Abstract
Background
Mycoplasma pneumoniae (M pneumoniae) is a common human etiology of respiratory infections. Nuclear acid sequence‐based amplification (NASBA) shows good value for the detection of M pneumoniae that surpasses PCR. However, the optimal detection technology still remains to be identified. The purpose of this meta‐analysis was to systematically evaluate the overall accuracy of NASBA for diagnosing M pneumoniae infections.
Methods
The databases PubMed, Cochrane Library, Google Scholar, CNKI, Wang Fang, and Baidu Scholar were comprehensively searched from their initiation date to December 2017 for NASBA in the diagnosis of M pneumoniae infection. Meta‐DiSc 1.4 statistical software was used to evaluate the sensitivity (SEN), specificity (SPE), negative likelihood ratio (−LR), positive likelihood ratio (+LR), diagnostic odds ratio (DOR), and summary receiver operating characteristic (SROC). RevMan 5.2 statistical software was used for quality evaluation of the included articles. Publication bias was evaluated by funnel plot.
Results
Six articles with high quality, including 10 studies, were finally included in this meta‐analysis. The combined statistics results for the diagnosis of M pneumoniae infection by NASBA were 0.77 (SEN, 95% CI: 0.71 to 0.82); 0.98 (SPE, 95% CI: 0.98 to 0.99); 0.22 (‐LR, 95% CI: 0.13 to 0.39); 50.38 (+ LR, 95% CI: 21.85 to 116.17); 292.72 (DOR, 95% CI: 95.02 to 901.75); and 0.9875 (the area under the curve of SROC).
Conclusion
Nuclear acid sequence‐based amplification is a reliable technique to diagnose M pneumoniae infection. However, whether it can replace PCR and serology need to be further studied.
Keywords: meta‐analysis, Mycoplasma pneumoniae, nuclear acid sequence‐based amplification, rapid diagnosis
1. INTRODUCTION
Mycoplasma pneumoniae is one of the primary pathogens resulting in both lower and upper respiratory infections in humans and all age groups,1, 2, 3, 4, 5 and is also the common respiratory system disease of pediatrics.6 M pneumoniae infections are responsible for more than 30% of the community‐acquired pneumonia (CAP) cases 7, 8, 9 and are also the main etiology of CAP in hospitalized patients, only ranking lower than Streptococcus pneumoniae.10, 11 The rates of upper respiratory infections differ across studies and may range by up to 50%.1 Although M pneumoniae can lead to infections in any epidemiologic setting, it shows a particularly significant burden in closed community settings, where outbreaks arise. Numerous outbreaks were recorded since the 1960s in different settings, for example, hospitals,12, 13 institutions,14, 15 military bases,16, 17 and religious communities.18, 19 In many patients, the symptomatic infections are mild, steady, and continue for weeks. However, severe cases requiring hospitalization and even death may happen, particularly among aging or immunocompromised people. Up to 25% of M pneumoniae patients presented extrapulmonary manifestations including the central nervous system, mucous membrane, and cardiovascular system.20 Some complications, for example, neurological manifestations, cause grave outcomes. Antibiotic treatment significantly moderates the symptoms and signs even before the thorough elimination of the bacteria. Early and rapid diagnosis of M pneumoniae infections has important clinical significance for the selection of correct antibiotics.
The clinical manifestation of patients with M pneumoniae infections shows no significant differences with other respiratory pathogen infections, such as Chlamydia pneumoniae; therefore, it is impossible to identify M pneumoniae infections only according to the clinical signs and symptoms, and laboratory tests for identifying M pneumoniae are especially important. However, there is lack of a definite standard test for diagnosis of M pneumoniae. 21
Mycoplasma pneumoniae culture is time‐consuming because it grows slowly (about 2 to 5 weeks for visible colonies appear).22 Serological assays are the most widely used technique in the laboratory diagnosis of M pneumoniae infections; nevertheless, the sensitivity of serology depends on the time phase when the first serum sample is collected after the M pneumoniae infections and the availability of the paired serum samples collected with a 2‐3‐week interval, as well as the sensitivity during the acute infectious stage.23 In addition, serological measurements of host immune responses did not directly measure organism load. Some infected patients have never found a detectable antibody response.24 Conversely, M pneumoniae IgM positive is also seen in healthy children.25 Thus, a nucleic acid amplification technique of was used,26 which includes polymerase chain reaction (PCR) to amplify the specific DNA fragment and the nuclear acid sequence‐based amplification (NASBA) using RNA templates to diagnose M pneumoniae infections. Compared with serology, PCR is rapid and sensitive with high specificity and is thus good for the early clinical diagnosis of M pneumoniae infection.27 Some experts recommend PCR as the gold standard for M pneumoniae diagnosis.
