Abstract
Contrast-induced acute kidney injury (CI-AKI) is a severe complication of intravascular applied radial contrast media, and recent progress in interventional therapy and angiography has revived interest in explaining detailed mechanisms and developing effective treatment. Recent studies have indicated a potential link between CI-AKI and microRNA (miRNA). However, the potential non-coding RNA-associated-competing endogenous RNA (ceRNA) pairs involved in CI-AKI still remain unclear. In this study, we systematically explored the circRNA or lncRNA-associated-ceRNA mechanism in a new rat model of CI-AKI through deep RNA sequencing. The results revealed that the expression of 38 circRNAs, 12 lncRNAs, 13 miRNAs and 127 mRNAs were significantly dysregulated. We performed Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway analyses for mRNAs with significantly different expression and then constructed comprehensive circRNA or lncRNA-associated ceRNA networks in kidney of CI-AKI rats. Thereafter, two constructed ceRNA regulatory pathways in this CI-AKI rat model—novel_circ_0004153/rno-miR-144-3p/Gpnmb or Naglu and LNC_000343/rno-miR-1956-5p/KCP—were validated by real-time qPCR. This study is the first one to provide a systematic dissection of non-coding RNA-associated ceRNA profiling in kidney of CI-AKI rats. The selected non-coding RNA-associated ceRNA networks provide new insight for the underlying mechanism and may profoundly affect the diagnosis and therapy of CI-AKI.
Keywords: non-coding RNA, ceRNA networks, CI-AKI, rat model
Introduction
Contrast-induced acute kidney injury (CI-AKI) is defined as acute renal dysfunction within 48–72 h after exposure to iodine contrast media (CM).1, 2 CI-AKI is a strong predictor of early and long-term adverse outcomes and often leads to prolonged hospitalization and increased medical costs.3, 4, 5 An exploration of both strategies and biomarkers for prevention of this disease is necessary and urgently needed.
Non-coding RNAs (ncRNAs) have become a research focus and have newly been demonstrated to play a significant role in multiple cellular physiological and pathological processes.6, 7, 8 These ncRNAs contain small ncRNAs (<200 nt long), such as microRNAs (miRNAs), and long ncRNAs (lncRNAs) (>200 nt long), as well as circular RNAs (circRNAs) with covalently closed loop structures.9, 10 Emerging evidence indicates that levels of miRNAs have been remarkably changed in different types of AKI animal models as well as in human biopsy samples.11, 12, 13 Functional studies also proved that restoration or knockdown of certain miRNAs was able to regulate the progression of AKI.14, 15 lncRNAs also play an important part in various biological processes, such as cell death, mitochondrial apoptosis, T cell differentiation, nuclear factor κB (NF-κB) signaling pathway in the progression of septic AKI, or ischemia-reperfusion (IR) AKI.16, 17, 18 Although the functional role of circRNAs in kidney diseases has not been systematically studied, they are highly expressed in kidney and have been reported to modulate cell-cycle progression and programmed cell death.19 This line of evidence indicates that ncRNAs may function in the progression of CI-AKI.
In most cases, miRNAs bind to the 3′ UTRs of their target gene to regulate expression and inhibit protein translation.20 Both lncRNAs and circRNAs can inhibit the function of miRNAs as miRNA sponges through the competing endogenous RNA (ceRNA) network.21, 22 The ncRNA-associated ceRNA networks may play a key role in inflammation, pyroptosis, apoptosis, or other biological activities. For example, the lncRNA MALAT1 directly repressed ELAVL1 and then decreased downstream NLRP3 expression via targeting miR-23c, thereby inhibiting cell pyroptosis and inflammation in diabetic nephropathy.23 It is of note that hsa_circ_0010729 inhibits apoptosis and promotes the cell proliferation and migration through targeting the miR-186/HIF-1α axis.24 This provides new evidence that ceRNA may underlie the mechanism of CI-AKI.
The potential link between CI-AKI and ncRNA-associated ceRNA networks shed new light on the research of CI-AKI. To date, only one study proved that circulating miRNA-188, -30a, and -30e were potential early biomarkers for CI-AKI.25 The potential role of ncRNAs and ncRNA-associated ceRNA networks in the pathogenesis of CI-AKI has not been fully understood. In this study, we developed whole-transcriptome sequencing (RNA sequencing [RNA-seq]) with the Illumina HiSeq 2000 and HiSeq 4000 in a control group and a new CI-AKI rat model26 to systematically identify differentially expressed lncRNAs, circRNAs, miRNAs, and mRNAs. This study is the first one to identify circRNA- or lncRNA-associated ceRNA networks in the CI-AKI Sprague-Dawley (SD) rat model and may provide new insight for diagnosis and therapeutic targets in CI-AKI.
Results
Evaluation of the CI-AKI SD Rat Model
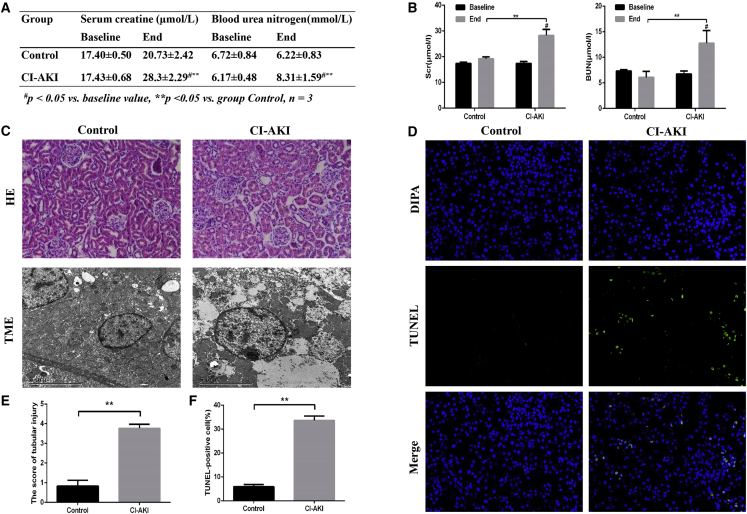
We used the change of serum creatine (SCr) to evaluate the renal function of SD rats. Twelve SD rats were divided into a CI-AKI group and a control group, and both were dehydrated for 48 h and then intramuscularly injected with furosemide (10 mL/kg) 30 min before iohexol (15 mL/kg) or 0.9% normal saline (15 mL/kg) intravenous injection. As shown in Figures 1A and 1B, SCr and blood urea nitrogen (BUN) were significantly increased after CM administration (p < 0.05), while no change was observed in the control group. Kidney histopathological changes were examined by H&E staining and transmission electron microscope examination (TME) (Figure 1C). H&E staining showed a small detachment of tubular cells in the control group (histologic scoring: 0.83 ± 0.29), while severe foamy degeneration and detachment of tubular cells occurred in the CI-AKI group (histologic scoring: 3.77 ± 0.21) (Figure 1E). Transmission electron microscope examination showed that normal mitochondria and few lysosomes and autophagosomes were presented in the control group. However, in the tubular cells of CI-AKI rats, karyopyknosis and chromatin set in edge with a marked accumulation of autophagosomes were observed, and more severe mitochondria ultrastructural changes were examined, including swelling, loss of cristae, and vacuolization in matrix. In the CI-AKI rats, apoptotic cells (percentage of TUNEL-positive cells: 33.64 ± 1.82) were significantly increased compared to those in the control group (percentage of TUNEL-positive cells: 5.91 ± 0.92; p < 0.05) (Figures 1D and 1F).
