Abstract
Background
The purpose of the current study was to assess the association between HOXA9 (homeobox A9) promoter methylation and head and neck squamous cell carcinoma (HNSCC) and its diagnostic value.
Methods
Quantitative methylation‐specific PCR (qMSP) was applied to measure HOXA9 promoter methylation levels in 145 paired HNSCC and corresponding normal tissue samples. Data from the Cancer Genome Atlas (TCGA) database (n = 578; 528 HNSCC and 50 normal) were also analyzed.
Results
Significantly higher levels of HOXA9 promoter methylation were detected in HNSCC, compared with normal, tissues (our cohort: P = 1.06E‐35; TCGA cohort: P = 3.06E‐39). Moreover, HOXA9 methylation was significantly increased in patients with advanced tumor (T) stage, lymph node metastasis, and advanced clinical stage. Areas under the receiver characteristic curves (AUCs) based on our cohort and TCGA data were 0.930 and 0.967, respectively.
Conclusion
In summary, our study reveals that HOXA9 promoter hypermethylation contributes to the risk of HNSCC and its progression and metastasis. Additionally, HOXA9 hypermethylation is a potential biomarker for the early diagnosis and screening of patients with HNSCC.
Keywords: diagnosis, HNSCC, HOXA9, metastasis, methylation
1. INTRODUCTION
Worldwide, head and neck cancer is the sixth most common malignancy and fifth leading cause of cancer‐related death.1 More than 90% of head and neck tumors are squamous cell carcinomas (HNSCC) arising from the epithelial mucosal membranes of the upper aerodigestive tract (oral and nasal cavity, oropharynx, hypopharynx, and larynx).2 Besides established risk factors (tobacco and alcohol abuse), high‐risk human papillomavirus (HPV) infection has recently been reported as an independent risk factor for a subset of HNSCC.3, 4 According to recently epidemiological data from the American Cancer Society, 64 690 new cases of HNSCC and at least 13 740 deaths are projected to occur in the United States in 2018,5 which is consistent with a continued rising trend over recent years.6, 7 As early‐stage HNSCC is often symptomless, the majority of HNSCC patients are diagnosed with advanced stage disease, including lymphatic metastasis and distant metastasis, and the 5‐year survival rate remains <50%,8 while outcomes and quality of life can be remarkably improved if HNSCC is detected at an early stage.9 In addition, biopsy by laryngoscope, which often requires general anesthesia, is considered the gold standard method for HNSCC diagnosis; the lack of a similarly valuable, non‐invasive and cost‐effective method for screening and early diagnosis of this disease is considered the major obstacle to improving the prognosis of HNSCC. Hence, identification of effective biomarkers for early HNSCC is an urgent priority for individual diagnosis and therapy.
Head and neck squamous cell carcinoma is a complex disease that can affect multiple sites and is caused by intricate interactions among genetic susceptibility, epigenetic modification, and environmental factors.10 Emerging evidence indicates that epigenetic inactivation of tumor suppressor genes (TSGs) resulting from promoter methylation is involved in the onset and progress of various cancers, including esophageal squamous cell carcinoma,11 cervical cancer,12 lung cancer,13 and breast cancer.14 Aberrant methylation events are frequent, chemically stable, and relatively early molecular changes during carcinogenesis,15, 16 which have potential as biomarkers for cancer screening and early diagnosis.17, 18
Homeobox (HOX) genes are a highly conserved family of 39 transcription factors that are grouped into four clusters, HOXA through HOXD.19, 20 HOX genes regulate and determine different cell types during embryonic development.20, 21 In addition, there is increasing evidence that HOX genes have important functions in regulation of the delicate balance between cell proliferation and differentiation during cancer development.22, 23 The HOXA9 gene, mapping to chromosome 7p15.2, is a member of this large family, and its abnormal expression is involved in the emergence of numerous solid and hematopoietic malignancies. HOXA9 is frequently activated in hematopoietic malignancies24; however, it can be downregulated in solid tumors,25, 26 particularly squamous cell carcinoma. Recent studies have revealed that hypermethylation of the HOXA9 promoter leads to its transcriptional inactivation in several of cancers, including those of the lung,28 breast,29 cervix,25 and bladder30; however, the relationship between HOXA9 and HNSCC remains unclear.
In the present study, we investigated the association between HOXA9 promoter methylation in 145 HNSCC patients and its potential diagnostic value. Furthermore, data from 578 samples available from the Cancer Genome Atlas (TCGA) database were analyzed to validate our findings.
