Summary
The neuronal gene repressor REST/NRSF recruits co-repressors, including CoREST, to modify histones and repress transcription. REST also functions as a tumor suppressor, but the mechanism remains unclear. We identified Chromodomain on Y-like (CDYL) as a REST co-repressor that physically bridges REST and the histone methylase G9a to repress transcription. Importantly, RNAi knockdown of REST, CDYL and G9a, but not CoREST, induced oncogenic transformation of immortalized primary human cells and derepression of the proto-oncogene TrkC. Significantly, transgenic expression of TrkC also induced transformation. This implicates CDYL-G9a, but not CoREST, in REST suppression of transformation, possibly by oncogene repression. CDYL knockdown also augments transformation in a cell culture model of cervical cancer, where loss of heterozygosity of the CDYL locus occurs. These findings demonstrate molecular strategies by which REST carries out distinct biological functions via different co-repressors, and provide critical insights into the role of histone modifying complexes in regulating cellular transformation.
Keywords: CDYL, G9a, REST, Chromatin, Histone, Transcription Regulation, Cellular Transformation, Cervical Cancer, HPV
Introduction
Regulated gene expression is critical for normal cellular function and plays a central role in developmental processes. This is achieved in large part by the activity of transcription factors that bind to gene regulatory elements, which recruit enzymatic complexes that render the chromatin environment of genes more or less favorable to transcription. Aberrations in either the transcription factors themselves, or the enzymatic complexes that mediate their activity, have been implicated in various diseases including cancer. However, the molecular mechanisms that link particular transcription regulatory complexes to specific biological and disease processes remain incompletely understood.
REST is a master negative regulator of neurogenesis that binds to conserved DNA sequences, RE1 sites, to repress gene transcription (Chong et al., 1995; Schoenherr and Anderson, 1995). Recently, unexpected roles for REST in regulating embryonic stem cell pluripotency and self-renewal and tumor suppression have also been described (Singh et al., 2008; Westbrook et al., 2005). However, it is unclear how this single transcription factor regulates apparently diverse biological processes. Gene repression by REST depends on the recruitment of multiple enzymatic co-repressor complexes that modify chromatin to repress transcription (Ballas and Mandel, 2005). These include mSin3A (Grimes et al., 2000; Huang et al., 1999), which recruits histone deacetylases (HDACs), and CoREST (Andres et al., 1999; Shi et al., 2003), which also recruits HDACs in addition to the histone demethylase, LSD1 (Shi et al., 2004). The H3K9 methyltransferase G9a is also associated with REST repression (Roopra et al., 2004), but it is not yet known how G9a is recruited to REST target promoters.
In the current study, we describe the purification of a Chromdomain on Y-like (CDYL) corepressor complex that contains REST. CDYL is a chromodomain protein that contains an unusual sequence homology with lipid metabolizing enzymes of the enoyl CoA hydratase family (Caron et al., 2003; Lahn et al., 2002). While the chromodomain of CDYL binds to methylated H3K9 residues on histones (Fischle et al., 2008 and unpublished observations), the function of the metabolic enzyme homology domain remains unclear, although it was shown to bind to HDAC1 (Caron et al., 2003). Previous work in our laboratory identified CDYL as a sub-stoichiometric component of the CtBP co-repressor supercomplex (Shi et al., 2003), and it was shown to repress heterologous gene transcription in reporter-based assays (Caron et al., 2003), consistent with a role in gene repression. However, by and large the function and mechanism of CDYL are unclear.
We have identified a CDYL complex that contains REST and the histone H3K9 methylases G9a, EuHMT1 and SETDB1, amongst others. We show that CDYL bridges the interaction between REST and G9a/EuHMT1 in vitro and in vivo, and functions as a REST co-repressor that facilitates G9a recruitment to REST target genes. Inhibition of CDYL and G9a, but not CoREST, leads to oncogenic transformation in vitro, suggesting a distinct role for both CDYL and G9a in REST suppression of cellular transformation. REST/CDYL/G9a directly represses transcription of the proto-oncogene TrkC, whose over-expression is sufficient to cause cellular transformation, suggesting that the mechanism of REST/CDYL/G9a transformation suppression may be linked to their ability to repress oncogene transcription. Finally, we provide evidence that transformation suppression by CDYL may be relevant to cervical cancer, where loss of heterozygosity (LOH) of the genetic locus encoding CDYL is frequent and correlates with poor prognosis (Arias-Pulido et al., 2004; Chatterjee et al., 2001; Krul et al., 1999). Together, these data provide new insights into CDYL function and mechanism, and support a specific role for CDYL and its associated histone methylase G9a in mediating REST tumor suppression function. These findings shed new light on molecular strategies utilized by a single eukaryotic DNA-binding transcription factor to regulate divergent biological functions.
