Abstract
Resistin is an adipokine produced in white adipose tissue that is thought to modulate insulin sensitivity in peripheral tissues (such as liver, skeletal muscle or adipose tissue). Human and murine resistin molecules share only about 60% sequence homology. [1] Contrary to humans, in which resistin is secreted mostly by macrophages, Park and Ahima 2013 resistin in rodents is produced primarily by the mature adipocytes of the white adipose tissue. Although resistin can bind to toll-like receptor 4 (TLF4) activating proinflammatory responses in human and rodents, [3], [4], [5], [6], [7], [8] the inflammatory actions of resistin in human monocytes were found to be mediated by resistin binding to adenylyl cyclase-associated protein 1 (CAP1). [9] In this study, we aimed to investigate the in vitro effects of resistin on the expression of various genes related to insulin resistance in mouse liver cells. Using BNL CL.2 cells, we investigated the effect of resistin in untransfected or CAP1 siRNA-transfected cells on the expression of 84 key genes involved in insulin resistance.
Keywords: Resistin, Adenylyl cyclase-associated protein 1 (CAP1), Insulin resistance, BNL CL.2 cells
| Subject area | Biology |
| More specific subject area | Inflammation |
| Type of data | Graphs, figures |
| How data was acquired | qRT-PCR array, QuantStudio 3 (Life Technologies) |
| Data format | Analyzed |
| Experimental factors | BNL CL.2 cells treated with resistin in the presence or absence of CAP1 siRNA transfection |
| Experimental features | Untransfected or CAP1 siRNA-transfected BNL CL.2 cells were treated with resistin, and mRNA expression levels of 84 genes related to insulin resistance were measured by using qRT-PCR array. |
| Data source location | New York, New York, USA |
| Data accessibility | Data is with this article |
| Related research article | Lee S, Lee H–C, Kwon Y–W, Lee SE, Cho Y, Kim J et al. Adenylyl cyclase-associated protein 1 is a receptor for human resistin and mediates inflammatory actions of human monocytes. Cell Metab. 2014 Mar 4; 19(3):484–97 |
Value of the data
|
1. Data
1.1. Optimization of CAP1 siRNA transfections
Our optimization experiments (data not shown) demonstrated that there was no statistically significant difference in CAP1 mRNA expression levels between untransfected and siRNA negative control-transfected cells. There was a significant decrease (86%) in CAP1 siRNA levels in CAP1 siRNA-transfected cells, compared to untransfected controls. There was no statistically significant difference in the cell viability between all of the treatment conditions.
1.2. Effect of resistin on the expression of genes related to insulin resistance in BNL CL.2 cells
Using quantitative RT-PCR array, we measured the expression levels of 84 key genes involved in the mechanisms behind type 2 diabetes mellitus (T2DM) in adipose tissue (the full list of genes is presented in Table 1, and list of genes grouped by function is provided in Table 2) in BNL CL.2 cells in the presence or absence of resistin (25 ng/ml for 24 hours) (Fig. 1).
Table 1.
List of genes examined. Table list of all genes measured by the PCR array, including the NCBI reference sequence database (RefSeq), gene abbreviations, full names and/or synonyms.
| RefSeq | Abbreviation | Full Name |
|---|---|---|
| NM_133360 | Acaca | Acetyl-Coenzyme A carboxylase alpha |
| NM_133904 | Acacb | Acetyl-Coenzyme A carboxylase beta |
| NM_007981 | Acsl1 | Acyl-CoA synthetase long-chain family member 1 |
| NM_019477 | Acsl4 | Acyl-CoA synthetase long-chain family member 4 |
| NM_009605 | Adipoq | Adiponectin, C1Q and collagen domain containing |
| NM_028320 | Adipor1 | Adiponectin receptor 1 |
| NM_197985 | Adipor2 | Adiponectin receptor 2 |
| NM_011785 | Akt3 | Thymoma viral proto-oncogene 3 |
| NM_009662 | Alox5 | Arachidonate 5-lipoxygenase |
| NM_009696 | Apoe | Apolipoprotein E |
| NM_009807 | Casp1 | Caspase 1 |
| NM_011331 | Ccl12 | Chemokine (C–C motif) ligand 12 |
| NM_009916 | Ccr4 | Chemokine (C–C motif) receptor 4 |
| NM_009917 | Ccr5 | Chemokine (C–C motif) receptor 5 |
| NM_009835 | Ccr6 | Chemokine (C–C motif) receptor 6 |
| NM_007643 | Cd36 | CD36 antigen |
| NM_007648 | Cd3e | CD3 antigen, epsilon polypeptide |
| NM_007678 | Cebpa | CCAAT/enhancer binding protein (C/EBP), alpha |
| NM_007700 | Chuk | Conserved helix-loop-helix ubiquitous kinase |
| NM_013493 | Cnbp | Cellular nucleic acid binding protein |
| NM_016715 | Crlf2 | Cytokine receptor-like factor 2 |
