Abstract
HIV infection has been associated with alterations in gut microbiota and related microbial metabolite production. However, the mechanisms of how these functional microbial metabolites may affect HIV immunopathogenesis and comorbidities, such as cardiovascular disease and other metabolic diseases, remain largely unknown. Here we review the current understanding of gut microbiota and related metabolites in the context of HIV infection. We focus on several bacteria-produced metabolites, including tryptophan catabolites, short-chain fatty acids and trimethylamine-N-oxide (TMAO), and discuss their implications in HIV infection and comorbidities. We also prospect future studies using integrative multiomics approaches to better understand host–microbiota–metabolites interactions in HIV infection, and facilitate integrative medicine utilizing the microbiota in HIV infection.
Keywords: : HIV, integrative omics, metabolites, metabolomics, microbiota
The human gastrointestinal microbiota, as a complex community of microorganisms, exerts a unique role in maintaining intestinal immune homeostasis, as well as in the induction, instruction and functionality of the whole host immune system. Interactions between host and bacteria occur primarily through evolutionarily conserved chemical dialogs that involve a multitude of metabolites and pathways [1–3]. In recent years, a lot of studies discovered the relationship between intestinal bacteria composition alternation (dysbiosis), bacterial metabolites, and human chronic diseases/disorders [4], such as diabetes [5–7], obesity [8,9], inflammatory bowel diseases (IBD) [10–12], periodontal disease [13], atherosclerosis, cardiovascular disease (CVD) [14–16], rheumatic diseases [17], and so on.
Previous studies suggested that HIV infection has an impact on the gut microbiome composition and then may affect microbial products/metabolites categories and concentrations. HIV infection has been linked with alterations of the enteric microbiome and distinct microbial metabolites, especially the increased bacterial populations in the intestinal tract [18,19]. Although the exact mechanisms remain unclear, the alterations of microbiota and microbial metabolites might be associated with the changes in the epithelium and mucosal immune system/landscape of the intestine caused in HIV infection, accompanying translocation of microbial products and possibly microorganisms themselves, which leads to dramatic alteration in the structural and immunological properties of this important organ system [18,20–25].
During the early stages of infection, HIV replicates in gut-related lymphoid tissue, resulting in rapid and substantial loss of lamina propria CD4+ T cells, including T helper (Th) 17 and Th22 cells, which play critical roles in modulating interaction with intestinal bacteria [25]. Rapid depletion of CD4+ T cells quickly leads to several immunological effects, mucosal barrier dysfunction, chronic inflammation, immune dysfunction, pronounced changes in the gut microbiome composition and subsequently altered microbial products, and then to disturbances of host–microbiome homeostasis [26–30]. It is proposed that microbial products also play important roles in HIV disease progression, because HIV preferentially infects activated T cells, while translocation of microbial products such as lipopolysaccharide to blood may result in systemic activation of T cells [31–33].
Due to the key interaction effect between features of HIV pathogenesis, gut microbiome and microbial metabolites, it is critical to thoroughly understand the impact of HIV infection on changes of gut microbiome diversity/composition and microbial metabolites; potential effect of treatment/clinical intervention on the gut microbiota and microbial metabolites; and integrative analysis of metabolome and microbiome.
Gut microbiome alteration in HIV infection
A number of studies have investigated gut microbiome alteration associated with HIV, although the relationship between HIV infection and the overall microbial community diversity (e.g., α- and β- diversity) remains uncertain [34]. Some studies reported loss of α-diversity in untreated HIV-infected patients, recent infected individuals, or those having special sexual performance (e.g., men who have sex with men [MSM]), compared with uninfected controls [35–37], while other studies did not find such results [29,38–40]. One very recent study which conducted a meta-analysis of 22 studies (n = 1032) indicated that HIV status was associated with a decrease in measures of α-diversity but not in MSM, and women displayed increased α-diversity compared with men who have sex with women (MSW) [41]. Differential clustering for HIV-positive patients have been reported in many studies using β-diversity indices, such as weighed/unweighted Unifrac distance and Bray–Curtis distance, compared with uninfected controls [28,38,42,43]. However, for β-diversity indices, the differential clustering trend has not been consistently observed, as several studies reported no clear separation by HIV infection status [40,44].
Lots of efforts have been made to investigate alternations in the gut microbiota taxonomic bacterial composition during HIV infection. At phyla level, several studies revealed that the relative abundances of Firmicutes in HIV-positive patients were lower than in HIV-negative individuals, while the relative abundances of Actinobacteria and Proteobacteria increased [42,43,45,46]. It is worth noting that these alternations lie on plenty of factors, not only the disease-specific characteristics (e.g., treated/untreated patients; short-term/long-term antiretroviral therapy [ART] use) but also other factors such as age, geography, sexual practices, diet, demographic background, exercise and so on [47–49]. A well-known example is the relationship between HIV infection and Prevotella. Earlier studies observed higher relative abundance of Prevotella in HIV-infected individuals compared with HIV-uninfected controls; however, several recent studies revealed that the enriched Prevotella was more strongly associated with sexual behaviors, MSM, rather than HIV infection itself [28,35,50,51]. Published studies on HIV infection and alteration in gut microbiome have been well-summarized in review articles by Ribeiro et al. and Liu et al. [19,52]
Gut microbiota-related metabolites in HIV infection
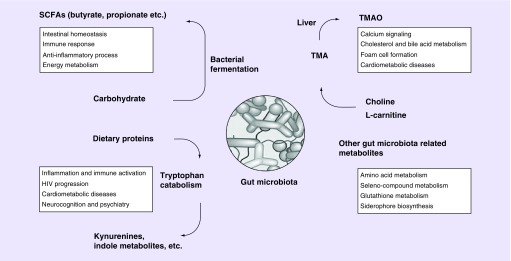
HIV infection elicits subsequent changes in enteric microbiota and alters microbial metabolites production. The alternation of microbial metabolites holds the ability to influence critical aspects of the host health status, HIV progression and multiple HIV comorbidities. Recent studies have reported that HIV infection may lead to metabolic changes associated with gut microbiota alterations, which is distinct from those induced in other diseases [53]. In this section, we focus on several well-documented microbial metabolites to sketch the overview of recent advances (Figure 1).
Figure 1. . Gut microbial metabolites in host physiology pathology.
SCFA: Short-chain fatty acid; TMA: Trimethylamine; TMAO: Trimethylamine-N-oxide.
