Abstract
This study was to investigate the expression correlation between long non-coding RNA metastasis-associated lung adenocarcinoma transcript 1 (lncRNA MALAT1), miR-200a-3p and programmed death-ligand 1 (PD-L1) in non-small cell lung cancer (NSCLC), and their roles in NSCLC. Real-time polymerase chain reaction (PCR) was performed to detect the expressions of MALAT1, miR-200a-3p and PD-L1 in NSCLC tissues and cells for the correlation analysis. The starBase and Targetscan databases were used to predict the binding sites between MALAT1 and miR-200a-3p, and miR-200a-3p and PD-L1, respectively. The targeting relationship between MALAT1 and miR-200a-3p, and miR-200a-3p and PD-L1 were further verified by real-time PCR and dual luciferase reporter gene assay. Cell proliferation was monitored by CCK8 and colony formation assays. The apoptosis was detected using flow cytometry. Wound healing assay and transwell assay were conducted to determine cell migration and invasion. In this study, we demonstrated that in NSCLC tissues, the expression level of MALAT1 was negatively correlated with that of miR-200a-3p, while positively correlated with PD-L1. Besides, MALAT1 promoted proliferation, mobility, migration, and invasion of NSCLC cells via sponging miR-200a-3p. PD-L1 was validated as a target of miR-200a-3p, and indirectly modulated by MALAT1. In conclusion, LncRNA MALAT1 facilitates the progression of NSCLC by modulating miR-200a-3p/PDL1 axis.
Keywords: lncRNA MALAT1, miR-200a-3p, NSCLC, PD-L1
Introduction
Lung cancer is one of the most common and deadly cancers worldwide, with an average of 228,150 new diagnoses and 147,510 deaths per year.1,2 Non-small cell lung cancer (NSCLC) accounts for about 85% of all cases of lung cancer.1,2 Many patients with NSCLC were diagnosed at the advanced stage.3 At present, the treatment methods are limited for patients with advanced and metastatic NSCLC. Obviously, it is urgent to discover indicators for early diagnosis and new therapeutic targets.
Long non-coding RNA (lncRNA) is a class of RNA with over 200 nucleotides in length. It is well known that lncRNA can regulate the expression of various genes, thus participating in the occurrence and development of tumors.4–7 For example, lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) promotes the progression of NSCLC by modulating miR-124 and miR-206.8,9 However, the mechanism of action of MALAT1 in NSCLC has not been fully elucidated.
In the tumorigenesis and progression of NSCLC, miRNA also plays an important regulatory role. For example, miR-433 inhibits the proliferation and invasion of NSCLC cells by directly targeting E2F3.10 And miR-182 promotes NSCLC cells by suppressing tumor suppressors FBXW7 and FBXW11.11 Mounting studies suggest that miR-200a-3p plays a tumor repressive role in multiple tumors, such as liver cancer and esophagus cancer.12–14 However, the detailed relationship between miR-200a-3p and NSCLC remains unclear.
Programmed death-ligand 1 (PD-L1, also called CD274, B7-H1), a 40-kDa transmembrane protein, is expressed on the surface of various cell types, including macrophages, dendritic cells, and endothelial cells.15 PD-L1 is also abundantly expressed in various cancer cells such as lung cancer, colon cancer, melanoma, and leukemia, and it participates in immune escape of tumor cells through the interaction with programmed cell death protein 1 (PD-1).16 PD-L1 is a critical “don’t find me” signal to the adaptive immune system.17 Tumor cells interact with receptors on the surface of antigen-presenting cells via PD-L1, to help cancer cells escape T-cell-mediated death and resist anti-tumor immune responses.18 Numerous studies have confirmed that PD-1/PD-L1 targeted drugs, like Opdivo and Keytruda, can help improve prognosis of patients with NSCLC.19–24 However, the upstream regulatory mechanism of PD-L1 has not been fully clarified.
Since the role of MALAT1, miR-200a-3p, and PD-L1 in NSCLC and the regulatory relationship have not been clarified clearly, this study aimed to explore the role of MALAT1/miR-200a-3p/PD-L1 axis in regulating the proliferation, apoptosis, and metastasis of NSCLC cells, providing theoretical basis for elucidating the molecular mechanism of NSCLC and discovery of new targets for NSCLC therapy.
Materials and methods
Clinical samples
Our study was approved by the Ethics Review Board of the Second Affiliated Hospital of South China University of Technology. And all patients signed written informed consents. Cancer samples were selected from 113 patients with NSCLC. The average age of patients was 58.77 ± 8.16 years. All patients had complete clinical and pathological data, and the pathological results of paraffin specimens were confirmed by professional pathologists. No patients received any radiotherapy or chemotherapy before surgery.
Cell lines and antibodies
Human NSCLC cell lines (A549 and CAL-12T) were purchased from Shanghai Sur Biotech Co., Ltd. All cells were cultured in RPMI-1640 medium (Gibco, Grand Island, NY, USA), containing 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA) and 1% double antibody (penicillin/streptomycin, Gibco, Thermo Fisher Scientific, USA) at 5% CO2 and 37°C. Overexpressed MALAT1 plasmid, MALAT1 shRNA, control plasmid, and control shRNA were designed and constructed by Shanghai Genechem Co., Ltd. Anti-PD-L1 antibody (Abcam, Cat. No. ab205921) was purchased from Abcam (Shanghai, China). Goat anti-rabbit secondary antibody (Santa, F030412) was purchased from Santa Cruz (CA, USA). The mouse anti-human β-actin monoclonal antibody was purchased from Kingsray (Beijing, China); the goat anti-mouse secondary antibody (colloidal gold label) was purchased from Santa Cruz (CA, USA).
