Abstract
Genetic profiling has been used to link mosquito bloodmeals to the individual humans, but this analysis has not been done for other mammalian bloodmeals. In this study, we describe a microsatellite-based method for identifying individual pigs in mosquito bloodmeals based on their unique multilocus genotypes. Eleven tetranucleotide microsatellites and a sex-specific marker were selected based on Smith-Waterman DNA sequence alignment scores from the reference genome and primers were designed with features that reduce primer dimers, promote complete adenylation, and enable fluorescent labeling of amplicons. A multiplex polymerase chain reaction (PCR) assay was optimized and validated by analyzing DNA of individual pigs from several nuclear families and breeds before it was used to analyze genomic DNA of pig-derived mosquito bloodmeals from villages of Papua New Guinea. Population analysis of the nuclear families showed high expected and observed heterozygosity. The probability of observing two unrelated or sibling individuals sharing the same genotype at a single microsatellite locus or a combination of loci was vanishingly low. Samples had unique genotypes and gender was accurately predicted. Analysis of 129 pig bloodmeals identified 19 unique genotypes, which varied greatly in frequency in the mosquito bloodmeal samples. The high allelic diversity of the microsatellite loci and low probability of false attribution of identity show that this genotyping method reliably distinguishes distantly and closely related pigs and can be used to identify individual pigs from genotyped mosquito bloodmeals.
Keywords: blood, microsatellite, mosquito, pig, genotype
Pig (Sus scrofa) is one of a number of domesticated mammalian species that live in close contact with humans (Frantz et al. 2015). While not competent for human malaria parasites, pigs are common hosts of Anopheles vectors of malaria (Russell et al. 2016, Keven et al. 2017) and of Culex vectors of Japanese encephalitis virus (JEV); pigs are also competent hosts of that virus (Hurk et al. 2001, 2003, 2008). When serving as hosts for Anopheles mosquitoes in malaria-endemic settings, pigs may contribute indirectly to malaria transmission by promoting population growth of mosquito vectors. In general, increasing domestic mammal density tends to favor malaria vector survival and increase potential for malaria parasite transmission when zoophilic malaria vectors are considered (Sota and Mogi 1989, Bouma and Rowland 1995), particularly by reducing the time required to locate any host for blood (Saul 2003). Further, the rate of feeding by Culex vectors of JEV on pigs increases when pigs are kept close to humans, a process that may enhance epizootic amplification of JEV and risk of infection in humans (Hurk et al. 2003). Similarly, presence of pigs reduces the human blood index of a mosquito population and spatial distribution of pigs relative to humans can cause heterogeneity in human exposure to Anopheles bites (Burkot et al. 1989); humans in areas with lower number of pigs may receive more bites than those in areas with higher number of pigs.
Evaluation of mosquito host selection involves sampling blood-fed mosquitoes and analyzing their bloodmeals to identify the source of the vertebrate blood, commonly achieved through PCR amplification of species-specific locus of the vertebrate mitochondrial cytochrome b gene (Kent and Norris 2005). Genotyping of human bloodmeals using forensic-based profiling systems allows identification of particular individual humans bitten by the mosquitoes (Chow-Shaffer et al. 2000, Michael et al. 2001, De Benedictis et al. 2003, Soremekun et al. 2004, Scott et al. 2006). It provides a means of assessing whether individuals or demographic groups contribute more bloodmeals than do others and whether the degree of heterogeneity of feeding on individual hosts or on particular groups of hosts has epidemiological significance (Dye and Hasibeder 1986, Hasibeder and Dye 1988, Bolzoni et al. 2015). Data of this type would be useful in targeting host-directed treatment strategies with mosquito-killing drugs to control mosquito vector populations (Omura and Crump 2017). Previous studies which evaluated the host selection behavior of a group of malaria-transmitting mosquito species in Papua New Guinea (PNG) found that individuals of all species fed on both humans and pigs with some species showing stronger fidelity to humans, others more to pigs, and others both hosts and also dogs depending upon their relative availability (Burkot et al. 1988, 1989; Keven et al. 2017). In on-going work, we are extending the human bloodmeal analysis to include identification of different human individuals with genotyping methods. In this article, we describe a method for genotyping pig bloodmeals. This will help us evaluate patterns of pig and human host choice by vectors of human malaria.
Materials and Methods
Samples
To validate the new genotyping method, DNA of 211 pigs (110 males, 101 females) collected in Michigan (Supp. Table S1) were analyzed. Of these, 22 were unrelated individuals of four different breeds: Landrace (n = 6), Duroc (n = 6), Hampshire (n = 5), and Yorkshire (n = 5). The other 189 were offspring of 18 different dams (or litters) and three different sires. Litters A (n = 12), B (n = 3), C (n = 4), E (n = 15), F (n = 11), G (n = 10), L (n = 14), M (n = 9), N (n = 9), O (n =12), P (n = 11), Q (n = 12), and R (n = 9) shared a common sire and were mixed breeds from crosses between Yorkshire (the dams) and Hampshire (the sire). Litters D (n = 13), I (n = 11), J (n = 14), and K (n = 7) shared a common sire and were pure Yorkshire breed. Individuals of litter H (n = 13) were pure Yorkshire breed and offspring of the third sire. Bloodmeals of female Anopheles mosquitoes (n = 129) confirmed by species-specific PCR (Kent and Norris 2005) to be imbibed from pigs were analyzed. The bloodmeal samples consisted of 20 Anopheles farauti sensu stricto from Mirap village and 109 An. farauti no. 4 from Kokofine village in the Madang province of PNG. We chose to analyze bloodmeal samples from these two villages because mosquitoes in these villages, particularly the two chosen species, were found to over utilize pigs compared to humans and other host species (Keven et al. 2017) and were thus likely to have multiple pig individuals in the bloodmeals. These mosquitoes were collected by barrier screen, a sampling method which uses vertically erected shade-cloth netting positioned between village houses and surrounding vegetation and acts as intercepting device for mosquitoes as they commute into and out of a village to seek blood hosts (Burkot et al. 2013, Keven et al. 2017). DNA was extracted (DNeasy Blood and Tissue Kit; Cat. No. 69506, Qiagen, Valencia, CA) from tail tissue (25 mg, routinely removed during standard management practices) of the 211 pigs, quantified using Qubit and Nanodrop machines, and adjusted to 3 ng/µl. DNA from mosquito bloodmeals was extracted as described elsewhere (Keven et al. 2017) and adjusted to 6 ng/µl.
