Abstract
Introduction
The implementation of spatial-covariance [18F]fluorodeoxyglucose positron emission tomography–based disease-related metabolic brain patterns as biomarkers has been hampered by intercenter imaging differences. Within the scope of the JPND-PETMETPAT working group, we illustrate the impact of these differences on Parkinson's disease–related pattern (PDRP) expression scores.
Methods
Five healthy controls, 5 patients with idiopathic rapid eye movement sleep behavior disorder, and 5 patients with Parkinson's disease were scanned on one positron emission tomography/computed tomography system with multiple image reconstructions. In addition, one Hoffman 3D Brain Phantom was scanned on several positron emission tomography/computed tomography systems using various reconstructions. Effects of image contrast on PDRP scores were also examined.
Results
Human and phantom raw PDRP scores were systematically influenced by scanner and reconstruction effects. PDRP scores correlated inversely to image contrast. A Gaussian spatial filter reduced contrast while decreasing intercenter score differences.
Discussion
Image contrast should be considered in harmonization efforts. A Gaussian filter may reduce noise and intercenter effects without sacrificing sensitivity. Phantom measurements will be important for correcting PDRP score offsets.
Keywords: Neuroimaging biomarker, FDG-PET, Harmonization, Hoffman 3D brain phantom, Principal component analysis
1. Introduction
The fundamentals of [18F]fluorodeoxyglucose positron emission tomography (FDG-PET) are well established and based on extensively explored molecular mechanisms. FDG-PET represents a unique tool for the in vivo assessment of resting-state cerebral metabolism, which is a proxy for neuronal activity and a direct index of synaptic function and density. Various events can contribute to synaptic dysfunction and consequent neurodegeneration captured by FDG-PET, such as altered intracellular signaling cascades and mitochondrial bioenergetics, impaired neurotransmitter release, and long-distance deafferentation effects [1].
The widespread availability of FDG-PET neuroimaging for the in vivo assessment of metabolic dysfunction in Alzheimer's disease (AD), Parkinson's disease (PD), and other neurodegenerative conditions has exposed important methodological and practical diagnostic issues. To date, a number of international working groups and consortia have advocated for the relevance of FDG-PET to the diagnostic workup of neurodegenerative diseases [2], [3], [4], [5], [6]. This is most likely due to methodological advances in FDG-PET data analysis, including univariate and multivariate methods, which considerably influence the accuracy of FDG-PET interpretation. Of note, several studies have demonstrated the diagnostic and prognostic value of PET techniques, in particular, when appropriate quantification methods are applied [5], [7], [8].
Accurate diagnosis of neurodegenerative diseases such as PD can be challenging, especially in the early stages, and visual interpretation of FDG-PET scans can be difficult in the absence of an expert reader [9]. An early, accurate diagnosis is essential for initiating earlier treatment to potentially alter disease course. In addition, it is important to be able to identify prodromal patients—such as those with idiopathic rapid eye movement sleep behavior disorder (RBD), considered to be prodromal for several parkinsonian disorders—for participation in drug trials. Therefore, interest in the development of stable progression imaging biomarkers for common neurodegenerative diseases has been steadily rising worldwide [6].
Many neurodegenerative conditions, such as PD, are characterized by distinct patterns of relative glucose hyper- and hypometabolism in the brain as assessed by FDG-PET imaging [10]. Using multivariate spatial covariance analysis methods such as Scaled Subprofile Model/Principal Component Analysis (SSM/PCA) [11], [12], neurodegenerative disease–related metabolic brain patterns for PD and other disorders have been identified by several research groups [13], [14], [15], [16], [17], [18]. The degree of pattern expression in individual FDG-PET scans can subsequently be quantified to get a disease-related pattern expression score.
The advantage of using such a method is that disease-related pattern expression can often be detected in presymptomatic, prodromal groups before structural changes occur [19], [20], [21]. In addition, they may be useful for discerning between differential diagnoses in early or atypical cases [10], [18], [22]. Subject scores generally increase with disease progression and decrease with effective treatment [16], [19], [23].
However, the widespread implementation of such SSM/PCA-derived disease-related metabolic brain patterns in multicenter collaborations and clinical practice has been hindered by differences between PET scanners as well as acquisition and reconstruction protocols. Variations in scanners and image reconstruction algorithms have been shown to systematically shift image quality and disease-related pattern subject scores [6], [24], [25], [26].
One way to resolve this is to apply a z-transformation to healthy control (HC) cohort data, such that the mean HC subject score is 0 with a standard deviation of 1. However, obtaining a HC cohort is not always feasible, especially for small and non-university-affiliated centers. In addition, z-scoring to HCs in different centers has the potential drawback of introducing additional factors of human variation, which could influence PD-related pattern (PDRP) expression score comparability between centers; a thorough, large-scale investigation into disease-related pattern score offsets in HCs with systematically differing ages, genders, ethnicities, and other factors has yet to be conducted.
Tomše et al. identified stable offsets in PDRP expression z-scores in HC and PD cohorts when adjusting image reconstruction factors such as time-of-flight (TOF) modeling and point spread function (PSF). It was found that PDRP topography was highly reproducible across FDG-PET reconstruction algorithms. In addition, within each type of reconstruction per scanner, discrimination between patients and HCs was not significantly impacted when using disease-related patterns derived using the same reconstruction method, whereas calibration with HCs was advised when different methods were used [25], [26]. However, the impact of additional variables on PDRP expression scores, such as histogram glucose uptake intensity distribution across voxels, as well as image contrast, has not yet been assessed. In addition, it is useful to examine raw PDRP expression scores, which are in principle represented in standard units independent of the center.
Ikari et al. suggested that the Hoffman 3D Brain Phantom (H3DBP) will likely represent the most promising approach to harmonizing image features and therefore to the implementation of disease-related patterns as biomarkers across centers [24]. To our knowledge, scanning the same H3DBP on different PET scanners to compare metabolic disease-related pattern expression differences has not been done before.
Comparison at a larger scale between univariate and multivariate analysis is also necessary, as this has been done almost exclusively for AD and related conditions, rather than for PD. There are currently very few univariate studies in classical PD [27]. Existing studies suggest that multivariate methods may be more accurate in early disease stages, but further investigation is necessary [28], [29], [30]. Validated univariate voxel-wise analysis methods such as statistical parametric mapping (SPM) have the advantage of being able to identify disease-related metabolic brain patterns at the single-subject level [31], [32], [33], [34], [35], [36], [37]. In addition, one study has suggested that SPM may be robust enough to withstand scanner and HC group differences between centers, a result which needs to be reproduced and explored on a pan-European scale [38].
