Candida auris is an emerging multidrug-resistant fungal pathogen that has been associated with nosocomial bloodstream and deep wound infections causing a high mortality rate mainly in intensive care unit (ICU) patients. Laboratories currently rely on phenotypic testing using commercial automated systems for identification of yeasts; however, this technique has often led to misidentification of C. auris to other closely related species.
KEYWORDS: BD Max, Candida auris, TaqMan probe, real-time PCR
ABSTRACT
Candida auris is an emerging multidrug-resistant fungal pathogen that has been associated with nosocomial bloodstream and deep wound infections causing a high mortality rate mainly in intensive care unit (ICU) patients. Laboratories currently rely on phenotypic testing using commercial automated systems for identification of yeasts; however, this technique has often led to misidentification of C. auris to other closely related species. We developed and validated a TaqMan-based real-time PCR assay on the BD Max platform targeting ribosomal DNA (rDNA) region nucleotide sequences to quickly and accurately test for C. auris infection from culture and clinical specimens. The assay is highly specific, reproducible, and sensitive, allowing detection of as low as 1 C. auris CFU per reaction within 3 h.
INTRODUCTION
Candida auris is a globally emerging multidrug-resistant fungal pathogen that has been associated with nosocomial bloodstream and deep wound infections causing a high mortality rate mainly in intensive care unit patients. C. auris infection has been reported in Asia, Africa, Europe, and the Americas (1–8). Various studies have reported that C. auris is resistant to many antifungal drugs, such as azoles, caspofungin, and amphotericin B (2, 9). The Centers for Disease Control and Prevention (CDC) has reported that C. auris can be spread through contact with contaminated surfaces and physical contact with a person who is infected or colonized (https://www.health.ny.gov/diseases/communicable/c_auris/). That poses a major challenge to clinicians and infection prevention groups.
C. auris is often misidentified as the closely related species Candida haemulonii by traditional microbiological methods, such as chromogenic agar, analytical profile index (API) strips, or automated systems such as the Vitek 2 (bioMérieux, Marcy l’Etoile, France) (2, 9, 10). Recently, matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) and techniques based on sequencing of the internal transcribed spacer (ITS) and D1/D2 segments of the ribosomal DNA (rDNA) have been used successfully to differentiate C. auris from other species (6, 9, 11–13). Those techniques, however, are laborious and expensive. The objective of this study was to develop and validate a new TaqMan-based real-time PCR assay targeting the rDNA region nucleotide sequences to quickly and accurately test for C. auris infection from culture or directly from clinical specimens using the BD Max system (BD Diagnostics, Sparks, MD), a fully integrated automated molecular platform combining specimen processing and real-time PCR processing.
MATERIALS AND METHODS
Primers and probe.
The primers and probe were designed based on the alignment of sequences currently available for the different Candida species from the National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov). Candida species were aligned using the multisequence alignment tools Clustal Omega version 1.2.4 (http://www.clustal.org/omega). The primer and probe sequences were designed to amplify genetic regions of the rRNA gene and the internal transcribed spacers (ITS) using the PrimerQuest primer design tool from Integrated DNA Technologies (Coralville, Iowa). Furthermore, they were analyzed using the OligoAnalyzer 3.1 tool (IDT) to ensure that they were at optimal conditions, and the Primer-BLAST tool from NCBI was used to confirm specificity before ordering from IDT (Coralville, IA).
Fungal and bacterial isolates and culture conditions.
