Accurate and timely identification of carbapenemase-producing Enterobacteriaceae (CPE) is critical for microbiology laboratories in order to facilitate infection prevention, reduce the spread of multiresistant Gram-negative bacilli, and reduce delays to effective antibiotic therapy. We undertook a study to compare the carbapenem inactivation method (CIM) against the modified carbapenem inactivation method (mCIM) on a broad range of CPE isolates from Australia, including a high proportion of blaIMP isolates.
KEYWORDS: Enterobacteriaceae, carbapenemase
ABSTRACT
Accurate and timely identification of carbapenemase-producing Enterobacteriaceae (CPE) is critical for microbiology laboratories in order to facilitate infection prevention, reduce the spread of multiresistant Gram-negative bacilli, and reduce delays to effective antibiotic therapy. We undertook a study to compare the carbapenem inactivation method (CIM) against the modified carbapenem inactivation method (mCIM) on a broad range of CPE isolates from Australia, including a high proportion of blaIMP isolates. Furthermore, we evaluated the performance of the mCIM with a reduced incubation time using automated incubation and digital plate reading in order to better facilitate quick confirmation of carbapenemases. The overall sensitivity of the mCIM was 98.2%, compared to 95.6% for the CIM. The minimum incubation time for the mCIM while maintaining its sensitivity was 12 hours. Both the CIM and mCIM perform well on a broad range of CPE isolates seen in Australia.
INTRODUCTION
The emergence and spread of resistant Gram-negative organisms represent significant threats to health care services and patient outcomes. Infections caused by carbapenemase-producing Enterobacteriaceae (CPE) result in increased patient mortality and morbidity (1, 2). This can result in significant excess health care-associated costs for patient management and also for outbreak control (3, 4). The rapid identification of transmissible resistance genes, particularly those that encode carbapenemases, is of critical importance to help prevent the spread of these organisms within health care settings, as well as to guide individual therapy and shape public health responses. Identification of genetic resistance mechanisms by nucleic acid amplification testing (NAAT) remains the gold standard for CPE confirmation and is generally used as a basis for epidemiological analysis and infection prevention and control intervention.
Despite the expanding role of molecular resistance detection, a significant role still exists for phenotypic detection of CPE prior to molecular confirmation. Phenotypic carbapenemase detection provides a cost-effective screening process that excludes carbapenem-resistant Enterobacteriaceae (CRE) that do not express carbapenemase enzymes and therefore do not require more expensive molecular testing. Phenotypic carbapenemase detection is also not limited by known genetic mutations or the number of molecular targets able to be assessed. The ideal phenotypic screening test therefore requires a high sensitivity for the detection of CPEs, a short turnaround time to facilitate rapid detection for infection prevention and outbreak investigation, and cost effectiveness for the microbiology laboratory. Additionally, the screening test needs to perform accurately over a range of carbapenem MICs, given that research has previously demonstrated that transmissible carbapenem resistance genes can be seen in organisms with low meropenem MICs, including those below the intermediate breakpoint (5).
A number of phenotypic carbapenemase screening methods have been utilized, including disc synergy testing, modified Hodge testing, acidimetric tests (including the Carba NP assay), and synergy-based gradient antibiotic susceptibility testing (6–8). All of these have limitations, including reduced sensitivity, long turnaround time, and variable performance across a range of phenotypes and genotypes (8–12). In 2015, van der Zwaluw et al. described a new method for carbapenemase detection based on the in vitro inactivation of meropenem by hydrolysis, the carbapenem inactivation method (CIM) (13). The CIM was shown to be rapid, inexpensive, simple to perform, and to have a sensitivity of 100% for CPE. Based on these results, the CIM was rapidly adopted into practice. However, in 2017, in response to apparent issues with test sensitivity found by some researchers, a modification of the CIM method (mCIM) was described and reported to have improved sensitivity compared to the CIM (14). For example, a comparison study of phenotypic CPE tests reported poor sensitivity of the CIM for the detection of blaKPC, blaNDM, blaOXA48, and blaIMP genotypes (15). Detection of these carbapenemases was better with the mCIM, particularly for blaKPC, blaNDM, and blaOXA48 genotypes, although apparently not for the blaIMP genotype. In contrast, Japanese researchers reported 100% sensitivity of the CIM test for detection of blaIMP-positive Enterobacteriaceae in their study (16). Therefore, the relative performance of the CIM and mCIM for blaIMP genotypes remains unclear. This is important in Australia, where blaIMP has to date been the most frequently detected CPE genotype (17–19).