We strongly support the implementation of these univariate and multivariate methods not only in academic research but also in routine clinical settings. The PETMETPAT project, under the umbrella of the EU Joint Programme–Neurodegenerative Disease Research (JPND), is currently addressing the issue of multivariate PDRP expression score harmonization specifically. Further exploration of factors which influence disease-related pattern scores in humans and the H3DBP is critical.
Here, we present a small cohort of HC, PD, and “intermediate” subjects with RBD scanned on one PET/computed tomography (CT) system, with images reconstructed in multiple ways, to illustrate the problem of large PDRP score expression ranges and the urgent need for harmonized imaging protocols. Prodromal patients in particular have a need for precise disease-related pattern score quantification for prognostic purposes. In addition, we have investigated PDRP raw score offsets in H3DBP scans performed on several different PET/CT systems with various reconstructions. Finally, we have attempted to pinpoint a unifying, underlying factor which influences image quality and resulting disease-related pattern scores, to suggest a starting point for the task of harmonization. To this end, we have investigated the effect of image contrast, expressed as gray-to-white matter ratios, in H3DBP and human subject scans.
2. Methods
2.1. Human subjects
2.1.1. [18F]FDG-PET imaging
As a proof of principle, we studied a retrospective cohort of five HCs (all male; median age 65 years, range 62–70); five nondemented, nonparkinsonian idiopathic RBD patients (all male; median age 64 years, range 50–67); and five nondemented subjects with early-stage PD (4 males; median age 65 years, range 62–78; average disease duration 2 years, range 0–5 years). HC subjects scored <5 on the RBD screening questionnaire [39] and did not have any first-degree family members with neurodegenerative disease. Exclusion criteria for all subjects included a history of (other) neurological diseases, diabetes mellitus, stroke, significant head trauma, or other relevant comorbidities.
All subjects underwent static [18F]FDG-PET imaging on a Siemens Biograph mCT64 PET/CT camera (Siemens, Munich, Germany) at the University Medical Center Groningen (UMCG) in the Netherlands. Central nervous system depressants were discontinued in all subjects for at least 24 hours before each scan. In patients with RBD, all RBD-related medications (e.g., melatonin or clonazepam) were discontinued for at least 24 hours before imaging. None of the patients with PD were medicated during the scan [19].
Images were reconstructed in 12 combinations of the following parameters: (a) Gaussian spatial smoothing with 0 mm (also known as unsmoothed/“all-pass”), 5 mm, and 10 mm full-width at half-maximum filters; (b) with and without both PSF and TOF modeling; and (c) with matrix sizes of 256 and 400, resulting in voxel sizes of 2 × 3.1819 × 3.1819 and 2 × 2.03642 × 2.03642, respectively. Reconstructions without TOF/PSF had 3 iterations and 24 subsets, whereas reconstructions with TOF/PSF had 3 iterations and 21 subsets.
2.1.2. Image processing and analysis
All images were spatially normalized onto an [18F]FDG-PET template [40] in Montreal Neurological Institute brain space using the SPM12 software (Wellcome Department of Imaging Neuroscience, Institute of Neurology, London, UK) in MATLAB (version R2017b; MathWorks, Natick, Massachusetts). Expression of the previously described PDRP [14] was calculated in the [18F]FDG-PET data as described by Spetsieris et al. [12].
In a nutshell, raw PDRP expression scores in new subject [18F]FDG-PET images were obtained by
-
1)
application of the same image transformations as were used in defining the PDRP (i.e., log transformation, within-subject demean, and subtraction of the existing grand mean profile),
-
2)
vectorization of image voxel values for both new [18F]FDG-PET data and the PDRP, and
-
3)
calculation of the inner products between these two vectors.
HCs tend to have lower (more negative) raw score values, and patients tend to express higher (more positive) score values. In general, the more advanced the disease is, the higher the corresponding disease-related pattern expression score will be.
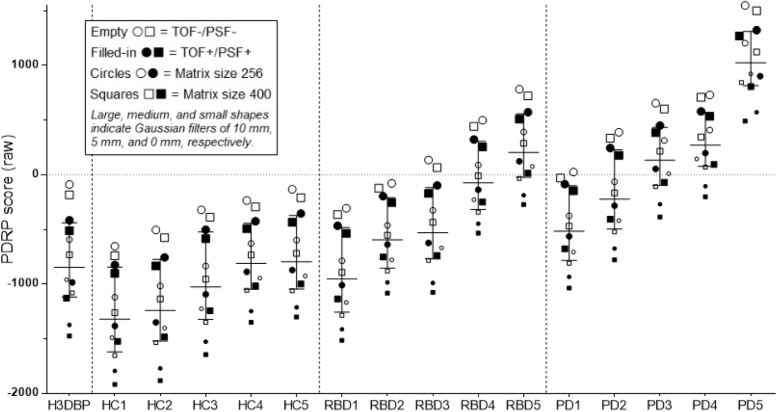
PDRP expression scores are presented for each individual subject/reconstruction (see Fig. 1). Given the small group size, the median and interquartile range (IQR) were used as markers of PDRP score expression and spread, respectively, for each group (HC, RBD, and PD) as a function of reconstruction (see Table 1).
Fig. 1.
The effects of Gaussian FWHM smoothing filter, TOF/PSF, and matrix size on raw, absolute PDRP expression scores in the H3DBP and five HC, RBD, and PD subjects scanned on the Siemens Biograph mCT64 PET/CT system at the UMCG. In all human subjects and the H3DBP, the reconstruction combination of 10 mm Gaussian filter, TOF−/PSF−, and matrix size of 256 consistently leads to the highest raw PDRP expression score, whereas the reconstruction combination of no Gaussian filter (0 mm/“all-pass”), and with TOF+/PSF+ and matrix size of 400, consistently leads to the lowest PDRP expression score. Per definition (based on how the PDRP is defined in this case)[14], [19], HCs tend to express more-negative raw PDRP values, and patients with PD tend to express more-positive raw PDRP values. Disease expression scores tend to increase with disease advancement. Abbreviations: FWHM, Full-width at half-maximum; TOF, time of flight; PSF, point spread function; PDRP, Parkinson's disease–related pattern; H3DBP, Hoffman 3D Brain Phantom; HC, healthy control; PD, Parkinson's disease; PET, positron emission tomography; CT, computed tomography; UMCG, University Medical Center Groningen.
Table 1.