| NM_026444 | Cs | Citrate synthase |
| NM_009910 | Cxcr3 | Chemokine (C-X-C motif) receptor 3 |
| NM_009911 | Cxcr4 | Chemokine (C-X-C motif) receptor 4 |
| NM_010130 | Adgre1 | EGF-like module containing, mucin-like, hormone receptor-like sequence 1 |
| NM_024406 | Fabp4 | Fatty acid binding protein 4, adipocyte |
| NM_007988 | Fasn | Fatty acid synthase |
| NM_030678 | Gys1 | Glycogen synthase 1, muscle |
| NM_013820 | Hk2 | Hexokinase 2 |
| NM_008337 | Ifng | Interferon gamma |
| NM_010512 | Igf1 | Insulin-like growth factor 1 |
| NM_010513 | Igf1r | Insulin-like growth factor I receptor |
| NM_010546 | Ikbkb | Inhibitor of kappaB kinase beta |
| NM_008365 | Il18r1 | Interleukin 18 receptor 1 |
| NM_008361 | Il1b | Interleukin 1 beta |
| NM_008362 | Il1r1 | Interleukin 1 receptor, type I |
| NM_144548 | Il23r | Interleukin 23 receptor |
| NM_001314054 | Il6 | Interleukin 6 |
| NM_010568 | Insr | Insulin receptor |
| NM_010570 | Irs1 | Insulin receptor substrate 1 |
| NM_001081212 | Irs2 | Insulin receptor substrate 2 |
| NM_008413 | Jak2 | Janus kinase 2 |
| NM_008493 | Lep | Leptin |
| NM_010704 | Lepr | Leptin receptor |
| NM_010719 | Lipe | Lipase, hormone sensitive |
| NM_008509 | Lpl | Lipoprotein lipase |
| NM_008517 | Lta4h | Leukotriene A4 hydrolase |
| NM_008927 | Map2k1 | Mitogen-activated protein kinase kinase 1 |
| NM_011952 | Mapk3 | Mitogen-activated protein kinase 3 |
| NM_016961 | Mapk9 | Mitogen-activated protein kinase 9 |
| NM_020009 | Mtor | Mechanistic target of rapamycin (serine/threonine kinase) |
| NM_021524 | Nampt | Nicotinamide phosphoribosyltransferase |
| NM_010907 | Nfkbia | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
| NM_145827 | Nlrp3 | NLR family, pyrin domain containing 3 |
| NM_138648 | Olr1 | Oxidized low density lipoprotein (lectin-like) receptor 1 |
| NM_011044 | Pck1 | Phosphoenolpyruvate carboxykinase 1, cytosolic |
| NM_011055 | Pde3b | Phosphodiesterase 3B, cGMP-inhibited |
| NM_133667 | Pdk2 | Pyruvate dehydrogenase kinase, isoenzyme 2 |
| NM_008814 | Pdx1 | Pancreatic and duodenal homeobox 1 |
| NM_008839 | Pik3ca | Phosphatidylinositol 3-kinase, catalytic, alpha polypeptide |
| NM_001024955 | Pik3r1 | Phosphatidylinositol 3-kinase, regulatory subunit, polypeptide 1 (p85 alpha) |
| NM_011144 | Ppara | Peroxisome proliferator activated receptor alpha |
| NM_011146 | Pparg | Peroxisome proliferator activated receptor gamma |
| NM_008904 | Ppargc1a | Peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| NM_011201 | Ptpn1 | Protein tyrosine phosphatase, non-receptor type 1 |
| NM_023258 | Pycard | PYD and CARD domain containing |
| NM_011255 | Rbp4 | Retinol binding protein 4, plasma |
| NM_009045 | Rela | V-rel reticuloendotheliosis viral oncogene homolog A (avian) |
| NM_022984 | Retn | Resistin |
| NM_028259 | Rps6kb1 | Ribosomal protein S6 kinase, polypeptide 1 |
| NM_009127 | Scd1 | Stearoyl-Coenzyme A desaturase 1 |
| NM_008871 | Serpine1 | Serine (or cysteine) peptidase inhibitor, clade E, member 1 |
| NM_011977 | Slc27a1 | Solute carrier family 27 (fatty acid transporter), member 1 |
| NM_009204 | Slc2a4 | Solute carrier family 2 (facilitated glucose transporter), member 4 |
| NM_007707 | Socs3 | Suppressor of cytokine signaling 3 |
| NM_011480 | Srebf1 | Sterol regulatory element binding transcription factor 1 |
| NM_033218 | Srebf2 | Sterol regulatory element binding factor 2 |
| NM_011486 | Stat3 | Signal transducer and activator of transcription 3 |
| NM_021297 | Tlr4 | Toll-like receptor 4 |
| NM_013693 | Tnf | Tumor necrosis factor |
| NM_011609 | Tnfrsf1a | Tumor necrosis factor receptor superfamily, member 1a |
| NM_011610 | Tnfrsf1b | Tumor necrosis factor receptor superfamily, member 1b |
| NM_009463 | Ucp1 | Uncoupling protein 1 (mitochondrial, proton carrier) |
| NM_013703 | Vldlr | Very low density lipoprotein receptor |
| NM_007393 | Actb | Actin, beta |
| NM_009735 | B2m | Beta-2 microglobulin |
| NM_010368 | Gusb | Glucuronidase, beta |
| NM_008084 | Gapdh | Glyceraldehyde-3-phosphate dehydrogenase |
| NM_008302 | Hsp90ab1 | Heat shock protein 90 alpha (cytosolic), class B member 1 |
| SA_00106 | MGDC | Mouse Genomic DNA Contamination |
| SA_00104 | RTC | Reverse Transcription Control |
| SA_00104 | RTC | Reverse Transcription Control |
| SA_00104 | RTC | Reverse Transcription Control |
| SA_00103 | PPC | Positive PCR Control |
| SA_00103 | PPC | Positive PCR Control |
| SA_00103 | PPC | Positive PCR Control |
Table 2.