Tryptophan catabolism
Tryptophan is a pivotal amino acid in the protein synthesis as well as the serotonin and melatonin biosynthesis, and also plays an important role in kynurenine pathway [54,55]. It is known that the rate of tryptophan catabolism, indicated by kynurenine-to-tryptophan (KYN/TRP) ratio, is increased in HIV infection and is linked to disease progression [56]. In previous studies, lots of efforts have been made to investigate the gut microbiota-associated tryptophan metabolism in HIV-infected patients. For example, Vujkovic-Cvijin et al. noted that progressive HIV-infected patients are enriched with bacterial communities that catabolize tryptophan by their capacity to produce the rate-limiting enzyme indoleamine 2,3-dioxygenase 1 (IDO-1) [29]. These HIV-associated microbiota alternations might influence immunity by emulation of the human kynurenine pathway. In the kynurenine pathway, tryptophan is catabolized into kynurenine and several downstream metabolites (e.g., kynurenic acid, anthranilic acid), mainly regulated by IDO-1, which is induced by Th1-type cytokines (e.g., interferon-γ) during inflammation and immune activation [57]. Vujkovic-Cvijin et al. [29] identified 140 genera that significantly contribute to tryptophan catabolism, a few of which encode the genetic machinery that performs the same tryptophan catabolism as human IDO-1. Kynurenine derivatives have been shown to impair mucosal immunity, resulting in increased bacterial translocation and higher mortality [58]. Increased levels of tryptophan catabolites, especially 3-hydroxyanthranilic acid, are directly interfaced with the biased balance of Th17 to Treg cells which further results in immune suppression, bacterial translocation and systemic inflammation [56]. These findings were further supported by metabolomics analysis in gut bacteria through detecting the kynurenine sub-product 3-hydroxyanthranilate [53]. Nevertheless, Vázquez-Castellanos et al. reported that the IDO1 gene or its expression was not found in the metagenomic and metatranscriptomic datasets from HIV-infected individuals [59]. Further studies are needed to address the mechanisms of tryptophan catabolism and host–microbiota interaction in the context of HIV infection.
The relationship between tryptophan catabolism and the comorbidities of HIV infection has also been reported. Recent data based on two large HIV cohorts indicated that HIV-infected individual had lower plasma tryptophan and higher kynurenic acid-to-tryptophan (KYNA/TRP) ratio compared with those without HIV infection; more interestingly, lower tryptophan and higher kynurenic acid-to-tryptophan (KYNA/TRP) ratio were associated with progression of carotid artery atherosclerosis in HIV infection [55]. Consistently, Siedner et al. found an association between the decreased plasma KYN/TRP ratio over 6 months of ART treatment and carotid artery intima media thickness (cIMT) [60]. The positive associations of KYN/TRP ratio with carotid artery subclinical atherosclerosis [61–63] and CVD risk [64–67] were also observed in non-HIV populations. In addition, among women with or at high risk for HIV infection, diabetes is associated with gut microbiota and plasma metabolites alteration; several metabolites in tryptophan catabolism, as well as the KYN/TRP ratio were higher in women with diabetes compared with those without diabetes [40]. Another study indicated that the combination of HIV infection and type 2 diabetes was associated with reduced gut microbiota diversity and increased plasma KYN/TRP ratio [68]. Nevertheless, for HIV comorbidities, the potential contributions and impacts of gut microbiota need to be further explored in large prospective studies powered for clinical end points.
The other interesting tryptophan catabolites worth noting are indole metabolites, a large group of gut bacteria-derived compounds. Ample evidence indicates that indoles derived from gut microbiota metabolism have significant biological effects that may contribute to the etiology of metabolic, cardiovascular and psychiatric diseases [69]. In progressive HIV infection, gut microbiota has a reduced ability to produce tryptophan-derived indole metabolites, which are associated with IL-22 produced by innate lymphocytes, and the loss of these lymphocytes increases the destruction of the epithelial barrier and exacerbates the overgrowth of pathogens [70–73].
Short-chain fatty acids
Short-chain fatty acids (SCFAs) are the main fermentation products of gut microbiota from dietary fibers. Acetate (two carbons), propionate (three carbons) and butyrate (four carbons) are three most abundantly produced SCFAs. Interesting links have been found between gut microbiota, SCFAs and the host’s physiology.
Butyrate is one of the most well-known predominant SCFAs and a high proportion of SCFA literature focused on this metabolite. Butyrate plays key roles in regulating intestinal homeostasis, an energy source for epithelial cells, a signaling molecule that modulates intestinal immune cell responses [74]. Several species under Firmicutes phylum are known as butyrate producers, which hold the ability to produce butyrate as a byproduct of fiber fermentation [74–76]. Previous studies indicated that, in both untreated [18,37,43] and treated [28,42,77] HIV-infected patients, a lot of the bacterial genera which related with butyrate producing (e.g., the well-known dominant butyrate producer: Roseburia, Coprococcus, Faecalibacterium, Eubacterium) were reduced, in association with altered SCFA profiles, compared with HIV-uninfected controls. However, not all of the species under Firmicutes phylum are decreased during HIV infection. For example, the increase of family Erysipelotrichaceae, associated with other inflammatory disorders, in the mucosa and stool samples of untreated and treated HIV-infected patients was observed in several studies [26,29,50]. Serrano-Villar et al. investigated the effects of prebiotics on microbial dysbiosis in the context of HIV infection and identified Faecalibacterium prausnitzii and Lachnospira were strongly correlated with butyrate abundance [78]. Dillon et al. found lower relative abundance of total butyrate-producing bacterial species in HIV-infected individuals compared with HIV-uninfected controls; and interestingly, Roseburia intestinalis which was inversely correlated with systemic markers of microbial translocation and immune activation, was depleted in the colonic mucosa of HIV-infected individuals [79]. In another study of HIV-infected women, one butyrate producer, Anaerococcus, was found to be inversely associated with tryptophan catabolism [40].
Besides butyrate, widely known gut microbiota-produced SCFA also include propionate and acetate, which have been also suggested to interact with host metabolism. Propionate can reduce the biosynthesis of cholesterol [80], providing protection against CVD [81]. Dialister is one of the most well-known propionate producers [82]. In animal models, butyrate and propionate have been documented to suppress weight gain with high fat diet-induced obesity, and acetate has been reported to reduce appetite via a central homeostatic mechanism [83,84]. Another study indicated that acetate acts on parasympathetic activity and support glucose-stimulated insulin secretion in a rodent model [85]. Peptococcus was reported as an acetate producer, and it also has the ability to produce butyrate [82]. Taken together, these findings have suggested a beneficial role of microbiota-produced SCFAs in gut, but most results were from animal studies or small human studies. Future studies with large sample sizes are needed to systemically investigate SCFAs in different human biospecimens (e.g., plasma, urine, and fecal) in relation to HIV infection. The mechanisms of how these SCFAs interact with HIV infection and their roles in gut microbiome–nutrition–physiology axis in HIV-infected patients remain largely unknown.