Compared with PCR, no DNA digestion enzyme pretreatment was necessary for NASBA to amplify RNAs. Additionally, NASBA does not need an expensive nucleic acid amplification reaction device when performing constant temperature amplification. NASBA can be completed with a regular constant temperature water bath because the entire process is carried out at 42°C instead of the thermal cycler and is thus very convenient.28 Meanwhile, only a few enzymatic cyclings are needed for NASBA to achieve the required target amount. For instance, to reach 106 amplification, PCR requires 20 cycles, while only 4‐5 cycles are needed for NASBA. Moreover, the mismatching rate is low and the cycle is shorter for NASBA than RT‐PCR.29 Therefore, it is worth exploring the diagnostic value of NASBA for M pneumoniae infections. NASBA has been shown to have a good diagnostic value for M pneumoniae infections compared with PCR as the gold standard; however, the evidence is not sufficient due to the relatively independent studies and small sample size. The aim of the current study was to perform a systematic review and meta‐analysis to evaluate the overall accuracy of NASBA to diagnose of M pneumoniae infections, as well as to provide powerful evidence for the possibility of using NASBA to diagnose M pneumoniae infections.
2. MATERIAL AND METHODS
2.1. Search strategy and selection criteria
Six electronic databases (PubMed, Cochrane Library, Google Scholar, CNKI, Wang Fang, and Baidu Scholar; three databases in English and three in Chinese) were comprehensively searched by two researchers independently from their initiation date to December 2017 with the following search terms: (i) NASBA OR Nucleic Acid Sequence‐Based Amplification; (ii) Mycoplasma pneumoniae OR M pneumoniae OR pneumonia mycoplasma. For the included studies, citation and reference lists were screened.
2.2. Inclusion criteria
Literature screening was conducted according to the inclusion criteria for the diagnostic test study from the Collaboration Screening and Diagnostic Test Method Group in the Cochrane Library.
Inclusion criteria: (i) studies in Chinese or English; (ii) patient samples; (iii) prospective or retrospective studies; (iv) sample size ≥30; (v) PCR used as the gold standard, and all the samples were detected with both PCR and NASBA; (vi) the data in the four grid tables could be directly obtained or indirectly calculated.
Exclusion criteria: (i) abstract, review, systematic review, or case report; (ii) duplicated publications; (iii) incomplete raw data; (iv) unavailable full text.
Data extraction: A predesigned data extraction form was used for data collection. Data were retrieved by two researchers independently according to the inclusion criteria and exclusion criteria from the reports. These data included (i) basic information of the included studies (including author, year of publication, region of the publication from); (ii) research design; (iii) data from the four grid table in the included studies, including true positive (TP), false positive (FP), false negative (FN), and true negative (TN); (iv) assessment of key points for risk of bias including nucleic acid extraction techniques and detection methods, etc Any disagreement needed the consensus of a third researcher.
Quality assessment of included studies: The methodological quality of the included studies was evaluated using the QUADAS‐2 tool recommended by the Cochrane Collaboration, and the RevMan 5.2 statistical software was used to display the quality of the study.
2.3. Statistical methods
The pooled analysis was performed using Meta‐DiSc 1.4 software. The ROC topographical plan was drawn, and the Spearman correlation coefficient between the logarithm of the sensitivity and logarithm of the (1‐specificity) was calculated to determine whether there was a threshold effect. There was threshold effect if P < 0.05. The random‐effects model was used to calculate the pooled parameters including SEN and SPE, +LR, −LR, DOR and their 95% CI for the forest plots to assess the probability of the accurate identification of both M pneumoniae‐infected and non‐infected individuals by NASBA. The heterogeneity of the eligible studies was evaluated by the I 2 test. The application of the effects model depends on the heterogeneity among the studies. If no significant heterogeneity was found (I 2 < 50%) in the included studies, a fixed‐effects model was used to analyze the results, whereas a random‐effects model was applied for the meta‐analysis if significant heterogeneity (I2 ≥ 50%) existed in the eligible studies. The SROC curve was created, and the area under the curve (AUC) was estimated. A value closer to one of the AUC indicates a higher value of NASBA for the diagnosis of M pneumoniae infections. The heterogeneity was analyzed and the meta‐regression analysis was performed using Meta‐DiSc 1.4 software to explore the source of heterogeneity. Funnel plots were made using Stata 12.0 software, and linear regression models were used to verify the symmetry of the funnel plots to identify publication bias. P < 0.05 was considered to indicate publication bias.