Genes by function. Table lists all of the genes examined, separated by function.
| Function | Gene |
|---|---|
| Insulin signaling | Akt3 |
| Gys1 | |
| Igf1 | |
| Igf1r | |
| Ikbkb(IKK2) | |
| Insr | |
| Irs1 | |
| Irs2 | |
| Map2k1(Mek1) | |
| Mapk3(Erk1) | |
| Mapk9(Jnk2) | |
| Mtor | |
| Pde3b | |
| Pik3ca(p110-alpha) | |
| Pik3r1(PI3KA, p85alpha) | |
| Ppargc1a(Pgc-1alpha, Ppargc1) | |
| Ptpn1(Ptp1b, Ptp) | |
| Rps6kb1 | |
| Slc2a4(Glut4) | |
| Socs3 | |
| Srebf1 | |
| Non-insulin dependent diabetes mellitus | Adipoq(Acrp30) |
| Hk2 | |
| Ikbkb(IKKbeta, IKK2) | |
| Insr | |
| Irs1 | |
| Irs2 | |
| Mapk3(Erk1) | |
| Mapk9(Jnk2) | |
| Mtor | |
| Pdx1(Ipf1) | |
| Pik3ca(p110-alpha) | |
| Pik3r1(PI3KA, p85-alpha) | |
| Slc2a4(Glut4) | |
| Socs3 | |
| Tnf | |
| Adipokine signaling | |
| Adipokines | Adipoq(Acrp30) |
| Il6 | |
| Lep | |
| Nampt | |
| Retn | |
| Serpine1(PAI-1) | |
| Tnf | |
| Receptors & transporters | Adipor1 |
| Adipor2 | |
| Cd36 | |
| Lepr | |
| Slc2a4(Glut4) | |
| Tnfrsf1a(Tnfr1) | |
| Tnfrsf1b | |
| Signaling downstream of adipokines | Akt3 |
| Chuk(Ikbka, Ikka) | |
| Ikbkb(IKK2) | |
| Irs1 | |
| Irs2 | |
| Jak2 | |
| Mapk9(Jnk2) | |
| Mtor | |
| Nfkbia(Iκb-alpha, Mad3) | |
| Ppara | |
| Ppargc1a(Ppargc1) | |
| Rela | |
| Socs3 | |
| Stat3 | |
| Innate immunity | Casp1(ICE) |
| Chuk(Ikbka, Ikka) | |
| Ikbkb(IKK2) | |
| Irs1 | |
| Irs2 | |
| Nlrp3 | |
| Nfkbia(Iκb-alpha, Mad3) | |
| Pycard(Tms1, Asc) | |
| Rela | |
| Tlr4 | |
| Inflammation | Alox5 |