Marker Selection and Primer Design
The pig reference genome (Sscrofa10.2) in the UCSC genome browser (https://genome.ucsc.edu/) was screened for tetranucleotide microsatellite sequences using the RepeatMasker track information via the Table browser feature. From a pool of candidate markers, 11 tetranucleotide microsatellite loci located on 10 different autosomal chromosomes were selected. Selection of the markers was based on Smith-Waterman sequence alignment (SW) scores (500 to 800), low GC content, and non-repetitive DNA in the flanking regions. The SW scores can provide useful estimates of heterozygosity and mutation rate for tetranucleotide microsatellites which allows the selection of markers with near maximal heterozygosity for a population while minimizing mutation rates (Venta et al. unpublished). Low GC content is important for PCR amplification efficiency, and all amplicons had GC percentages of less than 40%. All primers were designed to regions devoid of transposable elements to minimize spurious amplification elsewhere in the genome. All markers used also had a longest uninterrupted sequence (LUS; also known as the perfect repeat number) of at least eight in the pig reference genome, because shorter LUSs tend to have unacceptably low levels of polymorphism.
Using the web-based program Primer3 (http://bioinfo.ut.ee/primer3-0.4.0/), several candidate primer pairs with AA at the 3′ end of both forward and reverse sequence were generated for each marker. Primers with AA at 3′ end have been shown to reduce the probability of primer-dimer formation (Innis and Gelfand 1999). Of 28 primer pairs evaluated, 11 were chosen, one pair for each microsatellite marker. Four of these primer pairs had product sizes within 150–200 base pairs (bp) range, four had product sizes within 300–350 bp range and three had product sizes within 450–500 bp range. An amelogenin locus with published primer sequence (Lin et al. 2014) was included as sex-specific marker. Primers were also examined for internal matches using Autodimer (Vallone and Butler 2004). The markers, their chromosome location, repeat motif, SW score, primer sequences, and allele range are shown in Table 1. The two markers on chromosome 13 are 160 Mb apart and they are therefore likely to independently assort like markers on separate chromosomes under the assumption that this physical distance corresponds to a recombination distance of 50 cM.
Table 1.
Primer sequences and additional information of the 12 genetic markers
| Name | Chr | Repeat motif | SW | Primer sequences (5′–3′) | Allele range |
|---|---|---|---|---|---|
| SSchr13b | 13 | (TTTC)n | 522 |
CTCCAACTCACCTCCAAC AAACACAGGTAAAGAAAGGCCAAA GTTTCTTGAACCCTGGCAACCTTGAA |
494–561 |
| SSchr15 | 15 | (TTTC)n | 567 |
AAACCTCTCTCCACACCCAAA GACACATGGATGCTGTTACCTAAA GTTTCTTGATATGAGTA GAGGACAGGAAAGAA |
483–524 |
| SSchr1 | 1 | (TCTA)n | 531 |
AACTCCACCACTCCCACAA AACTGAGCCGTTTACAAACCAA GTTTCTTATGGGAATT CCGTTCAGGAAA |
353–509 |
| SSchr11 | 11 | (GAAA)n | 639 |
CTCCAACTCACCTCCAACAAA CAAATCTGCACCCAAGTGAA GTTTCTTCTGAAGCA GCATCTGTCTCAA |
339–369 |
| SSchr9 | 9 | (GAAA)n | 690 |
AAACCTCTCTCCACACCCAAA ACCACAGATTCCTTCTGCAA GTTTCTTCTGGGATTGC CTCCTTCAA |
336–463 |
| SSchr2 | 2 | (TTTC)n | 576 |
CTCACCTCCCACTCCACAAAAT CACTGCTCATTCCGCAAA GTTTCTTATGACATTCC TGTAGAAGGCTGAA |
328–389 |
| SSchr14 | 14 | (TTTC)n | 612 |
AACTCCACCACTCCCACAAA ATCCTTCTGCATTTTTCTATCAA GTTTCTTGTCAAAAGTACAT CCCCCTTCCTATAA |
332–368 |
| SSchr3 | 3 | (TAGA)n | 759 |
CTCCAACTCACCTCCAACAAA GGGTAGCCCCACCAAAGAA GTTTCTTATTCTGGGA TTAGTGATGCAA |
163–227 |
| SSchr8 | 8 | (TAAA)n | 500 |
AAACCTCTCTCCACACCCAA ATGCCTACTACCCCCTTCCAA GTTTCTTCTTGGCTCTT AGGAGGCATAA |
187–212 |
| SSchr13a | 13 | (TAGA)n | 558 |
CTCACCTCCCACTCCACAAAA AATTCTTGGGACTGAAACCAA GTTTCTTCCTCCTTAAT GGGGCTTCTAA |
179–203 |
| SSchr5 | 5 | (TAGA)n | 504 |
AACTCCACCACTCCCACAAAG ACCTGGCATCCAAAATCAA GTTTCTTAACCATCAAAAC CCCCTAAA |
154–184 |
| SS.Amel | X, Y | N/A | N/A |
AACTCCACCACTCCCACAAAG CAGGATCGGTCTGTTTTTC GTTTCTTATGCAAGCCCT CCGAGAA |
261, 264 |
Primer sequences for the 12 genetic markers shown along with their chromosome location (Chr), repeat motif, Smith-Waterman sequence alignment score (SW), and allele range in base pairs. For each marker, the top sequence is the forward primer and bottom sequence is the reverse primer. Section of the forward primers in bold font represents the tag sequence complementary to a universal primer in Table 2. The pigtail sequence GTTTCTT at the 5′ end of the reverse primer is shown in bold font.
To promote complete adenylation of PCR amplicons, each reverse primer was modified by adding to the 5′ end ‘pigtail’ sequence GTTTCTT (Brownstein et al. 1996). Each forward primer was modified by adding to the 5′ end a tag sequence complementary to one of four fluorescent-labeled universal primers (Table 2) which also end in AA. Of the 12 forward primers, three had a tag sequence complementary to universal primer 1 which was labeled at the 5′ end with the fluorescent dye 6-FAM, three had the tag sequence complementary to universal primer 2 which was labeled with the dye PET, two had the tag sequence complementary to universal primer 3 which was labeled with the dye NED, and four had the tag sequence complementary to universal primer 4 which was labeled with the dye VIC (Table 1).
