The enzymes that copy RNA genomes lack proofreading, and viruses that possess RNA genomes, such as West Nile virus, rapidly diversify into swarms of mutant lineages within a host. Intrahost variation of the primary genomic sequence of RNA viruses has been studied extensively because the extent of this variation shapes key virus phenotypes. However, RNA genomes also form complex secondary structures based on within-genome nucleotide complementarity, which are critical regulators of the cyclization of the virus genome that is necessary for efficient replication and translation. We sought to characterize variation in these secondary structures within populations of West Nile virus during serial passage in three bird species. Our study indicates that the intrahost population of West Nile virus is a diverse assortment of RNA secondary structures that should be considered in future analyses of intrahost viral diversity, but some regions that are critical for genome cyclization are conserved within hosts. Besides potential impacts on viral replication, structural diversity can influence the efficacy of small RNA antiviral therapies.
KEYWORDS: RNA virus, West Nile virus, evolution, genetic diversity, natural selection, quasispecies, secondary structure, untranslated region
ABSTRACT
RNA viruses are infamous for their high rates of mutation, which produce swarms of genetic variants within individual hosts. To date, analyses of intrahost genetic diversity have focused on the primary genome sequence. However, virus phenotypes are shaped not only by primary sequence but also by the secondary structures into which this sequence folds. Such structures enable viral replication, translation, and binding of small RNAs, yet within-host variation at the structural level has not been adequately explored. We characterized the structural diversity of the 5′ untranslated region (UTR) of populations of West Nile virus (WNV) that had been subject to five serial passages in triplicate in each of three bird species. Viral genomes were sampled from host serum samples at each passage (n = 45 populations) and subjected to next-generation sequencing. For populations derived from passages 1, 3, and 5 (n = 9 populations), we predicted the impact of each mutation occurring at a frequency of ≥1% on the secondary structure of the 5′ UTR. As expected, mutations in double-stranded (DS) regions of the 5′ UTR stem structures caused structural changes of significantly greater magnitude than did mutations in single-stranded (SS) regions. Despite the greater impact of mutations in DS regions, mutations in DS and SS regions occurred at similar frequencies, with no evidence of enhanced selection against mutation in DS regions. In contrast, mutations in two regions that mediate genome cyclization and thereby regulate replication and translation, the 5′ cyclization sequence and the UAR flanking stem (UFS), were suppressed in all three hosts.
IMPORTANCE The enzymes that copy RNA genomes lack proofreading, and viruses that possess RNA genomes, such as West Nile virus, rapidly diversify into swarms of mutant lineages within a host. Intrahost variation of the primary genomic sequence of RNA viruses has been studied extensively because the extent of this variation shapes key virus phenotypes. However, RNA genomes also form complex secondary structures based on within-genome nucleotide complementarity, which are critical regulators of the cyclization of the virus genome that is necessary for efficient replication and translation. We sought to characterize variation in these secondary structures within populations of West Nile virus during serial passage in three bird species. Our study indicates that the intrahost population of West Nile virus is a diverse assortment of RNA secondary structures that should be considered in future analyses of intrahost viral diversity, but some regions that are critical for genome cyclization are conserved within hosts. Besides potential impacts on viral replication, structural diversity can influence the efficacy of small RNA antiviral therapies.
INTRODUCTION
RNA viruses have exceptionally high mutation rates due to a lack of proofreading by viral RNA-dependent RNA polymerases (RdRp) (1, 2). As a consequence, RNA virus genomes rapidly diversify within individual hosts into a swarm of related mutant sequences (1–4). When this mutant swarm forms a unit upon which natural selection can act, it is termed a quasispecies (2, 5). Intrahost genetic diversity can shape viral phenotypes, including virulence and pathogenicity (6–15). To date, the characterization of virus genetic diversity has focused on quantifying variation in the linear RNA nucleotide sequence (6, 16–29). However, RNA genomes fold into complex secondary and tertiary structures, many located in untranslated regions (UTRs), which have essential functions in the viral life cycle (30–35). Alteration of the secondary structure per se has been shown to affect viral fitness (36–39). Additionally, for many RNA viruses, efficient genome replication and translation depend on long-range binding of sequences in the 5′ and 3′ UTRs that circularize the linear viral genome (40, 41).
Secondary structures in RNA virus genomes are generally highly conserved among consensus sequences of viruses isolated from separate hosts, reflecting their critical roles in the virus life cycle (42–47). The strength of selection is such that during passaging of viruses with altered secondary structures, compensatory mutations often arise that partially or completely reconstitute the wild-type structure (see, e.g., references 29 and 47). However, the vast majority of in silico analyses of viral genome structures to date (see, e.g., references 45 and 47–50) have relied on consensus sequences which do not reveal intrahost variation. An exception is a recent study by Ziv et al. (28), in which, using a novel cross-linking technique, at least five major alternative conformations of the Zika virus (ZIKV) genome were detected in vivo. Additionally, de Borba et al. (29) identified 10 single-nucleotide variants within secondary structures in the dengue virus (DENV) 3′ UTR that were under positive selection during replication in mosquitoes. Characterizing intrahost variation in RNA virus genome structure and the forces that shape it will provide deeper insight into the effects of population diversity on viral phenotypes, guide investigations of quasispecies dynamics, and reveal optimal targets for small RNA antiviral therapies (5, 51, 52).
In the current study, we tracked structural variation in the West Nile virus 5′ UTR over the course of serial passage in avian hosts. West Nile virus (WNV; genus Flavivirus) possesses a positive-sense RNA genome flanked by 5′ and 3′ UTRs (Fig. 1a). To facilitate replication and translation, secondary structures within the 5′ and 3′ UTRs of flaviviruses position the specific sequences (here referred to as binding sites) for long-range binding to achieve genome cyclization and also recruit and bind host and viral factors for replication and translation (43, 53–58). The linear and cyclized conformations of the genome occur at an approximately 1:1 ratio (59, 60). The transition between the linear and circular forms of the genome is mediated by the 5′ upstream of start codon (UAR) flanking stem (UFS) structure (61). The 5′ UTR was chosen for our analysis because it contains well-defined secondary structures of known function (Fig. 1a), stem-loops A and B (SLA and SLB, respectively), the UFS, and the capsid-coding region hairpin (cHP) (30, 62). Moreover, at 96 nucleotides, the 5′ UTR is small enough to be encompassed by a single 150-nucleotide Illumina sequencing read.
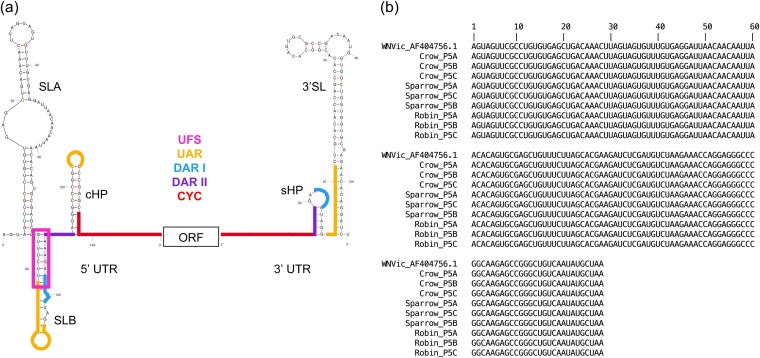
FIG 1.
(a) Representation of the WNV structural elements and binding sites of the 5′ and 3′ UTRs flanking the open reading frame (ORF). To initiate replication once cyclization has occurred, SLA binds to and directs the RdRp to the 3′ UTR (56–58). SLB contains the start codon for translation (55, 62), and cHP is a translation enhancer (67). The structure was generated using mFold online version 4.7 (74). Binding sites were identified by Brinton (62). (b) Alignment of the consensus sequences of the first 150 nucleotides of the WNV genome, via Geneious (105), shows that sequences of WNV after five passages in designated hosts are 100% identical with each other and with parental wild-type WNV infectious clone 3356 (WNVic).
WNV is maintained in birds and Culex mosquitoes and occasionally spills over to cause disease in horses and humans (63). In North America, the American crow, house sparrow, and American robin are known to propagate WNV outbreaks. These three species exhibit substantial differences in viremia and mortality. To explore host-specific impacts on the evolutionary dynamics of diversification of the open reading frame (ORF) sequence, WNV was previously passaged five times in triplicate in each of the three bird species, viral RNA was isolated from each infected bird, and viral genomes were sequenced (64). This study found that, in the ORF, all WNV populations were subject to strong purifying selection, but unique mutations and defective genomes were most frequent in virus passaged in crows, followed by those passaged in sparrows and then robins (64).
Here, we first sought to characterize structural diversity in each population by predicting the structure of the 5′ UTR carrying each mutation that occurred at a frequency of ≥1%, using two different structure prediction algorithms. We predicted only the secondary structures of the linear genome, because neither algorithm is capable of predicting pseudoknots and because we did not have complementary data on mutations in the 3′ UTR. While the WNV genome has three known pseudoknots, located in the capsid gene, NS1/2A genes, and the 3′ UTR (62), no pseudoknots have been identified in the 5′ UTR. Moreover, multiple studies have shown that mutations that distort the configuration of the secondary structures are deleterious for the fitness of WNV, as well as other flaviviruses (57, 58, 61, 65–68).
We then used our predicted structures to test the hypothesis that purifying selection acts to preserve secondary structures during replication within hosts, as it does during transmission between hosts. This hypothesis generated several testable predictions. First, it predicts that mutations with a disproportionately large impact on the secondary structure will be relatively infrequent in the population. Studies of HIV (45, 46, 69–72) and hepatitis C virus (73) indicate that changes in nucleotides in double-stranded (DS) regions of RNA secondary structures are more likely to induce structural changes than are changes in single-stranded (SS) regions. Moreover, using consensus sequences of HIV, Assis (45) showed that selection against mutations that break the base pairings that produce the DS structure was 2.7 times stronger than selection against mutations that did not cause such breakage. Consistent with these studies, we also detected a disproportionate impact of variation in DS regions on predicted WNV 5′ UTRs in the current study. We therefore predicted that variants in DS regions would be less common than would variants in SS regions. Additionally, we predicted that variants producing major structural changes would be less common than those producing minor changes. Finally, we predicted that variants in sequences or structures that enable or regulate the formation of the circularized genome, namely, binding sites and the UFS riboswitch (Fig. 1a), would be less common than variants outside the binding sites. While our data did not support the first two predictions, it did reveal a suppression of variation in the cyclization sequence (CYC) binding site and the UFS riboswitch for all three avian hosts.