| Casp1(Ice) | |
| Ccl12(MCP-5, Scya12) | |
| Ccr4 | |
| Ccr5 | |
| Chuk(Ikbka, Ikka) | |
| Cxcr4 | |
| Ifng | |
| Ikbkb(IKK2) | |
| Il1b | |
| Il23r | |
| Il6 | |
| Lta4h | |
| Nlrp3 | |
| Olr1 | |
| Pycard(Tms1, Asc) | |
| Rela | |
| Tnf | |
| Tnfrsf1a(Tnfr1) | |
| Tnfrsf1b | |
| Apoptosis | Pparg |
| Serpine1PAI-1) | |
| Tnf | |
| Casp1(Ice) | |
| Ikbkb(IKK2) | |
| Irs2 | |
| Mapk9(Jnk2) | |
| Nfkbia(Iκb-alpha, Mad3) | |
| Nlrp3 | |
| Pycard(Tms1, Asc) | |
| Rela | |
| Tnfrsf1a (Tnfr1) | |
| Jak2 | |
| Pik3ca (p110-alpha) | |
| Socs3 | |
| Rps6kb1 | |
| Tnfrsf1b | |
| Metabolic pathways | |
| Carbohydrate metabolism | Cs |
| Gys1 | |
| Hk2 | |
| Pck1 | |
| Pdk2 | |
| Lipid metabolism | Acaca |
| Acacb | |
| Acsl1 | |
| Acsl4 | |
| Apoe | |
| Cebpa | |
| Cnbp | |
| Fabp4 | |
| Fasn | |
| Lepr | |
| Lipe | |
| Lpl | |
| Ppara | |
| Pparg | |
| Ppargc1a(Ppargc1) | |
| Scd1 | |
| Srebf1 | |
| Srebf2 | |
| Metabolite transport | Apoe |
| Cd36 | |
| Fabp4 | |
| Rbp4 | |
| Slc2a4(Glut4) | |
| Slc27a1 | |
| Ucp1 | |
| Vldlr | |
| Infiltrating leukocyte markers | |
| Macrophages | Ccr5 |
| Cxcr4 | |
| Adgre1 | |
| Th1 cells | Ccr5 |
| Cd3e | |
| Cxcr3 | |
| Il18r1 | |
| Th2 cells | Ccr4 |
| Cd3e | |
| Crlf2 (Tslpr) | |
| Il1r1 | |
| Th17 cells | Ccr6 |
| Cd3e | |
| Il23r | |
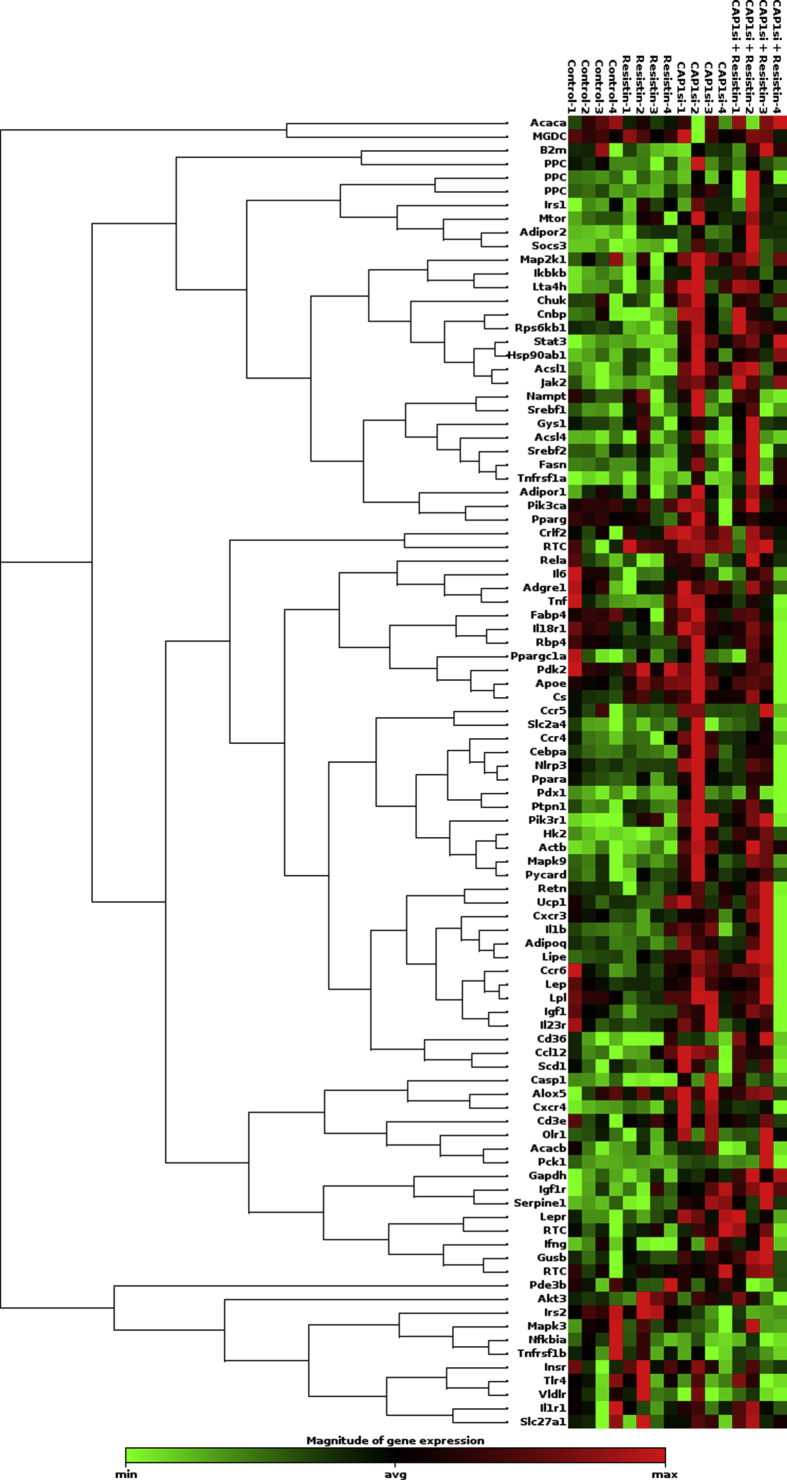
Fig. 1.
Gene expression–clustergram. Clustergram of the entire dataset displaying a heat map with dendrograms indicating co-regulated genes across groups or individual samples.