Immunohistochemical stain
The wax block containing the cervical cancer and the adjacent tissues was sliced, and the xylene was used for dewaxing and hydrating. They were incubated for 30 min at room temperature with a 0.3% H2O2 solution, overnight at 4°C with primary antibody (Anti-PD-L1 antibody, Abcam, ab205921, 1:100), for 1 h at 37°C with secondary antibody (goat anti-rabbit IgG). The sections were then rinsed with PBS buffer. DAB (Hubei Baiaosi Bioscience Co., Ltd.) was then used to stop the reaction after color development. The scoring criteria for immunohistochemical stain (IHC) results were completed by the pathologists from our hospital.
Quantitative reverse transcription polymerase chain reaction
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the instructions. And then the reverse transcription and amplification were conducted. Polymerase chain reaction (PCR) primers were purchased from Suzhou Genewiz Biotechnology Co., Ltd. The detection result was calculated by 2-ΔΔCt method. The primers used in this study were listed in the supplementary materials.
Cell transfection
We transfected A549 cells with the MALAT1 plasmid to successfully establish the MALAT1 overexpression models with LipofectamineTM 2000 (Invitrogen, USA), and CAL-12T cells with the MALAT1 shRNA to successfully construct MALAT1 knockdown models. The shRNA sequences used in this study were listed in the supplementary materials. MicroRNAs were also transfected into cells with LipofectamineTM 2000 (Invitrogen, USA).
Dual luciferase reporter assay
The target sequence of MALAT1 3’-UTR was predicted by StarBase. Wild type (WT) or mutant type (MUT) MALAT1 was subcloned into the pGL3 basic vector (Promega, Madison, WI, USA), and then transfected into A549 and CAL-12T cells. Afterward, cells were seeded in 24-well plates at 5000 cells/well. Luciferase activity was determined using dual luciferase system (Promega, Madison, WI, USA). MiR-200a-3p mimics or negative control (NC) microRNA were used to co-transfect A549 and CAL-12T cells with WT or mutant reporter vector. After 48 h of transfection, the luciferase activity was measured. Three duplicate holes were set in each group and the assay was repeated three times.
RNA-binding protein immunoprecipitation assay
The RNA-binding protein immunoprecipitation (RIP) assay was conducted to explore the interaction between MALAT1 and miR-200a-3p by using EZ-Magna RIP RNA-binding protein immunoprecipitation kit (Millipore, USA). Cells expressing ectogenic MALAT1 were lysed, and next the cell lysis was incubated with anti-Ago2 antibodies (Millipore, USA) coated on magnetic beads in RIP buffer. The precipitated RNAs were then isolated and reversely transcribed to cDNA. Then, quantitative reverse transcription PCR (qRT-PCR) was used to analyze miR-200a-3p level.
CCK-8 assay
A549 and CAL-12T cells in logarithmic growth phase were selected and seeded in 96-well plates at 1000 cells/well. A volume of 10 μL CCK8 solution (Hubei Biossci Biotechnology Co., Ltd.) was added to measure the absorbance at 450 nm (optical density (OD) value) using a microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Thereafter, the absorbance of cells was measured at 24 h, 48 h, 72 h, and 96 h.
Colony formation assay
Lung cancer cells A549 and CAL-12T were planted in six-well plates containing 1000 cells in each well. After 2 weeks of culture, the culture solution was discarded and carefully rinsed twice with PBS. Cells were then fixed using 4% paraformaldehyde for 10 min. A volume of 1 mL of crystal violet was added to each well, and the number of colony was recorded after staining.
Apoptosis assay
A549 and CAL-12T cells were collected, washed twice with PBS and resuspended in binding buffer. The cells were then stained with FITC Annexin V Apoptosis Detection Kit (Ruibo, Guangzhou, China) for 30 min at room temperature in the dark. Cell apoptosis was determined using flow cytometry (Becton Dickinson, Mountain View, USA).
Wound healing assay
The cells in each group were inoculated to 12-well plate with 2 × 105 per well. Subsequently, cells were cultured for 48 h. Once cells were paved, the water dropper of the pipette was used to make scratches along the bottom of the culture plate on the monolayer cultured cells. After that, the cells continued to be cultured for 24 h with serum-free medium. Ultimately, the scratches were observed.
Transwell assay
The migration experiment was carried out in culture chamber by using polycarbonate filter membrane with a specification of 8 μm. The cells of each group were made to be cell suspension with a density of 1 × 105/mL in a serum-free medium, and 200 μL cell suspension was inoculated into the upper chamber of the transwell chamber, 500 μL medium with 10% FBS was added to the lower chamber, and three compound wells were set in each group. After 24 h of culture, cells in the upper layer were gently wiped off by a cotton swab. Furthermore, 4% polyformaldehyde was used to fix cells for 10 min, and 0.1% crystal violet was added to stain cells for 10 min. The number of cells was then counted under the microscope. The small chamber covering invasive experiment was Matrigel, and the other steps were the same as the migration experiment.