A total of 113 fungal isolates and 12 bacteria species obtained from different sources were analyzed in this study. Of these isolates, 62 were from the Centers for Disease Control and Prevention (CDC) (Atlanta, GA) Candida auris and drug-resistant Candida panels, 45 were isolated from clinical specimens at Tampa General Hospital (Tampa, Florida), and 19 were acquired from the American Type Culture Collection (ATCC) (Manassas, VA). These included C. auris (n = 10), C. haemulonii (n = 4), Candida duobushaemulonii (n = 3), Candida lusitaniae (n = 2), Candida albicans (n = 21), Candida parapsilosis (n = 13), Candida glabrata (n = 36), Kodamaea ohmeri (n = 1), Candida krusei (n = 2), Candida tropicalis (n = 10), Candida guilliermondii (n = 1), Candida pelliculosa (n = 1), Candida rugosa (n = 1), Candida pararugosa (n = 1), Candida kefyr (n = 1), Saccharomyces cerevisiae (n = 4), Cryptococcus neoformans (n = 1), Cryptococcus albidus (n = 1), staphylococci (n = 2), streptococci (n = 2), enterococci (n = 2), Pseudomonas aeruginosa (n = 1), Burkholderia cepacia (n = 1), Klebsiella pneumoniae (n = 1), Escherichia coli (n = 1), Haemophilus influenzae (n = 1), and Neisseria meningitidis (n = 1). Fungal isolates were cultured on Sabouraud dextrose agar (Becton, Dickinson and Company, Sparks, MD), and the bacterial isolates were cultured on blood agar at 37°C for 24 h before testing.
Candida auris real-time PCR detection.
Fungal and bacterial cultures were resuspended in saline to a 0.5 McFarland standard using the bioMérieux DensiChek Plus colorimeter (bioMérieux). The DNA extraction and real-time PCR were carried out using the BD Max ExK DNA-3 extraction kit (BD Diagnostic Systems, Québec, Canada), along with the specific primers and probe designed to detect C. auris. An aliquot of 250 μl of sample (fungal suspension, bacterial suspension, spiked, or unspiked clinical specimen) was mixed with 250 μl of sorbitol buffer (2 M sorbitol, 200 mM EDTA, 29 mM β-mercaptoethanol added just before processing) and treated with 10 U of Zymolyase (Zymo Research, Irvine, CA) for 30 min at 30°C.
The mixture was then inoculated into the BD Max DNA-3 extraction kit sample buffer tube, which contains 750 μl of sample buffer, and placed on the extraction tray for automated extraction. Out of the 1.25 ml of buffer and sample mixture, the instrument transferred 937.5 μl (3/4 of the mixture) into the lysis tube. After the extraction, the DNA was eluted in 12.5 μl of neutralizing buffer in a Snap-1 tube. The PCR master mix was distributed in Snap-2 and Snap-3 tubes on the DNA-3 extraction strip. The Snap-2 tube contained the lyophilized PCR reagent mix and the primers and probe for the sample processing control (SPC). The Snap-3 tube contained the master mix that was prepared in-house consisting of 2 μl of DNA primer diluent (part of the BD Max kit), 1 μl of C. auris working primer mix (12.5 μM each primer C.aurFor and C.aurRev and 8.75μM C. auris probe [C.aurPb] in 1× Tris-EDTA [TE] buffer), and 9.5 μl of molecular grade water (Table 1). After extraction, the 12.5 μl of eluted DNA was mixed with the 12.5 μl of primers and probe in the Snap-3 tube. Of the 25 μl, only 4 μl of the mixture was transferred into the sealed BD Max PCR cartridge chamber where the PCR took place. Cycling conditions were as follows: 10 min hold at 98°C followed by 40 cycles of 30 s at 98°C and 30 s at 60°C. The channel settings for acquisition were 475/520 and 680/715 (excitation/emission) for C. auris and SPC, respectively. The PCR detector gain and threshold fluorescence for each channel were set at 50 and 100, respectively. Results were called positive if the C. auris target was amplified regardless of whether or not the SPC was amplified, negative if the C. auris target was not amplified and the SPC was amplified, and undetermined if the C. auris target was not amplified and the SPC was not amplified.
TABLE 1.
Primers and probes for real-time PCR for C. auris detection and sample processing control
| Primer name | Sequence (5′ to 3′)a | Target |
|---|---|---|
| C.auris | For: CGTGATGTCTTCTCACCAATCT | Partial sequence of ITS1 and 28S rRNA gene and complete sequence of 5.8S rRNA gene and ITS2 |
| Rev: TACCTGATTTGAGGCGACAAC | ||
| Pb:6-FAM/TTTGTGAAT/ZEN/GCAACGCCACCGC/IABkFQ | ||
| SPC | For: GGATCTAGCCGTGTGCCCGCT | Not disclosed by BD Max |
| Rev: GGCATGGAGGTTGTCCCATTTGTG | ||
| Pb:ATTO647N TGATGCCTCTTCACATTGCTCCACCTTTCCTBHQ3 |
For, forward; Rev, reverse; Pb, probe; SPC, sample processing control; ITS, internal transcribed spacer.