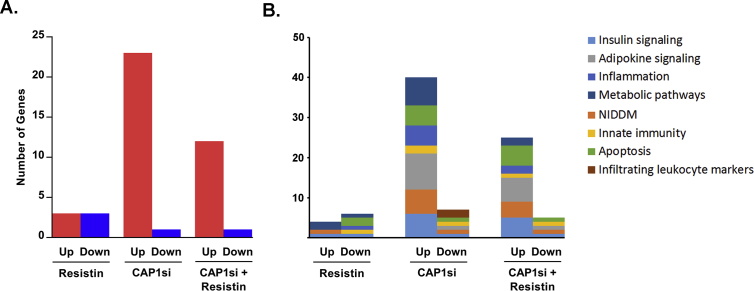
Resistin treatment resulted in statistically significant change in the expression of 6 genes (Table 3). Of these genes, 3 were significantly upregulated (Apoe, Cs, and Pik3rl) and 3 downregulated (Casp1, Rbp4, and Rps6kb1) (Fig. 2A). Functionally, these genes participate in metabolic pathways (Apoe, Cs, and Rbp4), insulin signaling (Pik3r1 and Rps6kb1), T2DM (Pik3r1), innate immunity (Casp1), inflammation (Casp1), and apoptosis (Casp1 and Rps6kb1) (Fig. 2B).
Table 3.
Fold regulation of gene expression (treatment vs. control). Fold change of gene expression of each treatment vs. control. p values <= 0.05 are marked in bold.
| Gene | Resistin |
CAP1 siRNA |
CAP1 siRNA + Resistin |
|||
|---|---|---|---|---|---|---|
| Fold Regulation | p-value | Fold Regulation | p-value | Fold Regulation | p-value | |
| Acaca | −1.14 | 0.169661 | −1.22 | 0.290632 | 1.00 | 0.878421 |
| Acacb | 1.01 | 0.906748 | 1.09 | 0.438927 | 1.06 | 0.643704 |
| Acsl1 | 1.02 | 0.724041 | 1.44 | 0.000491 | 1.36 | 0.018940 |
| Acsl4 | 1.04 | 0.542086 | 1.08 | 0.356823 | 1.12 | 0.206335 |
| Adipoq | −1.10 | 0.173396 | 1.32 | 0.041017 | 1.15 | 0.401499 |
| Adipor1 | −1.04 | 0.329231 | 1.02 | 0.640683 | 1.06 | 0.146953 |
| Adipor2 | 1.13 | 0.113931 | 1.17 | 0.028059 | 1.34 | 0.016324 |
| Akt3 | 1.07 | 0.126858 | 1.03 | 0.256181 | 1.01 | 0.782413 |
| Alox5 | 1.42 | 0.364008 | 1.75 | 0.099439 | 1.51 | 0.263775 |
| Apoe | 1.55 | 0.022577 | 1.29 | 0.046392 | −1.02 | 0.812274 |
| Casp1 | −1.89 | 0.003612 | 1.41 | 0.224487 | 1.24 | 0.252274 |
| Ccl12 | 1.22 | 0.306190 | 1.51 | 0.099450 | 1.33 | 0.142705 |
| Ccr4 | 1.00 | 0.925798 | 1.52 | 0.021117 | 1.17 | 0.271836 |
| Ccr5 | −1.05 | 0.612433 | 1.27 | 0.357504 | 1.07 | 0.791426 |
| Ccr6 | −1.07 | 0.570025 | 1.14 | 0.358616 | 1.07 | 0.661161 |
| Cd36 | −1.08 | 0.979498 | 1.17 | 0.480636 | 1.74 | 0.080825 |
| Cd3e | 1.30 | 0.367086 | 1.55 | 0.122220 | 1.28 | 0.416990 |
| Cebpa | −1.01 | 0.554566 | 1.16 | 0.045397 | 1.05 | 0.407643 |
| Chuk | −1.00 | 0.923392 | 1.07 | 0.230993 | 1.06 | 0.155313 |
| Cnbp | −1.08 | 0.148730 | 1.15 | 0.098214 | 1.12 | 0.158070 |
| Crlf2 | 1.13 | 0.152933 | 1.24 | 0.010318 | 1.10 | 0.290357 |
| Cs | 1.49 | 0.025871 | 1.28 | 0.023492 | 1.06 | 0.583900 |
| Cxcr3 | −1.16 | 0.143245 | 1.02 | 0.813042 | 1.05 | 0.617630 |
| Cxcr4 | 1.14 | 0.144379 | 1.66 | 0.013166 | 1.20 | 0.141848 |
| Adgre1 | −1.15 | 0.431959 | 1.20 | 0.273281 | −1.00 | 0.936820 |
| Fabp4 | −1.21 | 0.123579 | 1.02 | 0.702554 | −1.14 | 0.343194 |
| Fasn | −1.02 | 0.563222 | 1.05 | 0.370487 | 1.11 | 0.102888 |
| Gys1 | 1.00 | 0.891518 | 1.01 | 0.721490 | 1.05 | 0.204388 |
| Hk2 | −1.02 | 0.556124 | 1.39 | 0.009742 | 1.37 | 0.004545 |
| Ifng | −1.42 | 0.518183 | 1.76 | 0.147576 | 1.54 | 0.290151 |
| Igf1 | −1.20 | 0.063415 | 1.12 | 0.298831 | −1.00 | 0.909747 |
| Igf1r | 1.03 | 0.376609 | 1.12 | 0.009107 | 1.15 | 0.000795 |
| Ikbkb | 1.02 | 0.477848 | 1.10 | 0.031863 | 1.08 | 0.013828 |
| Il18r1 | −1.14 | 0.127650 | 1.07 | 0.378861 | −1.08 | 0.592830 |
| Il1b | 1.06 | 0.555982 | 1.26 | 0.122392 | 1.16 | 0.340112 |
| Il1r1 | −1.06 | 0.560278 | 1.01 | 0.957418 | 1.07 | 0.683408 |
| Il23r | −1.13 | 0.426607 | 1.16 | 0.476194 | −1.18 | 0.546692 |
| Il6 | −1.46 | 0.078072 | −1.21 | 0.351666 | −1.20 | 0.426350 |
