Helicobacter pylori antibiotic resistance is widespread and increasing worldwide. Routine detection of H. pylori mutations that invoke antimicrobial resistance may be a useful approach to guide antimicrobial therapy and possibly avert treatment failure.
KEYWORDS: DNA sequencing, Helicobacter pylori, antibiotic resistance, mutation
ABSTRACT
Helicobacter pylori antibiotic resistance is widespread and increasing worldwide. Routine detection of H. pylori mutations that invoke antimicrobial resistance may be a useful approach to guide antimicrobial therapy and possibly avert treatment failure. In this study, formalin-fixed, paraffin-embedded (FFPE) gastric biopsy specimens from a cohort of individuals from northern Ohio in the United States were examined using a next-generation sequencing (NGS) assay to detect H. pylori mutations that are known to confer resistance to clarithromycin, levofloxacin, and tetracycline. From January 2016 to January 2017, 133 H. pylori-infected gastric biopsy specimens were identified histologically and subsequently analyzed by NGS to detect mutations in gyrA, 23S rRNA, and 16S rRNA genes. The method successfully detected H. pylori in 126 of 133 cases (95% sensitivity). Mutations conferring resistance were present in 92 cases (73%), including 63 cases with one mutation (50%) and 29 cases with mutations in multiple genes (23%). Treatment outcomes were available in 58 cases. Sixteen of the 58 cases failed therapy (28%). Therapy failure correlated with the number of mutated genes: no failure in cases with no mutations (0/15), 19% (5/27) failure in cases with one gene mutation, and 69% (11/16) failure in cases with more than one mutated gene. Common 23S rRNA mutations (A2142G or A2413G) were present in 88% (14/16) of failed cases as opposed to in only 10% (4/42) of eradicated cases (P < 0.001). This NGS assay can be used on remnant specimens collected during standard-of-care testing to detect mutations that correlate with increased risk of treatment failure. A prospective study is needed to determine if the risk of treatment failure can be decreased by using this assay to guide antibiotic therapy.
INTRODUCTION
Helicobacter pylori infects close to one-half of the global population and is the main cause of peptic ulcer disease and a trigger for gastric cancer (1, 2). The current recommended empirical treatment for H. pylori eradication includes two or three antibiotics (typically clarithromycin and either amoxicillin or metronidazole) and one antisecretory drug for 14 days, with the eradication goal being higher than 80% (3–5). However, H. pylori acquires antibiotic resistance by mutation, which has dramatically increased over the past decades (6–8). The rate of clarithromycin resistance in the United States has increased from 10% to 24% in the 1990s to 24% to 70% in recent years (1, 3, 9). Increasing rates of resistance have also been reported in Europe and Asia (1, 2, 6, 10). This has caused a sharp global decline in the effectiveness of the recommended treatments that used to be >90% effective in the 1990s (5, 6, 11–13). Studies have shown that while a proton pump inhibitor (PPI)-clarithromycin-amoxicillin triple therapy has up to 88% eradication rate for clarithromycin-susceptible strains, the eradication rate drops to only 18% in clarithromycin-resistant strains (1, 14). Therefore, it is suggested to identify the H. pylori strains that are likely to fail empirical therapy and subsequently to choose a personalized therapeutic regimen with high likelihood of successful eradication. Several studies have reported that a treatment based on antimicrobial susceptibility testing is more effective than empirical treatment (15–17).
Susceptibility testing using bacterial culture followed by MIC phenotypic resistance testing is the gold standard technique to detect resistance. However, this practice is not widely used due to the fastidious nature of the organism and the long incubation time required. Enzyme immunoassay, fluorescence in situ hybridization (FISH), and several PCR-based methods have been described to detect H. pylori and the mutations related to clarithromycin resistance from biopsy specimens, gastric fluid, colonies, and even stool samples (12, 16). PCR methods based on the detection of point mutations offer high sensitivity and specificity and hence are alternatives to phenotypic testing, but PCR only allows for the detection of resistance mutations at a limited number of sites (16, 18). Currently, there is no simple and widely used method for predicting antimicrobial resistance, but there is opportunity to use molecular methods to develop a rapid and clinically useful approach to detect mutations associated with resistance. In this study, we investigated the ability of a novel next-generation sequencing (NGS) assay to detect resistance mutations in H. pylori by sequencing H. pylori DNA from formalin-fixed, paraffin-embedded (FFPE) gastric biopsy specimens. We also evaluated the correlation of these resistance mutations to clinical outcomes.
MATERIALS AND METHODS
Samples for NGS assay validation.