Results and Discussion
Identification of a CDYL repressor complex
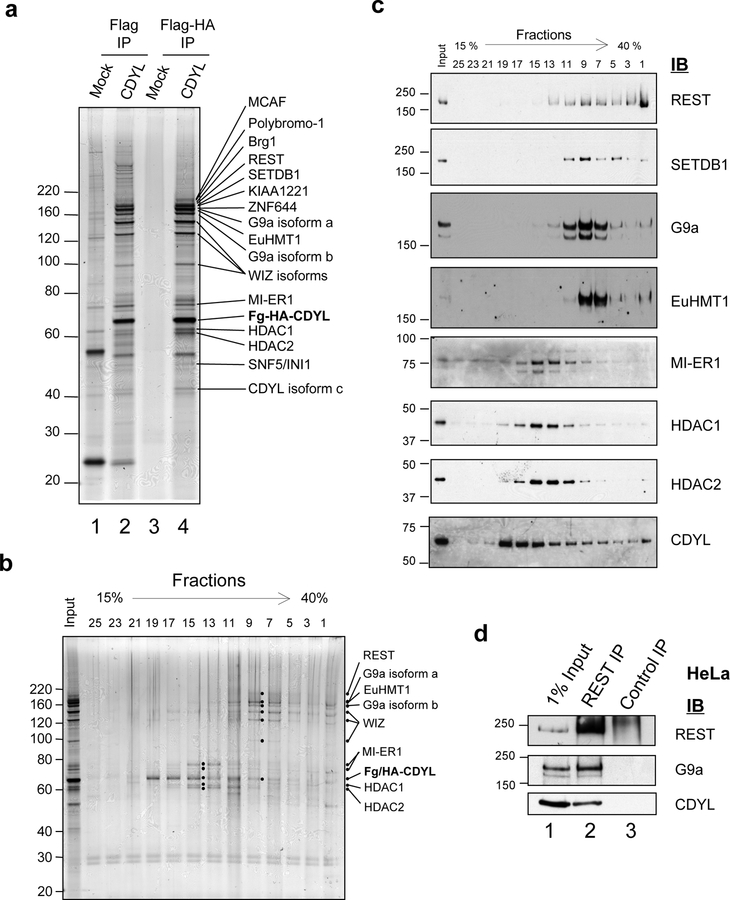
To gain insights into the mechanism and biological function of CDYL we carried out affinity purification of Flag-HA-tagged CDYL from Hela nuclear extracts to isolate CDYL-associated proteins (Fig. 1a, compare lanes 4 and 3). Mass spectrometry analysis identified over 22 associated proteins (Supplementary Table 1), the majority of which are involved in transcriptional repression. These include histone modifying enzymes, such as HDAC1 and HDAC2 (Taunton et al., 1996), G9a and EuHMT1 (Ogawa et al., 2002) and the H3K9 trimethyltransferase SETDB1 and its associated co-factor MCAF/hAM (Wang et al., 2003). Also identified was Mesoderm Induction Early Response 1 (MI-ER1) (Ding et al., 2003), a transcriptional co-repressor that binds HDAC1, is critical in Xenopus development and is aberrantly expressed in breast cancer (Ding et al., 2003), as well as its homolog MI-ER2, the function of which is unknown. Three large multi-zinc finger proteins, KIAA1221, its homolog ZNF644, and Widely Interspaced Zinc Fingers (WIZ) (Matsumoto et al., 1998), were also co-purified with CDYL. WIZ associates with G9a/EuHMT1, directly binds CtBP and represses E-cadherin gene expression (Ueda et al., 2006), while the functions of KIAA1221 and ZNF644 are unknown. We also identified the sequence-specific DNA binding factor RE1-binding Silencer of Transcription (REST)/Neuronal Specific Silencing Factor (NRSF) (Chong et al., 1995; Schoenherr and Anderson, 1995). REST is a silencer of neuronal gene transcription in non-neuronal cells and a master regulator of neuronal differentiation. Importantly, a recent report identified REST as a tumor suppressor (Westbrook et al., 2005), but the underlying mechanism is unclear.
Figure 1: CDYL forms a multiprotein complex with REST and histone H3K9 methyltransferases.
(a) Human CDYL tagged with both Flag and HA epitopes was sequentially immunoprecipitated from transduced HeLa nuclear extracts using Flag and HA antibody resins (lanes 2 and 4). Mock-transduced HeLa cells were used as a control (lanes 1 and 3). CDYL-associated proteins were resolved by SDS-PAGE and identified by mass spectrometry, as indicated on the silver stained gel shown. (b) Glycerol gradient ultracentrifugation was used to resolve CDYL-associated proteins into differentially sedimenting multiprotein complexes. Fractions were collected from the bottom of the gradient and analyzed by SDS-PAGE. Visualization of protein bands by silver staining revealed the presence of at least two CDYL sub-complexes, represented by fractions F15 and F9 respectively. The positions of CDYL, and major components of each sub-complex are indicated. (c) Immunoblotting of glycerol gradient fractions was used to confirm the identity of major components present in the two CDYL sub-complexes. (d) Antibodies against REST (lane 2), but not control IgG (lane 3), co-immunoprecipitated G9a and CDYL in the Flag-purified CDYL complex.
We next asked if REST, CDYL and the other co-purifying proteins form single or discrete sub-complexes. Glycerol gradient sedimentation analysis indicates that although a substantial fraction of purified Flag-HA-CDYL exists in the free form (Fig. 1b and c, fractions 19 – 25), at least two multiprotein sub-complexes appear to exist. A slower sedimenting complex, which peaks in fraction 15, contains MI-ER1/2, HDAC1 and HDAC2. The second, faster sedimenting complex peaks in fraction 9 and contains REST, WIZ and all three histone methyltransferases identified in the purification (Fig. 1b and c). This suggests that CDYL, REST and histone methyltransferases are likely to be in the same protein complex. We carried out co-immunoprecipitation to further examine this possibility. Importantly, both CDYL and G9a were co-immunoprecipitated with REST (Fig. 1d, compare lane 2 to lane 3) further supporting the idea that REST, CDYL and G9a are components of the same protein complex.