| Insr | 1.11 | 0.130969 | 1.04 | 0.591773 | 1.02 | 0.833929 |
| Irs1 | 1.12 | 0.305636 | 1.15 | 0.219709 | 1.20 | 0.177262 |
| Irs2 | 1.01 | 0.775771 | −1.13 | 0.017086 | −1.14 | 0.003924 |
| Jak2 | 1.03 | 0.424419 | 1.23 | 0.013603 | 1.27 | 0.009525 |
| Lep | −1.08 | 0.361070 | 1.16 | 0.211892 | −1.04 | 0.971677 |
| Lepr | 1.11 | 0.377068 | 1.59 | 0.005756 | 1.30 | 0.159982 |
| Lipe | 1.06 | 0.206055 | 1.17 | 0.012267 | 1.12 | 0.252451 |
| Lpl | −1.21 | 0.124427 | 1.10 | 0.365642 | −1.10 | 0.808003 |
| Lta4h | 1.02 | 0.531694 | 1.13 | 0.016815 | 1.11 | 0.022074 |
| Map2k1 | −1.06 | 0.445888 | 1.10 | 0.078819 | 1.06 | 0.321980 |
| Mapk3 | −1.02 | 0.680433 | −1.09 | 0.096201 | −1.04 | 0.580686 |
| Mapk9 | −1.00 | 0.917290 | 1.16 | 0.065311 | 1.13 | 0.132737 |
| Mtor | 1.03 | 0.684468 | 1.24 | 0.049218 | 1.19 | 0.080924 |
| Nampt | 1.01 | 0.846387 | 1.02 | 0.732144 | −1.02 | 0.896754 |
| Nfkbia | −1.09 | 0.318262 | −1.16 | 0.065727 | −1.17 | 0.069963 |
| Nlrp3 | −1.06 | 0.126789 | 1.24 | 0.126506 | 1.01 | 0.762575 |
| Olr1 | −1.38 | 0.504119 | 1.35 | 0.272188 | 1.33 | 0.287191 |
| Pck1 | −1.05 | 0.568571 | 1.31 | 0.077792 | 1.16 | 0.490821 |
| Pde3b | 1.02 | 0.825368 | 1.03 | 0.787814 | −1.15 | 0.190104 |
| Pdk2 | 1.14 | 0.394247 | 1.06 | 0.729603 | −1.23 | 0.473372 |
| Pdx1 | −1.01 | 0.710029 | 1.24 | 0.100216 | 1.04 | 0.657469 |
| Pik3ca | −1.03 | 0.399336 | −1.03 | 0.782751 | 1.01 | 0.744423 |
| Pik3r1 | 1.52 | 0.033282 | 1.25 | 0.000901 | 1.18 | 0.034911 |
| Ppara | −1.02 | 0.792466 | 1.21 | 0.080647 | 1.02 | 0.722852 |
| Pparg | −1.08 | 0.093212 | −1.03 | 0.865268 | −1.03 | 0.216785 |
| Ppargc1a | 1.01 | 0.998904 | 1.07 | 0.594209 | 1.00 | 0.998558 |
| Ptpn1 | 1.07 | 0.237874 | 1.29 | 0.032159 | 1.16 | 0.189144 |
| Pycard | 1.08 | 0.551026 | 1.42 | 0.036940 | 1.29 | 0.054659 |
| Rbp4 | −1.56 | 0.007272 | 1.06 | 0.482300 | −1.07 | 0.751105 |
| Rela | −1.04 | 0.598118 | 1.03 | 0.646466 | 1.07 | 0.279014 |
| Retn | −1.10 | 0.204038 | 1.11 | 0.099451 | 1.10 | 0.388516 |
| Rps6kb1 | −1.58 | 0.002976 | 1.06 | 0.144979 | 1.11 | 0.004902 |
| Scd1 | 1.05 | 0.821963 | 1.17 | 0.461504 | 1.18 | 0.410142 |
| Serpine1 | −1.03 | 0.820937 | 1.56 | 0.001029 | 1.37 | 0.043504 |
| Slc27a1 | 1.01 | 0.929373 | 1.04 | 0.805979 | 1.13 | 0.317341 |
| Slc2a4 | 1.04 | 0.761240 | 1.24 | 0.303759 | 1.15 | 0.331140 |
| Socs3 | −1.01 | 0.829376 | 1.27 | 0.021130 | 1.34 | 0.039503 |
| Srebf1 | 1.10 | 0.340675 | 1.18 | 0.186347 | 1.11 | 0.348476 |
| Srebf2 | −1.01 | 0.876772 | 1.02 | 0.732351 | 1.12 | 0.368694 |
| Stat3 | −1.01 | 0.653690 | 1.27 | 0.002095 | 1.29 | 0.001880 |
| Tlr4 | 1.03 | 0.737080 | −1.05 | 0.492373 | −1.03 | 0.761516 |
| Tnf | −1.22 | 0.480161 | 1.33 | 0.255783 | −1.14 | 0.607219 |
| Tnfrsf1a | 1.02 | 0.425885 | 1.05 | 0.359944 | 1.15 | 0.089583 |
| Tnfrsf1b | −1.06 | 0.567979 | −1.22 | 0.139907 | −1.17 | 0.161986 |
| Ucp1 | 1.01 | 0.881083 | 1.20 | 0.097531 | 1.05 | 0.631065 |
| Vldlr | 1.04 | 0.699680 | −1.11 | 0.501624 | −1.13 | 0.174714 |
| Actb | 1.01 | 0.777600 | 1.33 | 0.001409 | 1.33 | 0.001148 |
| B2m | −1.06 | 0.258822 | −1.02 | 0.642430 | 1.05 | 0.470054 |
| Gapdh | 1.01 | 0.814693 | 1.21 | 0.004777 | 1.45 | 0.001734 |
| Gusb | 1.10 | 0.303910 | 1.28 | 0.021514 | 1.38 | 0.038612 |
| Hsp90ab1 | 1.03 | 0.546044 | 1.19 | 0.049921 | 1.21 | 0.004717 |
Fig. 2.