The study was approved by the Institutional Research Board of the University Hospitals Cleveland Medical Center/Case Western Reserve University (UHCMC/CWRU). One hundred thirty-three gastric biopsy specimens positive for H. pylori (by morphology or immunohistochemistry) collected between January 2017 and January 2018 were selected by a retrospective search of the Department of Pathology archives database. Medical records were reviewed for each patient, including demographics, treatment of H. pylori infection, and treatment outcome. Only the first biopsy specimen obtained from a subject was analyzed in this study. Clinical eradication was defined as negative fecal antigen test, negative urea breath test, negative repeat biopsy, or clinical impression of cure (symptom relief) on follow-up assessments. Treatment failure was defined as a persistent positive fecal antigen test, urea breath test, repeat biopsy, or persistence of symptoms of dyspepsia in follow-up assessments.
DNA extraction and ion semiconductor-targeted NGS.
FFPE tissue blocks were retrieved, and 7-μm curls were prepared. DNA was extracted from the curls using the QIAamp FFPE DNA Isolation kit (catalog number 5110; Qiagen, Valencia, CA) according to the manufacturer’s instructions. The primers were designed to target the DNA regions that have been associated with antibiotic resistance. Clarithromycin resistance is due to single point mutations in the peptidyl transferase region of domain V of 23S rRNA (3). Tetracycline resistance is due to mutation in the binding site of tetracycline in 16S rRNA (14). Fluoroquinolone resistance is due to point mutations in the quinolone resistance determining region (QRDR) of gyrA at positions encoding amino acids 86, 87, 88, 91, or 97 (14, 19). We used 3 pairs of primers to amplify the three gene regions: gyrA, (forward) 5′-AAGGTTAGGCAGACGGCT-3′ and (reverse) 5′-TTAACCACCCCCATGGCGA-3′; 23S rRNA gene, (forward) 5′-GGTGGTATCTCAAGGATGGC-3′ and (reverse) 5′-GATCTAACCGCGGCAAGACG-3′(20); and 16S rRNA gene, (forward) 5′-TGGAGCATGTGGTTTAATTCGA-3′ and (reverse) 5′-TGCGGGACTTAACCCAACA-3′(21). An input of 30 ng of DNA was used for the multiplex PCR. The amplicons were then subjected to DNA end repair, deoxyribosyladenine (dA) tailing, and adapter ligation per the manufacture’s protocol (Thermo Fisher). The IonChef and Hi-Q View 200 kits were used for template amplification and enrichment, and sequencing was performed on either an Ion Torrent PGM with v318 Chip at approximately 1% of chip space per sample or an S5 GeneStudio with 510 Chip at approximately 5% of chip space.
Informatics pipeline and variant interpretation.
The low-quality bases from the end of the read (base < Q20) and raw reads (read lengths < 100 bp) were trimmed by Trimmomatic (version 0.38). The remaining clean reads were mapped with BWA (22) (version 0.7.5a-r405) against the database of 16S rRNA gene sequences compiled by Miao et al. (23). Reads that best mapped to H. pylori and had >99% similarity to H. pylori were extracted and aligned to the 16S rRNA gene of the reference genome (NC_000915). Single nucleotide variants (SNVs) and indel were called using SAMtools’s (24) (version 1.7) Mpileup module and Bcftools (minimum base and mapping quality of >10 and minimum coverage of >100 reads); finally, a series of mutations were generated. For determining variations in 23S rRNA and gyrA genes, all reads were aligned to the corresponding gene from the reference genome (NC_000915) followed by the identification of variants as described above for 16S. There is limited published data describing the clinical significance of a majority of variants for gyrA and 23S, and so only variants with a clear link to antibiotic resistance were considered clinically significant mutations (3, 8, 25–35).
Statistical analysis.
Clinical and demographic characteristics were compared using statistical tests (Fisher’s exact tests, chi-square, Mann-Whitney rank sum test, and Student’s t test) to assess differences between groups in categorical and nominal variables in the relationships between variations of multidrug resistance genes and the presence or absence of resistance mutations. A P value of ≤0.05 was considered statistically significant. Statistical analyses were performed using SigmaPlot 11.0.