Table 2.
Four universal primers and their assigned fluorescent dye
| Dye | Primer name | Primer sequence (5′–3′) |
|---|---|---|
| FAM | Universal 1 | CTCCAACTCACCTCCAACAAA |
| PET | Universal 2 | AAACCTCTCTCCACACCCAAA |
| NED | Universal 3 | CTCACCTCCCACTCCACAAA |
| VIC | Universal 4 | AACTCCACCACTCCCACAAA |
Each primer pair, without the tag sequence, was tested for nonspecific binding to untargeted regions of pig DNA and to non-porcine species by performing in silico PCR on all the genomes, including pig, in the UCSC genome database. As the UCSC genome database is limited in the number of different species, each primer pair was further tested for nonspecific binding against a wide range of nucleotide sequences in the National Center for Biotechnology Information database using Primer-BLAST tool (Ye et al. 2012). The in silico PCR can detect non-specific binding only when the primers have 100% nucleotide match to an untargeted region, but amplification of untargeted regions can still occur in vitro for primers with one or two nucleotide mismatches. For this reason, the primer pairs, with the tag sequence attached, were tested for species specificity by performing in vitro PCR on the genomic DNA of humans, dogs and five species of mosquitoes (An. farauti sensu stricto [n = 5], An. farauti no. 4 [n = 5], Anopheles punctulatus sensu stricto [n = 5] and Anopheles koliensis [n = 5] and Culex pipiens pipiens [n = 5]). These are organisms whose DNA is likely to be mixed with pig bloodmeal DNA of mosquitoes from PNG (Burkot et al. 1988, 1989; Logue et al. 2016; Keven et al. 2017). Unlabeled primers were obtained from Integrated DNA Technologies and the labeled universal primers were obtained from Applied Biosystems through ThermoFisher Scientific.
PCR Amplification and Genotyping
The 12 loci were coamplified in a single reaction (10 µl reaction volume) which consisted of 20 mM Tris–HCl, 50 mM KCl, 2 mM MgCl2, 0.2 mM of each dNTP, 0.08 μM of each universal primer, 0.08 μM of the reverse and 0.008 μM of the forward primers for the small amplicon size markers (SSchr3, SSchr8, SSchr13a, SSchr5 and SS.Amel), 0.2 μM reverse and 0.02 μM forward primers for the medium size markers (SSchr11, SSchr9, SSchr2 and SSchr14), 0.36 μM reverse and 0.036 μM forward primers for the large size markers (SSchr13b, SSchr15 and SSchr1), 0.4 units of AmpliTaq Gold and 3.0 ng (for pig samples) or 6.0 ng (for mosquito bloodmeals) of DNA template. Cycling condition consisted of one cycle of 94°C for 8 min followed by 40 cycles of 94°C for 1 min, 57°C for 2 min, and 72°C for 3 min, and one cycle of 72°C for 60 min. The PCR proceeded in two steps nested in a single reaction as described in details elsewhere (Schuelke 2000). PCR products were analyzed through the Michigan State University Genomics Core facility by capillary electrophoresis (ABI 3730 Genetic Analyzer, Applied Biosystems, Foster City, CA) with LIZ 500 (Applied Biosystems, Foster City, CA) as internal size standard. Genotypes were determined using Peak Scanner software version 1.0 (Applied Biosystems, Foster City, CA) and alleles were represented by fragment size in base pairs. True off-ladder alleles were distinguished from false off-ladder calls due to rounding errors in allele scoring (based on the local Southern algorithm) by overlaying the electropherogram of all samples and manually identifying those samples with true off-ladder peaks.
Data Analyses
The genotype data of the 211 pigs were analyzed for number of alleles (Na) and heterozygosity, both observed (Ho) and expected (He), for each microsatellite marker. Probability of identity of a genetic marker, defined as the probability that two individuals randomly drawn from the same population share the same genotype at that locus, was calculated for each microsatellite marker using the formula , for unrelated individuals (Paetkau and Strobeck 1994), and , for siblings (Taberlet and Luikart 1999). In these formulae, n is the number of alleles of a single-locus marker and pi is the frequency of ith allele of that marker. For multilocus markers, combined probability of identity was calculated by taking the product of the PI or PIsib of the individual locus constituting the multilocus marker (Taberlet and Luikart 1999). That is, and , where k is the number of loci in the multilocus marker. These values were computed in GenAlEx software version 6.5 (Peakall and Smouse 2012). A plot of mean values of cPI and cPIsib versus k was constructed. The means were estimated by performing random sampling (with replacement) on the 11 PI and PIsib values in Table 3. For the k = 1, n = 1,000 samples were randomly drawn and mean and 95% confidence interval (CI) were calculated. For k > 1, k samples of PI or PIsib were randomly drawn and the cPI or cPIsib were calculated. This was repeated for n = 1,000 replicates before the mean and 95% CI were calculated. Computation of the plot was implemented in R software (version 3.4.2). For both pig and bloodmeal samples, each DNA profile was searched against every other profile for potential matches using multilocus match function in GenAlEx. Using the software Population (Langella 1999), a distance matrix containing estimates of shared alleles (Chakraborty and Jin 1993) between 41 individuals (22 unrelated pigs of four different breeds from Michigan and 19 pigs of unknown breeds from PNG bloodmeals) was computed and used to construct a neighbor-joining network tree.
Table 3.
Allele diversity and discriminatory power of the microsatellite markers
| Marker | N | N a | Ho | He | PI | PIsib |
|---|---|---|---|---|---|---|
| SSchr5 | 211 | 6 | 0.68 | 0.73 | 0.11 | 0.41 |
| SSchr13a | 211 | 11 | 0.55 | 0.62 | 0.18 | 0.49 |
| SSchr8 | 211 | 6 | 0.67 | 0.67 | 0.16 | 0.46 |
| SSchr3 | 211 | 10 | 0.54 | 0.60 | 0.23 | 0.51 |
| SSchr14 | 211 | 8 | 0.68 | 0.68 | 0.15 | 0.45 |
| SSchr2 | 211 | 9 | 0.81 | 0.76 | 0.09 | 0.39 |
| SSchr9 | 211 | 10 | 0.56 | 0.69 | 0.14 | 0.44 |
| SSchr11 | 211 | 8 | 0.68 | 0.63 | 0.18 | 0.48 |
| SSchr1 | 211 | 11 | 0.79 | 0.81 | 0.06 | 0.36 |
| SSchr15 | 211 | 8 | 0.83 | 0.71 | 0.13 | 0.43 |
| SSchr13b | 211 | 8 | 0.76 | 0.73 | 0.12 | 0.42 |
Columns contain sample size (N), number of different alleles (Na), observed heterozygosity (Ho), expected heterozygosity (He) and probability of identity for unrelated individuals (PI) and siblings (PIsib) for each of the 11 microsatellite markers based on analysis of 211 pig genotypes.
