Abstract
South Korea was free of the Middle East Respiratory Syndrome (MERS) until 2015. The MERS outbreak in South Korea during 2015 was the largest outbreak of the Coronavirus outside the Middle East. The major characteristic of this outbreak is inter- or intra-hospital transmission. This recent MERS outbreak in South Korea is examined and assessed in this paper. The main objectives of the study is to characterize the pattern of the MERS outbreak in South Korea based on a basic reproductive ratio, the probability of ultimate extinction of the disease, and the spatio-temporal proximity of occurrence between patients. The survival function method and stochastic branching process model are adapted to calculate the basic reproductive ratio and the probability of ultimate extinction of the disease. We further investigate the occurrence pattern of the outbreak using a spatio-temporal autocorrelation function.
Keywords: Middle East Respiratory Syndrome, Basic reproductive ratio, Probability of ultimate extinction, Autocorrelation
1. Introduction
Mathematical epidemic models are crucial tools to understand, analyze, predict and control infectious diseases. However epidemic modeling has been a long standing challenge because of its complexity. The epidemiology of communicable diseases such as hepatitis B, tuberculosis, influenza and HIV involves an interplay between the nature of infectious organisms, evolving and mutating over time, and their transmission dynamics through direct or indirect individual contacts. Therefore, it requires both biological and sociological perspectives when mathematically modeling the outbreak.
1.1. Previous works
Early contributions to infectious disease modeling were pioneered mostly by public health physicians. The first known result in mathematical epidemiology was to appraise the effectiveness of a controversial inoculation against smallpox in 1760 by a mathematician, Daniel Bernoulli (1766). In 1889, P. D. En'ko contributed to the development of theoretical basis of epidemiology (Dietz, 1988), correlating a discrete time model with actual cases of a measles epidemic, and Hamer developed a discrete time model to understand the spread of the measles epidemic in 1906 (Hamer, 1906). Ross assumed that the rate of a new infection was proportional to the numbers of susceptible and infectious individuals, and developed the ordinary differential equation models of vector-borne diseases to first model malaria in 1911 (Ross, 1911). However, one of the early triumphs in mathematical epidemiology was the formulation of a simple model by Kermack and McKendrick in 1927 (Kermack & McKendrick, 1932; McKendrick, 1926). Their predictions explained the behavior of countless epidemics, including one of the worst epidemics in history, known as the Great Plague of London, that dicimated more than of London's population in 1665–1666. The Kermack and McKendrick epidemic model is a standard SIR model, consisting of three compartments that are Susceptible (S), Infected (I) and Recovered (R), and assumes that the sizes of the compartments are large enough so that each compartment is assumed to be homogeneous, or at least there is homogeneous mixing in each subgroup if the population is stratified by activity levels. The compartment model has been often used, modified and applied to different epidemic models (Brauer & Castillo-Chavez, 2011; Hethcote, 2000; Keeling & Rohani, 2008), as well as other phenomena in social networking, viral marketing, sensor networking, etc. However, at the beginning stage of a disease outbreak, there are a relatively small number of infected individuals, and the transmission of the infection is stochastic rather than homogeneous where the individual contacts between members of the population are distinguishable and traceable. To describe the early stage of an epidemic, stochastic branching process, first formulated by Galton and Watson in 1874, was used to model the reproduction of a population from generation to generation. It was also used to study the extinction of family names by Galton and Watson. Gets et al. (Lloyd-Smith and SchreiberGetz, 2006) introduced a natural generalization of the basic reproductive ratio that was critical in controlling disease outbreak. In the stochastic branching process model, the major interest is placed on finding the probability that the disease eventually dies out, i.e., the probability of ultimate extinction, during the transmission.
Besides the mathematical models to predict epidemics, the temporal and spatial proximity of subjects has been studied and used to characterize an epidemic that occurs through direct contacts at different places and times. Earlier, spatial patterns were analyzed to describe the geographical spread of plant and human diseases (Campbell & Noe, 1985; Nicot, Rouse, & Yandell, 1984; Snow, 1855). John Snow (Snow, 1855) greatly contributed to the early medical geography by mapping and analyzing the major cholera outbreak of London in 1854. However, it was later realized that the disease outbreak patterns needed to be explained not only spatially but also temporally. The techniques of spatio-temporal autocorrelation were introduced with an example of population diffusion in North-west England by Bennett (Bennett, 1975a, 1975b) and it was well summarized in subsequent literature (Reynolds & Madden, 1988). Recently, the spatio-temporal analysis and autocorrelation were used to analyze fatal infectious human diseases such as Dengue fever epidemics in Southern Vietnam (Cuong et al., 2013), and the spread of Severe Acute Respiratory Syndrome (SARS) in mainland China (Cao et al., 2016), etc. Cao, in the analysis of the spread of SARS in China (Cao et al., 2016), used Bayesian Maximum Entropy modeling and observed the empirical covariance based on a fitted theoretical covariance model with behavior of sine fluctuation in space and exponential decay in time.
1.2. Background of MERS outbreak in South Korea
In this section, we provide the introduction and statistical summary of the MERS outbreak in South Korea to have the general picture of the outbreak though it may be known or published earlier.