RESULTS
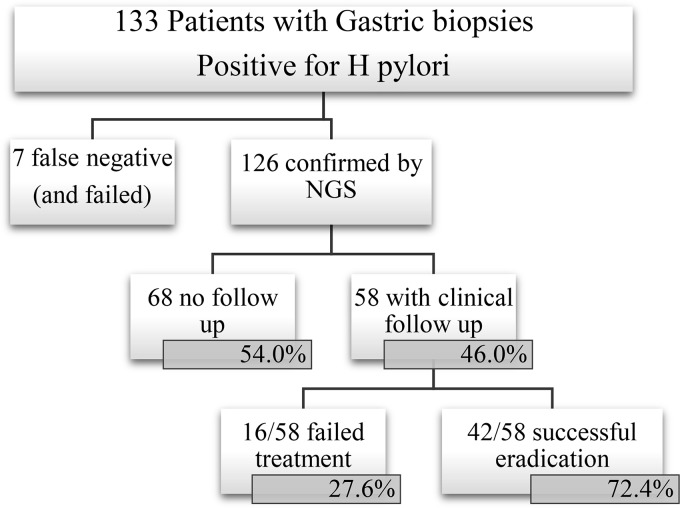
The NGS assay detected H. pylori in 126 of 133 histologically positive cases (Fig. 1), yielding 94.7% sensitivity. Table 1 shows the demographic data of the 126 NGS positive cases. Bacteria other than H. pylori were also detected using the 16S sequencing assay, and these bacteria and the frequency of their detection are reported in Table S1 in the supplemental material.
FIG 1.
Consort flow diagram of the study.
TABLE 1.
Demographic information (n = 126)
| Characteristics | Value(s) |
|---|---|
| Age (yr) | |
| Mean ± SD | 49.8 ± 20.0 |
| Range | 7–89 |
| Sex (n) | |
| Male | 39 |
| Female | 87 |
| Race (n) | |
| White | 50 |
| African-American | 55 |
| Other | 21 |
Prevalence of gene variants and resistance mutations.
The assay detected 11 different variants in 23S with at least one variant identified in 52 of 126 specimens, 28 different variants in gyrA in 75 specimens, and 4 different variants in 16S present in 12 specimens (see Tables S2 and S3). Of these variants, a subset has been described as invoking antibiotic resistance, and these included 6 mutations in 23S, 6 mutations in gyrA, and 4 mutations in 16S (Table 2). The prevalence of mutations known to cause antibiotic resistance was 48 (38.1%) in 23S, 66 (52.4%) in gyrA, and 12 (9.5%) in 16S (Table 3). The most common 23S mutations were A2143G, A2142G, and T2182C, and the most prevalent gyrA mutations were N87T and N87I (Table 2).
TABLE 2.
Prevalence of mutations
| Gene | No. among all positive cases (n = 126) | Positive cases with follow-up data |
|||
|---|---|---|---|---|---|
| Treatment failed (n = 16) |
Eradication (n = 42) |
||||
| No. | % | No. | % | ||
| 23S rRNA | |||||
| A2143G | 30 | 11 | 69 | 4 | 10 |
| A2142G | 10 | 4 | 25 | 1 | 2 |
| T2182C | 9 | 0 | 0 | 3 | 7 |
| C2195T | 8 | 1 | 6 | 3 | 7 |
| G2141A | 3 | 0 | 0 | 1 | 2 |
| A2223G | 1 | 1 | 6 | 0 | 0 |
| No mutations | 78 | 2 | 13 | 32 | 76 |
| gyrA | |||||
| N87T | 35 | 5 | 31 | 10 | 24 |
| N87I | 18 | 2 | 13 | 8 | 19 |
| D91N | 10 | 6 | 38 | 2 | 5 |
| N87K | 4 | 2 | 13 | 0 | 0 |
| D91G | 4 | 1 | 6 | 0 | 0 |
| D91Y | 3 | 2 | 13 | 0 | 0 |
| No mutations | 50 | 3 | 19 | 24 | 52 |
| 16S rRNA | |||||
| A928C | 6 | 2 | 13 | 3 | 7 |
| A926G | 5 | 1 | 6 | 1 | 2 |
| A928T | 1 | 1 | 6 | 0 | 0 |
| G927T | 0 | 0 | 0 | 0 | 0 |
| No mutations | 114 | 12 | 75 | 38 | 90 |
TABLE 3.
Clinically relevant mutation patterns and prevalence of H. pylori in the northeast Ohio cohort (n = 126)
| Mutation pattern | No. (%) of cases |
|---|---|
| No mutation | 34 (27.0) |
| Mutation in one gene | 63 (50.0) |
| 16S | 5 (4.0) |
| 23S | 20 (15.9) |
| gyrA | 38 (30.2) |
| Mutation in more than one gene | 29 (23.0) |
| 23S and 16S | 1 (0.8) |
| gyrA and 23S | 22 (17.5) |
| gyrA and 16S | 1 (0.8) |
| gyrA and 23S and 16S | 5 (4.0) |
Treatment outcomes.