CDYL bridges the interaction between REST and G9a
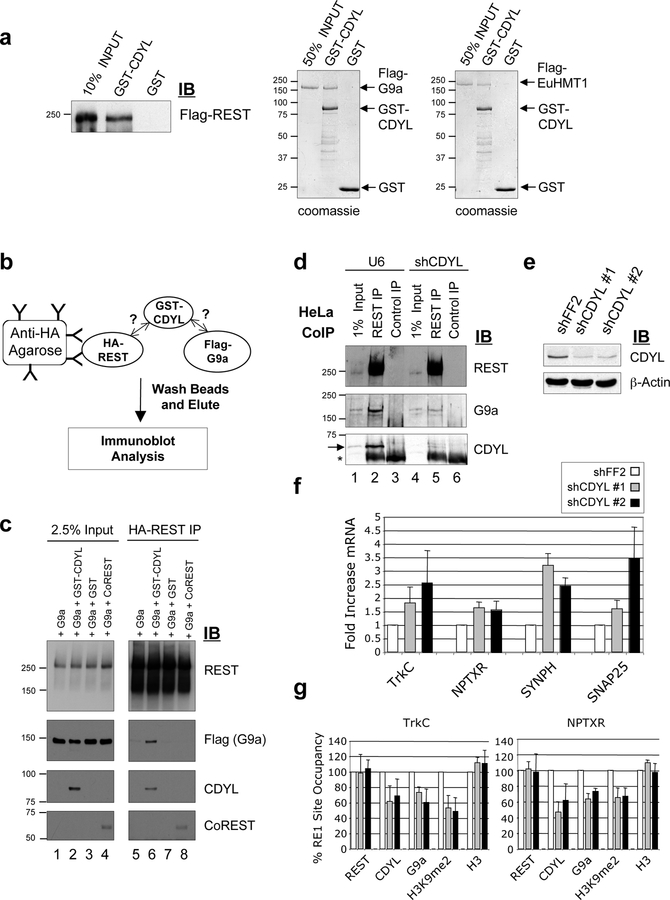
REST directly interacts with both the mSin3A and CoREST co-repressors for transcriptional repression (Andres et al., 1999; Grimes et al., 2000; Roopra et al., 2000). A recent study also showed that REST recruits G9a but it is unclear whether REST directly or indirectly interacts with G9a (Roopra et al., 2004). Interestingly, CoREST appears to be dispensable for G9a recruitment (Roopra et al., 2004). To determine whether CDYL may mediate the interaction between REST and G9a, we first carried out in vitro protein interaction assays. We found that purified GST-CDYL, but not GST alone, interacted directly with purified recombinant Flag-REST (Fig. 2a; Supp. Fig. 1). Purified recombinant G9a and EuHMT1 also interacted robustly with GST-CDYL in vitro (Fig. 2a). We then used an in vitro protein interaction assay (diagrammed in Fig. 2b) to ask if CDYL functions to bridge the interaction between REST and G9a. Purified HA-REST and Flag-G9a were incubated together in the presence or absence of GST-CDYL. The ability of HA-REST to interact with Flag-G9a was investigated by immunoprecipitating HA-REST and assaying for co-immunoprecipitation of Flag-G9a. As shown in Fig. 2c, purified REST and G9a do not interact in vitro (lane 5). However, GST-CDYL, but not GST alone or CoREST, restored binding of G9a (and EuHMT, data not shown) to REST (Fig. 2c, compare lane 6 with 7 and 8). These data strongly suggest that REST requires CDYL to bridge the interaction with G9a/EuHMT1.
Figure 2: CDYL bridges the interaction between REST and the histone methyltransferases G9a and EuHMT1 and is a co-repressor of REST target genes.
(a) Immunoblot analysis showing that purified GST-CDYL, but not GST, interacts with purified recombinant Flag-REST. Coomassie Blue stained SDS-PAGE gels showed that purified recombinant Flag-G9a and Flag-EuHMT1 also specifically interacted with GST-CDYL. The location of Flag-G9a and Flag-EuHMT1 on the gel is indicated. (b) Schematic diagram of the CDYL bridging assay to determine if REST interaction with G9a/EuHMT1 depends on CDYL. (c) CDYL bridging assay. Purified recombinant proteins were used to show that CDYL is required for the interaction of REST and G9a in vitro. Anti-HA resin was used to immunoprecipitate HA-REST and co-immunoprecipitation of purified Flag-G9a detected by immunoblot analysis. REST did not interact directly with G9a in vitro (lane 5). However, addition of GST-CDYL induced association of REST and G9a (lanes 6), but not GST alone (lane 7) or CoREST (lane 8). (d) REST was immunoprecipitated from control (U6) or CDYL RNAi HeLa nuclear extracts and assayed for co-immunoprecipitation of G9a by immunoblotting. Co-immunoprecipitation of G9a with REST was significantly diminished upon CDYL knockdown (compare lanes 2 and 5). Non-specific IgG did not immunoprecipitate REST, G9a or CDYL (lanes 3 and 6). The position of the CDYL band is indicated by an arrow and the IgG heavy chain by an asterisk. (e) Immunoblot showing knockdown of CDYL in TLM-HMEC by two independent shCDYL hairpins. RNAi directed against firefly luciferase, shFF2, was used as a control. (f) Knockdown of CDYL in TLM-HMEC derepresed of a number of REST target genes, as determined by quantitative RT-PCR. Samples were normalized to GAPDH expression levels and expressed as fold increase relative to control shRNA-treated cells (shFF2). Shown is the mean +/− s.d. of three independent assays. (g) Knockdown of CDYL in TLM-HMEC resulted in decreased occupancy of REST binding sites at the TrkC and NPTXR genes by CDYL, G9a and H3K9me2. Levels of REST and H3 occupancy were not affected. Samples were normalized to input chromatin and expressed as percentage occupancy relative to control shRNA-treated cells (shFF2). Shown is the mean +/− s.d. of three independent assays.
We then investigated whether CDYL mediates the interaction between REST and G9a in vivo. Although REST antibodies immunoprecipitated comparable amounts of REST from both control (U6) and shCDYL nuclear extracts (Fig. 2d and data not shown), co-immunoprecipitation of G9a was significantly diminished in the shCDYL extracts (Fig. 2d, compare lanes 2 and 5). This suggests that CDYL also mediates the interaction between endogenous REST and G9a in vivo. CDYL also contains a conserved N-terminal chromodomain that binds to methylated-H3K9 (Fischle et al. 2008 and unpublished observations). We speculate that CDYL chromodomain binding to methyl-H3K9 may stabilize CDYL recruitment to REST-binding sites, and/or facilitates spreading of the CDYL corepressor complex along the chromatin fiber from the initial point of recruitment. Such a model may explain why extended methyl-H3K9 domains are observed around certain REST binding sites (Roopra et al., 2004).