Results and Discussion
PCR Efficiency and Reliability
Both the in silico and in vitro PCR did not detect nonspecific binding or interspecies cross-reactivity for any of the primers. For the 340 PCR reactions performed (211 pigs and 129 bloodmeals), all 12 loci coamplified successfully in a full 12-plex with universal primers, with relatively few problems; only three microsatellites originally selected needed to be replaced due to technical difficulties (e.g., excess single base stutter due to a run of 15 As in one amplicon). A full 12-plex saves considerably on reagents and precious DNA samples, compared to smaller multiplexes or single marker amplifications. All of the 211 pig DNA samples had complete genotype profiles (Supp. Table S1). For the 129 bloodmeal samples, 8 (6.2%) had homozygous null alleles at locus SSchr9, but these bloodmeals were all taken from the same pig (Supp. Table S2). A mutation in the primer binding sites of the SSchr9 locus may have caused the null allele in this individual. Visualization of agarose gel electrophoresis under fluorescent light (Typhoon FLA900 Gel Imaging Scanner) showed that primer dimers were present but were threefold lower in intensity compared to the microsatellite-specific amplicons. This observation was consistent with an earlier one (Innis and Gelfand 1999) which showed that primers with AA at their 3′ end reduced primer dimers in multiplex reactions. Complete adenylation (>95% of the A+ peak) was observed for all markers and in all samples. This result was consistent with an earlier finding (Brownstein et al. 1996) that addition of the pigtail sequence to the 5′ end of reverse primers promoted complete adenylation, which is essential for preventing split peaks during allele scoring. Stutter peak heights varied among markers but were within the acceptable range of 0–15% of their true allele peak height. Low stutter peak height is important because stutter peaks are sometimes confused for true allele peaks if the stutter peaks are too tall. As expected, the amelogenin marker correctly predicted the sex of all 211 pigs. PCR analysis was replicated (n = 4) for 10 DNA samples (six pig tissues and four mosquito bloodmeals) and all showed 100% reproducibility in their DNA profile. This result indicates a high level of confidence in the assay to detect accurately and consistently the microsatellite genotypes of an individual pig.
Discriminatory Power of the Microsatellite Markers
Before the method was used to identify different pig individuals in mosquito bloodmeals based on unique DNA profiles, the power of the microsatellite markers to accurately distinguish between two individuals was assessed. The genotypes of the 211 pigs (Supp. Table S1) were evaluated for matching DNA profiles and no matches were found; all 211 pigs had unique DNA profiles and the sex-specific marker accurately predicted the sex of all the pigs. The absence of matching DNA profiles despite high degree of genetic relatedness among most of the pigs suggests high allele diversity of the markers. Indeed, high Na (mean: 8.64, min: 6, max: 11), Ho (mean: 0.69, min: 0.54, max: 0.83) and He (mean: 0.69, min: 0.60, max: 0.81) were observed for each of the microsatellite markers (Table 3). The observed Na were higher than expected, given that most (89.5%) of the pigs analyzed in this study were full or half siblings and therefore should be sharing the same few alleles. One factor for the high Na is the high SW score of the markers, which has been shown to be positively correlated with increased heterozygosity (Venta et al. unpublished). The heterozygosity for the microsatellites evaluated in this study was higher than those estimated for the pig microsatellites developed for forensic analyses by Lin and colleagues (Lin et al. 2014).
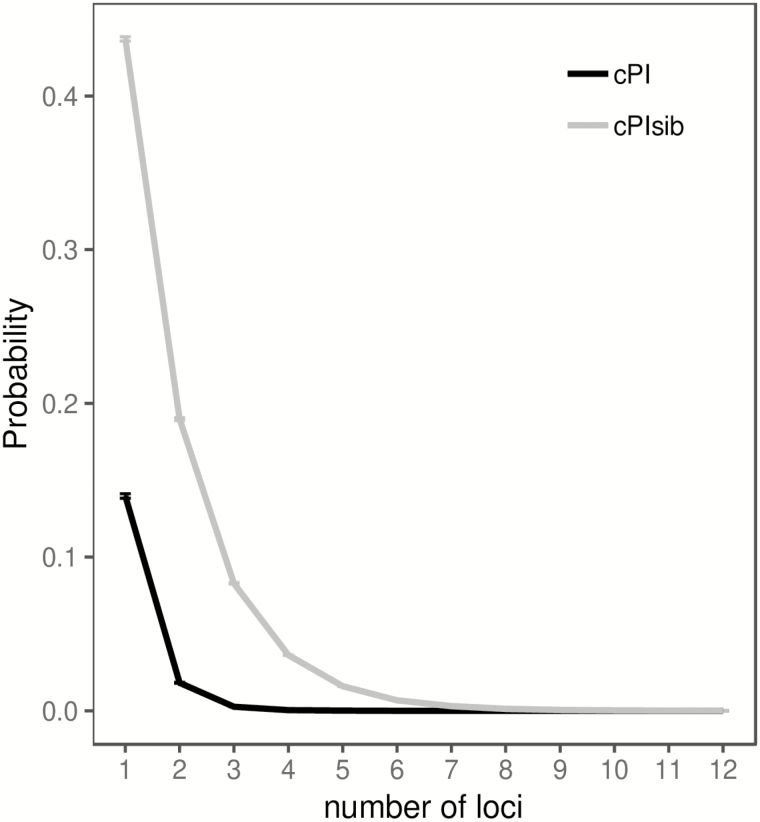
Highly polymorphic markers are useful for forensic identification because of their potential for distinguishing individuals. Analysis of the 211 pig genotypes gave very low per-marker PI (mean: 0.14, min: 0.06, max: 0.23) and PIsib (mean: 0.44, min: 0.36, max: 0.51) for all microsatellite markers (Table 3). This means that on average, the chance of observing two randomly drawn pigs sharing the same genotype for any of the microsatellite markers is 14% for unrelated individuals and 44% for siblings. As a multilocus marker (k = 11), extremely low combined probability of identity was calculated for both unrelated individuals (cPI = 2.5 × 10−10), and siblings (cPIsib = 1.1 × 10–4). These values were comparable to those reported for other pig multilocus markers (Caratti et al. 2010, Lin et al. 2014). From the plot of cPI and cPIsib versus multilocus markers with increasing k (Fig. 1), a minimum of four loci is sufficient to discriminate between unrelated individuals and at least nine loci are sufficient if siblings are involved. By using the full panel of 11 microsatellite markers, the power to accurately identify individual pigs or discriminate between different individuals based on their DNA profile is even higher.