Correlation with therapy outcomes was studied in a subset of 58 patients for which records of clinical follow-up were available. Clarithromycin (54 cases) and amoxicillin (47 cases) were the most commonly used antibiotics (Table 5). Of these 58 subjects, 16 (28%) failed treatment and 42 (72%) experienced successful eradication of H. pylori infection. Mutations were detected in all (16/16) in the failed therapy group, but only in 64% (27/42) in the successful eradication group (P < 0.01) (Table 4). There was no significant difference in the antibiotics prescribed when comparing the successfully eradicated and failed therapy groups, except for tetracycline, which was used more frequently in the failed group than in the eradicated group (P = 0.03) (Table 5).
TABLE 5.
Antibiotics used in cases with clinical follow-up
| Antibiotic(s) or characteristic | No. of cases treated (N = 58) | No. (%) of cases |
|
|---|---|---|---|
| Treatment failure (N = 16) | Eradication (N = 42) | ||
| Amoxicillin | 47 | 13 (81.2) | 34 (80.9) |
| Clarithromycin | 54 | 15 (93.7) | 39 (92.9) |
| Tetracycline | 8 | 5 (31.2) | 3 (7.1)a |
| Levofloxacin | 2 | 2 (12.5) | 0 (0) |
| Metronidazole | 12 | 6 (37.5) | 6 (14.3) |
| Combination antibiotic therapy | 56 | 16 (100) | 40 (95.2) |
| Mutation detected that invokes resistance to at least one of the prescribed antibiotics | 23 | 15 (93.75) | 8 (19.0)b |
P value <0.05.
P value <0.001.
TABLE 4.
Clinically relevant mutations and antibiotic therapy in cases with follow-up
aF, failed; S, successful eradication.
bShading indicates that the corresponding antibiotic was received.
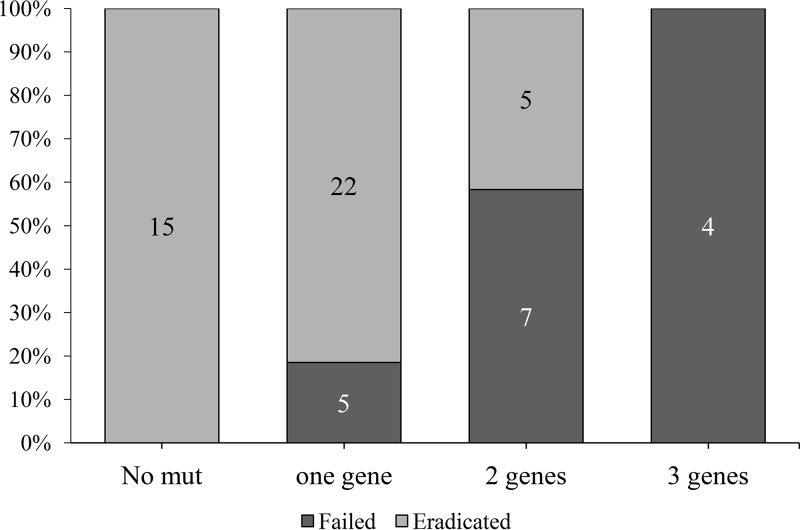
Of the 16 failed cases, 15 received clarithromycin, and 13 of these harbored 23S mutations associated with macrolide resistance. Of 42 eradicated cases, 39 had received clarithromycin, and only 8 of these 39 cases harbored 23S mutations. Regarding 16S mutations, of 16 failed cases, 5 received tetracycline, and 2 of these 5 cases harbored 16S mutations associated with tetracycline resistance. Of 42 eradicated cases, 3 had received tetracycline, and none of these 3 cases harbored mutations associated with tetracycline resistance (Tables 4 and 5). In the case of gyrA mutations and levofloxacin resistance, only 2 of 58 patients had received levofloxacin, and so we could not test a correlation between the therapy failure and mutation presence. Therapy failure positively correlated with the number of mutated genes. No therapy failures were reported in 15 specimens without mutations. The failure rate was 19% (5/27) in patients with 1 gene mutation and 69% (11/16) in specimens with mutations in more than one gene (Fig. 2). Total mutation burden (cumulative number of mutations of three genes) was greater in the failed treatment group than in the successful eradicated group (2.5 ± 1.0 versus 1.1 ± 1.0, respectively, P < 0.001).
FIG 2.
Failure rate based on the number of mutated genes. In our group of 58 patients with follow-up results, therapy failure increased with the number of gene mutations detected, while no therapy failure was reported in patients without mutations. Failure rate was 18.5% (5/27) in patients with 1 gene mutation and 68.8% (11/16) in patients with mutations in more than one gene.
