Abstract Abstract
Elbamycellaroseasp. nov., introduced in the new genus Elbamycella, was collected in the Mediterranean Sea in association with the seagrass Posidoniaoceanica and with the brown alga Padinapavonica. The affiliation of the new taxon to the family Juncigenaceae is supported by both morphology and phylogenetic inference based on a combined nrSSU and nrLSU sequence dataset. Maximum-likelihood and Bayesian phylogeny proved Elbamycellagen. nov. as a distinct genus within Juncigenaceae. The new genus has been compared with closely related genera and is characterised by a unique suite of characters, such as ascospores with polar appendages and peculiar shape and dimension of ascomata and asci.
Keywords: Marine fungi, new taxon, TBM clade
Introduction
Marine fungi are a considerable part of the huge diversity of microorganisms that inhabit the Oceans (Richards et al. 2012). These organisms, which are distributed worldwide, live on a broad range of biotic and abiotic substrates (e.g. algae, sponges, corals, sediments) (Jones and Pang 2012) and are divided in two major ecological categories, namely obligate and facultative marine fungi. The former grow and reproduce exclusively in the sea, the latter are terrestrial species that can actively grow and reproduce in marine environments. Those fungi whose obligate or facultative marine nature is undefined are called marine-derived (Raghukumar 2017).
The number of marine fungi has been estimated to exceed 10,000 taxa, but the most recent update in marine mycology listed only 1,206 species belonging to Ascomycota, Basidiomycota, Chytridiomycota, and Mucoromycota. Thus fungal diversity is largely undescribed (Pang and Jones 2017).
In an attempt to clarify the phylogeny of the genera Swampomyces Kohlm. & Volkm.-Kohlm. and Torpedospora Meyers, Sakayaroj et al. (2005) recognised a distinct lineage of marine Ascomycota within the class Sordariomycetes that was then named TBM (Torpedospora/Bertia/Melanospora) clade (Schoch et al. 2007). Following a re-evaluation of the marine fungi affiliated to the TBM clade, together with the terrestrial genus Falcocladium, new families were introduced to accommodate its four subclades: Juncigenaceae, Etheirophoraceae, Falcocladiaceae, and Torpedosporaceae, all belonging to the order Torpedosporales (Jones et al. 2014; Abdel-Wahab et al. 2018). Based on phylogeny and morphological data, Maharachchikumbura et al. (2015) introduced the order Falcocladiales (Falcocladiaceae) under the class Sordariomycetes.
Recently, during a survey focused on the fungal diversity in the Mediterranean Sea, two unidentified Sordariomycetes were isolated from the seagrass Posidoniaoceanica (L.) Delile (Panno et al. 2013) and from the brown alga Padinapavonica (L.) Thivy (Garzoli et al. 2018). The present paper provides a phylogenetic and morphological study of the two strains that turn out to represent a new genus within the family Juncigenaceae.
Material and methods
Fungal isolates
The fungal isolates investigated in this paper were previously retrieved from P.oceanica (MUT 4937 = CBS 130520) and P.pavonica (MUT 5443) from the coastal waters of Elba island, in the Mediterranean Sea (Panno et al. 2013; Garzoli et al. 2018) (Table 1). The two strains were originally isolated on corn meal agar medium supplemented with sea salts (CMASS; 3.4% w/v sea salt mix, Sigma-Aldrich, Saint Louis, USA, in ddH2O) and are preserved at the Mycotheca Universitatis Taurinensis (MUT), Italy, and CBS Collection of the Westerdijk Fungal Biodiversity Institute, the Netherlands.
Table 1.
Dataset used for phylogenetic analysis. Genbank sequences including newly generated ITS, LSU and SSU amplicons relative to Elbamycellarosea sp. nov. and TorpedosporaambispinosaMUT 3537.
