Abstract
Background/Aims:
The transcription factor Nrf2 is a master regulator of the antioxidant defense system, protecting cells from oxidative damage. We previously reported that the SUMO-targeted E3 ubiquitin ligase (STUbL), RING finger protein 4 (RNF4) accelerated the degradation rate of Nrf2 in promyelocytic leukemia-nuclear body (PML-NB)-enriched fractions and decreased Nrf2-mediated gene transcription. The mechanisms that regulate Nrf2 nuclear levels are poorly understood. In this study, we aim to explore the role of the second mammalian STUbL, Arkadia/RNF111 on Nrf2.
Methods:
Arkadia mediated ubiquitination was detected using co-immunoprecipitation assays in which whole cell lysates were immunoprecipated with anti-Nrf2 antibody and Western blotted with anti-hemagglutinin (HA) antibody or anti-Lys-48 ubiquitin-specific antibody. The half-life of Nrf2 was detected in whole cell lysates and promyelocytic leukemia-nuclear body enriched fractions by cycloheximide-chase. Reporter gene assays were performed using the antioxidant response element (ARE)-containing promoter Heme oxygenase-1 (HO-1).
Results:
We show that Arkadia/RNF111 is able to ubiquitinate Nrf2 resulting in the stabilization of Nrf2. This stabilization was mediated through Lys-48 ubiquitin chains, contrary to traditionally degradative role of Lys-48 ubiquitination, suggesting that Lys-48 ubiquitination of Nrf2 protects Nrf2 from degradation thereby allowing Nrf2-dependent gene transcription.
Conclusion:
Collectively, these findings highlight a novel mechanism to positively regulate nuclear Nrf2 levels in response to oxidative stress through Arkadia-mediated K48-linked ubiquitination of Nrf2.
Keywords: Nrf2, Nuclear degradation, Sumoylation, Promyelocytic leukemia-nuclear bodies, RNF111/Arkadia, K48-linked ubiquitination
Introduction
The transcription factor Nrf2 (nuclear factor erythroid 2-related factor 2) is a key oxidative response protein that enables cells to combat oxidative stress [1–6]. Under homeostatic conditions, its cellular level is kept low by Keap1-mediated Cul3/Rbx1-dependent ubiquitination, which targets it for proteasome-mediated degradation in the cytoplasm [7–10]. Upon oxidative stress, Nrf2 escapes from the Keap1-mediated repression in the cytoplasm to translocate to the nucleus where it forms heterodimers with small Maf proteins to bind to the antioxidant response element (ARE) in target genes to activate transcription [1, 2, 11]. It is becoming increasingly apparent that degradation of Nrf2 can also occur by Keap1-independent mechanisms [12–14]. For example, Hayes and his colleagues have reported that the Neh6 domain of Nrf2 contains two degrons that can mediate degradation of Nrf2 by the E3 ubiquitin ligase β-TrCP in a Keapl-independent manner [13]. However, the cellular locale of this activity has not been determined. Another Keap1-independent mechanism is exemplified by our recent finding that not only is Nrf2 a polysumoylatable protein, but Nrf2 can also be ubiquitinated by the polySUMO-specific E3 ubiquitin ligase known as RING finger protein 4 (RNF4), and subsequently degraded in promyelocytic leukemia-nuclear bodies (PML-NBs) [14].
PML-NBs are assemblies that are initiated by sumoylated PML protein within the nucleus, and typically contain PML protein, SUMO (small ubiquitin like-modifier), the nuclear antigen SP100, death domain-associated protein 6 (DAXX), and other proteins that dynamically localize to these domains [15–20]. There is much evidence for cross-talk within PML-NBs, between sumoylation and ubiquitination [21–23]. For example, RNF4 promotes polyubiquitination of polysumoylated PML in PML-NBs and the polyubiquitinated, polysumoylated PML adduct is subsequently degraded in these nuclear domains [24–26].
RING finger protein 111 (RNF111), also called Arkadia, was recently shown to ubiquitinate polysumoylated PML in PML-NBs, an event that also resulted in subsequent degradation of the polyubiquitinated PML product in PML-NBs [27]. Arkadia is the second of only two polySUMO-specific E3 ubiquitin ligases [also called SUMO-targeted ubiquitin ligases (STUbLs)] [27–32] so far identified in mammalian cells. Generally, STUbLs target sumoylated substrates for degradation by recognizing polysumoylated proteins through N-terminal SUMO-interacting motifs (SIMs) [31, 33]. Until the report by Erker et al. [27] that Arkadia ubiquitinates polysumoylated PML in PML-NBs, Arkadia was best known for being an E3 ubiquitin ligase that played a role in enhancing TGF-β signaling through ubiquitylating and subsequently degrading Smad7 [29], SnoN, and c-Ski [30] in a SUMO-independent manner.
STUbLs play important roles in regulating cellular processes by linking SUMO modification to the ubiquitin/proteasome system [33–36]. With RNF4 and Arkadia being the only two known human STUbLs, it is unclear how these enzymes coordinate their substrate modifications to regulate cellular activities. Each protein contains multiple SIM domains located in the N-terminus and a RING domain located in the C-terminus. The concerted action of these domains enables recognition of polysumoylated substrates with subsequent ubiquitination event/s, thereby creating a mark on the substrate. Generally, this mark has been associated with proteolytic degradation of the substrate, however recent evidence is demonstrating these marks can lead to the stabilization of the substrate with downstream effects such as, facilitating protein-protein interactions or increasing transcription [32, 37, 38].
Here, we report that Arkadia can ubiquitinate polysumoylated Nrf2, but unlike its action on polysumoylated PML its action on polysumoylated Nrf2 did not lead to proteolytic loss of the modified Nrf2, but rather to stabilization of Nrf2, presumably to enable continued transcriptional activity. Our study shows that the ubiquitination of polysumoylated Nrf2 by Arkadia occurs through lysine 48 of ubiquitin and results in stabilization of Nrf2.
Materials and Methods
Plasmids
3X Flag-tagged Arkadia (pBICEP-CMV2-3X-FLAG-ARK), pBICEP-CMV2-3X FLAG-ARK-C931A (RING domain mutant Flag-tagged Arkadia), and pBICEP-CMV2 (Flag vector) were obtained from Dr. Laurence Levy (INSERN, France). Flag-tagged Arkadia (S-FLAG-Strep-tagged RNF111), SFS-RNF111*RING expressing Arkadia mutated at its RING domain (W963A) and SFS-RNF111*SIM expressing Arkadia mutated at each of its three SIMs [VVVI(300–303)AAAA, VEIV(326–329)AAAA, and VVDL(382–385)AAAA] were obtained from Dr. Niels Mailand (University of Copenhagen, Denmark). Plasmids expressing HA-Ubiquitin (pRK5-HA-Ubiquitin) or mutant ubiquitins were purchased from Addgene (Cambridge, MA). HA-Ub-K63R was created by site-directed mutagenesis.