Figure 1.
Evaluation in the CI-AKI SD Rat Models
(A and B) Changes in the levels of Scr and BUN before dehydration and 24 h after saline or iohexol injection in the control and CI-AKI groups to evaluate renal function of SD rats. (C) Representative photomicrographs of tubular cell injuries in rat kidney tissues (H&E staining; original magnifications, ×200) and representative photomicrographs of mitochondrial ultrastructural changes in renal tubular epithelial cells (original magnifications, ×5000). (D) Immunofluorescent labeling for TUNEL in rat kidney tissue sections of the four groups (×200). Apoptotic cells in rat kidney tissue were stained for TUNEL (green). Nuclei were stained with DAPI (blue). (E) Quantitative analysis of histologic scoring. (F) Quantitative analysis of TUNEL-positive cells. More than 200 cells in each group were evaluated to determine the percentage of TUNEL-positive cells. The data are presented as the mean ± SEM; #p < 0.05 versus baseline value; **p < 0.05 versus group control.
Overview of circRNA-Seq and miRNA-Seq
After the removal of low-quality reads, as well as adapter-, poly-N-, and other contaminant-containing reads from the raw data, clean reads of circRNA- and miRNA sequencing (miRNA-seq) were obtained. We filtered reads based on length (21–22 nt) to select miRNAs, which were mapped to the rat reference sequence by Bowtie,27 and the miREvo28 and miRDeep229 software packages were integrated to predict the novel miRNAs. Finally, 746 matured miRNAs (628 known and 118 novel) were detected. Based on the theory of find_circ software,8 6,469 circRNAs were detected. These miRNAs and circRNAs were used for the subsequent analysis.
Overview of lncRNA-Seq and mRNA-Seq
After discarding the reads with adapters, poly-N > 10%, and any other possible contaminants, clean reads of lncRNAs were obtained. After mapped to the rat reference genome by using Bowtie and TopHat v2.0.9, a total of 1,789 matured lncRNAs were detected using five steps (step 1: number screening; step 2: length screening; step 3: annotation screening; step 4: expression level screening; and step 5: encoding potential screening) for systematic identification; these lncRNAs were used for the subsequent analysis. The cufflink results indicated that 25,701 protein coding transcripts were identified. These mRNAs were used for the subsequent analysis.
Differential Expression Analysis: Control versus CI-AKI Model
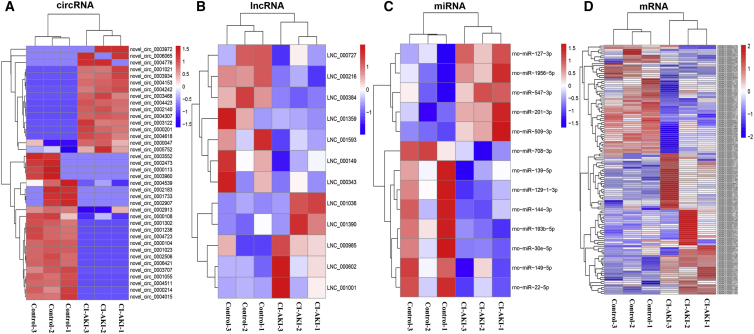
Differential expression ncRNAs and mRNAs in control rats and CI-AKI rats (n = 3) were indicated in a heatmap (Figure 2). We first identified 38 significantly dysregulated circRNA transcripts between the two groups based on transcripts per million (TPM) (p < 0.05; Table S1), with 16 upregulated and 22 downregulated transcripts in CI-AKI rats relative to those in control rats. A total of 13 significantly dysregulated miRNAs were also detected between the two groups based on TPM (p < 0.05; Table S2), with 5 upregulated in the CI-AKI model rat and 8 downregulated. The expression levels of the lncRNA and mRNA transcripts were estimated by fragments per kilobase of exons per million fragments mapped (FPKM). A total of 12 significantly dysregulated lncRNAs were identified, with 5 upregulated and 7 downregulated in the CI-AKI model rat (q < 0.05; p value was adjusted using q value30; Table S3). A total of 127 significantly dysregulated mRNA transcripts were identified, with 72 upregulated and 55 downregulated in the CI-AKI model rat (q < 0.05; Table S4).
Figure 2.
Expression Profiles of lncRNAs, circRNAs, miRNAs, and mRNAs
(A) Cluster analysis of differentially expressed circRNAs. (B) Cluster analysis of differentially expressed lncRNAs. (C) Cluster analysis of differentially expressed miRNAs. (D) Cluster analysis of differentially expressed mRNAs. Red indicates increased expression, and blue denotes decreased expression. The expression levels of miRNAs and circRNAs were quantified by TPMs, p < 0.05 was considered significantly different; the expression levels of lncRNAs and miRNAs were quantified by FPKMs, q < 0.05 was considered significantly different.
As displayed in Table 1, the most upregulated circRNA was a novel_circ_0004307 with 5.95 log2 fold change, and the most downregulated circRNA was a novel_circ_0002506 with −6.31-log2 fold change. The most upregulated lncRNA, miRNA, and mRNA were lnc_001001 (5.29 log2 fold change), rno-miR-201-3p (0.87 log2 fold change), and glycoprotein non-metastatic melanoma protein b (Gpnmb) (2.67 log2 fold change), respectively. The most downregulated lncRNA, miRNA, and mRNA were lnc_000384 (−3.03 log2 fold change), rno-miR-144-3p (−1.00 log2 fold change), and cyp27b1(−2.71 log2 fold change), respectively.
Table 1.