| Species | Strain Code | Source | ITS | SSU | LSU |
|---|---|---|---|---|---|
| HYPOCREALES | |||||
| Bionectriapityrodes (Mont.) Schroers | GJS95-26 | Bark | – | AY489696 | AY489728 |
| Clonostachysrosea (Link.) Schroers | GJS90-227 | Bark | – | AY489684 | AY489716 |
| Cordycepsmilitaris (L.) Fr. | NRRL 28021 | – | – | AF049146 | AF327374 |
| Fusariumsolani (Mart.) Sacc. | GJS89-70 | Bark | – | AY489697 | AY489729 |
| Trichodermadeliquescens (Sopp.) Jaklitsch | ATCC 208838 | Pine wood | – | AF543768 | AF543791 |
| MICROASCALES | |||||
| Cephalotrichumstemonitis (Pers.) Nees | AFTOL 1380 | Seed | – | DQ836901 | DQ836907 |
| Halosphaeriaappendiculata Linder | NTOU4004 | Driftwood | – | KX686781 | KX686782 |
| Lignincolalaevis Hohnk | JK5180A | Wooden stake | – | U46873 | U46890 |
| Microascustrigonosporus Emmons & Dodge | AFTOL 914 | – | – | DQ471006 | DQ470958 |
| Nimbosporaeffusa Kock | NTOU4018 | Intertidal wood | – | KX686793 | KX686794 |
| Noheaumiumi Kohlm. & Volkm. Kohlm. | NTOU4006 | Driftwood | – | KX686795 | KX686796 |
| Petriellasetifera (Schmidt) Curzi | AFTOL 956 | Wood panel in coastal water | – | DQ471020 | DQ470969 |
| TORPEDOSPORALES | |||||
| Etheirophoraceae | |||||
| Etheirophorablepharospora (Kohlm. & E. Kohlm.) Kohlm. & Volkm. Kohlm. | JK5397A | Bark on submerged proproots | – | EF027717 | EF027723 |
| E.unijubata Kohlm. & Volkm. Kohlm. | JK5443B | Submerged wood | – | EF027718 | EF027725 |
| Swampomycesarmeniacus Kohlm. & Volkm. Kohlm. | JK5059C | Mangroves | – | EF027721 | EF027728 |
| S.triseptatus Hyde & Nakagiri | CY2802 | Submerged wood | – | AY858942 | AY858953 |
| Juncigenaceae | |||||
| Juncigenaadarca Kohlm., Volkm. Kohlm. & Erikss | JK5548A | Juncus roemerianus | – | EF027720 | EF027727 |
| J.fruticosae (Abdel-Wahab, Abdel-Aziz & Nagah.) Mill. & Shearer | EF14 | Driftwood | – | GU252146 | GU252145 |
| IMI391650 | Driftwood | – | NG_061097 | NG_060791 | |
| Khaleijomycesmarinus Abdel-Wahab | MD1348 | Driftwood | – | MG717679 | MG717678 |
| Marinokulatichaetosa (Kohlm.) Jones & Panf | BCRC FU30271 | Driftwood | – | KJ866929 | KJ866931 |
| BCRC FU30272 | Driftwood | – | KJ866930 | KJ866932 | |
| Fulvocentrumaegyptiacum (Abdel-Wahab, El-Shar. & Jones) Jones & Abdel-Wahab | CY2973 | Mangroves | – | AY858943 | AY858950 |
| F.clavatisporum (Abdel-Wahab, El-Shar. & Jones) Jones & Abdel-Wahab | LP83 | Mangroves | – | AY858945 | AY858952 |
| Elbamycellarosea sp. nov. | MUT 4937 | P. oceanica | MK775496* | MK775501* | MK775499* |
| Elbamycellarosea sp. nov. | MUT 5443 | P. pavonica | MK775497* | MK775502* | MK775500* |
| Torpedosporaceae | |||||
| Torpedosporaambispinosa Kohlm. | CY3386 | Driftwood | – | AY858941 | AY858946 |
| BCC16003 | Driftwood | – | AY858940 | AY858949 | |
| MUT 3537 | Driftwood | MK775503* | MK775498* | MK775495* | |
| T.mangrovei (Abdel-Wahab & Nagah.) Jones & Abdel-Wahab | NBRC 105264 | Mangroves | NR_138418 | GU252150 | GU252149 |
| T.radiata Meyers | BCC11269 | Driftwood | – | AY858938 | AY858948 |
| PP7763 | Driftwood | – | AY858939 | AY858947 | |
| FALCOCLADIALES | |||||
| Falcocladiaceae | |||||
| Falcocladiummultivesiculatum Silveira, Alfenas, Crous & Wingf | CBS 120386 | Leaves | – | JF831928 | JF831932 |
| F.sphaeropedunculatum Crous & Alfenas | CBS 111292 | Leaves | – | JF831929 | JF831933 |
| F.thailandicum Crous & Himaman | CBS 121717 | Leaves | – | JF831930 | JF831934 |
| F.turbinatum Somrith., Sudhom, Tippawan & Jones | BCC22055 | Dead leaves | – | JF831931 | JF831935 |
| XYLARIALES | |||||
| Daldiniaconcentrica (Bolton) Ces. & De Not. | ATCC 36659 | Fraxinus sp. | – | U32402 | U47828 |
| Hypoxylonfragiforme (Pers.) Kickx | HKUCC 1022 | Bark | – | AY083810 | AY083829 |
| Xylariahypoxylon (L.) Grev. | AFTOL 51 | Rotting wood | – | AY544692 | AY544648 |
* = newly generated sequences
Morphological analysis
MUT 4937 and MUT 5443 were pre-grown on CMA-sea water (CMASW; 17 g corn meal agar in 1 L of sea water) for one month at 21 °C prior to inoculation in triplicate onto Petri dishes (9 cm Ø) containing CMASS, CMASW, Potato Dextrose Agar (PDA) SS or PDASW. Petri dishes were incubated at 10 °C and 21 °C. The colony growth, together with macroscopic and microscopic traits, were monitored for 28 days.
Reproductive structures were observed and captured using an optical microscope (Leica DM4500B, Leica microsystems GmbH, Germany) equipped with a camera (Leica DFC320, Leica microsystems GmbH, Germany). Macro- and microscopic features were compared with the available description of Juncigenaceae (Kohlmeyer et al. 1997; Abdel-Wahab et al. 2001; Jones et al. 2014; Abdel-Wahab et al. 2018).