Cell Culture and Transfection
HepG2 cells, obtained from Sigma-Aldrich (St. Louis, MO), were cultured in minimum essential medium (MEM) supplemented with 10% fetal bovine serum, 1 mM sodium pyruvate, 1X MEM nonessential amino acids (Invitrogen), and antibiotics (100 units of penicillin and 100 μg of streptomycin per ml) at 37 °C in 95% air:5% CO2 atmosphere [34]. HepG2 cells were seeded and grown to ~80% confluency and were then transfected with expression plasmids or empty vector using TransIT-2020 transfection reagent (Mirus Bio LLC, Madison, WI). When used, 48 h after transfection, 5 μM arsenic trioxide (As2O3) was added to cultures for 4 h. Transfection of siRNA was carried out using TransIT-X2 transfection reagent (Mirus Bio LLC, Madison, WI) according to manufacturer’s instructions for 48 h. Arkadia siRNA [CAU CUA GUG CUG UAA CGG A (sense) and UCC GUU ACA GCA CUA GAU G (anti-sense) was purchased from Sigma Life Science. Control siRNA was purchased from Dharmacon, Inc (Lafayette, CO).
Western Blot analysis
Proteins were resolved by SDS-PAGE (sodium-dodecyl sulfate poly-acrylamide gel electrophoresis) and were probed with the following antibodies: anti-Nrf2 (Abcam, Cambridge, MA; cat no: 62352), anti-GFP (ThermoFisher, Waltham, MA; cat no: A11122), anti-FLAG (Sigma Aldrich, St. Louis, MO; cat no: F1804 and F3165), anti-K48 ubiquitin specific (Cell Signaling Technology, Boston, MA; cat no: 8081S), anti-K63 ubiquitin specific (Abcam, Cambridge, MA; cat no: ab179434), anti-HA (Vanderbilt University Molecular Core, Nashville, TN), anti-RNF111 (Abnova, Taipei, Taiwan; cat no: H00054778), anti-β-actin-peroxidase (Sigma Aldrich, St. Louis, MO; cat no: A3854), anti-LDH (Epitomics, Burlingame, CA; cat no: 1980–1), anti-AKAP95 (Millipore, Burlington, MA; cat no: 06–417), and anti-Keap1 (Abcam, Cambridge, MA; cat no: ab31973) antibodies.
Co-Immunoprecipitation (Co-IP) Assays
Immunoprecipitation was performed with whole cell lysates of HepG2 cells. Briefly, 40 μl of Dynabeads® Protein G (ThermoFisher, Waltham, MA) were transferred to a 1.5 ml microcentrifuge tube (for each treatment) and incubated with 2 μg each of either normal rabbit IgG or indicated antibodies in 200 μl of wash buffer containing PBS, pH7.4, with 0.02% Tween-20 overnight at 4 °C for 1 h with gentle rotation. The microcentrifuge tube was placed on a magnetic rack to collect the Dynabeads®-antibody complex and the supernatant solution was removed. The Dynabeads®-antibody complex was then resuspended in 200 μl to 1 ml of whole cell lysate containing 500 μg to 1 mg protein of the antigen and incubated at 4 °C overnight with gentle rotation. The microcentrifuge tube was placed on a magnetic rack to collect the Dynabeads®-antibody-antigen complex, then washed with 200 μl of wash buffer containing PBS, pH 7.4, and 0.02% Tween-20. Each Dynabeads®-antibody-antigen complex was washed in 100 μl of PBS and transferred to a clean 1.5 ml microcentrifuge tube and then boiled for 10 min with 30 μl of 2X SDS sample buffer. The Dynabeads® were collected on a magnetic rack and the supernatant solution (containing the antigen-antibody complex) was resolved on 8% SDS-PAGE. Proteins were then analyzed by Western blotting using appropriate antibody as indicated in the legends to the various figures.
Reporter Gene Assay
Nrf2-dependent gene transcription was assessed by reporter gene assay using ARE-driven minimal promoter HO-1-ARE-luc. HepG2 cells (1 × 105 cells in 1 ml of medium per well) seeded in 24-well plates for 24 h were co-transfected with 0.2 μg of luciferase reporter plasmid (human minimal promoter Heme Oxygenase-1, HO-1-ARE-luc [14, 34] along with pCI-Nrf2, pCMV-SFS-Arkadia, and pRK5-HA-Ubiquitin or pRK5-HA-ubiquitin harboring mutations at lysine ubiquitination sites. The total amount of DNA was adjusted to 0.8 μg, if necessary, by adding the empty vector. The cells were harvested 20–24 h later, washed once with 1X PBS, pH 7.4, and lysed with 150 μl of 1X Cell Culture Lysis Reagent (Promega, Madison, WI) for 30 min at room temperature. Cell lysates were collected in 1.5-ml microcentrifuge tubes and centrifuged at 4 °C at 12, 000 × g (2 min) to remove cell debris. The luciferase activity and protein content of the cell extracts were then measured. For luciferase activity, 10 μl of the extracts were used to measure the integrated light units over 10 sec, using the luciferase assay system (Promega, Madison, WI) and a luminometer (Tropix, Inc., Bedford, MA), as recommended by the manufactures. Five μl of the extracts were used to determine protein content via the Bradford protein assay method (Bio-Rad, Hercules, CA), using bovine serum albumin as standard.
Preparation of Nuclear Matrix Fraction
Beginning with a partially purified nuclear pellet, the nuclear matrix fraction was prepared as previously described [14]. Briefly, HepG2 cells were pelleted and resuspended in 100 μl of buffer (Buffer A) containing 10 mM Hepes-KOH pH 7.9, 1.5 mM MgCl2, 10 mM KCl. Cells were then lysed by adding 1% NP-40. The lysate was then centrifuged and the resulting supernatant solution was collected as the cytoplasmic fraction. The resulting pellet (crude nuclear pellet) was resuspended in 100 μl of buffer A and lysed by adding NP-40. The lysed nuclear pellet fractions were re-washed with buffer A and the remaining partially-purified nuclear pellet was enriched for the nuclear matrix. The partially-purified nuclei were incubated with 100 μl of DNase I at a concentration of 1 μg/μl in buffer containing 10 mM PIPES pH 6.8, 300 mM sucrose, 50 mM NaCl, 3 mM MgCl2, 1 mM EGTA, 0.5% Triton-X 100 and mildly agitated at room temperature for 30 min. The DNase I-treated solution was then centrifuged and resuspended and washed, with a series of buffers as follows, to obtain the nuclear body (nuclear matrix) fraction. First, the pellet was resuspended in 100 μl buffer containing 250 mM (NH4)2SO4, 10 mM PIPES pH 6.8, 150 mM sucrose, 3 mM MgCl2, and 1 mM EGTA and incubated for 5 min at 4°C. The solution was then centrifuged and the process was repeated. The recovered pellet was washed by resuspending in a buffer containing 1 M NaCl, 10 mM PIPES pH 6.8, 150 mM sucrose, 3 mM MgCl2, and 1 mM EGTA and then washed by resuspending in buffer containing 2M NaCl, 10 mM PIPES pH 6.8, 150 mM sucrose, 3 mM MgCl2, and 1 mM EGTA. The resultant pellet (nuclear matrix) was resuspended in 50–100 μl of buffer containing 2X SDS, Tris-HCl pH 6.8.
