Campylobacter is a leading cause of foodborne diarrheal illness worldwide, with more than one million cases each year in the United States alone. The global emergence of antimicrobial resistance in this pathogen has become a growing public health concern. Florfenicol-resistant (FFNr) Campylobacter has been very rare in the United States. In this study, we employed whole-genome sequencing to characterize 16 multidrug-resistant Campylobacter coli isolates recovered from cattle in the United States. A gene [cfr(C)] was found to be responsible for resistance not only to florfenicol but also to several other antimicrobials, including linezolid, a critical drug for treating infections by Gram-positive bacteria in humans. The results showed that cfr(C) is located in a conjugative pTet MDR/virulence plasmid. This report highlights the power of antimicrobial resistance surveillance to uncover the intricacies of transmissible coresistance and provides information that is needed for accurate risk assessment and mitigation strategies.
KEYWORDS: Campylobacter, MDR, NARMS, WGS, florfenicol resistance, plasmid
ABSTRACT
Genomic analyses were performed on florfenicol-resistant (FFNr) Campylobacter coli isolates recovered from cattle, and the cfr(C) gene-associated multidrug resistance (MDR) plasmid was characterized. Sixteen FFNr C. coli isolates recovered between 2013 and 2018 from beef cattle were sequenced using MiSeq. Genomes and plasmids were found to be closed for three of the isolates using the PacBio system. Single nucleotide polymorphisms (SNPs) across the genome and the structures of MDR plasmids were investigated. Conjugation experiments were performed to determine the transferability of cfr(C)-associated MDR plasmids. The spectrum of resistance encoded by the cfr(C) gene was further investigated by agar dilution antimicrobial susceptibility testing. All 16 FFNr isolates were MDR and exhibited coresistance to ciprofloxacin, nalidixic acid, clindamycin, and tetracycline. All isolates shared the same resistance genotype, carrying aph (3′)-III, hph, ΔaadE (truncated), blaOXA-61, cfr(C), and tet(O) genes plus a mutation of GyrA (T86I). The cfr(C), aph (3′)-III, hph, ΔaadE, and tet(O) genes were colocated on transferable MDR plasmids ranging in size from 48 to 50 kb. These plasmids showed high sequence homology with the pTet plasmid and carried several Campylobacter virulence genes, including virB2, virB4, virB5, VirB6, virB7, virB8, virb9, virB10, virB11, and virD4. The cfr(C) gene conferred resistance to florfenicol (8 to 32 μg/ml), clindamycin (512 to 1,024 μg/ml), linezolid (128 to 512 μg/ml), and tiamulin (1,024 μg/ml). Phylogenetic analysis showed SNP differences ranging from 11 to 2,248 SNPs among the 16 isolates. The results showed that the cfr(C) gene located in the conjugative pTet MDR/virulence plasmid is present in diverse strains, where it confers high levels of resistance to several antimicrobials, including linezolid, a critical drug for treating infections by Gram-positive bacteria in humans. This report highlights the power of genomic antimicrobial resistance surveillance to uncover the intricacies of transmissible coresistance and provides information that is needed for accurate risk assessment and mitigation strategies.
IMPORTANCE Campylobacter is a leading cause of foodborne diarrheal illness worldwide, with more than one million cases each year in the United States alone. The global emergence of antimicrobial resistance in this pathogen has become a growing public health concern. Florfenicol-resistant (FFNr) Campylobacter has been very rare in the United States. In this study, we employed whole-genome sequencing to characterize 16 multidrug-resistant Campylobacter coli isolates recovered from cattle in the United States. A gene [cfr(C)] was found to be responsible for resistance not only to florfenicol but also to several other antimicrobials, including linezolid, a critical drug for treating infections by Gram-positive bacteria in humans. The results showed that cfr(C) is located in a conjugative pTet MDR/virulence plasmid. This report highlights the power of antimicrobial resistance surveillance to uncover the intricacies of transmissible coresistance and provides information that is needed for accurate risk assessment and mitigation strategies.
INTRODUCTION
Campylobacter is one of the leading bacterial causes of foodborne illness in the United States. Human infections are associated mainly with raw or undercooked chicken meat, but other sources such as beef, pork, lamb, water, and seafood also have been associated with Campylobacter infections (1). Antimicrobial resistance in Campylobacter is a public health concern (2–4). In 2013, the Centers for Disease Control and Prevention (CDC) classified drug-resistant Campylobacter as a serious threat in the United States (https://www.cdc.gov/drugresistance/threat-report-2013). The use of antimicrobials in animals and the potential contribution to generating resistance in foodborne bacteria have been important public health issues for many years. The U.S. National Antimicrobial Resistance Monitoring System (NARMS) was launched in 1996 to track changes in antimicrobial resistance in foodborne pathogens, including Campylobacter, isolated from food animals, retail meats, and humans. Currently, nine antimicrobials belonging to seven classes are included in the NARMS Campylobacter testing panel.