DNA extraction, PCR amplification, and data assembling
Genomic DNA was extracted from about 100 mg of mycelium carefully scraped from CMASS plates. Mycelium was transferred to a 2 mL Eppendorf tubes and disrupted in a MM400 tissue lyzer (Retsch GmbH, Haan, Germany). Extraction was accomplished using a NucleoSpin kit (Macherey Nagel GmbH, Duren, DE, USA) following the manufacturer’s instructions. The quality and quantity of DNA samples were measured spectrophotometrically with Infinite 200 PRO NanoQuant (TECAN, Switzerland) and stored at −20 °C.
The primer pairs ITS1/ITS4 (White et al. 1990), LROR/LR7 (Vilgalys and Hester 1990), and NS1/NS4 (White et al. 1990) were used to amplify the partial sequences of the internal transcribed spacers including the 5.8S rDNA gene (ITS), partial large ribosomal subunit (nrLSU), and partial small ribosomal subunit (nrSSU), respectively. Ribosomal genes were amplified in a T100 Thermal Cycler (Bio-Rad, Hercules, CA, USA), as previously described (Bovio et al. 2018). Reaction mixtures consisted of 60–80 ng DNA template, 10× PCR Buffer (15 mM MgCl2,500 mM KCl, 100 mM Tris-HCl, pH 8.3), 200 µM each dNTP, 1 μM each primer, 2.5 U Taq DNA Polymerase (Qiagen, Chatsworth, CA, USA), in 50 μL final volume. Following visualization of the amplicons on a 1.5% agarose gel stained with 5 mL 100 mL−1 ethidium bromide, PCR products were purified and sequenced at Macrogen Europe Laboratory (Madrid, Spain). The resulting ABI chromatograms were processed and assembled to obtain consensus sequences using Sequencer v. 5.0 (GeneCodes Corporation, Ann Arbor, Michigan, USA http://www.genecodes.com). Newly generated sequences were deposited in GenBank (Table 1).
Sequence alignment and phylogenetic analysis
A dataset consisting of nrLSU and nrSSU was assembled on the basis of BLASTn results and of a recent phylogenetic study focused on Torpedosporales (Abdel-Wahab et al. 2018). Reference sequences were retrieved from GenBank. Although nrITS regions were amplified for MUT 4937 and MUT 5443, they were not used for phylogenetic analyses, due to the lack of available ITS sequences for the strains present in the tree. Alignments were generated using MUSCLE (default conditions for gap openings and gap extension penalties), implemented in MEGA v. 7.0 (Molecular Evolutionary Genetics Analysis), visually inspected and trimmed by TrimAl v. 1.2 (http://trimal.cgenomics.org) to delimit and discard ambiguously aligned regions. nrITS alignment was not performed, due to the lack of reference sequences. Preliminary analyses suggested no incongruence among single-loci phylogenetic trees, as assessed through the Incongruence Length Difference (ILD) test (de Vienne et al. 2007). As a consequence, alignments were concatenated into a single data matrix with SequenceMatrix (Vaidya et al. 2011). The appropriate evolutionary model under the Akaike Information Criterion (AIC) was determined with jModelTest 2 (Darriba et al. 2012).
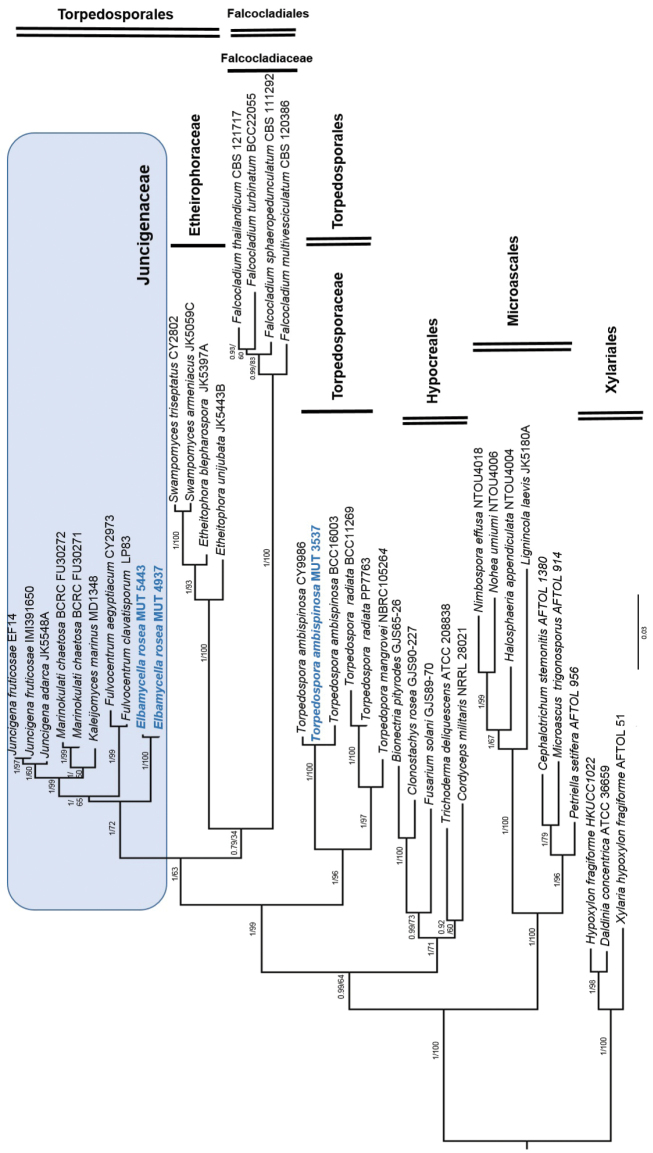
Phylogenetic inference was estimated using both Maximum Likehood (ML) and Bayesian Inference (BI). The ML analysis was performed using RAxML v. 8.1.2 (Stamatakis 2014) under GTR + I + G evolutionary model (best model) and 1,000 bootstrap replicates. Support values from bootstrapping runs (MLB) were mapped on the globally best tree using the “-f a” option of RAxML and “-x 12345” as a random seed to invoke the novel rapid bootstrapping algorithm. BI was performed with MrBayes 3.2.2 (Ronquist et al. 2012) with the same substitution model (GTR + I + G). The alignment was run for 10 million generations with two independent runs each containing four Markov Chains Monte Carlo (MCMC) and sampling every 100 iterations. The first quarter of the trees were discarded as “burn-in”. A consensus tree was generated using the “sumt” function of MrBayes and Bayesian posterior probabilities (BPP) were calculated. Consensus trees were visualized in FigTree v. 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree). Members of Xylariales (i.e. Xylariahypoxylon, Hypoxylonfragiforme, and Daldiniaconcentrica) were used as outgroup taxa. Due to topological similarity of the two resulting trees, only ML analysis with MLB and BPP values is reported (Fig. 1).