Western blot
The cells were lysed with radioimmunoprecipitation assay (RIPA) lysis buffer containing protease inhibitor. The supernatant was collected after high-speed centrifugation, and heated in water bath to denature the protein. After quantifying the protein by bicinchoninic acid (BCA) method, proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel electrophoresis, and incubated with Anti-PD-L1 antibody (Abcam, 1:500) at 4°C for 12 h. The polyvinylidene difluoride (PVDF) membrane was rinsed with the TBST solution, and then incubated with the secondary antibody (Hubei Biossci Biotechnology Co., Ltd., 1:2000) for 1 h at room temperature. Following that, the chemiluminescence was developed using a hypersensitive ECL (Hubei Biossci Biotechnology Co., Ltd.). β-actin was used as an internal reference.
Statistical analysis
GraphPad Prism 8 was used as the statistical processing software in this study. The t test was used to compare the data between the two groups. P < 0.05 was considered statistically different.
Results
MALAT1 expression in NSCLC was correlated with that of miR-200a-3p and PD-L1
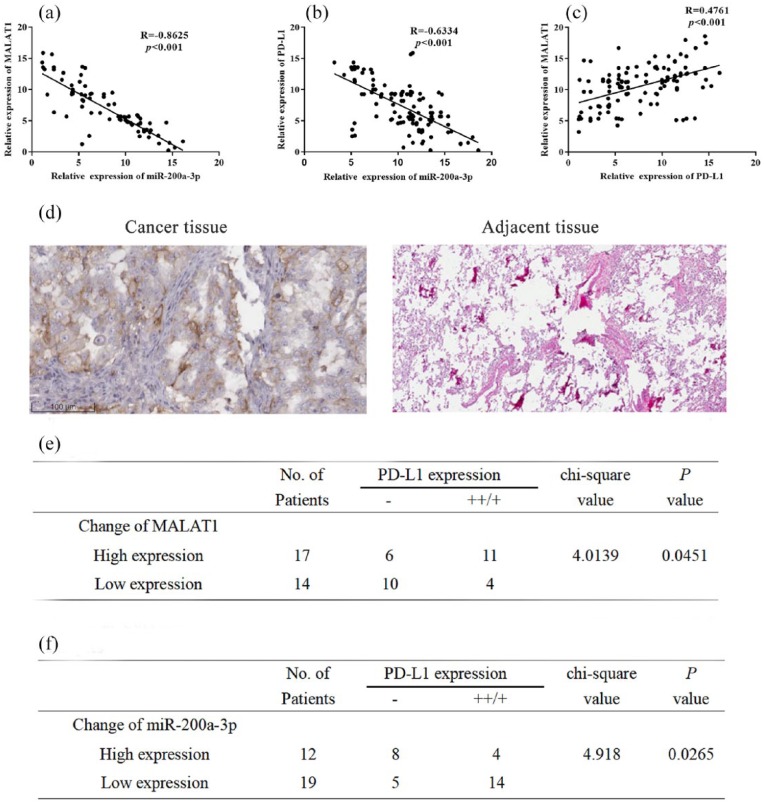
First, we detected the expression levels of MALAT1, miR-200a-3p, and PD-L1 in 113 NSCLC samples by qRT-PCR. Then, we conducted correlation analysis. The results showed that expression levels of MALAT1 and miR-200a-3p were inversely correlated (Figure 1(a), R = ‒0.8625, P < 0.001). Expression levels of miR-200a-3p and PD-L1 mRNA were also inversely correlated (Figure 1(b), R = ‒0.6334, P < 0.001), while expression levels of MALAT1 and PD-L1 mRNA were positively correlated (Figure 1(c), R = 0.4761, P < .001). In addition, higher PD-L1 immunohistochemical staining scores were negatively correlated with the expression level of miR-200a-3p, while positively correlated with the expression level of MALAT1 (Figure 1(a)–(f), chi-square test, P < 0.05). These data implied that there were potential regulatory relationships among MALAT1, miR-200a-3p, and PD-L1.
Figure 1.
Correlation among the expression levels of MALAT1, miR-200a-3p, and PD-L1: (a) The expression level of MALAT1 was negatively correlated with the expression level of miR-200a-3p in 113 NSCLC samples. (b) The expression level of miR-300a-3p was negatively correlated with the expression level of PD-L1 in 113 NSCLC samples. (c) The expression level of MALAT1 was positively correlated with the expression level of PD-L1 in 113 NSCLC samples. (d) IHC was used to detect the expression of PD-L1, and images of a pair of NSCLC tissues (left, ++) and adjacent tissues (right, ‒) were shown. (e) Correlation between IHC staining score of PD-L1 and MALAT1 in 31 NSCLC samples. (f) Correlation between IHC staining score of PD-L1 and miR-200a-3p in 31 NSCLC samples.