Florfenicol (FFN) belongs to a class of phenicol antimicrobials whose members are approved in the United States for treatment of bovine and swine respiratory infections (5, 6). Since 2004, NARMS has monitored resistance to florfenicol in Campylobacter and resistance has been very rare in human and food isolates, although resistance has been monitored in cecal samples only since 2013. The first florfenicol-resistant (FFNr) Campylobacter coli strains were detected in 2013 in cecal samples from beef cattle, accounting for 1.6% of the beef cattle isolates tested (n = 128), and the proportion increased to 4.4% resistant isolates in 2014 (n = 180). No FFNr C. coli isolates were detected in 2015 (n = 181), but such strains reappeared in 2016 (n = 200) and 2017 (n = 239), accounting for 2% and 0.8% of the resistance detected in the beef cattle C. coli isolates tested, respectively. All FFNr Campylobacter isolates were C. coli recovered from cecal contents collected from beef cattle postslaughter and prior to any intervention steps. Antimicrobial susceptibility testing (AST) showed that all of the FFNr C. coli isolates were multidrug resistant (MDR) and showed resistance to five of nine antimicrobials tested, including resistance to ciprofloxacin, clindamycin, florfenicol, nalidixic acid, and tetracycline.
The cfr(C) gene was first reported in 2017 by Tang et al. and was shown to be responsible for florfenicol resistance in C. coli (7). The cfr(C) gene encodes a protein that shares 55.1% and 54.9% amino acid identity with Cfr and Cfr(B), respectively (7). The cfr gene was first detected in Staphylococcus sciuri isolated from a bovine origin in 2000 (8) and was later detected in an Enterococcus faecium isolate collected from a human bloodstream infection in 2015 (9). The cfr(B) gene also has been detected in Clostridium difficile and E. faecium from humans (10, 11). Although the three cfr alleles show high sequence diversity, all of them confer resistance to members of five chemically unrelated antimicrobial classes, including phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramins (PhLOPSA phenotype) (7, 12). Previous studies showed that the cfr, cfr(B), and cfr(C) genes are located on various plasmids (7, 10, 11).
In the original study reported by Tang et al. (7), all FFNr C. coli isolates were recovered from cattle, with a 10% prevalence rate, but no FFNr Campylobacter jejuni isolates were detected. Their study showed that the cfr(C) gene located in the conjugative MDR plasmid also carried several other resistance genes, including tet(O), hph, and aphA-3, which conferred resistance to tetracycline, hygromycin, and kanamycin, respectively. The plasmid also carried a truncated aadE gene. Pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST) analysis showed that the increasing prevalence of cfr(C) in C. coli is due to clonal expansion (7). To further understand the mechanism of FFNr and the genetic context of its spread over time, we performed whole-genome sequencing (WGS) analyses of 16 FFNr C. coli isolates recovered between 2013 and 2018 to identify the resistance genotype and characterize FFNr MDR plasmids.
RESULTS AND DISCUSSION
Resistance phenotypes and genotypes.
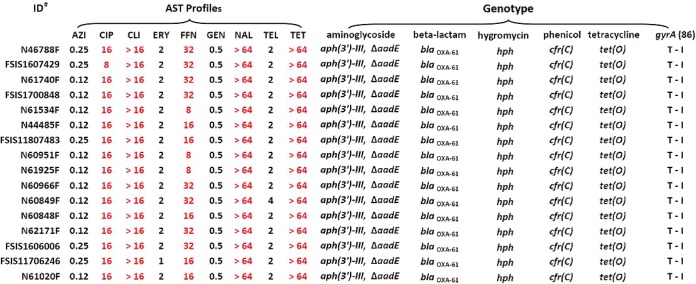
All 16 FFNr isolates were MDR and showed coresistance to ciprofloxacin (CIP), nalidixic acid (NAL), clindamycin (CLI), and tetracycline (TET) in testing using a NARMS Campylobacter panel. Additionally, they shared the same resistance genotype, carrying aph (3′)-III, hph, ΔaadE, blaOXA-61, cfr(C), and tet(O) genes plus the same mutation of GyrA T86I (Fig. 1). The cfr(C), tet(O), and gyrA T86I mutations are responsible for resistance to FFN/CLI, TET, and CIP/NAL, respectively, which showed a 100% correlation between resistance phenotype and genotype in the 16 isolates. aph (3′)-III, hph, and blaOXA-61 encode resistance to kanamycin, hygromycin, and β-lactam antibiotics, respectively, but these drugs were not included in the NAMRS testing panel. FFNr strain Tx40 reported by Tang was also MDR and showed resistance to CIP, TET, CLI, FFN, linezolid (LZD), tiamulin (TIA), chloramphenicol (CHL), and tedizolid (TED) (7).