Figure 1.
Phylogenetic inference of Elbamycellarosea sp. nov. based on a combined nrSSU and nrLSU dataset. The tree is rooted to Xylariahypoxylon. Branch numbers indicate BPP/MLB values; Bar = expected changes per site (0.03).
Sequence alignments and phylogenetic tree were deposited in TreeBASE (http://www.treebase.org, submission number 24426).
Results
Phylogenetic inference
Preliminary analyses were carried out individually with nrSSU and nrLSU. The topology of the single-locus trees was very similar and the ILD test confirmed the congruence between them (p = 0.001). The combined dataset consisted of an equal number of nrSSU and nrLSU sequences relative to 39 taxa (including MUT 4937 and MUT 5443) that represented 23 genera and 33 species (Table 1). Nine sequences (3 nrSSU, 3 nrLSU, and 3 nrITS) were newly generated while 72 were retrieved from GeneBank. SSU and LSU sequences relative to MUT 4937 and MUT 5443 displayed 100% and 99% similarity (3 bp substitutions). The combined dataset had an aligned length of 1676 characters, of which 1208 were constant, 92 were parsimony-uninformative and 376 parsimony informative (TL = 315, CI = 0.603715, RI = 0.802773, RC = 0.549296, HI = 0.396285).
The two isolates MUT 4937 and MUT 5443 clustered within the family Juncigenaceae together with Marinokulatichaetosa, Khaleijomycesmarinus, Juncigenaadarca, J.fruticosae, Fulvocentrumaegyptiacum, and F.clavatisporum (Fig. 1; BPP = 1; MLB = 72%) and formed a strongly supported monophyletic lineage (Fig. 1; BPP = 1; MLB = 100%) indicating that these strains are phylogenetically different from the other members of the family.
Taxonomy
Elbamycella
gen. nov. A. Poli, E. Bovio, V. Prigione & G.C. Varese
Mycobank: MB830648
Type species.
Elbamycellarosea sp. nov.
Etymology.
In reference to the geographic isolation site, Elba Island, Tuscany (Italy)
Phylogenetic placement.
Juncigenaceae, Sordariomycetes, Ascomycota. The genus Elbamycella gen. nov. clusters together with genera Marinokulati, Khaleijomyces, Juncigena, and Fulvocentrum (Fig. 1).
Description.
Ascomata superficial, erumpent or immersed, perithecial, scattered or gregarious, olivaceous-brown to black at maturity, globose, subglobose, ovoid or pyriform, glabrous; ostiolar neck long, pale-coloured; peridium of textura prismatica in the outer layers and textura globulosa in the inner layers. Asci evanescent, hyaline, cylindrical to clavate.
Ascopores cylindrical rounded at both ends, thin-walled, hyaline, straight or slightly curved, 3-septate, bearing subpolar, appendages.
Asexual morph unknown.