Middle East Respiratory Syndrome (MERS) is an infectious illness caused by a coronavirus that was first identified in Saudi Arabia in 2012. It resulted in a total of 2220 laboratory-confirmed cases including 790 related deaths with a case-fatality rate of as of May 2018 according to World Health Organization (WHO). South Korea was free of MERS until 2015. The outbreak of MERS in South Korea started in May 2015 and officially ended in December 2015. The index case of the outbreak in South Korea was a 68 year-old male who developed symptoms on May 11, 2015 and sought care at two outpatient clinics and two hospitals according to WHO. As a result, he exposed MERS to a number of medical staff, hospital patients, their family members and visitors, but was not diagnosed with MERS until May 20, 2015, nine days later. The last patient, diagnosed also with lymphoma, spent 172 days in quarantine after being diagnosed with MERS on June 7, 2015 and died on November 25, 2015. The Ministry of Health and Welfare (MoHW) in South Korea declared a formal end to the MERS virus outbreak on December 23, 2015, which was the 218th day since the confirmation of the first patient. The end date was achieved after 28 days without any new infection, or twice the maximum incubation period for the MERS virus. Among 186 infected patients, 39 died of MERS, equating the fatality rate of MERS in South Korea to . In this paper, we define a super-spreader as a laboratory-confirmed patient who infected the disease to more than five contacts.
The characteristics of the MERS outbreak in South Korea are that one index case who traveled to middle eastern countries introduced the disease to the population of South Korea, and the pattern of transmission is limited to inter-hospital or intra-hospital infection, except one possible case of household transmission. The size of the outbreak is much smaller than the size of the whole population in South Korea. It enables us to investigate the detailed transmission dynamics of the infectious disease. This paper is designed to make a situational assessment on the outbreak of MERS in South Korea based on the basic reproductive ratio and the probability of ultimate extinction. Furthermore, spatio-temporal proximity of the occurrence between patients is investigated using an autocorrelation function.
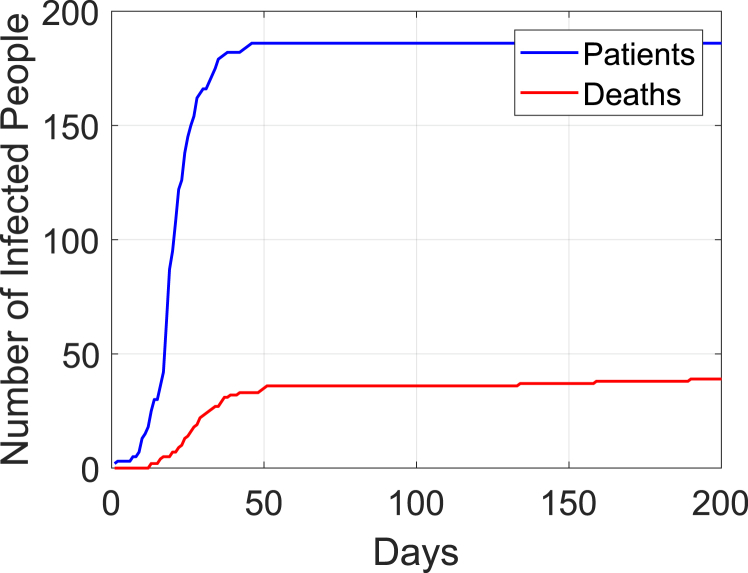
The data were publicly available by the Ministry of Health and Welfare, the Korea Centers for Disease Control and Prevention, and other press releases in South Korea. Each infected patient was numbered in chronological order based on the infection confirmation date. In this study, we count the period of the infection starting from Day 1 which corresponds to May 20, 2015. Since there is no change in epidemic status after Day 200, we use the outbreak period from Day 1 (May 20, 2015) to Day 200 (December 5, 2015) for figures in this paper. Fig. 1 displays the cumulative number of patients and deaths.
Fig. 1.
Cumulative number of infected patients each day from Day 1 to Day 200.
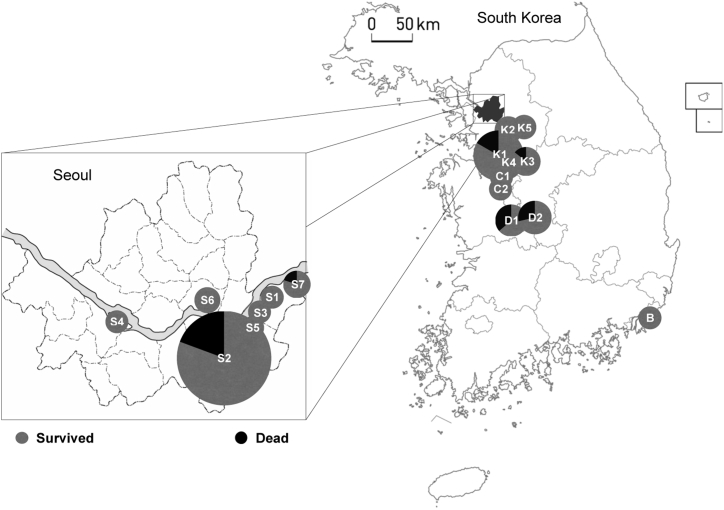
The generation is determined by how closely a patient is linked to the index case, Patient 1. The index case is the only member of Gen 0 (Generation 0) in this outbreak. Gen 1 is a group of people who were directly infected from Patient 1, Gen 2 is infected from the members of Gen 1, and so on. The MERS outbreak in South Korea ended with four generations from Gen 0 to Gen 3 with a total of 186 MERS patients. The outbreak started in Seoul, South Korea, the shaded region in Fig. 2, and spread out to the neighbor provinces. There were 17 hospitals where these 186 patients were confirmed with MERS. The hospitals are identified with the first letter of the province where each hospital is located and a number by random numbering within the province in Fig. 2. Table 1 provides the hospital identification and its corresponding location in latitude and longitude for geographic information, and it will be used for spatio-temporal analysis later in this paper.