Additional characteristics of the mCIM test can have impacts for the diagnostic laboratory. The mCIM prolongs the minimum incubation time for the test from 8 hours (the minimum incubation time for the CIM) to 22 hours (a result of increasing the disc incubation step from 2 hours to 4 hours and increasing the incubation step from a minimum of 6 hours to a minimum of 18 hours), potentially introducing clinically significant delays. Another major difference between the CIM and mCIM is that the mCIM includes an indeterminate category, which could increase the number of tests needed to proceed to additional phenotypic test or genotypic confirmatory testing.
Given the importance of rapid turnaround times and the high sensitivity for phenotypic testing, and given the uncertain impact of geographic variations in CPE phenotypes and genotypes on performance and workflow, we undertook a study to compare the performance of the CIM and mCIM on a range of local isolates and to validate the interpretation of the mCIM with a shorter incubation time.
MATERIALS AND METHODS
Enterobacteriaceae isolates and confirmation of MIC and carbapenemase genotype.
A range of clinical infection and infection control screening isolates with known carbapenemase genotypes from previous molecular testing were included in the study. Isolates were chosen to ensure a broad range of Enterobacteriaceae genera and species and a complete range of known locally prevalent CPE genotypes. Isolates were chosen from a collection of clinical isolates obtained between January 2008 and September 2017 and stored frozen at −80 degrees in the Department of Microbiology and Infectious Diseases (Liverpool Hospital) and the Centre of Infectious Diseases and Microbiology (Westmead Hospital). This collection was supplemented with prospectively collected clinical isolates from between September 2017 and March 2018. All isolates included were Enterobacteriaceae with meropenem MICs of ≥0.25 μg/ml as confirmed by Etest (bioMérieux, Mary-Etoile France). Stored isolates were subcultured from −80°C stock cultures in tryptic soy broth (TSB) with 5% horse blood and passaged twice prior to repeat phenotypic testing to confirm identity and susceptibility. Phenotypic testing was performed on 18- to 24-hour cultures. Meropenem MIC values were determined by gradient diffusion testing using Etest (bioMérieux, Mary-Etoile France).
All isolates included in the study were formally retested during the study to confirm the genetic mechanism of resistance (carbapenemase genotype) by NAAT using the AusDiagnostics CRE Panel (AusDiagnostics, Mascot, Australia). This multiplex panel included targets for blaVIM, blaIMP, blaKPC, blaNDM-1, blaIMI, blaSME, blaGES, blaOXA23-like, blaOXA48-like, blaOXA51-like, and blaOXA58-like carbapenemases.
Carbapenemase inhibition method.
The CIM test was performed per the methodology described by van der Zwaluw et al. (13). Briefly, a10-μl loop of test organism was suspended in 400 μl of sterile water and vortexed for 15 s. A 10-μg meropenem disc (Oxoid, Basingstoke, UK) was added to the suspension, and incubation was performed for 2 h ± 15 min. A Mueller-Hinton Agar (MHA) plate was inoculated by streaking a 0.5 MacFarland suspension of a meropenem-susceptible strain of Escherichia coli (ATCC strain 25922) in three directions over the entire surface of the plate. A sterile loop was used to transfer the meropenem disc onto the plate, and the plate was incubated for 18 hours at 37°C in aerobic conditions in the BD Kiestra Work Cell incubator. Plates were imaged at 2-hour intervals (commencing at 6 hours), without removing the plate from the incubator, using the BD Kiestra ReadA Browser software. Zones of inhibition were measured by the investigator using the zone diameter tool in the BD ReadA browser program, where zones are drawn over the zone of inhibition by the investigator and the diameter calculated by the software. The CIM was interpreted as positive (indicating the presence of a carbapenemase) when no zone of inhibition was seen around the meropenem disc and as negative (indicating the absence of a carbapenemase) when any zone of inhibition was measurable. The CIM can be read at any time after 6 hours of incubation, and for the purposes of this study the definitive reading time was set at 18 hours.