Elbamycella rosea
sp. nov. A. Poli, E. Bovio, V. Prigione & G.C. Varese
Mycobank: MB830649
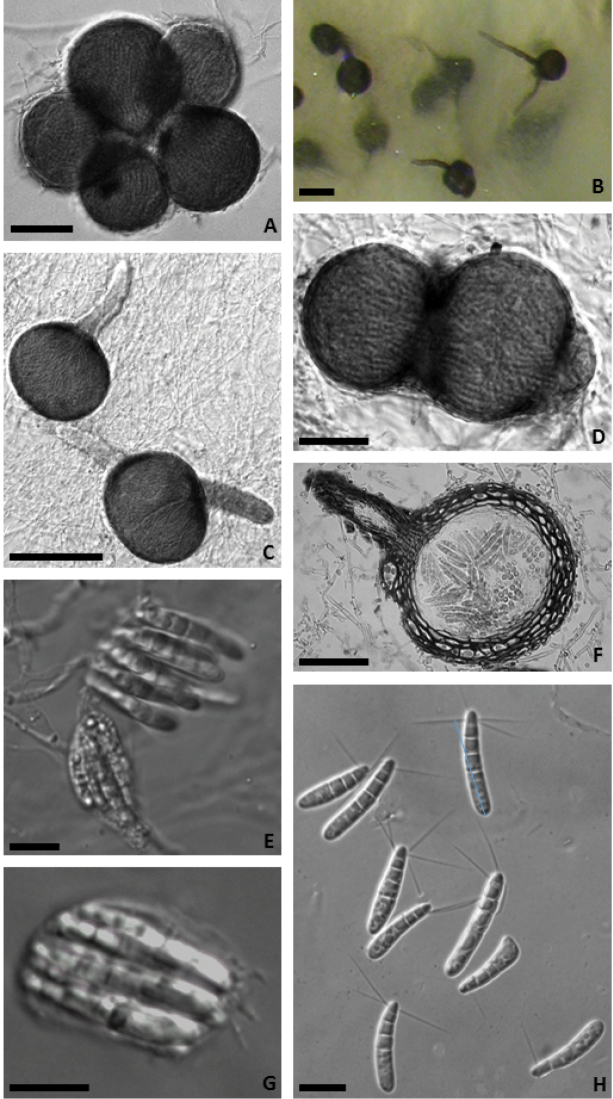
Figure 2.
Elbamycellarosea sp. nov. A, D group of young subglobose ascomata B, C globose ascomata with one or two necks E immature (bottom) and dehiscent (top) asci F ascoma in cross section G mature ascus with 8 ascospores H ascospores. Scale bars: 50 µm (A, D, F); 100 µm (B, C); 10 µm (E, G, H).
Figure 3.
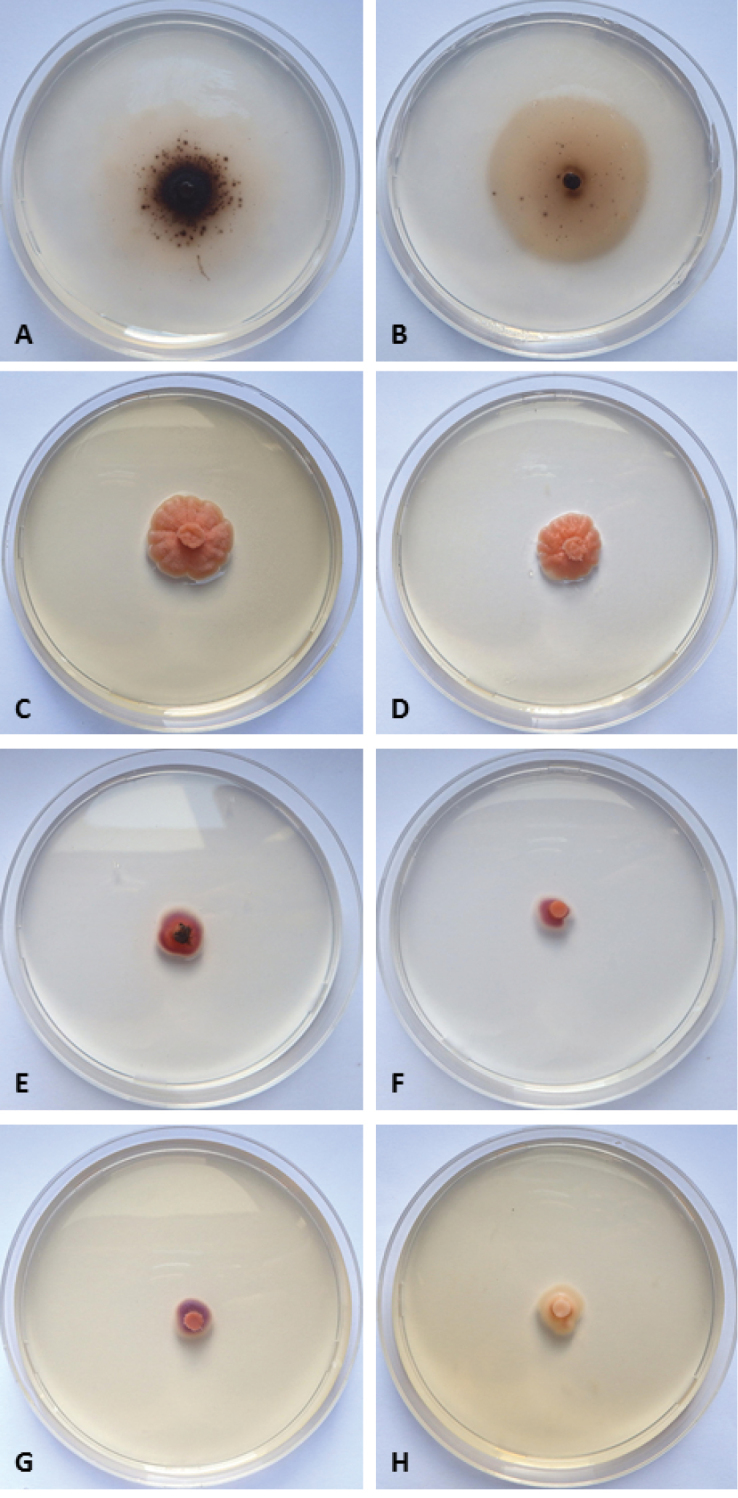
Elbamycellarosea sp. nov.: 28-days-old colonies at 21 °C on ACMASWBCMASSC PDASW D PDASS; 28-days-old colonies at 10 °C on ECMASWFCMASSG PDASW H PDASS.