23S rRNA mutations were significantly more common in treatment failure cases than in successful eradication cases (88% versus 24%, P < 0.001) (Table 4). Two 23S mutations of A2142G and A2143G were present in 88% of cases in the failed treatment group (14/16) as opposed to in only 10% of cases in the eradicated group (4/42) (P < 0.001) (Fig. 3).
FIG 3.
Failure rate based on A2143G and/or A2142G mutations. In our group of 58 patients with follow-up results, subjects with one of these mutations were significantly more likely to fail treatment than those with unmutated 23S rRNA (P < 0.001): 87.5% of patients who failed therapy, as opposed to only 9.5% in the eradicated group.
DISCUSSION
Our data demonstrated that targeted NGS testing of H. pylori from FFPE is a feasible approach to detect mutations associated with antimicrobial resistance and that the presence of these mutations correlated with treatment failure. In our cohort, clarithromycin resistance mutations were present in 38.1% (48/126), levofloxacin resistance mutations were present in 52.4% (66/126), and tetracycline resistance mutations were present in 9.5% (12/126) of specimens with detectable amounts of H. pylori DNA (Table 3). Amoxicillin resistance is attributed to penicillin binding protein mutations, and resistance is uncommon and was not investigated in this study. Metronidazole resistance is commonly encountered and has been associated with mutations in rdxA and/or frxA, but data regarding the value of detecting these mutations in predicting antibiotic resistance is limited; therefore, in this study, we did not investigate the mutations associated with metronidazole resistance (2, 36). Previous studies have shown that tailored treatments based on resistance data improve eradication rates and increase cost efficiency compared to the use of standard empirical triple therapy, especially in areas where clarithromycin resistance is higher than 15%, such as the United States (37, 38). In the future, this NGS assay could be used to tailor H. pylori therapy.
Clarithromycin was the most commonly used antibiotic in our cohort, and we identified a strong correlation between 23S mutation (which would invoke clarithromycin resistance) and treatment failure, even though these infections were treated with combination therapy in which one of the other antibiotics (e.g., amoxicillin) may have been active. Resistance to at least one of the prescribed antibiotics was detected in 93.7% of failed cases as opposed to in only 19% of eradicated cases (Table 5). This correlation highlights that resistance to even one antibiotic can significantly affect the clinical outcome of H. pylori eradication treatment.
When considering the results of 23S rRNA mutations alone, the presence of A2142G or A2143G alone correlated with treatment failure, and these mutations were previously reported to be associated with clarithromycin resistance (11, 32, 39). Of the 16 cases that failed therapy, 69% (11/16) had A2143G and 25% (4/16) had A2142G; cases with specimens containing one of these mutations were significantly more likely to fail treatment than those with unmutated 23S rRNA (P < 0.00) (Fig. 3).
Among the 6 common mutations in gyrA that invoke fluoroquinolone resistance, N87 and D91 are reported to be the most commonly encountered (14, 19, 40), and these mutations were the most frequent gyrA mutations in our cohort (Table 4). However, we did not observe a significant correlation between gyrA mutations and treatment success. Many subjects with gyrA mutations achieved successful eradication, and this may be because only three individuals in our study (3/58) received fluoroquinolone therapy. For similar reasons, 16S mutations also did not demonstrate a significant correlation with treatment success. In contrast, nearly all subjects in our study had received clarithromycin therapy (Table 5), and treatment failure correlated strongly with 23S mutations invoking macrolide resistance. However, despite no individual correlation between gyrA or 16S mutation and treatment success, we observed a direct correlation between the mutation burden across 16S, 23S, and gyrA genes and the likelihood of empirical treatment failure, regardless of the treatment regimen used (Table 5; see also Fig. S1 in the supplemental material). This finding suggests that H. pylori total mutation burden may be an independent risk factor of treatment failure, although additional studies are needed to confirm this possibility.
Our assay detected 11 different mutations in 23S and 28 mutations in gyrA, from which 6 mutations in 23S and 6 in gyrA have shown clinical correlation to antibiotic resistance. The rest of these mutations were regarded as variants of unknown significance. In vitro susceptibility testing of cultured isolates containing these variants of unknown significance may provide insight into whether these variants invoke antibiotic resistance. It is also worth noting that our assay was not designed to differentiate different strains of H. pylori or to identify mixed infections caused by more than one H. pylori strain.