MALAT1 sponges miR-200a-3p
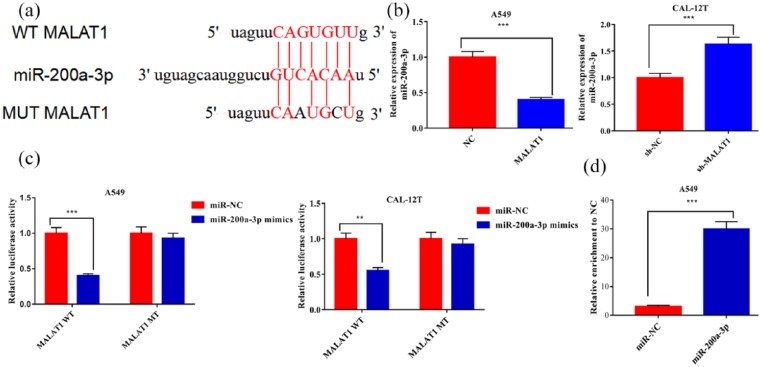
Then the target microRNAs of MALAT1 were predicted by starBase (http://starbase.sysu.edu.cn), and miR-200a-3p was found to be a candidate target of MALAT1 (Figure 2(a)). qRT-PCR demonstrated that overexpressed MALAT1 significantly decreased the expression level of miR-200a-3p in A549 cells, while knockdown of MALAT1 increased miR-200a-3p expression in CAL-12T cells (Figure 2(b)). In addition, luciferase reporter gene assay and RIP assay verified that MALAT1 had binding sites for miR-200a-3p, and could play a “sponge” role (Figure 2(c) and (d)).
Figure 2.
MALAT1 sponged miR-200a-3p and down-regulated its expression in NSCLC: (a) miR-200a-3p binding sequence of MALAT1 indicated that MALAT1 was a potential sponge of miR-200a-3p. (b) MALAT1 modulated the expression levels of miR-200a-3p in both A549 and CAL-12T cells. (c) miR-200a-3p significantly repressed the luciferase activity of wild type MALAT1 reporter, but did not change the luciferase activity of mutated MALAT1 reporter in A549 cells. (d) MALAT1 and miR-200a-3p simultaneously existed in the production precipitated by anti-AGO2.
**P < 0.01. ***P < 0.001.
MALAT1 promotes NSCLC cells via modulating miR-200a-3p
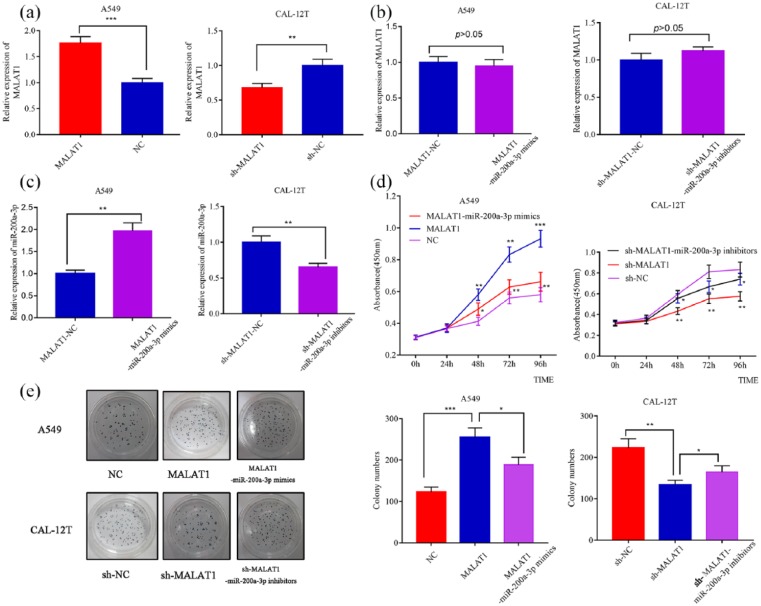
To further clarify the effect of MALAT1 and miR-200a-3p on the proliferation of NSCLC cells, we transfected MALAT1 plasmid into A549 cells to successfully establish MALAT1 overexpression models; MALAT1 shRNA was transfected into CAL-12T cells to successfully establish MALAT1 low expression models (Figure 3(a)). miR-200a-3p mimics or inhibitors were also transefected into NSCLC cells, but they did not change the expression level of MALAT1 (Figure 3(b) and (c)). Subsequently, CCK8 assay were conducted. The proliferation rate of CAL-12T cells with low-expressed MALAT1 significantly decreased than that of the control group, and the proliferation of CAL-12T cells with lowly expressed MALAT1 significantly increased after transfection of miR-200a-3p inhibitors (Figure 3(d)). Plate colony formation assays was performed to detect the proliferation of A549 and CAL-12T cells. The number of colony of A549 cells with overexpressed MALAT1 significantly increased than that of the control group, and it significantly decreased after transfection of miR-200a-3p mimics. The number of colony of CAL-12T cells with lowly expressed MALAT1 was significantly lower than that in the control group, and it significantly increased after transfection of miR-200a-3p inhibitors (Figure 3(e)).
Figure 3.
MALAT1 promotes proliferation of NSCLC cells via modulating miR-200a-3p. (a) Successful transfection of MALAT1 overexpression plasmid and shRNA into NSCLC cells was validated by qRT-PCR. (b) qRT-PCR showed that transfection of miR-200a-3p mimics or inhibitors did not change the expression level of MALAT1 in NSCLC cells. (c) qRT-PCR showed that transfection of miR-200a-3p mimics or inhibitors modulated the expression level of miR-200a-3p in NSCLC cells. (d) MALAT1 overexpression promoted proliferation of A549 cells and miR-200a-3p transfection partly reversed its function (left); knockdown of MALAT1 inhibited proliferation of CAL-12T cells and miR-200a-3p inhibitors neutralized its function (right). (e) MALAT1 overexpression promoted colony formation ability of A549 cells and miR-200a-3p transfection partly reversed its function (left); knockdown of MALAT1 inhibited colony formation of CAL-12T cells and miR-200a-3p inhibitors neutralized its function (right).