Fig. 2.
MERS outbreak in South Korea. S: Seoul, K: K(G) yeonggi Province, C: Chungcheong Province, D: Daejeon, and B: Busan.
Table 1.
Hospital Identification and its corresponding location in latitude and longitude.
| Area |
Seoul |
||||||
| Hospital ID | S1 | S2 | S3 | S4 | S5 | S6 | S7 |
| Latitude | 37.54 | 37.49 | 37.53 | 37.52 | 37.50 | 37.54 | 37.55 |
| Longitude |
127.13 |
127.09 |
127.11 |
126.94 |
127.09 |
127.07 |
127.16 |
| Area |
Gyeonggi |
Chungcheong |
|||||
| Hospital ID | K1 | K2 | K3 | K4 | K5 | C1 | C2 |
| Latitude | 37.01 | 37.22 | 36.99 | 36.99 | 37.23 | 36.93 | 36.78 |
| Longitude |
127.07 |
127.08 |
127.12 |
127.09 |
127.28 |
127.04 |
127.02 |
| Area |
Daejeon |
Busan |
|||||
| Hospital ID | D1 | D2 | B | ||||
| Latitude | 36.31 | 36.31 | 35.15 | ||||
| Longitude | 127.34 | 127.37 | 129.11 | ||||
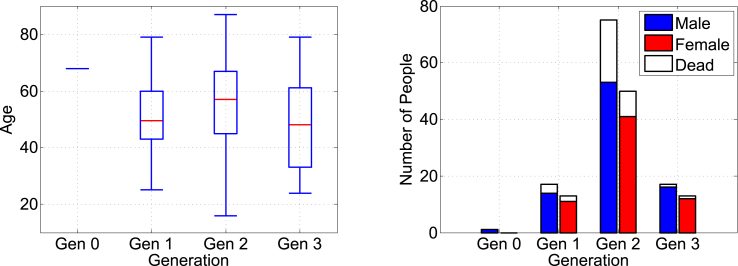
The box plot on the left in Fig. 3 displays the distribution of age, where the box represents the middle with the median in red. The median age of the entire population of patients in this MERS outbreak is 55, which is ten percent higher than the world median age (50) of MERS patients reported by the Centers for Disease Control and Prevention. The gender ratio of patients is 1.45:1, with 110 male and 76 female patients. The fatality rates are for male and for female, indicating male predominance with p-value = 0.0885 based on a one-sided test. Gen 2 has the highest rate of fatality, shown on the right in Fig. 3, with for male and for female.
Fig. 3.
Distributions of patient age (left) and gender (right) over generation.
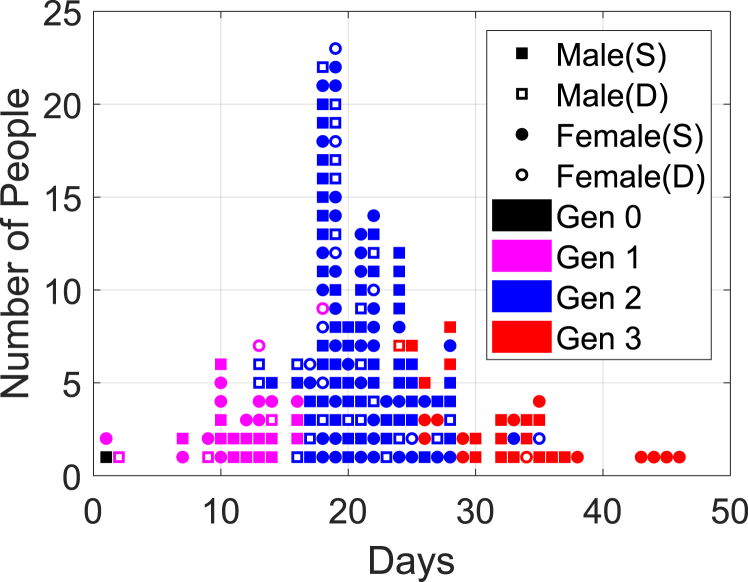
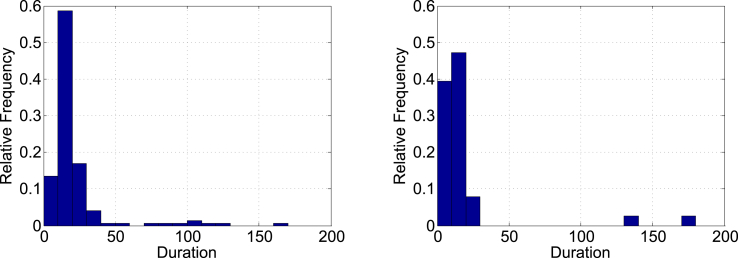
Fig. 4 summarizes the distribution of total new cases indicating gender and status where the square dots represent male and the round dots represent female, In the same figure the shaded dots indicate the survivors and the unshaded dots indicate the deceased. Each generation spanned about two weeks, which is close to the maximum incubation period for the MERS virus. The mode of the distribution in each generation is primarily determined by the time when the super-spreaders appear in the previous generation. The mode of the distribution in Fig. 4 is Day 19 in Gen 2. The distributions of infection period for both the survivors and the deceased are right-skewed as shown in Fig. 5. The median durations of infection are 15 days with the interquartile range for the survivors, and 11 days with the interquartile range for the deceased.