3. RESULTS
3.1. Literature identification and selection
The detailed process of the literature identification and selection is shown in Figure 1. A total of 26 publications were retrieved according to the search strategy described in the methods section (12 from PubMed, 0 from Cochrane Library, 5 from Google Scholar, 5 from CNKI, 2 from Wanfang, and 6 from Baidu Scholar), while nine duplicate studies, three reviews, four unrelated articles, and three articles without full text were excluded after reviewing the titles and abstracts. One article with incomplete original data and one article with data published repeatedly were further excluded after reading the full text. Finally, six articles 30, 31, 32, 33, 34, 35 meeting the inclusion criteria were included in this meta‐analysis.
Figure 1.
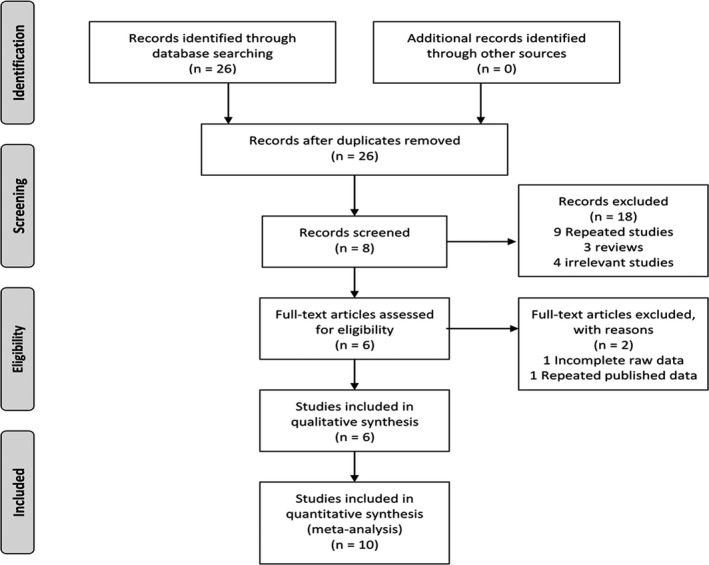
Flow diagram of study identification and inclusion
3.2. Characteristics and quality evaluation of the included studies
Ten groups of relatively independent data were included in the six included studies. The characteristics of the six included studies are shown in Table 1. The methodological quality of the articles was evaluated using the QUADAS‐2 tool. As shown in Figure 2, our results showed that the quality of the six included studies was high and that therefore the included studies were representative. Four studies were from Belgium, one study was from the Netherlands and one from France. Four studies were prospective and two studies did not report the study design method.
Table 1.
Characteristics of the included studies
| Author | Year | Region | Research methods | Instrument/Reagent | Type of NASBA | Gold standard | TP | FP | FN | TN |
|---|---|---|---|---|---|---|---|---|---|---|
| Templeton9 | 2003 | Netherlands | Prospective | NucliSens basic kit | RT‐NASBA | PCR | 12 | 0 | 0 | 94 |
| Loens10 | 2008 | Belgium | Prospective | Manual Boom | In‐house RT‐NASBA | PCR | 4 | 1 | 6 | 204 |
| Loens10 | 2008 | Belgium | Prospective | NucliSens MiniMAG | EasyQ RT‐NASBA | PCR | 9 | 0 | 1 | 205 |
| Loens10 | 2008 | Belgium | Prospective | Manual Boom | EasyQ RT‐NASBA | PCR | 7 | 0 | 3 | 205 |
| Loens11 | 2003 | Belgium | NA | NucliSens basic kit | RT‐NASBA | PCR | 16 | 0 | 1 | 100 |
| Loens11 | 2003 | Belgium | NA | NucliSens basic kit | Conventional NASBA | PCR | 17 | 0 | 0 | 100 |
| Bessede14 | 2010 | France | Retrospective | NucliSens EasyMAG | EasyQ RT‐NASBA | PCR and/or culture | 37 | 0 | 3 | 37 |
| Loens12 | 2008 | Belgium | Prospective | NucliSens basic kit | RT‐NASBA | RT‐PCR | 21 | 13 | 3 | 214 |
| Loens13 | 2008 | Belgium | NA | NucliSens basic kit | RT‐mono‐NASBA | RT‐PCR | 25 | 7 | 15 | 153 |
| Loens13 | 2008 | Belgium | NA | NucliSens basic kit | RT‐multiplex‐NASBA | RT‐PCR | 21 | 3 | 19 | 157 |
NA: Not Applicable
Figure 2.
