Abstract
Aristolochic acid (AA) is a potent human nephron-toxin and carcinogen. We previously reported that AA treatment resulted in DNA damage and mutation in the kidney and liver of rats. In this study, we have determined the DNA adducts and mutations induced by AA in rat spleen. Big Blue® transgenic rats were gavaged with 0, 0.1, 1.0, and 10.0 mg AA/kg body weight five-times/week for 3 months. Three DNA adducts, [7-(deoxyadenosin-N6-yl)-aristolactam I, 7-(deoxyadenosin-N6-yl)-aristolactam II and 7-(deoxyguanosin-N2-yl)-aris-tolactam I], were identified by 32P-postlabeling. Over the dose range studied, there were strong linear dose-responses for AA-DNA adduct formation in the treated rat spleens, ranging from 4.6 to 217.6 adducts/108 nucleotides. Spleen cII mutant frequencies also increased in a dose-dependent manner, ranging from 32.7 to 286.2 × 10−6 in the treated animals. Mutants isolated from the different treatment groups were sequenced; analysis of the resulting spectra indicated that there was a significant difference between the pattern of mutation in the 10 mg/kg AA-treated and the vehicle control rats. A:T → T:A transversion was the major type of mutation in AA-treated rats, whereas G:C → A:T transition was the main type of mutation in the vehicle controls. These results indicate that AA is genotoxic in the spleen of rats exposed under conditions that result in DNA adduct formation and mutation induction in kidney and liver.
Keywords: aristolochic acid, DNA adduct, mutagenicity, Big Blue® rat, rat spleen
INTRODUCTION
In the 1990s, patients in a Belgian weight loss program suffered an acute outbreak of progressive renal fibrosis, later called Chinese-herb nephropathy (CHN), and subsequently, urothelial carcinoma. The term CHN derived from evidence indicating that the toxicity was due to the accidental replacement of Stephania tetrandra by Aristo-lochia fangchi in a mixture of Chinese herbs ingested as part of the weight-loss program [Vanherweghem et al., 1993; Cosyns et al., 1994]. Subsequently, many other similar cases of CHN were reported worldwide [Cosyns, 2003; Debelle et al., 2008; Schmeiser et al., 2009]. As the source of the toxicity has been identified as the aristolochic acid (AA) contained in Aristolochia species [Debelle et al., 2008], the term CHN has been replaced by aristolochic acid nephropathy (AAN) [Gillerot et al., 2001]. Because herbal drugs derived from Aristolochia species have been used for medicinal purposes since antiquity and numerous ingredients containing AA are used regularly in some Asian countries, it is probable that the true incidence of AAN is underestimated [Debelle et al., 2008]. In addition, AA is an etiologic factor in the development of Balkan endemic nephropathy (BEN) [Arlt et al., 2007; Grollman et al., 2007], which was first described in the late 1950s. Thus, a high number of BEN cases are likely AAN cases.
Following the Belgian CHN/AAN incident and evidence indicating the etiopathological role of AA, the U.S. Food and Drug Administration issued warnings about the use and marketing of dietary supplements or other botanical-containing products that may contain AA [FDA, 2001]. Subsequent to this warning, however, products with possible AA content have been reported to be available on the internet [Gold and Slone, 2003] and such products continue to be used as traditional herbal medicines in some countries. FDA maintains an import alert restricting entry into the US of botanicals containing AA for use in dietary supplements [FDA, 2011].
AA is a family of structurally related nitrophenanthrene carboxylic acids, occurring in nature as a mixture of two major components, aristolochic acid I (AAI) and aristolochic acid II (AAII). AA is bioactivated by cytosolic nitroreductases [i.e., NAD(P)H:quinone oxidoreductase 1] or microsomal cytochromes P450 (i.e., CYP1A1 and CYP1A2) [Stiborova et al., 2011; Arlt et al., 2011a; Stiborova et al., 2012] and subsequently reacts with cellular proteins and DNA, leading to multiple forms of toxicity.
AA is mutagenic in bacterial and mammalian cell shortterm tests [Arlt et al., 2002] and carcinogenic in multiple tissues of both rats and mice. Rats treated orally with 0.1, 1.0, and 10.0 mg AA/kg body weight for 3 months developed squamous cell carcinomas in the forestomach and malignant tumors in the kidney and urinary tract, with incidences as high as 25%, 85%, and 100% at the three doses, respectively [Mengs et al., 1982]. Administration of 5 mg AA/kg body weight to mice for 3 weeks resulted in squamous cell carcinoma of the forestomach, adenocarcinoma of the glandular stomach, kidney adenomas, lung carcinomas, and uterine hemangiomas within 1 year of treatment [Mengs, 1988]. In 2002, the International Agency for Research on Cancer (IARC) concluded that there was limited evidence in humans and sufficient evidence in experimental animals for the carcinogenicity of AA, and that naturally occurring mixtures of AA are probably carcinogenic to humans (Group 2A) [IARC, 2002]. Based on new evidence, IARC recently reclassified AA as a Group 1 human carcinogen [Grosse et al., 2009].
In our previous studies, male Big Blue® transgenic rats were gavaged with 0, 0.1, 1.0, and 10.0 mg AA/kg body weight for 3 months. Levels of three major DNA adducts, 7-(deoxyadenosin-N6-yl)-aristolactam I (dA-AAI), 7-(deoxyadenosin-N6-yl)-aristolactam II (dA-AAII), and 7-(deoxyguanosin-N2-yl)-aristolactam I (dG-AAI), increased in both kidney and liver in a dose-responsive manner [Mei et al., 2006]. Similar dose-responses for cII mutant induction also were observed in both tissues, with A:T → T:A transversion identified as the predominant mutation in AA-treated rats [Mei et al., 2006; Chen et al., 2006a]. In addition, a significant dose-dependent induction of H-Ras codon 61 CAA → CTA mutation fraction was observed in both liver and kidney, which correlated significantly with AA-induced DNA adduct levels and cll mutant frequencies (MFs) [Wang et al., 2011]. Rat micro- array analysis detected more significantly altered genes involved in cancer-related pathways in kidney than in liver, and indicated that biological processes related to defense responses, apoptosis, and the immune response were altered significantly by AA exposure in kidney but not in liver [Chen et al., 2006b].
In vivo observations on the distribution of AA suggest that a number of tissues are potentially exposed. AA concentrations were measured in the kidney and other major organs (heart, liver, lung, and spleen), and also in the serum and urine of rats administrated 2 mg of AA orally, twice a day for 5 days [Liu et al., 2003]. The tissue AA concentration was highest in the lung and kidney, whereas liver and spleen AA concentrations were about 48% and 11% of kidney. DNA adduct levels were measured in a 34-year-old woman who had ingested an herbal drug containing AA for 9 months. The adenosine adduct of AAI was detected in her tissues at concentrations ranging from 1.0 to 21.9 adducts per 109 nucleotides. Spleen, at 21.2 adducts per 109 nucleotides, had the 2nd highest level of adducts after lung, followed by the adrenal gland, liver, and ureter, whereas only weak DNA binding was observed in bladder, brain, and kidney [Arlt et al., 2004].