Median and IQR of raw PDRP expression scores in the H3DBP/HC/RBD/PD scanned on the Siemens Biograph mCT64 PET/CT system at the UMCG
| Reconstructions | H3DBP | HC (all) | RBD (all) | PD (all) | |||
|---|---|---|---|---|---|---|---|
| 1 2 3 4 |
0 mm/all-pass |
TOF+/PSF+ TOF−/PSF− |
Matrix400 Matrix256 Matrix400 Matrix256 |
−1475 −1374 −1080 −960 |
−1645 ± 534 −1527 ± 522 −1348 ± 473 −1225 ± 457 |
−1076 ± 549 −984 ± 541 −781 ± 538 −668 ± 549 |
−385 ± 577 −270 ± 569 −107 ± 592 10 ± 564 |
| 5 6 7 8 |
5 mm |
TOF+/PSF+ TOF−/PSF− |
Matrix400 Matrix256 Matrix400 Matrix256 |
−1128 −984 −732 −592 |
−1243 ± 467 −1093 ± 462 −955 ± 399 −835 ± 390 |
−738 ± 503 −623 ± 502 −435 ± 542 −325 ± 553 |
−68 ± 501 54 ± 481 215 ± 512 315 ± 473 |
| 9 10 11 12 |
10 mm |
TOF+/PSF+ TOF−/PSF− |
Matrix400 Matrix256 Matrix400 Matrix256 |
−511 −416 −182 −89 |
−584 ± 339 −503 ± 330 −387 ± 282 −320 ± 269 |
−170 ± 510 −98 ± 517 66 ± 564 135 ± 577 |
385 ± 355 451 ± 334 602 ± 375 655 ± 342 |
Abbreviations: IQR, interquartile range; PDRP, Parkinson's disease–related pattern; H3DBP, Hoffman 3D Brain Phantom; HC, healthy control; RBD, idiopathic rapid eye movement sleep behavior disorder; PD, Parkinson's disease; PET, positron emission tomography; CT, computed tomography; UMCG, University Medical Center Groningen; TOF, time of flight; PSF, point spread function.
To investigate the impact of contrast on raw PDRP scores, gray-to-white matter recovery ratio values were obtained from [18F]FDG-PET images using in-house code created with MATLAB R2017b. The gray and white matter definition was extracted using SPM12's tissue probability map. A gray-to-white matter ratio was then calculated per scan using the average of the gray and white matter intensities, respectively.
Effects of spatial resolution on the gray-to-white matter ratios were also examined by comparing scans without a Gaussian spatial filter applied to them (0 mm/“all-pass”), and those with a 10 mm filter applied.
Owing to small group sizes, no rigorous statistical tests were performed. Instead, all individual data has been presented as-is.
2.2. Hoffman 3D Brain Phantom scans
2.2.1. [18F]FDG-PET imaging
To investigate the effect of different scanners and reconstruction settings on PDRP subject scores, the H3DBP underwent scanning on four PET/CT scanners from different hospitals. The following systems were used: the Siemens Biograph from the UMCG as described previously; a GE710 from the Catharina Hospital in Eindhoven, the Netherlands; a Philips Vereos from Philips Cleveland; and a Philips Ingenuity system from the VU University Medical Center (VUMC) in Amsterdam, the Netherlands. On each scanner, multiple clinically relevant reconstruction settings were applied, with and without TOF and PSF when possible.
2.2.2. Image processing and analysis
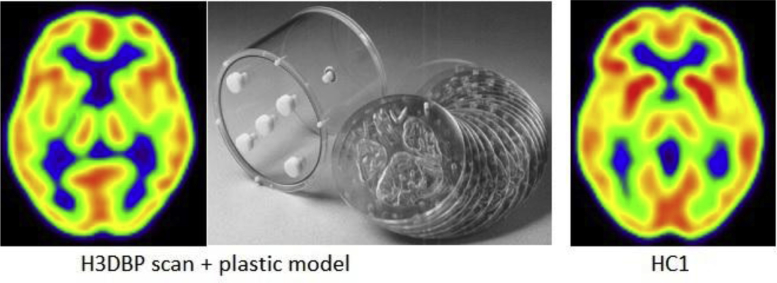
To obtain the raw PDRP expression scores, the steps as described in Section 2.1.2 were followed. An exception to this was the [18F]FDG-PET spatial normalization template used, which was modified for the H3DBP so as to exclude skull and soft tissue outside of the brain. To this end, one of the H3DBP data sets was spatially normalized to the template and correct spatial normalization was verified. Subsequently, this normalized scan was used as a spatial normalization template for all other H3DBP data, resulting in H3DBP scans taking up the same anatomical space as human subject scans (see Fig. 2).
Fig. 2.
Normalized H3DBP scan versus normalized HC1 scan [40] (Gaussian FWHM 8 mm filter, TOF+/PSF+, matrix 256; with approximately 50 MBq and 200 MBq [18F]FDG used in the H3DBP and HC1, respectively). Both scans were obtained from the Siemens Biograph mCT64 PET/CT System at the UMCG in the Netherlands. Abbreviations: FWHM, Full-width at half-maximum; TOF, time of flight; PSF, point spread function; H3DBP, Hoffman 3D Brain Phantom; FDG, fluorodeoxyglucose; UMCG, University Medical Center Groningen.
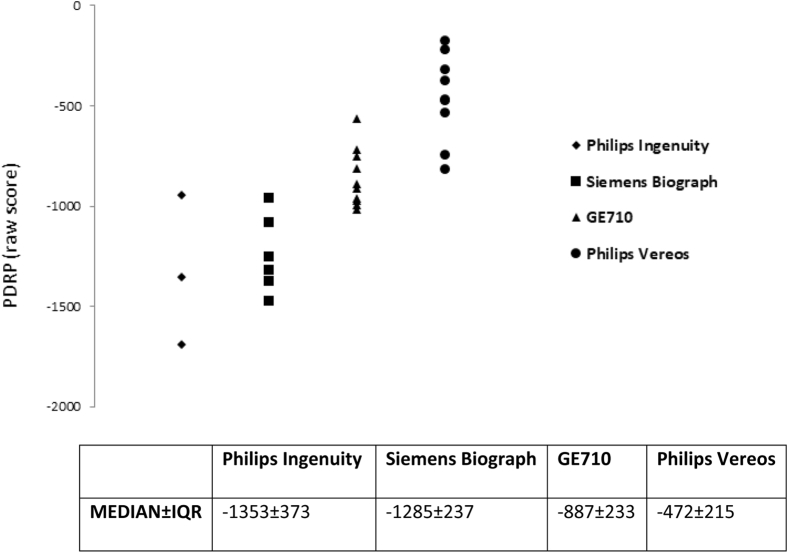
Once H3DBP PDRP scores were obtained for the Siemens Biograph mCT64 scanner, these scores were compared with the PDRP scores of the human subjects who underwent imaging on the same scanner (see Table 1 and Fig. 1). In addition, H3DBP PDRP scores, medians, and IQR were compared between the four scanners (see Fig. 3).
Fig. 3.
H3DBP PDRP expression scores on a Siemens, GE, and two Philips PET/CT systems with multiple image reconstructions each. Abbreviations: H3DBP, Hoffman 3D Brain Phantom; PDRP, Parkinson's disease–related pattern.