2. MATERIALS AND METHODS
2.1. Specimens and clinical data collection
Head and neck squamous cell carcinoma tissues (n = 145) and corresponding non‐tumor tissues were collected at the Department of Otolaryngology‐Head and Neck Surgery and Oral and Maxillofacial Surgery at Ningbo Lihuili Hospital between November 2012 and August 2017. Before surgery and tissue collection, all patients provided written informed consent. All specimens were freshly obtained and preserved in RNA‐fixer Reagent (Bioteke, Beijing, China) at −80°C until use. Final diagnoses were confirmed histopathologically. Histological grade was defined according to the National Comprehensive Cancer Network oncology guidelines. No patient received chemotherapy or radiotherapy before surgery. None of the patients had a history of hereditary cancer. Tumors were staged according to the TNM classification (7th edition) of the Union for International Cancer Control. Age, gender, smoking behavior, histological classification, tumor location, T classification, lymph metastasis, and tumor stage were extracted from medical records for all cases. The study was approved by the Human Research Ethical Committee of Ningbo Lihuili Hospital.
2.2. DNA extraction and bisulfite conversion
Genomic DNA was extracted from frozen tissues using a QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). A Nanodrop 2000 spectrophotometer (Thermal Scientific Co. Ltd., Wilmington, USA) was used to measure DNA concentration, and quality. extracted DNA was bisulfite‐converted subsequently using the EZ DNA Methylation‐Gold Kit, according to the manufacturer's protocol (Zymo Research, Orange, CA, USA). This procedure converts unmethylated cytosines to uracil, while the methylated cytosines are unaffected. Bisulfite‐converted DNA was stored in tris‐ethylenediaminetetraacetic acid buffer for subsequent methylation analysis.
2.3. Quantitative methylation‐specific polymerase chain reaction (qMSP)
Methylation levels of the HOXA9 promoter (chr7:27206577‐27206704) in 145 HNSCC and paired adjacent tissue samples were determined using a qMSP assay as described previously.31 ACTB was chosen as the internal control28 and human methylated DNA (Zymo Research, Orange, CA, USA) served as the positive control. The qMSP primer sequences were as follows: HOXA9, 5′‐TGATTATTTTTGTTTTAGGAGTCGT‐3′ (forward) and 5′‐TAAAAAAATTTATTTCTCACCCGTT‐3′ (reverse); ACTB, 5′‐TGGTGATGGAGGAGGTTTAGTAAGT‐3′ (forward) and 5′‐AACCAATAAAACCTACTCCTCCCTTAA‐3′ (reverse). PCR conditions for both methylated (M) and unmethylated (U) primer pairs comprised initial denaturation at 95°C for 10 minutes, followed by 45 cycles of 20‐seconds denaturation at 95°C, 20‐seconds annealing at 60°C, and 30‐seconds extension at 72°C. Products were stored at 4°C. The percentage of methylated reference (PMR) was calculated to determine the HOXA9 promoter methylation level.32
2.4. The Cancer Genome Atlas (TCGA) data mining study
The Cancer Genome Atlas (TCGA), supervised by the National Cancer Institute's Center for Cancer Genomics and the National Human Genome Research Institute, is a large‐scale cancer genome project which provides researchers with multi‐dimensional maps of the key genomic changes, clinicopathological information, and survival data in 33 types of cancer (http://cancergenome.nih.gov/)33 DNA methylation profiles (Illumina Human Methylation 450K) and details of the clinicopathological characteristic of patients providing the 528 HNSCC tissues and 50 non‐tumor tissues in TCGA cohort (Project Id: TCGA‐HNSC) were downloaded from the University of California Santa Cruz (UCSC) Xena browser (www.xena.ucsc.edu). The average β values of two Illumina Human Methylation 450K BeadChip probes (cg02643054 on chr7:27206544 and cg00905524 on chr7:27206907), close to the qMSP amplification fragment (chr7:27206577‐27206704), were used to evaluate HOXA9 methylation in this dataset.
2.5. Statistical analysis
All statistical analysis was performed using Statistical Program for Social Sciences (SPSS) 20.0 software (Chicago, IL, USA) and GraphPad Prism 6.0 (La Jolla, CA, USA), which were also used to generate figures. For comparisons between groups, independent Student's t test, paired Student's t test, and one‐way analysis of variance (one‐way ANOVA) tests were employed, as appropriate. Receiver operating characteristic (ROC) analysis was used to assess the diagnostic value of HOXA9 methylation for HNSCC. A two‐tailed P value <0.05 was defined as statistically significant.