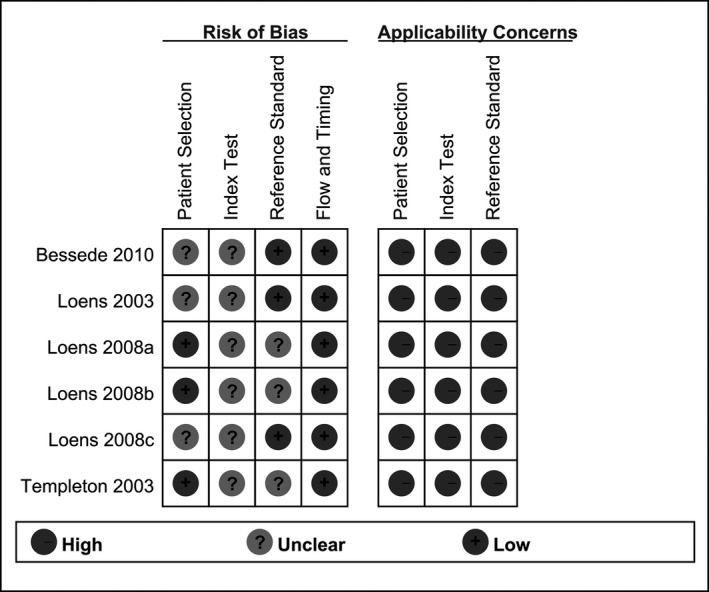
Quality evaluation of the included studies
3.3. Meta‐analysis and overall diagnostic accuracy
Threshold effect analysis: The scatter plot of the ROC topographical plan made from Meta‐Disc 1.4 software showed an atypical "shoulder‐arm shape." The Spearman correlation coefficient is r = 0.012 and P = 0.973, suggesting that Sen was negatively correlated with 1‐Spe, with no existing threshold effect.
The pooled results: a meta‐analysis was conducted for the pooled data extracted from the included studies using a random effects model. The results showed that SEN was 0.77 (95% CI: 0.71 to 0.82) (Figure S1); SPE was 0.98 (95% CI: 0.98 to 0.99) (Figure S2); + LR was 50.38 (95% CI: 21.85 to 116.17) (Figure S3); −LR was 0.22 (95% CI: 0.13 to 0.39) (Figure S4); DOR was 292.72 (95% CI: 95.02 to 901.75) (Figure S5); the area under the SROC curve (AUC) was 0.9875 and the Q* index was 0.9525 (Figure 3). The results of the pooled results suggest that NASBA shows a higher accuracy in the diagnosis of M pneumoniae infections, in which the potential to identify individuals without infections is higher than that to identify individuals with infections.
Figure 3.
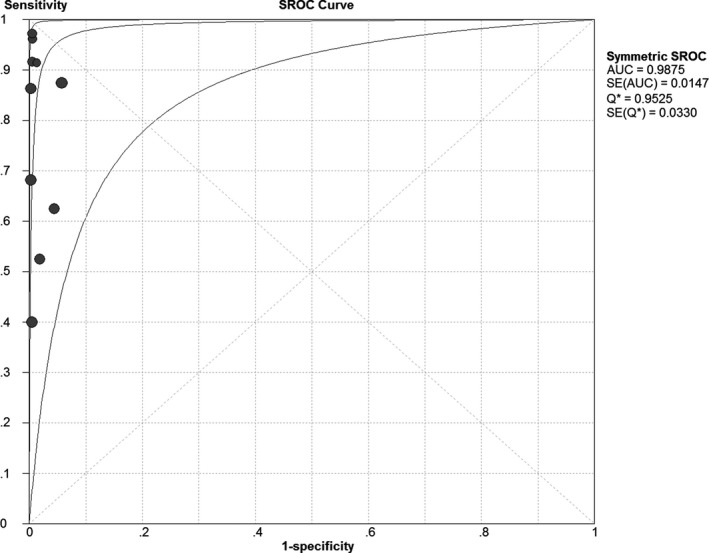
Summary receiver operating characteristic curves of Mycoplasma pneumoniae infections detected by NASBA
Our results showed that heterogeneity existed in the sensitivity, specificity, positive likelihood ratio, negative likelihood ratio, and diagnostic odds ratio (the I 2 was 82.3%, 80.6%, 62.8%, 79.5%, 62.4%, respectively). A meta‐regression analysis performed using the Meta‐DiSc 1.4 software for researchers, countries, nucleic acid extraction techniques, and detection methods showed that all P > 0.05, indicating that these factors are not able to explain the heterogeneity between the included studies. Other unknown factors led to the production of heterogeneity across the studies.
Publication bias assessment: Stale 12.0 software was used to make the Deeks’ funnel plot to identify publication bias for the included studies, and the linear regression model was used to test the symmetry of the funnel plot. Our result showed that P = 0.016, indicating the existence of publication bias (Figure 4).
Figure 4.