FIG 1.
Antimicrobial susceptibility testing profiles and resistance genotypes of FFNr Campylobacter coli strains isolated from cattle. ID, identifier; AZI, azithromycin; CIP, ciprofloxacin; CLI, clindamycin; ERY, erythromycin; FFN, florfenicol; GEN, gentamycin; NAL, nalidixic acid; TEL, telithromycin; TET, tetracycline.
Conjugation and cfr(C) coresistance to other antimicrobials.
Two FFNr C. coli strains, N61740F and N61925F, carrying the cfr(C) gene were used as donors for the conjugation experiment. The results showed that the cfr(C) gene was successfully transferred to a FFNS C. jejuni strain (N18880) based on species confirmation by PCR and the AST profiles of transconjugants. Two transconjugants (TCN61740F and TCN61925F) showed increasing MICs of CLI and FFN (≥4-fold and ≥8-fold, respectively) compared to the N18880 parent recipient strain (Table 1). Agar dilution antimicrobial sensitivity testing determined that the MICs of CLI, LZD, and TIA increased 32-fold to 64-fold, 8-fold to 32-fold, and 512-fold, respectively, in two transconjugants compared with the parent recipient strain (Table 1). Similar findings were reported from Tang’s study (7). Their results showed that the MICs of CLI, LZD, and TIA for transconjugant JL272/pTx40 or cloning/transformant C. jejuni 11168/pRY108-cfr(C) increased 32-fold, >16-fold, and >128-fold, respectively, compared with the parent strains (7). Both studies showed that differences in the genetic background of recipient/parent cells and the use of different susceptibility testing methods could result in variations in MICs.
TABLE 1.
Antimicrobial susceptibility of donors, recipients, and transconjugantsa
| CVM no. | MIC (μg/ml) |
Description | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Microbroth dilution |
Agar dilution |
||||||||||||
| AZI | CIP | CLI | ERY | FFN | GEN | NAL | TEL | TET | LZD | TIA | CLI | ||
| N61740F | 0.12 | 16 | >16 | 2 | 32 | 0.5 | >64 | 2 | >64 | 256 | 1,024 | 1,024 | Donor |
| N61925F | 0.12 | 16 | >16 | 2 | 16 | 0.5 | >64 | 2 | 64 | 256 | 1,024 | 1,024 | Donor |
| TCN61740F | >64 | 0.12 | >16 | 64 | 16 | 0.5 | ≤4 | 8 | 64 | 512 | 1,024 | 1,024 | Transconjugant |
| TCN61925F | >64 | 0.12 | 16 | 64 | 8 | 0.5 | ≤4 | 8 | >64 | 128 | 1,024 | 512 | Transconjugant |
| N18880R | >64 | 0.12 | 4 | >64 | 1 | 0.5 | ≤4 | 8 | 0.5 | 16 | 2 | 16 | Recipient |
CVM, Center for Veterinary Medicine; AZI, azithromycin; CIP, ciprofloxacin; CLI, clindamycin; ERY, erythromycin; FFN, florfenicol; GEN, gentamycin; NAL, nalidixic acid; TEL, telithromycin; TET, tetracycline; LZD, linezolid; TIA, tiamulin.
MDR virulence plasmids.