C. auris assay analytical sensitivity, specificity, and reproducibility.
The specificity and reproducibility of the assay were tested by different operators using 113 fungal isolates and 12 bacterial species. To determine the sensitivity of our new assay on the BD Max instrument, a series of seven 10-fold dilutions with a McFarland standard of 0.5 (2 × 106 CFU/ml) of C. auris AR0381, C. auris AR0385, and C. auris AR0389 from the CDC C. auris panel were tested in triplicate. The highest 10-fold dilution for which a threshold cycle CT value was observed was diluted further in a series of three 2-fold dilutions (1:2, 1:4, and 1:8) to find the lowest concentration at which a CT value was detected. The lowest concentration that produced a CT value was tested in triplicate to determine the limit of detection (LoD) of the assay on the BD Max.
Clinical sensitivity and specificity.
The clinical specificity and sensitivity of the assay was evaluated blindly in 50 leftover deidentified bronchoalveolar lavages, sputum, serum, urine, and wound swab samples. Fifteen of the samples were positive for C. haemulonii, C. parapsilosis, C. glabrata, C. albicans, C. tropicalis, and Curvularia spp. by the Vitek 2 (bioMérieux) or laboratory standard of care procedure for yeasts identification. The specificity of the assay was assessed on the 15 clinical specimens positive for yeasts other than C. auris and on 35 negative specimens spiked with 5 × 105 CFU of C. duobushaemulonii AR0391, C. haemulonii ATCC 22991, C. lusitaniae AR0398, and the C. auris AR0381 positive control. Clinical sensitivity and the LoD were tested on pooled specimens spiked with 500 CFU (high), 50 CFU (medium), and 5 CFU (low) of C. auris AR0381. In a separate experiment, 250 μl of pooled urine samples was inoculated with 500 CFU, 50 CFU, and 5 CFU of live or heat-killed C. auris to test the ability of the assay to assess C. auris infection or colonization over time. Unspiked specimens were used as controls. Cells were heat-killed at 95°C for 20 min and plated on Sabouraud dextrose agar; they were then incubated for 1 month to ascertain that the cells were actually not viable. Spiked and unspiked samples were either frozen at time of inoculation (time zero) or incubated at 37°C for up to 6 days. Samples were then tested as described above.
RESULTS
Primer design.
Primer-BLAST analysis showed that the primers/probe set was specific to C. auris isolates. They amplified a 111-bp fragment of the genomic regions of the 18S rRNA gene, the internal transcribed spacer 1, the 5.8S rRNA, the internal transcribed spacer 2, and partial sequence of the 28S rRNA of C. auris.
Assay specificity, sensitivity, and reproducibility.
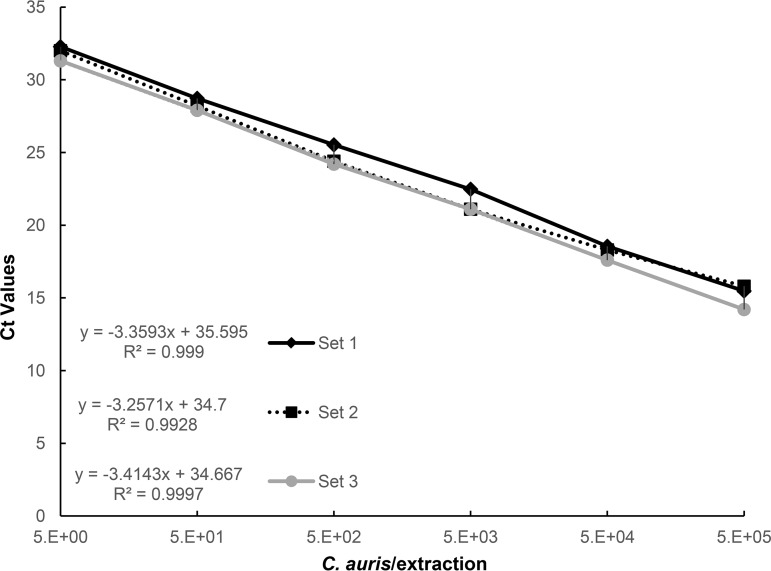
The data showed that the assay was highly sensitive with a PCR linearity of over 5 orders of magnitude with 99% efficiency and a limit of detection (LoD) as low as 1 CFU (CT = 34.3 ± 0.5); no CT value was observed at 5 × 10−1 CFU (Fig. 1). For the three 2-fold dilutions from the 5 × 100 dilution (highest 10-fold dilution for which a CT value was detected), we observed CT values of 33.4, 34.03, and 34.27 for 2.5 × 100 CFU, 1.3 × 100 CFU, and 0.6 × 100 CFU, respectively. Moreover, the specificity of the assay was determined by testing 113 fungal isolates, 10 of which were C. auris isolates, and 12 bacterial species. The data showed that the assay was highly specific and reproducible as C. auris isolates were consistently amplified while DNA from other species and organisms was not amplified (Table 2).