Type.
Italy, Tuscany, Mediterranean Sea, Elba Island (LI), Ghiaie ISL, 14–15m depth, 42°49’04”N, 10°19’20”E, on the brown alga Padinapavonica, 20 March 2010, R. Mussat-Sartor and N. Nurra, MUT 5443 holotype, living culture permanently preserved in metabolically inactively state by deep-freezing at Mycotheca Universitatis Taurinensis. A dried specimen of this culture grown on CMASS and CMASW has been deposited in the herbarium of the Department of Life Sciences and Systems Biology (TO Cryptogamia 3446).
Additional material examined.
Italy, Tuscany, Mediterranean Sea, Elba island (LI), Ghiaie ISL, 14–15m depth, 42°49’04”N, 10°19’20”E, on the seagrass Posidoniaoceanica, 20 March 2010, R. Mussat-Sartor and N. Nurra, MUT 4937 = CBS 130520.
Etymology.
In reference to the colour of the colony on the culture media.
Description.
Ascomata were produced on both CMASS and CMASW at 21 °C only, after 28 days of incubation. Mycelium hyaline to pale brown consisting of smooth-walled hyphae 2.5–4 µm wide (Fig. 2A–D, F).
Ascomata perithecial, scattered or gregarious (from 2 to 6-8), superficial, erumpent or immersed, olivaceous-brown to black at maturity, globose, subglobose, ovoid or pyriform, glabrous, up to 100–140 µm diameter; ostiolar neck, pale-coloured, single (sometimes 2, rarely 3), 55–70 µm long and 20–50 µm wide at the base; peridium 5–10 µm thick of textura prismatica in the outer layers and textura globulosa in the inner layers with cells with olivaceous-brown walls, in the neck consisting of hyaline, more elongated cells, from which numerous hyaline blunt hyphal projections 5–15 × 3–5 µm arise. Asci evanescent, hyaline, cylindrical to clavate 22–26 × 12–16 µm containing 8 spores; sterile elements not observed (Fig. 2E, G).
Ascopores cylindrical 23–28 × 4–5 µm, rounded at both ends, thin-walled, hyaline, straight or slightly curved, 3-septate, with a large basal cell 10–15 µm long and 3 shorter, upper cells, slightly constricted around the septa, the apical cell somewhat attenuated just below the blunt tip, bearing 3(4) subpolar, straight or slightly bent, acuminate, hyaline, smooth-walled cellular appendages 10–20 µm long (about 0.5–1 µm wide). In some spores the apical cell is divided by an additional septum; each cell of the spore contains a few oil-droplets 1.5–3.0 µm diameter (Fig. 2H).
Asexual morph not observed.
Colony description.
Colonies reaching 21–23 mm diameter on CMASW and 19–29 mm diameter on CMASS in 28 days at 21 °C, plane, thin, mycelium mainly submerged. Colonies pale pink in the centre becoming brown with age, colourless at the margins. Black spots due to ascomata groups in fruiting colonies. Reverse of the same colour of the surface (Fig. 3A, B).
Colonies on PDASW and PDASS reaching 10–14 mm diameter in 28 days at 21 °C, convolute, developing in height with irregular margins, salmon. Reverse of the same colour of the surface (Fig. 3C, D).
At 10 °C colony growth on all media very poor, attaining 5–8 mm diameter in 28 days. Colonies plane to slightly convolute with regular margins, pale pink to cyclamen. Reverse of the same colour of the surface (Fig. 3E–H).
Table 2.
Comparison of the main sexual morpholgical features of genera belonging to Juncigenaceae.