Repression of REST target genes by CDYL correlates with G9a recruitment and H3K9 dimethylation
We next asked whether the ability of CDYL to bridge the interaction between REST and G9a is important for gene repression in vivo. Quantitative RT-PCR showed that stable RNAi knockdown of CDYL de-repressed several REST target genes in TLM-HMEC (Fig. 2e and f). Chromatin immunoprecipitation (ChIP) revealed that the genomic regions inclusive of the RE1 sites at the TrkC and NPTXR genes were occupied by REST, CDYL and G9a and enriched in H3K9 dimethylation relative to the control GAPDH promoter (Supp. Fig. 2). Significantly, RNAi knockdown of CDYL also resulted in a diminished occupancy of CDYL, G9a and H3K9 dimethylation around the RE1 sites at both the TrkC and NPTXR genes (Fig. 2g). The moderate loss of occupancy by these factors may be due to the incomplete knockdown of CDYL in TLMHMEC resulting in a partial loss of CDYL function (Fig. 2e). In contrast, REST and histone H3 occupancy were not significantly diminished (Fig. 2g). Together, these data support the notion that CDYL is a REST co-repressor that facilitates G9a recruitment and H3K9 dimethylation at REST target genes for repression.
RNAi knockdown of CDYL and G9a induces transformation of TLM-HMEC
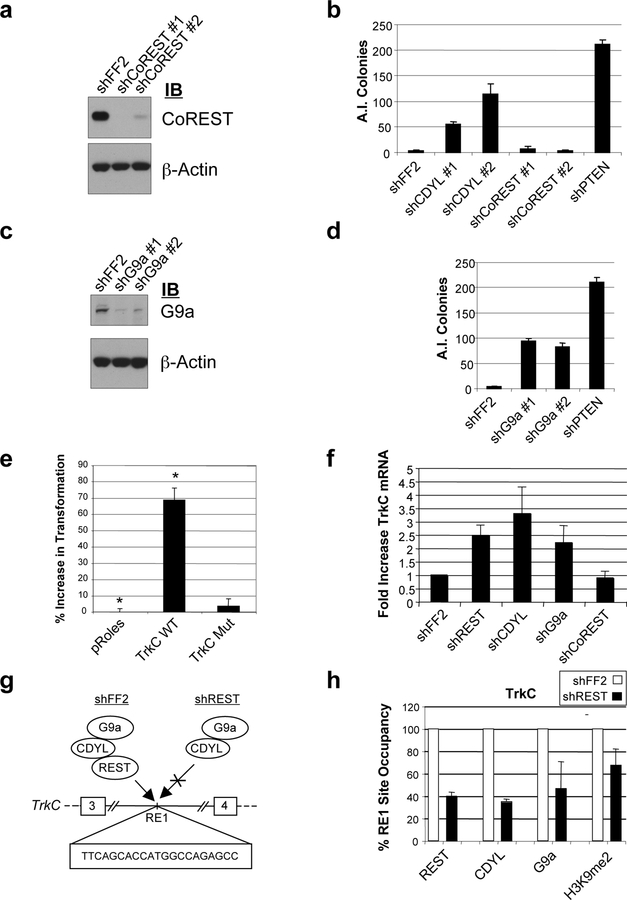
A recent finding described a somewhat unexpected function of REST as a tumor suppressor required for suppression of TLM-HMEC transformation in vitro (Westbrook et al., 2005). However, it remains unclear how REST mediates this transformation suppression activity. REST activity depends on the recruitment of co-repressor complexes, such as the CoREST complex, which modifies chromatin to repress transcription (Andres et al., 1999; Grimes et al., 2000). As discussed above, our evidence suggests that CDYL also functions as a REST co-repressor by recruiting H3K9 methyltransferases, including G9a. This prompted us to ask if CDYL may be involved in suppression of cellular transformation in TLM-HMEC. Using the same assay that identified REST as a tumor suppressor (Westbrook et al., 2005), we found that knockdown of CDYL, but not CoREST, is sufficient to induce transformation in these cells (Fig. 3a and b). These findings suggest that CDYL, but not CoREST, is required for suppression of TLM-HMEC transformation.
Figure 3: CDYL and G9a, but not CoREST, suppress transformation of TLM-HMEC and repress transcription of proto-oncogene TrkC.
(a) TLM-HMEC were stably transduced with two independent shCoREST constructs. Knockdown was confirmed by immunoblotting. CDYL knockdown is shown in Fig. 2e. (b) Transformation assays were performed by plating stable shRNA cells in semi-solid media and quantifying anchorage-independent (A.I.) colony formation after three weeks. shRNAs targeting firefly luciferase (shFF2) and PTEN tumor suppressor (shPTEN) were used as negative and positive controls for transformation respectively. (c, d) Similarly, two independent shRNA constructs were used to stably knockdown G9a expression in TLM-HMEC, and the resultant cells plated in semi-solid media to assay for transformation. (e) Transgenic expression of TrkC wild type (TrkC WT), but not a mutant TrkC lacking the extracellular and transmembrane domains (TrkC Mut), induced anchorage independent (A.I.) growth of TLM-HMEC relative to empty vector (pRoles). Shown is the mean +/− s.d. of triplicate samples (*p=0.004). (f) Quantitative RT-PCR demonstrating upregulated TrkC expression in TLMHMEC upon shRNA knockdown of REST, CDYL and G9a, but not shFF2 shCoREST. (g) Proposed regulation of TrkC by REST binding to a conserved RE1 site between exons 3 and 4, thereby recruiting CDYL and G9a. shREST is expected to cause loss of REST occupancy and failure to recruit CDYL and G9a. (h) Quantitative ChIP PCR analysis of the intronic TrkC RE1 site in TLM-HMEC. shREST induced diminished RE1 occupancy by REST, with a concomitant decrease in CDYL and G9a occupancy and H3K9me2 levels. shFF2 TLM-HMEC were used as a negative control.
The results of the transformation assays complement our biochemical observations that CDYL, but not CoREST, bridges the interaction between REST and G9a in vitro (Fig. 2c), and is important for repression mediated by REST (Fig. 2f and g). To determine whether transcriptional repression by REST-CDYL via the recruitment of histone methyltransferases may be important in transformation suppression, we investigated whether G9a also plays a role in suppressing anchorage-independent growth. As shown in Figure 3c and d, two independent shRNA plasmids that efficiently inhibited G9a expression induced transformation in TLM-HMEC. This suggests that suppression of TLM-HMEC transformation by REST may depend on its direct interaction with CDYL, which in turn recruits G9a to repress target genes.