FIG 1.
Real-time PCR assay analytical sensitivity on the BD Max. C. auris cell dilution series from a 0.5 McFarland standard containing 5 × 105 to 5 × 100 C. auris CFU/250 μl extraction reaction mixture with a slope of −3.35 and a correlation coefficient of R2 = 0.99. The assay is linear over 5 orders of magnitude with a detection limit of 1 C. auris CFU/250 μl of reaction mixture using 40 cycles. No CT value was observed for 5 × 10−1 CFU/250 μl.
TABLE 2.
Analytical specificity of C. auris assay on the BD Max
| Species | No. of isolates detected/no. tested | CT valuesa |
|---|---|---|
| C. auris | 10/10 | 15.0 ± 0.5 |
| C. haemulonii | 0/4 | ND |
| C. duobushaemulonii | 0/3 | ND |
| C. lusitaniae | 0/2 | ND |
| C. albicans | 0/21 | ND |
| C. tropicalis | 0/10 | ND |
| C. krusei | 0/2 | ND |
| C. glabrata | 0/36 | ND |
| C. guilliermondii | 0/1 | ND |
| C. parapsilosis | 0/13 | ND |
| C. kefyr | 0/1 | ND |
| S. cerevisiae | 0/4 | ND |
| C. pelliculosa | 0/1 | ND |
| C. rugosa | 0/1 | ND |
| C. pararugosa | 0/1 | ND |
| Kodamaea ohmeri | 0/1 | ND |
| C. neoformans | 0/1 | ND |
| C. albidus | 0/1 | ND |
| P. aeruginosa | 0/1 | ND |
| B. cepacia | 0/1 | ND |
| E. coli | 0/1 | ND |
| K. pneumoniae | 0/1 | ND |
| H. influenzae | 0/1 | ND |
| N. meningitidis | 0/1 | ND |
| S. epidermidis | 0/1 | ND |
| S. aureus | 0/1 | ND |
| S. pneumoniae | 0/1 | ND |
| S. pyogenes | 0/1 | ND |
| Enterococcus faecalis | 0/1 | ND |
| Enterococcus durans | 0/1 | ND |
| Humanb | 0/1 | ND |
ND, not detected; 5 × 105 CFU/250 μl reaction mixture was used.
Leftover pooled serum DNA control.
Detection of C. auris in clinical specimen.
Our data showed that it was possible to detect C. auris in all concentrations and clinical specimen types evaluated, including the following: sputum, urine, wound swabs, and serum. The only exception was in serum specimens spiked with 5 CFU/250 μl. The assay was as sensitive at detecting C. auris in clinical specimens as it was when the yeast was resuspended in saline control (Table 3). No DNA amplification was observed in unspiked specimen controls, specimens positive for yeast other than C. auris, or specimens that were spiked with C. haemulonii, C. duobushaemulonii, and C. lusitaniae (Table 4).
TABLE 3.