Trimethylamine-N-oxide
Trimethylamine-N-oxide (TMAO) is known as a gut microbiota-dependent choline metabolite. As the main precursor of TMAO, trimethylamine (TMA) is produced by certain intestinal bacteria such as Desulfovibrio spp., as a waste product of carnitine and choline metabolism [16]. It has been reported that TMAO production varied according to individual’s diversity of the gut microbiota and higher Firmicutes to Bacteroidetes abundance was observed accompanied with higher TMAO production [86]. Recent studies have revealed a strong relationship between TMAO and increased risk for atherogenesis and CVD [87–89]. At the mechanistic level, TMAO was found to contribute atherogenesis by changing calcium signaling, cholesterol and bile acid metabolism, fostering activation of inflammatory pathways and promoting foam cell formation [90].
A few studies have evaluated circulating TMAO levels among HIV-infected individuals but the results are controversial. Haissman et al. [91] reported a significant association between plasma TMAO and CD4 cell counts and a higher ratio of TMAO to its precursors, carnitine and betaine, in ART-treated patients compared with those without ART use, suggesting a potential role of ART in TMAO metabolism. However, many other studies did not find significant differences in TMAO levels between HIV-infected and uninfected individuals, and there were no significant associations of TMAO levels with HIV-related parameters (e.g., ART use, CD4 cell counts) [78,92,93]. One study indicated that there was no significant difference between the homolog gene of the cutC (Choline TMA Lyase) and cutD genes (Choline TMA−Lyase activating protein) in the metagenomes and metatranscriptomes of HIV- and HIV+ individuals [59]. Of note, these existing studies did not examine intestinal bacteria that can produce TMA, and the relationship between gut microbiota and circulating TMAO levels is unknown in the context of HIV infection. Future studies with gut microbiota data will help better understand the relationships among HIV infection, TMA-producing bacteria and TMAO.
Although the potential influences of HIV infection and ART on TMAO levels remain unclear, a number of recent studies have suggested potential associations between TMAO and various aspects of CVD in HIV-infected individuals [92–95]. In our prior work in two HIV cohorts, plasma TMAO levels were found to be positively correlated with serum biomarkers of monocyte activation and inflammation, and associated with progression of carotid atherosclerosis [96]. In addition, one earlier report showed an inverted U-shaped association between plasma TMAO levels and the presence of coronary artery stenosis in HIV-infected men [92]. In ART-experienced HIV-infected individuals, one study reported that circulating TMA levels, but not TMAO levels, were positively correlated with a number of coronary plaque features [93]. It has been suggested that TMAO could increase platelet hyper-reactivity, which is linked with cardiometabolic diseases and potential risks of thrombosis [97]. However, in both treated and untreated patients with HIV infection, there was no correlation between TMAO levels and platelet-hyperactivity [91]. Thus, further studies are needed to analyze the TMAO metabolic pathways, to clarify the relationship between TMAO and CVD risk in HIV-infected individuals.
Other gut microbiota-related metabolites
In the early stage of HIV epidemic, HIV wasting syndrome was characterized by the otherwise unexplained weight loss, malnutrition and diarrhea. Cunningham-Rundles et al. indicated that critical micronutrient deficiencies accompany with HIV disease, but the pathogenesis is still poorly understood [98]. Recent study revealed that the HIV-associated gut microbiota is unable to catabolize three amino acids, proline, phenylalanine and lysine, which is not evident in other infections or diseases [53]. This may contribute to nutritional deficits, including the wasting syndrome typically observed in advanced HIV-infected patients.
McHardy et al. investigated metagenomic functional capacity of rectal microbiota through 16S data between untreated HIV-infected patients and healthy control individuals [37]. There was an enrichment of genes and pathways involved in seleno compound metabolism, glutathione metabolism, siderophore biosynthetic, and folate biosynthesis in microbiome of HIV-infected individuals, and a depletion of genes involved in fructose/mannose metabolism, amino acid production and metabolism, and CoA biosynthesis. Notably, these functions did not appear in the HIV-infected individuals fully restored with ART.
In HIV-infected individuals, risk of bacterial pneumonia and tuberculosis is obviously increased, even in those with ART use. Cribbs et al. [99] investigated the metabolomic profile of bronchoalveolar lavages in HIV-infected patients as well as the correlation with lung bacteria, cystine, 3,5-dibromo-L-tyrosine and complex carbohydrates were significantly over-represented, indicated that HIV infection may change inflammatory and oxidative metabolic pathways in the lung associated with infections, such as the linoleate pathway. HIV infection also suppresses the Th1 T-cell subset or the glycerophospholipids synthesis pathway linked with alterations in surfactant [99]. Although, antiretroviral (ARV) treatment has improved the prognosis of HIV infection remarkably, it has also complicated the therapeutic management of TB, due to the interference between antituberculous drugs and ARV protease inhibitors/non-nucleoside reverse-transcriptase inhibitors, the accompanied immune restoration [100]. Further studies are needed to reveal the accompanied metabolomic alternations.
Potential effects of treatments & clinical interventions on gut microbiota & related metabolites
ART is combination of ARV drugs to maximally suppress the HIV virus and stop the progression of HIV disease. ART also prevents onward transmission of HIV. Vazquez-Castellanos et al. employed Shotgun metagenomics sequencing to compare stool samples from ART-treated, virally suppressed HIV-infected individuals with uninfected individuals, and suggested an altered functional profile: enrichment of genes encoding factors contributing to Lipopolysaccharide (LPS) biosynthesis, bacterial translocation and inflammation; loss of genes involved in amino acid metabolism and energy processes [42]. ART treatment, however, did not recover reduced α-diversity, whose bacterial richness is associated with immune dysfunction despite the fact that immune responders still displayed decreased intestinal bacterial richness [35]. This is in line with a recent study which found the continuous decrease of α-diversity in microbiome with ART-experienced patients [43].
Several studies have revealed that ART may have an impact on heme catabolism through inhibiting uridine glucuronyl transferases (UGT), hepatic enzymes, which are necessary for the disposal of bilirubin [45,101]. This might explain the lack of bilirubin in untreated HIV-infected patients, accumulation of bilirubin and biliverdinin gut microbiome from ART-experienced patients, and the link of ART with hyperbilirubinemia [45,102]. Biliverdin has been reported to reduce HIV viral infectivity and constitutes an important anti-inflammatory molecule [45,102,103]. Hence, immune recovery is suggested to be associated with accumulated biliverdin in the gut bacterial of HIV-infected patients [45].