The spleen functions in systemic circulation, sequestering cells from the bone marrow, thymus, and lymph nodes. It is also the largest secondary lymphoid organ, containing about one-fourth of the body’s lymphocytes. To provide additional information on the in vivo genotoxicity of AA, this study evaluated DNA adduct and mutation induction in rats treated with AA. Because these studies were conducted on the same Big Blue® rats used in our previous report of in vivo AA genotoxicity [Mei et al., 2006], we were able to compare AA adduct and mutation induction directly in kidney (a target tissue for AA toxicity), liver (a principle organ for xenobiotic metabolism), with spleen, an exposed tissue with no evidence for tumor induction.
MATERIALS AND METHODS
Chemical and Animals
AA was purchased from Sigma (St. Louis, MO). The AA content of the test agent was 96% (40% AAI and 56% AAII). Male Big Blue® transgenic rats were obtained from Taconic Laboratories (Germantown, NY). All animal procedures followed the recommendations of the NCTR Institutional Animal Care and Use Committee for the handling, maintenance, treatment, and sacrifice of the rats.
Treatments
Six-week-old Big Blue® rats were treated with 0.1, 1.0, or 10.0 mg AA/kg body weight by gavage, five times per week for 12 weeks; the treatment schedule was based on a previous carcinogenesis study [Mengs et al., 1982]. Vehicle control rats were gavaged with 0.9% sodium chloride using the same schedule as for the AA-treated rats. Six rats from each treatment group were sacrificed one day after the last treatment. Tissues, including the spleen, were isolated, frozen quickly in liquid nitrogen, and stored at −80°C.
DNA Adduct Analysis by 32P-Postlabeling
Spleen DNAs were isolated by a standard phenol extraction method. 32P-postlabeling analysis using the nuclease P1 enrichment and thin-layer chromatography (TLC) was performed as described previously [Phillips and Arlt, 2007; Arlt et al., 2011b]. Chromatographic conditions for TLC on polyethylenimine-cellulose plates (10 × 20 cm2; Macherey-Nagel, Düren, Germany) were: D1, 1.0 M sodium phosphate, pH 6.8; D3: 3.5 lithium-formate, 8.5 M urea, pH 4; D4, 0.8 M lithium chloride, 0.5 M Tris-HCl, 8.5 M urea, pH 9; and D5, 1.7 M sodium phosphate, pH 6. After chromatography, the TLC sheets were scanned using a Packard Instant Imager (Dowers Grove, IL). DNA adduct levels were calculated from the adduct cpm, the specific activity of [γ−32P]ATP, and the amount of DNA (pmol of DNA-P) used. Results are expressed as DNA adducts per 108 normal nucleotides. Enzymatic preparation of AA-DNA adduct reference compounds was performed as described previously [Arlt et al., 2001]. Urothelial DNA samples from AAN patients [Nortier et al., 2000] also were included in the analysis for comparison.
cII Mutation Assay
MFs were determined by the cII mutation assay as described previously [Mei et al., 2004a; Mei et al., 2004b]. High-molecular-weight genomic DNA was extracted from rat spleens using the RecoverEase DNA Isolation Kit (Stratagene; La Jolla, CA). The packaging of the phage, plating the packaged DNA samples, and determination of mutants were carried out following the manufacturer’s instructions for the Select-cII Mutation Detection System for Big Blue1 Rodents (Stratagene). The shuttle vector containing the cII target gene was rescued from total genomic DNA with phage packaging extract, and the resulting phage plated on Escherichia coli host strain G1250. To determine the total titer of packaged phages, G1250 bacteria were mixed with 1:3,000 dilutions of phage, plated on TB1 plates, and incubated overnight at 37°C (nonselective conditions). For mutant selection, the packaged phages were mixed with G1250, plated on TB1 plates, and incubated at 24°C for about 42 hr (conditions for cII selection). The cII MF is defined as the total number of mutant plaques (determined at 24°C) divided by the total number of plaques screened (determined at 37°C) and expressed as mutants per million plaque-forming units (pfus).
Sequence Analysis of the cII Mutants
All cII mutant plaques were isolated from the analyses conducted with spleen DNA and replated at low density to verify the mutant phenotype. Single, well-isolated plaques were transferred from these plates to individual wells of 96-well microplates containing 100 μl of sterile distilled water. The microplate with samples was heated at 100°C for 5 min and centrifuged at 12,000g for 3 min. For PCR amplification, 10 μl of the supernatant was added to 10 μl of Promega PCR Master Mix (Madison, WI) with cII primers [Slikker et al., 2004]. The PCR reaction was performed using a PCR System 9700 (Applied Biosystems, Foster City, CA), with the cycling parameters described previously [Slikker et al., 2004; Mei et al., 2005]. The PCR products were isolated using a QIA-Quick PCR product purification kit (Qiagen, Chatsworth, CA). The cII mutant DNA was sequenced with a CEQ Dye Terminator Cycle Sequencing Kit and a Beckman Coulter CEQ 8000 Genetic Analysis System (Brea, CA). The primer for cII mutation sequencing was the upstream primer used for the PCR.
Statistical Analyses
Analyses were performed using SigmaPlot 11.0 (Systat, Chicago, IL). All DNA adduct and MF data are expressed as the mean ± standard deviation from six rats per group. Statistical significance was determined by one-way analysis of variance followed by the Holm-Sidak method for pairwise comparisons. Because the variance increased with the magnitude of the MF, the MF data were log-transformed before conducting the analysis. Mutation spectra were compared using the computer program written by Cariello et al. [1994] for the Monte Carlo analysis developed by Adams and Skopek [1987].
RESULTS
AA-Induced DNA Adducts in the Spleen
Male Big Blue® transgenic rats were treated with AA 5 days a week for 12 weeks. At the end of the treatments, the mean body weights of the rats treated with 0.1, 1.0, and 10.0 mg/kg body weight AA were 5%, 7%, and 15% less than the vehicle controls [Mei et al., 2006]. DNA adducts formed in rat spleens were analyzed by 32P-post-labeling. The DNA adduct pattern induced by AA in the spleens was the same as previously observed in liver and kidney of AA-treated rats [Mei et al., 2006], consisting of dA-AAI (spot 1), dG-AAI (spot 2), and dA-AAII (spot 3), respectively (Table I). No DNA adducts were found in control rats. Levels of total AA-DNA adducts were 4.6, 29.4, and 217.6 per 108 nucleotides for the spleens of rats exposed to 0.1, 1.0, and 10.0 mg/kg body weight AA, respectively (Table I).
TABLE I.
DNA Adduct Formation in Spleen of Rats Treated With Different Doses of Aristolochic Acid
| Aristolochic acid (mg/kg body weight) | |||
|---|---|---|---|
| DNA adductab | 0.1 | 1.0 | 10.0 |
| dA-AAI | 1.06 ± 0.23 | 5.90 ± 0.59 | 55.69 ± 10.87 |
| dG-AAI | 1.07 ± 0.23 | 6.15 ± 0.64 | 44.28 ± 8.65 |
| dA-AAII | 2.49 ± 0.47 | 17.33 ± 1.31 | 117.67 ± 21.09 |
| Total | 4.62 ± 0.90 | 29.38 ± 2.17 | 217.64 ± 40.08 |
The values are the mean adducts ± SD per 108 nucleotides for each group of six rats.
In the control group of six rats, no AA-DNA adducts were found.