Statistical Analysis of All Differentially Expressed ncRNAs and mRNAs
| Differential Expression RNAs | Total No. | No. Upregulated | No. Downregulated | Most Upregulated (Log2 Fold Change) | Most Downregulated (Log2 Fold Change) |
|---|---|---|---|---|---|
| circRNA | 38 | 16 | 22 | novel_circ_0004307 (5.95) | novel_circ_0002506 (−6.31) |
| lncRNA | 12 | 5 | 7 | LNC_001001 (5.29) | LNC_000384 (−3.03) |
| miRNA | 13 | 5 | 8 | rno-miR-201-3p (0.87) | rno-miR-144-3p (−1.00) |
| mRNA | 127 | 72 | 55 | Gpnmb (2.67) | Cyp27b1 (−2.71) |
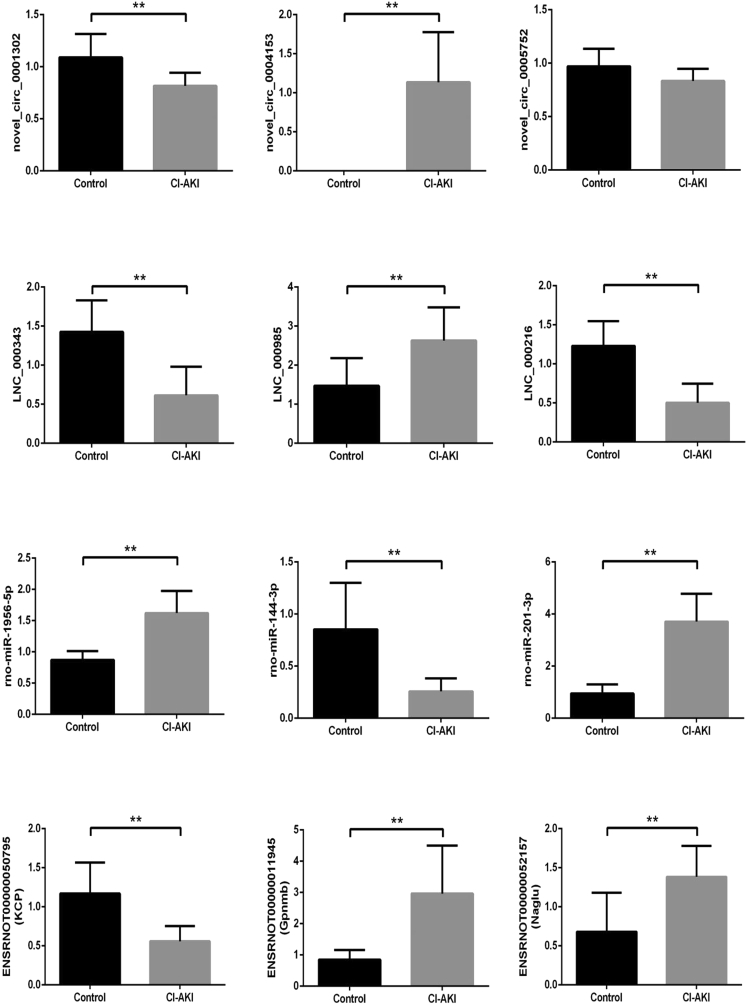
qRT-PCR Confirmation
qRT-PCR was used to confirm the differential expression identified in the RNA-seq data. We randomly selected 12 differentially expressed transcripts: three circRNAs, three lncRNAs, three miRNAs, and three mRNAs in an independent cohort of 5 CI-AKI model rats and 5 control rats. As shown in Figure 3, most selected transcripts were detected in control and CI-AKI model rat kidneys and exhibited significant differential expressions and relatively high verification rates. Only circRNA_005752 expressed opposite between microarray and qRT-PCR, but the difference was not statistically significant. In summary, qRT-PCR results confirmed the RNA-seq data.
Figure 3.
Validation of Transcript Expression by qRT-PCR between CI-AKI and Control Rats
Rat ACTB and U6 genes were used as housekeeping internal controls. Transcript expression was quantified relative to the expression level of ACTB using the comparative cycle threshold (ΔCt) method. The data are presented as the mean ± SEM (n = 5); **p < 0.05.
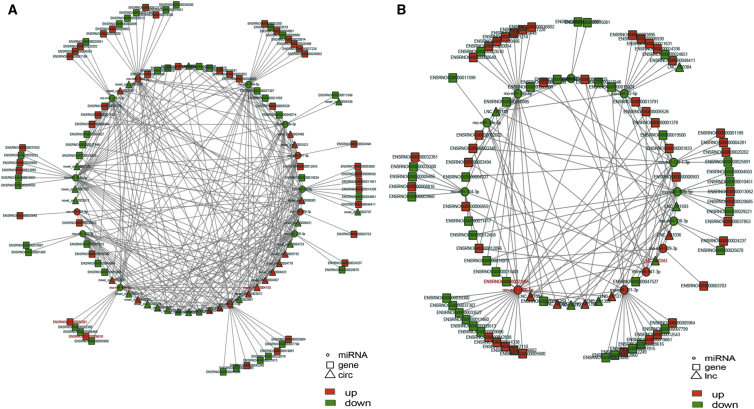
Construction of a circRNA- or lncRNA-Associated ceRNA Network
According to ceRNA hypothesis, the members of ceRNA (circRNAs, lncRNAs, and mRNAs) compete for the same miRNA response elements (MREs) to regulate each other. In this study, we found that the 38 circRNAs, 12 lncRNAs and 127 mRNAs with differential expression shared a common binding site of miRNA (13 remarkably dysregulated miRNAs). Two cases of the ceRNA network were covered (Figures 4A and 4B): one was circRNA or lncRNA (up in CI-AKI rats)-miRNA (down in CI-AKI rats)-mRNA (up in CI-AKI rats), and the other was circRNA or lncRNA (down in CI-AKI rats)-miRNA (up in CI-AKI rats)-mRNA (down in CI-AKI rats). These RNA interactions may serve as a novel perspective for exploring the underlying mechanism of CI-AKI. More details are listed in Tables S5 and S6.
Figure 4.
circRNA-Associated ceRNA Networks in CI-AKI Rats and lncRNA-Associated ceRNA Networks in CI-AKI Rats
(A) circRNA-associated ceRNA Networks in CI-AKI rats. (B) lncRNA-associated ceRNA networks in CI-AKI rats. The square nodes represent mRNAs, the triangle nodes indicate lncRNAs or circRNAs, the circular frames denote miRNAs, and the edges represent the competing interactions among them. Red represents upregulated expression, and green indicates downregulated expression.
Functional Annotation: GO and KEGG
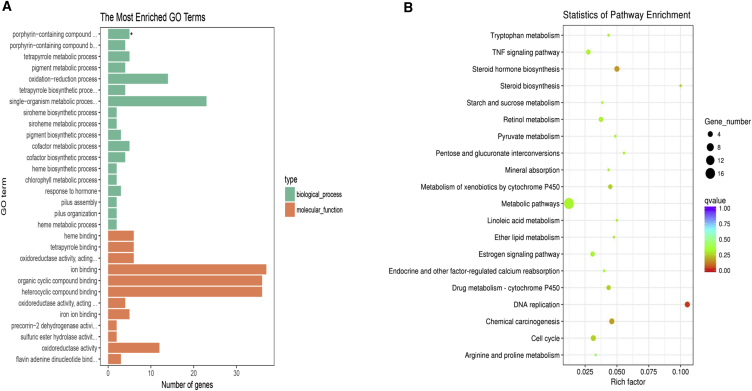
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed on the mRNAs involved in circRNA or lncRNA-associated ceRNA networks, respectively. As shown in Figure 5A, GO analysis showed 142 GO terms were markedly enriched (p < 0.05; Table S7), and the top three terms were porphyrin-containing compound metabolic process (GO: 0006778), porphyrin-containing compound biosynthetic process (GO: 0006779), and heme binding (GO: 0020037). Several kidney-injury-associated terms were also discovered, such as oxidation-reduction process (GO: 0055114) and oxidoreductase activity (GO: 0016491). KEGG pathway analysis showed that there were 17 pathways that were significantly enriched in these differential expression mRNAs involved in the circRNA or lncRNA-miRNA-mRNA networks (p < 0.05, Table S8). Of them, DNA replication, steroid hormone biosynthesis, and chemical carcinogenesis were the most enriched (Figure 5B). In short, the circRNA- or lncRNA-associated ceRNA network may participate in the physiological and pathological processes of CI-AKI from different aspects.