Fecal microbial transplantation is another clinical intervention using the transfer of fecal bacteria isolated from a healthy donor into the recipients in order to improve gut microbial diversity and healthier metabolic environment. This therapy displays significant effect in the treatment of patients with Clostridium difficile infection (CDI) [104]. Success of fecal microbial transplantation treatment in HIV infection is still lacking. However, pilot clinical trials (PROOV IT I and PROOV IT II) which are aimed to examine the impact of probiotics on the gut microbiome in HIV-infected, ART-naive or ART-experienced participants with poor CD4 recovery are currently in progress, and results from these studies will likely guide future avenues for microbiome studies in HIV-infected individuals with marked gut inflammation [105].
Integrative analysis of metabolome & microbiome
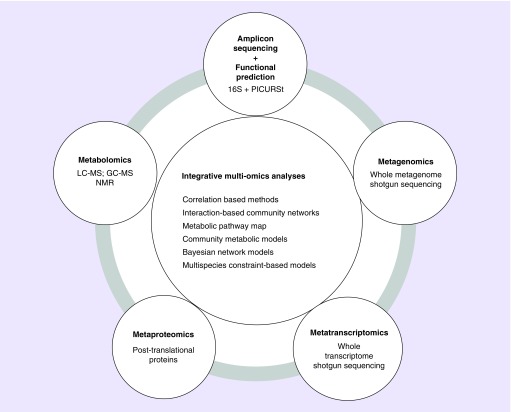
Developments in the next-generation high-throughput sequencing technologies, accompanied with progress in advanced multiomics bioinformatical/statistical algorithms, allow the rapid, cost-effective and comprehensive evaluation of gut microbiome and the functional perspective [106–108]. These efforts have dramatically advanced our understanding of host–microbiome homeostasis and the role of microbiota in human chronic diseases. Currently, the sequence based approaches include: marker gene amplicon (e.g., 16S rRNA) sequencing plus bioinformatics functional prediction pipeline and reference genomes database (e.g., PICURSt and KEGG) [109]; shotgun metagenomics sequencing, which allows one to directly profile the total gene content of the microbiota without amplification bias; meta-transcriptome (RNA-seq), which direct sequence the cDNA and have the potential to measure the active microorganisms and their effector molecules/genes/proteins [108]. However, as the final downstream products of genes and proteins, the metabolites are not applicable for sequence-based approaches. Targeted and untargeted metabolomics, based on gas chromatography mass spectrometry (GC–MS), liquid chromatography mass spectrometry (LC–MS) and nuclear magnetic resonance (NMR) have been widely used to identify metabolites that may play as functional mediators linking the microbiota and host chronic diseases. The integrated multiomics analyses, which combined complementary technologies together and connected direct measurements of the functional metabolites with the community structure of the microbiome as well as the genomic genes and functional pathways, hold the potential to improve our understanding of the complicated host–microbiota interactions, and provide deeper insight to mechanisms of functional changes during HIV progression (Figure 2).
Figure 2. . High-throughput omics approaches and integrative multiomics analyses.
GC–MS: Gas chromatography mass spectrometry; LC–MS: Liquid chromatography mass spectrometry; NMR: Nuclear magnetic resonance.
The integrated multiomics analysis is still in its developing stage, although some powerful bioinformatical/statistical algorithms and tools have showed promising potentials. The main stream approaches include Univariate and Multivariate Correlation-Based Approaches, Interaction-Based Community Networks, Topological Metabolic Models and Community Metabolic Models [110–113]. Other developing approaches include probabilistic graph models (e.g., Bayesian network models) and multispecies constraint-based models, which are complicated by determining the correct compartmentalization of species within a biological system, as well as choosing an optimal community objective function [114,115].
For integrated multiomics approaches applied in human cohort studies, the current trend is developing standard, powerful and visualizable bioinformatical/statistical algorithms and pipelines that could integrate the multiomics data accurately, realistically and effectively. Beside prolonged follow-up periods and increasing the study sample size, the study of bacterial microbiome could be extended to other microorganisms, such as fungal, parasitic, and viral (entero) pathogens. In addition, meta-transcriptome or proteomic approaches could also be applied, which may provide a better and clearer functional portrayal of influences and impacts of HIV infection, ART, and immune recovery on microbiota composition and related metabolites [19].
Few studies have used integrative approaches to investigate both gut microbiota and metabolites in HIV-infected individuals. Vesterbacka et al. found richer gut microbiota with distinct metabolic profile in HIV-infected elite controllers compared with progressor patients [73]. The distinct microbiota metabolic profile favored fatty acid metabolism, peroxisome proliferator-activated receptors-signaling and lipid biosynthesis protein pathways, combined with a decrease in carbohydrate metabolism and secondary bile acid synthesis [73]. These data suggested that HIV-induced changes in gut microbial communities are associated with metabolic alterations, which in turn, may contribute to clinical outcomes.
Conclusion & future perspective
In summary, during the past decade, the developments of high-throughput technologies and advanced bioinformatics/statistical algorithms provided us unprecedented resolution to track the alterations in microbiota metabolism and their impacts on host health. However, further integrated multiomics studies are still needed to improve our understanding of host–microbiota interactions, alterations in microbiota metabolism and possible mechanisms of functional changes during HIV infection.
One of the remaining challenges is to identify novel microbial metabolites, especially the discovery of key microbial metabolites that may indicate specific disease states, or that distinguish closely related disease conditions [71]. These metabolites could also be useful to identify individuals at risk for comorbidities (e.g., CVD) in HIV infection.
Furthermore, much work remains to fully characterize the impact of dietary components on changes in the microbial metabolites during HIV infection. Diet has a profound influence on the relationship between the intestinal microbiota and the host metabolite profiles [116]. The complex diet metabolism is dependent on multiple factors and could be affected by host genetic factors, host health status, diet quality and habits [117], as well as diverse microbial communities present in the gastrointestinal tract. Diet metabolism by gut microbiota produces small-molecule metabolites that could interfere with physiological processes such as immune homeostasis, energy metabolism, vascular function and neurological behavior [118,119]. However, the effects of dietary supplements on the gut microbial metabolites in HIV infection have been explored only to a limited extent.
Although our understanding of how HIV infection interacts with microbiota and microbial metabolites is advancing rapidly, there is only limited information available at the mechanism level or causation inference. As aforementioned, current evidence indicates that gut microbiota, microbiota-dependent metabolic pathways and microbial metabolites are not mere actors that are influenced by HIV infection [108,114], but rather they are influencing the whole health status and disease progression. In addition, the impact of HIV infection on these microbiota-related metabolites is also related to critical aspects of HIV comorbidities, such as CVD, diabetes, inflammation and lung infections [108].