*P < 0.05. **P < 0.01. ***P < 0.001.
MALAT1 inhibits apoptosis and promotes metastasis of NSCLC cells via modulating miR-200a-3p
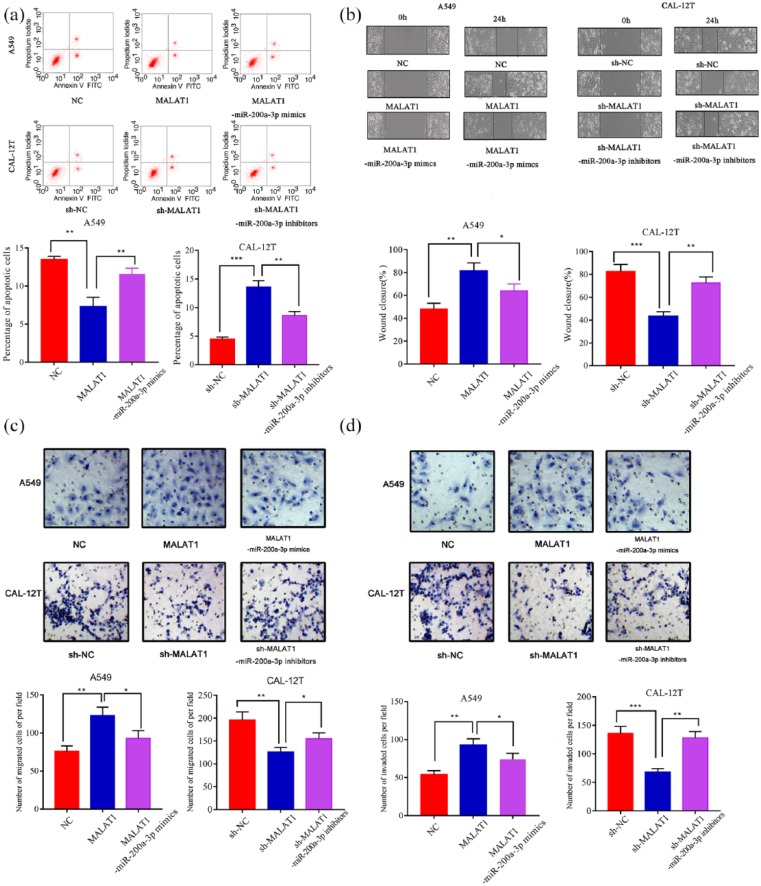
Next, we examined the effects of MALAT1 and miR-200a-3p on apoptosis and metastasis of NSCLC cells. Apoptosis experiments confirmed that the apoptotic rate of A549 cells with overexpressed MALAT1 was lower than that of control cells, and it was significantly increased after transfection of miR-200a-3p mimics; the apoptotic rate of CAL-12T cells with MALAT1 knockdown was higher than that of control group, and it is significantly decreased after transfection of miR-200a-3p inhibitors (Figure 4(a)). The wound healing assay indicated that the motility of A549 cells with overexpressed MALAT1 significantly increased compared with the control group, and it is significantly decreased after transfection of miR-200a-3p mimics; the motility of CAL-12T cells with MALAT1 knockdown was significantly inhibited, and transfection of miR-200a-3p inhibitors partly reversed this effect (Figure 4(b)). Subsequently, by transwell migration and invasion assays, we validated that the metastatic ability of A549 cells with overexpressed MALAT1 was significantly higher than that of the control group, and transfection of miR-200a-3p mimics partly neutralized the function of MALAT1; compared with that of the control group, the metastatic ability of CAL-12T cells with MALAT1 knockdown was blocked, but was increased after transfection of miR-200a-3p inhibitors (Figure 4(a) and (d)).
Figure 4.
MALAT1 inhibits apoptosis and promotes metastasis of NSCLC cells via modulating miR-200a-3p: (a) MALAT1 overexpression inhibited apoptosis of A549 cells and miR-200a-3p transfection induced apoptosis (left); knockdown of MALAT1 induced apoptosis of CAL-12T cells and miR-200a-3p inhibitors partly reversed it (right). (b) Wound healing assay showed that MALAT1 overexpression promoted motility of A549 cells and miR-200a-3p transfection partly reversed its function (left); knockdown of MALAT1 inhibited motility of CAL-12T cells and miR-200a-3p inhibitors neutralized its function (right). (c) Transwell assay showed MALAT1 overexpression promoted migration of A549 cells and miR-200a-3p transfection partly reversed its function (left); knockdown of MALAT1 inhibited migration of CAL-12T cells and miR-200a-3p inhibitors neutralized its function (right). (d) Transwell assay showed MALAT1 overexpression promoted invasion of A549 cells and miR-200a-3p transfection partly reversed its function (left); knockdown of MALAT1 inhibited invasion of CAL-12T cells and miR-200a-3p inhibitors neutralized its function (right).
*P < 0.05. **P < 0.01. ***P < 0.001.