Figure 5.
Bioinformatics Analysis in the ceRNA Network
(A and B) Data show bioinformatics analysis in the ceRNA network with GO (A) and KEGG (B) pathway analysis for the mRNAs regulated by the ncRNA-miRNA network.
Association Study
In this study, three limiting factors were applied to select circRNA- or lncRNA-associated ceRNA networks in CI-AKI. First, the selected pairs composed of ncRNAs and mRNAs should all be differentially and significantly expressed between the control and the CI-AKI rats (p or q < 0.05). Second, the levels of the selected ncRNAs and their target genes (mRNAs) in the rat kidney should be in a certain order of down (circRNAs or lncRNAs)-up (miRNAs)-down (mRNAs) or up (circRNAs or lncRNAs)-down (miRNAs)-up (mRNAs). Third, the selected pairs should be related to CI-AKI. For example, novel_circ_0004153, novel_circ_0004776, novel_circ_0006065, novel_circ_0004307, novel_circ_0003972, novel_circ_0000201, novel_circ_0000047, and novel_circ_0002140 (up in CI-AKI rats) were ceRNAs of rno-miR-144-3p (down in CI-AKI rats) targeting Naglu or Gpnmb (up in CI-AKI rats). lncRNA-000343, lncRNA-0001593, lncRNA-000727, lncRNA-0000149, and lncRNA-0001359 (down in CI-AKI rats) were ceRNAs of rno-miR-1956-3p (up in CI-AKI rats) targeting Kielin/chordin-like Protein (KCP) (down in CI-AKI rats). Therefore, we believe that the circRNA- or lncRNA-associated ceRNA networks mentioned earlier may take part in the pathogenesis and pathological process of CI-AKI. Additional results are listed in Table 2.
Table 2.
ncRNA-Associated ceRNA Networks Most Likely Involved in CI-AKI Pathogenesis
| circRNA or lncRNA | Log2 Fold Change | p Value | miRNA | Log2 Fold Change | p Value | Transcript ID | Gene ID | Gene Name | Log2 Fold Change | p Value |
|---|---|---|---|---|---|---|---|---|---|---|
| novel_circ_0004153 | 3.95 | 0.01934 | rno-miR-144-3p | −1.00 | 0.00115 | ENSRNOT00000011945 | ENSRNOG00000008816 | Gpnmb | 2.67 | 0.00005 |
| novel_circ_0004776 | 4.19 | 0.01249 | ||||||||
| novel_circ_0006065 | 4.21 | 0.01207 | ||||||||
| novel_circ_0004307 | 5.95 | 0.00006 | ||||||||
| novel_circ_0003972 | 4.09 | 0.01516 | ENSRNOT00000052157 | ENSRNOG00000032381 | Naglu | 0.64 | 0.00045 | |||
| novel_circ_0000201 | 3.77 | 0.02733 | ||||||||
| novel_circ_0000047 | 2.74 | 0.04435 | ||||||||
| novel_circ_0002140 | 3.49 | 0.04434 | ||||||||
| LNC_000343 | −2.49 | 0.00005 | rno-miR-1956-5p | 0.67 | 0.031072 | ENSRNOT00000050795 | ENSRNOG00000021855 | KCP | −1.57 | 0.00005 |
| LNC_001593 | −1.17 | 0.00005 | ||||||||
| LNC_000727 | −1.84 | 0.00005 | ||||||||
| LNC_000149 | −1.14 | 0.00005 | ||||||||
| LNC_001359 | −2.98 | 0.00020 |
Discussion
CI-AKI is a severe complication of intravascular applied radial CM. Intravenous hydration and use of low-or iso-osmolar iodine CM are recommended as prevention strategies for CI-AKI.31 However, recent progress in interventional therapy and angiography has revived interest in explaining detailed mechanisms and developing effective treatment, especially in patients suffering from hemodynamic instability, pre-existing decreased kidney function, diabetes, and so on.32 Many studies on CI-AKI focused on early biomarkers, the potential targets for therapy and prediction of early and late outcomes. Dr. Shi-qun Sun’s group proved that miRNAs were early potential biomarkers for CI-AKI, as plasma levels of miRNA-188, -30a, and -30e significantly distinguished patients with CI-AKI from those without CI-AKI.25 However, the role of circRNAs or lncRNAs in CI-AKI remains unknown.
Noncoding RNAs, a group of biomolecules that function at the RNA level, are involved in a wide range of physiological and pathological processes, including AKI. Function studies showed that miRNAs, such as miR-24, miR-126, miR-494, and miR-687, may regulate inflammation, programmed cell death, and cell cycle in the injury and repair stages of AKI by binding to target genes, which indicates their therapeutic potential.33 To date, the biological role of lncRNAs has become a hotspot in the research of AKI. Recent studies show that LINC00520 regulates the oncostatin M receptor (OSMR) expression level by targeting miR-27b-3 to promote renal IR injury development through the phosphatidylinositol 3-kinase/AKT signaling pathway.34 NEAT1 may serve as a vital therapeutic target and diagnostic marker in sepsis-induced AKI patients, for upregulation of NEAT1 activates the NF-κB pathway through regulating miR-204.35 In addition, the lncRNA MALAT1 and MEG3 in lipopolysaccharide-induced AKI are mediated by the miR-146a/NF-κB signaling pathway and miR-21/PDCD4 axis, respectively.35, 36 More importantly, circulating lncRNA transcript predicting survival in AKI (TapSAKI) can predict mortality in critically ill patients with AKI.37 In contrast, the functions of circRNAs in AKI are still unknown.38 However, their role in regulating programmed cell death39 and cell proliferation40 promises them as potential regulators in AKI. For example, mmu-circRNA-015947 was reported to have interacted with miRNAs (mmu-miR-188-3p, mmu-miR-329-5p, mmu-miR-3057-3p, mmu-miR-5098, and mmu-miR-683) to regulate downstream gene expression, thereby participating in apoptosis-correlated pathways involved in the pathogenesis of IR injury.41
Salmena et al.22 first proposed the “ceRNA hypothesis” describing lncRNAs or circRNAs could inhibit the function of miRNA to positively regulate targeted mRNA, functioning as miRNA sponges through the ceRNA networks. In the present study, we used RNA-seq to comprehensively analyze circRNA, lncRNA, miRNA, and mRNA profiles and ncRNA-associated ceRNA networks in the kidney of CI-AKI SD rats. The new CI-AKI rat model can well mimic CI-AKI in clinical practice. RNA-seq results showed that a total of 127 significantly dysregulated mRNA transcripts were identified. These mRNA transcripts are related to CI-AKI pathogenesis. For example, overexpression of stanniocalcin-1 inhibits reactive oxygen species (ROS) and renal IR injury via an AMP-activated protein kinase-dependent pathway.42, 43 Metallothionein 2A protects cells from toxic agents such as heavy metal ions or ROS and is a risk factor for chronic kidney disease and diabetes.44 According to the statistical data of RNA-seq results, 38 circRNA transcripts, 12 lncRNAs, and 13 miRNAs were significantly dysregulated in the CI-AKI rat model. These circRNAs, lncRNAs, and protein-coding mRNAs may function as ceRNAs to compete for the same MREs to regulate the expression of mRNA.