Deeper insights gained from integrated multiomics studies would presumably open the door to novel therapeutic approaches including probiotics, tailoring diet modulations and fecal microbiota transplantation, which hold the potential to modify gut microbiome composition toward a beneficial direction and to treat infectious diarrhea in the late stages of HIV infection [120,121]. For the known beneficial microbial metabolites, there will be more options such as selectively increasing the abundance of the specific gut microbiota that can produce beneficial metabolites, or by engineering endogenous gut microbiota to produce metabolites in high levels. These interventions are based on the modulation of either bacterial species or the bacterial biosynthetic enzymes related to producing the target metabolites [71]. These may inform future efforts and help work toward integrative medicine utilizing the microbiota in HIV infection.
Executive summary.
Gut microbiome alteration in HIV infection
HIV infection has been associated with alternations in gut microbiota, which may affect microbial metabolite production.
Gut microbiota-related metabolites in HIV infection
Alternations in tryptophan metabolism are observed during HIV infection, which have been associated with atherosclerosis and cardiovascular risk.
Short-chain fatty acids (SCFAs) are the main fermentation products of gut microbiota from dietary fibers, which may play beneficial roles in regulating intestinal homeostasis.
Trimethylamine-N-oxide (TMAO) is known as a gut microbiota-dependent choline metabolite. Recent studies have revealed potential associations between TMAO and various aspects of cardiovascular disease (CVD) in HIV-infected individuals.
Treatments such as antiretroviral therapy (ART) could affect many aspects of microbial functional profiles, while it did not recover reduced gut microbiome diversity.
Integrative analysis of metabolome & microbiome
The developing integrated multiomics analyses hold promising potentials which allow rapid, cost-effective, and comprehensive evaluation of gut microbiome and the functional perspective.
Future perspective
The remaining challenges include:
Identifying the novel biomarker microbial metabolites that may indicate specific disease states, or distinguish closely related disease condition.
Fully characterizing the impact of dietary components on changes in the microbial metabolites during HIV infection.
Understanding how HIV infection interacts with microbiota and microbial metabolites at mechanism levels.
Novel therapeutic approaches including probiotics, tailoring diet modulations and fecal microbial transplantation, etc.
Footnotes
Financial & competing interests disclosure
Q Qi is supported by the National Heart, Lung, and Blood Institute (NHLBI) K01HL129892, R01HL140976 and R01HL060712, and by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) R01DK119268. The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest
- 1.Martin FP, Sprenger N, Yap IK. et al. Panorganismal gut microbiome–host metabolic crosstalk. J. Proteom. Res. 8(4), 2090–2105 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Candela M, Guidotti M, Fabbri A, Brigidi P, Franceschi C, Fiorentini C. Human intestinal microbiota: cross-talk with the host and its potential role in colorectal cancer. Crit. Rev. Microbiol. 37(1), 1–14 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Wang Z, Zolnik CP, Qiu Y. et al. Comparison of fecal collection methods for microbiome and metabolomics studies. Front. Cell. Infect. Microbiol. 8, 301 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frank DN, Zhu W, Sartor RB, Li E. Investigating the biological and clinical significance of human dysbioses. Trends Microbiol. 19(9), 427–434 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alkanani AK, Hara N, Gottlieb PA. et al. Alterations in intestinal microbiota correlate with susceptibility to type 1 diabetes. Diabetes 64(10), 3510–3520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Markle JG, Frank DN, Mortin-Toth S. et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 339(6123), 1084–1088 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Mathis D, Benoist C. The influence of the microbiota on type-1 diabetes: on the threshold of a leap forward in our understanding. Immunol. Rev. 245(1), 239–249 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Brown K, DeCoffe D, Molcan E, Gibson DL. Diet-induced dysbiosis of the intestinal microbiota and the effects on immunity and disease. Nutrients 4(8), 1095–1119 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.John GK, Mullin GE. The gut microbiome and obesity. Curr. Oncol. Rep. 18(7), 45 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Frank DN, Robertson CE, Hamm CM. et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm. Bowel Dis. 17(1), 179–184 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl Acad. Sci. USA 104(34), 13780–13785 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li E, Hamm CM, Gulati AS. et al. Inflammatory bowel diseases phenotype, C. difficile and NOD2 genotype are associated with shifts in human ileum associated microbial composition. PLoS ONE 7(6), e26284 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, Qi J, Zhao H. et al. Metagenomic sequencing reveals microbiota and its functional potential associated with periodontal disease. Sci. Rep. 3, 1843 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koeth RA, Wang Z, Levison BS. et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 19(5), 576–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh V, Yeoh BS, Vijay-Kumar M. Gut microbiome as a novel cardiovascular therapeutic target. Curr. Opin. Pharmacol. 27, 8–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang WH, Wang Z, Levison BS. et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N. Engl. J. Med. 368(17), 1575–1584 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yeoh N, Burton JP, Suppiah P, Reid G, Stebbings S. The role of the microbiome in rheumatic diseases. Curr. Rheumatol. Rep. 15(3), 314-012-0314-y (2013). [DOI] [PubMed] [Google Scholar]
- 18.Dillon SM, Frank DN, Wilson CC. The gut microbiome and HIV-1 pathogenesis: a two-way street. AIDS 30(18), 2737–2751 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ribeiro ABDTM, Heimesaat MM, Bereswill S. Changes of the intestinal microbiome–host homeostasis in HIV-infected individuals – a focus on the bacterial gut microbiome. Eur. J. Microbiol. Immunol. 7(3), 158–167 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brenchley JM, Schacker TW, Ruff LE. et al. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J. Exp. Med. 200(6), 749–759 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chege D, Sheth PM, Kain T. et al. Sigmoid Th17 populations, the HIV latent reservoir, and microbial translocation in men on long-term antiretroviral therapy. AIDS 25(6), 741–749 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Sankaran S, George MD, Reay E. et al. Rapid onset of intestinal epithelial barrier dysfunction in primary human immunodeficiency virus infection is driven by an imbalance between immune response and mucosal repair and regeneration. J. Virol. 82(1), 538–545 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim CJ, Nazli A, Rojas OL. et al. A role for mucosal IL-22 production and Th22 cells in HIV-associated mucosal immunopathogenesis. Mucosal Immunol. 5(6), 670–680 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Kok A, Hocqueloux L, Hocini H. et al. Early initiation of combined antiretroviral therapy preserves immune function in the gut of HIV-infected patients. Mucosal Immunol. 8(1), 127–140 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Brenchley JM, Paiardini M, Knox KS. et al. Differential Th17 CD4 T-cell depletion in pathogenic and nonpathogenic lentiviral infections. Blood 112(7), 2826–2835 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dinh DM, Volpe GE, Duffalo C. et al. Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J. Infect. Dis. 211(1), 19–27 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellis CL, Ma ZM, Mann SK. et al. Molecular characterization of stool microbiota in HIV-infected subjects by panbacterial and order-level 16S ribosomal DNA (rDNA) quantification and correlations with immune activation. J. Acquir. Immune Defic. Syndr. 57(5), 363–370 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mutlu EA, Keshavarzian A, Losurdo J. et al. A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS Pathog. 10(2), e1003829 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vujkovic-Cvijin I, Dunham RM, Iwai S. et al. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci. Transl. Med. 5(193), 193ra91 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Describes the associations between altered bacterial genera and tryptophan catabolism in HIV progression. Highlighted the key enzymes.