Detection of C. auris in spiked clinical specimens on the BD Maxa
| CFU/250 μl specimen | Mean CT ± SD for: |
||||
|---|---|---|---|---|---|
| Saline | Serum | Urine | Sputum | Wound | |
| 5E + 02 | 26.93 ± 0.68 | 28.83 ± 1.63 | 25.15 ± 0.93 | 26.35 ± 0.92 | 26.78 ± 1.56 |
| 5E + 01 | 30.22 ± 1.65 | 32.55 ± 1.06 | 28.56 ± 1.72 | 29.05 ± 1.77 | 28.20 ± 0.14 |
| 5E + 00 | 33.27 ± 1.33 | NDb | 31.10 ± 3.49 | 32.70 ± 1.41 | 30.90 ± 2.05 |
C. auris was amplified by real-time PCR in spiked clinical specimen in 3 independent experiments.
ND, not detected; there was no detection in unspiked specimen control.
TABLE 4.
Clinical specificity of C. auris assay on the BD Max
| Specimen (no.) |
CT values for:a |
||||
|---|---|---|---|---|---|
| Serum | Urine | Sputum | Wound | BAL | |
| Spiked (5 × 105 CFU) | |||||
| C. auris AR0381 | 17.6 | 15.2 | 15.8 | 14.7 | 14.4 |
| C. haemulonii ATCC 22991 | ND | ND | ND | ND | ND |
| C. duobushaemulonii AR0391 | ND | ND | ND | ND | ND |
| C. lusitaniae AR0398 | ND | ND | ND | ND | ND |
| Positive on Vitek 2 | |||||
| C. haemulonii (1) | ND | ||||
| C. albicans (3) | ND | ND | ND | ||
| C. parapsilosis (2) | ND; ND | ||||
| C. glabrata (4) | ND; ND; ND; ND | ||||
| C. tropicalis (2) | ND | ND | |||
| Curvularia spp. (1) | ND | ||||
| Yeast (not C. neoformans) (2) | ND; ND | ||||
ND, not detected; BAL, bronchial lavages.
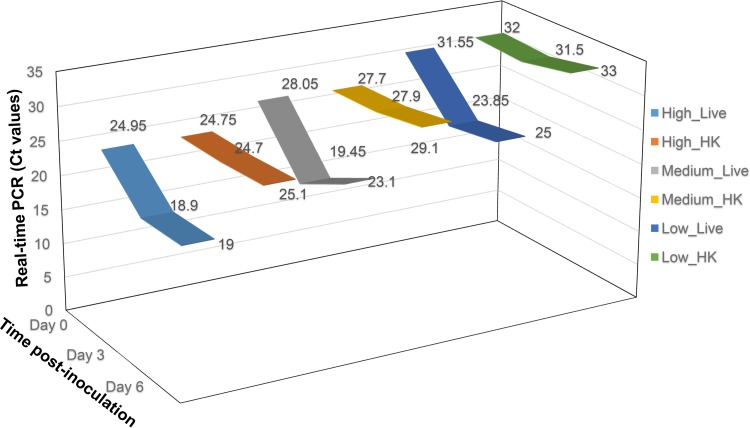
Moreover, we evaluated our assay in an ex vivo experiment mimicking C. auris infection in clinical specimens. A deidentified urine specimen was inoculated in triplicate with high, medium, and low CFU of live or heat-killed C. auris and incubated at 37°C for 6 days. The infection was assessed at time zero, day 3, and day 6 postspiking. The assay was capable of detecting an increase of 1,000-fold in C. auris burden at day 3 postinoculation (Fig. 2). The CT values for all three inoculating amounts decreased at day 3 for samples spiked with live cells. The decrease in CT values for the high CFU remained constant through 6 days. Although the CT values for the medium and low CFU went up by 1 to 2.5 CT from day 3, they were still lower than the CT values for the initial inoculation. On the other hand, CT values for the samples spiked with dead cells remained at inoculation level through 6 days. The constant CT values for heat-killed cells suggested that the DNA was not degraded.
FIG 2.
Detection of C. auris at different time points in clinical specimens. Urine samples inoculated with 500 CFU (high), 50 CFU (medium), and 5 CFU (low) of live or heat-killed C. auris. Real-time PCR data showed that the cells replicated at least 100-fold within 3 days and that the assay can detect not only DNA from live cells but also from dead cells at as low as 5 CFU, which persisted through the duration of the experiment (6 days). No amplification was observed in unspiked controls.