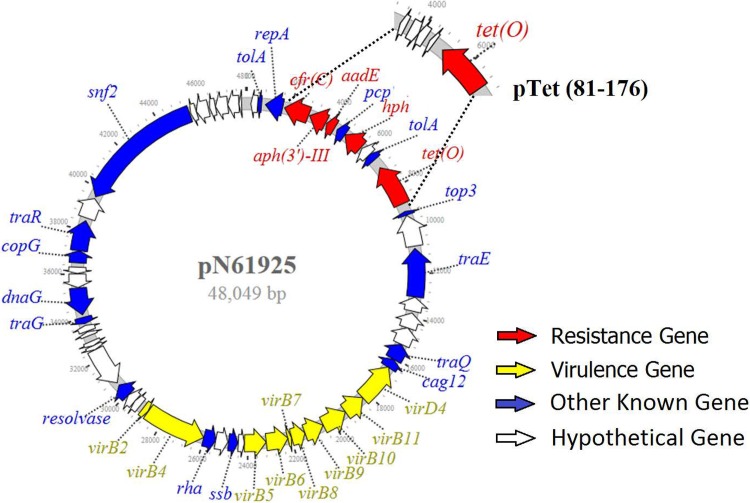
Three plasmids from strains N61925, N61740, and N46788F were closed using the PacBio long sequencing platform. Two plasmids, pN61925 and pN61740, were identical in size (48,049 bp), with 99.9% sequence identity. The third plasmid, pN46788F, consisted of 50,413 bp and showed >91% sequence identity with pN61925 and pN61740. All three plasmids were annotated to include 55 similar open reading frames (ORFs), including 22 encoding known function proteins and 23 encoding hypothetical proteins (Fig. 2). Among the genes with known functions, 4 resistance genes, namely, tet(O), hph, aph (3′)-III, and cfr(C), and 1 truncated resistance gene, ΔaadE, plus 10 virulence genes, virB2, virB4 (two copies), virB5, virB6, virB7, virB8, virB9, virB10, and virB11, were identified (Fig. 2). The structure and gene organization of pN61925, pN61740, and pN46788 were the same as those of the pTx40 plasmid reported previously by Tang et.al (7). We also compared the DNA sequences and overall gene organization characteristics of the pN61925 plasmid and the pTet 81-176 plasmid, the first pTet plasmid reported from Campylobacter (13). The two plasmids shared about 41 kb of sequence, and the only sequence difference was a region that encodes antimicrobial resistance genes. The pTet 81-176 plasmid carried only the tet(O) gene, whereas the pN61925 carried five resistance genes, namely, ΔaadE, hph, aph (3′)-III, cfr(C), and tet(O), in addition to pcp and tolA (Fig. 2). A plasmid with a structure similar to that of pTet 81-176 was also identified from C. coli isolated from NARMS retail chicken in early 2011 (14). The plasmid carried several antibiotic resistance genes, including a novel gentamicin resistance gene [aph(2’’)-Ig].
FIG 2.
Structure of multidrug resistance/virulence plasmid from FFNr Campylobacter coli.
The virulence factors encoded by pTet and cfr(C) plasmids were previously reported (13, 15, 16). These virulence factors are involved in bacterial pathogenesis, including adherence, invasion, motility, and immune evasion (15, 16). Some of these virulence factors are involved with the structure of type IV secretion systems (T4SSs), which have been found in both Gram-positive and Gram-negative bacteria, including Agrobacterium tumefaciens, Bordetella pertussis, several Brucella species, C. jejuni, Helicobacter pylori, and others (17, 18). T4SSs comprise a class of diverse transporters and secrete a wide range of substrates, ranging from single proteins to protein-protein and protein-DNA complexes, which are required for virulence in many pathogens (17, 18). More studies on Campylobacter pathogenesis associated with T4SSs and the pTet MDR virulence plasmid are needed.
Whole-genome SNP analysis.
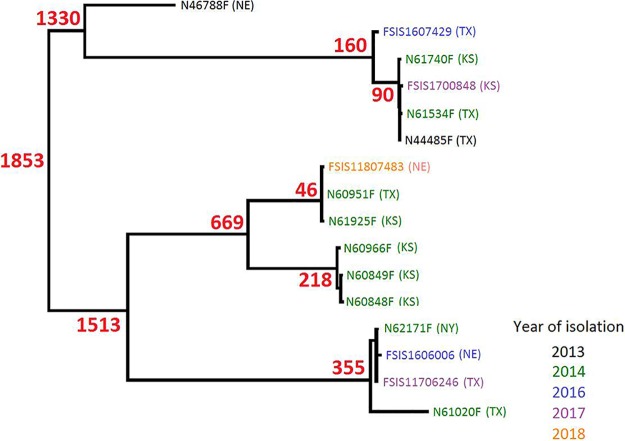
The SNP tree of 16 FFNr C. coli isolates showed that they were genetically diverse, and the 16 isolates were grouped into several distinct clusters. The number of whole-genome SNP (wgSNP) differences among 16 isolates ranged from 11 to 2,248 despite some of isolates having been collected in the same years from the same state (Fig. 3). For example, all 9 isolates recovered in 2014 (five from Kansas, three from Texas, and one from New York state) were scattered in all branches of the phylogenetic tree with a maximum 2,248 SNP differences, even though some isolates were from the same states and shared the same resistance phenotype and genotype (Fig. 1). This was most likely due to all isolates containing the same MDR plasmid carrying the same resistance genes, including aph (3′)-III, ΔaadE, hph, cfr(C), and tet(O). All isolates also carried a blaOXA-61 gene and had a mutation in GyrA (T86I), responsible for beta-lactam and quinolone resistance phenotypes, respectively (19). Previous studies showed that a blaOXA-61 gene is commonly present on the chromosome of C. jejuni and C. coli (20, 21).