Statistical Analysis
Statistical analyses were carried out using the Prism program (GraphPad Software, Inc, La Jolla, CA). Differences between multiple groups were tested using Tukey’s corrected one-way ANOVA. The data are presented as means ± S.E.M.
Results
Arkadia promotes Nrf2 ubiquitination
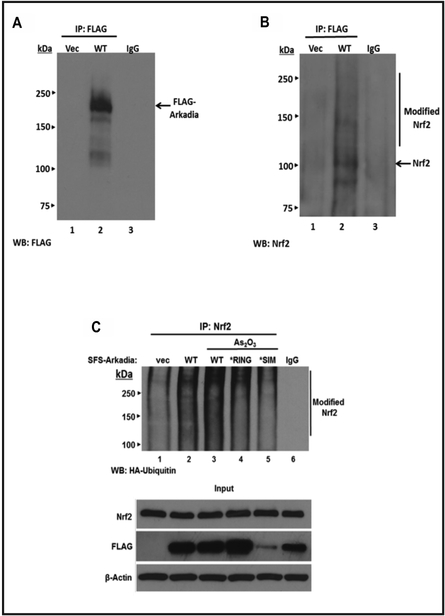
We have previously shown in vitro and in vivo that Nrf2 is a polysumoylatable protein and that it is polyubiquitinated by RNF4 thereby targeting it for degradation in PML-NBs [14]. To determine if Arkadia would have a similar effect on Nrf2 function, we investigated if Arkadia could associate with Nrf2 by transfecting HepG2 cells with a plasmid encoding FLAG-Arkadia and performed Co-immunoprecipitation (Co-IP) assays. The expression of Flag-Arkadia is verified in Fig. 1A. The results show that under normal growth conditions Arkadia co-precipitates with modified Nrf2, which is presumably sumoylated due to the presence of high molecular weight bands (Fig. 1B). Next, we wanted to determine if this association resulted in ubiquitination of Nrf2. To do this, we co-expressed Flag-Arkadia with HA-tagged Ubiquitin. We then immunoprecipated lysates with anti-Nrf2 antibody and blotted with anti-HA antibody to detect ubiquitination. Compared to empty vector control, expression of HA-Ubiquitin led to a significant increase in the level of HA-Ubiquitinated Nrf2 (Fig. 1C, lanes 1 and 2). To confirm that Arkadia was in fact mediating the formation of the HA-Ub-Nrf2 species, HepG2 cells were transfected with either Arkadia WT, an Arkadia*RING mutant (a mutation that renders Arkadia incapable of catalyzing ubiquitination), or with an Arkadia*SIM mutant plasmid (a mutation in which all three SIMs of Arkadia are mutated to disrupt recognition of sumoylated substrates) [32] followed by treatment with arsenic trioxide (As203). Arsenic trioxide is a known inducer of the oxidative stress pathway and results in increased expression of Nrf2, an increase of SUMO-2/3 attachment to proteins, as well as induces the formation of PML-NBs [39–41]. Our results show an increase in the levels of Nrf2 ubiquitination in cells expressing WT Arkadia and as expected, a decrease in the levels of Nrf2 ubiquitination in cells expressing both the RING and SIM mutants. This suggests that Nrf2 ubiquitination was mediated by exogenously expressed Arkadia. The input panel in Fig. 1C shows the presence of the FLAG-Arkadia proteins. It should be noted that the expression of FLAG-Arkadia*SIM was not as high as that of WT FLAG-Arkadia and FLAG-Arkadia*RING. This phenomenon was also reported by Poulsen et al. [32]. Taken together, we demonstrate that Nrf2 is a substrate for Arkadia and that Arkadia is able to ubiquitinate polysumoylated Nrf2 in response to oxidative stress.
Fig. 1.
Arkadia interacts with and ubiquitylates endogenous Nrf2. HepG2 cells were transfected with 5 μg of plasmid expressing FLAG vector or FLAG-Arkadia. After 48 h, cells were harvested and whole-cell lysates were immunopreciptated with 1 μg of anti-FLAG antibody (Sigma) and Western blotted with anti-FLAG antibody (A) or with anti-Nrf2 antibody (B). In panel C, HepG2 cells were transfected with 2 μg of pRK5-HA-ubiquitin and vector (pFLAG) alone, plasmid encoding wild-type Arkadia, Arkadia*RING (Arkadia-W963A) that cannot mediate ubiquitylation, or Arkadia*SIM [VWI(300–303)AAAA, VEIV(326–329)AAAA, and VVDL(382–385)AAAA] which has all three SIMs of Arkadia mutated and therefore can not recognize sumoylated substrates. After 48 h, the cells were incubated with/without As203 (5 μM) for 4 h and then harvested. Whole cell lysates were subjected to Co-IP assays in which cells were immunoprecipitated with anti-Nrf2 antibody and Western-blotted with anti-HA antibody to detect ubiquitylation of Nrf2. Non-immune serum (IgG) was used as control. The same whole cell lysates were immunoblotted with anti-Nrf2 antibody, anti-FLAG antibody to detect the presence of FLAG-Arkadia and anti β-actin as a loading control.