TrkC/NTRK3 induces cellular transformation and is repressed by REST, CDYL and G9a
We considered the possibility that CDYL and G9a may suppress transformation in TLMHMEC by repressing the transcription of an oncogene. The proto-oncogene TrkC/NTRK3 is a receptor tyrosine kinase that contains an intronic RE1 site and is de-repressed by CDYL RNAi in TLM-HMEC (Fig. 2f). TrkC plays a critical role in neurogenesis and cancers of the neural lineage (Nakagawara, 2001), and other types of cancers (Jin et al., 2007; McGregor et al., 1999). We found that stable transgenic expression of TrkC in TLM-HMEC induced cellular transformation, identifying TrkC as a candidate oncogene in these cells (Fig. 3e). Expression of a TrkC mutant lacking the extracellular and transmembrane domains failed to induce transformation (Fig. 3e). Importantly, TrkC is up-regulated in TLM-HMEC upon shRNA knockdown of REST, CDYL and G9a, but not in shCoREST or shFF2 TLM-HMEC (Fig. 3f). We then investigated if REST knockdown in the shREST TLM-HMEC in which transformation induction was previously reported (Westbrook et al., 2005), affected the occupancy of CDYL, G9a and H3K9me2 at the TrkC REST binding site. We found that knockdown of REST in TLM-HMEC not only results in diminished REST occupancy, but also diminished occupancy of CDYL, G9a and H3K9me2 (Fig. 3g and h). These findings identify TrkC as a direct target of the REST/CDYL/G9a repressor complex and suggest that TrkC repression may play a role in REST/CDYL/G9a-mediated suppression of cellular transformation. Future studies will determine whether the REST repressor complex also regulates additional oncogenes and tumor suppressor genes to accomplish tumor suppression.
CDYL RNAi augments cellular transformation of human squamous epithelial cells expressing HPV16 oncoproteins E6 and E7
Given the role of CDYL in suppressing cellular transformation (Fig. 3b), we next investigated a potential role of CDYL loss of function in clinical cancer. Several studies have reported frequent loss of herterozygosity (LOH) of 6p25, which encompasses the CDYL locus, in cervical carcinoma (CC) (42.6 – 70.4% LOH of 6p25 in invasive CC) (Arias-Pulido et al., 2004; Chatterjee et al., 2001; Krul et al., 1999; Mazurenko et al., 2006). The major risk factor for CC is infection by specific “high-risk” HPV types, which express the E6 and E7 oncoproteins that target tumor suppressor pathways and contribute directly to genomic instability (Howley, 2007). However, only a small percentage of HPV-infected women develop CC (Munoz et al., 2003), suggesting that whereas HPV infection might be necessary for CC, it is not sufficient. Furthermore, specific cytogenetic abnormalities are commonly observed in CCs, suggesting that mutations in specific cellular genes might contribute to carcinogenic progression. Significantly, 6p25 LOH was observed in the earliest stages of CC, and is more prevalent in precancerous lesions that have a high risk of progressing to invasive CC, compared to low- or neutral-risk precancerous lesions (Arias-Pulido et al., 2004; Chatterjee et al., 2001; Krul et al., 1999). This suggests that mutation of candidate tumor suppressor genes in this region may play a critical role in CC progression. However, the identity of such tumor suppressor genes has remained elusive.
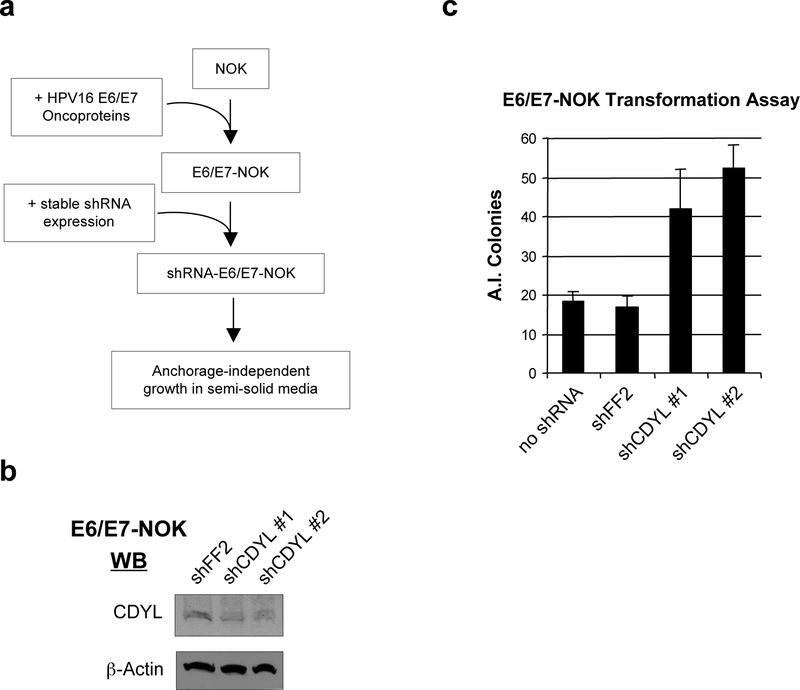
We used an in vitro model to ask if inhibition of CDYL cooperates with high-risk HPV16 E6 and E7 oncoproteins to transform squamous epithelial cells, the normal host cell of HPV. Low passage primary human squamous epithelial cells (Normal Oral Keratinocytes, NOK), immortalized by expression of exogenous hTERT, were stably transduced with HPV16 oncoproteins E6 and E7 (E6/E7-NOK), or an empty viral vector (Fig. 4a). CDYL was then stably knocked-down in these cells by shRNA, with shFF2 used as a negative control (Fig. 4b). The cells were plated in semi-solid media and assayed for anchorage independent growth of colonies four weeks later. As has previously been demonstrated, the expression of HPV16 E6 and E7 oncoproteins alone was sufficient for keratinocyte transformation (Munger et al., 1989). Significantly, knockdown of CDYL in E6/E7-NOK by two independent shRNAs resulted in a consistent 2.5 to 3-fold increase in colony formation relative to controls (Fig. 4c). This indicates that knockdown of CDYL augmented the transformation phenotype of E6/E7-expressing NOK. Importantly, NOK that were not transduced with HPV16 E6 and E7 did not form colonies, even upon CDYL knockdown (data not shown). Together, these data suggest that CDYL mutation, though insufficient to transform squamous epithelial cells alone, may enhance the transformation phenotype of an HPV E6/E7 expressing cell.