FIG 3.
High-quality SNP (hqSNP) core genome tree of FFNr Campylobacter coli strains isolated from cattle.
Tang et al. characterized 34 cfr(C)-positive C. coli isolates by PFGE and MLST and suggested that clonal expansion was involved in the spread of cfr(C)-positive C. coli isolates in feedlot cattle in the United States (7). The phylogenetic tree showed that the 16 isolates were grouped into several distinct clusters, suggesting that these isolates belong to several clones. Within each cluster, there were isolates from different states and years, suggesting that clonal expansion also played a role in the dissemination of the cfr(C)-positive C. coli isolates in different feedlots. Tang’s study showed that all 34 positive C. coli isolates belong to a single clone based on PFGE and MLST. In contrast, the SNP tree showed that 16 cfr(C)-positive C. coli isolates belong to several clones, indicating that a MDR cfr(C) plasmid could also be disseminated through horizontal transfer. The differences between these two studies may be due to the sampling interval. The isolates in Tang’s study were isolated earlier, presumably closer to the original emergence event during which cfr(C) spread on United States cattle farms, mainly through an ascendant clone, followed by plasmid dissemination. The second possible factor accounting for the differences could the different subtyping methods used to define clonality. WGS has more discriminatory power than PFGE and MLST and can provide better confirmation for clonality. The cfr(C) gene encodes resistance to several antimicrobials, including the oxazolidinone class, whose members represent the last resort for treating MDR Gram-positive bacterial infections in humans. So far, the cfr(C) gene has not been reported in Campylobacter isolates recovered from humans, U.S. Department of Agriculture Food Safety and Inspection Service (USDA FSIS) regulatory samples, or retail meat, and neither has the cfr(C) gene been detected in C. jejuni in the United States. Continued monitoring of cfr(C) transmission is needed to understand the spread of the gene and of the MDR plasmids to different bacterial pathogens.
MATERIALS AND METHODS
Bacterial strains.
All 16 FFNr C. coli isolates were recovered from beef cattle cecal contents between 2013 and 2018 as part of the US NARMS program (Table 2). The isolates were obtained by the USDA FSIS. The isolates were grown on sheep blood agar plates (Thermo Fisher Scientific/Remel, Lenexa, KS) at 42°C under microaerobic conditions (85% N2, 10% CO2, and 5% O2). WGS data for the FFNr C. coli isolates recovered from cattle cecal contents at slaughter were generated by FSIS or FDA. One florfenicol-susceptible (FFNs), erythromycin-resistant (ERYr) C. jejuni isolate (N18880) recovered from retail chicken was used as the recipient strain for conjugation experiments. The resistance phenotypes of all 16 isolates were previously determined by the broth microdilution method using the NARMS Campylobacter panel (22).
TABLE 2.
Florfenicol-resistant Campylobacter coli strains in this studya
| Strain ID | Month | Yr | Source(s) | State | NCBI accession no. |
|---|---|---|---|---|---|
| N44485F | April | 2013 | Heifer | TX | SRR7821186 |
| N46788Fb | July | 2013 | Steer | NE | SRR7821185/MK541987 |
| N60848F | February | 2014 | Heifer | KS | SRR7821188 |
| N60849F | February | 2014 | Beef cows | KS | SRR7821187 |
| N60951F | February | 2014 | Steer | TX | SRR7821190 |
| N60966F | February | 2014 | Beef cows | KS | SRR7821189 |
| N61020F | March | 2014 | Heifer | TX | SRR7821192 |
| N61534F | May | 2014 | Steer | TX | SRR7821191 |
| N61740Fb | July | 2014 | Steer | KS | SRR7821184/MK541988 |
| N61925Fb | August | 2014 | Steer | KS | SRR7821183/MK541989 |
| N62171F | October | 2014 | Steer | NY | SRR7821182 |
| FSIS1606006 | February | 2016 | Heifer | NE | SRR3214652 |
| FSIS1607429 | July | 2016 | Heifer | TX | SRR4175492 |
| FSIS1700848 | March | 2017 | Heifer | KS | SRR5517169 |
| FSIS11706246 | November | 2017 | Heifer | TX | SRR6495259 |
| FSIS11807483 | January | 2018 | Heifer | NE | SRR6743237 |
KS, Kansas; NE, Nebraska; TX, Texas; NY, New York.
The indicated isolates were sequenced by the use of both the MiSeq and PacBio platforms.
Genome sequencing, assembly, and annotation.