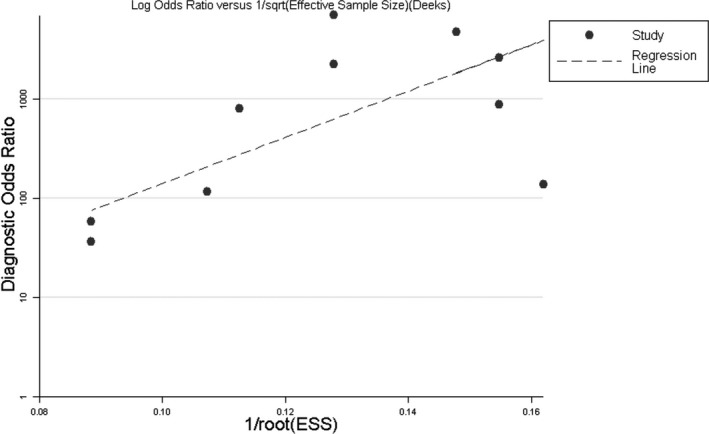
Deeks’ funnel plot asymmetry test evaluating the publication bias based on the diagnostic odds ratio for NASBA detection of Mycoplasma pneumoniae infections
4. DISCUSSION
A clear diagnosis of M pneumoniae infections as soon as possible is of great significance to guide medication, prevent complications, and control disease development. Traditional methods for detecting M pneumoniae infections include classical culture, serological tests, and nucleic acid detection.
However, M pneumoniae is difficult to culture with a low positive isolation rate and long duration; thus, M pneumoniae culture is not suitable for rapid clinical diagnosis.36 Therefore, the diagnosis of M pneumoniae infections essentially relied on serology.37 Unfortunately, the serological test has low sensitivity, poor specificity, and cross‐reactivity with other respiratory mycoplasmas or viruses.38, 39, 40 Nucleic acid detection is rapid and sensitive with high specificity and is thus good for the rapid clinical diagnosis of M pneumoniae infection.
The nucleic acid detection methods that have been reported include traditional PCR, NASBA, LAMP, and others.29 PCR technology has been used to detect M pneumoniae infections for approximately 20 years with quite a few limitations, such as the fact that the PCR inhibitors in samples may result in false‐negative results; contamination can easily lead to false positives; it is relative difficult to obtain high quality samples; and the time point for sampling impacts results. The diagnostic accuracy of PCR has been reported to decrease over 7 days after disease onset vs the serology.39, 40, 41, 42
Nuclear acid sequence‐based amplification has been reported to show a high sensitivity and specificity in the detection of M pneumoniae infection, as well as an even higher value than PCR in the detection of certain microbial infections, such as invasive fungal infections.43 In addition, NASBA also has the advantage of no thermal cycling instruments required with high amplification efficiency,28 which warrants further development and utilization. However, different diagnostic studies in this field have generated inconsistent diagnostic accuracy due to the small sample size, and the exact diagnostic accuracy of NASBA for M pneumoniae is difficult to establish. There are currently no systematic reviews or meta‐analyses assessing NASBA for the detection of M pneumoniae; therefore, this systematic review and meta‐analysis were conducted with the intention to provide more powerful evidence for the feasibility of NASBA in the diagnosis of M pneumoniae infections.
The results of ourmeta‐analysis showed that the pooled specificity was 0.98 (95% CI: 0.98 to 0.99), suggesting that NASBA had a very low omission diagnosis rate for M pneumoniae infections. The pooled sensitivity was 0.77 (95% CI: 0.71 to 0.82), suggesting that NASBA may have a misdiagnosis rate for diagnosing M pneumoniae infections. The misdiagnosis rate may be related to the longer sample freezing and the degradation of RNA in the samples.44 The area under the SROC curve is less than 0.5, which indicates that there is no diagnostic value for the tested technology. The area under the SROC curve and the Q * index are closer to 1, indicating higher diagnostic values. Our results showed that the area under the curve of SROC (AUC) was 0.99 and Q * index was 0.9525. +LR was 50.38 (95% CI: 21.85 to 116.17), suggesting that the positive results in M pneumoniae‐infected individuals was 50.38‐fold that of non‐infected individuals; additionally, the −LR was 0.22 (95% CI: 0.13 to 0.39), suggesting a 0.22‐fold greater chance of negative results in M pneumoniae‐infected individuals than in non‐infected individuals.
The DOR was 292.72 (95%CI: 95.02 to 901.75). A greater value led to a better distinguishing effect of the diagnostic tests when the DOR value is >1. Our results showed a higher value indicating a better distinguishing diagnostic effect of NASBA for M pneumoniae infection.
Based on the above comprehensive meta‐analysis, it can be concluded that NASBA may be a reliable tool for diagnosing M pneumoniae infection.