AA-Induced MFs in the Spleen cII Gene
DNA from each spleen was packaged two-three times either to confirm the MF or to obtain a minimum of 2 × 105 pfus for mutant detection. The spleen MFs for the vehicle control Big Blue® rats ranged from 17 to 53 × 10−6, with an average of 35 ± 14 × 10−6, which is slightly higher than the MFs determined for liver and kidney of control rats [Mei et al., 2006]. The results from the cII mutation assay indicated that there was a dose- dependent increase in MF for the spleens of AA-treated rats (Table II). The MFs for the middle- and high-dose groups (62 ± 28 × 10−6 and 286 ± 82 × 10−6) were significantly higher than the MFs for control and low- dose groups (P < 0.05 and P < 0.001, respectively). There were also significant differences between the MFs of the high- and middle-dose groups (P < 0.001).
TABLE II.
The cII Mutant Frequencies in Spleen of Rats Treated With Different Doses of Aristolochic Acid (mg/kg Body Weight)
| Group (mg/kg body weight) | Total plaques screened (×103) | Mutant plaques | Mutant Frequency (×10−6) | Mean ± SD (X10−6) |
|---|---|---|---|---|
| 0 | 391 | 11 | 28.1 | |
| 148 | 5 | 33.8 | ||
| 231 | 4 | 17.3 | ||
| 300 | 15 | 50.0 | ||
| 223 | 12 | 53.8 | ||
| 204 | 6 | 29.4 | ||
| 35.4 ± 13.9 | ||||
| 0.1 | 125 | 3 | 24.0 | |
| 421 | 15 | 35.6 | ||
| 233 | 5 | 21.5 | ||
| 384 | 19 | 49.5 | ||
| 299 | 9 | 30.1 | ||
| 452 | 16 | 35.4 | ||
| 32.7 ± 10.1 | ||||
| 1.0 | 229 | 6 | 26.2 | |
| 236 | 16 | 67.8 | ||
| 234 | 20 | 85.5 | ||
| 210 | 11 | 52.4 | ||
| 288 | 12 | 41.7 | ||
| 70 | 7 | 100.0 | ||
| 62.3 ± 27.6a,b | ||||
| 10.0 | 285 | 85 | 298.8 | |
| 196 | 44 | 224.5 | ||
| 109 | 37 | 339.4 | ||
| 135 | 56 | 414.8 | ||
| 173 | 42 | 242.8 | ||
| 122 | 24 | 196.7 | ||
| 286.2 ± 81.5c,d |
P < 0.05 (significantly higher than the control group).
Significantly higher than the group treated with 0.1 mg/kg (P < 0.05).
P < 0.001 (significantly higher than the control group).
Significantly higher than the groups treated with 0.1 and 1.0 mg/kg (P < 0.001).
Mutation Spectra in the Spleen cII Gene
All cII mutant plaques obtained from vehicle control and AA-treated spleens were isolated and replated at low density to verify the mutants, and then analyzed for sequence alterations in the cII gene. We successfully sequenced 52 mutants from the six control rats (about 98% of mutant plaques) and 41, 61, and 196 mutants from six rats each that were treated with either 0.1, 1.0, or 10.0 mg/kg body weight AA, respectively (about 61%, 85%, and 68% of mutant plaques in these three groups). Mutations that were found more than once among the mutants isolated from a single animal were assumed to be siblings and to represent only one independent mutation. As shown in Table III, we identified 47 independent cII mutations for control rats (90% of mutants with sequence) and 31, 54, and 173 independent mutations for the low-, middle-, and high-dose-treated rats, respectively (76%, 89%, and 88% of mutants with sequence in these three groups). The mutations were distributed throughout the cII gene in both the control and AA-treated groups. Of the six control rats, four of them had G:C base substitutions at base pairs 25, 34, 103, and 196 of the cII gene. In high-dose-treated group, three or four rats had A:T base substitutions at base pairs 2, 26, 127, 139, 161, 178, and 294, and these mutations were not found in vehicle control rats (Table III).
TABLE III.
Mutations in the cII Gene of Spleen From AA-Treated and Control Male Big Blue® Rats
| Number of mutations (independent) | |||||||
|---|---|---|---|---|---|---|---|
| AA (mg/kg body weight) | |||||||
| Positiona | Mutationb | Amino acid change | Sequence context 5′ → 3′c | Control | 0.1 | 1.0 | 10.0 |
| −18 | C → G | N/A | tatCTAagg | 1 | |||
| −13 | G → A | N/A | ctaAGGaaa | 1 | |||
| −1 | T → C | N/A | ttaCATatg | 1 | |||
| 1 | A → G | Met → Val | catATGgtt | 1 | 4 (2) | ||
| A → T | Met → Leu | catATGgtt | 1 | ||||
| 2 | T → A | Met → Lys | catATGgtt | 1 d | 1 | ||
| T → C | Met → Thr | catATGgtt | 2 d | ||||
| 3 | G → A | Met → Ile | catATGgtt | 1 | 1 | ||
| G → C | Met → Ile | catATGgtt | 1 | ||||
| G → T | Met → Ile | catATGgtt | 2(1) | 1 | |||
| 4 | G → C | Val → Leu | atgGTTcgt | 1 | |||
| 14 | A → T | Asn → Ile | gcaAACaaa | 1 | |||
| 15 | C → A | Asn → Lys | gcaAACaaa | 1 | |||
| 16 | A → T | Lys → Stop | aacAAAcgc | 2 | |||
| 19 | C → T | Arg → Cys | aaaCGCaac | 1 | |||
| 24 | C → G | Asn → Lys | cgcAACgag | 1 | |||
| 25 | G → A | Glu → Lys | aacGAGgct | 3 | 2 | ||
| G → T | Glu → Stop | aacGAGgct | 1 | 1 | |||
| 26 | A → G | Glu → Gly | aacGAGgct | 1 | |||
| A → T | Glu → Val | aacGAGgct | 3 | ||||
| 28 | G → A | Ala → Thr | gagGCTcta | 1 | |||
| G → C | Ala → Pro | gagGCTcta | 1 | ||||
| 31 | C → G | Leu → Val | gctCTAcga | 1 | 1 | ||
| 32 | T → C | Leu → Pro | gctCTAcga | 1 | |||
| 34 | C → G | Arg → Gly | ctaCGAatc | 1 | |||
| C → T | Arg → Stop | ctaCGAatc | 4 | 1 | 1 | ||
| 35 | G → A | Arg → Gln | ctaCGAatc | 1 | 1 | 1 | |
| G → C | Arg → Pro | ctaCGAatc | 1 | ||||
| G → T | Arg → Leu | ctaCGAatc | 1 | ||||