3. RESULTS
In the current study, 145 HNSCC and corresponding non‐tumor tissue samples were collected to investigate the association of HOXA9 promoter methylation and HNSCC. Illumina Human Methylation 450K data from 528 patients with histologically confirmed HNSCC, including tumor tissues in all cases and matched adjacent normal tissues in 50 cases, were available from TCGA project, and two Methylation 450K CpG sites (cg02643054 and cg00905524) located near the tested fragment (chr7:27206577‐27206704) were chosen to verify our findings (Figure 1).
Figure 1.
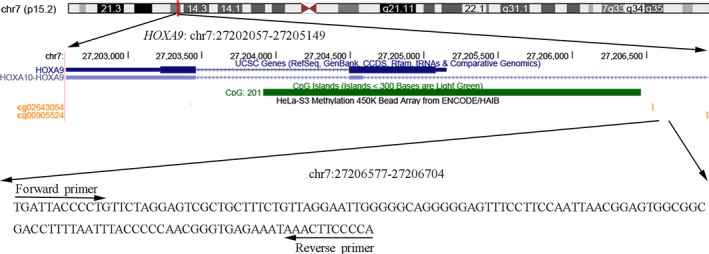
Genomic position of the quantitative methylation‐specific PCR (qMSP) amplified fragment shown in the UCSC genome browser (human 2009 assembly; GRCh37/hg19)). Two available CpG probes (cg00905524 and cg02643054) in the Illumina Human Methylation 450K also map to the HOXA9 promoter, close to the qMSP amplified fragment
In our study cohort, we found that the HOXA9 methylation levels in tumor tissues were significantly higher than those in paired adjacent normal tissues: median PMR with interquartile range, 0.359 (0.221, 0.556) vs 0.075 (0.043, 0.116), P = 1.06E‐35 (Figure 2A). Subsequently, we determined HOXA9 methylation levels using data from TCGA database. As shown in Figure 2B, this analysis validated our finding that there was a significant difference in HOXA9 methylation levels in tumor compared with adjacent non‐tumor tissues(median β value with interquartile range, 0.529 (0.457, 0.594) vs 0.226 (0.186, 0.268), P = 3.06E‐39).
Figure 2.
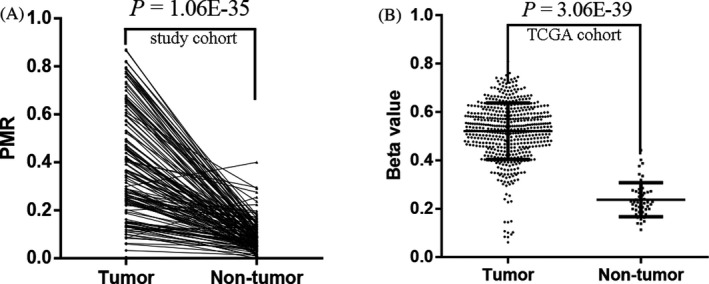
Comparisons of HOXA9 promoter methylation levels in HNSCC tumor and adjacent non‐tumor tissues. A, our study cohort: P = 1.06E‐35; B, TCGA cohort: P = 3.06E‐39
Based on these findings, we analyzed the association between HOXA9 methylation levels, its expression levels, and the clinicopathological characteristics of patients with HNSCC, including age, gender, smoking behavior, histological classification, tumor location, T classification, lymph metastasis, and tumor stage. As shown in Table 1, HOXA9 hypermethylation in human HNSCC tissues was associated with T classification (P = 0.008), lymph metastasis (P = 0.012), and tumor stage (P = 0.004). Importantly, these findings were replicated by analysis of TCGA data. HOXA9 was significantly hypermethylated in patients with advanced pathologic tumors (P = 0.041), advanced pathologic nodal category (P = 0.047), and advanced pathologic stage (P = 0.009) (Table 2). Additionally, a borderline significant difference in HOXA9 methylation status was identified between male and female patients in TCGA data (P = 0.06). This may be explained in part by differences in smoking habits, with female patients mainly being non‐smokers; however, no statistically significant correlation was identified with any other clinicopathological characteristic in either our study cohort or the TCGA cohort.
Table 1.