The methodological qualities of the six articles included in this meta‐analysis were assessed using the QUADAS‐2 tool, which showed a low bias risk of the included case selection, index test, gold standard utility, good case‐flow, good progression status, and high clinical applicability. Ten of the six included articles showed no threshold effect but did show heterogeneity. A meta‐regression analysis was performed to explore the heterogeneous sources, and the results showed that researchers, countries, nucleic acid extraction techniques, and detection methods were not the main sources of heterogeneity, indicating that there are other unknown factors to produce heterogeneity across the included studies. Existence of the potential problem may somehow reduce the stability of the research. Possible causes of clinical heterogeneity or methodological heterogeneity include: (i) the pathogenic conditions of the patients providing the samples is different; (ii) collection, transportation, and storage of samples are different; (iii) different experimental environments.
Deeks’ funnel plots were made for the included studies. The linear regression model was used to test the symmetry of the funnel plots, and our results suggested the existence of publication bias, which may be due to some negative study results not yet having been published, which may impact our results.
There are some limitations to this study, such as: (i) unpublished data were not searched. There may be some negative results, that is, studies without statistical significance in the unpublished literature; this may result in more significant publication bias. (ii) The included studies were all from Europe, and the findings from other regions and countries are absent. (iii) The included documents are relatively old. The most recent article included in this meta‐analysis was published in 2010. The development and innovation of NASBA technology in recent years has not been reflected in our research. The diagnostic value of NASBA will also increase as it is developed. (iv) The number of included studies is relatively small. (v) There are deficiencies in using PCR positive as a gold standard. For example, high sensitivity of PCR may lead to false positives.24, 45
In conclusion, the present systematic review and meta‐analysis suggest an important diagnostic value of NASBA for M pneumoniae infections based on the available evidence. The rapid clinical diagnosis of M pneumoniae infections using NASBA is feasible, and it can be used as an effective supplement for laboratory diagnosis of M pneumoniae. However, the above conclusion still needs to be further verified with a larger sample size and more involved areas.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest.
Supporting information
ACKNOWLEDGMENTS
This work was supported by College students' scientific research project of the third affiliated hospital of Guangzhou medical university (No. 2018A007)
Huang C, Huang P‐T, Yao J‐Y, Li Z‐W, Weng L‐B, Guo X-G. Pooled analysis of nuclear acid sequence‐based amplification for rapid diagnosis of Mycoplasma pneumoniae infection. J Clin Lab Anal. 2019;33:e22879 10.1002/jcla.22879
REFERENCES
- 1. Waites KB, Talkington DF. Mycoplasma pneumoniae and its role as a human pathogen. Clin Microbiol Reviews. 2004;17(4):697–728, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Principi N, Esposito S, Blasi F, Allegra L. Role of Mycoplasma pneumoniae and Chlamydia pneumoniae in children with community‐acquired lower respiratory tract infections. Clin Infect Dis. 2001;32(9):1281–1289. [DOI] [PubMed] [Google Scholar]
- 3. Esposito S, Cavagna R, Bosis S, Droghetti R, Faelli N, Principi N. Emerging role of Mycoplasma pneumoniae in children with acute pharyngitis. Eur J Clin Microbiol Infect Dis. 2002;21(8):607–610. [DOI] [PubMed] [Google Scholar]
- 4. Foy HM. Infections caused by Mycoplasma pneumoniae and possible carrier state in different populations of patients. Clin Infect Dis. 1993;17(Suppl 1):S37–S46. [DOI] [PubMed] [Google Scholar]
- 5. Marston BJ, Plouffe JF, File TM Jr, et al. Incidence of community‐acquired pneumonia requiring hospitalization. Results of a population‐based active surveillance Study in Ohio. The Community‐Based Pneumonia Incidence Study Group. Arch Intern Med. 1997;157(15):1709–1718. [PubMed] [Google Scholar]
- 6. Han MS, Yun KW, Lee HJ, et al. Contribution of co‐detected respiratory viruses and patient age to the clinical manifestations of Mycoplasma pneumoniae pneumonia in children. Pediatric Infect Dis J. 2017; 37: 531‐536. [DOI] [PubMed] [Google Scholar]