Overexpression of Arkadia stabilizes Nrf2
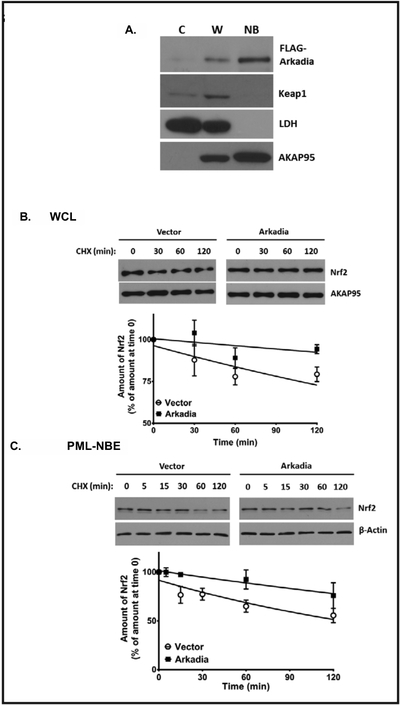
To determine if the Arkadia-mediated ubiquitination of polysumoylated Nrf2 occurred within PML-NB domains, we isolated PML-NBs from As2O3--treated HepG2 cells and blotted for exogenously expressed Arkadia. FLAG-Arkadia was most prominent in the nuclear body (NB) fraction (Fig. 2A), which was also enriched for AKAP95, a nuclear matrix protein, and was free of the cytoplasmic marker lactate dehydrogenase (LDH). The NB fraction was also enriched for Nrf2 [14] and was free of biologically active Keap1, the inhibitory protein for Nrf2. This suggests that the effects of Arkadia-mediated ubiquitination of Nrf2 may occur in PML-NBs. Based on the reports that RNF4 [24–26] and Arkadia [27] target polysumoylated PML for degradation in PML-NBs, we postulated that like RNF4, Arkadia would also target polysumoylated Nrf2 for degradation in PML-NBs. To test this, we conducted cycloheximide chase assays in HepG2 cells overexpressing Arkadia to monitor and compare the degradation of Nrf2 in untreated whole cell lysates (Fig. 2B) and in the PML-NB fraction of As2O3-treated HepG2 cells (Fig. 2C). Interestingly, the results show nominal degradation of Nrf2 in the whole cell lysate fraction and the PML-NB fraction suggesting that trafficking of Arkadia to PML-NBs enables ubiquitination of Nrf2 and an increase in its stability. Having shown that RNF4 shortens the half-life (t1/2) of Nrf2 [14], these results demonstrate the antagonistic activity of human STUbLs in regulating Nrf2 protein levels, particularly in PML-NBs.
Fig. 2.
Arkadia-stabilized Nrf2 in whole-cell lysates and PML-NB enrichments. In panel A, whole-cell lysate (W) of HepG2 cells grown to 80% confluency and incubated with As203 (2 μM) for 4 h was separated into cytoplasmic (C) and nuclear body (NB) fractions, as previously described (13). The fractions were resolved on SDS-PAGE gel (8%) and immunoblotted for transfected Arkadia (FLAG-Arkadia), Keapl, LDH, and AKAP95. LDH and AKAP95 were used as markers for the cytoplasm and nuclear matrix, respectively. B, C, HepG2 cells were transfected with vector alone or plasmid encoding wild-type FLAG-Arkadia. Forty-eight hours after transfection, HepG2 cells were incubated with cycloheximide (100 μg/μl), then harvested at the indicated time points thereafter. The lysates were then immunoblotted for Nrf2. Representative blots are shown. The Western blot analyses were quantified densitometrically using UN-SCAN-IT software (Silk Scientific, Inc., Orem, UT). The values plotted are means ± S.E. for 5 experiments and are plotted on a semi-log scale. The data are presented as percentage plots after normalizing against the loader (actin for whole cell lysate or AKAP95 for NB fraction), taking the values for no treatment with cycloheximide (zero time) as 100%, representative blots ares shown. B, cycloheximide-chase using whole cell lysates to measure degradation of endogenous Nrf2. C, cycloheximide-chase using PML-NB fraction from As2O3-treated HepG2 cells (prepared as in A) to measure degradation of endogenous Nrf2.
Arkadia ubiquitinates Nrf2 through Lys48 ubiquitin chains
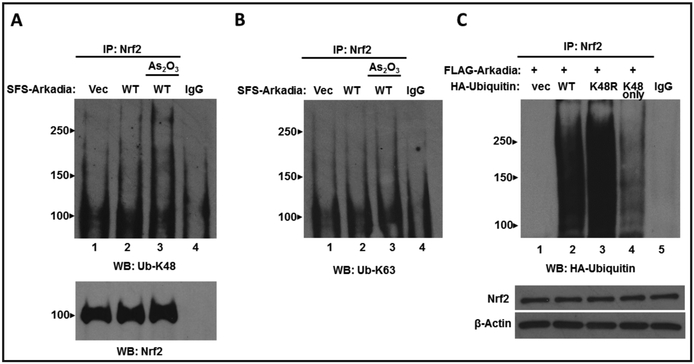
Ubiquitination processes generating K48-linked ubiquitin chains traditionally target such products for degradation by the 26S proteasome, whereas K63-linked polyubiquitination generally results in protein stabilization or facilitation of protein-protein interactions [32, 42–44, review 45]. These reports combined with our observation that Nrf2 was not degraded by Arkadia, led us to the hypothesis that Arkadia mediates K63-linked ubiquitination of Nrf2. To address this, HepG2 cells were co-transfected with plasmids encoding FLAG-Arkadia and wild-type HA-Ubiquitin followed by treatment with As2O3. Whole cell lysates were prepared and immunoprecipitated using anti-Nrf2 antibody, then probed using anti-K63 or anti-K48 ubiquitin specific antibodies. The results intriguingly show that when probed with anti-Ub-K48 antibody we observe an increase in K48-linked ubiquitination of Nrf2 (Fig. 3A) However, when probed with anti-Ub-K63 antibody there is no change in K63-linked ubiquitination of Nrf2 (Fig. 3B). This provides evidence that Arkadia mediates K48-linked ubiquitination of Nrf2.
Fig. 3.
Arkadia ubiquitylates polysumoylated Nrf2 through K48-linked ubiquitin chains. HepG2 cells were transfected with 2 μg of plasmid expressing empty vector (pFLAG) or FLAG-Arkadia and pRK5-HA-Ubiquitin. After 48 hr the cells were incubated with or without As2O3 (5 μM) for 4 hrs and then harvested. Whole-cell lysates were generated and immunoprecipitated with anti-Nrf2 antibody and Western blotted with (A) anti-K48-specific ubiquitin antibody or (B) anti-K63-specific ubiquitin antibody. Control IPs were blotted with anti-Nrf2 antibody (C). HepG2 cells were transfected with FLAG-Arkadia and wild-type HA-Ubiquitin or mutant plasmids expressing HA-Ub-K48R or HA-UbK48 only. Whole cell lysates were generated and immunoprecipitated with anti-Nrf2 antibody and Western blotted with anti-HA antibody. The same whole cell lysates were immunoblotted with anti-Nrf2 antibody and anti-β-actin as a loading control.
To further ascertain Arkadia mediated K48-linked ubiquiylation of Nrf2, an in vivo ubiquitination assay was performed using two different K48 ubiquitin mutant plasmids (Fig. 3C). HepG2 cells were co-transfected with FLAG-Arkadia and either plasmids expressing HA empty vector, wild-type Ub, Ub-K48R (in which Lys-48 is mutated to Arg) or Ub-K48 only (in which all Lys residues are mutated to Arg except K48). Whole cell lysates were generated and immunoprecipited using anti-Nrf2 antibody and then probed with anti-HA antibody to detect ubiquitination. The results show that as compared to wild-type Ub, there was a slight increase in ubiquitination in cells expressing Ub-K48R. We speculate that there is an increase in ubiquitination because the lack of Lys-48 allows for an increase in ubiquitination through the six remaining Lys ubiquitination sites. In lysates expressing Ub-K48 only, Nrf2 was indeed ubiquitinated via Ub-K48 only however the level of ubiquitination was decreased as compared to wild-type ubiquitin and Ub-K48R. This phenomenon is most likely due to the Ub-K48 only plasmid only harboring one site for ubiquitination while wild-type ubiquitin and Ub-K48R contain seven and six Lys sites for ubiquitination, respectively, therefore it is expected that the level of ubiquitination will be less with the Ub-K48 only plasmid containing only one site for ubiquitination.