- 30.Lozupone CA, Rhodes ME, Neff CP, Fontenot AP, Campbell TB, Palmer BE. HIV-induced alteration in gut microbiota: driving factors, consequences, and effects of antiretroviral therapy. Gut Microbes 5(4), 562–570 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Brenchley JM, Douek DC. HIV infection and the gastrointestinal immune system. Mucosal Immunol. 1(1), 23–30 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brenchley JM, Price DA, Schacker TW. et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat. Med. 12(12), 1365–1371 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Moriyama K, Ando C, Tashiro K. et al. Polymerase chain reaction detection of bacterial 16S rRNA gene in human blood. Microbiol. Immunol. 52(7), 375–382 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Liu J, Williams B, Frank D, Dillon SM, Wilson CC, Landay AL. Inside out: HIV, the gut microbiome, and the mucosal immune system. J. Immunol. 198(2), 605–614 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Noguera-Julian M, Rocafort M, Guillen Y. et al. Gut microbiota linked to sexual preference and HIV infection. EBioMedicine 5, 135–146 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang L, Poles MA, Fisch GS. et al. HIV-induced immunosuppression is associated with colonization of the proximal gut by environmental bacteria. AIDS 30(1), 19–29 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McHardy IH, Li X, Tong M. et al. HIV infection is associated with compositional and functional shifts in the rectal mucosal microbiota. Microbiome 1(1), 26-2618-1-26 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dinh DM, Volpe GE, Duffalo C. et al. Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J. Infect. Dis. 211(1), 19–27 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dillon SM, Lee EJ, Kotter CV. et al. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol. 7(4), 983–994 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moon JY, Zolnik CP, Wang Z. et al. Gut microbiota and plasma metabolites associated with diabetes in women with, or at high risk for, HIV infection. EBioMedicine 37, 392–400 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tuddenham SA, Koay WLA, Zhao N. et al. The impact of HIV infection on gut microbiota alpha-diversity: an individual level meta-analysis. Clin. Infect. Dis. pii:ciz258 (2019) (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vazquez-Castellanos JF, Serrano-Villar S, Latorre A. et al. Altered metabolism of gut microbiota contributes to chronic immune activation in HIV-infected individuals. Mucosal Immunol. 8(4), 760–772 (2015). [DOI] [PubMed] [Google Scholar]; • Shotgun metagenomics based study that provide valuable information for gut microbiome alteration and functional contents in HIV infection.
- 43.Nowak P, Troseid M, Avershina E. et al. Gut microbiota diversity predicts immune status in HIV-1 infection. AIDS 29(18), 2409–2418 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Monaco CL, Gootenberg DB, Zhao G. et al. Altered virome and bacterial microbiome in human immunodeficiency virus-associated acquired immunodeficiency syndrome. Cell. Host Microbe 19(3), 311–322 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Serrano-Villar S, Rojo D, Martinez-Martinez M. et al. Gut bacteria metabolism impacts immune recovery in HIV-infected individuals. EBioMedicine 8, 203–216 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Villar-Garcia J, Guerri-Fernandez R, Moya A. et al. Impact of probiotic saccharomyces boulardii on the gut microbiome composition in HIV-treated patients: a double-blind, randomised, placebo-controlled trial. PLoS ONE 12(4), e0173802 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Filippo C, Cavalieri D, Di Paola M. et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl Acad. Sci. USA 107(33), 14691–14696 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yatsunenko T, Rey FE, Manary MJ. et al. Human gut microbiome viewed across age and geography. Nature 486(7402), 222–227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clarke SF, Murphy EF, O’Sullivan O. et al. Exercise and associated dietary extremes impact on gut microbial diversity. Gut 63(12), 1913–1920 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Lozupone CA, Li M, Campbell TB. et al. Alterations in the gut microbiota associated with HIV-1 infection. Cell. Host Microbe 14(3), 329–339 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Armstrong AJS, Shaffer M, Nusbacher NM. et al. An exploration of Prevotella-rich microbiomes in HIV and men who have sex with men. Microbiome 6, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu J, Williams B, Frank D, Dillon SM, Wilson CC, Landay AL. Inside out: HIV, the gut microbiome, and the mucosal immune system. J. Immunol. 198(2), 605–614 (2017). [DOI] [PubMed] [Google Scholar]; • Reviews paper well-summarized microbiome alterations associated with HIV-1 infection.