Previous studies have shown a correlation between the presence of H. pylori mutations and in vitro antibiotic resistance (3), and the present study demonstrates that mutation screening directly from biopsy specimens can predict treatment failure. The NGS assay described in this study can be performed with FFPE specimens, which are routinely obtained as part of standard of care. This NGS assay has prognostic value, and it could potentially be used for personalized therapy to avoid the use of antibiotics that are likely to be ineffective. Our study corroborates others’ findings that specific mutations in 23S rRNA (A2142G or A2413G) correlate with increased likelihood of treatment failure when clarithromycin is used. This study is the first to suggest that total mutation burden across 16S rRNA, 23S rRNA, and gyrA genes correlates with increased likelihood of treatment failure; further investigation should be performed to test the validity of this finding.
Supplementary Material
ACKNOWLEDGMENT
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01834-18.
REFERENCES
- 1.Malfertheiner P, Megraud F, O'Morain CA, Atherton J, Axon AT, Bazzoli F, Gensini GF, Gisbert JP, Graham DY, Rokkas T, El-Omar EM, Kuipers EJ, European Helicobacter Study Group . 2012. Management of Helicobacter pylori infection–the Maastricht IV/Florence consensus report. Gut 61:646–664. doi: 10.1136/gutjnl-2012-302084. [DOI] [PubMed] [Google Scholar]
- 2.Teh X, Khosravi Y, Lee WC, Leow AH, Loke MF, Vadivelu J, Goh KL. 2014. Functional and molecular surveillance of Helicobacter pylori antibiotic resistance in Kuala Lumpur. PLoS One 9:e101481. doi: 10.1371/journal.pone.0101481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen D, Cunningham SA, Cole NC, Kohner PC, Mandrekar JN, Patel R. 2017. Phenotypic and molecular antimicrobial susceptibility of Helicobacter pylori. Antimicrob Agents Chemother 61:e02530-16. doi: 10.1128/AAC.02530-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghotaslou R, Leylabadlo HE, Asl YM. 2015. Prevalence of antibiotic resistance in Helicobacter pylori: a recent literature review. World J Methodol 5:164–174. doi: 10.5662/wjm.v5.i3.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chey WD, Leontiadis GI, Howden CW, Moss SF. 2017. ACG clinical guideline: treatment of Helicobacter pylori infection. Am J Gastroenterol 112:212–239. doi: 10.1038/ajg.2016.563. [DOI] [PubMed] [Google Scholar]
- 6.Megraud F. 2004. H. pylori antibiotic resistance: prevalence, importance, and advances in testing. Gut 53:1374–1384. doi: 10.1136/gut.2003.022111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fennerty MB, Kovacs TO, Krause R, Haber M, Weissfeld A, Siepman N, Rose P. 1998. A comparison of 10 and 14 days of lansoprazole triple therapy for eradication of Helicobacter pylori. Arch Intern Med 158:1651–1656. doi: 10.1001/archinte.158.15.1651. [DOI] [PubMed] [Google Scholar]
- 8.Kim JM, Kim JS, Kim N, Jung HC, Song IS. 2005. Distribution of fluoroquinolone MICs in Helicobacter pylori strains from Korean patients. J Antimicrob Chemother 56:965–967. doi: 10.1093/jac/dki334. [DOI] [PubMed] [Google Scholar]
- 9.Shiota S, Reddy R, Alsarraj A, El-Serag HB, Graham DY. 2015. Antibiotic resistance of Helicobacter pylori among male United States veterans. Clin Gastroenterol Hepatol 13:1616–1624. doi: 10.1016/j.cgh.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park JY, Dunbar KB, Mitui M, Arnold CA, Lam-Himlin DM, Valasek MA, Thung I, Okwara C, Coss E, Cryer B, Doern CD. 2016. Helicobacter pylori clarithromycin resistance and treatment failure are common in the USA. Dig Dis Sci 61:2373–2380. doi: 10.1007/s10620-016-4091-8. [DOI] [PubMed] [Google Scholar]
- 11.Megraud F, Benejat L, Ontsira Ngoyi EN, Lehours P. 2015. Molecular approaches to identify Helicobacter pylori antimicrobial resistance. Gastroenterol Clin North Am 44:577–596. doi: 10.1016/j.gtc.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Dong F, Ji D, Huang R, Zhang F, Huang Y, Xiang P, Kong M, Nan L, Zeng X, Wu Y, Bao Z. 2015. Multiple genetic analysis system-based antibiotic susceptibility testing in Helicobacter pylori and high eradication rate with phenotypic resistance-guided quadruple therapy. Medicine (Baltimore) 94:e2056. doi: 10.1097/MD.0000000000002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graham DY, Fischbach L. 2010. Helicobacter pylori treatment in the era of increasing antibiotic resistance. Gut 59:1143–1153. doi: 10.1136/gut.2009.192757. [DOI] [PubMed] [Google Scholar]