Fig. 1.
Plot of mean combined probability of identity of multilocus markers with increasing number of loci for both unrelated individuals (cPI) and siblings (cPIsib). The 95% CI bars are too small to be seen.
Mosquito Bloodmeal Profiles
After confirming that the marker panel has high discriminatory power, the pig genotypes analyzed from the 129 mosquito bloodmeals (Supp. Table S2) were evaluated and 19 unique DNA profiles were identified; nine were identified in mosquitoes from Mirap and 10 in mosquitoes from Kokofine. In both villages, these unique DNA profiles were not equally represented in the mosquito bloodmeals. Some DNA profiles were found in a single bloodmeal sample whereas others occurred in multiple samples (Table 4). Given the high confidence that a unique DNA profile based on this marker panel represents an individual pig, those mosquitoes with matching DNA profiles all fed on the same pig. The heterogeneity in the frequency of mosquito bloodmeals taken from these individuals may be due to stochastic effects of the sampling procedure, but it may also be the result of underlying ecological or spatial factors—a hypothesis which this assay was developed to investigate.
Table 4.
Sex and frequency of the unique DNA profiles identified in the mosquito bloodmeals
| Unique profile | Site | Sex | Frequency |
|---|---|---|---|
| profile_01 | Mirap | Female | 1 |
| profile_02 | Mirap | Male | 1 |
| profile_03 | Mirap | Female | 3 |
| profile_04 | Mirap | Female | 8 |
| profile_05 | Mirap | Female | 1 |
| profile_06 | Mirap | Female | 1 |
| profile_07 | Mirap | Male | 3 |
| profile_08 | Mirap | Male | 1 |
| profile_09 | Mirap | Male | 1 |
| profile_10 | Kokofine | Female | 53 |
| profile_11 | Kokofine | Male | 8 |
| profile_12 | Kokofine | Male | 16 |
| profile_13 | Kokofine | Female | 19 |
| profile_14 | Kokofine | Male | 2 |
| profile_15 | Kokofine | Male | 2 |
| profile_16 | Kokofine | Female | 1 |
| profile_17 | Kokofine | Female | 6 |
| profile_18 | Kokofine | Male | 1 |
| profile_19 | Kokofine | Female | 1 |
Genetic Clusters
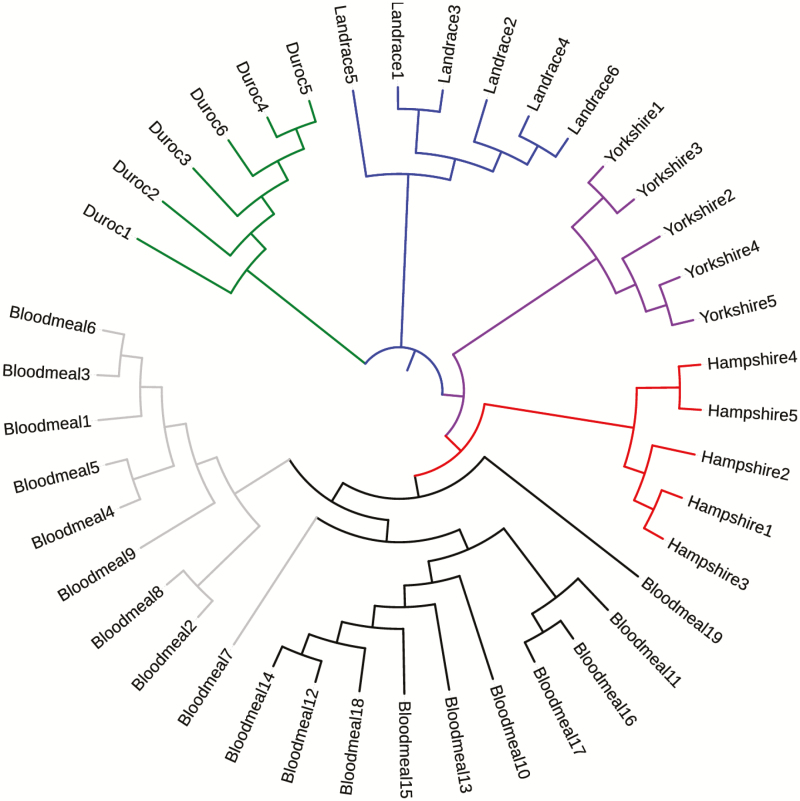
In some cases, information about the breed of a pig is needed. However, in PNG, the breed of pigs is not usually known to the owners of the animals and predictions may need to be made based on their genotypes. The ability of the microsatellite markers to cluster individual pigs based on genetic relatedness, or predict the breed of an unknown pig, was evaluated based on the neighbor-joining network tree of the 41 individuals (Fig. 2). The tree showed that these genetic markers are capable of grouping individuals according to their genetic similarities as all 22 individuals with known breeds were assigned correctly to the cluster representing their breed. The bloodmeal-identified individuals with unknown breeds were grouped in two clusters separate from the four breeds, indicating that these individuals do not belong to any of the four breeds. The two clusters correspond to the two villages; the pigs from Mirap clustered separately from those of Kokofine, suggesting local inbreeding.
Fig. 2.
Neighbor-joining network tree based on proportion of shared alleles showing genetic similarities of 41 pigs (22 unrelated individuals of 4 different breeds (Duroc, Landrace, Yorkshire and Hampshire) and 19 bloodmeal-derived individuals of unknown breeds from Mirap [grey tree branches] and Kokofine [black tree branches] villages in Papua New Guinea).