Effect of knockdown of Arkadia on Nrf2 Ubiquitination
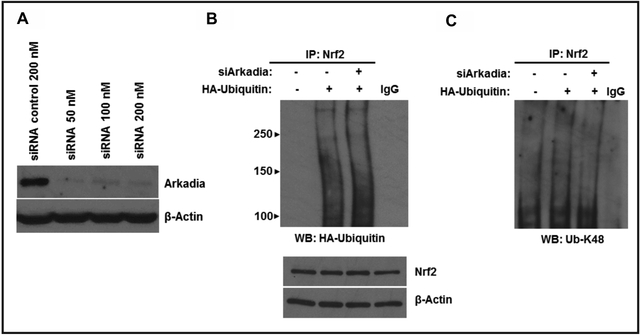
To further demonstrate the role of Arkadia on Nrf2 ubiquitination, we used control siRNAs or siRNAs targeting Arkadia to knock down Arkadia in HepG2 cells. We then evaluated the levels of Arkadia in whole cell lysates by Western blotting. This analysis showed that siArkadia did indeed knockdown endogenous Arkadia with 50 nM of siArkadia yielding the best knockdown (Fig. 4A). HepG2 cells were then transfected with siArkadia and wild-type Ub. Wh ole cell lysates were generated and immunoprecipitated with anti-Nrf2 antibody and probed with either anti-HA antibody (Fig. 4B) or anti-Ub-K48 antibody (Fig. 4C). The results show that there was a slight decrease in overall ubiquitination when Arkadia was knocked down (Fig. 4B) indicating that Arkadia only contributes marginally to the overall ubiquitination status of Nrf2. Interestingly, there was also a slight decrease in K48-linked ubiquitination of Nrf2 when Arkadia was knocked down (Fig. 4C) confirming that Arkadia does in fact mediate K48-linked ubiquitination of Nrf2. These results reveal a novel mechanism for STUbL-mediated ubiquitination of target proteins. To our knowledge, this is the first report of K48-linkages by Arkadia resulting in protein stabilization.
Fig. 4.
Effect of Arkadia knockdown on Nrf2 ubiquitylation. A, HepG2 cells were transfected with 200 nM of control siRNA or 50 nM, 100 nM or 200 nM ofsiArkadiato detect the effeciency of Arkadia knockdown. B, C, HepG2 cells were transfected with 50 nM of control siRNA or siArkadia along with plasmid encoding wild-type HA-Ub, HA-Ub-K48R or HA-Ub-K48 only. After 48 h, cells were treated with As203 (5 μM) for 4h and harvested. Whole cell lysates were generated and immunoprecipitated with anti-Nrf2 antibody and blotted with anti-HA antibody (B) or anti-K48-specific ubiquitin antibody (C). The same whole cell lysates were immunoblotted with anti-Nrf2 antibody and anti-β-actin as a loading control.
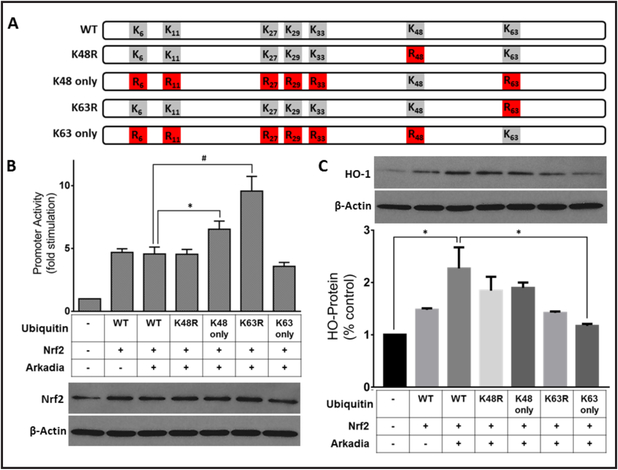
Impact ofK48-linked ubiquitination on Nrf2-dependentgene transcription
To determine how human STUbLs regulate Nrf2 function, we assessed Nrf2 transcriptional activity by performing reporter genes assays. HepG2 cells were co-transfected with the prototypic ARE-driven minimal promoter Heme Oxygenase-1-ARE-luc, a plasmid expressing Arkadia, and a series of ubiquitin mutant plasmids (Fig. 5B). In this experiment, four ubiquitin mutant plasmids (illustrated in Fig. 5A) harboring mutations at various lysine positions were used in individual gene reporter assays to determine the preferred linkage pattern required for Arkadia to regulate Nrf2-mediated transcriptional activity. The results show comparable levels of gene activity following expression of Ub-WT or Ub-K48R (Fig. 5B). Intriguingly, we observe a significant increase in gene activity when the ubiquitin plasmid containing only one available Lys residue (Ub-K48 only) was used, suggesting a preference for K48-mediated linkages. Additionally, when the ubiquitin mutant plasmid with only Lys 63 mutated to Arg (Ub-K63R) was expressed, there was a more pronounced increase in transcriptional activity suggesting mechanisms that utilize Ub-K63 are no longer functional, and all available substrate is shifted for other ubiquitin linkages, presumably K48 (Fig. 5B). To evaluate the effects of Arkadia mediated stabilization of Nrf2 on the ability to activate the endogenous HO-1 gene, HepG2 cells were prepared as in panel 5B and Western blotted with anti-HO-1 antibody and quantified (Fig. 5C). HO-1 protein levels were significantly increased in cells expressing Arkadia and Nrf2 as compared to control. These results show that in the presence of Arkadia, Nrf2 regulated the endogenous HO-1 promoter and consequently HO-1 protein levels in a similar manner. These results demonstrate that Arkadia uses K48-linked ubiquitination to stabilize Nrf2.
Fig. 5.
Arkadia induces Nrf2-dependent ARE-driven gene transcription. A, Schematic of pRK5-HA-Ubiquitin mutant plasmids. Five HA-ubiquitin mutation plasmids were used; HA-ubiquitin harboring a single K→R mutation at position K48 (Ub-K48R) or at position K63 (Ub-K63R), HA-ubiquitin bearing a single Lys at either position 48 (Ub-K48 only) or position 63 only (Ub-K63 only) with the other six Lys residues in ubiquitin mutated to Arg. B, C, HepG2 cells were co-transfected with 0.2 μg each of HO-1-ARE-luc reporter gene construct and heterologous Nrf2 (pCI-Nrf2) as inducer, pRK5-HA-ubiquitin or its 5 mutants described in A, and FLAG-Arkadia. Representative blots are shown, B. C, HepG2 cells were prepared as in B were blotted for the endogenous target gene hemeoxygenase-1. Representative blots are shown. Values plotted are means ± S.E.M. for duplicate assays from 10 different experiments analyzed by one-way ANOVA. * Statistically different (p<0.05), # statistically different (p<0.0001).