Association of HOXA9 promoter methylation with clinicopathological characteristics of HNSCC patients in our study cohort
| Characteristics | N | Mean ± SD | P value |
|---|---|---|---|
| Gender | |||
| Female | 30 | 0.407 ± 0.228 | 0.698 |
| Male | 115 | 0.389 ± 0.222 | |
| Age | |||
| <60 y | 78 | 0.397 ± 0.235 | 0.786 |
| ≥ 60 y | 67 | 0.387 ± 0.209 | |
| Smoking behavior | |||
| No | 28 | 0.359 ± 0.233 | 0.376 |
| Yes | 117 | 0.400 ± 0.221 | |
| Histological classification | |||
| Well and Moderately | 126 | 0.379 ± 0.220 | 0.063 |
| Poorly | 19 | 0.481 ± 0.228 | |
| Tumor location | |||
| Oral cavity | 21 | 0.360 ± 0.224 | 0.881 |
| Oropharynx | 6 | 0.393 ± 0.273 | |
| Hypopharynx | 26 | 0.413 ± 0.120 | |
| Larynx | 92 | 0.394 ± 0.228 | |
| T classification | |||
| T1 + 2 | 82 | 0.350 ± 0.214 | 0.008a |
| T3 + 4 | 63 | 0.448 ± 0.223 | |
| Lymph metastasis | |||
| No | 97 | 0.360 ± 0.213 | 0.012a |
| Yes | 48 | 0.458 ± 0.231 | |
| Tumor stage | |||
| Stage I + II | 64 | 0.333 ± 0.211 | 0.004a |
| Stage III +IV | 81 | 0.439 ± 0.222 | |
The difference in HOXA9 promoter methylation between these groups was significant.
Table 2.
The association of HOXA9 promoter methylation with clinicopathological characteristics of HNSCC patients in TCGA cohort
| Characteristics | N | Mean ± SD | P value |
|---|---|---|---|
| Gender | |||
| Female | 142 | 0.506 ± 0.107 | 0.06 |
| Male | 386 | 0.526 ± 0.120 | |
| Age | |||
| <60 y | 260 | 0.524 ± 0.120 | 0.602 |
| ≥60 y | 267 | 0.518 ± 0.114 | |
| Smoking behavior | |||
| No | 122 | 0.528 ± 0.107 | 0.44 |
| Yes | 393 | 0.519 ± 0.120 | |
| Alcohol history | |||
| No | 165 | 0.511 ± 0.134 | 0.225 |
| Yes | 352 | 0.526 ± 0.108 | |
| Histological grade | |||
| G1 + 2 | 374 | 0.520 ± 0.111 | 0.779 |
| G3 + 4 | 132 | 0.523 ± 0.125 | |
| Tumor site | |||
| Oral cavity | 392 | 0.526 ± 0.109 | 0.241 |
| Oropharynx | 9 | 0.547 ± 0.076 | |
| Hypopharynx | 10 | 0.494 ± 0.168 | |
| Larynx | 117 | 0.504 ± 0.138 | |
| HPV status | |||
| Positive | 41 | 0.534 ± 0.142 | 0.119 |
| Negative | 74 | 0.495 ± 0.117 | |
| Pathologic tumor category | |||
| Tis/T1/T2 | 190 | 0.504 ± 0.120 | 0.041a |
| T3/T4 | 276 | 0.527 ± 0.116 | |
| Pathologic nodal category | |||
| No | 180 | 0.504 ± 0.119 | 0.047a |
| Yes | 240 | 0.528 ± 0.117 | |
| Pathologic stage | |||
| StageI + II | 104 | 0.490 ± 0.116 | 0.009a |
| StageIII + IV | 352 | 0.525 ± 0.118 | |
The difference of HOXA9 promoter methylation between these groups was significant.
Sensitivity and specificity are objective and easy to understand; however, they are often affected by the use of different threshold values. We examined the diagnostic value of HOXA9 promoter methylation in HNSCC using ROC curve analysis, which is a synthesized index that reflects the accuracy of diagnostic test. An area under the ROC curve (AUC) close to 1.0 signifies that the test has almost perfect discrimination. The maximum Youden index was used as a cutoff point. In our study cohort, HOXA9 hypermethylation yielded an AUC of 0.930 (95% CI: 0.901‐0.959), a sensitivity of 79.3%, and a specificity of 93.8%, with a cutoff value of 0.185 (Figure 3A). In the TCGA cohort, HOXA9 hypermethylation yielded an AUC of 0.967 (95% CI: 0.952‐0.983), a sensitivity of 93.6%, and a specificity of 92.0% with a cutoff value of 0.343 (Figure 3B).
Figure 3.