DISCUSSION
Candida species are among the leading causes of nosocomial bloodstream infection contributing significantly to morbidity and mortality. According to the CDC, as of December 2018, there have been 520 clinical cases of C. auris reported in 12 States in the United States, with New York, Illinois, and New Jersey having the most cases. An additional 975 patients have been found to be colonized with C. auris by targeted screening in 6 out of the 12 states with reported clinical cases (https://www.cdc.gov/fungal/candida-auris/tracking-c-auris.html). Most of those cases are individuals who have recently traveled to places outside of the United States where many cases of C. auris have been reported. In fact, strains of C. auris found in the United States have been linked to those parts of the world (https://www.cdc.gov/fungal/candida-auris/tracking-c-auris.html). As time goes by, we expect those numbers to rise. Therefore, there has been an urgent need for the development of reliable molecular assays for the detection of C. auris and for the implementation of infection control and prevention measures.
Recently, PCR-based methods for identification of C. auris infection have started to surface. Kordalewska et al. described a PCR assay based on melt curve analysis for the detection of C. auris (14). Most recently, while our assay was being developed, a number of molecular assays have been documented. Leach et al. described a TaqMan-based real-time PCR assay targeting the ITS2 region of the ribosomal gene for detection of C. auris from surveillance samples (15). Arastehfar et al. developed two multiplex PCR assays targeting the rDNA gene to detect and differentiate C. auris from closely related species as follows: an endpoint PCR assay validated using the mouse model and a fluorescent dyes-based reverse transcriptase PCR (RT-PCR) assay validated in serum samples spiked with DNA from the target species (16, 17). Although those assays are different in some ways, they share one common aspect in that DNA extraction is done manually, which can be time-consuming, labor-intensive, and in some cases limit the number of samples that can be processed at once. Ahmad et al. (18) validated a high-throughput DNA extraction method using dermal swabs samples for detection of C. auris using the assay developed by Leach et al. However, the use of a fully automated and integrated system for rapid molecular detection of C. auris is still lacking.
We developed an assay that is not only based on the TaqMan probe chemistry but also targets both the ITS1 and ITS2 of the ribosomal gene capable of rapidly detecting C. auris both from culture as well as directly from different types of patient specimens on the BD Max automated platform. This region has previously been targeted to not only show discrimination among Candida species but also to compare variations among closely related species (1, 19). As opposed to the previously described assays, in which DNA extraction is either done manually or separate from the PCR amplification platform, our assay is carried out on a fully integrated system that performs both nucleic acid extraction and real-time PCR providing results for as many as 24 samples within 3 h after culture or directly from clinical specimens. On the BD Max sample-to-answer platform, minimal hands-on time is required, reducing potential human error and contamination. Based on culture and evaluation on the Vitek 2, our assay has a 100% clinical sensitivity and specificity in a variety of specimen types, although with a smaller pool of samples compared to the other assays.
Whether from culture or directly from clinical specimens, our C. auris assay is highly specific, sensitive, and reproducible. The analytical specificity of the assay was established using a wide range of Candida isolates from different species as well as other organisms, including Gram-positive and Gram-negative bacteria. No cross-reactivity, false positives, or false negatives were observed within the parameters of the assay, even when closely related species to C. auris were tested. The assay was shown to be very sensitive with a limit of detection as low as 1 CFU/extraction and to have high PCR efficiency.
Fortunately, we have not found a C. auris-positive specimen in our hospital since this study started, and attempts to acquire C. auris-positive specimens have been unsuccessful so far. Therefore, we mimicked an infection environment by spiking different types of clinical specimens with known amounts of yeast and assessing the clinical sensitivity and specificity of our assay. The assay proved to be able to accurately detect C. auris in clinical specimens. However, the assay seemed to be more sensitive in detecting C. auris in the other specimen types than it was in serum, as the detection in serum was 2 CT values higher than in the other specimen types with the same inoculation dose. That and the inability to detect 5 CFU/250 μl of serum specimen may be due to some inhibitory effects, as the CT values for the sample processing control were higher for both serum and sputum compared to that of the other specimen types. As with any PCR-based assays, our assay can also detect DNA from dead cells. This confirms the shortfall of any PCR-based assay that DNA from dead cells can get amplified and a positive result does not necessarily mean an active infection. These data, nevertheless, show how highly sensitive this assay is and that an actual infection with C. auris can be monitored by running the assay on samples collected at different time points.