Discussion
In this study, we reveal a novel role for Arkadia in the ubiquitination of the transcription factor Nrf2. First, we show that Arkadia is able to ubiquitinate Nrf2. Second, we showed that Arkadia is able to stabilize Nrf2 and lastly, we show that Arkadia ubiquitinates Nrf2 through ubiquitin K48 linkages. To our knowledge, this is this the first study showing that Arkadia can stabilize a protein through ubiquitin K48 linkages.
Since the discovery of Nrf2, a major quest in the field has been to understand the mechanisms that regulate the levels of this master antioxidant transcription factor. It is now known that there are several layers of Nrf2 regulation [7–10, 12–14]. The most characterized regulatory mode is the Keap 1-dependent mechanism in unstressed cells, in which Nrf2 activity is suppressed by constant ubiquitination of Nrf2, marking it for proteasomal degradation in the cytosol [7–10]. This led to questions as to whether Nrf2 could be degraded in the nucleus. It was postulated that Nrf2 could be degraded in the nucleus in oxidatively stressed cells through the Neh6 degron, although the E3 ligases mediating this degradation is not known [46]. While Keap1 has been implicated in nuclear degradation, the mechanism by which Keap1 enters the nucleus was not delineated because it lacks a nuclear localization sequence [6]. These studies have provided much insight into Nrf2 regulation, yet they fail to completely address the influence of post-translational modifications (PTMs) on nuclear Nrf2 levels. There have been extensive studies on the PTMs of Keap1 and/or Nrf2 that lead to the activation of Nrf2 [1–5, 8–10]. Emerging areas of interest now seem to focus on PTMs of Nrf2 after it is activated. In that context, reports have surfaced on four types of PTMs: phosphorylation [12, 13, 47–49], acetylation [34, 50], ubiquitination [12, 13], and sumoylation [14]. The demonstration that Nrf2 is a target for sumoylation is a recent finding; therefore, the potential consequences of this modification have not yet been fully explored. In our initial report that Nrf2 is a sumoylatable protein, we showed that RNF4 ubiquitinated polysumoylated Nrf2, resulting in negative regulation of the modified Nrf2 within PML-NBs [14].
Our present work has uncovered two aspects of Nrf2 function. First, we show that polysumoylated Nrf2 can also be ubiquitinated by the STUbL RNF111/Arkadia (Fig. 1). However, this Arkadia-induced ubiquitination of polysumoylated Nrf2 did not result in proteolytic degradation of the modified Nrf2 as seen with RNF4 (Fig 2, ref. 14). Instead, it stabilized Nrf2. Second, the Arkadia-induced stabilization of Nrf2 was specifically mediated by Ub-K48 linkages (Figs. 3 and 4), and allowed ARE-driven gene transcription (Fig. 5). We initially predicted that co-expression of Arkadia with Ub-WT would result in an increase of transcriptional activity, while co-expression with Ub-K48R would result in decreased activity of the HO-1 promoter. Strikingly, the levels of both were comparable (Fig. 5).
It has only been a decade since the discovery of STUbLs, which provided the first direct link between sumoylation and ubiquitination [36, 51, 52]. In this context, STUbLs were reported to utilize their SIMs to recognize polysumoylated substrates resulting in subsequent polyubiquitination with proteolytic degradation [31, 33]. Arkadia has previously been reported to induce ubiquitination of sumoylated PML [27] leading to proteolytic loss of PML, which is similar to the effect mediated by RNF4 [24–26]. As the newest member of the human STUbL family, it is not surprising that Arkadia shares similar substrates as RNF4. What is unclear about STUbLs, is how they mediate these varying outcomes. Emerging evidence is providing insight that extends the capabilities of these enzymes to establish ubiquitin marks, which can promote the stabilization of proteins. For example, Arkadia was demonstrated to promote K63-linked ubiquitination of xeroderma pigmentosum C (XPC) protein [32]. This effect resulted in stabilization, rather than proteolysis of XPC. Thus far, RNF4 and Arkadia appear to share similar functions as the activity of both have been shown to stabilize proteins, facilitate protein-protein interactions, influence the DNA damage response as well as increase transcription [32, 37, 38]. Interestingly, these activities have been mediated by protein ubiquitination through both K48-linked and K63-linked ubiquitin chains [24, 27, 32, 37]. It would be interesting to determine which ubiquitin linkage is involved in RNF4-mediated ubiquitination of Nrf2.
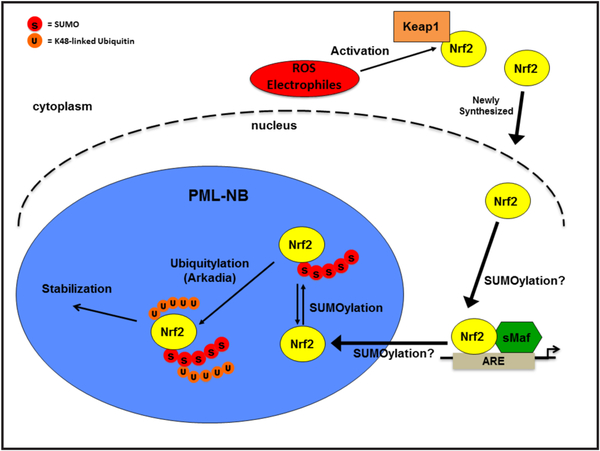
We show here that Arkadia associates with polysumoylated Nrf2 and that this association is increased in response to oxidative stress resulting in Nrf2 ubiquitination and stabilization in PML-NBs (Figs. 1 and 2). More importantly, the Arkadia-induced stabilization of the polysumoylated Nrf2 substrate involved the attachment of K48-linked ubiquitin chain(s), rather than the predicted K63-linked ubiquitin chains [Fig. 3]. We speculate K48 polyubiquitination by Arkadia could be a protective mark that prevents degradation of polysumoylated Nrf2. It can be postulated that the effects of Arkadia on Nrf2 during oxidative stress is a means of protecting Nrf2 from degradation until Nrf2-mediated transcription is complete. RNF4 is then able to ubiquitinate Nrf2 and target it for degradation. In certain cancers somatic mutations in Keap1 or Nrf2 disrupt the regulation of Nrf2 by Keap1, resulting in increased levels of Nrf2. In such cells, excessively high levels of Nrf2 have been reported to sustain cell proliferation and carcinogenesis, and make such cells resistant to chemotherapeutic drugs. This study has the potential to provide understanding as to how sumoylation-dependent stabilization of Nrf2 by Arkadia (or degradation by RNF4) in such circumstances would be beneficial or detrimental to decreasing proliferation and chemoresistance by such cells. Based on the data presented in this study, we propose a model depicted in Fig. 6, where upon activation, newly synthesized Nrf2 translocates to the nucleus and subsequently into PML-NBs where it is ubiquitinated. This ubiquitination marks Nrf2 for stabilization and thus enables gene transcription, which is mediated by Arkadia. While our study unfolds a new target for Arkadia, Arkadia has not been extensively researched as a STUbL and the effects of Arkadia-mediated SUMO-targeted ubiquitination are still unfolding. It would be interesting to determine which SUMO sites in Nrf2 are responsible for SUMO-targeted ubiquitination of Nrf2 by Arkadia.