Modified carbapenemase inhibition method.
The mCIM was set up as described in the Clinical and Laboratory Standards Institute (CLSI) M100 Performance Standards for Antimicrobial Susceptibility Testing (20). Briefly, a calibrated 1-μl loop of test organism was suspended in 2 ml of tryptic soy broth (TSB) and vortexed for 15 s. A 10-μg meropenem disc (Oxoid, Basingstoke, UK) was added to the suspension and incubated for 4 hours ± 15 min at 35°C. A Mueller-Hinton Agar (MHA) plate was inoculated by streaking a 0.5 MacFarland suspension of a meropenem-susceptible strain of Escherichia coli (ATCC strain 25922) in three directions over the entire surface of the plate. A sterile loop was used to transfer the meropenem disc onto the plate, and the plate was incubated for 18 hours at 37°C in aerobic conditions in the BD Kiestra Work Cell incubator. Plates were imaged at 2-hour intervals (commencing at 6 hours), without removing the plate from the incubator, using the BD Kiestra ReadA Browser software. Zones of inhibition were measured by the investigator using the zone diameter tool in the BD ReadA browser program. The mCIM was interpreted as positive (indicating the presence of a carbapenemase) when the zone of inhibition around the meropenem disc was ≤15 mm; negative (indicating the absence of carbapenemase) when the zone of inhibition was ≥19 mm; and indeterminate when the zone of inhibition was between 16 and 18 mm. The definitive reading time for the mCIM was after 18 hours incubation in accordance with the CLSI guideline.
For the purposes of the study, the plate incubation time for the mCIM was modified to be interpreted after 6 hours, 8 hours, 10 hours, 12 hours, 14 hours, and 16 hours of incubation to see if the incubation time of the test could be reduced while maintaining accuracy.
RESULTS
A total of 160 isolates were identified as Enterobacteriaceae with a meropenem MIC of ≥0.25μg/ml and underwent genotyping by NAAT, as well as phenotypic carbapenemase testing by CIM and mCIM. A total of 107 retrospectively and prospectively collected isolates were included from the Department of Microbiology and Infectious Diseases (Liverpool Hospital), which comprised 61 clinical isolates and 46 infection control screening isolates, as well as 53 isolates from the Centre of Infectious Diseases and Microbiology (Westmead Hospital). NAAT confirmed the presence of one or more carbapenemase genes in 137 isolates and the absence of a carbapenemase gene in 23 isolates. The 137 CPE organisms included 55 blaIMP isolates, 29 blaOXA48-like isolates, 23 blaKPC isolates, 13 blaVIM isolates, 9 blaNDM-1 isolates, 6 combined blaNDM-1 and blaOXA48-like isolates, 1 combined blaNDM-1 and blaKPC isolate, and 1 combined blaIMP and blaOXA23 isolate. Details of the isolates can be seen in Table 1.
TABLE 1.
Isolate characteristics and results of CIM and mCIM at 18 h of plate incubation
| Carbapenemase gene | Species | No. of isolates | Meropenem MIC (μg/ml) | CIM (no. of results) |
mCIM (no. of results) |
% susceptibilitya |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Indeterminate | Negative | S | I | R | ||||
| blaIMP | All blaIMP species | 55 | 0.25 to ≥32 | 54 | 1 | 54 | 1 | 49.1 | 18.2 | 32.7 | |
| Citrobacter braakii | 1 | 1 | 1 | 1 | |||||||
| Citrobacter farmeri | 1 | 1 | 1 | 1 | |||||||
| Citrobacter freundii | 11 | 1 to 8 | 1 | 11 | |||||||
| Citrobacter koseri | 1 | 1 | 1 | 1 | |||||||
| Enterobacter aerogenes | 2 | 1 to 8 | 2 | 2 | |||||||
| Enterobacter cloacae | 14 | 0.25 to 4 | 13 | 1 | 13 | 1 | |||||