| Fungus | Ascomata | Periphyses/ Paraphyses | Asci | Ascospores | Reference |
| Marinokulati chaetosa | Immersed to superficial, dark brown, ostiolate, papillate; neck 20 × 70 µm | Both present, septate, wide | 102−135 × 12−18 µm; cylindrical to clavate, attenuate at the base, thick-walled at the apex, containing 8 spores | 25.5−36.5 × 7.5−11.5 µm; 3-septate, hyaline, fusiform to ellipsoidal with polar and equatorial appendages | Jones et al. 2014 |
| Khaleijomyces marinus | Superficial to immersed, hyaline to yellow-orange to reddish brown, ostiolate; 110−175 × 100−115 µm; neck 120-175 × 40-50 µm | Periphyses present in the neck | 60−98 × 12−16 µm; cymbiform to fusiform, thin-walled, with no apical apparatus, containing 8 spores | 12−26 × 6-8 µm; 1-4-septate; ellipsoidal to fusiform; hyaline, smooth-walled | Abdel-Wahab et al. 2018 |
| Juncigena adarca | Immersed, ostiolate, papillate, 225−400 × 135-200 µm; neck 85−170 × 50−85 µm | Both present; Paraphyses thin, branched, septate | 115−140 × 10−13 µm; fusiform to cylindrical, short pedunculate, apical apparatus with a ring, containing 8 spores | 26.5−34.5 × 6−7 µm; 3-septate, hyaline, fusiform to ellipsoidal, no appendages, smooth wall, constricted | Kohlmeyer et al. 1997 |
| Fulvocentrum aegyptiacum | Immersed, dark brown, ostiolate; 240−280 × 170−190 µm; neck 70−80 µm diameter | Both present; Paraphyses numerous, in a gel, unbranched | 145−155 × 9−10 µm; Short pedicellate, apically thickened, containing 8 spores | 15−20 × 6−8 µm; 3-septate, ellipsoidal, hyaline | Abdel-Wahab et al. 2001 |
| Fulvocentrum clavatisporum | Immersed, dark brown, ostiolate; 160−170 × 160−190 µm; neck 50 µm long | Both present; Paraphyses numerous, in a gel, unbranched | 80−96 × 10−13µm; Pedicellate, apically thickened, containing 8 spores | 25−28 × 5−6 µm; 3-septate, clavate, hyaline | Abdel-Wahab et al. 2001 |
| Fulvocentrum rubrum | Erumpent to superficial, olive-brown to dark brown, ostiolate; 145−270 µm; neck 310−390 × 50−55 µm | Both present | Fusiform or obclavate, 95−130 × 13−19 µm; persistent, thin-walled, containing 8 spores | Ellipsoidal to clavate, no appendages, 25−33 × 6−9 µm; hyaline to faint apricot, smooth walled, 3−5-septate | Abdel-Wahab et al. 2019 |
| Elbamycellarosea sp. nov. | Superficial, erumpent or immersed; 100–140 µm diam; olivaceous-brown to black; ostiolar neck 55−70 × 20–50 µm | Cylindrical to clavate 22−26 × 12−16 µm containing 8 spores | Cylindrical 23−28 × 4−5 µm; hyaline, generally 3-septate, bearing 3(4) subpolar appendages 10–20 × 0.5–1 µm | This study |
Discussion
The novel genus Elbamycella is introduced in this study and has been compared to the closest genera. Herein, the two strains MUT 4937 and MUT 5443 represented a new species that formed a well-supported cluster phylogenetically distant from the related genera of Juncigenaceae.
From a morphological point of view, the relatedness with the other species belonging to Juncigenaceae is confirmed by i) 3-septate spores (1–4 only in K.marinus), ii) 8-spored asci, and iii) ascomata with an elongated neck (Kohlmeyer et al. 1997; Abdel-Wahab et al. 2001; Abdel-Wahab et al. 2010; Jones et al. 2014; Abdel-Wahab et al. 2018). Elbamycellarosea sp. nov. is furthermore characterised by the presence of polar appendages on the ascospores. Marinokulatichaetosa displays this feature too, although it can be distinguished from E.rosea sp. nov. by additional, equatorially placed appendages. Additonally, in the new species, spores are cylindrical, not fusiform-ellipsoidal as in M.chaetosa (Jones et al. 2014). Khaleijomycesmarinus, Juncigenaadarca, Fulvocentrumaegyptiacum, F.clavatisporum, and the recently described F.rubrum differ in the shape and dimensions of the ascospores (Jones et al. 2014; Abdel-Wahab et al. 2018; Abdel-Wahab et al. 2019); generally asci and ascomata are larger than those observed in E.rosea sp. nov.
As no sexual form is known for J.fruticosae, the comparison with E.rosea sp. nov. is not possible. However, the similarity or identity to this species is excluded by the phylogenetical distance.
Ecologically, the described Juncigenaceae are species having a marine origin. So far, they have all been retrieved from driftwood in the intertidal of salt marshes (Kohlmeyer et al. 1997; Jones et al. 2014). The new species was found for the first time underwater, in association with the seagrass P.oceanica and the brown alga P.pavonica, two different organisms that were sampled in close proximity. This could be related to a successful spore dispersal; indeed polar appendages are known to facilitate floatation and attachment (Overy et al. 2019).
Supplementary Material
Acknowledgements
This work was funded by Fondazione CRT, Turin, Italy. The authors are grateful to Pelagosphera s.c.r.l. for harvesting algal and seagrasses samples.