RESULTS
Overview of WNV 5′ UTR variants.
In order to assess the impact of different avian hosts on WNV diversity and fitness, WNV infectious clone 3356 (WNVic) was passaged five times in each of three different bird species in triplicate (64). Each viral population was then sequenced using an Illumina platform. Data from the open reading frame (ORF) were analyzed previously (64). In the current study, we analyzed the secondary structure of the 5′ UTR in populations from the first, third, and fifth passages in order to capture rapid, intermediate, and gradual selection, respectively. The total number of 5′ UTR paired sequenced reads per bird and replicate for all passages varied from 3,198 (robin passage 5, replicate B) to 3,155,764 (sparrow passage 3, replicate B). The average number of paired reads by bird across all 3 passages and replicates were 726,362 for crows, 868,899 for sparrows, and 522,048 for robins.
We analyzed only variants occurring at a frequency of ≥1.0%. This sample included a total of 133 variants from passage 1 (15 crow, 30 sparrow, and 88 robin), 178 variants from passage 3 (62 crow, 62 sparrow, and 54 robin), and 192 variants from passage 5 (44 crow, 74 sparrow, and 74 robin). A summary file of variant frequencies was used to generate this set of variants; consequently, linkages of variants within haplotypes could not be determined. Most of these variants occurred at frequencies that ranged from 1.0 to 38.2% (Table 1), but at passage 3, crow replicate C and sparrow replicate B contained two and five mutations, respectively, that reached consensus (i.e., occurred in more than 50% of the reads) but not fixation. However, high frequency mutations were not detected by passage 5, and the consensus sequences of virus populations from each replicate (A, B, and C) in each of the three bird hosts show 100% identity of the 5′ UTR sequence with each other and with parental WNVic (Fig. 1b).
TABLE 1.
Summary of paired reads and variants with frequencies of ≥1.0% from the 5′ UTR
| Passage no. | Replicate | Total no. of 5′ UTR reads | Range in frequency of variants occurring at ≥1.0 (%) | No. of unique variants ≥1.0% |
|---|---|---|---|---|
| 1 | Crow | |||
| A | 1,105,842 | 1.0 | 1 | |
| B | 1,003,674 | 1.1–1.8 | 6 | |
| C | 1,429,252 | 1.2–2.1 | 8 | |
| Sparrow | ||||
| A | 1,163,411 | 1.0–16.4 | 3 | |
| B | 1,146,719 | 2.3–13.5 | 5 | |
| C | 366,481 | 1.0–2.6 | 22 | |
| Robin | ||||
| A | 70,153 | 1.0–6.5 | 39 | |
| B | 1,462,416 | 1.0–4.4 | 26 | |
| C | 93,296 | 1.0–3.9 | 23 | |
| 3 | Crow | |||
| A | 898,274 | 1.0–4.2 | 30 | |
| B | 821,125 | 1.0–3.6 | 27 | |
| C | 1,189,599 | 1.0–52.3 | 5 | |
| Sparrow | ||||
| A | 728,521 | 1.5–14.4 | 9 | |
| B | 3,155,764 | 1.0–65.7 | 18 | |
| C | 1,195,437 | 1.0–17.1 | 35 | |
| Robin | ||||
| A | 622,342 | 1.0–1.9 | 8 | |
| B | 2,142,138 | 1.0–3.1 | 26 | |
| C | 541,686 | 1.0–2.9 | 20 | |
| 5 | Crow | |||
| A | 29,162 | 1.1–1.3 | 4 | |
| B | 29,682 | 1.0–3.9 | 24 | |
| C | 30,649 | 1.0–6.4 | 27 | |
| Sparrow | ||||
| A | 23,897 | 1.0–2.3 | 19 | |
| B | 14,837 | 1.0–13.0 | 36 | |
| C | 25,027 | 1.0–6.1 | 22 | |
| Robin | ||||
| A | 23,898 | 1.0–38.2 | 35 | |
| B | 3,198 | 1.0–3.0 | 25 | |
| C | 9,308 | 1.0–2.9 | 24 |
To assess the impact of each variant, the corresponding wild-type (wt) nucleotide was replaced in silico with the mutated nucleotide within a sequence encompassing the first 150 nucleotides of the wt WNVic sequence. The secondary structure for all mutated sequences was then predicted using the mFold application version 4.7 (http://unafold.rna.albany.edu/ [74]). The resulting structures with the lowest Gibbs free energy were compared to the wt structure, and changes between the two were categorized as major if six or more changes in strandedness occurred between the wt and the mutated structure (e.g., SS to DS or DS to SS), minor if fewer than six such changes occurred, and none if no structure change was predicted. As our categorization of major and minor was arbitrary, the analyses that compared major versus minor versus no structure change were repeated to compare any structure change versus no structure change. After the final passage, 21 variants occurred in all three hosts, and 8 variants occurred in two hosts (Table 2). Of the variants that occurred in all three species, 57.1% caused a minor structure change and 42.9% caused no structure change. Of the variants that occurred in two hosts, 50% resulted in minor structure change, while 37.5% resulted in a major structure change, and the remaining 12.5% did not alter the structure.
TABLE 2.
Twenty-nine variants from passage 5 that were found in more than one avian host
| Variant | Structure change | Passage 5 replicate(s) |
||
|---|---|---|---|---|
| Crow | Sparrow | Robin | ||
| A124T | None | B, C | A, B, C | A, B, C |
| A125G | None | B, C | A, B, C | A, B, C |
| A127C | None | B, C | A, B, C | A, B, C |
| A127T | None | B, C | A, B, C | A, B, C |
| C118T | None | B, C | A, B, C | A, B, C |
| C120G | Minor | B, C | A, B, C | A, B, C |
| C123T | None | B, C | A, B, C | A, B, C |
| C130A | Minor | B, C | A, B, C | A, B, C |
| G116A | None | B, C | A, B, C | A, B, C |
| G121T | Minor | B, C | A, B, C | A, B, C |
| G122A | Minor | B, C | A, B, C | A, B, C |
| G126T | None | B, C | A, B, C | A, B, C |
| G128C | Minor | B, C | A, B, C | A, B, C |
| G131T | Minor | B, C | A, B, C | A, B, C |
| G132C | Minor | B, C | A, B, C | A, B, C |
| G132T | Minor | B, C | A, B, C | A, B, C |
| G117A | Minor | B, C | B | A, C |
| G115A | None | B, C | B | A |
| A114C | Minor | C | B | A, C |
| G113A | Minor | C | B | A, C |
| C123G | Minor | C | B | A |
| A34T | Major | A, B | B | |
| A50G | Major | C | B | |
| A26T | Major | B | A | |
| G2A | None | C | A | |
| G121A | Minor | B | B | |
| G131A | Minor | B | A | |
| T3C | Minor | C | A, B | |
| T7C | Minor | A, C | B, C | |
Overview of WNV 5′ UTR structures.
To predict the secondary structure, we initially folded all variant 5′ UTR sequences using mFold with the temperature set to 37°C. All resulting unique 5′ UTR structures (n = 80) are shown in Table S1 in the supplemental material; 41.8% of all variants resulted in no change, 49.5% resulted in a minor change, and 8.6% resulted in a major change. We next tested whether our choice of mFold had a large impact on our predicted structures by randomly selecting 20% of the unique structures (n = 16) and predicting their structures in RNAstructure version 6.0.1 (https://rna.urmc.rochester.edu/RNAstructureWeb/) at 37°C (Table S2). Eighty-one percent of the outputs from mFold and RNAstructure were identical; of those that differed, none shifted from no change to major change. The distribution of major, minor, or no structural changes did not differ significantly among the structures predicted in mFold and RNAstructure (χ2 = 1.7, df = 2, P = 0.43). Next, we tested whether folding sequences at 40°C, the average body temperature of birds, rather than at 37°C affected the structure predictions. The same 16 structures were refolded in RNAstructure version 6.0.1 at 40°C (Table S2); 100% were identical in structure classification (e.g., major, minor, or no change from the wild type). We also tested the potential effect of high fever in crows on the predicted secondary structures of passage 5 variants and found that 96.0% of the structures that folded at 37°C showed the same structure change classification when the variants were folded at 42°C; there was also not a significant difference in the distribution of major, minor, and no structure changes between the structures predicted at 37°C and 42°C (χ2 = 0.17, df = 2, P = 0.92).
5′ UTR structures. Download Table S1, DOCX file, 2.1 MB (2.1MB, docx) .
Copyright © 2019 Scroggs et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Structure comparison from mFold and RNAstructure at 37°C and 40°C. Download Table S2, DOCX file, 2.0 MB (2MB, docx) .
Copyright © 2019 Scroggs et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To assess the impact of selecting the first (lowest free energy) structure predicted by mFold rather than the second (i.e., next lowest free energy) structure on our analysis, we compared the first and second predicted structures from mFold for the 16 mutations described above and found that the prediction of major, minor, and no structure change was identical for 15 out of the 16 mutations (Table S3).
Comparison of first and second predicted structures and ΔG values (kcal/mol) by mFold. Download Table S3, DOCX file, 0.9 MB (910.8KB, docx) .
Copyright © 2019 Scroggs et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Impact of SS or DS variant location on structure change.