Fig. 4.
Summary distribution of infected cases.
Fig. 5.
Distributions of infection periods for survivors (left) and non-survivors (right).
2. Methods
2.1. Reproductive ratio and probability of extinction
The basic reproductive ratio is important since a disease outbreak is classified as a minor outbreak or a major outbreak depending on this number. A minor outbreak ultimately extinguishes itself with the probability of 1 when , while a major outbreak defines itself by the boundless spread of the disease with a positive probability when . There are more than one possible interpretations for the definition of the basic reproductive rate (R). We adopt the survival function method that is introduced in (Heesterbeek & Dietz, 1996), known as the gold standard determination of R. In this approach, the reproductive ratio is defined as
| (1) |
where is the average number of newly infected individuals whom one infectious patient will produce per unit time when infected for total time t, and is the survival function, i.e., the probability that a newly infected individual remains infectious for at least time t. The function is called the reproduction function. This approach allows us to use a time dependent instead of a fixed constant rate β.
The stochastic branching process model is often used to describe the beginning stage of a disease outbreak when the number of infected patients remains significantly smaller than the entire population. The network of individual contacts and transmission is more focused in this approach, and a network diagram of patients, consisting of two major objects: vertices and edges, is often used to visualize it. Vertices represent infected patients in our study, and edges are contacts between the members that cause transmission of the disease. We assume that every contact through each edge leads to an infection. The orientation of transmission always flows from a generation to a higher generation but never flows reversely, which means that the previous generation cannot be infected again by any subsequent generation. The degree of a vertex is the number of edges connected to the vertex in the network. For a random node with a vertex degree k, the excess degree of the vertex is defined as the number of contacts infected by the current vertex. Since the current vertex has the k connected neighbor vertices including the source node who infected the disease to the current node, and a patient cannot be infected again from its descendants as we assumed earlier, then the excess degree of the current node is . The distribution of vertex degree is essential in the description of disease transmission since it determines how fast or slow the disease spreads or dies out.
We start with the probability generating function of the neighbor distribution in a power series representation:
| (2) |
where is the probability density function for a discrete random variable X. The power series (2) converges for since this series is differentiable and . The interval is taken as the domain for since z represents probability. Let be the probability that a vertex has excess degree and consider the probability generating function of the descendent distribution in terms of :
| (3) |
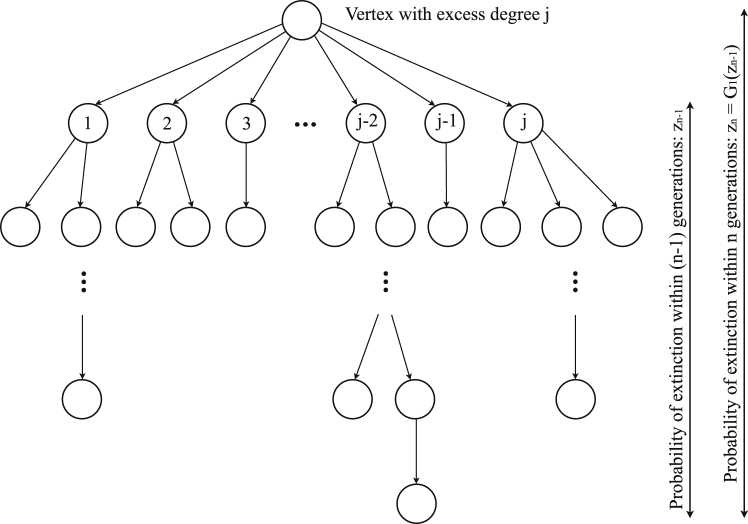
To derive the probability of ultimate extinction, let be the probability that the infection will die out within n generations. We assume that is positive, otherwise the infection will never die out, and that is non-negative for all . For a random vertex with excess degree j, the probabilities and are compared and depicted in Fig. 6. The sequence is monotonically increasing since the probability that an infection dies out within n generations is generally larger than or equal to the probability that it dies out within generations. For a certain j, the probability of ultimate extinction of the infection within n generations is the product of and . Thus the probability is the summation over all possible non-negative integers , which is the expected value of where :
| (4) |
Fig. 6.
Probability that a disease dies out within n generations from a vertex with excess degree j.
Since is a Cauchy sequence, then it converges. Let be the limit value of as n goes to infinity, . By taking the limit to Equation (4), then we have:
| (5) |
which is a fixed-point problem. It is important to know where the zeros of Equation (5) are located since it provides the probability for equilibrium when the infection eventually dies out. If is 1, then the infection will certainly extinguish, otherwise it may manifest into a major outbreak. The detailed calculation and derivation can be found in (Brauer & Castillo-Chavez, 2011).
2.2. Spatio-temporal analysis: auto-correlation
While the stochastic branching process model gathers the information of the individual transmission in each generation and predicts the probability of the ultimate extinction of the disease, it does not include both spatial and temporal information of the transmission. Autocorrelation analysis is the most straightforward way to establish direct links between two series. In this section, we adapt the autocorrelation function to analyze the spatio-temporal proximity of occurrence during a disease outbreak.