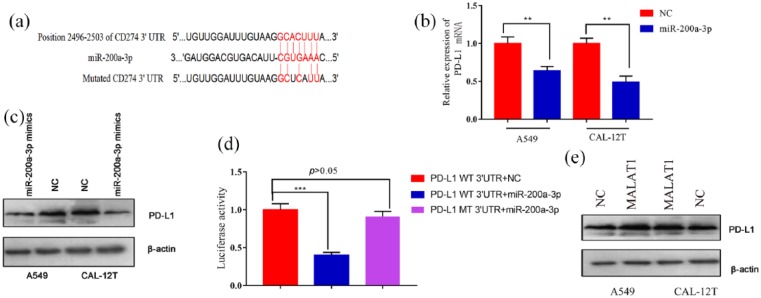
MiR-200a-3p directly targets 3’-UTR of PD-L1
Next, the target gene of miR-200a-3p was predicted by TargetScan (www.targetscan.org), and PD-L1 was found to be a candidate target gene of miR-200a-3p (Figure 5(a)). qRT-PCR and Western blot results showed that there was a significant decrease in PD-L1 mRNA and protein after the transfection of miR-200a-3p into both A549 and CAL-12T cells (Figure 5(b) and (c)). Subsequently, luciferase reporter gene assay revealed that miR-200a-3p could specifically bind to 3’-UTR of PD-L1 (Figure 5(d)). Western blot also showed that PD-L1 expression was significantly up-regulated after MALAT1 plasmid transfection (Figure 5(e)). These results indicated that PD-L1 was a downstream gene of miR-200a-3p, and MALAT1 might indirectly regulate its expression.
Figure 5.
PD-L1 was a target gene of miR-200a-3p and could be indirectly up-regulated by MALAT1: (a) miR-200a-3p binding sequence of PD-L1 3’-UTR indicated that PD-L1 was a potential target of miR-200a-3p. (b) qRT-PCR showed that transfection of miR-200a-3p mimics significantly attenuated the mRNA levels of endogenous PD-L1 in both A549 and CAL-12T cells. (c) Western blot showed that transfection of miR-200a-3p mimics significantly attenuated the protein levels of endogenous PD-L1 in both A549 and CAL-12T cells. (d) miR-200a-3p significantly repressed the luciferase activity of wide type 3’-UTR of PD-L1, but did not change the luciferase activity of mutated 3’-UTR of PD-L1 in A549 cells. (e) Western blot showed that overexpression of MALAT1 significantly increased the protein levels of endogenous PD-L1 in both A549 and CAL-12T cells.
**P < 0.01. ***P < 0.001.
Discussion
NSCLC is one of the most common malignant tumors. However, the current diagnosis and treatment methods for NSCLC are limited, and the therapeutic effect is still far from satisfactory.25–28 MALAT1 and PD-L1 promote NSCLC progression, while multiple miR-200 family members play a role in fighting it.29–36 Consistent with previous reports, we found that MALAT1 and PD-L1 were highly expressed in NSCLC, while miR-200a-3p was significantly downregulated. The starBase and TargetScan suggested that miR-200a-3p has possible binding sites of MALAT1, and PD-L1 was also one of the downstream targets of miR-200a-3p. These findings prompted us to further investigate the role of MALAT1/miR-200a-3p/PD-L1 axis in the tumorigenesis and progression of NSCLC.
MALAT1 is a unfavorable biomarker in a variety of tumors, such as prostate cancer, breast cancer, oral squamous cell carcinoma, NSCLC, and other cancers.37–41 But interestingly, MALAT1 has tumor suppressive function by reducing the expression of matrix metalloproteinase 2 in glioma.42 This suggested that the function of MALAT1 may differ depending on the type of tumor cell and its specific molecular interaction. In recent years, competing endogenous RNA (ceRNA) hypothesis has come into prominence in cancer researches. LncRNAs can act as miRNA sponges by sequestering their target miRNAs and affect gene expression indirectly.43–45 In this study, we demonstrated that the up-regulation of MALAT1 in NSCLC cells caused a significant decrease in the expression of miR-200a-3p. Furthermore, after transfecting miR-200a-3p mimics into the above NSCLC cells with overexpressed MALAT1, the cancer-promoting effect of MALAT1 was reversed. Knockdown of MALAT1 caused the up-regulation of miR-200a-3p, and the malignant phenotypes of cancer cells were promoted by transfecting miR-200a-3p inhibitors into NSCLC cells with low-expressed MALAT1. The direct binding sites of miR-200a-3p to MALAT1 were also confirmed by the dual luciferase reporter gene assay. Our study showed that lncRNA MALAT1 elicited its biological effects partly by acting as a sponge for miR-200a-3p, which affected the ability of miR-200a-3p to bind to its targets.
In recent years, several studies have indicated that the miR-200 family played a tumor suppressive role in a variety of tumors.46–49 For example, miR-200c inhibits the growth of breast cancer cells by modulating the HIPK1/β-Catenin axis;47 miR-200a-3p impedes the proliferation of renal cell carcinoma and induces apoptosis by targeting sperm-associated antigen 9.49 In this study, we demonstrated that transfection of miR-200a-3p into NSCLC cells caused a significant decrease in the expression of PD-L1 and arrested the proliferation and metastasis of NSCLC cells. The direct binding of miR-200a-3p to PD-L1 was confirmed by the dual luciferase reporter gene assay. Thus, we confirmed that in NSCLC cells, miR-200a-3p can negatively regulate the expression of PD-L1, thereby inhibiting the immune escape of tumor cells.