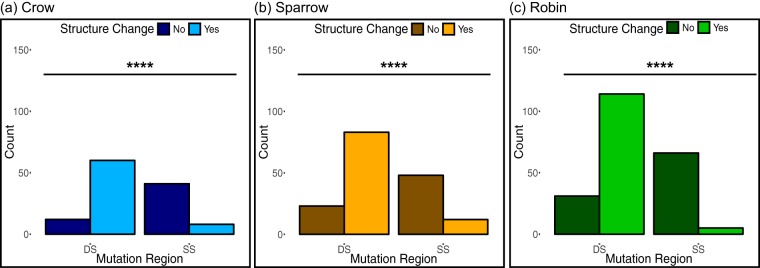
We first tested whether variation from the wild-type sequence in DS regions would be more likely to cause predicted structure change than would variation in SS regions. This assumption was borne out for all three bird-passaged WNV populations, as shown in Fig. 2a to c. Next, we evaluated the degree of predicted structure change and found that variants in DS regions were significantly more likely to cause minor structure changes than were variants in SS regions, while variants in SS regions tended not to alter structure (Fig. 3a to c). However, variants in DS and SS regions were equally likely to cause major structure change.
FIG 2.
(a to c) Variants in DS regions of the WNV 5′ UTR sequence were more likely to cause predicted structure changes than were variants in SS regions in virus passaged in crows (a), sparrows (b), and robins (c). The tendencies of variants from wild-type sequence in DS and SS regions to cause any change in secondary structure were compared using Fisher’s exact test for virus populations pooled across passages 1, 3, and 5 for each bird species (n = 121 variants for crow, n = 166 for sparrow, and n = 216 for robin). ****, P < 0.0001.
FIG 3.
Variants in DS regions of the WNV 5′ UTR were more likely to cause minor structure change than were variants in SS regions, while variants in the DS and SS regions were equally likely to cause major structure change. (a to c) Variants in SS regions were more likely to cause no structure change than were variants in DS regions in viruses passaged in crows (a), sparrows (b), and robins (c). The degrees of predicted structure change as a result of variants in SS or DS regions were compared using Fisher’s exact test for samples from each bird species pooled across passages 1, 3, and 5 (n = 121 variants for crow, n = 166 for sparrow, and n = 216 for robin). ****, P < 0.0001.
Distribution of variants in DS and SS regions.
To determine whether the disproportionate impact of variation in DS regions led to selection for the suppression of such variants, we tested whether variants in DS regions occurred less frequently than expected based on the percentage of nucleotides in DS regions in the wild-type WNV 5′ UTR structure. The distribution of variants in the DS and SS regions of the linear genome at each passage was compared to the expected distribution of the first 150 nucleotides (nt) of wild-type WNV, which is 60.7% DS and 39.3% SS. When stratified by passage, WNV populations showed no significant variation from the expected distribution of variants occurring in DS and SS regions after correction for multiple comparisons, as shown in Fig. 4 and Table S4. Moreover, there was no indication of a decrease or increase in the percentage of variants occurring in DS regions over the course of the five passages, with the exception of viruses passaged once in crows, wherein more variants than expected occurred in SS regions (adjusted P = 0.02) (Fig. 4 and Table S4). As an additional analysis of selection on mutations in DS versus SS regions, we followed the approach used by Assis (45) in which an exact binomial test was used to detect differences in mutation saturation levels between DS and SS sites. In crows, DS and SS sites were equally saturated with mutations, except for passage 1, which has 6.1% saturation at SS nucleotides compared to 1.5% for DS nucleotides (adjusted P = 0.002; see Table 3 for full statistics). The saturations of mutations at DS and SS nucleotides in WNV passaged in sparrows and robins were most often equal, but the saturation of mutations at DS nucleotides was greater than that at SS nucleotides at passages 1 and 5 for robins and passage 5 for sparrows (adjusted P < 0.05 for robin passages 1 and 5 and sparrow passage 5; see Table 3 for full statistics). The average difference was an increase of 6.3% in saturation at DS sites.
FIG 4.
Variants were not less common in DS regions than expected in the WNV 5′ UTR for virus passaged in crows, sparrows, and robins, with the exception of the variants for WNV passaged in crows once, which were more likely to occur in SS regions. Positive values indicate more variants than expected in a DS region, and negative values indicate more variants than expected in an SS region. Using a chi-square test with Bonferroni’s correction for multiple comparisons, only passage 1 variants from crow-passaged WNV differed from the expected (χ2 = 7.3, df: 1, P = 0.007, adjusted P = 0.02); see Table S4 for complete statistics. *, adjusted P < 0.05.
TABLE 3.
Saturation of mutations in DS and SS sites
| Passage no. | Bird | No. of mutations: |
Saturation (%) of: |
P value | Adjusted P valuea | ||||
|---|---|---|---|---|---|---|---|---|---|
| In DS regions | In SS regions | Per DS site | Per SS site | DS sites | SS sites | ||||
| 1 | Crow | 4 | 11 | 0.04 | 0.18 | 1.5 | 6.1 | 0.0005 | 0.002 |
| Sparrow | 17 | 13 | 0.19 | 0.22 | 6.3 | 7.2 | 0.6 | 1.00 | |
| Robin | 59 | 29 | 0.66 | 0.48 | 21.9 | 16.1 | 0.01 | 0.03 | |
| 3 | Crow | 40 | 22 | 0.44 | 0.37 | 14.8 | 12.2 | 0.2 | 0.57 |
| Sparrow | 38 | 24 | 0.42 | 0.40 | 14.1 | 13.3 | 0.7 | 1.00 | |
| Robin | 34 | 20 | 0.38 | 0.33 | 12.6 | 11.1 | 0.4 | 1.00 | |
| 5 | Crow | 28 | 16 | 0.80 | 0.82 | 10.4 | 8.9 | 0.4 | 1.00 |
| Sparrow | 51 | 23 | 0.57 | 0.38 | 18.9 | 12.8 | 0.004 | 0.01 | |
| Robin | 52 | 22 | 0.58 | 0.37 | 19.3 | 12.2 | 0.0007 | 0.002 | |
Adjusted with Bonferroni’s correction for multiple comparisons.
χ2 test results for differences between expected and observed distributions of DS and SS mutations from Fig. 4. Download Table S4, DOCX file, 0.01 MB (13.5KB, docx) .
Copyright © 2019 Scroggs et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Frequency of predicted structure change in the WNV 5′ UTR among different host species.
Grubaugh et al. (64) reported significant differences in WNV fitness and in the distribution and frequency of mutations in virus populations passaged in different hosts. However, in the current study, there was no difference among species in the total number of variants that resulted in predicted structure change (e.g., major and minor structure change combined versus no structure change; Kruskal-Wallis χ2 = 2, df = 2, P = 0.37) or even a correspondence of the ranking of the total number of change-driving variants (crow > robin > sparrow) with the relative fitness reported by Grubaugh et al. (64) (robin > sparrow > crow). The same patterns held for variants that caused major structure changes (robin > crow > sparrow; Kruskal-Wallis χ2 = 2, df = 2, P = 0.37). The frequency of variants in DS regions, irrespective of predicted structure change, did not differ among hosts (59.5% variants in DS regions for crows, 53.9% for sparrows, and 67.1% for robins; χ2 = 2.0, df = 2, P = 0.4).
Viral variants in the sequences and structures that enable and regulate genome circularization.
As the WNV binding sites and the UFS together regulate transitions between the linear and circular genome, we predicted that selection should act to purge variation within these sites, in particular relative to the remainder of the 5′ UTR sequence. To test this prediction, we compared the proportion of variants within and outside specific elements to the proportion of nucleotides within and outside these elements to assess whether variants within them occur more or less often than expected by chance. Additionally, we used an exact binomial test (45) to compare mutation saturation levels within and outside the specific elements.
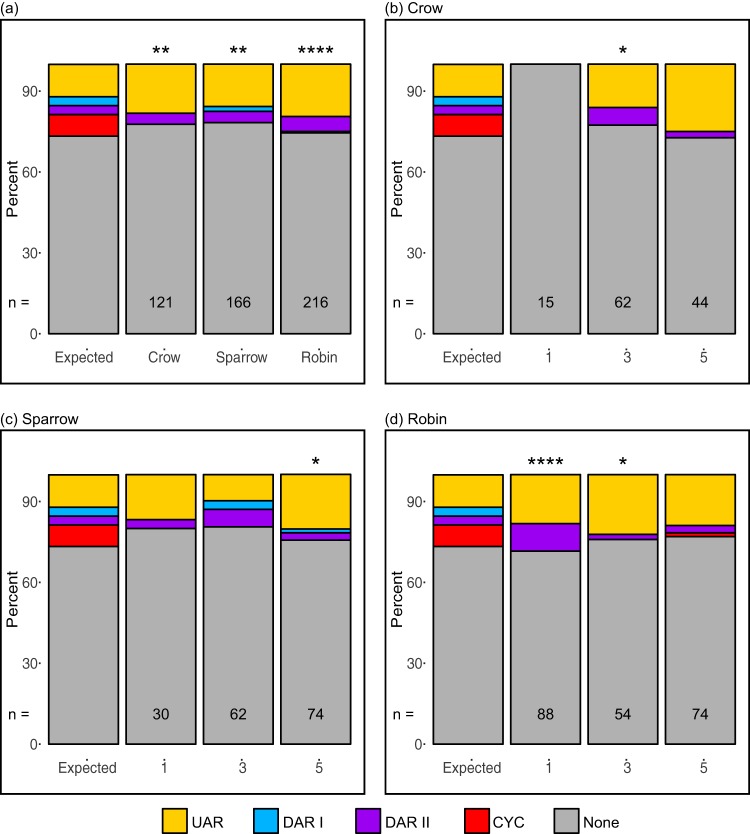
Figure 5a depicts the expected distribution of nucleotides within and outside the binding sites (UAR, DAR I, DAR II, and CYC), as well as the host-specific distributions of variants in these regions, pooled across passages 1, 3, and 5. Fewer variants occurred in the CYC binding site sequence than expected in each of the three bird species. The frequency of variants in the DAR II binding site was not significantly different from the expected frequency in any bird species.
FIG 5.
The frequency of variants within designated binding sites in the WNV 5′ UTR differed from the expected distribution. (a) When the variants were pooled by passage (numbers 1, 3, and 5 on the bottom of each graph), all bird-passaged WNV populations had fewer variants than expected in the CYC binding site. (b to d) Variants located in binding site sequences by passage for viruses passaged in crows (b), sparrows (c), and robins (d) were less common in all binding sites except the UAR. Full statistics from χ2 tests are in Table S5. The UFS riboswitch was also analyzed, and results are described in the text; however, as this sequence overlaps binding sequences, it could not be illustrated in this figure. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
χ2 test results for differences between expected and observed mutations within and outside binding sites from Fig. 5b to d. Download Table S5, DOCX file, 0.01 MB (13.3KB, docx) .