- 14.Megraud F, Lehours P. 2007. Helicobacter pylori detection and antimicrobial susceptibility testing. Clin Microbiol Rev 20:280–322. doi: 10.1128/CMR.00033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.López-Góngora S, Puig I, Calvet X, Villoria A, Baylina M, Muñoz N, Sanchez-Delgado J, Suarez D, García-Hernando V, Gisbert JP. 2015. Systematic review and meta-analysis: susceptibility-guided versus empirical antibiotic treatment for Helicobacter pylori infection. J Antimicrob Chemother 70:2447–2455. doi: 10.1093/jac/dkv155. [DOI] [PubMed] [Google Scholar]
- 16.Arslan N, Yılmaz Ö, Demiray-Gürbüz E. 2017. Importance of antimicrobial susceptibility testing for the management of eradication in Helicobacter pylori infection. World J Gastroenterol 23:2854–2869. doi: 10.3748/wjg.v23.i16.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith SM, O'Morain C, McNamara D. 2014. Antimicrobial susceptibility testing for Helicobacter pylori in times of increasing antibiotic resistance. World J Gastroenterol 20:9912–9921. doi: 10.3748/wjg.v20.i29.9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thung I, Aramin H, Vavinskaya V, Gupta S, Park JY, Crowe SE, Valasek MA. 2016. Review article: the global emergence of Helicobacter pylori antibiotic resistance. Aliment Pharmacol Ther 43:514–533. doi: 10.1111/apt.13497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zerbetto De Palma G, Mendiondo N, Wonaga A, Viola L, Ibarra D, Campitelli E, Salim N, Corti R, Goldman C, Catalano M. 2017. Occurrence of mutations in the antimicrobial target genes related to levofloxacin, clarithromycin, and amoxicillin resistance in Helicobacter pylori isolates from Buenos Aires City. Microb Drug Resist 23:351–358. doi: 10.1089/mdr.2015.0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Zhao F, Kong M, Wang S, Nan L, Hu B, Olszewski MA, Miao Y, Ji D, Jiang W, Fang Y, Zhang J, Chen F, Xiang P, Wu Y, Zhao H. 2016. Validation of a high-throughput multiplex genetic detection system for Helicobacter pylori identification, quantification, virulence, and resistance analysis. Front Microbiol 7:1401. doi: 10.3389/fmicb.2016.01401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang S, Lin S, Kelen GD, Quinn TC, Dick JD, Gaydos CA, Rothman RE. 2002. Quantitative multiprobe PCR assay for simultaneous detection and identification to species level of bacterial pathogens. J Clin Microbiol 40:3449–3454. doi: 10.1128/JCM.40.9.3449-3454.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miao J, Han N, Qiang Y, Zhang T, Li X, Zhang W. 2017. 16SPIP: a comprehensive analysis pipeline for rapid pathogen detection in clinical samples based on 16S metagenomic sequencing. BMC Bioinformatics 18:568. doi: 10.1186/s12859-017-1975-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nonaka S, Matsuzaki K, Kazama T, Nishiyama H, Ida Y, Koyano S, Sonobe K, Okamura N, Saito R. 2014. Antimicrobial susceptibility and mechanisms of high-level macrolide resistance in clinical isolates of Moraxella nonliquefaciens. J Med Microbiol 63:242–247. doi: 10.1099/jmm.0.061788-0. [DOI] [PubMed] [Google Scholar]
- 26.Posteraro P, Branca G, Sanguinetti M, Ranno S, Cammarota G, Rahimi S, De Carlo M, Posteraro B, Fadda G. 2006. Rapid detection of clarithromycin resistance in Helicobacter pylori using a PCR-based denaturing HPLC assay. J Antimicrob Chemother 57:71–78. doi: 10.1093/jac/dki406. [DOI] [PubMed] [Google Scholar]
- 27.Agudo S, Pérez-Pérez G, Alarcón T, López-Brea M. 2011. Rapid detection of clarithromycin resistant Helicobacter pylori strains in Spanish patients by polymerase chain reaction-restriction fragment length polymorphism. Rev Esp Quimioter 24:32–36. [PMC free article] [PubMed] [Google Scholar]