Although the assay uses mainly the BD Max platform, it can be adapted to any system capable of real-time PCR detection. For laboratories that do not have a sample-to-answer instrument, the yeast DNA can be extracted using an automated instrument or manually, and the template DNA can be used for amplification using the same cycling conditions as mentioned above in a thermocycler capable of real-time PCR detection. We will also continue developing the assay in different clinical samples as C. auris-positive specimen become available.
In all, the emergence of this nosocomial and multidrug-resistant Candida species has heightened concerns among many health care providers and infection control and prevention groups around the world and in the United States. This assay provides a fast and direct detection method from clinical specimens that takes no more than 15 min hands-on time and less than 2.5 h for results compared to culture methods, which take days and may result in misidentification of C. auris. This assay will provide health care groups and providers with a much needed tool to help with C. auris infection detection, control, and prevention.
ACKNOWLEDGMENTS
We thank the CDC for providing the Candida auris panel and the drug-resistant Candida panel.
REFERENCES
- 1.Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H. 2009. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol 53:41–44. doi: 10.1111/j.1348-0421.2008.00083.x. [DOI] [PubMed] [Google Scholar]
- 2.Chowdhary A, Sharma C, Duggal S, Agarwal K, Prakash A, Singh PK, Jain S, Kathuria S, Randhawa HS, Hagen F, Meis JF. 2013. New clonal strain of Candida auris, Delhi, India. Emerg Infect Dis 19:1670–1673. doi: 10.3201/eid1910.130393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Magobo RE, Corcoran C, Seetharam S, Govender NP. 2014. Candida auris-associated candidemia, South Africa. Emerg Infect Dis 20:1250–1251. doi: 10.3201/eid2007.131765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borman AM, Szekely A, Johnson EM. 2017. Isolates of the emerging pathogen Candida auris present in the UK have several geographic origins. Med Mycol 55:563–567. doi: 10.1093/mmy/myw147. [DOI] [PubMed] [Google Scholar]
- 5.Ruiz Gaitan AC, Moret A, Lopez Hontangas JL, Molina JM, Aleixandre Lopez AI, Cabezas AH, Mollar Maseres J, Arcas RC, Gomez Ruiz MD, Chiveli MA, Canton E, Peman J. 2017. Nosocomial fungemia by Candida auris: first four reported cases in continental Europe. Rev Iberoam Micol 34:23–27. doi: 10.1016/j.riam.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Morales-Lopez SE, Parra-Giraldo CM, Ceballos GA, Martinez HP, Rodriguez GJ, Alvarez-Moreno CA, Rodriguez JY. 2017. Invasive infections with multidrug-resistant yeast Candida auris, Colombia. Emerg Infect Dis 23:162–164. doi: 10.3201/eid2301.161497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCarthy M. 2016. Hospital transmitted Candida auris infections confirmed in the US. BMJ 355:i5978. doi: 10.1136/bmj.i5978. [DOI] [PubMed] [Google Scholar]
- 8.Vallabhaneni S, Kallen A, Tsay S, Chow N, Welsh R, Kerins J, Kemble SK, Pacilli M, Black SR, Landon E, Ridgway J, Palmore TN, Zelzany A, Adams EH, Quinn M, Chaturvedi S, Greenko J, Fernandez R, Southwick K, Furuya EY, Calfee DP, Hamula C, Patel G, Barrett P, Lafaro P, Berkow EL, Moulton-Meissner H, Noble-Wang J, Fagan RP, Jackson BR, Lockhart SR, Litvintseva AP, Chiller TM. 2017. Investigation of the first seven reported cases of Candida auris, a globally emerging invasive, multidrug-resistant fungus—United States, May 2013–August 2016. Am J Transplant 17:296–299. doi: 10.1111/ajt.14121. [DOI] [PubMed] [Google Scholar]