| Escherichia coli | 1 | >32 | 1 | 1 | |||||||
| Klebsiella oxytoca | 5 | 0.25 to 2 | 5 | 5 | |||||||
| Klebsiella pneumoniae | 16 | 0.25 to >32 | 16 | 16 | |||||||
| Morganella morganii | 2 | 0.25 | 2 | 2 | |||||||
| Proteus mirabilis | 1 | 16 | 1 | 1 | |||||||
| blaIMP blaOXA23 | Citrobacter koseri | 1 | 0.5 | 1 | 1 | 100 | 0 | 0 | |||
| blaKPC | All blaKPC species | 23 | 0.5 to >32 | 23 | 23 | 17.4 | 4.3 | 78.3 | |||
| Escherichia coli | 2 | 0.5 to 1 | 2 | 2 | |||||||
| Klebsiella pneumoniae | 21 | 1 to >32 | 21 | 21 | |||||||
| blaKCP blaNDM-1 | Klebsiella pneumoniae | 1 | >32 | 1 | 1 | 0 | 0 | 100 | |||
| blaNDM-1 | All blaNDM-1 species | 9 | 8 to >32 | 9 | 9 | 0 | 0 | 100 | |||
| Escherichia coli | 4 | 9 to >32 | 4 | 4 | |||||||
| Klebsiella pneumoniae | 5 | 16 to >32 | 5 | 5 | |||||||
| blaNDM-1 blaOXA48-like | All blaNDM-1 blaOXA48-like species | 6 | 16 to >32 | 6 | 6 | 0 | 0 | 100 | |||
| Escherichia coli | 1 | >32 | 1 | 1 | |||||||
| Klebsiella pneumoniae | 5 | 16 to >32 | 5 | 5 | |||||||
| blaOXA48-like | All blaOXA48 species | 29 | 0.25 to >32 | 25 | 4 | 28 | 1 | 41.4 | 6.9 | 51.7 | |
| Enterobacter aerogenes | 1 | 2 | 1 | 1 | |||||||
| Escherichia coli | 14 | 0.25 to 4 | 11 | 3 | 14 | ||||||
| Klebsiella pneumoniae | 14 | 1 to >32 | 13 | 1 | 13 | 1 | |||||
| blaVIM | All blaVIM species | 13 | 0.25 to 32 | 12 | 1 | 13 | 23.1 | 30.8 | 46.2 | ||
| Enterobacter cloacae | 3 | 2 to 32 | 3 | 3 | |||||||
| Escherichia coli | 3 | 0.25 to 2 | 3 | 3 | |||||||
| Klebsiella pneumoniae | 6 | 0.5 to 16 | 6 | 6 | |||||||
| Proteus mirabilis | 1 | 2 | 1 | 1 | |||||||
| Non-CPE | All non-CPE species | 23 | 0.25 to >32 | 56.5 | 13.0 | 30.4 | |||||
| Enterobacter aerogenes | 3 | 0.25 to >32 | 3 | 1 | 2 | ||||||
| Enterobacter asburiae | 1 | 0.25 | 1 | 1 | |||||||
| Enterobacter cloacae | 5 | 0.5 to >32 | 5 | 1 | 4 | ||||||
| Escherichia coli | 2 | 0.25 to 2 | 2 | 2 | |||||||
| Klebsiella pneumoniae | 9 | 0.25 to >32 | 9 | 9 | |||||||
| Morganella morganii | 2 | 0.25 | 2 | 2 | |||||||
| Proteus mirabilis | 1 | 0.25 | 1 | 1 | |||||||
Susceptibility breakpoints per CLSI Performance Standards for Antimicrobial Susceptibility Testing (20). S, susceptible; I, intermediate; R, resistant.
After 18 hours of disc incubation, the CIM correctly identified 131 of the 137 carbapenemase-producing isolates (95.6% agreement), and 23 of 23 non-carbapenemase-producing isolates (100% agreement). The mCIM correctly identified 135 of the 137 CPE isolates (98.5% agreement), and 20 of the 23 non-CPE isolates (87.0% agreement), with the 3 incorrectly classified isolates returning indeterminate results after 18 hours of disc incubation. None of the carbapenemase-negative isolates were classified as positive by the mCIM, and none of the carbapenemase-positive isolates were classified as indeterminate. The percent agreement to molecular testing of the CIM and mCIM for different genotypes is shown in Table 2. Both tests correctly identified all blaIMP blaOXA23, blaKPC, blaKCP blaNDM-1, and blaNDM-1 blaOXA48-like carbapenemases. One blaIMP Enterobacter cloacae isolate with an MIC of 0.25 μg/ml tested negative in both the CIM and mCIM assays. The mCIM performed better than the CIM for blaOXA48-like CPE (agreement, 96.6% versus 86.2%) and for blaVIM CPE (agreement, 100% versus 92.3%) isolates. Discrepant isolates are detailed in Table 3. The median zone of inhibition diameter for non-CPE isolates with the CIM test was 25 mm, while the median zone of inhibition diameter with the mCIM was 20 mm.