Fig. 6.
Proposed model for Nrf2 nuclear regulation. Upon activation, newly synthesized Nrf2 is translocated to the nucleus where it presumably undergoes sumoylation. Within PML-NBs polysumoylated Nr f2 is ubiquitylated by Arkadia with K48-linked ubiquitin chains. Ubiqutylation by Arkadia marks Nrf2 for stabilization, which enales transcription.
Acknowledgements
We thank Dr. Laurence Levy (INSERM, Paris, France) for providing us with expression plasmid for wild-type Arkadia and Arkadia mutated at its RING domain and Dr. Niels Mailand (University of Copenhagen, Denmark) for providing us with plasmids expressing Arkadia mutated at either its RING domain or its SIMs. *This work was supported by NIH grant SC1CA143985 and by CTSA award No. ULITR000445 from the National Center for Advancing Translational Sciences. +Supported by NIH grant # 2 S21MD000104. ++Supported by NIH grant # 5 R25GM0599994.
Abbreviations
- AKAP95
A-kinase-anchoring (protein 95)
- ARE
antioxidant (response element)
- HO1-ARE-luc
heme (oxygenase 1-luciferase reporter gene construct)
- Keap1
Kelch-like (ECH-associated protein 1)
- Nrf2
nuclear (factorerythroid 2-related factor 2)
- PML
promyelocytic (leukemia protein)
- PML-NB
promyelocytic (leukemia-nuclear body)
- SIM
SUMO-interaction (motif)
- SUMO
small (ubiquitin-like modifier)
- STUbL
SUMO-targeted (ubiquitin ligase)
Footnotes
Disclosure Statement
The authors declare that they have no conflicts of interest with the contents of this article.
References
- 1.Nguyen T, Sherratt PJ, Pickett CB: Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu Rev Pharmacol Toxicol 2003;43:233–260. [DOI] [PubMed] [Google Scholar]
- 2.Motohashi H, Yamamoto M: Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol Med 2004;10:549–557. [DOI] [PubMed] [Google Scholar]
- 3.Nguyen T, Sherratt PJ, Nioi P, Yang CS, Pickett CB: Nrf2 controls constitutive and inducible expression of ARE-driven genes through a dynamic pathway involving nucleocytoplasmic shuttling by Keap1. J Biol Chem 2005;280:32485–32492. [DOI] [PubMed] [Google Scholar]
- 4.Kobayashi M, Yamamoto M: Molecular mechanisms activating the Nrf2-Keap1 pathway of antioxidant gene regulation. Antiox Redox Signal 2005;7:385–395. [DOI] [PubMed] [Google Scholar]
- 5.Kensler TW, Wakabayashi N, Biswal S: Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol 2007;47:89–116. [DOI] [PubMed] [Google Scholar]
- 6.Nguyen T, Nioi P, Pickett, CB: The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J Biol Chem 2009;284:13291–13295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M: Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev 1999;13:76–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang DD, Lo S-C, Cross JV, Templeton DJ, Hannik M: Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol 2004;24:10941–10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kobayashi A, Kang M, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M: Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol 2004;24:7130–7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furukawa M, Xiong Y: BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin3-Roc1 ligase. Mol Cell Biol 2005;25:162–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Motohashi H, O’Connor T, Katsuoka F, Engel JD, Yamamoto M: Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene 2002;294:1–12. [DOI] [PubMed] [Google Scholar]
- 12.Rada P, Rojo AI, Chowdhry S, McMahon M, Hayes JD, Cuadrado A: SCF/β-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol Cell Biol 2011;31:1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chowdhry S, Zhang Y, McMahon M, Sutherland C, Cuadrado A, Hayes JD: Nrf2 is controlled by two distinct beta-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene 2013;32:3765–3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malloy MT, McIntosh DJ, Walters TS, Flores A, Goodwin JS, Arinze IJ: Trafficking of the transcription factor Nrf2 to promyelocytic leukemia-nuclear bodies: Implications for degradation of Nrf2 in the nucleus. J Biol Chem 2013;288:14569–14583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sternsdorf T, Jensen K, Will H: Evidence for covalent modification of the nuclear dot-associated proteins PML and SP100 by PIC1/SUMO-1. J Cell Biol 1997;139:1621–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishov AM, Sotnikov AG, Negorev D, Vladimirova OV, Neff N, Kamitani T, Yeh ET, Strauss JF III, Maul GG: PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J Cell Biol 1999;147: 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duprez E, Saurin AJ, Desterro JM, Lallemand-Breitenbach V, Howe K, Boddy MN, Solomon E, de Thé H, Hay RT, Freemont PS: SUMO-1 modification of the acute promyelocytic leukemia protein PML: implications for nuclear localisation. J Cell Sci 1999;112:381–393. [DOI] [PubMed] [Google Scholar]
- 18.Zhong S, Müller S, Ronchetti S, Freemont PS, Dejean A, Pandolfi PP: Role of SUMO-1-modified PML in nuclear body formation. Blood 2000;95:2748–2753. [PubMed] [Google Scholar]
- 19.Shen TH, Lin HK, Scaglioni PP, Yung TM, Pandolfi PP: The mechanism of PML-nuclear body formation. Mol Cell 2006;24:331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lallemand-Breitenbach V, de Thé H: PML nuclear bodies. Cold Spring Harbor Perspect Biol 2010;2:a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernardi R, Pandolfi PP: Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol 2007;8:1006–1016. [DOI] [PubMed] [Google Scholar]
- 22.Bailey D, O’Hare P: Comparison of the SUMO1 and ubiquitin conjugation pathways during inhibition of proteasome activity with evidence of SUMO1 recycling. Biochem J 2005;392:271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hay RT: Decoding the SUMO signal. Biochem Soc Transactions 2013;41:463–473. [DOI] [PubMed] [Google Scholar]
- 24.Tatham MH, Geoffroy M-C, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT: RNF4 is a poly-SUMO-specific ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol 2008;10:538–546. [DOI] [PubMed] [Google Scholar]