| 38 | T → A | Ile → Asn | cgaATCgag | 1 | |||
| 39 | C → G | Ile → Met | cgaATCgag | 1 | |||
| 40 | G → A | Glu → Lys | atcGAGagt | 1 | 2 | ||
| 43 | A → G | Ser → Gly | gagAGTgcg | 1 | |||
| 47 | C → G | Ala → Gly | agtGCGttg | 1 | |||
| −C | Frameshift | agtGCGttg | 1 | ||||
| 49 | T → A | Leu → Met | gcgTTGctt | 1 | |||
| 50 | T → C | Leu → Ser | gcgTTGctt | 1 | |||
| 51 | G → T | Leu → Phe | gcgTTGctt | 1 | |||
| 53 | T → C | Leu → Pro | ttgCTTaac | 1 | |||
| 55 | A → C | Asn → His | cttAACaaa | 1 | |||
| 58 | A → T | Lys → Stop | aacAAAatc | 1 | |||
| A → G | Lys → Glu | aacAAAatc | 1 | ||||
| 60 | +AATCGC | Frameshift | aacAAAatc | 1 | |||
| 60 | A → T | Lys → Lys | aacAAAatc | 1 | |||
| 63 | C → G | Ile → Met | gcaATCgca | 1 | |||
| 64 | G → A | Ala → Thr | atcGCAatg | 1 | 4 | ||
| 68–69 | −TG | Frameshift | gcaATGctt | 1 | |||
| 71 | T → C | Leu → Pro | atgCTTgga | 1 | 1 | ||
| 73 | G → A | Gly → Arg | cttGGAact | 1 | 1 | ||
| 74 | G → T | Gly → Val | cttGGAact | 1 | |||
| 77 | C → A | Thr → Asn | ggaACTgag | 1 | |||
| 82 | A → G | Lys → Glu | gagAAGaca | 1 | |||
| 86 | C → G | Thr → Arg | aagACAgcg | 2 | |||
| 89 | C → A | Ala → Glu | acaGCGgaa | 2(1) | |||
| C → T | Ala → Val | acaGCGgaa | 1 | 2 | 1 | ||
| 90–91 | −G | Frameshift | acaGCGGAAgct | 1 | |||
| 94 | G → A | Ala → Thr | gaaGCTgtg | 1 | |||
| G → T | Ala → Ser | gaaGCTgtg | 1 | ||||
| 99–101 | −G | Frameshift | gctGTGGGCgtt | 1 | |||
| 100 | G → A | Gly → Ser | gtgGGCgtt | 1 | |||
| G → T | Gly → Cys | gtgGGCgtt | 1 | ||||
| 101 | G → T | Gly → Val | gtgGGCgtt | 1 | |||
| 103 | G → A | Val → Ile | ggcGTTgat | 3 (2) | 3 (2) | 5 (2) | 3 |
| G → C | Val → Leu | ggcGTTgat | 1 | 1 | |||
| G → T | Val → Phe | ggcGTTgat | 1 | ||||
| 104 | T → A | Val → Asp | ggcGTTgat | 1 | 1 | ||
| 108 | T → A | Asp → Glu | gttGATaag | 1 | |||
| 109 | A → G | Lys → Glu | gatAAGtcg | 1 | |||
| A → T | Lys → Stop | gatAAGtcg | 1 | ||||
| 110 | A → T | Lys → Met | gatAAGtcg | 2 | |||
| 111 | G → T | Lys → Asn | gatAAGtcg | 1 | |||
| 113 | C → T | Ser → Leu | aagTCGcag | 1 | 1 | 1 | |
| 115 | C → T | Gln → Stop | tcgCAGatc | 1 | |||
| 116 | A → T | Gln → Leu | tcgCAGatc | 1 | |||
| 118 | A → T | Ile → Phe | cagATCagc | 1 | |||
| 119 | T → A | Ile → Asn | cagATCagc | 1 | |||
| 120 | C → G | Ile → Met | cagATCagc | 1 | |||
| 124 | A → T | Arg → Trp | agcAGGtgg | 2 | |||
| 125 | G → T | Arg → Met | agcAGGtgg | 1 | |||
| 127 | T → A | Trp → Arg | aggTGGaag | 3 | |||
| 129–132 | +A | Frameshift | aggTGGAAGagg | 1 | |||
| 129 | G → A | Trp → Stop | aggTGGaag | 1 | |||
| G → C | Trp → cys | aggTGGaag | 1 | ||||
| 130 | A → T | Lys → Stop | tggAAGagg | 1 d | |||
| 135 | G → C | Arg → Ser | aagAGGgac | 1 | |||
| 139 | T → A | Trp → Arg | gacTGGatt | 4 (3) | |||
| 141 | G → T | Trp → Cys | gacTGGatt | 1 | 1 | ||
| 145 | C → A | Pro → Thr | attCCAaag | 1 | |||
| 146 | C → A | Pro → Gln | attCCAaag | 1 | |||
| 150 | G → T | Lys → Asn | ccaAAGttc | 1 | 1 | 1 | |
| 158 | T → C | Met → Thr | tcaATGctg | 1 | |||
| 159 | G → A | Met → Ile | tcaATGctg | 1 | |||
| 161 | T → A | Leu → Gln | atgCTGctt | 2 | |||
| T → C | Leu → Pro | atgCTGctt | 1 | ||||
| 164 | T → A | Leu → His | ctgCTTgct | 2 | |||
| 167 | C → G | Ala → Gly | cttGCTgtt | 1 | |||
| 172 | C → A | Leu → Ile | gttCTTgaa | 1 | |||
| C → T | Leu → Phe | gttCTTgaa | 1 | ||||
| −C | Frameshift | gttCTTgaa | 1 | ||||
| 175 | G → C | Glu → Gln | cttGAAtgg | 1 | |||
| G → T | Glu → Stop | cttGAAtgg | 1 | ||||
| 176 | A → T | Glu → Val | cttGAAtgg | 1 | 2 | ||
| 178/185 | +G | Frameshift | gaaTGGGGGGTCgtt | 4 (2) | 4 (3) | 5 (2) | 5 (4) |
| 178 | T → A | Trp → Arg | gaaTGGggg | 3 | |||
| 179 | G → A | Trp → Stop | gaaTGGggg | 1 | 3 | ||
| 179–184 | −G | Frameshift | gaaTGGGGGGTCgtt | 1 | 2 (1) | 3 | 6 (3) |
| 181 | G → T | Gly → Trp | tggGGGgtc | 1 | |||
| 182 | G → T | Gly → Val | tggGGGgtc | 3 | |||
| 185 | T → A | Val → Asp | gggGTCgtt | 1 | 2 | ||
| T → C | Val → Ala | gggGTCgtt | 1 | ||||
| T → G | Val → Gly | gggGTCgtt | 1 | 1 | |||
| 190 | G → C | Asp → Gln | gttGACgac | 1 | |||
| G → T | Asp → Tyr | gttGACgac | 1 | ||||
| 190–192 | −GAC | Frameshift | gttGACgac | 1 | |||
| 191 | A → G | Asp → Gly | gttGACgac | 1 | |||
| 191–192 | AC → TT | Asp → Val | gttGACgac | 1 | |||
| 193 | G → A | Asp → Asn | gacGACgac | 1 | 1 | 5 (3) | |
| G → C | Asp → His | gacGACgac | 4(1) | ||||
| G → T | Asp → Tyr | gacGACgac | 1 | 2 | |||
| 193–194 | GA → TT | Asp → Tyr | gacGACgac | 1 | |||
| 194 | A → T | Asp → Val | gacGACgac | 1 | |||
| 196 | G → A | Asp → Asn | gacGACatg | 4 (3) | 2 | 1 | 3 |
| G → C | Asp → His | gacGACatg | 2 | ||||
| G → T | Asp → Tyr | gacGACatg | 1 | 1 | |||
| 199 | A → G | Met → Val | gacATGgct | 4 (3) | |||
| 200 | T → A | Met → Lys | gacATGgct | 1 | |||
| T → C | Met → Thr | gacATGgct | 1 | ||||
| 201–202 | −G | Frameshift | gacATGGCTcga | 1 | |||
| 202 | G → C | Ala → Pro | atgGCTcga | 1 | |||
| 206 | G → A | Arg → Gln | gctCGAttg | 2 (1) | 1 | ||
| G → C | Arg → Pro | gctCGAttg | 1 | 1 | |||
| 208 | T → A | Leu → Met | cgaTTGgcg | 2(1) | |||
| 209 | T → A | Leu → Stop | cgaTTGgcg | 2 | |||