Copyright © 2019 Scroggs et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
When stratified by passage and host species (Fig. 5b to d and Table S5), variants in the CYC binding site sequence remained less common than expected, although many of the differences were no longer significant due to small sample size. For all three hosts, at passage 1, variants were less common in the CYC binding site, although the difference was only significant for the robin-passaged virus and were more common in the UAR binding site, except for the 15 crow-passaged variants, which were all located outside a binding site. By passage 3, variants were still less common in the CYC binding site, but the difference was only significant for crow- and robin-passaged viruses. At passage 5, variants were less common in the DAR I, DAR II, and CYC binding sites than expected; however, they were more common in the UAR binding site across all host species; these differences were only significant for sparrow-passaged virus.
A binomial analysis of mutation saturation within and outside binding sites relative to other regions of the 5′ UTR revealed that, with the exception of the UAR in robin-passaged virus, when significant differences were detected, they reflected lower mutation saturation in the binding sites (Table 4). Mutation saturation inside the CYC binding site was significantly lower than that outside the binding site for virus passaged in each of the three host species.
TABLE 4.
Saturation of mutations within and outside binding sites and the UFS element
| Bird | Binding sitea | No. of mutations: |
Saturation (%) of: |
P value | Adjusted P valueb | ||||
|---|---|---|---|---|---|---|---|---|---|
| Inside | Outside | Per inside site | Per outside site | Inside sites | Outside sites | ||||
| Crow | UAR | 20 | 94 | 1.11 | 0.29 | 37.04 | 28.48 | 0.18 | 0.70 |
| DAR I | 0 | 94 | 0.00 | 0.29 | 0.00 | 28.48 | 0.009 | 0.04 | |
| DAR II | 5 | 94 | 1.00 | 0.29 | 33.33 | 28.48 | 0.78 | 1.00 | |
| CYC | 0 | 94 | 0.00 | 0.29 | 0.00 | 28.48 | 7.98E−06 | 0 | |
| All BS | 25 | 94 | 0.63 | 0.29 | 20.83 | 28.48 | 0.07 | NA | |
| UFS | 0 | 121 | 0.00 | 0.90 | 0.00 | 30.10 | 7.30E−08 | NA | |
| Sparrow | UAR | 26 | 130 | 1.44 | 0.29 | 48.15 | 39.39 | 0.21 | 0.84 |
| DAR I | 3 | 130 | 0.60 | 0.29 | 20.00 | 39.39 | 0.19 | 0.74 | |
| DAR II | 7 | 130 | 1.40 | 0.29 | 46.67 | 39.39 | 0.60 | 1.00 | |
| CYC | 0 | 130 | 0.00 | 0.29 | 0.00 | 39.39 | 2.47E−08 | 0 | |
| All BS | 36 | 130 | 0.90 | 0.29 | 30.00 | 39.39 | 0.04 | NA | |
| UFS | 0 | 166 | 0.00 | 1.24 | 0.00 | 41.29 | 1.18E−11 | NA | |
| Robin | UAR | 42 | 161 | 2.33 | 0.29 | 77.78 | 48.79 | 1.60E−05 | 0 |
| DAR I | 0 | 161 | 0.00 | 0.29 | 0.00 | 48.79 | 6.48E−05 | 0 | |
| DAR II | 12 | 161 | 2.40 | 0.29 | 80.00 | 48.79 | 1.88E−02 | 0.08 | |
| CYC | 1 | 161 | 0.08 | 0.29 | 2.78 | 48.79 | 1.45E−09 | 0 | |
| All BS | 55 | 161 | 1.38 | 0.29 | 45.83 | 48.79 | 0.52 | NA | |
| UFS | 1 | 215 | 0.06 | 1.60 | 2.08 | 53.48 | 6.26E−15 | NA | |
BS, binding sites.
Adjusted with Bonferroni’s correction for multiple comparisons. NA, not applicable.
In the UFS, only a single mutation was detected from passages 1, 3, and 5 at a frequency over 1.0%. This mutation was identified from virus passaged once in robins at a frequency of 1.2% and did not alter the structure of the 5′ UTR. The difference in the distributions of mutations inside and outside the UFS compared to the expected distribution was significantly different for all hosts pooled by each passage (P < 0.0001 for all three). The saturation of mutations in the UFS site of virus passaged in robins was significantly lower, at 2.1% compared to 53.5% mutation saturation outside the UFS site (P = 6.26E−15, Table 4).
Sensitivity of structure of 3′ and 5′ UTR cyclization binding sites to nucleotide variation.
The sequencing coverage of the WNV 3′ UTR in this study was not adequate for our analysis because the reads did not consistently cover to the 3′ terminus of the 3′ UTR. To understand the potential impact of variants in the 3′ UTR binding sites on structure, we systematically altered the sequences of the 3′ binding sites of the wild-type WNV genome in silico (UAR, nt 10951 to 10962 [83% DS]; DAR I, nt 10942 to 10946 [20% DS]; DAR II, nt 10935 to 10939 [100% DS], and CYC, nt 10923 to 10934 [0% DS]) to reflect all three possible alternative nucleotides at each position. For example, if the original nucleotide was an A, the three mutated sequences contained a C, G, or T at that position. This analysis was implemented on the 5′ UTR binding sites as well (UAR, nt 82 to 95 and 124 to 127 [55% DS]; DAR I, nt 99 to 103 [80% DS]; DAR II, nt 109 to 113 [40% DS]; and CYC, nt 135 to 146 [25% DS]). The sequences were folded using mFold. For both the 5′ and 3′ UTR, variants in DS regions caused more structure change than did variants in SS regions, which most often did not cause structure change (Fig. 6a and b).
FIG 6.
Analysis of the impact of all possible variants within 5′ and 3′ UTR binding sites on predicted structure. (a and b) Variants in DS regions in both the 5′ UTR and the 3′ UTR tend to cause structure change (minor and major), while variants in SS regions do not, as detected by Fisher’s exact tests. (c and d) In the 5′ UTR, variants in each of four binding sites are equally likely to result in structure change or no structure change; in the 3′ UTR, variants in different binding sites differ significantly in their tendency to cause structure change, as detected by Fisher’s exact tests. ****, P < 0.0001.
When stratified by binding site, structure change and no structure change were equally likely in response to variants at each of the four binding sites in the 5′ UTR (Fig. 6c, Fisher’s exact P = 0.3). In the 3′ UTR, variants in the DAR II and UAR cause structure changes more often than did variants that occur in the CYC and DAR I binding sites (Fig. 6d).
DISCUSSION
This study characterized intrahost variation in the secondary structures of the untranslated regions of the WNV genome. Specifically, we tested the hypothesis that purifying selection, which acts to conserve secondary structures of many viruses during transmission between hosts (42, 43, 45, 47, 75), would also act to conserve secondary structures of the linear genome as well as binding sites and structures that promote and regulate formation of the circularized form of the genome. We focused on the WNV 5′ UTR, which folds into several structures that are critical for virus fitness (57, 58, 65–68) and are highly conserved among different WNV isolates (55, 62) and across the mosquito-borne flaviviruses generally (30, 44). Further, we leveraged an experimental evolution data set to track variation and the overall evolution of WNV genome structures across serial passages in different avian hosts (64).
Using in silico structure predictions, we first demonstrated that, as in other viruses (42, 45, 76, 77), nucleotide variation in DS regions of the 5′ UTR was more likely to alter the folded structure of the 5′ UTR than was variation in SS regions. Unfortunately, due to insufficient read coverage, we were not able to perform the same analyses of variant distribution in the WNV 3′ UTR. To overcome this limitation, we instead analyzed the impacts of all possible variants within the four binding sites in both the 5′ UTR and 3′ UTR known to mediate genome cyclization by binding to each other. This analysis also demonstrated that the variants in DS regions caused changes in predicted structure significantly more often than did variants in SS regions.
While the majority of mutant genomes are likely inviable, they are not irrelevant because these genomes provide the pool of variability upon which selection can act when the environment of the virus changes due, for example, to stimulation of the immune response, a host shift, or drug treatment. Despite the inordinately large impact of variants in DS regions on predicted secondary structure, such variants were detected as frequently as expected by chance, contradicting our initial prediction that they would be subject to detectable purifying selection. Indeed, an analysis of mutation saturation revealed that three out of nine viral populations were more saturated with mutations in DS sites than in SS sites. Moreover, there was no evidence that the frequency of variants resulting in predicted structural changes, irrespective of DS or SS location, changed in a concerted way over the course of experimental passage, or covaried with fitness, as measured by Grubaugh et al. (64). In contrast, intrahost purifying selection on the amino acid sequence has been documented for WNV (17, 78–83), as well as arthropod-borne viruses, like DENV and bluetongue virus, and for directly transmitted viruses, like HIV and hepatitis C virus (HCV) (84–89). Grubaugh et al. (64) assessed intrahost selection on the ORF of the WNV populations analyzed in this study and found that the proportion of variants that resulted in nonsynonymous changes decreased after five passages (0.7 at input to 0.18 to 0.25 at passage 5), although the frequency of unique intrahost single nucleotide variants in the ORF increased (64). Grubaugh et al. (64) detected lower nucleotide variation in the ORF at passage 5 than we did in the 5′ UTR, yet they were able to identify selection across the ORF. This indicates that the detection of selection in the 5′ UTR is possible, although the region is smaller and could experience more stochastic variation. Thus, while purifying selection on the amino acid sequence of within-host WNV populations was detectable after five passages in the selected bird species (64), selection to preserve UTR secondary structures in the same populations was not.