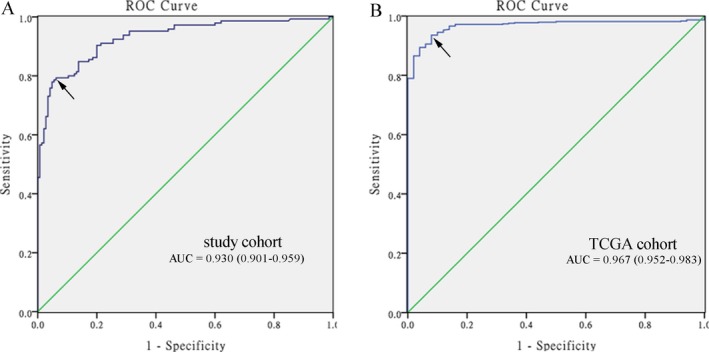
Receiver operating characteristic (ROC) curves to assess the diagnostic value of HOXA9 promoter methylation in HNSCC patients. A, in our study cohort, the area under the curve (AUC) was 0.930. B, in TCGA cohort, the AUC was 0.967. The arrows indicate cutoff points
4. DISCUSSION
Hypermethylation, causing the transcriptional silencing of the promoters of tumor suppressor genes (TSGs), occurs in various malignancies as part of the process of carcinogenesis.34 Compared with other molecular markers, DNA hypermethylation is a common and early event during the progression of various tumors and it is chemically and biologically more stable than RNA or the majority of proteins.35 Given these advantages and the development of technology for their detection, methylation biomarkers have great potential for use in early screening and diagnosis of cancer.36 HOXA9 functions as a tumor suppressor gene that suppresses breast tumor growth and metastasis.23 Furthermore, methylation of the HOXA9 promoter is associated with progression and prognosis in numerous cancers. 28, 29
In the current study, we recruited 145 HNSCC patients to investigate the association of HOXA9 methylation with HNSCC and its potential for use in detection of HNSCC. Our results showed that methylation levels of the HOXA9 promoter were significantly higher in HNSCC than adjacent non‐tumor tissues. Similarly, further bioinformatics analyses of TCGA data confirmed that HOXA9 methylation was higher in HNSCC compared with normal tissues. Taken together, these findings suggest that HOXA9 methylation is a risk factor for HNSCC and has potential value for its diagnosis.
Subsequently, we also determined the association between HOXA9 promoter methylation and the clinicopathological characteristics of patients with HNSCC. Tumor invasion and clinical stage are vital factors for assessing prognosis in patients with cancer and are among the most common tools used for that purpose.37, 38 In our qMSP study, we demonstrated a significantly elevated frequency of HOXA9 promoter methylation in patients with advanced tumor stage and advanced clinical stage, which was confirmed by analysis of TCGA data. Overall, these results suggest that HOXA9 methylation may be involved in the progression and metastasis of HNSCC. Given the well‐developed lymphatic network in the neck region, HNSCC has a high propensity to undergo lymph node metastasis40; however, because of the high incidence of occult lymph node metastasis,41 the accurate diagnosis of lymph node metastases remains challenging. Analyses of both our cohort and TCGA data showed that HOXA9 promoter methylation levels were significantly higher in HNSCC patients with lymph node metastasis compared with those without, providing a potential means to distinguish HNSCC patients with lymphatic metastasis. Additionally, an almost statistically significant difference in HOXA9 methylation status was found between male and female patients in TCGA data (P = 0.06), which may be partly attributable to differences in smoking habits, with female patients mainly being non‐smokers.
Therapeutic procedures and prognosis differ substantially between early‐ and late‐stage HNSCC. Early‐stage HNSCC patients receive minimally invasive surgery or irradiation alone, with good outcomes, while late‐stage patients receive aggressive therapy, such as expanded surgery and/or concomitant chemoradiotherapy, resulting in dismal survival rates and poor quality of life.42, 43 Screening for HNSCC depends on clinical symptoms and imaging examinations (laryngoscopy, computed tomography, magnetic resonance imaging, and positron emission tomography) and histopathological examination44; however, owing to the non‐specificity of symptoms in early‐stage disease and ineffective conventional cancer‐related biomarkers,45 the early detection of HNSCC remains unsatisfactory. As they occur early in carcinogenesis and have other advantageous characteristics, abnormal methylation patterns represent potential markers for early detection of cancer and can even be non‐invasively detected in various body fluids (blood, bronchial aspirates, brushing, saliva, and urine).46, 47 In the present study, we constructed ROC curves and calculated the AUC to determine the diagnostic value of HOXA9 methylation for HNSCC. The AUC values were close to 1.0, signifying near perfect diagnostic power. The AUC for our study was 0.930 and that based on TCGA data was even more encouraging at 0.967, indicating that HOXA9 promoter methylation has excellent diagnostic accuracy for HNSCC.