Figure 4: RNAi knockdown of CDYL augments transformation of immortalized primary human squamous epithelial cells (NOK) by HPV16 oncoproteins E6 and E7.
(a) Normal Oral Keratinocyte (NOK) transformation assay. hTERT-immortalized primary human squamous epithelial cells (NOK) were engineered to stably express HPV16 oncoproteins E6 and E7 using retroviral transduction (E6/E7-NOK). These cells were then stably transduced with two independent shRNA vectors targeting knockdown of CDYL. As a negative control, cells were stably transduced with shRNA vectors targeting firefly luciferase (shFF2). Cells were plated in semi-solid media to assay for anchorage-independent (A.I.) growth. (b) Immunoblotting confirmed shRNA knockdown of CDYL in E6/E7-NOK. (c) Transformation assay showing that shCDYL augments A.I. colony formation of E6/E7-NOK. Shown is the mean of triplicate samples +/− s.d.
Taken together our findings support a model whereby CDYL bridges the interaction between REST and G9a to repress transcription of genes important for suppression of cellular transformation in TLM-HMEC, including the proto-oncogene TrkC. As REST binding sites have been mapped to some 1946 sites in the genome (Johnson et al., 2007; Otto et al., 2007), we speculate that other target genes may also participate in suppression of transformation by the REST/CDYL/G9a repressor complex. The failure of RNAi knockdown of CoREST to transform TLM-HMEC suggests that different REST co-repressors mediate different functions, with CDYL specifically implicated in REST suppression of transformation. The fact that CDYL facilitates the interaction between REST and G9a, and that G9a is also required for suppression of transformation in TLMHMEC, suggest that the ability of CDYL to bridge REST-G9a interaction is important for its mechanism of transformation suppression. Importantly, our findings also suggest a role for CDYL in suppression of cellular transformation by HPV16 oncoproteins in squamous epithelial cells (Fig. 4), the normal host cells of HPV infection. Given the frequent loss of heterozygosity (LOH) of the locus encoding CDYL in cervical cancer, and its correlation with poor prognosis, this suggests that CDYL may contribute to suppression of transformation in cervical cancer associated with HPV infection. By demonstrating that RNAi inhibition of CDYL leads to cellular transformation in two independent cell culture models, TLM-HMEC and E6/E7-NOK, our findings suggest that CDYL may play a general role in suppression of cellular transformation, and potentially, clinical cancer. Taken together, these findings cast new light on the mechanism of REST and CDYL activities, and provide new insights into the molecular mechanisms that link a particular histone-modifying complex with a specific biological process.
Experimental Procedures
Cell Culture, Viral Transduction and RNAi
HeLa cells were maintained as per Shi et al. (2003), TLM-HMEC cells were maintained as per Westbrook et al. (2005), and NOK cells were maintained as per Piboonniyom et al. (2003). Retroviral vectors expressing Flag-HA-CDYL mRNA isoform 2 were constructed as described previously (Shi et al., 2003). Retroviral and lentiviral shRNA vectors were obtained from Open Biosystems (clone Id: shCDYL #1, V2HS_68448; shCDYL #2, V2HS_68451; shCoREST #1, V2HS_87301; shCoREST #2, V2HS_87304; shG9a #1, TRCN0000115667; shG9a #2, TRCN0000115668), or as previously described (Westbrook et al., 2005). pMSCV-neo (Clontech) expressing HPV16-E6/E7 cDNA p1321 (Munger et al., 1989) was a gift from Agata Smogorzewska. Preparation of viruses and cell transduction was as described previously (Westbrook et al., 2005). HeLa cells were transfected with pBluescript-U6 or pBluescript-U6-shRNA plasmid constructed to target CDYL sequence AAGGTACATCTCCGTTCATGG, as previously described (Sui et al., 2002). Cells were co-transfected with pBABE-puro and selected with 1 ug/mL puromycin for 48 h before preparation of nuclear extracts.
CDYL Tandem Affinity Purification and Mass Spectrometry
CDYL-associated proteins were purified from HeLa cell nuclear extracts an identified by mass spectrometry as previously described (Shi et al., 2003).
Glycerol Gradient Sedimentation
Flag-HA-CDYL tandem-affinity purified material was analyzed as previously described (Shi et al., 2003), using 4 mL, 15 – 40% glycerol gradients in Buffer A (20 mM Tris-HCl, pH 7.9, 5 mM MgCl2, 10% glycerol, 1 mM phenylmethylsulphonyl fluoride (PMSF), 0.1% Nonidet P40, 10 mM 2-mercaptoethanol) containing 100 mM KCl, and centrifuged for 15 h at 55,000 r.p.m. Fractions were collected from the bottom of the gradient, resolved by SDS-PAGE and analyzed by silver staining or immunoblotting.
REST Immunoprecipitation
Flag-purified Flag-HA-CDYL-associated proteins or HeLa nuclear extracts were diluted three-fold in IP buffer containing 50 mM Tris (pH 7.3), 150 mM NaCl, 0.1% NP-40, 1 mM EDTA (pH 8), 1 mM PMSF, 5 mM 2-Mercaptoethanol and 10 mM MG132 (Sigma) and incubated overnight with REST IgG (gift of Gail Mandel) or non-specific IgG at 4°C. Protein G agarose beads (Sigma) were added for 1 h at 4°C, then washed four times with IP buffer. Beads were boiled in SDS-PAGE loading buffer and eluates analyzed by Immunoblotting.