Relevance to the Study of Mosquito-Borne Disease Ecology
The development of immunological and PCR-based assays to identify vertebrate host species in mosquito bloodmeals has greatly enabled our ability to study important aspects of the ecology of disease vectors, particularly vertebrate host range (i.e., the different species of hosts utilized by a mosquito population) and host selection tendency (i.e., the propensity to feed more on a particular host species than others) of the vectors (Tempelis 1975; Charlwood et al. 1985; Burkot et al. 1988, 1989; Sousa et al. 2001; Apperson et al. 2002; Basseri et al. 2005; Elizondo-Quiroga et al. 2006; Molaei et al. 2006; Oshaghi et al. 2006; Tirados et al. 2006; Zimmerman et al. 2006; Abbasi et al. 2009; Hamer et al. 2009; Chaves et al. 2010; Molaei et al. 2010; Kek et al. 2014; Logue et al. 2016; Keven et al. 2017). In addition to the species identification methods, molecular methods for identifying particular individuals in mosquito bloodmeals are important for investigating vector-host contact patterns and their consequences on disease transmission. For example, the basic reproductive number—a measure of persistence and spread of communicable diseases—increases when few individuals in a community are bitten more frequently than others (i.e., nonrandom or clustered feeding pattern) (Dye and Hasibeder 1986, Hasibeder and Dye 1988). Although DNA profiling methods are well developed and routinely used in human and wildlife forensic investigations, only a few studies have applied them to human-derived mosquito bloodmeals (Chow-Shaffer et al. 2000, Michael et al. 2001, De Benedictis et al. 2003, Soremekun et al. 2004, Scott et al. 2006) and none for pig-derived bloodmeals. To the best of our knowledge, our study is probably the first to develop and apply DNA profiling to animal-derived mosquito bloodmeals. In the same way analysis of human-derived bloodmeals can be used to determine vector-human contact pattern and its consequences on human disease transmission, this pig profiling method can be used to determine vector-pig contact pattern and how it affects the transmission of pig pathogens such as JEV. A more useful application of the pig bloodmeal profiling is to apply it in parallel with human bloodmeal profiling to confirm whether the observed distribution of bloodmeals taken on human individuals in the village is the result of anthropogenic factors such as bed nets (in which case the distribution of bites is most likely to differ between the two host species) or environmental or spatial factors (in which case the distribution of bites is likely to co-vary for both host species). Such knowledge is useful for guiding disease control programs, particularly targeted interventions.
Conclusion
In this paper, we describe the development and validation of a microsatellite multiplex assay for profiling pig DNA extracted from pig tissues and mosquito bloodmeals. We show, based on the low probabilities of identity as well as the absence of matching DNA profiles among highly related individuals, that the assay can accurately determine different pig individuals in a sample of blood-fed mosquitoes based on unique DNA profiles. This method can be used in studies that seek to link mosquito bloodmeals to individual pigs. Although a standard DNA profiling method (Lin et al. 2014) for forensic testing of pigs is available and can be used in mosquito bloodmeal studies, it requires fluorescent labeling of all primers and is therefore expensive for analyzing large number of samples. By using fluorescent-labeled universal primers, the assay described here can analyze thousands of mosquito bloodmeals and pig DNA samples with relatively less expense. Also, by selecting microsatellites based on the SW scores, the markers described here are likely to be more polymorphic and thus have higher discriminatory power. DNA profiling by sequencing of hypervariable region of mitochondrial genome has been used for human bloodmeals (Logue et al. 2016) and can be extended to pig bloodmeals. However, this approach has low discriminatory power (Parson et al. 1998), especially for distinguishing between closely related individuals or highly inbred populations. Unlike the standard method (Lin et al. 2014), the assay described here is not intended for use in forensic analysis cases and, at this point, does not meet all the ISFG requirements (Linacre et al. 2011) but it is nevertheless extremely useful for the study of mosquito vectors. Further improvements of this method such as the inclusion of additional microsatellite markers and development of allelic ladder are possible.
Supplementary Material
Acknowledgments
We thank Kevin Turner (manager of the Michigan State University Swine Farm) for giving us access to pig tail tissues and Dr. Catherine Ernst and Nancy Raney from the Michigan State University Department of Animal Science for providing us with pig DNA samples during the early stage of this project. The mosquitoes used in this study were collected with the help of entomology field technicians of the Papua New Guinea Institute of Medical Research. This study was funded by grants from the National Institute of Allergy and Infectious Diseases (grant number: U19AI089686) and Fogarty International Center (grant number: 2D43TW007377) of the US National Institute of Health.
References Cited
- Abbasi I., Cunio R., and Warburg A.. . 2009. Identification of blood meals imbibed by phlebotomine sand flies using cytochrome b PCR and reverse line blotting. Vector Borne Zoonotic Dis. 9: 79–86. [DOI] [PubMed] [Google Scholar]
- Apperson C. S., Harrison B. A., Unnasch T. R., Hassan H. K., Irby W. S., Savage H. M., Aspen S. E., Watson D. W., Rueda L. M., Engber B. R., . et al. 2002. Host-feeding habits of Culex and other mosquitoes (Diptera: Culicidae) in the Borough of Queens in New York City, with characters and techniques for identification of Culex mosquitoes. J. Med. Entomol. 39: 777–785. [DOI] [PubMed] [Google Scholar]
- Basseri H. R., Moosakazemi S. H., Yosafi S., Mohebali M., Hajaran H., and Jedari M.. . 2005. Anthropophily of malaria vectors in Kahnouj district, south of Kerman, Iran. Iranian J. Publ. Health 34: 27–35. [Google Scholar]
- Bolzoni L., Pugliese A., and Rosà R.. . 2015. The role of heterogeneity on the invasion probability of mosquito-borne diseases in multi-host models. J. Theor. Biol. 377: 25–35. [DOI] [PubMed] [Google Scholar]