The current study had some limitations. A number of methods have been developed to assess biomarkers in biological fluids to non‐invasively identify early HNSCC.49, 50 Due to a lack of blood and/or saliva samples, we were unable to explore the diagnostic value of non‐invasive detection of HNSCC methylation. In addition, emerging evidence indicates that a panel of several methylation genes could improve cancer diagnosis,51 while our study focused on a single gene, which may not completely satisfy the requirements for clinical application. Therefore, future investigation is needed to determine whether a panel for analysis of the HOXA9 promoter combined with other epigenetic biomarkers will be of higher diagnostic value, particularly using liquid biopsies.
5. CONCLUSION
In conclusion, HOXA9 promoter hypermethylation is associated with the risk for HNSCC and its progression and metastasis. Additionally, HOXA9 hypermethylation has potential for use as a biomarker for the early diagnosis and screening of patients with HNSCC.
ACKNOWLEDGMENTS
This research was supported by grants from the Zhejiang Provincial Natural Science Foundation of China (No. LY14H160003), the Medical and Health Research Project of Zhejiang Province (No. 2012ZDA042), the Medical and Health Training Project of Zhejiang Province (No. 2014PYA017), the Scientific Innovation Team Project of Ningbo (No. 2012B82019), the Ningbo Natural Science Foundation (No. 2015A610221 and No. 2017A610236), and Ningbo Health Branding Subject Fund (PPXK2018‐02).
Zhou C, Li J, Li Q, et al. The clinical significance of HOXA9 promoter hypermethylation in head and neck squamous cell carcinoma. J Clin Lab Anal. 2019;33:e22873 10.1002/jcla.22873
Contributor Information
Zhenhua Wu, Email: zhenhuawu75@163.com.
Zhisen Shen, Email: szs7216@163.com.
Hongxia Deng, Email: xixizb@163.com.
REFERENCES
- 1. Vokes EE, Weichselbaum RR, Lippman SM, Hong WK. Head and neck cancer. N Engl J Med. 1993;328(3):184‐194. [DOI] [PubMed] [Google Scholar]
- 2. Thompson L. World Health Organization classification of tumours: pathology and genetics of head and neck tumours. Ear Nose Throat J. 2006;85(2):74. [PubMed] [Google Scholar]
- 3. Jung AC, Briolat J, Millon R, et al. Biological and clinical relevance of transcriptionally active human papillomavirus (HPV) infection in oropharynx squamous cell carcinoma. Int J Cancer. 2010;126(8):1882‐1894. [DOI] [PubMed] [Google Scholar]
- 4. Sturgis EM, Cinciripini PM. Trends in head and neck cancer incidence in relation to smoking prevalence: an emerging epidemic of human papillomavirus‐associated cancers? Cancer. 2007;110(7):1429‐1435. [DOI] [PubMed] [Google Scholar]
- 5. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7‐30. [DOI] [PubMed] [Google Scholar]
- 6. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7‐30. [DOI] [PubMed] [Google Scholar]
- 7. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7‐30. [DOI] [PubMed] [Google Scholar]
- 8. Laramore GE, Scott CB, al‐Sarraf M,et al. Adjuvant chemotherapy for resectable squamous cell carcinomas of the head and neck: report on Intergroup Study 0034 . Int J Radiat Oncol Biol Phys. 1992;23(4):705‐713. [DOI] [PubMed] [Google Scholar]
- 9. Miller KD, Siegel RL, Lin CC, et al. treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66(4):271‐289. [DOI] [PubMed] [Google Scholar]
- 10. Mroz EA, Tward AD, Hammon RJ, Ren Y, Rocco JW. Intra‐tumor genetic heterogeneity and mortality in head and neck cancer: analysis of data from the Cancer Genome Atlas. PLoS Med. 2015;12(2):e1001786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Luo M, Li Y, Shi X, et al. Aberrant methylation of EYA4 promotes epithelial‐mesenchymal transition in esophageal squamous cell carcinoma. Cancer Sci. 2018;109(6):1811‐1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu D, Zhang J, Fan P,et al. Methylation in the promoter regions of WT1, NKX6‐1 and DBC1 genes in cervical cancer tissues of Uygur women in Xinjiang. Genet Mol Biol. 2018;41(1):9‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Feng Y, Wang L, Wang M. Alteration of DACH1 methylation patterns in lung cancer contributes to cell proliferation and migration. Biochem Cell Bio. 2018;96(5):602‐609. [DOI] [PubMed] [Google Scholar]