Genomic DNA was extracted by the use of a QIAamp 96 DNA QIA cube HT kit (Qiagen, Gaithersburg, MD, USA) and an automated high-throughput DNA extraction machine (QIAcube HT) per the manufacturer’s instructions. WGS was performed on an Illumina MiSeq platform using v3 reagent kits (Illumina, San Diego, CA, USA) and the 2 × 300 paired-end option. Assembly was performed de novo for each isolate using CLC Genomics Workbench version 8.0 (CLC bio, Aarhus, Denmark). Three isolates (N46788F, N61740F, and N61925F) were selected to close the genomes and plasmids by the use of a Pacific Biosciences (PacBio) RS II sequencer (PacBio, Menlo Park, CA. USA). The continuous-long reads were assembled by the use of the PacBio Hierarchical Genome Assembly Process (HGAP3.0) program. Genomes were annotated using the RAST annotation server (http://rast.nmpdr.org/). Among the 16 FFNr C. coli isolates sequenced on the MiSeq, there was a median of 70 contigs (ranging from 23 to 573) and 103-fold coverage (ranging from 26 to 138) per genome.
Identification of antimicrobial resistance genotypes.
Antimicrobial resistance genes were identified by using Perl scripts to perform local BLAST with ResFinder (https://cge.cbs.dtu.dk/services/ResFinder/) with at least 85% nucleotide identity and 50% sequence length corresponding to known resistance gene sequences. Sequences showing less than 100% identity and/or sequence length were examined by additional BLAST analysis to identify the appropriate resistance genes. Mutations in the gyrA gene were identified using an in-house pipeline (19).
Plasmid analysis.
The pTet (81-176) plasmid sequences were downloaded from NCBI GenBank under accession number AY394561. Our cfr(C) MDR plasmid sequences were subjected to a BLAST search against the pTet plasmid sequences to determine sequence homology. The pTet plasmid and our cfr(C) MDR plasmids were also annotated using the RAST annotation server to compare the annotated genes among these plasmids.
Whole-genome phylogenetic analysis.
Single nucleotide polymorphism (SNP) analysis of 16 FFNr C. coli isolates was performed using the Food and Drug Administration Center for Food Safety and Applied Nutrition (CFSAN) SNP Pipeline (http://snp-pipeline.readthedocs.io/en/latest/). The complete genome of C. coli strain MG1116 (NCBI accession number CP017868) was used as a reference genome. VarScan (23) was used to detect SNPs. Plasmid sequences were excluded from the SNP analysis. SNP redundancy by linkage disequilibrium (LD) was reduced and the phylogenetic tree was constructed with the maximum likelihood algorithm using the SNPhylo package (24).
Conjugation and antimicrobial susceptibility testing.
Two FFNr C. coli isolates (N61740F and N61925F) that carried the cfr(C) gene were used as donor strains. C. jejuni N18880, susceptible to florfenicol (FFNs) but resistant to erythromycin (ERYr), was chosen as a recipient strain based on its antimicrobial susceptibility profile (Table 1). C. jejuni N18880 was isolated from chicken breast in 2008 and was previously sequenced (accession number SRR9072097). The method used for the agar plate mating experiments was previously described by Chen et al. (14). Briefly, to prepare donor and recipient strains, one loopful of bacteria grown overnight on a sheep blood agar plate was resuspended in 200 μl LB broth; 10 μl of each donor strain was spotted separately onto each of seven 10-μl spots of recipient strain on a fresh sheep blood agar plate. Plates were incubated overnight at 42°C under microaerobic conditions. Each coculture was scraped from the plate and resuspended in 500 μl LB broth. A 100 μl volume of each of 1:10 and 1:100 dilutions of the resuspension was plated on agar plates supplemented with appropriate selective agents. Florfenicol (8 μg/ml) and erythromycin (16 μg/ml) were used as selecting markers for the conjugation experiment. Successful transconjugants were confirmed by comparing the resistant phenotypes of donors, recipients, and transconjugants by the use of a Sensititre automated antimicrobial susceptibility system in accordance with the instructions of the manufacturer (Thermo Fisher Scientific; Trek Diagnostics, Cleveland, OH) and the NARMS Campylobacter panel (catalog no. CAMPY). Nine antimicrobial agents were tested, including azithromycin (AZI), ciprofloxacin (CIP), clindamycin (CLI), erythromycin (ERY), florfenicol (FFN), gentamicin (GEN), nalidixic acid (NAL), telithromycin (TEL), and tetracycline (TET). C. jejuni ATCC 33560 was used as the quality control organism according to guidelines of the Clinical and Laboratory Standards Institute (CLSI). Interpretation of susceptibility testing results was based on the European Committee on Antimicrobial Susceptibility Testing (EUCAST) epidemiological cutoff values (http://www.eucast.org/). PCR analysis was used to confirm that the transconjugants were C. jejuni (25).