- 7. Chiang C‐H, Huang C‐C, Chan W‐L, et al. Association between mycoplasma pneumonia and increased risk of ischemic stroke: a nationwide study. Stroke. 2011;42(10):2940–2943. [DOI] [PubMed] [Google Scholar]
- 8. File TM. Community‐acquired pneumonia. Lancet. 2003;362(9400):1991–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Don M, Fasoli L, Paldanius M, et al. Aetiology of community‐acquired pneumonia: serological results of a paediatric survey. Scand J Infect Dis. 2005;37(11–12):806–812. [DOI] [PubMed] [Google Scholar]
- 10. Porath A, Schlaeffer F, Lieberman D. The epidemiology of community‐acquired pneumonia among hospitalized adults. J Infect. 1997;34(1):41–48. [DOI] [PubMed] [Google Scholar]
- 11. Lauderdale T‐L, Chang F‐Y, Ben R‐J, et al. Etiology of community acquired pneumonia among adult patients requiring hospitalization in Taiwan. Respir Med. 2005;99(9):1079–1086. [DOI] [PubMed] [Google Scholar]
- 12. Kashiwagi S, Hayashi J, Nomura H, et al. An outbreak of Mycoplasma pneumoniae infections in a hospital in Japan. Kurume Med J. 1985;32(4):293–296. [DOI] [PubMed] [Google Scholar]
- 13. Kleemola M, Jokinen C. Outbreak of Mycoplasma pneumoniae infection among hospital personnel studied by a nucleic acid hybridization test. J Hosp Infect. 1992;21(3):213–221. [DOI] [PubMed] [Google Scholar]
- 14. Dorigo‐Zetsma JW, de Wit M, Szabo JS, Schneeberger PM. Epidemic of respiratory tract infections by Mycoplasma pneumoniae in an institute for mentally disabled, investigated with polymerase chain reaction of a throat swab specimen. Ned Tijdschr Geneeskd. 1999;143(24):1261–1265. [PubMed] [Google Scholar]
- 15. Klausner JD, Passaro D, Rosenberg J, et al. Enhanced control of an outbreak of Mycoplasma pneumoniae pneumonia with azithromycin prophylaxis. J Infect Dis. 1998;177(1):161–166. [DOI] [PubMed] [Google Scholar]
- 16. Klement E, Talkington Df, Wasserzug O, et al. Identification of risk factors for infection in an outbreak of Mycoplasma pneumoniae respiratory tract disease. Clin Infect Dis. 2006;43(10):1239–1245. [DOI] [PubMed] [Google Scholar]
- 17. Mogabgab WJ. Mycoplasma pneumoniae and adenovirus respiratory illnesses in military and university personnel, 1959–1966. Am Rev Respirat Dis. 1968;97(3):345–358. [DOI] [PubMed] [Google Scholar]
- 18. Muldoon RL, Raucci J, Kowalski J, Rajashekaraiah K. An outbreak of Mycoplasma pneumoniae respiratory illness in a semi‐closed religious commune. Ann Emerg Med. 1982;11(11):613–615. [DOI] [PubMed] [Google Scholar]
- 19. Waring AL, Halse TA, Csiza CK, Carlyn CJ, Arruda Musser K, Limberger RJ. Development of a genomics‐based PCR assay for detection of Mycoplasma pneumoniae in a large outbreak in New York State. J Clin Microbiol. 2001;39(4):1385–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Copete AR, Aguilar YA, Rueda ZV, Velez LA. Genotyping and macrolide resistance of Mycoplasma pneumoniae identified in children with Community‐Acquired Pneumonia (CAP) in Medellin‐Colombia. Int J Infect Dis. 2017; 66: 113‐120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Montagnani F, Rossetti B, Vannoni A, Cusi MG, De Luca A. Laboratory diagnosis of Mycoplasma pneumoniae infections: data analysis from clinical practice. New Microbiol. 2018;41(3):203–207. [PubMed] [Google Scholar]
- 22. Loens K, Ursi D, Goossens H, Ieven M. Molecular diagnosis of Mycoplasma pneumoniae respiratory tract infections. J Clin Microbiol. 2003;41(11):4915–4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chaudhry R, Sharma S, Javed S, Passi K, Dey AB, Malhotra P. Molecular detection of Mycoplasma pneumoniae by quantitative real‐time PCR in patients with community acquired pneumonia. Indian J Med Res. 2013;138:244–251. [PMC free article] [PubMed] [Google Scholar]
- 24. Waites KB, Xiao L, Liu Y, Balish MF, Atkinson TP. Mycoplasma pneumoniae from the respiratory tract and beyond. Clinical Microbiol Rev. 2017;30(3):747–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Loens K, Ieven M. Mycoplasma pneumoniae: current knowledge on nucleic acid amplification techniques and serological diagnostics. Front Microbiol. 2016;7:448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Song M, Zhang Y, Li S, et al. A sensitive and rapid immunoassay for Mycoplasma pneumoniae in children with pneumonia based on single‐walled carbon nanotubes. Sci Rep. 2017;7(1):16442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dash S, Chaudhry R, Dhawan B, Dey AB, Kabra SK, Das BK. Clinical spectrum and diagnostic yields of Mycoplasma pneumoniae as a causative agent of community‐acquired pneumonia. J Lab Physicians. 2018;10(1):44–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lu X, Shi X, Wu G, Wu T, Qin R, Wang Y. Visual detection and differentiation of Classic Swine Fever Virus strains using nucleic acid sequence‐based amplification (NASBA) and G‐quadruplex DNAzyme assay. Sci Rep. 2017;7:44211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maffert P, Reverchon S, Nasser W, Rozand C, Abaibou H. New nucleic acid testing devices to diagnose infectious diseases in resource‐limited settings. Eur J Clin Microbiol Infect Dis. 2017;36(10):1717–1731. [DOI] [PubMed] [Google Scholar]