Gray-to-white matter recovery ratios were also obtained in H3DBP data as described in Section 2.1.2 and compared with human subject data. Ratio differences between unsmoothed H3DBP scans and scans with a 10 mm Gaussian full-width at half-maximum spatial smoothing filter applied to them were also examined.
In addition, the potential impact of differences between voxel-wide glucose uptake value distributions (histograms) on H3DBP raw PDRP scores across scanners and reconstructions was also explored. To this end, in-house code was created with MATLAB R2017b to re-scale the image intensity of H3DBP data obtained from the GE710, Philips Vereos, and Philips Ingenuity scanners to match the intensity range of the H3DBP data obtained from the UMCG's Siemens Biograph mCT64 scanner, reconstructed with TOF+/PSF+, matrix size 256. Raw PDRP expression scores were then calculated in the scans with transformed histograms (results not shown).
2.3. Other
An overview of JPND-PETMETPAT working group proceedings can be found in Supplementary Material A. In addition, an anonymous survey was conducted to identify practical variations in scanning protocols between centers—see Supplementary Material B.
3. Results
3.1. PDRP scores in HC, RBD, and PD subjects scanned on the UMCG Siemens Biograph mCT64 system
As expected, raw PDRP expression scores tended to follow an upward trend from HC to RBD subjects and from RBD to PD subjects who underwent FDG-PET brain imaging on the Siemens Biograph mCT64 PET scanner at the UMCG (see Table 1).
The most obvious impact on PDRP expression was due to Gaussian smoothing filter variations, with higher smoothing filters associated with increased expression of the PDRP (Fig. 1). Concurrently, the PDRP score IQRs tended to decrease in HC and PD subject scans as the smoothing filter increased. Score changes were most prominent among the HC subject scans, which have the lowest intrinsic raw PDRP scores.
Next, the TOF/PSF status had a moderate impact on PDRP score expression, with TOF−/PSF− reconstruction being associated with increased raw PDRP scores as compared with TOF+/PSF+ (Fig. 1). Similarly to the Gaussian filter offsets, the most prominent differences between TOF+/PSF+ and TOF−/PSF− were observed in scans obtained from HCs. In addition, as the Gaussian smoothing filter increased, the median score differences between the TOF+/PSF+ and TOF−/PSF− reconstructed scans decreased for all subjects.
Finally, the matrix size had a small but consistent impact on PDRP expression, with a matrix size of 256 being associated with slightly increased raw PDRP scores compared with a matrix size of 400 (Fig. 1). Once again, the biggest difference in PDRP scores as a function of matrix size was seen in scans from HCs. In addition, as the Gaussian smoothing filter increased, the score differences between the scans reconstructed with matrix size 256 versus 400 decreased for all subjects.
Overall, HC scans, which have intrinsically lower PDRP expression scores, had greater score offsets as a result of reconstruction differences than PD subject scans. The median PDRP score-spread in scans reconstructed with 0 mm, TOF+/PSF+, and matrix size 400 (representing the lowest PDRP score across all subjects) to scans reconstructed with 10 mm, TOF−/PSF−, and matrix size 256 (representing the highest PDRP score across all subjects) was 1325 points in HC, versus 1211 points in RBD, and 1040 points in PD.
3.2. H3DBP PDRP scores
3.2.1. UMCG Siemens Biograph mCT64 reconstructions
The H3DBP raw PDRP expression scores on the UMCG Siemens Biograph mCT64 scanner fell within the IQR for both the HC and RBD subjects, irrespective of the reconstruction used. Overall, the influence of the Gaussian filter, TOF/PSF reconstruction, and matrix size followed the same pattern in H3DBP PDRP scores as seen in human subjects (see Figs. 1 and 2). Between 0 and 5 mm Gaussian smoothing filter, a median raw PDRP expression score offset of +358 was observed across all reconstructions (i.e., all combinations of matrix sizes and TOF/PSF). This is close to the +368 median offset seen in HCs and higher than the offset seen in the RBD and PD subjects. From 5 to 10 mm Gaussian smoothing filter, a median raw PDRP score offset of +559 was observed, which is higher than the +532 median offset seen in the HCs and higher than in the other subjects.
From TOF+/PSF+ to the TOF−/PSF− reconstructions, a median PDRP expression score offset of +394 was observed in the H3DBP scans, which is somewhat higher than the offset seen from 0 to 5 mm smoothing filters and higher than the offsets seen in human subjects. This is also in contrast to the trend seen in human subjects, where greater median score offsets were seen from 0 to 5 mm Gaussian smoothing filters relative to TOF/PSF reconstruction differences (+343 and +245, respectively).
From matrix size of 400 to 256, a median score offset of +130 was seen in the H3DBP scans across all reconstructions (i.e. all combinations of 0/5/10 mm Gaussian smoothing filter and TOF/PSF). This was slightly higher than the +115 offset seen in HCs.
Overall, the H3DBP had a greater score spread between the reconstruction representing its lowest PDRP expression score (0 mm, TOF+/PSF+, matrix size 400) and the reconstruction representing its highest score (10 mm, TOF−/PSF−, matrix size 256), 1386 points, than that seen in human subjects.
3.2.2. Comparing Siemens, GE, and two Philips PET/CT systems at different centers
In addition, we found large differences in H3DBP raw PDRP scores on different PET/CT scanners (Fig. 3). The Philips Ingenuity PET scanner had the lowest (i.e. most negative) H3DBP PDRP scores across all variations of its standard reconstruction protocols, followed by the Siemens Biograph scanner; the score ranges for the reconstructions between these two scanners largely overlapped. Most of the GE710 PDRP expression scores were significantly higher (i.e. less negative) than those of the Philips Ingenuity or Siemens Biograph, with only the lowest GE710 scores and the highest Philips Ingenuity and Siemens Biograph scores overlapping. Median Philips Vereos scanner PDRP expression scores were higher than the GE710 scores, mostly overlapping with the GE710 range but showing no overlap with either the Philips Ingenuity or the Siemens Biograph scanner H3DBP PDRP score range. The lowest raw PDRP score attainable on the Philips Vereos was −818, whereas the highest (i.e. least negative) PDRP scores attainable on the Philips Ingenuity and Siemens Biograph were −945 and −960, respectively.
Similarly to the human subjects scanned on the Siemens Biograph mCT64 PET/CT system, increasing Gaussian smoothing filter from 0 to 5 mm and from 5 to 10 mm was found to systematically increase H3DBP PDRP scores in the Siemens Biograph, GE710, and the Philips Vereos and Ingenuity scanners. The offset from 5 to 10 mm full-width at half-maximum smoothing filter was also greater than between 0 and 5 mm across all systems.