Number of genes significantly affected by treatment. A. The graph represents the number of statistically significant (p < 0.05) up- or down-regulated genes. B. Graph represents genes significantly up- or down-regulated within each treatment group, grouped by function.
1.3. Role of CAP1 in mediating insulin sensitivity actions of resistin in BNL Cl.2 cells
Transfection of the BNL CL.2 cells with CAP1 siRNA resulted in significant change in the expression levels of 24 genes. 23 of these were upregulated (Acsl1, Adipoq, Adipor2, Apoe, Ccr4, Cebpa, Crlf2, Cs, Cxcr4, Hk2, Igflr, Ikbkb, Jak2, Lepr, Lipe, Lta4h, Mtor, Pik3r1, Ptpn1, Pycard, Serpine1, Socs3, and Stat3), and one (Irs2) was downregulated (Fig. 1A). Divided by function, these genes participate in metabolic pathways (Acsl1, Apoe1, Cebpa, Cs, Hk2, Lepr, and Lipe), insulin signaling (Igf1r, Ikbkb, Mtor, Pik3r1, Ptpn1, Socs3, and Irs2), T2DM (Adipoq, Hk2, Ikbkb, Mtor, Pik3r1, Socs3, and Irs2), adipokine signaling (Adipoq, Adipor2, Ikbkb, Jak2, Lepr, Mtor, Serpine1, Socs3, Stat3, and Irs2), innate immunity (Ikbkb, Pycard, and Irs2), inflammation (ccr4, Cxcr4, Ikbkb, Lta4h, and Pycard), apoptosis (Ikbkb, Jak2, Pycard, Serpine1, Socs3, and Irs2), or are markers of infiltrated leukocytes (Ccr4 and Crlf2) (Fig. 1, Fig. 2B).
When CAP1 siRNA-transfected cells were treated with resistin, the expression of 13 genes was significantly affected: 12 genes (Acsl1, Adipor2, Hk2, Igf1r, Ikbkb, Jak2, Lta4h, Pik3r1, Rps6kb1, Serpine1, Socs3, and Stat3) were upregulated, and 1 gene (Irs2) was downregulated (Table 3 and Fig. 2A). Divided by function, these genes were involved in metabolic pathways (Acsl1 and Hk2), insulin signaling (Igf1r, Ikbkb, Pik3r1, Rps6kb1, Socs3, and Irs2), T2DM (Hk2, Ikbkb, Pik3r1, Socs3, and Irs2), adipokine signaling (Adipor2, Ikbkb, Jak2, Serpine1, Socs3, Stat3, and Irs2), innate immunity (Ikbkb and Irs2), inflammation (Ikbkb and Lta4h), or apoptosis (Ikbkb, Jak2, Rps6kb1, Serpine1, Socs3, and Irs2) (Table 3 and Fig. 1, Fig. 2B).
2. Experimental design, materials and methods
2.1. Reagents
Mouse recombinant resistin (Sigma-Aldrich, Cat. # SRP4560) was resuspended in water to a stock concentration of 100 μg/ml and further diluted to 25 μg/ml before cell treatment.
2.2. Cell culture
BNL CL.2 mouse liver cells were purchased from American Type Culture Collection (ATCC) (Cat. # TIB-73) and grown in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (VWR International, Cat. # 89510–186) and antibiotic/antimycotic solution (Penicillin, Streptomycin, Amphotericin B) (Corning, Cat. # 30-004-Cl), and incubated at 37 °C with 10% CO2.
2.3. Experiment design
BNL CL.2 cells were seeded in 6-well tissue culture plates with 2 ml tissue culture medium, in a density of 0.5 × 10 [6] cells and grown for one day (to approximately 60% confluency). Resistin treatment was performed by adding 2 μl (25 μg/ml) resistin to the appropriate wells.