In Vitro Protein Interaction Assays
Recombinant proteins were prepared as described in Supplemental Experimental Procedures. For GST pull-down assays, GST-CDYL (2 μg) was incubated with Flag-REST (200 ng) or Flag-G9a/Flag-EuHMT1 (1 ug), overnight at 4°C in 400 μL Buffer A containing 150 mM NaCl and 0.1% BSA. Beads were washed four times with Buffer A supplemented with 100 mM NaCl and boiled in sample buffer to elute bound proteins. For CDYL bridging assays, HA-REST (200 ng) and Flag-G9a/Flag-EHMT1 (500 ng) were incubated overnight at 4°C with either GST-CDYL (190 ng), GST alone (50 ng) or His6-CoREST (150 ng) in 200 μl Buffer A containing 100 mM NaCl and 0.1% BSA. Complexes were purified using anti-HA resin (Santa Cruz), washed four times in Buffer A supplemented with 100 mM NaCl, eluted by boiling in sample buffer and analyzed by immunoblotting.
RT-PCR and Chromatin Immunoprecipitation
RNA was extracted using Trizol reagent (Invitrogen) as per manufacturer’s protocol. Quantitative RT-PCR was performed using the Lightcycler 480 kit (Roche). RT-PCR primer pairs were as previously described (Bruce et al., 2004; Shi et al., 2004). ChIP was performed according to the Upstate protocol, as previously described (Shi et al. 2003). ChIP PCR primers are listed in Supplemental Experimental Procedures.
Transformation Assays
TLM-HMEC transformation assays were performed as previously described (Westbrook et al., 2005). Normal Oral Keratinocytes (NOK) cells were described previously (Piboonniyom et al., 2003). Stable shRNA cells were constructed as described in Supplemental Experimental Procedures. For NOK transformation assays, 105 cells were plated in triplicate on 6 cm plates. Cells were seeded in a final volume of 1.5 mL of 0.3 % noble agar (Sigma), on 0.6 % bottom agar plates. Each week 0.5 mL of fresh complete medium was added to the plates. Colonies of 100 μm or greater were counted by microscopy after 4 weeks.
Supplementary Material
Acknowledgements:
We gratefully acknowledge the assistance of Benedikt Kessler and Eric Spooner of the Harvard Medical School PFPC in completing the mass spectrometric analysis. We thank Gail Mandel for providing REST antibody and reagents, Grace Gill and Jian Ouyang for His-CoREST, Agata Smogorzewska for pMSCV-neo-HPV16-E6/E7, Karl Munger for NOK cells and Yoshihiro Nakatani and Bijan Sobhian for G9a antibody and expression plasmid. We also thank Grace Gill, Keith Blackwell, Wade Harper and their laboratories for critical discussion of the project. This work was supported by NIH grants GM071004 (to Y.S.) and PO1 CA050661 (to P.M.H.). S.J.E is an investigator with the Howard Hughes Medical Institute, and T.F.W. is funded by grant PDF0403175 from the Susan G. Komen Breast Cancer Foundation. Yang Shi is a cofounder of Constellation Pharmaceuticals and member of its scientific advisory board.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication.As a service to our customers we are providing this early version of the manuscript.The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andres ME, Burger C, Peral-Rubio MJ, Battaglioli E, Anderson ME, Grimes J, Dallaman J, Ballas N, and Mandel G (1999). CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc. Natl. Acad. Sci. USA 96, 9873–9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias-Pulido H, Joste N, and Wheeler CM (2004). Loss of heterozygosity on chromosome 6 in HPV-16 positive cervical carcinomas carrying the DRB1*1501-DQB1*0602 haplotype. Genes Chromosomes Cancer 40, 277–284. [DOI] [PubMed] [Google Scholar]
- Ballas N, and Mandel G (2005). The many faces of REST oversee epigenetic programming of neuronal genes. Current opinion in neurobiology 15, 500–506. [DOI] [PubMed] [Google Scholar]
- Bruce AW, Donaldson IJ, Wood IC, Yerbury SA, Sadowski MI, Chapman M, Gottgens B, and Buckley NJ (2004). Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc Natl Acad Sci U S A 101, 10458–10463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron C, Pivot-Pajot C, van Grunsven LA, Col E, Lestrat C, Rousseaux S, and Khochbin S (2003). Cdyl: a new transcriptional co-repressor. EMBO Rep 4, 877–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Pulido HA, Koul S, Beleno N, Perilla A, Posso H, Manusukhani M, and Murty VV (2001). Mapping the sites of putative tumor suppressor genes at 6p25 and 6p21.3 in cervical carcinoma: occurrence of allelic deletions in precancerous lesions. Cancer Res 61, 2119–2123. [PubMed] [Google Scholar]
- Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, and Mandel G (1995). REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell 80, 949–957. [DOI] [PubMed] [Google Scholar]
- Ding Z, Gillespie LL, and Paterno GD (2003). Human MI-ER1 alpha and beta function as transcriptional repressors by recruitment of histone deacetylase 1 to their conserved ELM2 domain. Mol Cell Biol 23, 250–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Franz H, Jacobs SA, Allis CD, and Khorasanizadeh S (2008). Specificity of the Chromodomain Y Chromosome Family of Chromodomains for Lysine-methylated ARK(S/T) Motifs. J Biol Chem 283, 19626–19635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes JA, Nielsen SJ, Battaglioli E, Miska EA, Speh JC, Berry DL, Atouf F, Holdener BC, Mandel G, and Kouzarides T (2000). The co-repressor mSin3A is a functional component of the REST-CoREST repressor complex. J Biol Chem 275, 9461–9467. [DOI] [PubMed] [Google Scholar]
- Howley PM, and Lowy DR. (2007). Papillomaviruses In Fields Virology, Howley D.M.K.a.P.M., ed. (Philadelphia, PA: Lippincott Williams and Wilkins; ), pp. 2299–2354. [Google Scholar]