TABLE 2.
Positive percent agreement of CIM and mCIM with NAAT after 18 h of plate incubation by carbapenemase genotype
| CPE gene | No. of isolates | CIM |
mCIM |
||
|---|---|---|---|---|---|
| No. positive | % positive | No. positive | % positive | ||
| All CPE genes | 137 | 131 | 95.6 | 135 | 98.5 |
| blaIMP | 55 | 54 | 98.2 | 54 | 98.2 |
| blaIMP blaOXA23 | 1 | 1 | 100 | 1 | 100 |
| blaKPC | 23 | 23 | 100 | 23 | 100 |
| blaKCP blaNDM-1 | 1 | 1 | 100 | 1 | 100 |
| blaNDM-1 | 9 | 9 | 100 | 9 | 100 |
| blaNDM-1 blaOXA48-like | 6 | 6 | 100 | 6 | 100 |
| blaOXA48-like | 29 | 25 | 86.2 | 28 | 96.6 |
| blaVIM | 13 | 12 | 92.3 | 13 | 100 |
TABLE 3.
CPE isolates with discrepant results between CIM and mCIM
| Organism | Meropenem MIC (μg/ml) | Carbapenemase gene | CIM |
mCIM |
||
|---|---|---|---|---|---|---|
| Zone (mm) | Result | Zone (mm) | Result | |||
| Proteus mirabilis | 2 | blaVIM | 21 | Negative | 6 | Positive |
| Escherichia coli | 0.5 | blaOXA48-like | 17 | Negative | 6 | Positive |
| Escherichia coli | 0.25 | blaOXA48-like | 14 | Negative | 6 | Positive |
| Escherichia coli | >32 | blaOXA48-like | 15 | Negative | 15 | Positive |
For the evaluation of CIM and mCIM at reduced incubation times, the zones of inhibition were read every 2 hours from 6 to 18 hours. There was no change in the interpretation of the CIM at any time after 6 hours of incubation. The CIM correctly identified 95.6% of CPE isolates and 100% of non-CPE isolates at 6 hours. In contrast, the mCIM missed 1 CPE isolate at 6 hours that was subsequently correctly classified after 8 hours of incubation (Fig. 1). Therefore, the mCIM reached maximum agreement (98.2%) after 8 hours incubation (Table 4). When the indeterminate mCIM results were included as positives (as they would be when this test is used as a screening test prior to NAAT confirmation), 98.2% of CPE isolates and 87.0% of non-CPE isolates were correctly identified after 8 hours incubation with the mCIM test.
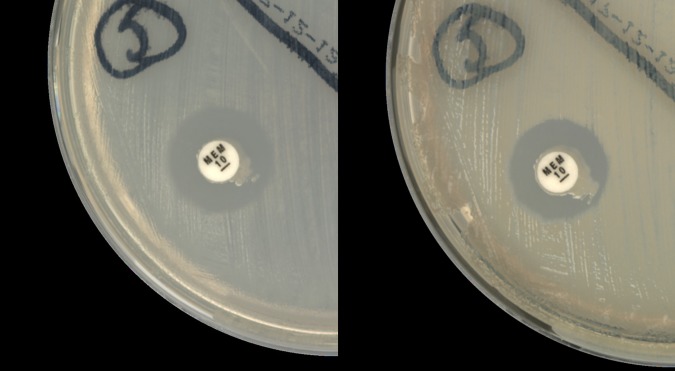
FIG 1.
Comparison of indeterminate mCIM result at 6 h with a zone size of 16 mm (left) and positive result at 8 h with a zone size of 15 mm (right). The isolate was a blaIMP Klebsiella pneumoniae strain with an MIC of >32 μg/ml.