However, our study did reveal evidence of purifying selection on the sequence of the 5′ CYC binding site and the UFS structural element. In all three bird species, fewer variants occurred in the CYC binding site sequence and the UFS than was expected. The 5′ CYC is the first binding site to bind the 3′ UTR during cyclization and is thus considered the most important site for replication (30), and the UFS is critical for viral replication and genome cyclization (61). Viral replication is suppressed when mutations that break the UFS double bonds are introduced but is partially restored with compensatory mutations (61). Point mutations in the WNV 5′ CYC binding site have been shown to abolish 5′ and 3′ genomic interactions (54) resulting in decreased replication and fitness (90, 91). Mutations in the 5′ CYC site may have prevented replication or may, by preventing cyclization, have accelerated genome degradation by exoribonucleases (92). Either effect may enhance the strength of selection against variants in the 5′ CYC binding site relative to other cyclization sites. Previous analyses of mutations in the 5′ CYC (54, 90, 91) did not include any of the variants identified in our study, so the impact of these specific variants on cyclization is not yet known. Recently, human microRNA required for viral propagation was found to bind the 5′ CYC of the ZIKV genome (28), demonstrating the importance of the 5′ CYC beyond intragenomic long-range viral RNA-RNA binding.
We acknowledge three important caveats to the conclusions above. First, all of our inferences are based on predicted structures, which are not a perfect representation of the actual structures adopted by the virus genome (see, e.g., reference 93). Because we sought to analyze individual variants rather than consensus populations, we were necessarily limited to analyzing predicted structures. In particular, the programs we used to predict structure cannot predict pseudoknots, the presence of which could certainly influence the fitness impacts of variants. While the WNV pseudoknot in the capsid gene, called the downstream cyclization sequence (dCS), spans nt 171 to 438 and is located near the 5′ UTR, there are no documented pseudoknots within the 5′ UTR (94). Recent empirical structural characterizations of the complete genomes of DENV (95) and ZIKV (28) confirmed the presence of the dCS pseudoknot and did not identify any pseudoknots within the 5′ UTR. While unlikely, future analyses may reveal WNV-specific pseudoknots not previously identified, which would then prompt us to reexamine our findings. Additionally, our analysis focused on the structures present in the linear genome, which can be different than the cyclized genome. For example, in the cyclized ZIKV genome, cHP remains mostly intact, but SLB opens completely and binds to the 3′ UTR (28).
Second, because folding and analyzing each structure were laborious, we analyzed only the variants that occurred at a frequency of ≥1.0%, an arbitrary cutoff.
Third, we focused only on avian hosts, but WNV undergoes an alternating cycle of replication in hosts and vectors, and the diversity of the WNV population depends whether the virus is replicating in host or vector. In the avian host, strong purifying pressure restricts genetic diversity, whereas this selection pressure is relaxed in the mosquito vector, resulting in increased diversity (17, 78, 79, 81, 96, 97). Thus, structural diversification may follow different patterns in the mosquito vector relative to the patterns found in the current study.
The structural variation detected in this study could influence the efficacy of antisense oligomer (98, 99) and small interfering RNA (siRNA) antiviral therapies (100) against WNV and contribute to the evolution of resistance to these agents. Many antisense agents are designed to bind sequences in the UTRs, and their binding relies upon perfect complementarity to the target sequence (101, 102). As appealing as antisense antiviral therapies may be, they must be considered in the context of the rapidly evolving virus populations. Using the same WNV infectious clone as that used in this study, Deas et al. (101) demonstrated that translation of WNV was suppressed by a phosphorodiamidate morpholino oligomer (PMO) that targeted the first 20 nucleotides of the 5′ UTR (located at the base of stem-loop A; Fig. 1). Resistance to the PMO evolved quickly (within two passages) as a result of two or three variants in the PMO binding sequence (103). A total of five mutations (G9U, G19A, C8G, C11G, and A23G) were identified in the resistant WNV 5′ UTRs, two of which were shared across at least two of the three replicates (G9U and G19A) (103). We identified G9U three times in our data set (robin passage 1 replicate A and sparrow passage 3 replicates A and B) at low frequencies (<2.3%). Additionally, we found that G9U occurs in a DS region and causes minor structure change in SLA. These results indicate that simply targeting oligomers to structures that are conserved across hosts will not prevent the evolution of resistance within hosts; instead, regions that are subject to purifying selection within hosts must be identified. To this end, the CYC binding site and UFS element may be ideal targets.
In sum, our study suggests that the intrahost population of WNV comprises a high degree of variation not just in primary RNA sequence but in RNA structure as well. Moreover, we detected no evidence of purifying selection acting to sieve out mutations that mediate structural change, even major structural changes, within hosts, save in the UFS stem structure. In their recent review of quasispecies theory, Domingo and Perales (5) expanded the mechanisms of quasispecies variation beyond point mutations to also include recombination, reassortment, gene duplication, and gene transfers. Our study suggests that, when considering the results of such mutations, variations in secondary structures should be considered in tandem with variations in the primary sequence.
MATERIALS AND METHODS
Serial passage of WNV and whole-genome sequencing.
As previously described in detail by Grubaugh et al. (64) WNVic (infectious clone 3356) (104) was injected into wild-caught American crows (Corvus brachyrhynchos), house sparrows (Passer domesticus), and American robins (Turdus migratorius). Three days postinfection, serum samples were collected and used to infect a new set of naive birds for a total of 5 passages, resulting in 45 virus populations sampled (five passages in three bird species each replicated in triplicate). Viral RNA was isolated and sequenced on the Illumina HiSeq 2000 platform (Beckman Coulter Genomics, Danvers, MA), and the data are available at the NCBI under BioProject number PRJNA281547 (64).
Identification of variants.
Part of SLB and all of cHP, as well as three of the binding sites (Fig. 1a), extend from the 5′ UTR into the capsid gene (62). Thus, for this analysis, we included the whole 5′ UTR and the first 54 nucleotides of the ORF to incorporate all of these elements, for a total region of 150 nucleotides. We compiled the variants that occurred at a frequency of ≥1.0% for passages 1, 3, and 5 for all three bird hosts. All variant frequencies were calculated using Geneious version 9.0.5 (105).
Prediction of secondary structure and classification of structure changes.
The secondary structure of the first 150 nt of wild-type WNVic (GenBank accession no. AF404756.1) was predicted using the online mFold application version 4.7 (http://unafold.rna.albany.edu/ [74]). The default folding parameters were utilized for a linear RNA sequence, which includes a fixed folding temperature of 37°C, 1 M NaCl, 5% suboptimality, maximum interior loop size of 30 nt, and maximum interior loop asymmetry of 30 nt. The resulting structure was identical to previously documented secondary structures for the WNV 5′ UTR (55, 62).
To determine the structure of variant haplotypes, each specific nucleotide variation from the wild-type sequence was identified, and the corresponding change was made to a template sequence comprising the 150 nt at the 5′ end of the wild-type WNV genome (Fig. 1a). Each variant sequence was folded using mFold, as described above. Two properties of each nucleotide variant were noted, as follows: (i) whether the nucleotide occurred in a DS or SS region of the wild-type structure, and (ii) whether the variant occurred in one of four sites known to bind sequences in the 3′ UTR, namely, UAR (upstream of start codon), DAR I, DAR II (downstream of start codon), and CYC (cyclization sequence) (Fig. 1a) (62). Variants of the haplotypes that contained two or three mutations were folded individually and in combination. Unless otherwise noted, only the RNA structures resulting from individual variants were used for analyses. To test the sensitivity of our results to our choice of folding program, we assigned a numerical code to each of the unique structures and used a random number generator to create a subset of 20% of total structures, which we refolded in RNAstructure version 6.0.1 at 37°C. Additionally, these sequences were also folded using RNAstructure version 6.0.1 at 40°C, the average bird body temperature (106). RNAstructure was used to test the effect of folding temperature because at 40°C, mFold version 2.3 does not recapitulate the wild-type WNV 5′ UTR structure (62). As crows experience a high fever during WNV infection (107), the crow viral variants from passage 5 were also folded using RNAstructure at 42°C.
Based on the folded structure, each variant was classified as causing a major structure change, a minor structure change, or no structure change (i.e., the structure was identical to the wild type). A major structure change was defined as six or more changes in strandedness, i.e., SS to DS or DS to SS, while a minor structure change was fewer than six but more than zero changes in strandedness.
Statistical analysis.
The distribution of variants causing structure change (major or minor) and variant location (DS or SS) was analyzed using contingency analyses to obtain a χ2 statistic or Fisher’s exact P value, with Bonferroni’s correction where applicable. The Kruskal-Wallis rank sum test was used to detect correlations between host-specific viral fitness and structure change. As described by Assis (45), the proportion of mutations in DS or SS sites (number of mutations in DS or SS sites divided by total number of DS or SS nucleotides) and the proportion of DS or SS sites that are saturated with mutations (calculated as the [number of mutations in DS or SS sites divided by the total number of DS or SS nucleotides multiplied by 3] × 100) were calculated for each bird and passage. An exact binomial test was then used to detect differences in mutation saturations in the DS and SS sites. Exact binomial tests were also conducted for each binding site and the UFS sequence. Statistics were conducted using R (https://www.r-project.org/ [108]).
Data availability.
The sequencing data utilized for this project have been deposited in the NCBI Sequence Read Archive database and can be accessed with the BioProject identifier PRJNA281547.
ACKNOWLEDGMENTS
This work was funded by grants from the National Institutes of General Medical Sciences New Mexico INBRE (grant 8P20GM103451-12 to K.A.H., S.L.P.S., J.A.S., A.S., and F.D.S.) and from the National Institute of Allergy and Infectious Disease, National Institutes of Health (grants 1R15AI113628-01 to K.A.H. and A1067380 to G.D.E.).
An NSF-RCN on Infectious Disease Evolution Across Scales contributed significantly to the ideas underpinning this study.
S.L.P.S. and K.A.H. conceived and designed the structure analyses; G.D.E., N.D.G., and D.R.S. performed the in vivo experiment and sequencing; S.L.P.S. conducted all in silico analyses; S.L.P.S. and K.A.H. conducted the statistical analysis; J.A.S., A.S., and F.D.S. contributed to the bioinformatics analyses; and S.L.P.S. and K.A.H. wrote the manuscript.