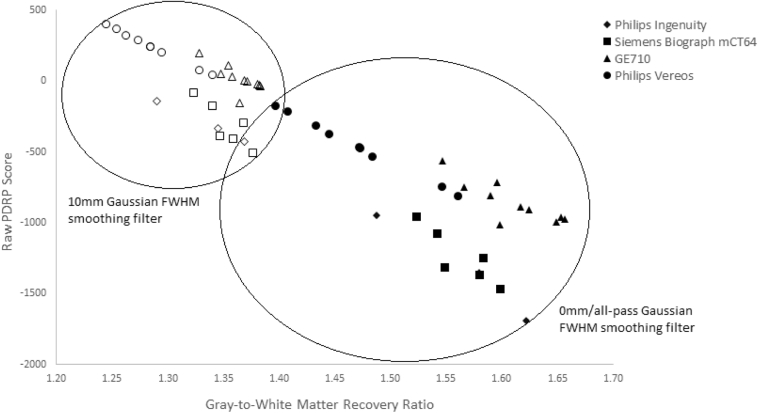
3.2.3. Gray-to-white matter recovery ratios in different PET systems
The gray-to-white matter ratios were calculated for the H3DBP on the four scanners. Independent of the scanner or reconstruction, raw PDRP score expression was inversely correlated with the gray-to-white matter ratio (Fig. 4). Both Philips scanners showed the highest correlations between gray-to-white matter ratios and the H3DBP raw PDRP scores, with both having R2 of over 0.97, whereas the GE scanner showed the lowest correlation, with an R2 of approximately 0.69. Nonetheless, the GE710 had a similar gray-to-white matter ratio range to the Philips Ingenuity and Siemens Biograph scanners while having significantly higher median raw PDRP scores. The Philips Vereos had lower gray-to-white matter ratios than the other scanners, regardless of reconstruction.
Fig. 4.
H3DBP gray-to-white matter recovery ratios versus raw PDRP expression scores on four different PET/CT systems. Multiple data points per scanner correspond to multiple reconstructions (i.e. TOF+/PSF+, TOF−/PSF−, etc.), just as inFig. 3. Abbreviations: H3DBP, Hoffman 3D Brain Phantom; PDRP, Parkinson's disease–related pattern; PET, positron emission tomography; CT, computed tomography.
In addition, it was found that when a 10 mm Gaussian smoothing filter was applied to the scans, the gray-to-white matter contrast systematically decreased. We also observed that the spread of gray-to-white matter ratios and PDRP scores decreased while maintaining intercenter distinctions (Fig. 4).
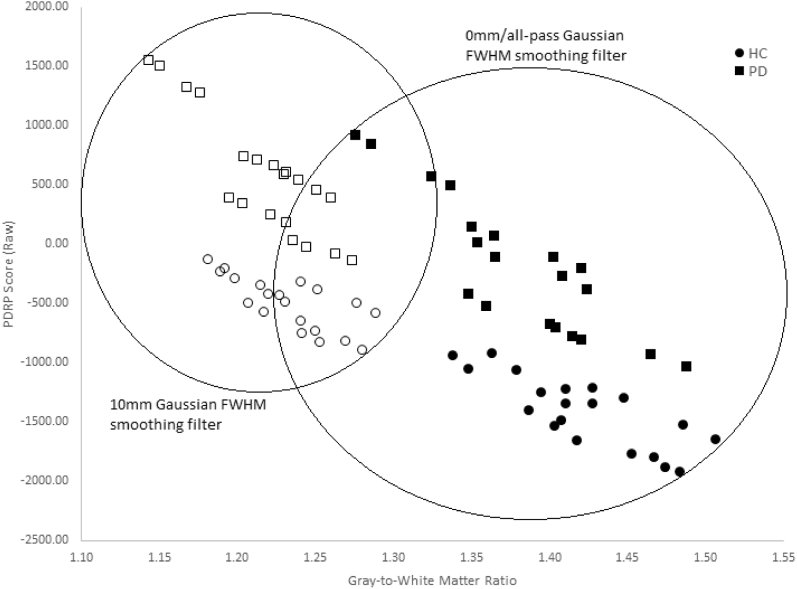
A similar trend among the HC and PD subjects from Section 3.1 was observed (Fig. 5); the highest gray-to-white matter ratios and lowest raw PDRP scores corresponded to the HC subjects, whereas the lowest gray-to-white matter ratios and highest raw PDRP scores corresponded to the patients with PD. Furthermore, application of a 10 mm Gaussian smoothing filter decreased gray-to-white matter contrast systematically. The spread of the gray-to-white matter ratios and raw PDRP scores was also reduced while maintaining a separation between the HC and PD subjects.
Fig. 5.
HC and PD gray-to-white matter ratios versus raw PDRP expression scores. Five HCs and 5 PD patients in four reconstructions each (TOF+/PSF+ with matrix size 256, TOF+/PSF+ with matrix size 400, TOF−/PSF− with matrix size 256, and TOF−/PSF− with matrix size 400) on the Siemens Biograph mCT64 at the UMCG in the Netherlands. Abbreviations: PDRP, Parkinson's disease–related pattern; TOF, time of flight; PSF, point spread function; HC, healthy control; PD, Parkinson's disease.
4. Discussion
To the best of our knowledge, no other study has directly investigated raw PDRP expression score differences in human subjects and the H3DBP caused by reconstruction and PET scanner differences. Our study showed that PDRP raw scores are systematically influenced by a variety of PET/CT system and reconstruction-related factors. On the UMCG Siemens Biograph mCT64 scanner, offsets in human subject and H3DBP raw PDRP expression scores occurred as a result of differences in reconstruction protocols with regard to the Gaussian smoothing filter, TOF/PSF reconstruction, and matrix (or voxel) size. In addition, significant PDRP score offsets were seen in H3DBP scans on different PET systems with a variety of reconstructions, with the Philips Ingenuity and Siemens Biograph scanners having the lowest median raw PDRP scores and the Philips Vereos system having the highest.
Higher Gaussian smoothing filters consistently raised PDRP expression scores across all scanner and reconstruction settings tested; increasing the smoothing filter from 5 to 10 mm led to larger score offsets relative to increasing the filter from 0 to 5 mm. Furthermore, subject scans with intrinsically lower (i.e. negative) raw PDRP expression scores, such as HC scans, showed greater score offsets as a result of reconstruction setting differences than PD subject scans with higher intrinsic (i.e. less negative, or positive) raw PDRP expression scores. The H3DBP, being within the HC range, followed this trend as well. This suggests that simple linear score corrections may not be an adequate solution when compensating for large intercenter differences.