We modify the traditional definition of autocorrelation function (Reynolds & Madden, 1988) for our analysis. Let a realization indicate occurrence of a disease where if a vertex infects the disease to another vertex , otherwise . We note that the direction of transmission is always from one generation to the next generation but never in reverse. Let be the index set of vertices and be the index set of vertices that are infected by a vertex in the neighborhood of vertex for .
We define indicator functions, in space and in time, respectively, such that
where and are the spatial distance and serial interval of occurrence between two vertices and , respectively. For the sake of simplifying notation, we adopt:
where plays as an indicator if the distance between the two serial vertices and is s or not, and plays as an indicator whether the serial interval between these two vertices is t or not. For all and , we define the spatio-temporal autocorrelation function of occurrence as:
| (6) |
which describes how likely the distance and time difference between two connected vertices vary together when one vertex, , transmitted the disease to the neighbor vertices for . The autocorrelation in Equation (6) is defined differently from traditional definitions where only the fluctuation parts of variables are considered. However, we use the full variable term instead of the fluctuation part since we are interested in the number of occurrences rather than the fluctuation of occurrence.
3. Results and discussion
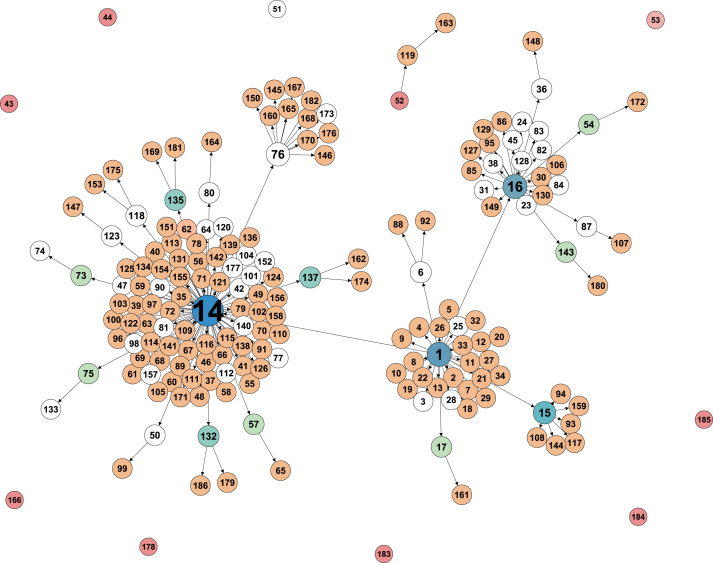
The network map for the transmission of MERS in South Korea during 2015 is shown in Fig. 7. The vertices without any shade represent the deceased. It starts from the index case (Gen 0), numbered as 1 in the map, who transmitted MERS to 29 people in the next generation, Gen 1, and so on. Since the sources of infection for Patients 43, 44, 51, 53, 166, 178, 183, 184 and 185 are unknown, we assume that their excess degree of vertex is 0. The source of infection for Patient 52 is also unknown but the excess degree of vertex is 1 as Patient 52 infected MERS to Patient 119. In Fig. 7, there are a few super-spreaders, Patients 1, 14, 15, 16 and 76, who contributed to of all active cases.
Fig. 7.
Transmission diagram of the MERS outbreak in South Korea. The nodes without shade represent the deceased. The different shades represent different out-degrees, i.e., the excess vertex degrees.
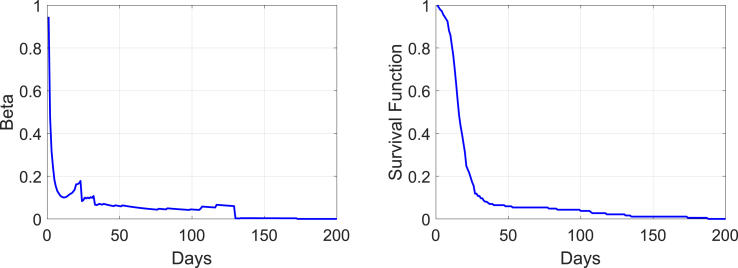
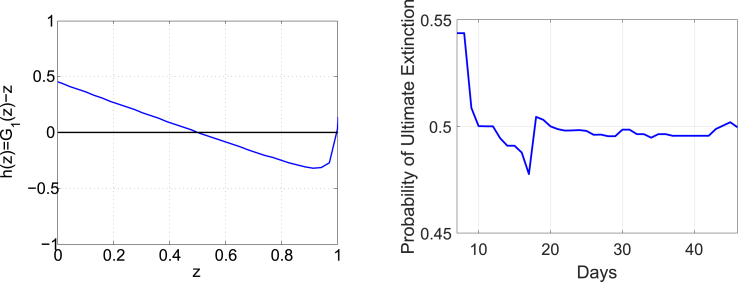
To calculate the reproductive rate for the MERS outbreak in South Korea, we use Equation (1) in a discrete form. The average number of new patients from an infectious patient per unit time (one day) and the survival function at time t are graphed in Fig. 8. The calculated reproductive ratio based on these two functions is which is considerably larger than 1 and this outbreak is classified as a major outbreak. For the probability of ultimate extinction that is the root of Equation (5), we define . The graph on the left in Fig. 9 shows and the probability of ultimate extinction for , i.e., the zero of the function less than 1, is located at . The graph on the right shows the profile of the probability of ultimate extinction over the outbreak. The probability drops but immediately increases after the appearance of super-spreaders on Days 14–16, which indicates how important successful quarantine of super-spreaders is during an epidemic.