Recently, PD-L1 is a hot topic in cancer research, and drugs targeting PD-L1 have been shown to help patients improve their prognosis. For example, in pancreatic cancer, interferon-γ promotes epithelial mesenchymal transition and expression of PD-L1, whereas targeting PD-1/PD-L1 immunotherapy improves prognosis.50 Studies have found that in patients with NSCLC, survival time after treatment with anti-PD-1 therapy is significantly prolonged.51 In this experiment, we up-regulated the expression of PD-L1 in NSCLC and promoted the mobility, proliferation, migration, and invasion of NSCLC cells. While knocking down the expression of PD-L1, the above biological behaviors were inhibited. Our findings are consistent with current reports.
In summary, we found that LncRNA MALAT1 accelerated the progression of NSCLC by modulating the miR-200a-3p/PDL1 axis, and had a positive significance for the selection of new targeted drugs and enrichment of therapeutic methods in the future. However, we need to further confirm that LncRNA MALAT/miR-200a-3p/PDL1 can regulate the biological behavior of NSCLC cells via animal experiments.
Supplemental Material
Supplemental material, Supplementary_Materials for LncRNA MALAT1 contributes to non-small cell lung cancer progression via modulating miR-200a-3p/programmed death-ligand 1 axis by Shuquan Wei, Kangwei Wang, Xiaomei Huang, Zhuxiang Zhao and Ziwen Zhao in International Journal of Immunopathology and Pharmacology
Footnotes
Author contributions: S.Q.W. and Ziwen Zhao conceived and designed the experiments; S.Q.W., K.W.W., X.M.H., and Zhuxiang Zhao performed the experiments; S.Q.W. contributed to statistic analysis; S.Q.W. and Ziwen Zhao wrote the article; and all authors read and approved the final article.
Data availability: The data used to support the findings of this study are available from the corresponding author upon request.
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Ziwen Zhao  https://orcid.org/0000-0002-4374-9914
https://orcid.org/0000-0002-4374-9914
Supplemental material: Supplemental material for this article is available online.
References
- 1. Siegel RL, Miller KD, Jemal A. (2019) Cancer statistics, 2019. CA: A Cancer Journal for Clinicians 69(1): 7–34. [DOI] [PubMed] [Google Scholar]
- 2. Bray F, Ferlay J, Soerjomataram I, et al. (2018) Cancer statistics, 2018. CA: A Cancer Journal for Clinicians 68(6): 394–424. [DOI] [PubMed] [Google Scholar]
- 3. Du Q, Li E, Liu Y, et al. (2018) CTAPIII/CXCL7: A novel biomarker for early diagnosis of lung cancer. Cancer Medicine 7: 325–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhou Q, Tang X, Tian X, et al. (2018) LncRNA MALAT1 negatively regulates MDSCs in patients with lung cancer. Journal of Cancer 9(14): 2436–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lin L, Li H, Zhu Y, et al. (2018) Expression of metastasis-associated lung adenocarcinoma transcript 1 long non-coding RNA in vitro and in patients with non-small cell lung cancer. Oncology Letters 15(6): 9443–9449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jin Y, Feng SJ, Qiu S, et al. (2017) LncRNA MALAT1 promotes proliferation and metastasis in epithelial ovarian cancer via the PI3K-AKT pathway. European Review for Medical and Pharmacological Sciences 21(14): 3176–3184. [PubMed] [Google Scholar]
- 7. Yang YR, Zang SZ, Zhong CL, et al. (2014) Increased expression of the lncRNA PVT1 promotes tumorigenesis in non-small cell lung cancer. International Journal of Clinical and Experimental Pathology 7(10): 6929–6935. [PMC free article] [PubMed] [Google Scholar]
- 8. Li S, Mei Z, Hu HB, et al. (2018) The lncRNA MALAT1 contributes to non-small cell lung cancer development via modulating miR-124/STAT3 axis. Journal of Cellular Physiology 233(9): 6679–6688. [DOI] [PubMed] [Google Scholar]
- 9. Tang Y, Xiao G, Chen Y, et al. (2018) LncRNA MALAT1 promotes migration and invasion of non-small-cell lung cancer by targeting miR-206 and activating Akt/mTOR signaling. Anti-cancer Drugs 29(8): 725–735. [DOI] [PubMed] [Google Scholar]
- 10. Liu N, Liu Z, Zhang W, et al. (2018) MicroRNA433 reduces cell proliferation and invasion in nonsmall cell lung cancer via directly targeting E2F transcription factor 3. Molecular Medicine Reports 18(1): 1155–1164. [DOI] [PubMed] [Google Scholar]
- 11. Chang H, Liu YH, Wang LL, et al. (2018) MiR-182 promotes cell proliferation by suppressing FBXW7 and FBXW11 in non-small cell lung cancer. American Journal of Translational Research 10(4): 1131–1142. [PMC free article] [PubMed] [Google Scholar]
- 12. Gong Y, Mao J, Wu D, et al. (2018) Circ-ZEB1.33 promotes the proliferation of human HCC by sponging miR-200a-3p and upregulating CDK6. Cancer Cell International 18: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tak H, Kang H, Ji E, et al. (2018) Potential use of TIA-1, MFF, microRNA-200a-3p, and microRNA-27 as a novel marker for hepatocellular carcinoma. Biochemical and Biophysical Research Communications 497(4): 1117–1122. [DOI] [PubMed] [Google Scholar]
- 14. Zang Y, Tai Y, Wan B, et al. (2016) miR-200a-3p promotes the proliferation of human esophageal cancer cells by post-transcriptionally regulating cytoplasmic collapsin response mediator protein-1. International Journal of Molecular Medicine 38(5): 1558–1564. [DOI] [PubMed] [Google Scholar]