Gray-to-white matter ratios were shown to be inversely correlated to raw PDRP expression scores on different PET scanners, with the strongest association seen in the two Philips scanners. The results suggest that 0 mm (“all-pass”) Gaussian smoothing with TOF+/PSF+ reconstruction and matrix size of 400, all factors which lead to lower PDRP expression, also lead to higher image contrast. Conversely, the 10 mm Gaussian smoothing filter with TOF−/PSF− reconstruction and matrix size of 256 lead to higher expression of the PDRP together with lower image contrast. A similar trend in gray-to-white matter ratios was observed in our human subjects, with HCs (with the lowest, most negative PDRP expression scores) displaying the highest gray-to-white matter ratios and patients with PD (with the highest, least negative or positive PDRP expression scores) exhibiting the lowest gray-to-white matter ratios.
Our results illustrate that a substantial problem exists, which prevents raw PDRP scores derived from FDG-PET scans produced in different centers from being directly comparable with each other at face-value. This impedes efforts to properly implement the PDRP and other validated disease-related patterns as robust imaging biomarkers for disease progression in multicenter and international studies. As shown by Tomše et al., accurate and comparable differential diagnosis among multiple subject groups is possible, provided that either the PDRP used is derived using the same methods as the subject groups tested or that appropriate calibration to a local HC cohort is applied [25], [26].
The advantage of this study in comparison to the ones by Tomše et al. is that raw PDRP expression scores were used. This way, scores can be directly compared and extraneous human variations in age, gender, ethnicity, and other potential factors, which may exist between different HC groups—and which may impede getting comparable z-scores from different centers—are eliminated. Once there is a standardized protocol in place to harmonize raw PDRP expression scores between different centers, in principle, it will be possible to z-score the raw PDRP scores of one center to the HC cohort of another. This is a necessary step as many centers, particularly small or non-university affiliated hospitals, may have practical barriers to assembling their own HC cohorts.
Finally, we were able to rule out histogram scaling differences between scanners as a cause of PDRP score shifts between centers (results not shown). This is due to the SSM process removing scan scaling differences with the use of a log transformation followed by a within-subject centering of image intensity values before the application of a principal component analysis [12]. As a result, we were able to narrow in on image noise and especially contrast differences as a major culprit in PDRP score shifts.
We found that applying a higher Gaussian smoothing filter (thereby masking image noise and reducing overall contrast levels in the scan) systematically decreased overall H3DBP PDRP score variability between centers while maintaining intercenter distinctions (Fig. 4), something which has not been demonstrated previously. An analogous result was shown in HC and PD subjects as well (Fig. 5). However, it also resulted in an overall PDRP expression increase across all H3DBP and human subject scans. This is logical considering that in the neurodegenerative disease process, a decrease in global glucose uptake is to be expected over time. Because the majority of glucose uptake occurs in the gray matter as opposed to the white, the gray-to-white matter contrast ratio will naturally decrease while disease-related pattern expression score increases with disease progression.
Some of the limitations of this study include the small size of the HC, RBD, and PD groups, presented here for proof of concept. In addition, all of the raw PDRP expression scores calculated in this study were based on the UMCG's established PDRP, which was determined from 17 HC and 19 PD subject scans [14], [19]. We did not study how PDRPs derived from different HC and PD cohorts scanned at different centers would impact the PDRP expression scoring. Furthermore, we only examined a limited selection of image reconstructions. We did not investigate clinical scanning protocol differences between centers, which may introduce additional variations in scan data, such as uptake time, injected activity, and factors which would not be possible to test on a H3DBP, for instance blood glucose levels or whether eyes are open or closed during patient preparation for the scan (See Supplementary Material B). Efforts to harmonize PDRP expression scores in different centers will have to produce a standardized clinical scanning protocol as well.
The H3DBP had scores within the HC (and RBD) range when tested for PDRP expression, but it is not clear which level of expression the H3DBP will have for other neurodegenerative disease-related patterns or compared to other age groups/demographics. Perhaps it will be of value to consider developing classic “diseased” brain phantoms based on validated disease-related patterns to refine score offset corrections. In addition, there is a high likelihood that disease-related pattern scores derived from HC and patient scans are influenced by additional factors such as age, gender, and perhaps other factors as well. It may be necessary to examine a critical mass of HCs in order to be able to derive “gold standard” benchmarks of disease-related pattern expression based on these variables.
Based on the above, simple linear PDRP score correction does not appear to offer a satisfactory solution. Consequently, we have begun applying machine learning techniques to overcome scan variability between centers. In a preliminary investigation by the University of Groningen–Department of Mathematics, HC cohorts from three European centers were compared: virtually all of the subjects could be classified on the basis of scanner origin using brain FDG spatial covariance information [41]. We are hopeful that it will be possible to use the distinctive features detected in scans from different PET systems to correct for intercenter variability.
In addition, minimizing some PET/CT system and image reconstruction-related differences in PDRP scores could also be achieved by implementing a multicenter calibration and quality-control program aimed at harmonizing image quality and contrast. This could further improve the performance of SSM/PCA analysis and assessment of disease-related pattern expression without loss of spatial detail. This may be of importance for disease-related patterns in certain neurodegenerative diseases which affect relatively small brain structures such as the hippocampus.
5. Conclusion
Raw PDRP expression scores in human subjects and the H3DBP were systematically influenced by variations in PET scanners and reconstruction parameters, impeding the ability to compare scores from different centers at face-value. These results reaffirm the need for the development of a harmonization protocol. One common underlying factor between various scanners and reconstructions which consistently, inversely correlated to raw PDRP expression scores was image contrast. Therefore, we believe that image contrast will be an important factor to take into account in the future development of a harmonization protocol. In addition, employing an optimal Gaussian smoothing filter may help mask noise and diminish intercenter differences without sacrificing sensitivity. Because we see a similar pattern of reconstruction effects in both human subject and H3DBP scans, the H3DBP will in all likelihood represent the most practical and standardized way forward to correct for PDRP score offsets between different scanners and reconstruction protocols. This would also circumvent the need for each center to collect their own HC cohorts for z-scoring. Furthermore, we recommend examining machine learning methods as a possible way to correct for intrinsic scanner differences.
Research in Context.
-
1.
Systematic review: In May 2017, an EU Joint Programme - Neurodegenerative Disease Research (JPND) symposium was held in Madrid to address the lack of standardized [18F]FDG-PET neuroimaging protocols and its effect on uni- and multivariate disease-related brain pattern expression quantification. Previous studies have recognized the impact of certain factors on image quality and resulting biomarker expression; however, a more thorough examination was still needed.
-
2.
Interpretation: Different PET/CT scanners and reconstruction algorithms systematically influenced multivariate Parkinson’s disease-related pattern (PDRP) expression in human subjects and the Hoffman 3D Brain Phantom (H3DBP). Image contrast inversely correlated to PDRP expression, and a Gaussian spatial filter reduced contrast while decreasing intercenter differences.