- Huang Y, Myers SJ, and Dingledine R (1999). Transcriptional repression by REST: recruitment of Sin3A and histone deacetylase to neuronal genes. Nat Neurosci 2, 867–872. [DOI] [PubMed] [Google Scholar]
- Jin W, Yun C, Kwak MK, Kim TA, and Kim SJ (2007). TrkC binds to the type II TGF-beta receptor to suppress TGF-beta signaling. Oncogene 26, 7684–7691. [DOI] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, and Wold B (2007). Genome-wide mapping of in vivo protein-DNA interactions. Science 316, 1497–1502. [DOI] [PubMed] [Google Scholar]
- Krul EJ, Kersemaekers AM, Zomerdijk-Nooyen YA, Cornelisse CJ, Peters LA, and Fleuren GJ (1999). Different profiles of allelic losses in cervical carcinoma cases in Surinam and The Netherlands. Cancer 86, 997–1004. [PubMed] [Google Scholar]
- Lahn BT, Tang ZL, Zhou J, Barndt RJ, Parvinen M, Allis CD, and Page DC (2002). Previously uncharacterized histone acetyltransferases implicated in mammalian spermatogenesis. Proc Natl Acad Sci U S A 99, 8707–8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K, Ishii N, Yoshida S, Shiosaka S, Wanaka A, and Tohyama M (1998). Molecular cloning and distinct developmental expression pattern of spliced forms of a novel zinc finger gene wiz in the mouse cerebellum. Brain Res Mol Brain Res 61, 179–189. [DOI] [PubMed] [Google Scholar]
- Mazurenko NN, Bliev A, Bidzhieva BA, Peskov D, Snigur NV, Savinova EB, and Kiselev FL (2006). [Loss of heterozygosity at chromosome 6 as a marker of early genetic alterations in cervical intraepithelial neoplasias and microinvasive carcinomas]. Molekuliarnaia biologiia 40, 436–447. [PubMed] [Google Scholar]
- McGregor LM, McCune BK, Graff JR, McDowell PR, Romans KE, Yancopoulos GD, Ball DW, Baylin SB, and Nelkin BD (1999). Roles of trk family neurotrophin receptors in medullary thyroid carcinoma development and progression. Proc Natl Acad Sci U S A 96, 4540–4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger K, Phelps WC, Bubb V, Howley PM, and Schlegel R (1989). The E6 and E7 genes of human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J. Virol 63, 4417–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz N, Bosch FX, de Sanjose S, Herrero R, Castellsague X, Shah KV, Snijders PJ, and Meijer CJ (2003). Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med 348, 518–527. [DOI] [PubMed] [Google Scholar]
- Nakagawara A (2001). Trk receptor tyrosine kinases: a bridge between cancer and neural development. Cancer Lett 169, 107–114. [DOI] [PubMed] [Google Scholar]
- Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, and Nakatani Y (2002). A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 296, 1132–1136. [DOI] [PubMed] [Google Scholar]
- Otto SJ, McCorkle SR, Hover J, Conaco C, Han JJ, Impey S, Yochum GS, Dunn JJ, Goodman RH, and Mandel G (2007). A new binding motif for the transcriptional repressor REST uncovers large gene networks devoted to neuronal functions. J Neurosci 27, 6729–6739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piboonniyom SO, Duensing S, Swilling NW, Hasskarl J, Hinds PW, and Munger K (2003). Abrogation of the retinoblastoma tumor suppressor checkpoint during keratinocyte immortalization is not sufficient for induction of centrosome-mediated genomic instability. Cancer Res 63, 476–483. [PubMed] [Google Scholar]
- Roopra A, Qazi R, Schoenike B, Daley TJ, and Morrison JF (2004). Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol Cell 14, 727–738. [DOI] [PubMed] [Google Scholar]
- Roopra A, Sharling L, Wood IC, Briggs T, Bachfischer U, Paquette AJ, and Buckley NJ (2000). Transcriptional repression by neuron-restrictive silencer factor is mediated via the Sin3-histone deacetylase complex. Mol Cell Biol 20, 2147–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenherr CJ, and Anderson DJ (1995). The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science 267, 1360–1363. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, and Shi Y (2004). Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953. [DOI] [PubMed] [Google Scholar]
- Shi Y, Sawada J, Sui G, Affarel B, Whetstine JR, Lan F, Ogawa H, Luke MP, and Nakatani Y (2003). Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422, 735–738. [DOI] [PubMed] [Google Scholar]
- Singh SK, Kagalwala MN, Parker-Thornburg J, Adams H, and Majumder S (2008). REST maintains self-renewal and pluripotency of embryonic stem cells. Nature 453, 223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui G, Soohoo C, Affar el B., Gay F, Shi Y, Forrester WC and Shi Y (2002). A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America 99, 5515–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taunton J, Hassig CA and Schreiber SL (1996). A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272, 408–11. [DOI] [PubMed] [Google Scholar]
- Ueda J, Tachibana M, Ikura T, and Shinkai Y (2006). Zinc finger protein Wiz links G9a/GLP histone methyltransferases to the co-repressor molecule CtBP. J Biol Chem 281, 20120–20128. [DOI] [PubMed] [Google Scholar]
- Wang H, An W, Cao R, Xia L, Erdjument-Bromage H, Chatton B, Tempst P, Roeder RG, and Zhang Y (2003). mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol Cell 12, 475–487. [DOI] [PubMed] [Google Scholar]
- Westbrook TF, Martin ES, Schlabach MR, Leng Y, Liang AC, Feng B, Zhao JJ, Roberts TM, Mandel G, Hannon GJ, et al. (2005). A genetic screen for candidate tumor suppressors identifies REST. Cell 121, 837–848. [DOI] [PubMed] [Google Scholar]
- Zhao JJ, Gjoerup OV, Subramanian RR, Cheng Y, Chen W, Roberts TM, and Hahn WC (2003). Human mammary epithelial cell transformation through the activation of phosphatidylinositol 3-kinase. Cancer Cell 3, 483–495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.