- Bouma M., and Rowland M.. . 1995. Failure of passive zooprophylaxis: cattle ownership in Pakistan is associated with a higher prevalence of malaria. Trans. R. Soc. Trop. Med. Hyg. 89: 351–353. [DOI] [PubMed] [Google Scholar]
- Brownstein M. J., Carpten J. D., and Smith J. R.. . 1996. Modulation of non-templated nucleotide addition by Taq DNA polymerase: primer modifications that facilitate genotyping. Biotechniques. 20: 1004–1006, 1008. [DOI] [PubMed] [Google Scholar]
- Burkot T. R., Graves P. M., Paru R., and Lagog M.. . 1988. Mixed blood feeding by the malaria vectors in the Anopheles punctulatus complex (Diptera: Culicidae). J. Med. Entomol. 25: 205–213. [DOI] [PubMed] [Google Scholar]
- Burkot T. R., Dye C., and Graves P. M.. . 1989. An analysis of some factors determining the sporozoite rates, human blood indexes, and biting rates of members of the Anopheles punctulatus complex in Papua New Guinea. Am. J. Trop. Med. Hyg. 40: 229–234. [DOI] [PubMed] [Google Scholar]
- Burkot T. R., Russell T. L., Reimer L. J., Bugoro H., Beebe N. W., Cooper R. D., Sukawati S., Collins F. H., and Lobo N. F.. . 2013. Barrier screens: a method to sample blood-fed and host-seeking exophilic mosquitoes. Malar. J. 12: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caratti S., Rossi L., Sona B., Origlia S., Viara S., Martano G., Torre C., and Robino C.. . 2010. Analysis of 11 tetrameric STRs in wild boars for forensic purposes. Forensic Sci. Int. Genet. 4: 339–342. [DOI] [PubMed] [Google Scholar]
- Chakraborty R., and Jin L.. . 1993. Determination of relatedness between individuals using DNA fingerprinting. Hum. Biol. 65: 875–895. [PubMed] [Google Scholar]
- Charlwood J. D., Dagoro H., and Paru R.. . 1985. Blood-feeding and resting behaviour in the Anopheles punctulatus Donitz complex (Diptera: Culicidae) from coastal Papua New Guinea. Bull. Entomol. Res. 75: 463–475. [Google Scholar]
- Chaves L. F., Harrington L. C., Keogh C. L., Nguyen A. M., and Kitron U. D.. . 2010. Blood feeding patterns of mosquitoes: random or structured? Front. Zool. 7: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow-Shaffer E., Sina B., Hawley W. A., De Benedictis J., and Scott T. W.. . 2000. Laboratory and field evaluation of polymerase chain reaction-based forensic DNA profiling for use in identification of human blood meal sources of Aedes aegypti (Diptera: Culicidae). J. Med. Entomol. 37: 492–502. [DOI] [PubMed] [Google Scholar]
- De Benedictis J., Chow-Shaffer E., Costero A., Clark G. G., Edman J. D., and Scott T. W.. . 2003. Identification of the people from whom engorged Aedes aegypti took blood meals in Florida, Puerto Rico, using polymerase chain reaction-based DNA profiling. Am. J. Trop. Med. Hyg. 68: 437–446. [PubMed] [Google Scholar]
- Dye C., and Hasibeder G.. . 1986. Population dynamics of mosquito-borne disease: effects of flies which bite some people more frequently than others. Trans. R. Soc. Trop. Med. Hyg. 80: 69–77. [DOI] [PubMed] [Google Scholar]
- Elizondo-Quiroga A., Flores-Suarez A., Elizondo-Quiroga D., Ponce-Garcia G., Blitvich B. J., Contreras-Cordero J. F., Gonzalez-Rojas J. I., Mercado-Hernandez R., Beaty B. J., and Fernandez-Salas I.. . 2006. Host-feeding preference of Culex quinquefasciatus in Monterrey, northeastern Mexico. J. Am. Mosq. Control Assoc. 22: 654–661. [DOI] [PubMed] [Google Scholar]
- Frantz L. A., Schraiber J. G., Madsen O., Megens H. J., Cagan A., Bosse M., Paudel Y., Crooijmans R. P., Larson G., and Groenen M. A.. . 2015. Evidence of long-term gene flow and selection during domestication from analyses of Eurasian wild and domestic pig genomes. Nat. Genet. 47: 1141–1148. [DOI] [PubMed] [Google Scholar]
- Hamer G. L., Kitron U. D., Goldberg T. L., Brawn J. D., Loss S. R., Ruiz M. O., Hayes D. B., and Walker E. D.. . 2009. Host selection by Culex pipiens mosquitoes and West Nile virus amplification. Am. J. Trop. Med. Hyg. 80: 268–278. [PubMed] [Google Scholar]
- Hasibeder G., and Dye C.. . 1988. Population dynamics of mosquito-borne disease: persistence in a completely heterogeneous environment. Theor. Popul. Biol. 33: 31–53. [DOI] [PubMed] [Google Scholar]
- Hurk A. F. V. D., Nisbet D. J., Johansen C. A., Foley P. N., Ritchie S. A., and Mackenzie J. S.. . 2001. Japanese encephalitis on Badu Island, Australia: the first isolation of Japanese encephalitis virus from Culex gelidus in the Australasian region and the role of mosquito host-feeding patterns in virus transmission cycles. Trans. R. Soc. Trop. Med. Hyg. 95: 595–600. [DOI] [PubMed] [Google Scholar]
- Hurk A. F. V. D., Nisbet D. J., Hall R. A., Kay B. H., Mackenzie J. S., and Ritchie S. A.. . 2003. Vector competence of Australian mosquitoes (Diptera: Culicidae) for Japanese Encephalitis Virus. J. Med. Entomol. 40: 82–90. [DOI] [PubMed] [Google Scholar]
- Hurk A. F. V. D., Ritchie S. A., Johansen C. A., Mackenzie J. S., and Smith G. A.. . 2008. Domestic pigs and Japanese Encephalitis Virus infection, Australia. Emerg. Infect. Dis. 14: 1736–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innis M., and Gelfand D.. . 1999. Optimization of PCR: conversations between Michael and David, pp. 3–22. InInnis M. A., Gelfand D. H., and Sninsky J. J. (eds.), PCR applications. Academic Press, Cambridge. [Google Scholar]
- Kek R., Hapuarachchi H. C., Chung C. Y., Humaidi M. B., Razak M. A., Chiang S., Lee C., Tan C. H., Yap G., Chong C. S., . et al. 2014. Feeding host range of Aedes albopictus (Diptera: Culicidae) demonstrates its opportunistic host-seeking behavior in rural Singapore. J. Med. Entomol. 51: 880–884. [DOI] [PubMed] [Google Scholar]
- Kent R. J., and Norris D. E.. . 2005. Identification of mammalian blood meals in mosquitoes by a multiplexed polymerase chain reaction targeting cytochrome B. Am. J. Trop. Med. Hyg. 73: 336–342. [PMC free article] [PubMed] [Google Scholar]
- Keven J. B., Reimer L., Katusele M., Koimbu G., Vinit R., Vincent N., Thomsen E., Foran D. R., Zimmerman P. A., and Walker E. D.. . 2017. Plasticity of host selection by malaria vectors of Papua New Guinea. Parasit. Vectors 10: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langella O. 1999. Populations 1.2.30 http://bioinformatics.org/populations/ computer program, version By Langella, O.