- 28.Hwang TJ, Kim N, Kim HB, Lee BH, Nam RH, Park JH, Lee MK, Park YS, Lee DH, Jung HC, Song IS. 2010. Change in antibiotic resistance of Helicobacter pylori strains and the effect of A2143G point mutation of 23S rRNA on the eradication of H. pylori in a single center of Korea. J Clin Gastroenterol 44:536–543. doi: 10.1097/MCG.0b013e3181d04592. [DOI] [PubMed] [Google Scholar]
- 29.Binh TT, Shiota S, Suzuki R, Matsuda M, Trang TT, Kwon DH, Iwatani S, Yamaoka Y. 2014. Discovery of novel mutations for clarithromycin resistance in Helicobacter pylori by using next-generation sequencing. J Antimicrob Chemother 69:1796–1803. doi: 10.1093/jac/dku050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim JM, Kim JS, Kim N, Kim YJ, Kim IY, Chee YJ, Lee CH, Jung HC. 2008. Gene mutations of 23S rRNA associated with clarithromycin resistance in Helicobacter pylori strains isolated from Korean patients. J Microbiol Biotechnol 18:1584–1589. [PubMed] [Google Scholar]
- 31.Agudo S, Perez-Perez G, Alarcon T, Lopez-Brea M. 2010. High prevalence of clarithromycin-resistant Helicobacter pylori strains and risk factors associated with resistance in Madrid, Spain. J Clin Microbiol 48:3703–3707. doi: 10.1128/JCM.00144-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Versalovic J, Osato MS, Spakovsky K, Dore MP, Reddy R, Stone GG, Shortridge D, Flamm RK, Tanaka SK, Graham DY. 1997. Point mutations in the 23S rRNA gene of Helicobacter pylori associated with different levels of clarithromycin resistance. J Antimicrob Chemother 40:283–286. doi: 10.1093/jac/40.2.283. [DOI] [PubMed] [Google Scholar]
- 33.Bachir M, Allem R, Benejat L, Tifrit A, Medjekane M, Drici AE, Megraud F, Douidi KT. 2018. Molecular detection of mutations involved in Helicobacter pylori antibiotic resistance in Algeria. J Antimicrob Chemother 73:2034–2038. doi: 10.1093/jac/dky167. [DOI] [PubMed] [Google Scholar]
- 34.Toledo H, López-Solís R. 2010. Tetracycline resistance in Chilean clinical isolates of Helicobacter pylori. J Antimicrob Chemother 65:470–473. doi: 10.1093/jac/dkp457. [DOI] [PubMed] [Google Scholar]
- 35.Lawson AJ, Elviss NC, Owen RJ. 2005. Real-time PCR detection and frequency of 16S rDNA mutations associated with resistance and reduced susceptibility to tetracycline in Helicobacter pylori from England and Wales. J Antimicrob Chemother 56:282–286. doi: 10.1093/jac/dki199. [DOI] [PubMed] [Google Scholar]
- 36.Lauener FN, Imkamp F, Lehours P, Buissonniere A, Benejat L, Zbinden R, Keller PM, Wagner K. 2019. Genetic determinants and prediction of antibiotic resistance phenotypes in Helicobacter pylori. J Clin Med 8:E53. doi: 10.3390/jcm8010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romano M, Marmo R, Cuomo A, De Simone T, Mucherino C, Iovene MR, Montella F, Tufano MA, Del Vecchio Blanco C, Nardone G. 2003. Pretreatment antimicrobial susceptibility testing is cost saving in the eradication of Helicobacter pylori. Clin Gastroenterol Hepatol 1:273–278. doi: 10.1016/S1542-3565(03)00131-9. [DOI] [PubMed] [Google Scholar]
- 38.Cosme A, Montes M, Martos M, Gil I, Mendarte U, Salicio Y, Pineiro L, Recasens MT, Ibarra B, Sarasqueta C, Bujanda L. 2013. Usefulness of antimicrobial susceptibility in the eradication of Helicobacter pylori. Clin Microbiol Infect 19:379–383. doi: 10.1111/j.1469-0691.2012.03844.x. [DOI] [PubMed] [Google Scholar]
- 39.van Doorn LJ, Glupczynski Y, Kusters JG, Megraud F, Midolo P, Maggi-Solca N, Queiroz DM, Nouhan N, Stet E, Quint WG. 2001. Accurate prediction of macrolide resistance in Helicobacter pylori by a PCR line probe assay for detection of mutations in the 23S rRNA gene: multicenter validation study. Antimicrob Agents Chemother 45:1500–1504. doi: 10.1128/AAC.45.5.1500-1504.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsuzaki J, Suzuki H, Nishizawa T, Hirata K, Tsugawa H, Saito Y, Okada S, Fukuhara S, Hibi T. 2012. Efficacy of sitafloxacin-based rescue therapy for Helicobacter pylori after failures of first- and second-line therapies. Antimicrob Agents Chemother 56:1643–1645. doi: 10.1128/AAC.05941-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.