GO and KEGG pathway analyses were constructed to the target genes (mRNAs) involved in circRNA- or lncRNA-associated ceRNA networks. GO analyses revealed that the pathological process of CI-AKI may be regulated by these networks from different angles, including porphyrin-containing compound metabolic (GO: 0006778) and biosynthetic (GO: 0006779) processes, heme binding (GO: 0020037), and so on. KEGG pathway analysis showed that these dysregulated molecules mainly related to DNA replication, steroid hormone biosynthesis, and chemical carcinogenesis. For instance, flavin-containing monooxigenases (FMOs) (GO: 0006778; GO: 0006779) are a polymorphic family of drug- and pesticide-metabolizing enzymes. Rodriguez-Fuentes et al.45 reported that the expression and activities of FMOs may be influenced by hyperosmotic conditions in the kidney of rats. It has also been proved that renal collectrin (GO: 0006778; GO: 0006779) protects against salt-sensitive hypertension and is downregulated by angiotensin II. Increasing the abundance or activity of collectrin may have therapeutic benefits in the treatment of hypertension and salt sensitivity.46 In addition, phospholipase A2 group VII (Pla2g7) (GO: 0006778), the only known phospholipases that specifically select these oxidized and/or short-chained phospholipids as substrates, were purified and cloned as platelet-activating factor acetylhydrolases. Foulks et al.47 proved that selective hydrolysis of phospholipid oxidation products was an early and critical way to overcome oxidative membrane damage and oxidant-induced cell death. Renal oxidative stress level characterized by elevated heme oxygenase 1 (HO-1), cytochrome P450 E1 (CYP2E1) (GO: 0020037, steroid hormone biosynthesis, chemical carcinogenesis) in cisplatin-induced renal dysfunction,48 and inhibition of CYP2E1 coupled with the induction of Nrf2 can effectively reduce CYP2E1-mediated rhabdomyolysis-induced acute kidney injury.49 What is more, previous studies have shown that UDP-glucuronosyltransferases (UGTs) catalyze phase II biotransformation reactions in which lipophilic substrates are conjugated with glucuronic acid to increase water solubility and enhance excretion. Ugt2b35 (steroid hormone biosynthesis and chemical carcinogenesis) was expressed in kidney, potentially influencing drug metabolism and pharmacokinetics.50 Although the correlations between the aforementioned genes and CI-AKI have not yet been reported, to our knowledge, we speculate that development of CI-AKI in this rat model may be related to hyperosmotic conditions, angiotensin, oxidative stress, drug metabolism, and pharmacokinetics. Further research is necessary.
In this study, strict constraints were applied to select the most possible circRNA- or lncRNA-associated ceRNA networks that are involved in the pathogenesis and development of CI-AKI. For example, we selected circRNA-associated ceRNA networks that participated in the control of the Naglu gene or Gpmnb gene. Novel_circ_0004153 circRNAs were ceRNAs of rno-miR-144-3p targeting Naglu or Gpnmb. The Naglu gene is an indicator of renal tubulointerstitial damage and associated with inflammation, such as nuclear factor kB (NF-kB) activation.51 It can be used to differentiate AKI types and predict AKI development, especially prerenal AKI.52 Gpnmb serves as a negative regulator of inflammation in macrophages and plays a protective role against the AKI.53 Another one is the lncRNA-associated ceRNA network that participates in regulation of the KCP gene. lncRNA_000343 were ceRNAs of rno-miR-1956-5p targeting KCP. KCP transgenic mice were significantly more resistant to interstitial fibrosis and renal injury via inhibiting both transforming growth factor β (TGF-β) and activin signals while enhancing bone morphogenetic protein signaling, which can attenuate both acute and chronic renal injury.54, 55, 56 The circRNA- or lncRNA-associated ceRNA networks in CI-AKI are highly complex and diverse. Our ongoing efforts will provide basic information to understand these ncRNA-associated ceRNA networks. There exists an enormous challenge to understand this regulatory mechanism in CI-AKI, and further exploration is needed.
We elucidated the circRNA- or lncRNA-associated ceRNA profiles of control and CI-AKI rats using deep RNA-seq analysis. To the best of our knowledge, this is the first report examining the expression of lncRNAs, circRNAs, miRNAs, and mRNAs in CI-AKI. These findings further expanded our understanding of ceRNA networks and help us to explore their regulatory functions in the pathogenesis and pathological process of CI-AKI. These novel networks may be potential biomarkers or therapeutic targets in CI-AKI. Taken together, this strategy provides new insight for the underlying mechanism and may profoundly affect the diagnosis and therapy of CI-AKI.
Materials and Methods
Preparation of Tissues
Male SD rats were acclimated to the environment for 7 days before starting the study, weighing approximately 250–300 g. All the operations were handled in accordance with the guidelines on animal care of the Second Xiangya Hospital of Central South University. Twelve SD rats were enrolled and randomly assigned to the CI-AKI group or control group (n = 6). Animals were dehydrated for 48 h and then underwent furosemide (10 mL/kg; Harvest Pharmaceutical, Shanghai, China) injection 30 min before administration of iohexol (15 mL/kg, group CI-AKI; iodine, 350 mg/mL; GE Healthcare, Shanghai, China) or 0.9% normal saline (15 mL/kg, control group). We randomly selected three rats from each group for the following tests.
Blood Biomarker Assay
SCr and BUN levels were measured with an automatic biochemical analyzer (Hitachi 7170A, Tokyo, Japan).
H&E Staining
Renal tissue was fixed in 10% neutral-buffered formalin for a minimum of 24 h and embedded in paraffin; then, 4-μm-thick tissue sections were cut using a microtome; and, finally, histopathological evaluation was conducted with H&E staining. For semiquantitative analysis of the frequency and severity of renal lesions, we randomly selected 10 high-magnification (×200) cortical regions and the outer stripe of the outer medulla. The specimens were scored according to a semiquantitative scale based on the extent of foamy degeneration and detachment of tubular cells:57 no injury (0); mild: <25% (1); moderate: <50% (2); severe: <75% (3); and very severe: >75% (4).
Transmission Electron Microscope Examination
2.5% glutaraldehyde-fixed kidney samples were dehydrated, osmosed, embedded, ultrathin sectioned, and stained in uranyl acetate and lead citrate conventionally. The ultrastructure of the tubular cells was observed under a Hitachi H7700 electron microscope.
dUTP Nick-End Labeling Assay
According to the manufacturer’s protocol, a commercial kit (in situ cell death assay kit; Roche, Basel, Switzerland) for terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay was used on paraffin sections of the renal cortical medulla border region. All cells were counted in five different views per section. TUNEL-positive cells were expressed as percentage of total cells.