- Lin Y. C., Hsieh H. M., Lee J. C., Hsiao C. T., Lin D. Y., Linacre A., and Tsai L. C.. . 2014. Establishing a DNA identification system for pigs (Sus scrofa) using a multiplex STR amplification. Forensic Sci. Int. Genet. 9: 12–19. [DOI] [PubMed] [Google Scholar]
- Linacre A., Gusmão L., Hecht W., Hellmann A. P., Mayr W. R., Parson W., Prinz M., Schneider P. M., and Morling N.. . 2011. ISFG: recommendations regarding the use of non-human (animal) DNA in forensic genetic investigations. Forensic Sci. Int. Genet. 5: 501–505. [DOI] [PubMed] [Google Scholar]
- Logue K., Keven J. B., Cannon M. V., Reimer L., Siba P., Walker E. D., Zimmerman P. A., and Serre D.. . 2016. Unbiased characterization of Anopheles mosquito blood meals by targeted high-throughput sequencing. PLoS Negl. Trop. Dis. 10: e0004512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael E., Ramaiah K. D., Hoti S. L., Barker G., Paul M. R., Yuvaraj J., Das P. K., Grenfell B. T., and Bundy D. A.. . 2001. Quantifying mosquito biting patterns on humans by DNA fingerprinting of bloodmeals. Am. J. Trop. Med. Hyg. 65: 722–728. [DOI] [PubMed] [Google Scholar]
- Molaei G., Andreadis T. G., Armstrong P. M., Anderson J. F., and Vossbrinck C. R.. . 2006. Host feeding patterns of Culex mosquitoes and West Nile virus transmission, northeastern United States. Emerg. Infect. Dis. 12: 468–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molaei G., Cummings R. F., Su T., Armstrong P. M., Williams G. A., Cheng M. L., Webb J. P., and Andreadis T. G.. . 2010. Vector-host interactions governing epidemiology of West Nile virus in Southern California. Am. J. Trop. Med. Hyg. 83: 1269–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura S., and Crump A.. . 2017. Ivermectin and malaria control. Malar. J. 16: 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshaghi M. A., Chavshin A. R., and Vatandoost H.. . 2006. Analysis of mosquito bloodmeals using RFLP markers. Exp. Parasitol. 114: 259–264. [DOI] [PubMed] [Google Scholar]
- Paetkau D., and Strobeck C.. . 1994. Microsatellite analysis of genetic variation in black bear populations. Mol. Ecol. 3: 489–495. [DOI] [PubMed] [Google Scholar]
- Parson W., Parsons T. J., Scheithauer R., and Holland M. M.. 1998. Population data for 101 Austrian Caucasian mitochondrial DNA d-loop sequences: application of mtDNA sequence analysis to a forensic case. Int. J. Legal Med. 111: 124–132. [DOI] [PubMed] [Google Scholar]
- Peakall R., and Smouse P. E.. . 2012. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research–an update. Bioinformatics. 28: 2537–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell T. L., Beebe N. W., Bugoro H., Apairamo A., Cooper R. D., Collins F. H., Lobo N. F., and Burkot T. R.. . 2016. Determinants of host feeding success by Anopheles farauti. Malar. J. 15: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saul A. 2003. Zooprophylaxis or zoopotentiation: the outcome of introducing animals on vector transmission is highly dependent on the mosquito mortality while searching. Malar. J. 2: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuelke M. 2000. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 18: 233–234. [DOI] [PubMed] [Google Scholar]
- Scott T. W., Githeko A. K., Fleisher A., Harrington L. C., and Yan G.. . 2006. DNA profiling of human blood in anophelines from lowland and highland sites in western Kenya. Am. J. Trop. Med. Hyg. 75: 231–237. [PubMed] [Google Scholar]
- Soremekun S., Maxwell C., Zuwakuu M., Chen C., Michael E., and Curtis C.. . 2004. Measuring the efficacy of insecticide treated bednets: the use of DNA fingerprinting to increase the accuracy of personal protection estimates in Tanzania. Trop. Med. Int. Health 9: 664–672. [DOI] [PubMed] [Google Scholar]
- Sota T., and Mogi M.. . 1989. Effectiveness of zooprophylaxis in malaria control: a theoretical inquiry, with a model for mosquito populations with two bloodmeal hosts. Med. Vet. Entomol. 3: 337–345. [DOI] [PubMed] [Google Scholar]
- Sousa C. A., Pinto J., Almeida A. P., Ferreira C., do Rosário V. E., and Charlwood J. D.. . 2001. Dogs as a favored host choice of Anopheles gambiae sensu stricto (Diptera: Culicidae) of São Tomé West Africa. J. Med. Entomol. 38: 122–125. [DOI] [PubMed] [Google Scholar]
- Taberlet P., and Luikart G.. . 1999. Non-invasive genetic sampling and individual identification. Biol. J. Linn. Soc. 68: 41–55. [Google Scholar]
- Tempelis C. H. 1975. Host-feeding patterns of mosquitoes, with a review of advances in analysis of blood meals by serology. J. Med. Entomol. 11: 635–653. [DOI] [PubMed] [Google Scholar]
- Tirados I., Costantini C., Gibson G., and Torr S. J.. . 2006. Blood-feeding behaviour of the malarial mosquito Anopheles arabiensis: implications for vector control. Med. Vet. Entomol. 20: 425–437. [DOI] [PubMed] [Google Scholar]
- Vallone P. M., and Butler J. M.. . 2004. AutoDimer: a screening tool for primer-dimer and hairpin structures. Biotechniques. 37: 226–231. [DOI] [PubMed] [Google Scholar]
- Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., and Madden T. L.. . 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 13: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman R. H., Galardo A. K., Lounibos L. P., Arruda M., and Wirtz R.. . 2006. Bloodmeal hosts of Anopheles species (Diptera: Culicidae) in a malaria-endemic area of the Brazilian Amazon. J. Med. Entomol. 43: 947–956. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.