- 30. Templeton KE, Scheltinga SA, Graffelman AW, et al. Comparison and evaluation of real‐time PCR, real‐time nucleic acid sequence‐based amplification, conventional PCR, and serology for diagnosis of Mycoplasma pneumoniae . J Clin Microbiol. 2003;41(9):4366–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Loens K, Ursi D, Goossens H, Ieven M. Evaluation of the NucliSens miniMAG RNA extraction and real‐time NASBA applications for the detection of Mycoplasma pneumoniae and Chlamydophila pneumoniae in throat swabs. J Microbiol Methods. 2008;72(2):217–219. [DOI] [PubMed] [Google Scholar]
- 32. Loens K, Ieven M, Ursi D, et al. Detection of Mycoplasma pneumoniae by real‐time nucleic acid sequence‐based amplification. J Clin Microbiol. 2003;41(9):4448–4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Loens K, Beck T, Ursi D, et al. Development of real‐time multiplex nucleic acid sequence‐based amplification for detection of Mycoplasma pneumoniae, Chlamydophila pneumoniae, and Legionella spp. in respiratory specimens. J Clin Microbiol. 2008;46(1):185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Loens K, Beck T, Ursi D, et al. Evaluation of different nucleic acid amplification techniques for the detection of M pneumoniae, C pneumoniae and Legionella spp. in respiratory specimens from patients with community‐acquired pneumonia. J Microbiol Methods. 2008;73(3):257–262. [DOI] [PubMed] [Google Scholar]
- 35. Bessede E, Renaudin H, Clerc M, de Barbeyrac B, Bebear C, Pereyre S. Evaluation of the combination of the NucliSENS easyMAG and the EasyQ applications for the detection of Mycoplasma pneumoniae and Chlamydia pneumoniae in respiratory tract specimens. Eur J Clin Microbiol Infect Dis. 2010;29(2):187–190. [DOI] [PubMed] [Google Scholar]
- 36. Ou L, Lv Q, Wu C, Hao H, Zheng Y, Jiang Y. Development of a lateral flow immunochromatographic assay for rapid detection of Mycoplasma pneumoniae‐specific IgM in human serum specimens. J Microbiol Methods. 2016;124:35–40. [DOI] [PubMed] [Google Scholar]
- 37. Chan YR, Morris A. Molecular diagnostic methods in pneumonia. Curr Opin Infect Dis. 2007;20(2):157–164. [DOI] [PubMed] [Google Scholar]
- 38. Li W, Liu Y, Zhao Y, Tao R, Li Y, Shang S. Rapid diagnosis of Mycoplasma pneumoniae in children with pneumonia by an immuno‐chromatographic antigen assay. Sci Rep. 2015;5:15539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tjhie JH, van Kuppeveld FJ, Roosendaal R, et al. Direct PCR enables detection of Mycoplasma pneumoniae in patients with respiratory tract infections. J Clin Microbiol. 1994;32(1):11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Haaheim H, Vorland L, Gutteberg TJ. Laboratory diagnosis of respiratory diseases: PCR versus serology. Nucleosides Nucleotides Nucleic Acids. 2001;20(4–7):1255–1258. [DOI] [PubMed] [Google Scholar]
- 41. Nilsson AC, Bjorkman P, Persson K. Polymerase chain reaction is superior to serology for the diagnosis of acute Mycoplasma pneumoniae infection and reveals a high rate of persistent infection. BMC Microbiol. 2008;8:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kashyap B, Kumar S, Sethi GR, Das BC, Saigal SR. Comparison of PCR, culture & serological tests for the diagnosis of Mycoplasma pneumoniae in community‐acquired lower respiratory tract infections in children. Indian J Med Res. 2008;128(2):134–139. [PubMed] [Google Scholar]
- 43. Du L, Xia Y, He Y, Pu Q, Hua R, Wu W. Development and evaluation of enzyme‐linked immunosorbent assay of nucleic acid sequence‐based amplification for diagnosis of invasive aspergillosis. AMB Express. 2016;6(1):91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhao Y, Paderu P, Railkar R, et al. Blood Aspergillus RNA is a promising alternative biomarker for invasive aspergillosis. Med Mycol. 2016;54(8):801–807. [DOI] [PubMed] [Google Scholar]
- 45. Lin LJ, Chang FC, Chi H, et al. The diagnostic value of serological studies in pediatric patients with acute Mycoplasma pneumoniae infection. J Microbiol Immunol Infect. 2018. 10.1016/j.jmii.2018.09.001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