REFERENCES
- 1.Eigen M. 1993. Viral quasispecies. Sci Am 269:42–49. doi: 10.1038/scientificamerican0793-42. [DOI] [PubMed] [Google Scholar]
- 2.Lauring AS, Andino R. 2010. Quasispecies theory and the behavior of RNA viruses. PLoS Pathog 6:e1001005. doi: 10.1371/journal.ppat.1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steinhauer DA, Holland JJ. 1987. Rapid evolution of RNA viruses. Annu Rev Microbiol 41:409–431. doi: 10.1146/annurev.mi.41.100187.002205. [DOI] [PubMed] [Google Scholar]
- 4.Biebricher CK, Eigen M. 2006. What is a quasispecies?, p 1–31. In Domingo E. (ed), Quasispecies: concept and implications for virology. Springer Berlin Heidelberg, Berlin, Heidelberg, Germany. [Google Scholar]
- 5.Domingo E, Perales C. 2018. Quasispecies and virus. Euro Biophys 47:443–457. doi: 10.1007/s00249-018-1282-6. [DOI] [PubMed] [Google Scholar]
- 6.Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. 2006. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 439:344–348. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arnold JJ, Vignuzzi M, Stone JK, Andino R, Cameron CE. 2005. Remote site control of an active site fidelity checkpoint in a viral RNA-dependent RNA polymerase. J Biol Chem 280:25706–25716. doi: 10.1074/jbc.M503444200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfeiffer JK, Kirkegaard K. 2005. Increased fidelity reduces poliovirus fitness and virulence under selective pressure in mice. PLoS Pathog 1:e11. doi: 10.1371/journal.ppat.0010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coffey LL, Beeharry Y, Borderia AV, Blanc H, Vignuzzi M. 2011. Arbovirus high fidelity variant loses fitness in mosquitoes and mice. Proc Natl Acad Sci U S A 108:16038–16043. doi: 10.1073/pnas.1111650108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng J, Wang H, Xie X, Li C, Zhou G, Yang D, Yu L. 2014. Ribavirin-resistant variants of foot-and-mouth disease virus: the effect of restricted quasispecies diversity on viral virulence. J Virol 88:4008–4020. doi: 10.1128/JVI.03594-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lloyd SB, Lichtfuss M, Amarasena TH, Alcantara S, De Rose R, Tachedjian G, Alinejad-Rokny H, Venturi V, Davenport MP, Winnall WR, Kent SJ. 2016. High fidelity simian immunodeficiency virus reverse transcriptase mutants have impaired replication in vitro and in vivo. Virol 492:1–10. doi: 10.1016/j.virol.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 12.Van Slyke GA, Arnold JJ, Lugo AJ, Griesemer SB, Moustafa IM, Kramer LD, Cameron CE, Ciota AT. 2015. Sequence-specific fidelity alterations associated with West Nile virus attenuation in mosquitoes. PLoS Pathog 11:e1005009. doi: 10.1371/journal.ppat.1005009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griesemer SB, Kramer LD, Van Slyke GA, Pata JD, Gohara DW, Cameron CE, Ciota AT. 2017. Mutagen resistance and mutation restriction of St. Louis encephalitis virus. J Gen Virol 98:201–211. doi: 10.1099/jgv.0.000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukuhara T, Yamamoto S, Ono C, Nakamura S, Motooka D, Mori H, Kurihara T, Sato A, Tamura T, Motomura T, Okamoto T, Imamura M, Ikegami T, Yoshizumi T, Soejima Y, Maehara Y, Chayama K, Matsuura Y. 2017. Quasispecies of hepatitis C virus participate in cell-specific infectivity. Sci Rep 7:45228. doi: 10.1038/srep45228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willett BJ, Kraase M, Logan N, McMonagle E, Varela M, Hosie MJ. 2013. Selective expansion of viral variants following experimental transmission of a reconstituted feline immunodeficiency virus quasispecies. PLoS One 8:e54871. doi: 10.1371/journal.pone.0054871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herring BL, Tsui R, Peddada L, Busch M, Delwart EL. 2005. Wide range of quasispecies diversity during primary hepatitis C virus infection. J Virol 79:4340–4346. doi: 10.1128/JVI.79.7.4340-4346.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jerzak G, Bernard KA, Kramer LD, Ebel GD. 2005. Genetic variation in West Nile virus from naturally infected mosquitoes and birds suggests quasispecies structure and strong purifying selection. J Gen Virol 86:2175–2183. doi: 10.1099/vir.0.81015-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salemi M. 2013. The intrahost evolutionary and population dynamics of human immunodeficiency virus type 1: a phylogenetic perspective. Infect Dis Rep 5:3. doi: 10.4081/idr.2013.s1.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romano CM, Lauck M, Salvador FS, Lima CR, Villas-Boas LS, Araújo ESA, Levi JE, Pannuti CS, O’Connor D, Kallas EG. 2013. Inter- and intra-host viral diversity in a large seasonal DENV2 outbreak. PLoS One 8:e70318. doi: 10.1371/journal.pone.0070318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simmonds P. 2004. Genetic diversity and evolution of hepatitis C virus–15 years on. J Gen Virol 85:3173–3188. doi: 10.1099/vir.0.80401-0. [DOI] [PubMed] [Google Scholar]
- 21.Rambaut A, Pybus OG, Nelson MI, Viboud C, Taubenberger JK, Holmes EC. 2008. The genomic and epidemiological dynamics of human influenza A virus. Nature 453:615–619. doi: 10.1038/nature06945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y, Sharp PM, Fowkes M, Kocher O, Joseph JT, Koralnik IJ. 2004. Analysis of 15 novel full-length BK virus sequences from three individuals: evidence of a high intra-strain genetic diversity. J Gen Virol 85:2651–2663. doi: 10.1099/vir.0.79920-0. [DOI] [PubMed] [Google Scholar]
- 23.Morelli MJ, Wright CF, Knowles NJ, Juleff N, Paton DJ, King DP, Haydon DT. 2013. Evolution of foot-and-mouth disease virus intra-sample sequence diversity during serial transmission in bovine hosts. Vet Res 44:12. doi: 10.1186/1297-9716-44-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneider WL, Roossinck MJ. 2001. Genetic diversity in RNA virus quasispecies is controlled by host-virus interactions. J Virol 75:6566–6571. doi: 10.1128/JVI.75.14.6566-6571.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cuevas JM, Willemsen A, Hillung J, Zwart MP, Elena SF. 2015. Temporal dynamics of intrahost molecular evolution for a plant RNA virus. Mol Biol Evol 32:1132–1147. doi: 10.1093/molbev/msv028. [DOI] [PubMed] [Google Scholar]
- 26.Moya A, Holmes EC, González-Candelas F. 2004. The population genetics and evolutionary epidemiology of RNA viruses. Nat Rev Microbiol 2:279–288. doi: 10.1038/nrmicro863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duffy S, Shackelton LA, Holmes EC. 2008. Rates of evolutionary change in viruses: patterns and determinants. Nat Rev Genet 9:267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- 28.Ziv O, Gabryelska MM, Lun ATL, Gebert LFR, Sheu-Gruttadauria J, Meredith LW, Liu ZY, Kwok CK, Qin CF, MacRae IJ, Goodfellow I, Marioni JC, Kudla G, Miska EA. 2018. COMRADES determines in vivo RNA structures and interactions. Nat Methods 15:785–788. doi: 10.1038/s41592-018-0121-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Borba L, Villordo SM, Marsico FL, Carballeda JM, Filomatori CV, Gebhard LG, Pallarés HM, Lequime S, Lambrechts L, Sánchez Vargas I, Blair CD, Gamarnik AV. 2019. RNA structure duplication in the dengue virus 3′ UTR: redundancy or host specificity? mBio 10:e02506-18. doi: 10.1128/mBio.02506-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernández-Sanlés A, Ríos-Marco P, Romero-López C, Berzal-Herranz A. 2017. Functional information stored in the conserved structural RNA domains of flavivirus genomes. Front Microbiol 8:546. doi: 10.3389/fmicb.2017.00546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Witwer C, Rauscher S, Hofacker IL, Stadler PF. 2001. Conserved RNA secondary structures in Picornaviridae genomes. Nucleic Acids Res 29:5079–5089. doi: 10.1093/nar/29.24.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thurner C, Witwer C, Hofacker IL, Stadler PF. 2004. Conserved RNA secondary structures in flaviviridae genomes. J Gen Virol 85:1113–1124. doi: 10.1099/vir.0.19462-0. [DOI] [PubMed] [Google Scholar]
- 33.Chapman EG, Costantino DA, Rabe JL, Moon SL, Wilusz J, Nix JC, Kieft JS. 2014. The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production. Science 344:307–310. doi: 10.1126/science.1250897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Logan G, Newman J, Wright CF, Lasecka-Dykes L, Haydon DT, Cottam EM, Tuthill TJ. 2017. Deep sequencing of foot-and-mouth disease virus reveals RNA sequences involved in genome packaging. J Virol 92:e01159-17. doi: 10.1128/JVI.01159-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tuplin A. 2015. Diverse roles and interactions of RNA structures during the replication of positive-stranded RNA viruses of humans and animals. J Gen Virol 96:1497–1503. doi: 10.1099/vir.0.000066. [DOI] [PubMed] [Google Scholar]
- 36.Macadam AJ, Ferguson G, Burlison J, Stone D, Skuce R, Almond JW, Minor PD. 1992. Correlation of RNA secondary structure and attenuation of Sabin vaccine strains of poliovirus in tissue culture. Virology 189:415–422. doi: 10.1016/0042-6822(92)90565-7. [DOI] [PubMed] [Google Scholar]
- 37.Kirkpatrick BD, Whitehead SS, Pierce KK, Tibery CM, Grier PL, Hynes NA, Larsson CJ, Sabundayo BP, Talaat KR, Janiak A, Carmolli MP, Luke CJ, Diehl SA, Durbin AP. 2016. The live attenuated dengue vaccine TV003 elicits complete protection against dengue in a human challenge model. Sci Transl Med 8:330ra36. doi: 10.1126/scitranslmed.aaf1517. [DOI] [PubMed] [Google Scholar]