Fig. 8.
Graphs of and the survival function in Equation (1).
Fig. 9.
The function on the left and the probability of ultimate extinction in terms of days on the right.
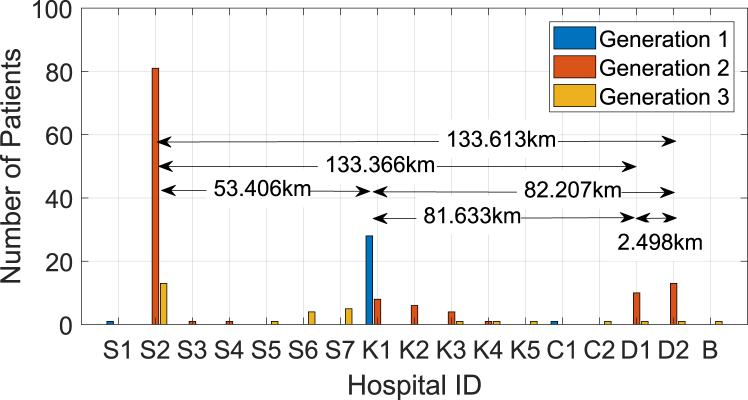
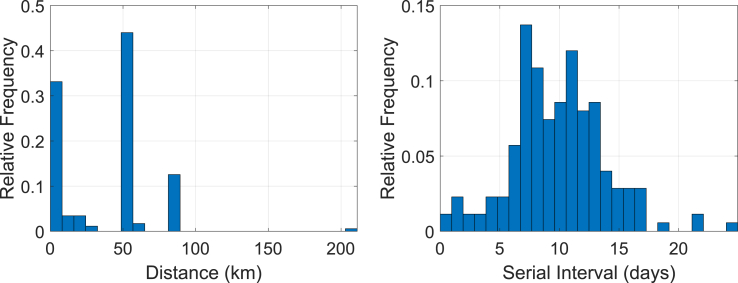
The demographical distribution of each generation over the 17 hospitals in Fig. 2 as well as in Table 1 is depicted in Fig. 10. One may expect a strong association between the size (bed counts) of the hospital and the number of MERS cases. However, we found the association to be very weak with a coefficient of determination close to zero for this particular outbreak. This explains the importance of quarantine regardless of hospital size. Our interest lies in the spatio-temporal proximity of occurrence between two patients where one patient infected MERS to the other, i.e. the two connected vertices in Fig. 7. For this purpose, we use the geographical distance and the difference of the confirmation dates (serial interval) between these paired patients. The distance is measured by the haversine formula between two hospitals in latitude and longitude in Table 1. While this Euclidean distance may not give the exact travel distance in a metropolis such as Seoul in South Korea, we assume that it is sufficient to characterize the geometrical distances between hospitals or the travel distances of patients. The distributions of the distance and the serial interval between all connected vertices in Fig. 7 are depicted in Fig. 11. As we observe in the graph on the left in Fig. 11, there are three major distances, approximately 0 km, 53 km and 81 km, that are well associated with the major distances in Fig. 10, characterizing inter-hospital or intra-hospital transmission. Most of the patients in Gen 1 are located at Hospital K1 in Fig. 10, where the index case in Gen 0 was also confirmed, contributing the highest probability at the distance of 0 km in the left graph of Fig. 11. The major hospitals in Gen 2 are S2, K1-K3 and D1-D2. Considering the three prominent distances in the graph on the left in Fig. 11, we infer that the disease was transmitted intensely between S2 and K1-K3 (about 53 km apart), and between K1-K3 and D1-D2 (about 81 km apart), rather than transmitted directly between S2 and D1-D2 (about 133 km apart), unless otherwise intra-hospital (0 km apart) transmission in Gen 2. Lastly, the major hospitals in Gen 3 are located mostly within 25 km from each other in Seoul, and the total number of patients in Gen 3 is not as high as those in other generations. The mean serial time interval is 9.8057 days with a confidence interval of (9.1975, 10.414) in the graph on the right in Fig. 11.
Fig. 10.
Number of patients confirmed at each hospital over each generation.
Fig. 11.
Distributions of the distance and serial interval between two connected vertices (patients) in Fig. 7.