- 9.Kathuria S, Singh PK, Sharma C, Prakash A, Masih A, Kumar A, Meis JF, Chowdhary A. 2015. Multidrug-resistant Candida auris misidentified as Candida haemulonii: characterization by matrix-assisted laser desorption ionization-time of flight mass spectrometry and DNA sequencing and its antifungal susceptibility profile variability by Vitek 2, CLSI broth microdilution, and Etest method. J Clin Microbiol 53:1823–1830. doi: 10.1128/JCM.00367-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizusawa M, Miller H, Green R, Lee R, Durante M, Perkins R, Hewitt C, Simner PJ, Carroll KC, Hayden RT, Zhang SX. 2017. Can multidrug-resistant Candida auris be reliably identified in clinical microbiology laboratories? J Clin Microbiol 55:638–640. doi: 10.1128/JCM.02202-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghosh AK, Paul S, Sood P, Rudramurthy SM, Rajbanshi A, Jillwin TJ, Chakrabarti A. 2015. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry for the rapid identification of yeasts causing bloodstream infections. Clin Microbiol Infect 21:372–378. doi: 10.1016/j.cmi.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 12.Girard V, Mailler S, Chetry M, Vidal C, Durand G, van Belkum A, Colombo AL, Hagen F, Meis JF, Chowdhary A. 2016. Identification and typing of the emerging pathogen Candida auris by matrix-assisted laser desorption ionisation time of flight mass spectrometry. Mycoses 59:535–538. doi: 10.1111/myc.12519. [DOI] [PubMed] [Google Scholar]
- 13.Prakash A, Sharma C, Singh A, Kumar Singh P, Kumar A, Hagen F, Govender NP, Colombo AL, Meis JF, Chowdhary A. 2016. Evidence of genotypic diversity among Candida auris isolates by multilocus sequence typing, matrix-assisted laser desorption ionization time-of-flight mass spectrometry and amplified fragment length polymorphism. Clin Microbiol Infect 22:277.e1–227.e9. doi: 10.1016/j.cmi.2015.10.022. [DOI] [PubMed] [Google Scholar]
- 14.Kordalewska M, Zhao Y, Lockhart SR, Chowdhary A, Berrio I, Perlin DS. 2017. Rapid and accurate molecular identification of the emerging multidrug-resistant pathogen Candida auris. J Clin Microbiol 55:2445–2452. doi: 10.1128/JCM.00630-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leach L, Zhu Y, Chaturvedi S. 2018. Development and validation of a real-time PCR assay for rapid detection of Candida auris from surveillance samples. J Clin Microbiol 56:e01223-17. doi: 10.1128/JCM.01223-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arastehfar A, Fang W, Badali H, Vaezi A, Jiang W, Liao W, Pan W, Hagen F, Boekhout T. 2018. Low-cost tetraplex PCR for the global spreading multi-drug resistant fungus, Candida auris and its phylogenetic relatives. Front Microbiol 9:1119. doi: 10.3389/fmicb.2018.01119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arastehfar A, Fang W, Daneshnia F, Al-Hatmi AM, Liao W, Pan W, Khan Z, Ahmad S, Rosam K, Lackner M, Lass-Florl C, Hagen F, Boekhout T. 2019. Novel multiplex real-time quantitative PCR detecting system approach for direct detection of Candida auris and its relatives in spiked serum samples. Future Microbiol 14:33–45. doi: 10.2217/fmb-2018-0227. [DOI] [PubMed] [Google Scholar]
- 18.Ahmad A, Spencer JE, Lockhart SR, Singleton S, Petway DJ, Bagarozzi DA Jr, Herzegh OT. 2019. A high-throughput and rapid method for accurate identification of emerging multidrug-resistant Candida auris. Mycoses 23:12907. doi: 10.1111/myc.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Groenewald M, Robert V, Smith MT. 2011. The value of the D1/D2 and internal transcribed spacers (ITS) domains for the identification of yeast species belonging to the genus Yamadazyma. Persoonia 26:40–46. doi: 10.3767/003158511X559610. [DOI] [PMC free article] [PubMed] [Google Scholar]