RNA Extraction and Qualification
Total RNA from each renal tissue sample was isolated using TRIzol Reagent (Invitrogen). Briefly, RNA degradation and contamination were monitored with 1% agarose gels. RNA purity was checked using the NanoPhotometer spectrophotometer (IMPLEN, Los Angeles, CA, USA). RNA concentration and integrity were measured using the Qubit RNA Assay Kit with the Qubit 2.0 Flurometer (Life Technologies, CA, USA) and the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 System (Agilent Technologies, CA, USA), respectively.
RNA-Seq
Strand-specific cDNA libraries were constructed and sequenced on an Illumina HiSeq 2000/4000 platform (Novogene Bioinformatic Technology, Beijing, China). The methods were previously described, and more details are described in Methods S1 and S2. All subsequent analyses were performed using clean reads after removing the adaptor reads and low-quality tags.
Expression Analysis
FPKMs of both lncRNAs and coding genes in each sample were calculated by Cuffdiff (v2.1.1).58 Gene FPKMs were the sum of the FPKMs of transcripts in each gene group. FPKMs were calculated based on the length of the fragments and read counts mapped to this fragment. The expression levels of miRNAs and circRNAs were quantified by TPM using the following normalization formula:59 normalized expression = mapped read count/total reads ⋅ 1,000,000.
Differential Expression Analysis
For the samples with biological replicates, differential expression analysis of the control and CI-AKI model groups was performed using the DESeq R package (1.8.3). We adjusted the p value as a q value using the Benjamini & Hochberg method.30 A p or q value < 0.05 was considered significantly different.
ceRNA Network Analysis
We analyzed the significant difference expression levels of ncRNAs and mRNAs between the control group and the CI-AKI model. The sequences of circRNAs, lncRNAs, and mRNAs were screened to search the potential MREs. MiRanda (http://www.microrna.org/microrna/home.do) was used to predict miRNA-binding seed sequence sites and target genes. Protein-protein interaction analysis of differentially expressed genes was based on the STRING database using Cytoscape 3.4.0.
GO and KEGG Enrichment Analyses
The circRNA or lncRNA-miRNA-enriched genes were analyzed by using the GO database and KEGG database. The GO categories (http://geneontology.org) cover defined terms that describe gene product attributes. KEGG60 is a database resource applied to understand high-level functions and utilities of the biological system (https://www.genome.jp/kegg/). We used KOBAS61 software to test the statistical enrichment of the target gene candidates in KEGG pathways.
Real-Time qPCR Validation
Total RNA was isolated from kidney tissues using TRIzol Reagent (Invitrogen), and RNA (1 μg) was reverse transcribed to cDNA using the Reverse Transcriptase M-MLV (Takara, Japan) according to manufacturer’s instructions. The real-time qPCR reaction was performed using the SYBR Green assay (Takara, Japan) in the CFX96 Real-Time PCR Detection System (Bio-Rad) with the following conditions: 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. The specific quantitative primers were designed and synthesized by Sangon Biotech (Sangon Biotech, Shanghai, China) and listed in Table S9. The primers of ACTB (for circRNA, lncRNA, and mRNA) and U6 (for miRNA) were synthesized as endogenous controls.
Statistical Analysis
All statistical analysis was conducted by using SPSS 22.0. The data of normal distribution were presented with mean values ± SD. The mean comparison between the two groups was conducted with Student’s t test. The enumeration data were expressed by rate and analyzed by chi-square test. The expression level of each mRNA, miRNA, lncRNA, and circRNA was represented as fold change using the 2−ΔΔCt method on real-time qPCR analysis. p values or q values < 0.05 were considered statistically significant.
Author Contributions
W.C. and X.-W.L. established the CI-AKI rat model and sample collection. W.C., X.-W.L., and Y.-Q.X. carried out the identification experiments. W.C. and S.-B.D. participated in the design of the study and performed the statistical analysis. W.C. drafted the manuscript, and S.-B.D. helped to correct it.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (nos. 81570618 and 81873607) and the National Science and Technology Support Program of China (no. 2013BAH05F02).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.05.011.
Supplemental Information
References
- 1.Kellum J.A., Lameire N., KDIGO AKI Guideline Work Group Diagnosis, evaluation, and management of acute kidney injury: a KDIGO summary (Part 1) Crit. Care. 2013;17:204. doi: 10.1186/cc11454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomsen H.S., Morcos S.K. Contrast media and the kidney: European Society of Urogenital Radiology (ESUR) guidelines. Br. J. Radiol. 2003;76:513–518. doi: 10.1259/bjr/26964464. [DOI] [PubMed] [Google Scholar]
- 3.Seeliger E., Sendeski M., Rihal C.S., Persson P.B. Contrast-induced kidney injury: mechanisms, risk factors, and prevention. Eur. Heart J. 2012;33:2007–2015. doi: 10.1093/eurheartj/ehr494. [DOI] [PubMed] [Google Scholar]
- 4.Azzalini L., Spagnoli V., Ly H.Q. Contrast-induced nephropathy: from pathophysiology to preventive strategies. Can. J. Cardiol. 2016;32:247–255. doi: 10.1016/j.cjca.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 5.Luo M., Yang Y., Xu J., Cheng W., Li X.W., Tang M.M., Liu H., Liu F.Y., Duan S.B. A new scoring model for the prediction of mortality in patients with acute kidney injury. Sci. Rep. 2017;7:7862. doi: 10.1038/s41598-017-08440-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dou C., Cao Z., Yang B., Ding N., Hou T., Luo F., Kang F., Li J., Yang X., Jiang H. Changing expression profiles of lncRNAs, mRNAs, circRNAs and miRNAs during osteoclastogenesis. Sci. Rep. 2016;6:21499. doi: 10.1038/srep21499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 8.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 9.Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hombach S., Kretz M. Non-coding RNAs: classification, biology and functioning. Adv. Exp. Med. Biol. 2016;937:3–17. doi: 10.1007/978-3-319-42059-2_1. [DOI] [PubMed] [Google Scholar]