- 38.Whitehead SS, Durbin AP, Pierce KK, Elwood D, McElvany BD, Fraser EA, Carmolli MP, Tibery CM, Hynes NA, Jo M, Lovchik JM, Larsson CJ, Doty EA, Dickson DM, Luke CJ, Subbarao K, Diehl SA, Kirkpatrick BD. 2017. In a randomized trial, the live attenuated tetravalent dengue vaccine TV003 is well-tolerated and highly immunogenic in subjects with flavivirus exposure prior to vaccination. PLoS Negl Trop Dis 11:e0005584. doi: 10.1371/journal.pntd.0005584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shan C, Muruato AE, Nunes BTD, Luo H, Xie X, Medeiros DBA, Wakamiya M, Tesh RB, Barrett AD, Wang T, Weaver SC, Vasconcelos PFC, Rossi SL, Shi P-Y. 2017. A live-attenuated Zika virus vaccine candidate induces sterilizing immunity in mouse models. Nat Med 23:763–767. doi: 10.1038/nm.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Villordo SM, Gamarnik AV. 2009. Genome cyclization as strategy for flavivirus RNA replication. Virus Res 139:230–239. doi: 10.1016/j.virusres.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Villordo SM, Alvarez DE, Gamarnik AV. 2010. A balance between circular and linear forms of the dengue virus genome is crucial for viral replication. RNA 16:2325–2335. doi: 10.1261/rna.2120410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mauger DM, Golden M, Yamane D, Williford S, Lemon SM, Martin DP, Weeks KM. 2015. Functionally conserved architecture of hepatitis C virus RNA genomes. Proc Natl Acad Sci U S A 112:3692–3697. doi: 10.1073/pnas.1416266112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gritsun TS, Venugopal K, de A Zanotto PM, Mikhailov MV, Sall AA, Holmes EC, Polkinghorne I, Frolova TV, Pogodina VV, Lashkevich VA, Gould EA. 1997. Complete sequence of two tick-borne flaviviruses isolated from Siberia and the UK: analysis and significance of the 5′ and 3′-UTRs. Virus Res 49:27–39. doi: 10.1016/S0168-1702(97)01451-2. [DOI] [PubMed] [Google Scholar]
- 44.Ng WC, Soto-Acosta R, Bradrick SS, Garcia-Blanco MA, Ooi EE. 2017. The 5ʹ and 3ʹ untranslated regions of the flaviviral genome. Viruses 9:137. doi: 10.3390/v9060137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Assis R. 2014. Strong epistatic selection on the RNA secondary structure of HIV. PLoS Pathog 10:e1004363. doi: 10.1371/journal.ppat.1004363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Le S-Y, Chen J-H, Chatterjee D, Maizel JV. 1989. Sequence divergence and open regions of RNA secondary structures in the envelope regions of the 17 human immunodeficiency virus isolates. Nucleic Acids Res 17:3275–3288. doi: 10.1093/nar/17.8.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watters KE, Choudhary K, Aviran S, Lucks JB, Perry KL, Thompson JR. 2018. Probing of RNA structures in a positive sense RNA virus reveals selection pressures for structural elements. Nucleic Acids Res 46:2573–2584. doi: 10.1093/nar/gkx1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JWJ, Swanstrom R, Burch CL, Weeks KM. 2009. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu K, Heng X, Garyu L, Monti S, Garcia EL, Kharytonchyk S, Dorjsuren B, Kulandaivel G, Jones S, Hiremath A, Divakaruni SS, Lacotti C, Barton S, Tummillo D, Hosic A, Edme K, Albrecht S, Telesnitsky A, Summers MF. 2011. NMR detection of structures in the HIV-1 5ʹ-leader RNA that regulate genome packing. Science 334:242–245. doi: 10.1126/science.1210460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pollom E, Dang KK, Potter EL, Gorelick RJ, Burch CL, Weeks KM, Swanstrom R. 2013. Comparison of SIV and HIV-1 genomic RNA structures reveals impact of sequence evolution on conserved and non-conserved structural motifs. PLoS Pathog 9:e1003294. doi: 10.1371/journal.ppat.1003294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bavia L, Mosimann ALP, Aoki MN, Duarte dos Santos CN. 2016. A glance at subgenomic flavivirus RNAs and microRNAs in flavivirus infections. Virol J 13:84. doi: 10.1186/s12985-016-0541-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tycowski KT, Guo YE, Lee N, Moss WN, Vallery TK, Xie M, Steitz JA. 2015. Viral noncoding RNAs: more surprises. Genes Dev 29:567–584. doi: 10.1101/gad.259077.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khromykh AA, Meka H, Guyatt KJ, Westaway EG. 2001. Essential role of cyclization sequences in flavivirus RNA replication. J Virol 75:6719–6728. doi: 10.1128/JVI.75.14.6719-6728.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang B, Dong H, Stein DA, Iversen PL, Shi P-Y. 2008. West Nile virus genome cyclization and RNA replication require two pairs of long-distance RNA interactions. Virology 373:1–13. doi: 10.1016/j.virol.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 55.Brinton MA, Dispoto JH. 1988. Sequence and secondary analysis of the 5ʹ-terminal region of flavivirus genome RNA. Virology 162:290–299. doi: 10.1016/0042-6822(88)90468-0. [DOI] [PubMed] [Google Scholar]
- 56.Filomatori CV, Iglesias NG, Villordo SM, Alvarez DE, Gamarnik AV. 2011. RNA sequences and structures required for the recruitment and activity of the dengue virus polymerase. J Biol Chem 286:6929–6939. doi: 10.1074/jbc.M110.162289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li X-F, Jiang T, Yu X-D, Deng Y-Q, Zhao H, Zhu Q-Y, Qin E-D, Qin C-F. 2010. RNA elements within the 5ʹ untranslated region of the West Nile virus genome are critical for RNA synthesis and virus replication. J Gen Virol 91:1218–1223. doi: 10.1099/vir.0.013854-0. [DOI] [PubMed] [Google Scholar]
- 58.Dong H, Zhang B, Shi P-Y. 2008. Terminal structures of West Nile virus genomic RNA and their interactions with viral NS5 protein. Virology 381:123–135. doi: 10.1016/j.virol.2008.07.040. [DOI] [PubMed] [Google Scholar]
- 59.Alvarez DE, Lodeiro MF, Ludueña SJ, Pietrasanta LI, Gamarnik AV. 2005. Long-range RNA-RNA interactions circularize the dengue virus genome. J Virol 79:6631–6643. doi: 10.1128/JVI.79.11.6631-6643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ferreira GP, Trindade GS, Vilela JMC, Da Silva MIN, Andrade MS, Kroon EG. 2008. Climbing the steps of viral atomic force microscopy: visualization of dengue virus particles. J Microsc 231:180–185. doi: 10.1111/j.1365-2818.2008.02028.x. [DOI] [PubMed] [Google Scholar]
- 61.Liu ZY, Li XF, Jiang T, Deng YQ, Ye Q, Zhao H, Yu JY, Qin CF. 2016. Viral RNA switch mediates the dynamic control of flavivirus replicase recruitment by genome cyclization. Elife 5:e17636. doi: 10.7554/eLife.17636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brinton MA. 2013. Replication cycle and molecular biology of the West Nile virus. Viruses 6:13–53. doi: 10.3390/v6010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kramer LD, Styer LM, Ebel GD. 2008. A global perspective on the epidemiology of West Nile virus. Annu Rev Entomol 53:61–81. doi: 10.1146/annurev.ento.53.103106.093258. [DOI] [PubMed] [Google Scholar]
- 64.Grubaugh ND, Smith DR, Brackney DE, Bosco-Lauth AM, Fauver JR, Campbell CL, Felix TA, Romo H, Duggal NK, Dietrich EA, Eike T, Beane JE, Bowen RA, Black WC, Brault AC, Ebel GD. 2015. Experimental evolution of an RNA virus in wild birds: evidence for host-dependent impacts on population structure and competitive fitness. PLoS Pathog 11:e1004874. doi: 10.1371/journal.ppat.1004874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Filomatori CV, Lodeiro MF, Alvarez DE, Samsa MM, Pietrasanta L, Gamarnik AV. 2006. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev 20:2238–2249. doi: 10.1101/gad.1444206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lodeiro MF, Filomatori CV, Gamarnik AV. 2009. Structural and functional studies of the promoter element for dengue virus RNA replication. J Virol 83:993–1008. doi: 10.1128/JVI.01647-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Clyde K, Harris E. 2006. RNA secondary structure in the coding region of dengue virus type 2 directs translation start codon selection and is required for viral replication. J Virol 80:2170–2182. doi: 10.1128/JVI.80.5.2170-2182.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Clyde K, Barrera J, Harris E. 2008. The capsid-coding region hairpin element (cHP) is a critical determinant of dengue virus and West Nile virus RNA synthesis. Virology 379:314–323. doi: 10.1016/j.virol.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Le S-Y, Chen J-H, Braun MJ, Gonda MA, Maizel JV. 1988. Stability of RNA stem-loop structure and distribution of non-random structure in the human immunodeficiency virus (HIV-I). Nucleic Acids Res 16:5153–5168. doi: 10.1093/nar/16.11.5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yoshida K, Nakamura M, Ohno T. 1997. Mutations of the HIV type 1 V3 loop under selection pressure with neutralizing monoclonal antibody NM-01. AIDS Res Hum Retroviruses 13:1283–1290. doi: 10.1089/aid.1997.13.1283. [DOI] [PubMed] [Google Scholar]
- 71.Sanjuán R, Bordería AV. 2011. Interplay between RNA structure and protein evolution in HIV-1. Mol Biol Evol 28:1333–1338. doi: 10.1093/molbev/msq329. [DOI] [PubMed] [Google Scholar]