- 53.Serrano-Villar S, Rojo D, Martinez-Martinez M. et al. HIV infection results in metabolic alterations in the gut microbiota different from those induced by other diseases. Sci. Rep. 6, 26192 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu G, Chen S, Zhong J, Teng K, Yin Y. Crosstalk between tryptophan metabolism and cardiovascular disease, mechanisms, and therapeutic implications. Oxid. Med. Cell. Longev. 2017, 1602074 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qi Q, Hua S, Clish CB. et al. Plasma tryptophan-kynurenine metabolites are altered in human immunodeficiency virus infection and associated with progression of carotid artery atherosclerosis. Clin. Infect. Dis. 67(2), 235–242 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Favre D, Mold J, Hunt PW. et al. Tryptophan catabolism by indoleamine 2,3-dioxygenase 1 alters the balance of TH17 to regulatory T cells in HIV disease. Sci. Transl. Med. 2(32), 32ra36 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taylor MW, Feng GS. Relationship between interferon-gamma, indoleamine 2,3-dioxygenase, and tryptophan catabolism. FASEB J. 5(11), 2516–2522 (1991). [PubMed] [Google Scholar]
- 58.Byakwaga H, Hunt PW, Laker-Oketta M. et al. The kynurenine pathway of tryptophan catabolism and AIDS-associated Kaposi sarcoma in Africa. J. Acquir. Immune Defic. Syndr. 70(3), 296–303 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vazquez-Castellanos JF, Serrano-Villar S, Jimenez-Hernandez N. et al. Interplay between gut microbiota metabolism and inflammation in HIV infection. ISME J. 12(8), 1964–1976 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Siedner MJ, Kim JH, Nakku RS. et al. Persistent immune activation and carotid atherosclerosis in HIV-infected ugandans receiving antiretroviral therapy. J. Infect. Dis. 213(3), 370–378 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pertovaara M, Raitala A, Juonala M. et al. Indoleamine 2,3-dioxygenase enzyme activity correlates with risk factors for atherosclerosis: the Cardiovascular Risk in Young Finns study. Clin. Exp. Immunol. 148(1), 106–111 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Niinisalo P, Raitala A, Pertovaara M. et al. Indoleamine 2,3-dioxygenase activity associates with cardiovascular risk factors: the Health 2000 study. Scand. J. Clin. Lab. Invest. 68(8), 767–770 (2008). [DOI] [PubMed] [Google Scholar]
- 63.Kato A, Suzuki Y, Suda T. et al. Relationship between an increased serum kynurenine/tryptophan ratio and atherosclerotic parameters in hemodialysis patients. Hemodial. Int. 14(4), 418–424 (2010). [DOI] [PubMed] [Google Scholar]
- 64.Wirleitner B, Rudzite V, Neurauter G. et al. Immune activation and degradation of tryptophan in coronary heart disease. Eur. J. Clin. Invest. 33(7), 550–554 (2003). [DOI] [PubMed] [Google Scholar]
- 65.Pawlak K, Domaniewski T, Mysliwiec M, Pawlak D. The kynurenines are associated with oxidative stress, inflammation and the prevalence of cardiovascular disease in patients with end-stage renal disease. Atherosclerosis 204(1), 309–314 (2009). [DOI] [PubMed] [Google Scholar]
- 66.Sulo G, Vollset SE, Nygard O. et al. Neopterin and kynurenine-tryptophan ratio as predictors of coronary events in older adults, the Hordaland Health Study. Int. J. Cardiol. 168(2), 1435–1440 (2013). [DOI] [PubMed] [Google Scholar]
- 67.Pedersen ER, Midttun O, Ueland PM. et al. Systemic markers of interferon-gamma-mediated immune activation and long-term prognosis in patients with stable coronary artery disease. Arterioscler. Thromb. Vasc. Biol. 31(3), 698–704 (2011). [DOI] [PubMed] [Google Scholar]
- 68.Hoel H, Hove-Skovsgaard M, Hov JR. et al. Impact of HIV and type 2 diabetes on gut microbiota diversity, tryptophan catabolism and endothelial dysfunction. Sci. Rep. 8(1), 6725 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Konopelski P, Ufnal M. Indoles – gut bacteria metabolites of tryptophan with pharmacotherapeutic potential. Curr. Drug Metab. 19(10), 883–890 (2018). [DOI] [PubMed] [Google Scholar]
- 70.Zelante T, Iannitti RG, Cunha C. et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 39(2), 372–385 (2013). [DOI] [PubMed] [Google Scholar]
- 71.Zhang LS, Davies SS. Microbial metabolism of dietary components to bioactive metabolites: opportunities for new therapeutic interventions. Genome Med. 8(1), (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Romani L, Zelante T, De Luca A. et al. Microbiota control of a tryptophan-AhR pathway in disease tolerance to fungi. Eur. J. Immunol. 44(11), 3192–3200 (2014). [DOI] [PubMed] [Google Scholar]
- 73.Vesterbacka J, Rivera J, Noyan K. et al. Richer gut microbiota with distinct metabolic profile in HIV infected elite controllers. Sci. Rep. 7(1), 6269 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Aliment. Pharmacol. Ther. 27(2), 104–119 (2008). [DOI] [PubMed] [Google Scholar]
- 75.MacFarlane S, MacFarlane GT. Regulation of short-chain fatty acid production. Proc. Nutr. Soc. 62(1), 67–72 (2003). [DOI] [PubMed] [Google Scholar]
- 76.Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 294(1), 1–8 (2009). [DOI] [PubMed] [Google Scholar]
- 77.Sun Y, Ma Y, Lin P. et al. Fecal bacterial microbiome diversity in chronic HIV-infected patients in China. Emerg. Microbes Infect. 5, e31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Serrano-Villar S, Vazquez-Castellanos JF, Vallejo A. et al. The effects of prebiotics on microbial dysbiosis, butyrate production and immunity in HIV-infected subjects. Mucosal Immunol. 10(5), 1279–1293 (2017). [DOI] [PubMed] [Google Scholar]; • Identified short-chain fatty acids correlated bacteria and investigated the effects of prebiotics on microbial dysbiosis in the context of HIV infection.
- 79.Dillon SM, Kibbie J, Lee EJ. et al. Low abundance of colonic butyrate-producing bacteria in HIV infection is associated with microbial translocation and immune activation. AIDS 31(4), 511–521 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wolever TM, Spadafora PJ, Cunnane SC, Pencharz PB. Propionate inhibits incorporation of colonic [1,2-13C]acetate into plasma lipids in humans. Am. J. Clin. Nutr. 61(6), 1241–1247 (1995). [DOI] [PubMed] [Google Scholar]
- 81.Scott KP, Duncan SH, Flint HJ. Dietary fibre and the gut microbiota. Nutr. Bull. 33(3), 201–211 (2008). [Google Scholar]
- 82.Aldunate M, Srbinovski D, Hearps AC. et al. Antimicrobial and immune modulatory effects of lactic acid and short chain fatty acids produced by vaginal microbiota associated with eubiosis and bacterial vaginosis. Front. Physiol. 6, 164 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Frost G, Sleeth ML, Sahuri-Arisoylu M. et al. The short-chain fatty acid acetate reduces appetite via a central homeostatic mechanism. Nat. Commun. 5, 3611 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vallianou NG, Stratigou T, Tsagarakis S. Microbiome and diabetes: where are we now? Diabetes Res. Clin. Pract. 146, 111–118 (2018). [DOI] [PubMed] [Google Scholar]
- 85.Perry RJ, Peng L, Barry NA. et al. Acetate mediates a microbiome-brain-beta-cell axis to promote metabolic syndrome. Nature 534(7606), 213–217 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cho CE, Taesuwan S, Malysheva OV. et al. Trimethylamine-N-oxide (TMAO) response to animal source foods varies among healthy young men and is influenced by their gut microbiota composition: a randomized controlled trial. Mol. Nutr. Food Res. 61(1), (2017). [DOI] [PubMed] [Google Scholar]
- 87.Brown JM, Hazen SL. The gut microbial endocrine organ: bacterially derived signals driving cardiometabolic diseases. Annu. Rev. Med. 66, 343–359 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Romano KA, Vivas EI, Amador-Noguez D, Rey FE. Intestinal microbiota composition modulates choline bioavailability from diet and accumulation of the proatherogenic metabolite trimethylamine-N-oxide. MBio 6(2), e02481-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tang WH, Hazen SL. The contributory role of gut microbiota in cardiovascular disease. J. Clin. Invest. 124(10), 4204–4211 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Velasquez MT, Ramezani A, Manal A, Raj DS. Trimethylamine N-oxide: the good, the bad and the unknown. Toxins 8(11), pii:E326 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Haissman JM, Haugaard AK, Ostrowski SR. et al. Microbiota-dependent metabolite and cardiovascular disease marker trimethylamine-N-oxide (TMAO) is associated with monocyte activation but not platelet function in untreated HIV infection. BMC Infect. Dis. 17(1), 445 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Miller PE, Haberlen SA, Brown TT. et al. Brief report: intestinal microbiota-produced trimethylamine-N-oxide and its association with coronary stenosis and HIV serostatus. J. Acquir. Immune Defic. Syndr. 72(1), 114–118 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Srinivasa S, Fitch KV, Lo J. et al. Plaque burden in HIV-infected patients is associated with serum intestinal microbiota-generated trimethylamine. AIDS 29(4), 443–452 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Haissman JM, Knudsen A, Hoel H. et al. Microbiota-dependent marker TMAO is elevated in silent ischemia but is not associated with first-time myocardial infarction in HIV infection. J. Acquir. Immune Defic. Syndr. 71(2), 130–136 (2016). [DOI] [PubMed] [Google Scholar]
- 95.Knudsen A, Christensen TE, Thorsteinsson K. et al. Microbiota-dependent marker TMAO is not associated with decreased myocardial perfusion in well-treated HIV-infected patients as assessed by 82Rubidium PET/CT. J. Acquir. Immune Defic. Syndr. 72(4), e83–e85 (2016). [DOI] [PubMed] [Google Scholar]
- 96.Shan Z, Clish CB, Hua S. et al. Gut microbial-related choline metabolite trimethylamine-N-oxide is associated with progression of carotid artery atherosclerosis in HIV infection. J. Infect. Dis. 218(9), 1474–1479 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Linked plasma trimethylamine-N-oxide levels with monocyte activation, inflammation and progression of carotid atherosclerosis in two HIV cohorts.