TABLE 4.
Proportion of CIM and mCIM tests classified correctly at different plate incubation times
| Enterobacteriaceae group | Test | Incubation time |
||||||
|---|---|---|---|---|---|---|---|---|
| 6 h (%) |
8 h (%) |
10 h (%) |
12 h (%) |
14 h (%) |
16 h (%) |
18 h (%) |
||
| CPE | CIM | 95.6 | 95.6 | 95.6 | 95.6 | 95.6 | 95.6 | 95.6 |
| mCIM | 97.8 | 98.5 | 98.5 | 98.5 | 98.5 | 98.5 | 98.5 | |
| Non-CPE | CIM | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| mCIM | 91.3 | 87.0 | 87.0 | 87.0 | 87.0 | 87.0 | 87.0 | |
DISCUSSION
The findings presented here confirm that the phenotypic carbapenem inactivation method is a sensitive method of screening for carbapenemase production in Enterobacteriaceae. The test performs well across a broad range of phenotypes and genotypes, and, in particular, it performs well for local Australian isolates and can therefore be recommended as an effective component of local CPE screening protocols. In contrast to previously reported reduced detection of blaKPC, blaNDM-1, and blaIMP genotypes by the original CIM test, we found that the test performed well for these genotypes. But we found that the CIM may not detect all blaOXA48-like carbapenemase-producing organisms. On the other hand, the mCIM detected more blaOXA48-like CPE isolates than did the CIM. The increased percent agreement to molecular testing of the mCIM in CPE isolates (98.5% versus 95.6%) was, however, associated with a reduced percentage agreement (87% versus 100%) in non-CPE isolates, and this loss of specificity resulted from several non-CPE isolates (3 isolates) producing indeterminate results. For a laboratory that does not confirm mCIM-positive isolates by NAAT, there could be misclassification for these indeterminate organisms, or the status of these organisms would not be resolved. However, this would not be a problem in a lab that routinely confirms all positive or indeterminate isolates by NAAT or that accepts positive results as tested and confirms any indeterminate results by NAAT.
Our findings also support reduced incubation time for the mCIM in the setting of automated incubation and digital plate reading. No changes in category call were seen between incubation times of 8 hours and 18 hours, allowing significantly reduced overall turnaround time of the test from a minimum of 22 hours to 12 hours (8 hours of incubation plus 4 hours of disc setup and hydrolysis), potentially aiding patient care and infection control measures. This reduction in incubation time may also facilitate better laboratory testing workflow, particularly when NAAT confirmation is required before reporting the result to the clinician. However, our study suggests that reducing the incubation time of the mCIM to 6 hours (to align with the CIM minimum incubation time) would not be advisable, as this may lead to some false-negative results.
The relative strengths of this study are the large number and broad variety of genotypes and phenotypes tested, including those of organisms less well covered in previous studies. The results for mCIM sensitivity are similar to findings from Yamada et al. (21), and both the larger number of isolates and range of genotypes we have included in our study make these data robust. In particular, our study included a large number of blaIMP-positive isolates and demonstrated that both the CIM and mCIM have a high percent agreement with molecular methods for blaIMP, in contrast to previous research, which had shown reduced sensitivity of carbapenemase inactivation for blaIMP (21). The large number of blaIMP isolates included in our study, and good correlation with previous research that demonstrated a high sensitivity for CIM with blaIMP, confirms the utility of this test in detecting blaIMP (16). The limitations of the study are that many of the isolates were collected from a relatively small geographical area, increasing the chance that the isolates might be epidemiologically linked and therefore of reduced genetic variability. The use of retrospective isolates also favors Enterobacteriaceae that had already been identified as CPE, which thus may be more likely to test positive on phenotypic testing. However, we attempted to overcome this limitation by also using stored Enterobacteriaceae isolates that had previously tested negative for carbapenemase activity and by prospective enrollment of organisms that would enter the CPE confirmation pathway. As the isolates used in the study were not prospectively collected for the purposes of screening, definite conclusions about the use of CIM and mCIM for screening cannot be drawn, but rather this serves as a comparability study for carbapenem inactivation against genotypic testing. Furthermore, our study included the use of automated incubation and digital plate reading in the BD Kiestra Work Cell incubator, which may have more stable environmental conditions, as the incubator is not opened during incubation and imaging. As automated incubation is not currently available in many laboratories, our results may not be generalizable to laboratories that do not use these technologies.