-
3.
Future directions: Further investigation into optimal harmonized data acquisition and reconstruction methods is necessary to achieve adequate intercenter data comparability. The H3DBP will be important for assessing and correcting for these intercenter effects. Machine-learning methods for applying such corrections should also be explored.
Acknowledgments
This study was funded by Dutch “Stichting ParkinsonFonds” and the EU Joint Program – Neurodegenerative Disease Research (JPND): Project PETMETPAT.
Footnotes
The authors report no conflicts of interest.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.dadm.2019.04.002.
Supplementary Data
References
- 1.Perani D. FDG-PET and amyloid-PET imaging: The diverging paths. Curr Opin Neurol. 2014;27:405–413. doi: 10.1097/WCO.0000000000000109. [DOI] [PubMed] [Google Scholar]
- 2.Albert M.S., DeKosky S.T., Dickson D., Dubois B., Feldman H.H., Fox N.C. The diagnosis of mild cognitive impairment due to Alzheimer's disease: Recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7:270–279. doi: 10.1016/j.jalz.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McKeith I.G., Dickson D.W., Lowe J., Emre M., O'Brien J.T., Feldman H. Diagnosis and management of dementia with lewy bodies: Third report of the DLB consortium. Neurology. 2005;65:1863–1872. doi: 10.1212/01.wnl.0000187889.17253.b1. [DOI] [PubMed] [Google Scholar]
- 4.Garibotto V., Herholz K., Boccardi M., Picco A., Varrone A., Nordberg A. Clinical validity of brain fluorodeoxyglucose positron emission tomography as a biomarker for Alzheimer's disease in the context of a structured 5-phase development framework. Neurobiol Aging. 2017;52:183–195. doi: 10.1016/j.neurobiolaging.2016.03.033. [DOI] [PubMed] [Google Scholar]
- 5.Meles S.K., Teune L.K., de Jong B.M., Dierckx R.A., Leenders K.L. Metabolic imaging in parkinson disease. J Nucl Med. 2017;58:23–28. doi: 10.2967/jnumed.116.183152. [DOI] [PubMed] [Google Scholar]
- 6.Nobili F., Westman E., Kogan R.V., Pereira J.B., Massa F., Grazzini M. Clinical utility and research frontiers of neuroimaging in movement disorders. Q J Nucl Med Mol Imaging. 2017;61:372–385. doi: 10.23736/S1824-4785.17.03018-7. [DOI] [PubMed] [Google Scholar]
- 7.Perani D., Schillaci O., Padovani A., Nobili F.M., Iaccarino L., Della Rosa P.A. A survey of FDG- and amyloid-PET imaging in dementia and GRADE analysis. Biomed Res Int. 2014;2014:785039. doi: 10.1155/2014/785039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frisoni G.B., Bocchetta M., Chetelat G., Rabinovici G.D., de Leon M.J., Kaye J. Imaging markers for Alzheimer disease: which vs how. Neurology. 2013;81:487–500. doi: 10.1212/WNL.0b013e31829d86e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyer P.T., Frings L., Rucker G., Hellwig S. (18)F-FDG PET in parkinsonism: differential diagnosis and evaluation of cognitive impairment. J Nucl Med. 2017;58:1888–1898. doi: 10.2967/jnumed.116.186403. [DOI] [PubMed] [Google Scholar]
- 10.Eckert T., Tang C., Ma Y., Brown N., Lin T., Frucht S. Abnormal metabolic networks in atypical parkinsonism. Mov Disord. 2008;23:727–733. doi: 10.1002/mds.21933. [DOI] [PubMed] [Google Scholar]
- 11.Eidelberg D. Metabolic brain networks in neurodegenerative disorders: a functional imaging approach. Trends Neurosci. 2009;32:548–557. doi: 10.1016/j.tins.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spetsieris P., Ma Y., Peng S., Ko J.H., Dhawan V., Tang C.C. Identification of disease-related spatial covariance patterns using neuroimaging data. J Vis Exp. 2013 doi: 10.3791/50319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma Y., Tang C., Spetsieris P.G., Dhawan V., Eidelberg D. Abnormal metabolic network activity in Parkinson's disease: test-retest reproducibility. J Cereb Blood Flow Metab. 2007;27:597–605. doi: 10.1038/sj.jcbfm.9600358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teune L.K., Renken R.J., de Jong B.M., Willemsen A.T., van Osch M.J., Roerdink J.B. Parkinson's disease-related perfusion and glucose metabolic brain patterns identified with PCASL-MRI and FDG-PET imaging. Neuroimage Clin. 2014;5:240–244. doi: 10.1016/j.nicl.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meles S.K., Tang C.C., Teune L.K., Dierckx R.A., Dhawan V., Mattis P.J. Abnormal metabolic pattern associated with cognitive impairment in Parkinson's disease: a validation study. J Cereb Blood Flow Metab. 2015;35:1478–1484. doi: 10.1038/jcbfm.2015.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niethammer M., Eidelberg D. Metabolic brain networks in translational neurology: concepts and applications. Ann Neurol. 2012;72:635–647. doi: 10.1002/ana.23631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teune L.K., Strijkert F., Renken R.J., Izaks G.J., de Vries J.J., Segbers M. The Alzheimer's disease-related glucose metabolic brain pattern. Curr Alzheimer Res. 2014;11:725–732. doi: 10.2174/156720501108140910114230. [DOI] [PubMed] [Google Scholar]
- 18.Teune L.K., Renken R.J., Mudali D., De Jong B.M., Dierckx R.A., Roerdink J.B. Validation of Parkinsonian disease-related metabolic brain patterns. Mov Disord. 2013;28:547–551. doi: 10.1002/mds.25361. [DOI] [PubMed] [Google Scholar]
- 19.Meles S.K., Vadasz D., Renken R.J., Sittig-Wiegand E., Mayer G., Depboylu C. FDG PET, dopamine transporter SPECT, and olfaction: combining biomarkers in REM sleep behavior disorder. Mov Disord. 2017;32:1482–1486. doi: 10.1002/mds.27094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meles S.K., Renken R.J., Janzen A., Vadasz D., Pagani M., Arnaldi D. The metabolic pattern of idiopathic REM sleep behavior disorder reflects early-stage Parkinson disease. J Nucl Med. 2018;59:1437–1444. doi: 10.2967/jnumed.117.202242. [DOI] [PubMed] [Google Scholar]
- 21.Meles S.K., Pagani M., Arnaldi D., De Carli F., Dessi B., Morbelli S. The Alzheimer's disease metabolic brain pattern in mild cognitive impairment. J Cereb Blood Flow Metab. 2017;37:3643–3648. doi: 10.1177/0271678X17732508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schindlbeck K.A., Eidelberg D. Network imaging biomarkers: Insights and clinical applications in Parkinson's disease. Lancet Neurol. 2018;17:629–640. doi: 10.1016/S1474-4422(18)30169-8. [DOI] [PubMed] [Google Scholar]