- 97.Zhu W, Gregory JC, Org E. et al. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell 165(1), 111–124 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cunningham-Rundles S, Ahrne S, Johann-Liang R. et al. Effect of probiotic bacteria on microbial host defense, growth, and immune function in human immunodeficiency virus type-1 infection. Nutrients 3(12), 1042–1070 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cribbs SK, Uppal K, Li S. et al. Correlation of the lung microbiota with metabolic profiles in bronchoalveolar lavage fluid in HIV infection. Microbiome 4, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Aaron L, Saadoun D, Calatroni I. et al. Tuberculosis in HIV-infected patients: a comprehensive review. Clin. Microbiol. Infect. 10(5), 388–398 (2004). [DOI] [PubMed] [Google Scholar]
- 101.McPhee F, Caldera PS, Bemis GW, McDonagh AF, Kuntz ID, Craik CS. Bile pigments as HIV-1 protease inhibitors and their effects on HIV-1 viral maturation and infectivity in vitro. Biochem. J. 320(Pt 2), 681–686 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wegiel B, Otterbein LE. Go green: the anti-inflammatory effects of biliverdin reductase. Front. Pharmacol. 3, 47 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fowler CJ. Oleamide: a member of the endocannabinoid family? Br. J. Pharmacol. 141(2), 195–196 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gianotti RJ, Moss AC. Fecal microbiota transplantation: from Clostridium difficile to inflammatory bowel disease. Gastroenterol. Hepatol. 13(4), 209–213 (2017). [PMC free article] [PubMed] [Google Scholar]
- 105.Kim CJ, Walmsley SL, Raboud JM. et al. Can probiotics reduce inflammation and enhance gut immune health in people living with HIV: study designs for the Probiotic Visbiome for Inflammation and Translocation (PROOV IT) pilot trials. HIV Clin. Trials 17(4), 147–157 (2016). [DOI] [PubMed] [Google Scholar]
- 106.Di Bella JM, Bao Y, Gloor GB, Burton JP, Reid G. High throughput sequencing methods and analysis for microbiome research. J. Microbiol. Methods 95(3), 401–414 (2013). [DOI] [PubMed] [Google Scholar]
- 107.Frank DN, Pace NR. Gastrointestinal microbiology enters the metagenomics era. Curr. Opin. Gastroenterol. 24(1), 4–10 (2008). [DOI] [PubMed] [Google Scholar]
- 108.Serrano-Villar S, Moreno S, Ferrer M. The functional consequences of the microbiome in HIV: insights from metabolomic studies. Curr. Opin. HIV AIDS 13(1), 88–94 (2018). [DOI] [PubMed] [Google Scholar]
- 109.Langille MG, Zaneveld J, Caporaso JG. et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31(9), 814–821 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Theriot CM, Koenigsknecht MJ, Carlson PE Jr. et al. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat. Commun. 5, 3114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.El Aidy S, Derrien M, Merrifield CA. et al. Gut bacteria–host metabolic interplay during conventionalisation of the mouse germfree colon. ISME J. 7(4), 743–755 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sigurdsson MI, Jamshidi N, Steingrimsson E, Thiele I, Palsson BO. A detailed genome-wide reconstruction of mouse metabolism based on human Recon 1. BMC Syst. Biol. 4, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Borenstein E, Kupiec M, Feldman MW, Ruppin E. Large-scale reconstruction and phylogenetic analysis of metabolic environments. Proc. Natl Acad. Sci. USA 105(38), 14482–14487 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chong J, Xia J. Computational approaches for integrative analysis of the metabolome and microbiome. Metabolites 7(4), (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.McGeachie MJ, Sordillo JE, Gibson T. et al. Longitudinal prediction of the infant gut microbiome with dynamic bayesian networks. Sci. Rep. 6, 20359 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature 474(7351), 327–336 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ding M, Ellervik C, Huang T. et al. Diet quality and genetic association with body mass index: results from 3 observational studies. Am. J. Clin. Nutr. 108(6), 1291–1300 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.El-Far M, Tremblay CL. Gut microbial diversity in HIV infection post combined antiretroviral therapy: a key target for prevention of cardiovascular disease. Curr. Opin. HIV AIDS. 13(1), 38–44 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rowland I, Gibson G, Heinken A. et al. Gut microbiota functions: metabolism of nutrients and other food components. Eur. J. Nutr. 57(1), 1–24 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rabe SM. Treatment of recurrent Clostridium difficile infection with fecal transplantation. Gastroenterol. Nurs. 37(2), 156–163; quiz 164–165 (2014). [DOI] [PubMed] [Google Scholar]
- 121.Newman KM, Rank KM, Vaughn BP, Khoruts A. Treatment of recurrent Clostridium difficile infection using fecal microbiota transplantation in patients with inflammatory bowel disease. Gut Microbes 8(3), 303–309 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]