Carbapenem inactivation remains a simple and accurate method for phenotypically detecting carbapenemases. When assessing whether the original CIM or the mCIM is appropriate for laboratories, the local CPE epidemiology should be considered, as a high local prevalence of blaOXA48-like enzymes would favor the use of mCIM over CIM and may make the use of CIM inappropriate. Other factors to be considered include the required turnaround time for the test, as well as the ease of disc reading. When interpreting the test, we found that interpretation of the mCIM was more difficult, as the zone of inhibition for many isolates was close to the zone diameter cutoffs. The median zone of inhibition diameter for non-CPE isolates with the CIM test was 25 mm, while the median zone of inhibition diameter with the mCIM was 20 mm, with many isolates sitting within 1 to 2 mm of the indeterminate zone cutoff. More recently, CLSI has also addressed the issues of microcolonies, which can affect the reading of the mCIM but which also increase the complexity of interpretation (20). For laboratories using mCIM, the plate incubation step can safely be reduced to 8 hours, reducing the overall turnaround time of the test.
ACKNOWLEDGMENTS
We acknowledge the contributions of the Centre for Infectious Diseases & Microbiology (CIDM) at the Institute of Clinical Pathology & Medical Research (ICPMR) Westmead, Australia, for the contribution of many of the isolates used in this study.
Funding was provided by the Department of Infectious Diseases and Microbiology at Liverpool Hospital.
REFERENCES
- 1.Tumbarello M, Viale P, Viscoli C, Trecarichi EM, Tumietto F, Marchese A, Spanu T, Ambretti S, Ginocchio F, Cristini F, Losito AR, Tedeschi S, Cauda R, Bassetti M. 2012. Predictors of mortality in bloodstream infections caused by Klebsiella pneumoniae carbapenemase-producing K. pneumoniae: importance of combination therapy. CID 55:943–950. doi: 10.1093/cid/cis588. [DOI] [PubMed] [Google Scholar]
- 2.Borer A, Saidel-Odes L, Riesenberg K, Eskira S, Peled N, Nativ R, Schlaeffer R, Sherf M. 2009. Attributable mortality rate for carbapenem-resistant Klebsiella pneumoniae bacteraemia. Infect Control Hosp Epidemiol 30:972–976. doi: 10.1086/605922. [DOI] [PubMed] [Google Scholar]
- 3.Zilberberg MD, Nathanson BH, Sulham K, Weihong F, Shorr AF. 2017. Carbapenem resistance, inappropriate empiric treatment and outcomes among patients hospitalized with Enterobacteriaceae urinary tract infection, pneumonia and sepsis. BMC Infect Dis 17:279. doi: 10.1186/s12879-017-2383-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Otter JA, Burgess P, Davies F, Mookerjee S, Singleton J, Gilchrist M, Parsons D, Brannigan ET, Robotham J, Holmes AH. 2017. Counting the cost of an outbreak of carbapenemase-producing Enterobacteriaceae: and economic evaluation from a hospital perspective. Clin Microbiol Infec 23:188–196. doi: 10.1016/j.cmi.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Fattouh R, Tijet N, McGeer A, Poutanen SM, Melano RG, Patel SN. 2016. What is the appropriate meropenem MIC for screening of carbapenemase-producing Enterobacteriaceae in low-prevalence settings? Antimicrob Agents Chemother 60:1556–1559. doi: 10.1128/AAC.02304-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nordmann P, Poirel L, Dortet L. 2012. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 18:1503–1507. doi: 10.3201/eid1809.120355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hrabal J, Chudackova E, Papagiannnitsis CC. 2014. Detection of carbapenemases in Enterobacteriaceae: a challenge for diagnostic microbiological laboratories. Clin Microbiol Infect 20:839–853. doi: 10.1111/1469-0691.12678. [DOI] [PubMed] [Google Scholar]
- 8.Nordmann P, Poirel L. 2013. Strategies for identification of carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother 68:487–489. doi: 10.1093/jac/dks426. [DOI] [PubMed] [Google Scholar]