2.4. siRNA transfection
CAP1 siRNA transfection was performed using Opti-MEM Reduced Serum Medium (Gibco, Cat. # 31985–070), Lipofectamine RNAiMAX transfection reagent (Invitrogen, Cat. # 13778–075), and mouse CAP1 Silencer Select siRNA (Life Technologies, Cat. # 4390771, siRNA ID# s63297) following manufacturer's protocol. Transfection was performed for 6 hours; the cell culture medium was then replaced with complete medium for overnight cell growth.
2.5. RNA extraction and reverse transcription
After completing the experiments, the cells were washed one time with ice-cold PBS and RNA was extracted using TRIzol Reagent (Ambion, Cat. # 15596018), chloroform, and iso-propanol. Total RNA concentration was quantified using NanoDrop One spectrophotometer (Thermo Scientific). All of the samples were normalized to 1 mg/ml of total RNA. Reverse transcriptase (RT) reaction was performed using qScript cDNA SuperMix kit (QuantaBio, Cat. # 95048). RT reaction was performed using the following conditions: 25 °C/5 min, 42 °C/30 min, 85 °C/5 min, 4 °C/∞. After RT reaction, the cDNA samples were diluted 10 times with water.
2.6. Quantitative RT-PCR array
Quantitative RT-PCR array was performed using RT [2] SYBR Green ROX qPCR Mastermix (Qiagen) and RT [2] ProfilerTM PCR Array Mouse Insulin Resistance kit (QIAGEN, Cat. # PAMM-156ZA). Master mix for each plate consisted of: 1350 μl 2× RT2 SYBR Green Master Mix, 102 μl of cDNA, and 1248 μl of PCR grade water, and 25 μl of it was pipetted into each PCR plate well. Quantitative PCR was performed using the following conditions: hold stage: 50 °C/2 min, 95 °C/10 min; PCR stage: 95 °C/15 sec, 60 °C/1 min (40 cycles); melt curve stage: 60 °C/1 min, 95 °C/1 sec.
2.7. Data analysis
Results were obtained from 4 separate experiments. Data analysis was performed using QIAGEN web-based software (https://dataanalysis.qiagen.com). Fold change values were calculated using ΔΔCt method (2ˆ(-ΔΔCt)). p-values were calculated based on Student's t-test of the replicate 2ˆ(-ΔCt) values for each gene in the control group and the treatment groups.
Acknowledgements
This project was supported by a grant from Gerald J. and Dorothy Friedman New York Foundation for Medical Research.
Conflict of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Park H.K., Ahima R.S. Resistin in rodents and humans. Diabetes Metab. J. 2013 Dec;37(6):404–414. doi: 10.4093/dmj.2013.37.6.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel L., Buckels A.C., Kinghorn I.J., Murdock P.R., Holbrook J.D., Plumpton C. Resistin is expressed in human macrophages and directly regulated by PPAR gamma activators. Biochem. Biophys. Res. Commun. 2003 Jan 10;300(2):472–476. doi: 10.1016/s0006-291x(02)02841-3. [DOI] [PubMed] [Google Scholar]
- 3.Tarkowski A., Bjersing J., Shestakov A., Bokarewa M.I. Resistin competes with lipopolysaccharide for binding to toll-like receptor 4. J. Cell Mol. Med. 2010 Jun;14(6B):1419–1431. doi: 10.1111/j.1582-4934.2009.00899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benomar Y., Gertler A., De Lacy P., Crépin D., Ould Hamouda H., Riffault L. Central resistin overexposure induces insulin resistance through Toll-like receptor 4. Diabetes. 2013 Jan;62(1):102–114. doi: 10.2337/db12-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benomar Y., Amine H., Crépin D., Al Rifai S., Riffault L., Gertler A. Central resistin/TLR4 impairs adiponectin signaling, contributing to insulin and FGF21 resistance. Diabetes. 2016;65(4):913–926. doi: 10.2337/db15-1029. [DOI] [PubMed] [Google Scholar]
- 6.Jiang Y., Lu L., Hu Y., Li Q., An C., Yu X. Resistin induces hypertension and insulin resistance in mice via a TLR4-dependent pathway. Sci. Rep. 2016 Feb 26;6:22193. doi: 10.1038/srep22193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jang J.C., Li J., Gambini L., Batugedara H.M., Sati S., Lazar M.A. Human resistin protects against endotoxic shock by blocking LPS-TLR4 interaction. Proc. Natl. Acad. Sci. U.S.A. 2017;114(48):E10399–E10408. doi: 10.1073/pnas.1716015114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miao J., Benomar Y., Al Rifai S., Poizat G., Riffault L., Crépin D. Resistin inhibits neuronal autophagy through Toll-like receptor 4. J. Endocrinol. 2018 Jul;238(1):77–89. doi: 10.1530/JOE-18-0096. [DOI] [PubMed] [Google Scholar]
- 9.Lee S., Lee H.-C., Kwon Y.-W., Lee S.E., Cho Y., Kim J. Adenylyl cyclase-associated protein 1 is a receptor for human resistin and mediates inflammatory actions of human monocytes. Cell Metabol. 2014 Mar 4;19(3):484–497. doi: 10.1016/j.cmet.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]