- 11.Bhatt K., Kato M., Natarajan R. Mini-review: emerging roles of microRNAs in the pathophysiology of renal diseases. Am. J. Physiol. Renal Physiol. 2016;310:F109–F118. doi: 10.1152/ajprenal.00387.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lorenzen J.M. Vascular and circulating microRNAs in renal ischaemia-reperfusion injury. J. Physiol. 2015;593:1777–1784. doi: 10.1113/JP270318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilflingseder J., Sunzenauer J., Toronyi E., Heinzel A., Kainz A., Mayer B., Perco P., Telkes G., Langer R.M., Oberbauer R. Molecular pathogenesis of post-transplant acute kidney injury: assessment of whole-genome mRNA and miRNA profiles. PLoS ONE. 2014;9:e104164. doi: 10.1371/journal.pone.0104164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X.Y., Zhang K., Jiang Z.Y., Cai L.H. MiR-204/miR-211 downregulation contributes to candidemia-induced kidney injuries via derepression of Hmx1 expression. Life Sci. 2014;102:139–144. doi: 10.1016/j.lfs.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 15.Liu X.J., Hong Q., Wang Z., Yu Y.Y., Zou X., Xu L.H. MicroRNA-34a suppresses autophagy in tubular epithelial cells in acute kidney injury. Am. J. Nephrol. 2015;42:168–175. doi: 10.1159/000439185. [DOI] [PubMed] [Google Scholar]
- 16.Mimura I., Hirakawa Y., Kanki Y., Kushida N., Nakaki R., Suzuki Y., Tanaka T., Aburatani H., Nangaku M. Novel lnc RNA regulated by HIF-1 inhibits apoptotic cell death in the renal tubular epithelial cells under hypoxia. Physiol. Rep. 2017;5:e13203. doi: 10.14814/phy2.13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chun-Mei H., Qin-Min G., Shu-Ming P., Xiang-Yang Z. Expression profiling and ontology analysis of circulating long non-coding RNAs in septic acute kidney injury patients. Clin. Chem. Lab. Med. 2016;54:e395–e399. doi: 10.1515/cclm-2015-1281. [DOI] [PubMed] [Google Scholar]
- 18.Ding Y., Guo F., Zhu T., Li J., Gu D., Jiang W., Lu Y., Zhou D. Mechanism of long non-coding RNA MALAT1 in lipopolysaccharide-induced acute kidney injury is mediated by the miR-146a/NF-κB signaling pathway. Int. J. Mol. Med. 2018;41:446–454. doi: 10.3892/ijmm.2017.3232. [DOI] [PubMed] [Google Scholar]
- 19.Xu T., Wu J., Han P., Zhao Z., Song X. Circular RNA expression profiles and features in human tissues: a study using RNA-seq data. BMC Genomics. 2017;18(Suppl 6):680. doi: 10.1186/s12864-017-4029-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim I., Kwak H., Lee H.K., Hyun S., Jeong S. β-Catenin recognizes a specific RNA motif in the cyclooxygenase-2 mRNA 3′-UTR and interacts with HuR in colon cancer cells. Nucleic Acids Res. 2012;40:6863–6872. doi: 10.1093/nar/gks331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tay Y., Rinn J., Pandolfi P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X., Zeng L., Cao C., Lu C., Lian W., Han J., Zhang X., Zhang J., Tang T., Li M. Long noncoding RNA MALAT1 regulates renal tubular epithelial pyroptosis by modulated miR-23c targeting of ELAVL1 in diabetic nephropathy. Exp. Cell Res. 2017;350:327–335. doi: 10.1016/j.yexcr.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 24.Dang R.Y., Liu F.L., Li Y. Circular RNA hsa_circ_0010729 regulates vascular endothelial cell proliferation and apoptosis by targeting the miR-186/HIF-1α axis. Biochem. Biophys. Res. Commun. 2017;490:104–110. doi: 10.1016/j.bbrc.2017.05.164. [DOI] [PubMed] [Google Scholar]
- 25.Sun S.Q., Zhang T., Ding D., Zhang W.F., Wang X.L., Sun Z., Hu L.H., Qin S.Y., Shen L.H., He B. Circulating microRNA-188, -30a, and -30e as early biomarkers for contrast-induced acute kidney injury. J. Am. Heart Assoc. 2016;5:e004138. doi: 10.1161/JAHA.116.004138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng W., Zhao F., Tang C.Y., Li X.W., Luo M., Duan S.B. Comparison of iohexol and iodixanol induced nephrotoxicity, mitochondrial damage and mitophagy in a new contrast-induced acute kidney injury rat model. Arch. Toxicol. 2018;92:2245–2257. doi: 10.1007/s00204-018-2225-9. [DOI] [PubMed] [Google Scholar]
- 27.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wen M., Shen Y., Shi S., Tang T. miREvo: an integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinformatics. 2012;13:140. doi: 10.1186/1471-2105-13-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedländer M.R., Mackowiak S.D., Li N., Chen W., Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang N., Qian P., Yan T.D., Phan K. Periprocedural effects of statins on the incidence of contrast-induced acute kidney injury: A systematic review and trial sequential analysis. Int. J. Cardiol. 2016;206:143–152. doi: 10.1016/j.ijcard.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 32.Su X., Xie X., Liu L., Lv J., Song F., Perkovic V., Zhang H. Comparative effectiveness of 12 treatment strategies for preventing contrast-induced acute kidney injury: a systematic review and Bayesian network meta-analysis. Am. J. Kidney Dis. 2017;69:69–77. doi: 10.1053/j.ajkd.2016.07.033. [DOI] [PubMed] [Google Scholar]
- 33.Ren G.L., Zhu J., Li J., Meng X.M. Noncoding RNAs in acute kidney injury. J. Cell. Physiol. 2019;234:2266–2276. doi: 10.1002/jcp.27203. [DOI] [PubMed] [Google Scholar]
- 34.Tian X., Ji Y., Liang Y., Zhang J., Guan L., Wang C. LINC00520 targeting miR-27b-3p regulates OSMR expression level to promote acute kidney injury development through the PI3K/AKT signaling pathway. J. Cell Physiol. 2019;234:14221–14233. doi: 10.1002/jcp.28118. [DOI] [PubMed] [Google Scholar]
- 35.Chen Y., Qiu J., Chen B., Lin Y., Chen Y., Xie G., Qiu J., Tong H., Jiang D. Long non-coding RNA NEAT1 plays an important role in sepsis-induced acute kidney injury by targeting miR-204 and modulating the NF-κB pathway. Int. Immunopharmacol. 2018;59:252–260. doi: 10.1016/j.intimp.2018.03.023. [DOI] [PubMed] [Google Scholar]
- 36.Yang R., Liu S., Wen J., Xue L., Zhang Y., Yan D., Wang G., Liu Z. Inhibition of maternally expressed gene 3 attenuated lipopolysaccharide-induced apoptosis through sponging miR-21 in renal tubular epithelial cells. J. Cell. Biochem. 2018;119:7800–7806. doi: 10.1002/jcb.27163. [DOI] [PubMed] [Google Scholar]
- 37.Lorenzen J.M., Schauerte C., Kielstein J.T., Hubner A., Martino F., Fiedler J., Gupta S.K., Faulhaber-Walter R., Kumarswamy R., Hafer C. Circulating long noncoding RNATapSaki is a predictor of mortality in critically ill patients with acute kidney injury. Clin. Chem. 2015;61:191–201. doi: 10.1373/clinchem.2014.230359. [DOI] [PubMed] [Google Scholar]