- 9.Literacka E, Herda M, Baraniak A, Żabicka D, Hryniewicz W, Skoczyńska A, Gniadkowski M. 2017. Evaluation of the Carba NP test for carbapenemase detection in Enterobacteriaceae, Pseudomonas spp. and Acinetobacter spp., and its practical use in the routine work of a national reference laboratory for susceptibility testing. Eur J Clin Microbiol Infect Dis 36:2281–2287. doi: 10.1007/s10096-017-3062-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tijet N, Boyd D, Patel SN, Mulvey MR, Melano RG. 2013. Evaluation of the Carba NP test for rapid detection of carbapenemase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother 57:4578–4580. doi: 10.1128/AAC.00878-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galani I, Rekatsina PD, Hatzaki D, Plachouras D, Souli M, Giamarellou H. 2008. Evaluation of different laboratory tests for the detection of metallo-β-lactamases in Enterobacteriaceae. J Antimicrob Chemother 61:548–553. doi: 10.1093/jac/dkm535. [DOI] [PubMed] [Google Scholar]
- 12.Girlich D, Poirel L, Nordmann P. 2012. Value of the modified Hodge test for detection of emerging carbapenemases in Enterobacteriaceae. J Clin Microbiol 50:477–479. doi: 10.1128/JCM.05247-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van der Zlauw K, de Haan A, Pluister GN, Bootsma HJ, de Neeling AJ, Schouls LM. 2015. The carbapenem inactivation method (CIM), a simple and low-cost alternative for the Carba NP test to assess phenotypic carbapenemase activity in Gram-negative rods. PLoS One 10:e0123690. doi: 10.1371/journal.pone.0123690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pierce VM, Simner PJ, Lonsway DR, Roe-Carpenter DE, Johnson JK, Brasso WB, Bobenchik AM, Lockett ZC, Charnot-Katsikas A, Ferraro MJ, Thomson RB Jr, Jenkins SG, Limbago BM, Das S. 2017. Modified carbapenem inactivation method for phenotypic detection of carbapenemase production among Enterobacteriaceae. J Clin Microbiol 55:2321–2333. doi: 10.1128/JCM.00193-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamma PD, Opene BNA, Gluck A, Chambers KK, Carroll KC, Simner PJ. 2017. Comparison of 11 phenotypic assays for accurate detection of carbapenemase-producing Enterobacteriaceae. J Clin Microbiol 55:1046–1055. doi: 10.1128/JCM.02338-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saito K, Nakano R, Suzuki Y, Nakano A, Ogawa Y, Yonekawa S, Endo S, Mizuno F, Kasahara K, Mikasa K, Kaku M, Yano H. 2017. Suitability of carbapenem inactivation method (CIM) for detection of IMP metallo-β-lactamase-producing Enterobacteriaceae. J Clin Microbiol 55:1220–1222. doi: 10.1128/JCM.02275-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Australian Commission on Safety and Quality in Health Care. 2017. CARALert: the national alert system for critical antibiotic resistance; first annual report, March 2016–March 2017. Australian Commission on Safety and Quality in Health Care, Sydney, Australia. [Google Scholar]
- 18.Bell JM, Turnidge JD, Coombs GW, Daley DM, Gottlieb T, Robson J, George N. 2016. Australian Group on Antimicrobial Resistance Australian Enterobacteriaceae Sepsis Outcome Program Annual Report, 2014. Commun Dis Intell Q Rep 40:E229–E235. [DOI] [PubMed] [Google Scholar]
- 19.Logan LK, Weinstein RA. 2017. The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis 215:S28–S36. doi: 10.1093/infdis/jiw282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clinical and Laboratory Standards Institute. 2018. Performance standards for antimicrobial susceptibility testing, 28th ed. CLSI supplement M100 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 21.Yamada K, Kashiwa M, Arai K, Nagano N, Saito R. 2017. Evaluation of the modified carbapenem inactivation method and sodium mercaptoacetate-combination method for the detection of metallo-β-lactamase production by carbapenemase-producing Enterobacteriaceae. J Microbiol Methods 137:112–115. doi: 10.1016/j.mimet.2016.11.013. [DOI] [PubMed] [Google Scholar]