- 72.Berkhout B. 1992. Structural features in TAR RNA of human and simian immunodeficiency viruses: a phylogenetic analysis. Nucleic Acids Res 20:27–31. doi: 10.1093/nar/20.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tuplin A, Wood J, Evans DJ, Patel AH, Simmonds P. 2002. Thermodynamic and phylogenetic prediction of RNA secondary structures in the coding region of hepatitis C virus. RNA 8:824–841. doi: 10.1017/S1355838202554066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zuker M. 2003. mFold Web server for nucleic acid folding and hybridization predicition. Nucleic Acids Res 31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Patiño-Galindo JÁ, González-Candelas F. 2017. Comparative analysis of variation and selection in the HCV genome. Infect Genet Evol 49:104–110. doi: 10.1016/j.meegid.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 76.Moss WN, Priore SF, Turner DH. 2011. Identification of potential conserved RNA secondary structure throughout influenza A coding regions. RNA 17:991–1011. doi: 10.1261/rna.2619511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cloete LJ, Tanov EP, Muhire BM, Martin DP, Harkins GW. 2014. The influence of secondary structure, selection and recombination on rubella virus nucleotide substitution rate estimates. Virol J 11:166. doi: 10.1186/1743-422X-11-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jerzak GVS, Bernard K, Kramer LD, Shi PY, Ebel GD. 2007. The West Nile virus mutant spectrum is host-dependant and a determinant of mortality in mice. Virology 360:469–476. doi: 10.1016/j.virol.2006.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Grubaugh ND, Fauver JR, Ruckert C, Weger-Lucarelli J, Garcia-Luna S, Murrieta RA, Gendernalik A, Smith DR, Brackney DE, Ebel GD. 2017. Mosquitoes transmit unique West Nile virus populations during each feeding episode. Cell Rep 19:709–718. doi: 10.1016/j.celrep.2017.03.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ehrbar DJ, Ngo KA, Campbell SR, Kramer LD, Ciota AT. 2017. High levels of local inter- and intra-host genetic variation of West Nile virus and evidence of fine-scale evolutionary pressures. Infect Genet Evol 51:219–226. doi: 10.1016/j.meegid.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 81.Jerzak GVS, Brown I, Shi PY, Kramer LD, Ebel GD. 2008. Genetic diversity and purifying selection in West Nile virus populations are maintained during host switching. Virology 374:256–260. doi: 10.1016/j.virol.2008.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ciota AT, Kramer LD. 2013. Vector-virus interactions and transmission dynamics of West Nile virus. Viruses 5:3021–3047. doi: 10.3390/v5123021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ciota AT, Kramer LD. 2010. Insights into arbovirus evolution and adaptation from experimental studies. Viruses 2:2594–2617. doi: 10.3390/v2122594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Parameswaran P, Charlebois P, Tellez Y, Nunez A, Ryan EM, Malboeuf CM, Levin JZ, Lennon NJ, Balmaseda A, Harris E, Henn MR. 2012. Genome-wide patterns of intrahuman dengue virus diversity reveal associations with viral phylogenetic clade and interhost diversity. J Virol 86:8546–8558. doi: 10.1128/JVI.00736-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Holmes EC. 2003. Patterns of intra- and interhost nonsynonymous variation reveal strong purifying selection in dengue virus. J Virol 77:11296–11298. doi: 10.1128/jvi.77.20.11296-11298.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lequime S, Fontaine A, Ar Gouilh M, Moltini-Conclois I, Lambrechts L. 2016. Genetic drift, purifying selection and vector genotype shape dengue virus intra-host genetic diversity in mosquitoes. PLoS Genet 12:e1006111. doi: 10.1371/journal.pgen.1006111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bonneau KR, Mullens BA, MacLachlan NJ. 2001. Occurrence of genetic drift and founder effect during quasispecies evolution of the VP2 and NS3/NS3A genes of bluetongue virus upon passage between sheep, cattle, and Culicoides sonorensis. J Virol 75:8298–8305. doi: 10.1128/jvi.75.17.8298-8305.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kils-Hütten L, Cheynier R, Wain-Hobson S, Meyerhans A. 2001. Phylogenetic reconstruction of intrapatient evolution of human immunodeficiency virus type 1: predominance of drift and purifying selection. J Gen Virol 82:1621–1627. doi: 10.1099/0022-1317-82-7-1621. [DOI] [PubMed] [Google Scholar]
- 89.Geller R, Estada Ú, Peris JB, Andreu I, Bou JV, Garijo R, Cuevas JM, Sabariegos R, Mas A, Sanjuán R. 2016. Highly heterogeneous mutation rates in the hepatitis C virus genome. Nat Microbiol 1:16045. doi: 10.1038/nmicrobiol.2016.45. [DOI] [PubMed] [Google Scholar]
- 90.Lo MK, Tilgner M, Bernard KA, Shi P-Y. 2003. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3ʹ untranslated region of West Nile virus by use of a reporting replicon that differentiates between viral translation and RNA replication. J Virol 77:10004–10014. doi: 10.1128/jvi.77.18.10004-10014.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Basu M, Brinton MA. 2011. West Nile virus (WNV) genome RNAs with up to three adjacent mutations that disrupt long distance 5ʹ-3ʹ cyclization sequence basepairs are viable. Virology 412:220–232. doi: 10.1016/j.virol.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jinek M, Coyle SM, Doudna JA. 2011. Coupled 5′ nucleotide recognition and processivity in Xrn1-mediated mRNA decay. Mol Cell 41:600–608. doi: 10.1016/j.molcel.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Romero TA, Tumban E, Jun J, Lott WB, Hanley KA. 2006. Secondary structure of dengue virus type 4 3ʹ untranslated region: impact of deletion and substitution mutations. J Gen Virol 87:3291–3296. doi: 10.1099/vir.0.82182-0. [DOI] [PubMed] [Google Scholar]
- 94.Friebe P, Peña J, Pohl MOF, Harris E. 2012. Composition of the sequence downstream of the dengue virus 5′ cyclization sequence (dCS) affects viral RNA replication. Virol 422:346–356. doi: 10.1016/j.virol.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dethoff EA, Gokhale NS, Boerneke MA, Muhire BM, Martin DP, Sacco MT, McFadden MJ, Weinstein JA, Messer WB, Horner SM, Weeks KM. 2018. Pervasive tertiary structure in the dengue virus RNA genome. Proc Natl Acad Sci U S A 115:11513–11518. doi: 10.1073/pnas.1716689115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brackney DE, Beane JE, Ebel GD. 2009. RNAi targeting of West Nile virus in mosquito midguts promotes virus diversification. PLoS Pathog 5:e1000502. doi: 10.1371/journal.ppat.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Deardorff ER, Fitzpatrick KA, Jerzak GVS, Shi P-Y, Kramer LD, Ebel GD. 2011. West Nile virus experimental evolution in vivo and the trade-off hypothesis. PLoS Pathog 7:e1002335. doi: 10.1371/journal.ppat.1002335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Warren TK, Shurtleff AC, Bavari S. 2012. Advanced morpholino oligomers: a novel approach to antiviral therapy. Antiviral Res 94:80–88. doi: 10.1016/j.antiviral.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stein DA, Shi P-Y. 2008. Nucleic acid-based inhibition of flavivirus infections. Front Biosci 13:1385–1395. doi: 10.2741/2769. [DOI] [PubMed] [Google Scholar]
- 100.Beloor J, Maes N, Ullah I, Uchil P, Jackson A, Fikrig E, Lee SK, Kumar P. 2018. Small interfering RNA-mediated control of virus replication in the CNS is therapeutic and enables natural immunity to West Nile virus. Cell Host Microbe 23:549–556.e3. doi: 10.1016/j.chom.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Deas TS, Binduga-Gajewska I, Tilgner M, Ren P, Stein DA, Moulton HM, Iversen PL, Kauffman EB, Kramer LD, Shi P-Y. 2005. Inhibition of flavivirus infections by antisense oligomers specifically suppressing viral translation and RNA replication. J Virol 79:4599–4609. doi: 10.1128/JVI.79.8.4599-4609.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kanda T, Steele R, Ray R, Ray RB. 2007. Small interfering RNA targeted to hepatitis C virus 5ʹ nontranslated region exerts potent antiviral effect. J Virol 81:669–676. doi: 10.1128/JVI.01496-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Deas TS, Bennett CJ, Jones SA, Tilgner M, Ren P, Behr MJ, Stein DA, Iversen PL, Kramer LD, Bernard KA, Shi PY. 2007. In vitro resistance selection and in vivo efficacy of morpholino oligomers against West Nile virus. Antimicrob Agents Chemother 51:2470–2482. doi: 10.1128/AAC.00069-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shi P-Y, Tilgner M, Lo MK, Kent K. a, Bernard K. a. 2002. Infectious cDNA clone of the epidemic West Nile virus from New York City. J Virol 76:5847–5856. doi: 10.1128/jvi.76.12.5847-5856.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Prinzinger R, Preßmar A, Schleucher E. 1991. Body temperature in birds. Comp Biochem Physiol Part A Mol Integr Physiol 99:499–506. doi: 10.1016/0300-9629(91)90122-S. [DOI] [Google Scholar]
- 107.Nemeth NM, Thomsen BV, Spraker TR, Benson JM, Bosco-Lauth AM, Oesterle PT, Bright JM, Muth JP, Campbell TW, Gidlewski TL, Bowen RA. 2011. Clinical and pathologic responses of American crows (Corvus brachyrhynchos) and fish crows (C. ossifragus) to experimental West Nile virus infection. Vet Pathol 48:1061–1074. doi: 10.1177/0300985811398249. [DOI] [PubMed] [Google Scholar]
- 108.R Core Team. 2016. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
5′ UTR structures. Download Table S1, DOCX file, 2.1 MB (2.1MB, docx) .
Copyright © 2019 Scroggs et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Structure comparison from mFold and RNAstructure at 37°C and 40°C. Download Table S2, DOCX file, 2.0 MB (2MB, docx) .
Copyright © 2019 Scroggs et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Comparison of first and second predicted structures and ΔG values (kcal/mol) by mFold. Download Table S3, DOCX file, 0.9 MB (910.8KB, docx) .
Copyright © 2019 Scroggs et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
χ2 test results for differences between expected and observed distributions of DS and SS mutations from Fig. 4. Download Table S4, DOCX file, 0.01 MB (13.5KB, docx) .
Copyright © 2019 Scroggs et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
χ2 test results for differences between expected and observed mutations within and outside binding sites from Fig. 5b to d. Download Table S5, DOCX file, 0.01 MB (13.3KB, docx) .
Copyright © 2019 Scroggs et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The sequencing data utilized for this project have been deposited in the NCBI Sequence Read Archive database and can be accessed with the BioProject identifier PRJNA281547.