| 210 | G → T | Leu → Phe | cgaTTGgcg | 3 | |||
| 211 | G → C | Ala → Pro | ttgGCGcga | 1 | |||
| 212 | C → A | Ala → Glu | ttgGCGcga | 2 | |||
| C → G | Ala → Gly | ttgGCGcga | 1 | ||||
| C → T | Ala → Val | ttgGCGcga | 2 | 7 (2) | 3 | 6 (5) | |
| 214 | C → A | Arg → Arg | gcgCGAcaa | 1 | |||
| C → T | Arg → Stop | gcgCGAcaa | 3 | 3 (2) | 3 (2) | 10(5) | |
| 215 | G → T | Arg → Leu | gcgCGAcaa | 1 | |||
| 217 | C → T | Gln → Stop | cgaCAAgtt | 1 | |||
| 218 | A → T | Gln → Leu | cgaCAAgtt | 3 | |||
| 220 | G → A | Val → Ile | caaGTTgct | 1 | |||
| G → T | Val → Phe | caaGTTgct | 1 | ||||
| 224 | C → A | Ala → Asp | gttGCTgcg | 1 | |||
| 226 | G → C | Ala → Pro | gctGCGatt | 1 | |||
| 228 | −G | Frameshift | gctGCGatt | 1 | 3 (1) | ||
| 230 | T → A | Ile → Asn | gcgATTctc | 1 | |||
| 232 | C → T | Leu → Phe | attCTCacc | 1 | |||
| 233 | T → A | Leu → His | attCTCacc | 1 | |||
| T → C | Leu → Pro | attCTCacc | 1 | ||||
| 240 | −T | Frameshift | accAATaaa | 1 | |||
| 241 | A → T | Lys → Stop | aatAAAaaa | 1 | |||
| 241–246 | −A | Frameshift | aatAAAAAAcgc | 1 | |||
| 253 | −G | Frameshift | ccgGCGgca | 1 | |||
| 274 | C → A | Gln → Lys | gaaCAAatc | 1 | |||
| C → G | Gln → Glu | gaaCAAatc | 1 | ||||
| 275 | A → T | Gln → Leu | gaaCAAatc | 1 | |||
| 276 | A → T | Gln → His | gaaCAAatc | 1 | |||
| 287 | A → T | Glu → Val | atgGAGttc | 1 | |||
| 292 | T → A | Stop → Arg | ttcTGAggt | 1 | |||
| T → C | Stop → Arg | ttcTGAggt | 1 | ||||
| 293 | G → C | Stop → Ser | ttcTGAggt | 1 | |||
| G → T | Stop → Leu | ttcTGAggt | 1 | ||||
| 294 | A → G | Stop → Trp | ttcTGAggt | 1 | |||
| A → T | Stop → Cys | ttcTGAggt | 1 | 3 | |||
| Total | 52 (47) | 41 (31) | 61 (54) | 196 (173) | |||
Abbreviations: −, deletion; +, insertion
Position 1 is the first base of the start codon in the cII coding sequence.
Presented in term of sequence change on nontranscribed DNA strand.
Uppercase indicates target codon and target bases are underlined.
Complex mutation.
Table IV summarizes the types of independent mutations in the spleen cII gene from low-, middle-, and high- dose-treated rats and the vehicle control rats. The overall pattern of mutations in the spleen of rats treated with the middle and high doses of AA differed significantly from the mutation spectrum of the vehicle control rats (P = 0.036 and P = 0.003, respectively), whereas the mutation spectrum in the spleens of low-dose-treated rats was not significantly different from the control group (P = 0.846). A:T → T:A transversion (27%) was the major type of spleen mutation in the high-dose-treated group (Table IV), and the percentages of this type of mutation also were increased in the low- (10%) and middle-dose (17%) groups when compared with the control group (4%). The mutation spectrum for the control rat spleens was dominated by G:C → A:T transition (51%), and this mutation type was decreased in the middle-dose (26%) and high- dose (25%) groups.
TABLE IV.
Summary of Independent Mutations in the cII Gene of Spleen From Control and Aristolochic Acid-Treated Rats
| Aristolochic acid (mg/kg body weight) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Control | 0.1 | 1.0a | 10.0b | |||||
| Type of mutation | Number | % | Number | % | Number | % | Number | % |
| G:C → C:G | 4 | 8.5 | 3 | 9.7 | 8 | 14.8 | 18 | 10.4 |
| G:C → A:T | 24 | 51.1 | 17 | 54.8 | 14 | 25.9 | 43 | 24.9 |
| G:C → T:A | 10 | 21.3 | 3 | 9.7 | 9 | 16.7 | 24 | 13.9 |
| A:T → T:A | 2 | 4.3 | 3 | 9.7 | 9 | 16.7 | 47 | 27.2 |
| A:T → C:G | 1 | 2.1 | 0 | 0 | 0 | 0 | 2 | 1.1 |
| A:T → G:C | 1 | 2.1 | 1 | 3.2 | 4 | 7.4 | 20 | 11.6 |
| Frameshift | 4 | 8.5 | 4 | 12.9 | 9 | 16.7 | 16 | 9.2 |
| Tandem base substitution | 1 | 2.1 | 0 | 0 | 0 | 0 | 1 | 0.6 |
| Complex mutation | 0 | 0 | 0 | 0 | 1 | 1.8 | 2 | 1.1 |
| Total mutants screened | 47 | 31 | 54 | 173 | ||||
Spectrum for AA-treated spleens was significantly different from the control.
P = 0.0364.
P = 0.0029.
DISCUSSION
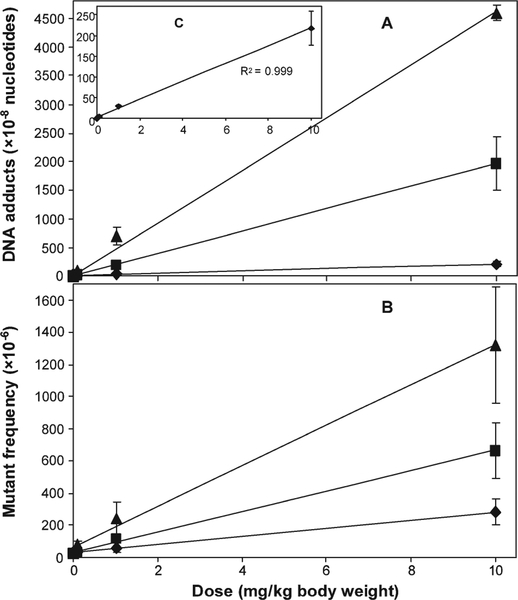
We previously reported on DNA adduct formation and mutation induction in kidney and liver of Big Blue® transgenic rats treated with 0, 0.1, 1.0, or 10.0 mg AA per kg body weight for 3 months [Chen et al., 2006a; Mei et al., 2006]. The results of this study indicate that, as was the case with kidney and liver, AA induced a dose- dependent increase of DNA damage and cII MF in rat spleens (Tables I and II). The DNA adduct levels in the kidneys and livers, however, were ~20-fold and ~5 to 9-fold higher, respectively, than those in the spleens (Fig. 1). Based on the three data points per tissue derived from AA-treated rats, the total DNA binding in all three tissues increased in a linear dose-dependent manner (Figs. 1A and 1C). Comparison of the MFs detected in the three tissues indicated that the spleen cII MFs were about half those of liver and one-fourth those of kidney (Fig. 1B). Over the dose range studied, the two endpoints for AA genotoxicity, DNA adduction formation and mutation induction, were linearly correlated in the spleen. Similar correlations were observed previously for the liver and kidney samples.