- 14. Cao X, Tang Q, Holland‐Letz T, et al. Evaluation of promoter methylation of RASSF1A and atm in peripheral blood of breast cancer patients and healthy. Int J Mol Sci. 2018;19(3):E900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Forest M, O'Donnell KJ, Voisin G, et al. Agreement in DNA methylation levels from the Illumina 450K array across batches, tissues, and time. Epigenetics. 2018;13(1):19‐32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou C, Li J, Li Q. CDKN2A methylation in esophageal cancer: a meta‐analysis. Oncotarget. 2017;8(30):50071‐50083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Delpu Y, Cordelier P, Cho WC, Torrisani J. DNA methylation and cancer diagnosis. Int J Mol Sci. 2013;14(7):15029‐15058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fece de la Cruz F, Corcoran RB Methylation in cell‐free DNA for early cancer detection. Ann Oncol. 2018;29(6):1351‐1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Apiou F, Flagiello D, Cillo C, Malfoy B, Poupon MF, Dutrillaux B. Fine mapping of human HOX gene clusters. Cytogenet Cell Genet. 1996;73(1–2):114‐115. [DOI] [PubMed] [Google Scholar]
- 20. Lappin TR, Grier DG, Thompson A, Halliday HL. HOX genes: seductive science, mysterious mechanisms. Ulster Med J. 2006;75(1):23‐31. [PMC free article] [PubMed] [Google Scholar]
- 21. Castelli‐Gair J. Implications of the spatial and temporal regulation of Hox genes on development and evolution. Int J Dev Biol. 1998;42(3):437‐444. [PubMed] [Google Scholar]
- 22. Grier DG, Thompson A, Kwasniewska A, McGonigle GJ, Halliday HL, Lappin TR. The pathophysiology of HOX genes and their role in cancer. J Pathol. 2005;205(2):154‐171. [DOI] [PubMed] [Google Scholar]
- 23. Sun M, Song C‐X, Huang H, et al. HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis. Proc Natl Acad Sci USA. 2013;110(24):9920‐9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dorsam ST, Ferrell CM, Dorsam GP, et al. The transcriptome of the leukemogenic homeoprotein HOXA9 in human hematopoietic cells. Blood. 2004;103(5):1676‐1684. [DOI] [PubMed] [Google Scholar]
- 25. Alvarado‐Ruiz L, Martinez‐Silva MG, Torres‐Reyes LA, et al. HOXA9 is Underexpressed in cervical cancer cells and its restoration decreases proliferation, migration and expression of epithelial‐to‐mesenchymal transition genes. Asian Pac J Cancer Prev. 2016;17(3):1037‐1047. [DOI] [PubMed] [Google Scholar]
- 26. Popovic R, Erfurth F, Zeleznik‐Le N. Transcriptional complexity of the HOXA9 locus. Blood Cells Mol Dis. 2008;40(2):156‐159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reinert T, Modin C, Castano Fm, et al. Comprehensive genome methylation analysis in bladder cancer: identification and validation of novel methylated genes and application of these as urinary tumor markers. Cli Cancer Res. 2011;17(17):5582‐5592. [DOI] [PubMed] [Google Scholar]
- 28. Wrangle J, Machida Eo, Danilova L, et al. Functional identification of cancer‐specific methylation of CDO1, HOXA9, and TAC1 for the diagnosis of lung cancer. Cli Cancer Res. 2014;20(7):1856‐1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li M, Li X, Zhuang Y, et al. Induction of HOXA9 expression in three‐dimensional organotypic culture of the Claudin‐low breast cancer cells. Oncotarget. 2016;7(32):51503‐51514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim YJ, Yoon HY, Kim JS, et al. HOXA9, ISL1 and ALDH1A3 methylation patterns as prognostic markers for nonmuscle invasive bladder cancer: array‐based DNA methylation and expression profiling. Int J Cancer. 2013;133(5):1135‐1142. [DOI] [PubMed] [Google Scholar]