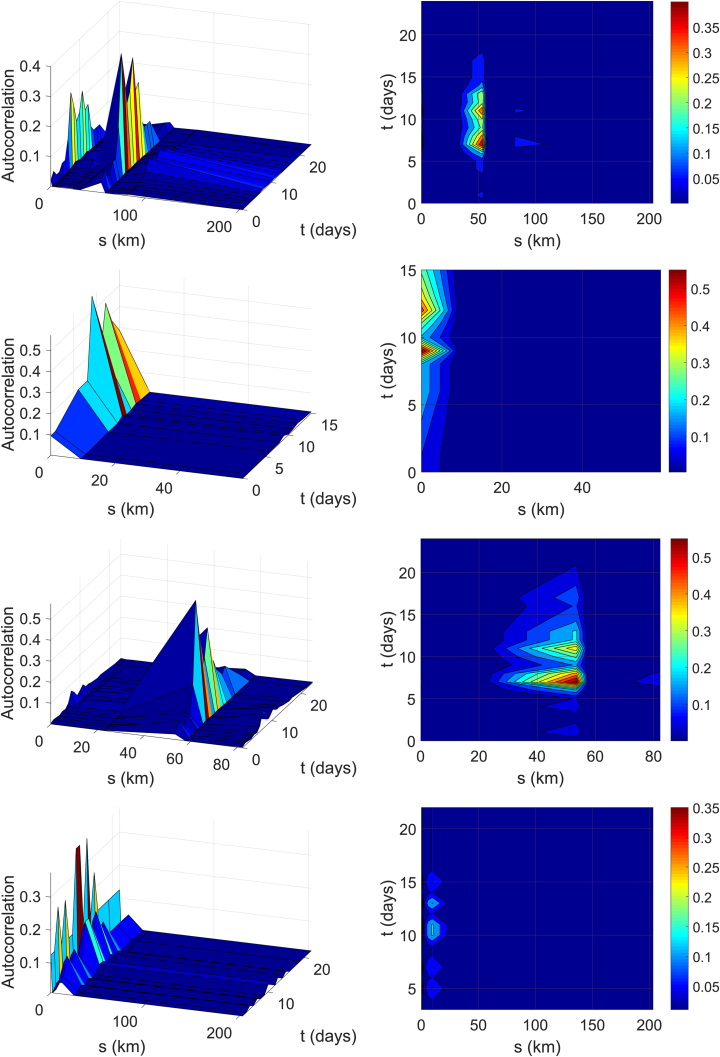
Fig. 12 demonstrates the spatio-temporal autocorrelation results of the MERS outbreak in South Korea, based on Equation (6). The first row in Fig. 12 is the profile of over the entire transmission period in the three-dimensional presentation in the left and the corresponding contour plots in -plane in the right. The second to the fourth rows in Fig. 12 are the autocorrelations for Gens 1, 2 and 3, respectively. The autocorrelation over the full domain has a few distinct spikes at certain s- and t-lags. The peaks in the spatial and temporal lag domain explains that there are a few hospitals mainly involved in the transmission, that characterizes a typical pattern of inter- and intra-hospital transmission. These patterns of the autocorrelation in spatial and temporal directions can be closely associated with Fig. 11. The transmissions in Gen 1 were mainly intra-hospital as there is very little spatial distribution. The index case was confirmed with MERS in Hospital K1 and most of Gen 1 were located at the same hospital, which creates a strong autocorrelation at as depicted in the second row in Fig. 12. In Fig. 10, we can infer that most of Gen 2 in Hospital S2 were infected by patients from Hospital K1 in Gen 1 that were located about 53 km apart. This explains the peak in the autocorrelation in Gen 2. The patients in Gen 3 were mostly infected from patients at the same hospital or nearby hospitals. To understand these graphs better, we note that the projections of the autocorrelation in the first row in Fig. 12 onto s-axis and t-axis have similar patterns with the left graph and the right graph in Fig. 11, respectively.
Fig. 12.
Spatio-temporal Autocorrelation functions for the entire period of transmission (first row), Gen 1 (second row), Gen 2 (third row) and Gen 3 (fourth row) from the top. The 3-dimensional presentation on the left and the corresponding contour figures in -plane on the right.
4. Conclusion
In this paper, we examine and assess the outbreak of MERS in South Korea based on the reproductive ratio and the probability of ultimate extinction as well as the spatio-temporal autocorrelation of occurrence.
The basic reproductive ratio is the expected number of secondary infections arising from an infectious individual over the entire epidemic. Since the estimation of the reproductive ratio is based on sufficient data acquired from the fully-completed or well established epidemic, the prediction of further development of the epidemic is somewhat limited (Mills et al., 2004). Though there are different approaches for estimating the reproductive ratio (Hefferman, Smith, & Wahl, 2005), we chose to use the survival function method that is widely applicable as there is no assumption of a constant transmission rate. To calculate the reproductive ratio, the average contact rate as a function of t is directly simulated from the data instead of using a constant rate in other works (Chang, 2017; Choi, Jung, Choi, Hur, & Ki, 2018), and the survival function is also examined from the outbreak data. The resulting reproductive ratio for the MERS outbreak in South Korea is , and the MERS outbreak in South Korea is classified as a major outbreak.
The stochastic branching process model fits well with this scenario of the MERS outbreak in South Korea since it involves a relatively small number of patients compared to the whole population of South Korea, allowing us to trace the individual contacts and gain the detailed dynamics of the transmission. Based on the transmission data, we estimate the probability of ultimate extinction by using the probability generating function and assuming the probability of extinction eventually reaches the equilibrium state. The probability of ultimate extinction of the MERS outbreak in South Korea is calculated as 0.4996.
While the reproductive ratio and the probability of ultimate extinction are useful to classify the outbreak and predict the extinction of the disease in a long term, it fails to provide any spatial or spatio-temporal aspects of the transmission. The spatial and temporal proximity of the transmission between occurrences is measured based on the autocorrelation function that describes how likely the spatial and temporal differences between patients vary together. The autocorrelation profiles the entire transmission period, as well as over each generation period, and provides good insight in understanding and characterizing the patterns of inter-hospital or intra-hospital transmission of the MERS outbreak in South Korea. We observed a few distinct peaks of autocorrelation values in the spatial and temporal domain, clearly displaying the characteristics of the MERS outbreak in South Korea.