Fig. 1.
Total DNA adduct levels (A) and cII mutant frequencies (B) in spleen, liver, and kidney of Big Blue rats treated with different doses (0.1, 1.0, and 10.0 mg/kg body weight) of aristolochic acid for 12 weeks. The data represent the mean ± SD of groups of six rats. Spleen data for total DNA adduct levels (C, inner figure of A) and mutant frequencies are from Table II, whereas liver data are from Mei et al [2006] and kidney data are from Chen et al [2006a]. (♦) spleen; (■) liver; and (▲) kidney.
To determine if there were any differences between vehicle control and AA-treated rats in terms of clonal expansion or mutational spectra, we sequenced the cII mutants isolated from all four groups in this study. Since the cell turnover time in rat spleen is about 15 days [Cameron, 1971], the number of sibling mutants in the animal will increase and result in a higher MF if a mutated clone is disproportionately expanded. As shown in Table III, 90% of the mutants sequenced from the control rats and 88% of the mutants from the high-dose-treated rats contained independent mutations, suggesting minimum clonal expansion. Although mutations in AA-treated rats were distributed throughout the cII gene, A:T base substitutions at base pairs 2, 26, 127, 139, 161, 178, and 294 occurred more than three times (i.e., in three or more different rats; Table III), indicating that certain base pairs may be mutated preferentially.
Comparison of the vehicle control mutation spectra in spleen, liver, and kidney revealed no significant differences (Table V). In contrast, the overall pattern of mutations induced by the high-dose of AA in spleen was significantly different from kidney and liver, whereas there was no difference between the liver and kidney mutation spectra [Mei et al., 2006]. This may be explained by the relatively modest increases in MF produced by AA in spleens when compared with those in kidney and liver (Fig. 1) and by the differences in DNA adduct formation. The low-, middle-, and high-dose AA treatments resulted in 10%, 17%, and 27% A:T → T:A transversion, respectively, while this mutation accounted for only 4% of the mutations in the spleens from control animals (Table IV). The percentage of A:T → G:C transition was also increased from 2 to 12% with increasing AA dose. Combining all mutations at A:T sites results in a larger difference in the types of mutations in the high-dose-treated and control groups, i.e., 40% vs. 9% mutations at A:T base pairs (Table IV). AA also resulted in increases of A:T → T:A transversion and A:T → G:C transition in rat kidney and liver [Chen et al. 2006a; Mei et al. 2006]. It was reported that AA induced mainly A:T → T:A trans-version in the kidney, bladder, and forestomach of Muta mice [Kohara et al., 2002]. Collectively, these results suggest that reaction with deoxyadenine is the major mutagenic pathway for AA. Based on the preponderance of A → T transversion in the mutation spectra, the adenine adducts formed by AA (dA-AAI and dA-AAII) appear to have greater miscoding potential than the guanine adducts [Broschard et al., 1994; Stiborova et al., 1994].
TABLE V.
Statistical Pairwise Multiple Comparisons of Mutational Spectra in Spleen, Liver, and Kidney From Rats Treated With 10 mg/kg Body Weight Aristolochic Acid (AA) or the Vehicle Control
| Comparisona | P value | Statistically significant difference |
|---|---|---|
| Spleen-control vs. liver-control | 0.43176 | No |
| Spleen-control vs. kidney-control | 0.73764 | No |
| Spleen-AA vs. liver-AA | 0.00000 | Yes |
| Spleen-AA vs. kidney-AA | 0.00059 | Yes |
Liver data are from Mei et al. [2006] and kidney data are from Chen et al. [2006a].
As to the significance of these mutations for cancer, a high frequency of A → T transversion has been reported in codon 61 of the Ha-ras oncogene in tumors from rodents treated with AAI [Schmeiser et al., 1991] and in TP53 mutations from AAN and BEN patients [Lord et al., 2004; Grollman et al., 2007; Moriya et al., 2011]. A high frequency of A → T transversion mutation also has been observed in the human TP53 gene of the Hupki (human TP53 knock-in) mouse model [Nedelko et al. 2009; Kucab et al., 2010]. Thus, AA-DNA adducts at adenine residues appear to be the critical premutagenic lesions in the carcinogenic process [Arlt et al., 2000; Arlt et al., 2007].
The genotoxicity of AA has been studied extensively in vitro and in vivo. A large body of evidence suggests that AA-induced DNA adduct formation, followed by cellular proliferation and fixation of mutations, is responsible for cancer development in AA-treated animals [Arlt et al., 2002; Arlt et al., 2007]. However, AA-DNA adducts in AAN patients have been found in several organs in addition to the urinary tract, including the liver and spleen, but AAN-associated tumors have been observed only in urothelial tissue [Nortier et al., 2003; Arlt et al., 2004; Lord et al., 2004]. In our previous study, we found that treatment with 10 mg/kg body weight AA for 3 months resulted in a relatively high DNA adduct levels and MFs in liver as well as kidney [Mei et al., 2006]; in this study we found adducts and mutations in spleen (Fig. 1). These observations suggest that additional factors other than DNA damage and mutations may be critical for the high incidence of urothelial tumors. In female mice, oral AA exposure caused malignant lymphoma (50%), as well as tumors of the forestomach (100%), lung (100%), kidney (75%), and uterus (37%) [Mengs, 1988]. A single AA-treated male rat was reported to have tumors of the hematopoietic (blood-producing) system [NTP, 2008]. It has been reported that AA treatment of Muta mouse resulted in significantly higher lacZ and cII MFs in several different tissues, including spleen and bone marrow (1.6- to 2.6-fold) [Kohara et al., 2002]. In addition, AA increased micronuclei in bone marrow cells of mice [Mengs and Klein, 1988], induced sister chromatid exchanges [Abel and Schimmer, 1983] and micronucleus formation [Kevekordes et al., 2001] in cultured human peripheral lymphocytes. Taken together, it is possible that a longer follow-up of the Belgian cohort may indicate that AA exposure has an increased risk of malignancy in hematopoietic tissues (lymphomas and leukemia). However, it should be noted that additional studies should be performed to understand genotoxic activities of AA in spleen.
In summary, using chronic treatment conditions comparable to those known to produce tumors in rats [Mengs et al., 1982], AA exposure resulted in dose-responsive increases in the total DNA adducts and the cII MF in rat spleen. The MF and levels of adducts in spleen were lower than those previously reported for kidney and liver [Mei et al., 2006]. Sequence analysis of the cII mutants from the spleen indicated that there was a significant difference in the mutation spectra in 10 mg/kg AA-treated and vehicle control rats. This study provides additional support to a growing body of evidence for a mutagenic mechanism for AA carcinogenesis in rodents.
ACKNOWLEDGMENTS
The authors thank Dr. Robert H. Heflich for helpful discussions and comments. This research was partly supported by appointment (ERE) to the Summer Student Research Program at the NCTR administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the U.S. Food and Drug Administration (FDA). The views presented in this article do not necessarily reflect those of the U.S. FDA. LPM and ERE performed the cII mutation assay and sequence analysis. XG was involved in the sequence analysis of the mutants and data analyses. VMA performed the DNA adduct analysis. TC and NM designed the study. NM performed animal treatment and prepared the manuscript with important intellectual input from VMA and TC. All authors approved the final version of manuscript.