- 38.Cheng X., Joe B. Circular RNAs in rat models of cardiovascular and renal diseases. Physiol. Genomics. 2017;49:484–490. doi: 10.1152/physiolgenomics.00064.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nan A., Chen L., Zhang N., Liu Z., Yang T., Wang Z., Yang C., Jiang Y. A novel regulatory network among LncRpa, CircRar1, MiR-671 and apoptotic genes promotes lead-induced neuronal cell apoptosis. Arch. Toxicol. 2017;91:1671–1684. doi: 10.1007/s00204-016-1837-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shan K., Liu C., Liu B.H., Chen X., Dong R., Liu X., Zhang Y.Y., Liu B., Zhang S.J., Wang J.J. Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation. 2017;136:1629–1642. doi: 10.1161/CIRCULATIONAHA.117.029004. [DOI] [PubMed] [Google Scholar]
- 41.Lin S.P., Ye S., Long Y., Fan Y., Mao H.F., Chen M.T., Ma Q.J. Circular RNA expression alterations are involved in OGD/R-induced neuron injury. Biochem. Biophys. Res. Commun. 2016;471:52–56. doi: 10.1016/j.bbrc.2016.01.183. [DOI] [PubMed] [Google Scholar]
- 42.Huang L., Belousova T., Chen M., DiMattia G., Liu D., Sheikh-Hamad D. Overexpression of stanniocalcin-1 inhibits reactive oxygen species and renal ischemia/reperfusion injury in mice. Kidney Int. 2012;82:867–877. doi: 10.1038/ki.2012.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pan J.S., Huang L., Belousova T., Lu L., Yang Y., Reddel R., Chang A., Ju H., DiMattia G., Tong Q., Sheikh-Hamad D. Stanniocalcin-1 inhibits renal ischemia/reperfusion injury via an AMP-activated protein kinase-dependent pathway. J. Am. Soc. Nephrol. 2015;26:364–378. doi: 10.1681/ASN.2013070703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lenhard D.C., Frisk A.L., Lengsfeld P., Pietsch H., Jost G. The effect of iodinated contrast agent properties on renal kinetics and oxygenation. Invest. Radiol. 2013;48:175–182. doi: 10.1097/RLI.0b013e31827b70f9. [DOI] [PubMed] [Google Scholar]
- 45.Rodríguez-Fuentes G., Coburn C., Currás-Collazo M., Guillén G., Schlenk D. Effect of hyperosmotic conditions on flavin-containing monooxygenase activity, protein and mRNA expression in rat kidney. Toxicol. Lett. 2009;187:115–118. doi: 10.1016/j.toxlet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chu P.L., Gigliotti J.C., Cechova S., Bodonyi-Kovacs G., Chan F., Ralph D.L., Howell N., Kalantari K., Klibanov A.L., Carey R.M. Renal collectrin protects against salt-sensitive hypertension and is downregulated by angiotensin II. J. Am. Soc. Nephrol. 2017;28:1826–1837. doi: 10.1681/ASN.2016060675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Foulks J.M., Weyrich A.S., Zimmerman G.A., McIntyre T.M. A yeast PAF acetylhydrolase ortholog suppresses oxidative death. Free Radic. Biol. Med. 2008;45:434–442. doi: 10.1016/j.freeradbiomed.2008.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y.Z., Ren S., Yan X.T., Li H.P., Li W., Zheng B., Wang Z., Liu Y.Y. Improvement of cisplatin-induced renal dysfunction by Schisandra chinensis stems via anti-inflammation and anti-apoptosis effects. J. Ethnopharmacol. 2018;217:228–237. doi: 10.1016/j.jep.2018.01.033. [DOI] [PubMed] [Google Scholar]
- 49.Wang Z., Shah S.V., Liu H., Baliga R. Inhibition of cytochrome P450 2E1 and activation of transcription factor Nrf2 are renoprotective in myoglobinuric acute kidney injury. Kidney Int. 2014;86:338–349. doi: 10.1038/ki.2014.65. [DOI] [PubMed] [Google Scholar]
- 50.Buckley D.B., Klaassen C.D. Tissue- and gender-specific mRNA expression of UDP-glucuronosyltransferases (UGTs) in mice. Drug Metab. Dispos. 2007;35:121–127. doi: 10.1124/dmd.106.012070. [DOI] [PubMed] [Google Scholar]
- 51.Mise K., Hoshino J., Ueno T., Hazue R., Hasegawa J., Sekine A., Sumida K., Hiramatsu R., Hasegawa E., Yamanouchi M. Prognostic value of tubulointerstitial lesions, urinary N-acetyl-β-d-glucosaminidase, and urinary β2-microglobulin in patients with type 2 diabetes and biopsy-proven diabetic nephropathy. Clin. J. Am. Soc. Nephrol. 2016;11:593–601. doi: 10.2215/CJN.04980515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Doi K., Katagiri D., Negishi K., Hasegawa S., Hamasaki Y., Fujita T., Matsubara T., Ishii T., Yahagi N., Sugaya T., Noiri E. Mild elevation of urinary biomarkers in prerenal acute kidney injury. Kidney Int. 2012;82:1114–1120. doi: 10.1038/ki.2012.266. [DOI] [PubMed] [Google Scholar]
- 53.Zhou L., Zhuo H., Ouyang H., Liu Y., Yuan F., Sun L., Liu F., Liu H. Glycoprotein non-metastatic melanoma protein b (Gpnmb) is highly expressed in macrophages of acute injured kidney and promotes M2 macrophages polarization. Cell. Immunol. 2017;316:53–60. doi: 10.1016/j.cellimm.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 54.Soofi A., Wolf K.I., Emont M.P., Qi N., Martinez-Santibanez G., Grimley E., Ostwani W., Dressler G.R. The kielin/chordin-like protein (KCP) attenuates high-fat diet-induced obesity and metabolic syndrome in mice. J. Biol. Chem. 2017;292:9051–9062. doi: 10.1074/jbc.M116.771428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soofi A., Zhang P., Dressler G.R. Kielin/chordin-like protein attenuates both acute and chronic renal injury. J. Am. Soc. Nephrol. 2013;24:897–905. doi: 10.1681/ASN.2012070759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bradford S.T.J., Ranghini E.J., Grimley E., Lee P.H., Dressler G.R. High-throughput screens for agonists of bone morphogenetic protein (BMP) signaling identify potent benzoxazole compounds. J. Biol. Chem. 2019;294:3125–3136. doi: 10.1074/jbc.RA118.006817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Racusen L.C., Solez K. Nephrotoxic tubular and interstitial lesions: morphology and classification. Toxicol. Pathol. 1986;14:45–57. doi: 10.1177/019262338601400106. [DOI] [PubMed] [Google Scholar]
- 58.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou L., Chen J., Li Z., Li X., Hu X., Huang Y., Zhao X., Liang C., Wang Y., Sun L. Integrated profiling of microRNAs and mRNAs: microRNAs located on Xq27.3 associate with clear cell renal cell carcinoma. PLoS ONE. 2010;5:e15224. doi: 10.1371/journal.pone.0015224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., Katayama T., Kawashima S., Okuda S., Tokimatsu T., Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mao X., Cai T., Olyarchuk J.G., Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21:3787–3793. doi: 10.1093/bioinformatics/bti430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.