Finally, we observe that a few super-spreaders greatly contributed to the MERS epidemic in South Korea, infecting of all active cases, and elevated the reproductive ratio. To reduce the number of super-spreaders and consequently the reproductive ratio, more thorough quarantine can be ordered. However, Lipsitch et al. (Lipsitch et al., 2003) also argued the trade-off between reducing the reproductive ratio and stressing the population with excessive quarantine, suggesting that the reproductive ratio may not always be the best measure. As a future endeavor, modeling super-spreading events with non-diffusive terms in mathematical equations will be investigated and a better quarantine plan will be suggested.
Sources of financial support
None.
Declarations of conflicts of interest
No conflict of interest.
Acknowledgements
None.
Handling Editor: J. Wu
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.idm.2019.06.002.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Bennett R.J. The representation and identification of spatio-temporal systems; an example of population diffusion in north-west england. Transactions of the Institute of British Geographers. 1975;66:73–94. [Google Scholar]
- Bennett R.J. Pion Ltd; 1975. Spatial time series. [Google Scholar]
- Bernoulli D. 1766. Essai d'une nouvelle analyse de la mortalite causée par la petite vérole, et des avantages de l’inoculation pour la prévenir; p. 1. Mém. Math. Phys. Acad. Roy. Sci., Paris. [Google Scholar]
- Brauer F., Castillo-Chavez C. Springer Science and Business Media; 2011. Mathematical models in population biology and epidemiology. [Google Scholar]
- Campbell C.L., Noe J.P. The spatial analysis of soilborne pathogens and root diseases. Annual Review of Phytopathology. 1985;23:129–148. [Google Scholar]
- Cao C., Chen W., Zheng S., Zhao J., Wang J., Cao W. Analaysis of spatiotemporal characteristics of pandemic sars spread in mainland China. BioMed Research International. 2016;(7247983):1–12. doi: 10.1155/2016/7247983. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H. Estimation of basic reproduction number of the middle east respiratory syndrome coronavirus (mers-cov) during the outbreak in South Korea. BioMedical Engineering Online. 2017;16(1) doi: 10.1186/s12938-017-0370-7. (79) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S., Jung E., Choi B.Y., Hur Y.J., Ki M. High reproduction number of middle east respiratory syndrome coronavirus in nosocomial outbreaks: Mathematical modelling in Saudi Arabia and South Korea. Journal of Hospital Infection. 2018;99(2):162–168. doi: 10.1016/j.jhin.2017.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuong H.Q., Vu N.T., Cazelles B., Boni M.F., Thai K.T.D., Rabaa M.A. Spatiotemporal dynamics of dengue epidemics, southern vietnam. Emerging Infectious Diseases. 2013;19(6):945–953. doi: 10.3201/eid1906.121323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz K. The first epidemic model: A historical note on P. D. En'ko. Australian Journal of Statistics. 1988;30(1):56–65. [Google Scholar]
- Hamer W.H. Bedford Press; 1906. Epidemic disease in England. [Google Scholar]
- Heesterbeek J.A.P., Dietz K. The concept of in epidemic theory. Statistica Neerlandica. 1996;50(1):89–110. [Google Scholar]
- Hefferman J.M., Smith R.J., Wahl L.M. Perspectives on the basic reproductive ratio. Journal of the Royal Society Interface. 2005;2(4):281–293. doi: 10.1098/rsif.2005.0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hethcote H.W. The mathematics of infectious diseases. Society for Industrial and Applied Mathematics. 2000;42(4):599–653. [Google Scholar]
- Keeling M.J., Rohani P. Princeton University Press; 2008. Modeling infectious diseases in humans and animals. [Google Scholar]
- Kermack W.O., McKendrick A.G. Contributions to the mathematical theory of epidemics. ii. the problem of endemicity. Proceedings of the Royal Society of London Series A. 1932;138(834):55–83. [Google Scholar]
- Lipsitch M., Cohen T., Cooper B., Robins J.M., Ma S., James L. Transmission dynamics and control of severe acute respiratory syndrome. Science. 2003;300(5627):1966–1970. doi: 10.1126/science.1086616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Smith J.O., Schreiber S.J., Getz W.M. Moving beyond averages: Individual-level variation in disease transmission. Contemporary Mathematics. 2006;410:235–258. [Google Scholar]
- McKendrick A.G. Applications of mathematics to medical problems. Proceedings of the Edinburgh Mathematical Society (Revised Edition) 1926;44:98–130. [Google Scholar]
- Mills C.E., Robins J.M., Lipsitch M. Transmissibility of 1918 pandemic influenza. Nature. 2004;432(7019):904–906. doi: 10.1038/nature03063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicot P.C., Rouse D.I., Yandell B.S. Comparison of statistical methods for studying spatial patterns of soilborne plant pathogens in the field. Phytopathology. 1984;74:1399–1402. [Google Scholar]
- Reynolds K.M., Madden L.V. Analaysis of epidemics using spatio-temporal autocorrelation. Phytopathology. 1988;78:240–246. [Google Scholar]
- Ross R. John Murray; London: 1911. The prevention of malaria. [Google Scholar]
- Snow J. 2nd ed. John Churchill; London: 1855. On the mode of communication of Cholera. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.