- 31. Li J, Zhou C, Ni S, et al. Methylated claudin‐11 associated with metastasis and poor survival of colorectal cancer. Oncotarget. 2017;8(56):96249‐96262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shen Z, Lin L, Cao B, Zhou C, Hao W, Ye D. LZTS2 promoter hypermethylation: a potential biomarker for the diagnosis and prognosis of laryngeal squamous cell carcinoma. World J surg oncol. 2018;16(1):42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Weinstein JN, Collisson EA, Mills GB, et al. The Cancer Genome Atlas Pan‐Cancer analysis project. Nat Genet. 2013;45(10):1113‐1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shen Z, Zhou C, Li J, et al. SHISA3 promoter methylation is a potential diagnostic and prognostic biomarker for laryngeal squamous cell carcinoma. BioMed Res Int. 2017;2017:9058749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3(4):253‐266. [DOI] [PubMed] [Google Scholar]
- 36. Wittenberger T, Sleigh S, Reisel D, et al. DNA methylation markers for early detection of women's cancer: promise and challenges. Epigenomics. 2014;6(3):311‐327. [DOI] [PubMed] [Google Scholar]
- 37. Feng M, Wang W, Fan Zi, et al. Tumor volume is an independent prognostic indicator of local control in nasopharyngeal carcinoma patients treated with intensity‐modulated radiotherapy. Radiat Oncol. 2013;8:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huang SH, Xu W, Waldron J, et al. Refining American Joint Committee on Cancer/Union for International Cancer Control TNM stage and prognostic groups for human papillomavirus‐related oropharyngeal carcinomas. J Clin Oncol. 2015;33(8):836‐845. [DOI] [PubMed] [Google Scholar]
- 39. Talmi YP, Takes RP, Alon EE, et al. Prognostic value of lymph node ratio in head and neck squamous cell carcinoma. Head Neck. 2018;40(5):1082‐1090. [DOI] [PubMed] [Google Scholar]
- 40. Dogan E, Cetinayak HO, Sarioglu S, Erdag TK, Ikiz AO. Patterns of cervical lymph node metastases in oral tongue squamous cell carcinoma: implications for elective and therapeutic neck dissection. J Laryngol Otol. 2014;128(3):268‐273. [DOI] [PubMed] [Google Scholar]
- 41. Leusink FK, van Es RJ, de Bree R, et al. Novel diagnostic modalities for assessment of the clinically node‐negative neck in oral squamous‐cell carcinoma. Lancet Oncol. 2012;13(12):e554‐561. [DOI] [PubMed] [Google Scholar]
- 42. Choong N, Vokes E. Expanding role of the medical oncologist in the management of head and neck cancer. CA Cancer J Clin. 2008;58(1):32‐53. [DOI] [PubMed] [Google Scholar]
- 43. Savvides PP. The role of chemotherapy in the management of patients with head and neck cancer. Semin Plast Surg. 2010;24(2):137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jovanovic MB. Diagnosis of laryngeal carcinoma. Med pregl. 2008;61(11–12):591–595. [DOI] [PubMed] [Google Scholar]
- 45. Barak V, Meirovitz A, Leibovici V, et al. The diagnostic and prognostic value of tumor markers (CEA, SCC, CYFRA 21–1, TPS) in head and neck cancer patients. Anticancer Res. 2015;35(10):5519–5524. [PubMed] [Google Scholar]
- 46. Ni S, Ye M, Huang T. Short stature homeobox 2 methylation as a potential noninvasive biomarker in bronchial aspirates for lung cancer diagnosis. Oncotarget. 2017;8(37):61253–61263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ye M, Huang T, Ni C, Yang P, Chen S. Diagnostic capacity of RASSF1A promoter methylation as a biomarker in tissue, brushing, and blood samples of nasopharyngeal carcinoma. EBioMedicine. 2017;18:32–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ye M, Huang T, Ying Y, et al. Detection of 14‐3‐3 sigma (sigma) promoter methylation as a noninvasive biomarker using blood samples for breast cancer diagnosis. Oncotarget. 2017;8(6):9230–9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ahn SM, Chan J, Zhang Z, et al. Saliva and plasma quantitative polymerase chain reaction‐based detection and surveillance of human papillomavirus‐related head and neck cancer. JAMA Otolaryngol Head Neck Surg. 2014;140(9):846–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sun Y, Du W, Zhou C, et al. A computational method for prediction of saliva‐secretory proteins and its application to identification of head and neck cancer biomarkers for salivary diagnosis. IEEE Trans Nanobioscience. 2015;14(2):167–174. [DOI] [PubMed] [Google Scholar]
- 51. Liu S, Chen X, Chen R, et al. Diagnostic role of Wnt pathway gene promoter methylation in non small cell lung cancer. Oncotarget. 2017;8(22):36354–36367. [DOI] [PMC free article] [PubMed] [Google Scholar]