Citation
Poli A, Bovio E, Verkley G, Prigione V, Varese GC (2019) Elbamycella rosea gen. et sp. nov. (Juncigenaceae, Torpedosporales) isolated from the Mediterranean Sea. MycoKeys 55: 15–28. https://doi.org/10.3897/mycokeys.55.35522
Funding Statement
Fondazione CRT - Torino, Italy
References
- Abdel-Wahab MA, El-Sharouney H, Jones EBG. (2001) Two new intertidal lignicolous Swampomyces species from Red Sea mangroves in Egypt. Fungal Diversity 8: 35–40. [Google Scholar]
- Abdel-Wahab MA, Pang KL, Nagahama T, Abdel-Aziz FA, Jones EBG. (2010) Phylogenetic evaluation of anamorphic species of Cirrenalia and Cumulospora with the description of eight new genera and four new species. Mycological Progress 9: 537–558. 10.1007/s11557-010-0661-x [DOI] [Google Scholar]
- Abdel-Wahab MA, El-Samawaty AMA, El Gorban AM, Yassin MA, Alsaadi MH. (2018) Khaleijomycesmarinus gen. et sp nov. (Juncigenaceae, Torpedosporales) a new lignicolous marine fungus from Saudi Arabia. Phytotaxa 340: 277–285. 10.11646/phytotaxa.340.3.8 [DOI] [Google Scholar]
- Abdel-Wahab MA, Jones EBG, Bahkali AHA, El-Gorban AM. (2019) Marine fungi from Red Sea mangroves in Saudi Arabia with Fulvocentrumrubrum sp. nov. (Torpedosporales, Ascomycota). Nova Hedwigia 108: 365–377. 10.1127/nova_hedwigia/2018/0511 [DOI] [Google Scholar]
- Bovio E, Garzoli L, Poli A, Prigione V, Firsova D, McCormack G, Varese G. (2018) The culturable mycobiota associated with three Atlantic sponges, including two new species: Thelebolusbalaustiformis and T.spongiae. Fungal Systematics and Evolution 1: 141–167. 10.3114/fuse.2018.01.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772–772. hhttps://doi.org/ 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed]
- de Vienne DM, Giraud T, Martin OC. (2007) A congruence index for testing topological similarity between trees. Bioinformatics 23: 3119–3124. 10.1093/bioinformatics/btm500 [DOI] [PubMed] [Google Scholar]
- Garzoli L, Poli A, Prigione V, Gnavi G, Varese GC. (2018) Peacock’s tail with a fungal cocktail: first assessment of the mycobiota associated with the brown alga Padinapavonica. Fungal Ecology 35: 87–97. 10.1016/j.funeco.2018.05.005 [DOI] [Google Scholar]
- Jones EG, Pang K-L. (2012) Marine fungi and fungal-like organisms. Walter de Gruyter. 10.1515/9783110264067 [DOI]
- Jones EBG, Suetrong S, Cheng WH, Rungjindamai N, Sakayaroj J, Boonyuen N, Somrothipol S, Abdel-Wahab MA, Pang KL. (2014) An additional fungal lineage in the Hypocreomycetidae (Falcocladium species) and the taxonomic re-evaluation of Chaetosphaeriachaetosa and Swampomyces species, based on morphology, ecology and phylogeny. Cryptogamie Mycologie 35: 119–138. 10.7872/crym.v35.iss2.2014.119 [DOI] [Google Scholar]
- Kohlmeyer J, Volkmann-Kohlmeyer B, Eriksson OE. (1997) Fungi on Juncusroemerianus .9. New obligate and facultative marine ascomycotina. Botanica Marina 40: 291–300. 10.1515/botm.1997.40.1-6.291 [DOI] [Google Scholar]
- Maharachchikumbura SSN, Hyde KD, Jones EBG, McKenzie EHC, Huang SK, Abdel-Wahab MA, Daranagama DA, Dayarathne M, D’Souza MJ, Goonasekara ID, Hongsanan S, Jayawardena RS, Kirk PM, Konta S, Liu JK, Liu ZY, Norphanphoun C, Pang KL, Perera RH, Senanayake IC, Shang QJ, Shenoy BD, Xiao YP, Bahkali AH, Kang JC, Somrothipol S, Suetrong S, Wen TC, Xu JC. (2015) Towards a natural classification and backbone tree for Sordariomycetes. Fungal Diversity 72: 199–301. 10.1007/s13225-015-0331-z [DOI] [Google Scholar]
- Overy DP, Rama T, Oosterhuis R, Walker AK, Pang KL. (2019) The neglected marine fungi, sensu stricto, and their isolation for natural products’ discovery. Marine Drugs 17: 20. 10.3390/md17010042 [DOI] [PMC free article] [PubMed]
- Pang KL, Jones EBG. (2017) Recent advances in marine mycology. Botanica Marina 60: 361–362. 10.1515/bot-2017-0048 [DOI] [Google Scholar]
- Panno L, Bruno M, Voyron S, Anastasi A, Gnavi G, Miserere L, Varese GC. (2013) Diversity, ecological role and potential biotechnological applications of marine fungi associated to the seagrass Posidoniaoceanica. New Biotechnology 30: 685–694. 10.1016/j.nbt.2013.01.010 [DOI] [PubMed] [Google Scholar]
- Raghukumar S. (2017) Fungi in Coastal and Oceanic Marine Ecosystems: Marine Fungi Springer, Cham, 1–36. 10.1007/978-3-319-54304-8_1 [DOI]
- Richards TA, Jones MD, Leonard G, Bass D. (2012) Marine fungi: their ecology and molecular diversity. Annual Review of Marine Science 4: 495–522. 10.1146/annurev-marine-120710-100802 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakayaroj J, Pang KL, Jones EBG, Phongpaichit S, Vrijmoed LLP, Abdel-Wahab MA. (2005) A systematic reassessment of the marine ascomycetes Torpedospora and Swampomyces. Botanica Marina 48: 395–406. 10.1515/bot.2005.053 [DOI] [Google Scholar]
- Schoch CL, Sung GH, Volkmann-Kohlmeyer B, Kohlmeyer J, Spatafora JW. (2007) Marine fungal lineages in the Hypocreomycetidae. Mycological Research 111: 154–162. 10.1016/j.mycres.2006.10.005 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidya G, Lohman DJ, Meier R. (2011) SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27: 171–180. 10.1111/j.1096-0031.2010.00329.x [DOI] [PubMed] [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: a Guide to Methods and Applications 18: 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.