- 25.Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, Zhou J, Zhu J, Raught B, de Thé H: Arsenic degrades PML or PML-RARα through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol 2008;10:547–555. [DOI] [PubMed] [Google Scholar]
- 26.Geoffroy M-C, Jaffray EG, Walker KJ, Hay RT: Arsenic-induced SUMO-dependent recruitment of RNF4 into PML-nuclear bodies. Mol Biol Cell 2010;21:4227–4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erker Y, Neyret-Khan H, Seeler JS, Dejean A, Atfi A, Levy L: Arkadia, a novel SUMO-targeted ubiquitin ligase involved in PML degradation. Mol Cell Biol 2013;33:2163–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Episkopou V, Arkell R, Timmons PM, Walsh JJ, Andrew RL, Swan D: Induction of the mammalian node requires Arkadia function in the extraembryonic lineages. Nature 2001;410:825–830. [DOI] [PubMed] [Google Scholar]
- 29.Koinuma D, Shinozaki M, Komura A, Goto K, Saitoh M, Hanyu A, Ebina M, Nukiwa T, Miyazawa K, Imamura T, Miyazono K: Arkadia amplifies TGF-β superfamily signaling through degradation of Smad7. EMBO J 2003;22:6458–6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagano Y, Mavrakis K, Lee KL, Fujii T, Koinuma D, Sase H, Yuki K, Isogaya K, Saitoh M, Imamura T, Episkopou V, Miyazono K, Miyazawa K: Arkadia induces degradation of SnoN and c-Ski to enhance transforming growth factor-β signaling. J Biol Chem 2007;282:20492–20501. [DOI] [PubMed] [Google Scholar]
- 31.Sun H, Hunter T: Poly-small ubiquitin-like modifier (polySUMO)-binding proteins identified through a string search. J Biol Chem 2012;287:42071–42083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poulsen SL, Hansen RK, Wagner SA, von Cuijk L, von Belle GJ, Streicher W, Wikström M, Choudhary C, Houtsmuller AB, Marteijn JA, Bekker-Jensen S, Mailand N: RNF111/Arkadia is a SUMO-targeted ubiquitin ligase that facilitates the DNA damage response. J Cell Biol 2013;201:797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sriramachandran A, Dohmen R: SUMO-targeted ubiquitin ligases. Bioch Biophys Acta 2014;1843:75–85. [DOI] [PubMed] [Google Scholar]
- 34.Kawai Y, Garduño K, Theodore M, Yang J, Arinze IJ: Acetylation-deacetylation of the transcription factor Nrf2 (nuclear factor erythroid 2-related factor 2) regulates its transcriptional activity and nucleocytoplasmic localization. J Biol Chem 2011;286:7629–7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perry JJ, Tainer JA, Boddy MN: A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem Sci 2008;33:201–208. [DOI] [PubMed] [Google Scholar]
- 36.Uzunova K, Göttsche K, Miteva M, Weisshaar SR, Glanemann C, Schnellhardt M, Niessen M, Scheel H, Hofmann K, Johnson ES, Praefcke GJ, Dohmen RJ: Ubiquitin-dependent proteolytic control of SUMO conjugates. J Biol Chem 2007;282:34167–34175. [DOI] [PubMed] [Google Scholar]
- 37.Yin Y, Seifert A, Chua JS, Maure JF, Golebiowski F, Hay RT: SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev 2012;26:1196–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas J, Abed M, Heuberger J, Novak R, Zohar Y, Lopez A, Trausch-Azar J, Ilagan M, Benhamou D, Dittmar G: RNF4-dependent oncogene activation by protein stabilization. Cell Rep 2016;16:3388–3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weisshaar SR, Keusekotten K, Krause A, Horst C, Springer HM, Göttsche K, Dohmen RJ, Praefcke GJ: Arsenic trioxide stimulates SUMO-2/3 modification leading to RNF4-dependent proteolytic targeting of PML. FEBS Lett 2008;582:3174–3178. [DOI] [PubMed] [Google Scholar]
- 40.He X, Chen MG, Lin GX, Ma Q: Arsenic induces NAD(P)H-quinone oxidoreductase 1 by disrupting the Nrf2-Keap1-Cul3 complex and recruiting Nrf2-Maf to the antioxidant response element enhancer. J Biol Chem 2006;281:23620–23631. [DOI] [PubMed] [Google Scholar]
- 41.Sahin U, Ferhi O, Jeanne M, Benhenda S, Berthier C, Jollivet F, Niwa-Kawakita M, Faklaris O, Setterblad N, de Thé H, Lalleman-Breitenbach V: Oxidative stress-induced assembly of PML nuclear bodies controls sumoylation of partner proteins. J Cell Biol 2014;204:931–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pickart CM, Fushman D: Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol 2004;8:610–616. [DOI] [PubMed] [Google Scholar]
- 43.Erpapazoglou Z, Walker O, Haguenauer-Tsapis R: Versatile roles of K63-linked ubiquitin in trafficking. Cells 2014;3:1027–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kommander D, Rape M: The ubiquitin code. Annu Rev Biochem 2012;81:203–229. [DOI] [PubMed] [Google Scholar]
- 45.Hershko A, Ciechanover A: The ubiquitin system. Annu Rev Biochem 1998; 67:425–479. [DOI] [PubMed] [Google Scholar]
- 46.McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD: Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J Biol Chem 2004;279:31556–31567. [DOI] [PubMed] [Google Scholar]
- 47.Cullinan SB, Zhang D, Hannink M, Arvisais E, Kaufman RJ, Dieh JA: Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol Cell Biol 2003;23:7198–7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Apopa PL, He X, Ma Q: Phosphorylation of Nrf2 in the transcriptional activation domain by casein kinase 2 (CK2) is critical for the nuclear translocation and transcriptional function of Nrf2 in IMR-32 neuroblastoma cells. J Biochem Mol Toxicol 2008;22:63–76. [DOI] [PubMed] [Google Scholar]
- 49.Sun Z, Huang Z, Zhang DD: Phosphorylation of Nrf2 at multiple sites by MAP kinases has a limited contribution in modulating the Nrf2-dependent antioxidant response. PLoS ONE 2009;4:e6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun Z, Chin YE, Zhang DD: Acetylation of Nrf2 by p300/CBP augments promoter-specific DNA binding of Nrf2 during the antioxidant response. Mol Cell Biol 2009;29:2658–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prudden J, Pebernard S, Raffa G, Slavin D, Perry J, Trainer J, McGowan C, Boddy M: SUMO-targeted ubiquitin ligases in genome stability. EMBO J 2007;26:4089–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun H, Leverson J, Hunter T: Conserved function of RNF4 family of proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J 2007;26:4102–4112. [DOI] [PMC free article] [PubMed] [Google Scholar]