To measure the contribution of cfr(C) to linezolid (LZD), tiamulin (TIA), and clindamycin (CLI) resistance, MICs were measured for the donors, recipients, and transconjugants by agar dilution as described previously after transmission of the cfr(C)-containing plasmid (26, 27). Briefly, agar plates were prepared with four drugs with ranges of concentrations from 0.125 μg/ml to 1,024 μg/ml for each antimicrobial. MICs were determined based on CLSI guidelines (28) and were recorded as the lowest concentration of antimicrobial agent that completely inhibited the visible growth of the organism on the agar surface after incubation at 42°C for 24 h.
ACKNOWLEDGMENTS
We are grateful to Maureen Davidson for helpful comments and manuscript review. We also acknowledge Claudia Lam for handling all WGS submissions.
This work was supported by internal funds of the U.S. Food and Drug Administration.
The views expressed in this article are ours and do not necessarily reflect the official policy of the Department of Health and Human Services, the U.S. Food and Drug Administration, and Centers for Disease Control and Prevention or the U.S. government. Mention of trade names or commercial products in this publication is solely for providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture or Food and Drug Administration.
We declare that we have no conflicts of interest.
REFERENCES
- 1.Zhao S, Young SR, Tong E, Abbott JW, Womack N, Friedman SL, McDermott PF. 2010. Antimicrobial resistance of Campylobacter isolates from retail meat in the United States between 2002 and 2007. Appl Environ Microbiol 76:7949–7956. doi: 10.1128/AEM.01297-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scallan E, Griffin PM, Angulo FJ, Tauxe RV, Hoekstra RM. 2011. Foodborne illness acquired in the United States–unspecified agents. Emerg Infect Dis 17:16–22. doi: 10.3201/eid1701.091101p2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luangtongkum T, Jeon B, Han J, Plummer P, Logue CM, Zhang Q. 2009. Antibiotic resistance in Campylobacter: emergence, transmission and persistence. Future Microbiol 4:189–200. doi: 10.2217/17460913.4.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen X, Naren GW, Wu CM, Wang Y, Dai L, Xia LN, Luo PJ, Zhang Q, Shen JZ. 2010. Prevalence and antimicrobial resistance of Campylobacter isolates in broilers from China. Vet Microbiol 144:133–139. doi: 10.1016/j.vetmic.2009.12.035. [DOI] [PubMed] [Google Scholar]
- 5.Schwarz S, Kehrenberg C, Doublet B, Cloeckaert A. 2004. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol Rev 28:519–542. doi: 10.1016/j.femsre.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 6.White DG, Piddock LJ, Maurer JJ, Zhao S, Ricci V, Thayer SG. 2000. Characterization of fluoroquinolone resistance among veterinary isolates of avian Escherichia coli. Antimicrob Agents Chemother 44:2897–2899. doi: 10.1128/aac.44.10.2897-2899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang Y, Dai L, Sahin O, Wu Z, Liu M, Zhang Q. 2017. Emergence of a plasmid-borne multidrug resistance gene cfr(C) in foodborne pathogen Campylobacter. J Antimicrob Chemother 72:1581–1588. doi: 10.1093/jac/dkx023. [DOI] [PubMed] [Google Scholar]
- 8.Schwarz S, Werckenthin C, Kehrenberg C. 2000. Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob Agents Chemother 44:2530–2533. doi: 10.1128/aac.44.9.2530-2533.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morroni G, Brenciani A, Antonelli A, D'Andrea MM, Di Pilato V, Fioriti S, Mingoia M, Vignaroli C, Cirioni O, Biavasco F, Varaldo PE, Giovanetti E. 2018. Characterization of a multiresistance plasmid carrying the optrA and cfr resistance genes from an Enterococcus faecium clinical isolate. Front Microbiol 9:2189. doi: 10.3389/fmicb.2018.02189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen LH, Vester B. 2015. A cfr-like gene from Clostridium difficile confers multiple antibiotic resistance by the same mechanism as the cfr gene. Antimicrob Agents Chemother 59:5841–5843. doi: 10.1128/AAC.01274-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deshpande LM, Ashcraft DS, Kahn HP, Pankey G, Jones RN, Farrell DJ, Mendes RE. 2015. Detection of a new cfr-like gene, cfr(B), in Enterococcus faecium isolates recovered from human specimens in the United States as part of the SENTRY Antimicrobial Surveillance Program. Antimicrob Agents Chemother 59:6256–6261. doi: 10.1128/AAC.01473-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Long KS, Poehlsgaard J, Kehrenberg C, Schwarz S, Vester B. 2006. The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob Agents Chemother 50:2500–2505. doi: 10.1128/AAC.00131-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Batchelor RA, Pearson BM, Friis LM, Guerry P, Wells JM. 2004. Nucleotide sequences and comparison of two large conjugative plasmids from different Campylobacter species. Microbiology 150:3507–3517. doi: 10.1099/mic.0.27112-0. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y, Mukherjee S, Hoffmann M, Kotewicz ML, Young S, Abbott J, Luo Y, Davidson MK, Allard M, McDermott P, Zhao S. 2013. Whole-genome sequencing of gentamicin-resistant Campylobacter coli isolated from U.S. retail meats reveals novel plasmid-mediated aminoglycoside resistance genes. Antimicrob Agents Chemother 57:5398–5405. doi: 10.1128/AAC.00669-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bacon DJ, Alm RA, Burr DH, Hu L, Kopecko DJ, Ewing CP, Trust TJ, Guerry P. 2000. Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect Immun 68:4384–4390. doi: 10.1128/iai.68.8.4384-4390.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bacon DJ, Alm RA, Hu L, Hickey TE, Ewing CP, Batchelor RA, Trust TJ, Guerry P. 2002. DNA sequence and mutational analyses of the pVir plasmid of Campylobacter jejuni 81-176. Infect Immun 70:6242–6250. doi: 10.1128/iai.70.11.6242-6250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ward DV, Draper O, Zupan JR, Zambryski PC. 2002. Peptide linkage mapping of the Agrobacterium tumefaciens vir-encoded type IV secretion system reveals protein subassemblies. Proc Natl Acad Sci U S A 99:11493–11500. doi: 10.1073/pnas.172390299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fronzes R, Christie PJ, Waksman G. 2009. The structural biology of type IV secretion systems. Nat Rev Microbiol 7:703–714. doi: 10.1038/nrmicro2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao S, Tyson GH, Chen Y, Li C, Mukherjee S, Young S, Lam C, Folster JP, Whichard JM, McDermott PF. 2016. Whole-genome sequencing analysis accurately predicts antimicrobial resistance phenotypes in Campylobacter spp. Appl Environ Microbiol 82:459–466. doi: 10.1128/AEM.02873-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alfredson DA, Korolik V. 2007. Antibiotic resistance and resistance mechanisms in Campylobacter jejuni and Campylobacter coli. FEMS Microbiol Lett 277:123–132. doi: 10.1111/j.1574-6968.2007.00935.x. [DOI] [PubMed] [Google Scholar]
- 21.Whitehouse CA, Young S, Li C, Hsu CH, Martin G, Zhao S. 2018. Use of whole-genome sequencing for Campylobacter surveillance from NARMS retail poultry in the United States in 2015. Food Microbiol 73:122–128. doi: 10.1016/j.fm.2018.01.018. [DOI] [PubMed] [Google Scholar]
- 22.FDA. 2015. National antimicrobial resistance monitoring system. https://www.fda.gov/animal-veterinary/national-antimicrobial-resistance-monitoring-system/2015-narms-integrated-report.
- 23.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. 2012. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee TH, Guo H, Wang X, Kim C, Paterson AH. 2014. SNPhylo: a pipeline to construct a phylogenetic tree from huge SNP data. BMC Genomics 15:162. doi: 10.1186/1471-2164-15-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang G, Clark CG, Taylor TM, Pucknell C, Barton C, Price L, Woodward DL, Rodgers FG. 2002. Colony multiplex PCR assay for identification and differentiation of Campylobacter jejuni, C. coli, C. lari, C. upsaliensis, and C. fetus subsp. fetus. J Clin Microbiol 40:4744–4747. doi: 10.1128/JCM.40.12.4744-4747.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao S, Mukherjee S, Li C, Jones SB, Young S, McDermott PF. 21 December 2018, posting date. Cloning and expression of novel aminoglycoside phosphotransferase genes from Campylobacter and their role in the resistance to six aminoglycosides. Antimicrob Agents Chemother doi: 10.1128/AAC.01682-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao S, Mukherjee S, Chen Y, Li C, Young S, Warren M, Abbott J, Friedman S, Kabera C, Karlsson M, McDermott PF. 2015. Novel gentamicin resistance genes in Campylobacter isolated from humans and retail meats in the USA. J Antimicrob Chemother 70:1314–1321. doi: 10.1093/jac/dkv001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.CLSI. 2016. Performance standards for antimicrobial susceptibility testing, 26th ed CLSI supplement M100-S26 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]