Grant sponsors:
Association for International Cancer Research (AICR), Cancer Research UK.
REFERENCES
- Abel G, Schimmer O. 1983. Induction of structural chromosome aberrations and sister chromatid exchanges in human lymphocytes in vitro by aristolochic acid. Hum Genet 64:131–133. [DOI] [PubMed] [Google Scholar]
- Adams WT, Skopek TR. 1987. Statistical test for the comparison of samples from mutational spectra. J Mol Biol 194:391–396. [DOI] [PubMed] [Google Scholar]
- Arlt VM, Wiessler M, Schmeiser HH. 2000. Using polymerase arrest to detect DNA binding specificity of aristolochic acid in the mouse H-ras gene. Carcinogenesis 21:235–242. [DOI] [PubMed] [Google Scholar]
- Arlt VM, Pfohl-Leszkowicz A, Cosyns J, Schmeiser HH. 2001. Analyses of DNA adducts formed by ochratoxin A and aristolochic acid in patients with Chinese herbs nephropathy. Mutat Res 494: 143–150. [DOI] [PubMed] [Google Scholar]
- Arlt VM, Stiborova M, Schmeiser HH. 2002. Aristolochic acid as a probable human cancer hazard in herbal remedies: A review. Mutagenesis 17:265–277. [DOI] [PubMed] [Google Scholar]
- Arlt VM, Alunni-Perret V, Quatrehomme G, Ohayon P, Albano L, Gaid H, Michiels JF, Meyrier A, Cassuto E, Wiessler M, Schmeiser HH, Cosyns JP. 2004. Aristolochic acid (AA)-DNA adduct as marker of AA exposure and risk factor for AA nephropathy-associated cancer. Int J Cancer 111:977–980. [DOI] [PubMed] [Google Scholar]
- Arlt VM, Stiborova M, vom Brocke J, Simoes ML, Lord GM, Nortier JL, Hollstein M, Phillips DH, Schmeiser HH. 2007. Aristolochic acid mutagenesis: Molecular clues to the aetiology of Balkan endemic nephropathy-associated urothelial cancer. Carcinogenesis 28:2253–2261. [DOI] [PubMed] [Google Scholar]
- Arlt VM, Levova K, Barta F, Shi Z, Evans JD, Frei E, Schmeiser HH, Nebert DW, Phillips DH, Stiborova M. 2011a. Role of P450 1A1 and P450 1A2 in bioactivation versus detoxication of the renal carcinogen aristolochic acid I: Studies in Cyp1a1(−/−), Cyp1a2(−/−), and Cyp1a1/1a2(−/−) mice. Chem Res Toxicol 24:1710–1719. [DOI] [PubMed] [Google Scholar]
- Arlt VM, Zuo J, Trenz K, Roufosse CA, Lord GM, Nortier JL, Schmeiser HH, Hollstein M, Phillips DH. 2011b. Gene expression changes induced by the human carcinogen aristolochic acid I in renal and hepatic tissue of mice. Int J Cancer 128:21–32. [DOI] [PubMed] [Google Scholar]
- Broschard TH, Wiessler M, von der Lieth CW, Schmeiser HH. 1994. Translesional synthesis on DNA templates containing site-specifically placed deoxyadenosine and deoxyguanosine adducts formed by the plant carcinogen aristolochic acid. Carcinogenesis 15: 2331–2340. [DOI] [PubMed] [Google Scholar]
- Cameron IL. 1971Cell proliferation and renewal in the mammalian body In: Cameron IL, Thrasher JD, editors. Cellular and Molecular Renewal in the Mammalian Body. New York:Academic Press; pp 45–85. [Google Scholar]
- Cariello NF, Piegorsch WW, Adams WT, Skopek TR. 1994. Computer program for the analysis of mutational spectra: Application to p53 mutations. Carcinogenesis 15:2281–2285. [DOI] [PubMed] [Google Scholar]
- Chen L, Mei N, Yao L, Chen T. 2006a. Mutations induced by carcinogenic doses of aristolochic acid in kidney of Big Blue transgenic rats. Toxicol Lett 165:250–256. [DOI] [PubMed] [Google Scholar]
- Chen T, Guo L, Zhang L, Shi L, Fang H, Sun Y, Fuscoe JC, Mei N. 2006b. Gene expression profiles distinguish the carcinogenic effects of aristolochic acid in target (kidney) and non-target (liver) tissues in rats. BMC Bioinformatics 7(Suppl 2):S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosyns JP. 2003. Aristolochic acid and ‘Chinese herbs nephropathy’: A review of the evidence to date. Drug Saf 26:33–48. [DOI] [PubMed] [Google Scholar]
- Cosyns JP, Jadoul M, Squifflet JP, De Plaen JF, Ferluga D, van Ypersele de Strihou C. 1994. Chinese herbs nephropathy: A clue to Balkan endemic nephropathy? Kidney Int 45:1680–1688. [DOI] [PubMed] [Google Scholar]
- Debelle FD, Vanherweghem JL, Nortier JL. 2008. Aristolochic acid nephropathy: A worldwide problem. Kidney Int 74:158–169. [DOI] [PubMed] [Google Scholar]
- FDA. 2001. FDA concerned about botanical products, including dietary supplements, containing aristolochic acid. USFDA, Center for Food Safety and Applied Nutrition. Available at: http://www.fda.gov/Food/DietarySupplements/Alerts/ucm095272.htm [Accessed 28 March 2012].