- 23.Huang C., Tang C., Feigin A., Lesser M., Ma Y., Pourfar M. Changes in network activity with the progression of Parkinson's disease. Brain. 2007;130:1834–1846. doi: 10.1093/brain/awm086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ikari Y., Akamatsu G., Nishio T., Ishii K., Ito K., Iwatsubo T. Phantom criteria for qualification of brain FDG and amyloid PET across different cameras. EJNMMI Phys. 2016;3:23. doi: 10.1186/s40658-016-0159-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomse P., Jensterle L., Rep S., Grmek M., Zaletel K., Eidelberg D. The effect of 18F-FDG-PET image reconstruction algorithms on the expression of characteristic metabolic brain network in Parkinson's disease. Phys Med. 2017;41:129–135. doi: 10.1016/j.ejmp.2017.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomse P., Peng S., Pirtosek Z., Zaletel K., Dhawan V., Eidelberg D. The effects of image reconstruction algorithms on topographic characteristics, diagnostic performance and clinical correlation of metabolic brain networks in parkinson's disease. Phys Med. 2018;52:104–112. doi: 10.1016/j.ejmp.2018.06.637. [DOI] [PubMed] [Google Scholar]
- 27.Pilotto A., Premi E., Paola Caminiti S., Presotto L., Turrone R., Alberici A. Single-subject SPM FDG-PET patterns predict risk of dementia progression in Parkinson disease. Neurology. 2018;90:e1029–e1037. doi: 10.1212/WNL.0000000000005161. [DOI] [PubMed] [Google Scholar]
- 28.Habeck C., Foster N.L., Perneczky R., Kurz A., Alexopoulos P., Koeppe R.A. Multivariate and univariate neuroimaging biomarkers of Alzheimer's disease. Neuroimage. 2008;40:1503–1515. doi: 10.1016/j.neuroimage.2008.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Habeck C., Stern Y., Alzheimer's Disease Neuroimaging Initiative Multivariate data analysis for neuroimaging data: Overview and application to Alzheimer's disease. Cell Biochem Biophys. 2010;58:53–67. doi: 10.1007/s12013-010-9093-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scarmeas N., Habeck C.G., Zarahn E., Anderson K.E., Park A., Hilton J. Covariance PET patterns in early Alzheimer's disease and subjects with cognitive impairment but no dementia: utility in group discrimination and correlations with functional performance. Neuroimage. 2004;23:35–45. doi: 10.1016/j.neuroimage.2004.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friston K.J., Holmes A.P., Worsley K.J., Poline J.-P., Frith C.D., Frackowiak R.S.J. Statistical parametric maps in functional imaging: A general linear approach. Hum Brain Mapp. 1994;2:189–210. [Google Scholar]
- 32.Perani D., Cerami C., Caminiti S.P., Santangelo R., Coppi E., Ferrari L. Cross-validation of biomarkers for the early differential diagnosis and prognosis of dementia in a clinical setting. Eur J Nucl Med Mol Imaging. 2016;43:499–508. doi: 10.1007/s00259-015-3170-y. [DOI] [PubMed] [Google Scholar]
- 33.Perani D., Della Rosa P.A., Cerami C., Gallivanone F., Fallanca F., Vanoli E.G. Validation of an optimized SPM procedure for FDG-PET in dementia diagnosis in a clinical setting. Neuroimage Clin. 2014;6:445–454. doi: 10.1016/j.nicl.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iaccarino L., Chiotis K., Alongi P., Almkvist O., Wall A., Cerami C. A cross-validation of FDG- and amyloid-PET biomarkers in mild cognitive impairment for the risk prediction to dementia due to Alzheimer's disease in a clinical setting. J Alzheimers Dis. 2017;59:603–614. doi: 10.3233/JAD-170158. [DOI] [PubMed] [Google Scholar]
- 35.Caminiti S.P., Alongi P., Majno L., Volonte M.A., Cerami C., Gianolli L. Evaluation of an optimized [(18) F]fluoro-deoxy-glucose positron emission tomography voxel-wise method to early support differential diagnosis in atypical parkinsonian disorders. Eur J Neurol. 2017;24:687.e26. doi: 10.1111/ene.13269. [DOI] [PubMed] [Google Scholar]
- 36.Caminiti S.P., Ballarini T., Sala A., Cerami C., Presotto L., Santangelo R. FDG-PET and CSF biomarker accuracy in prediction of conversion to different dementias in a large multicentre MCI cohort. Neuroimage Clin. 2018;18:167–177. doi: 10.1016/j.nicl.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caminiti S.P., Sala A., Iaccarino L., Beretta L., Pilotto A., Gianolli L. Brain glucose metabolism in lewy body dementia: Implications for diagnostic criteria. Alzheimers Res Ther. 2019;11 doi: 10.1186/s13195-019-0473-4. 20-2019-0473-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Presotto L., Ballarini T., Caminiti S.P., Bettinardi V., Gianolli L., Perani D. Validation of 18F-FDG-PET single-subject optimized SPM procedure with different PET scanners. Neuroinformatics. 2017;15:151–163. doi: 10.1007/s12021-016-9322-9. [DOI] [PubMed] [Google Scholar]
- 39.Stiasny-Kolster K., Mayer G., Schafer S., Moller J.C., Heinzel-Gutenbrunner M., Oertel W.H. The REM sleep behavior disorder screening questionnaire–a new diagnostic instrument. Mov Disord. 2007;22:2386–2393. doi: 10.1002/mds.21740. [DOI] [PubMed] [Google Scholar]
- 40.Della Rosa P.A., Cerami C., Gallivanone F., Prestia A., Caroli A., Castiglioni I. A standardized [18F]-FDG-PET template for spatial normalization in statistical parametric mapping of dementia. Neuroinformatics. 2014;12:575–593. doi: 10.1007/s12021-014-9235-4. [DOI] [PubMed] [Google Scholar]
- 41.Van Veen R., Talavera Martinez L., Kogan R.V., Meles S.K., Mudali D., Roerdink J.B.T.M. Machine learning based analysis of FDG-PET image data for the diagnosis of neurodegenerative diseases. In: Petkov N., Strisciuglio N., Travieso-González C., editors. Applications of intelligent systems - proceedings of the 1st international APPIS conference 2018, frontiers in artificial intelligence and applications 310. IOS Press; Amsterdam: 2018. pp. 280–289. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.