- FDA. 2011. Detention without physical examination of bulk/finished dietary supplements products containing aristolochic acid. US: FDA; Available at: http://www.accessdata.fda.gov/cms_ia/importalert_141.html [Accessed 28 March 2012]. [Google Scholar]
- Gillerot G, Jadoul M, Arlt VM, van Ypersele De Strihou C, Schmeiser HH, But PP, Bieler CA, Cosyns JP. 2001. Aristolochic acid nephropathy in a Chinese patient: Time to abandon the term “Chinese herbs nephropathy”? Am J Kidney Dis 38:E26. [DOI] [PubMed] [Google Scholar]
- Gold LS, Slone TH. 2003. Aristolochic acid, an herbal carcinogen, sold on the Web after FDA alert. N Engl J Med 349:1576–1577. [DOI] [PubMed] [Google Scholar]
- Grollman AP, Shibutani S, Moriya M, Miller F, Wu L, Moll U, Suzuki N, Fernandes A, Rosenquist T, Medverec Z, Jakovina K, Brdar B, Slade N, Turesky RJ, Goodenough AK, Rieger R, Vukelic M, Jelakovic B. 2007. Aristolochic acid and the etiology of endemic (Balkan) nephropathy. Proc Natl Acad Sci USA 104:12129–12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse Y, Baan R, Straif K, Secretan B, El Ghissassi F, Bouvard V, Benbrahim-Tallaa L, Guha N, Galichet L, Cogliano V. 2009. A review of human carcinogens-Part A: Pharmaceuticals. Lancet Oncol 10:13–14. [DOI] [PubMed] [Google Scholar]
- IARC. 2002. Some traditional herbal medicines, some mycotoxins, nephthalene and styrene IARC Monographs on the Evaluation of Carcinogenic Risks to Humans 82 Lyon, France: Available at: http://monographs.iarc.fr/ENG/Monographs/vol82/mono82.pdf [Accessed 28 March 2012]. [PMC free article] [PubMed] [Google Scholar]
- Kevekordes S, Spielberger J, Burghaus CM, Birkenkamp P, Zietz B, Paufler P, Diez M, Bolten C, Dunkelberg H. 2001. Micronucleus formation in human lymphocytes and in the metabolically competent human hepatoma cell line Hep-G2: Results with 15 naturally occurring substances. Anticancer Res 21:461–469. [PubMed] [Google Scholar]
- Kohara A, Suzuki T, Honma M, Ohwada T, Hayashi M. 2002. Mutagenicity of aristolochic acid in the lambda/lacZ transgenic mouse (MutaMouse). Mutat Res 515:63–72. [DOI] [PubMed] [Google Scholar]
- Kucab JE, Phillips DH, Arlt VM. 2010. Linking environmental carcinogen exposure to TP53 mutations in human tumours using the human TP53 knock-in (Hupki) mouse model. FEBS J 277:2567–2583. [DOI] [PubMed] [Google Scholar]
- Liu MC, Maruyama S, Mizuno M, Morita Y, Hanaki S, Yuzawa Y, Matsuo S. 2003. The nephrotoxicity of Aristolochia manshuriensis in rats is attributable to its aristolochic acids. Clin Exp Nephrol 7:186–194. [DOI] [PubMed] [Google Scholar]
- Lord GM, Hollstein M, Arlt VM, Roufosse C, Pusey CD, Cook T, Schmeiser HH. 2004. DNA adducts and p53 mutations in a patient with aristolochic acid-associated nephropathy. Am J Kidney Dis 43:e11–e17. [DOI] [PubMed] [Google Scholar]
- Mei N, Chou MW, Fu PP, Heflich RH, Chen T. 2004a. Differential mutagenicity of riddelliine in liver endothelial and parenchymal cells of transgenic Big Blue rats. Cancer Lett 215:151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei N, Heflich RH, Chou MW, Chen T. 2004b. Mutations induced by the carcinogenic pyrrolizidine alkaloid riddelliine in the liver cII gene of transgenic Big Blue rats. Chem Res Toxicol 17:814–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei N, Heflich RH, Moore MM, Chen T. 2005. Age-dependent sensitivity of Big Blue transgenic mice to the mutagenicity of N-ethyl-N-nitrosourea (ENU) in liver. Mutat Res 572:14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei N, Arlt VM, Phillips DH, Heflich RH, Chen T. 2006. DNA adduct formation and mutation induction by aristolochic acid in rat kidney and liver. Mutat Res 602:83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengs U 1988. Tumour induction in mice following exposure to aristolochic acid. Arch Toxicol 61:504–505. [DOI] [PubMed] [Google Scholar]
- Mengs U, Klein M. 1988. Genotoxic effects of aristolochic acid in the mouse micronucleus test. Planta Med 54:502–503. [DOI] [PubMed] [Google Scholar]
- Mengs U, Lang W, Poch JA. 1982. The carcinogenic action of aristolochic acid in rats. Arch Toxicol 51:107–119. [Google Scholar]
- Moriya M, Slade N, Brdar B, Medverec Z, Tomic K, Jelakovic B, Wu L, Truong S, Fernandes A, Grollman AP. 2011. TP53 Mutational signature for aristolochic acid: An environmental carcinogen. Int J Cancer 129:1532–1536. [DOI] [PubMed] [Google Scholar]
- Nedelko T, Arlt VM, Phillips DH, Hollstein M. 2009. TP53 mutation signature supports involvement of aristolochic acid in the aetiology of endemic nephropathy-associated tumours. Int J Cancer 124:987–990. [DOI] [PubMed] [Google Scholar]
- Nortier JL, Martinez MC, Schmeiser HH, Arlt VM, Bieler CA, Petein M, Depierreux MF, De Pauw L, Abramowicz D, Vereerstraeten P, Vanherweghem JL. 2000. Urothelial carcinoma associated with the use of a Chinese herb (Aristolochia fangchi). N Engl J Med 342:1686–1692. [DOI] [PubMed] [Google Scholar]
- Nortier JL, Schmeiser HH, Muniz Martinez MC, Arlt VM, Vervaet C, Garbar CH, Daelemans P, Vanherweghem JL. 2003. Invasive urothelial carcinoma after exposure to Chinese herbal medicine containing aristolochic acid may occur without severe renal failure. Nephrol Dial Transplant 18:426–428. [DOI] [PubMed] [Google Scholar]
- NTP. 2008. Report on carcinogens background document for aristolochic acids. National Toxicology Program; Available at: http://ntp.niehs.nih.gov/files/Aristolochic_Acids_(FINAL-02Sep08)_Redo02[03].pdf [Accessed 28 March 2012]. [PubMed] [Google Scholar]
- Phillips DH, Arlt VM. 2007. The 32P-postlabeling assay for DNA adducts. Nat Protoc 2:2772–2781. [DOI] [PubMed] [Google Scholar]
- Schmeiser HH, Scherf HR, Wiessler M. 1991. Activating mutations at codon 61 of the c-Ha-ras gene in thin-tissue sections of tumors induced by aristolochic acid in rats and mice. Cancer Lett 59:139–143. [DOI] [PubMed] [Google Scholar]
- Schmeiser HH, Stiborova M, Arlt VM. 2009. Chemical and molecular basis of the carcinogenicity of Aristolochia plants. Curr Opin Drug Discov Devel 12:141–148. [PubMed] [Google Scholar]
- Slikker W III, Mei N, Chen T. 2004. N-ethyl-N-nitrosourea (ENU) increased brain mutations in prenatal and neonatal mice but not in the adults. Toxicol Sci 81:112–120. [DOI] [PubMed] [Google Scholar]
- Stiborova M, Fernando RC, Schmeiser HH, Frei E, Pfau W, Wiessler M. 1994. Characterization of DNA adducts formed by aristolochic acids in the target organ (forestomach) of rats by 32P-postlabelling analysis using different chromatographic procedures. Carcinogenesis 15:1187–1192. [DOI] [PubMed] [Google Scholar]
- Stiborova M, Mareis J, Frei E, Arlt VM, Martinek V, Schmeiser HH. 2011. The human carcinogen aristolochic acid i is activated to form DNA adducts by human NAD(P)H:quinone oxidoreductase without the contribution of acetyltransferases or sulfotransferases. Environ Mol Mutagen 52:448–459. [DOI] [PubMed] [Google Scholar]
- Stiborova M, Levova K, Barta F, Shi Z, Frei E, Schmeiser HH, Nebert DW, Phillips DH, Arlt VM. 2012. Bioactivation versus detoxication of the urothelial carcinogen aristolochic acid I by human cytochrome P450 1A1 and 1A2. Toxicol Sci 125:345–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanherweghem JL, Depierreux M, Tielemans C, Abramowicz D, Dratwa M, Jadoul M, Richard C, Vandervelde D, Verbeelen D, Vanhaelen-Fastre R. 1993. Rapidly progressive interstitial renal fibrosis in young women: Association with slimming regimen including Chinese herbs. Lancet 341:387–391. [DOI] [PubMed] [Google Scholar]
- Wang Y, Meng F, Arlt VM, Mei N, Chen T, Parsons BL. 2011. Aristolochic acid-induced carcinogenesis examined by ACB-PCR quantification of H-Ras and K-Ras mutant fraction. Mutagenesis 26:619–628. [DOI] [PubMed] [Google Scholar]