- 15. Xie K, Wang C, Qin N, et al. (2016) Genetic variants in regulatory regions of microRNAs are associated with lung cancer risk. Oncotarget 7(30): 47966–47974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shi L, Chen S, Yang L, et al. (2013) The role of PD-1 and PD-L1 in T-cell immune suppression in patients with hematological malignancies. Journal of Hematology and Oncology 6(1): 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ohaegbulam KC, Assal A, Lazar-Molnar E, et al. (2015) Human cancer immunotherapy with antibodies to the PD-1 and PD-L1 pathway. Trends in Molecular Medicine 21(1): 24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen T, Li Q, Liu Z, et al. (2019) Peptide-based and small synthetic molecule inhibitors on PD-1/PD-L1 pathway: A new choice for immunotherapy? European Journal of Medicinal Chemistry 161: 378–398. [DOI] [PubMed] [Google Scholar]
- 19. Lian S, Xie R, Ye Y, et al. (2019) Simultaneous blocking of CD47 and PD-L1 increases innate and adaptive cancer immune responses and cytokine release. EBioMedicine 42: 281–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu Y, Gu W, Li J, et al. (2019) Silencing PD-1 and PD-L1 with nanoparticle-delivered small interfering RNA increases cytotoxicity of tumor-infiltrating lymphocytes. Nanomedicine 14: 0237. [DOI] [PubMed] [Google Scholar]
- 21. Liao LC, Hu B, Zhang SP. (2019) Macrophages participate in the immunosuppression of condyloma acuminatum through the PD-1/PD-L1 signalling pathway. Journal of the Chinese Medical Association 82: 413–418. [DOI] [PubMed] [Google Scholar]
- 22. Jiang Y, Chen M, Nie H, et al. (2019) PD-1 and PD-L1 in cancer immunotherapy: Clinical implications and future considerations. Human Vaccines and Immunotherapeutics 15: 1111–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. DiTucci C, Capone C, Galati G, et al. (2019) Immunotherapy in endometrial cancer: New scenarios on the horizon. Journal of Gynecologic Oncology 30(3): e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bylicki O, Barazzutti H, Paleiron N, et al. (2019) First-line treatment of non-small-cell lung cancer (NSCLC) with immune checkpoint inhibitors. Biodrugs: Clinical Immunotherapeutics, Biopharmaceuticals and Gene Therapy 33(2): 159–171. [DOI] [PubMed] [Google Scholar]
- 25. Passiglia F, Galvano A, Rizzo S, et al. (2018) Looking for the best immune-checkpoint inhibitor in pre-treated NSCLC patients: An indirect comparison between nivolumab, pembrolizumab and atezolizumab. Inter-national Journal of Cancer 142(6): 1277–1284. [DOI] [PubMed] [Google Scholar]
- 26. Yang Z, Tam KY. (2018) Combination strategies using EGFR-TKi in NSCLC therapy: Learning from the gap between pre-clinical results and clinical outcomes. International Journal of Biological Sciences 14(2): 204–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schrank Z, Chhabra G, Lin L, et al. (2018) Current molecular-targeted therapies in NSCLC and their mechanism of resistance. Cancers 10(7): piiE224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gotfrit J, Jonker C, Zhang T, et al. (2019) Inpatients versus outpatients with advanced non-small cell lung cancer: Characteristics and outcomes. Cancer Treatment and Research Communications 19: 100130. [DOI] [PubMed] [Google Scholar]
- 29. Wang JZ, Xiang JJ, Wu LG, et al. (2017) A genetic variant in long non-coding RNA MALAT1 associated with survival outcome among patients with advanced lung adenocarcinoma: A survival cohort analysis. BMC Cancer 17(1): 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang R, Xia Y, Wang Z, et al. (2017) Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochemical and Biophysical Research Communications 490(2): 406–414. [DOI] [PubMed] [Google Scholar]
- 31. Zhang CG, Yin DD, Sun SY, et al. (2017) The use of lncRNA analysis for stratification management of prognostic risk in patients with NSCLC. European Review for Medical and Pharmacological Sciences 21: 115–119. [PubMed] [Google Scholar]
- 32. Bravaccini S. (2019) What impacts the cost-effectiveness of PD-L1 testing in non-small cell lung cancer? Lung Cancer 132: 152–153. [DOI] [PubMed] [Google Scholar]
- 33. Vigliar E, Malapelle U, Iaccarino A, et al. (2019) PD-L1 expression on routine samples of non-small cell lung cancer: Results and critical issues from a 1-year experience of a centralised laboratory. Journal of Clinical Pathology 72(6): 412–417. [DOI] [PubMed] [Google Scholar]
- 34. Liu C, Hu W, Li LL, et al. (2018) Roles of miR-200 family members in lung cancer: More than tumor suppressors. Future Oncology 14(27): 2875–2886. [DOI] [PubMed] [Google Scholar]
- 35. Nishijima N1, Seike M1, Soeno C1, et al. (2016) miR-200/ZEB axis regulates sensitivity to nintedanib in non-small cell lung cancer cells. International Journal of Oncology 48(3): 937–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pacurari M, Addison JB, Bondalapati N, et al. (2013) The microRNA-200 family targets multiple non-small cell lung cancer prognostic markers in H1299 cells and BEAS-2B cells. International Journal of Oncology 43(2): 548–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Malakar P, Stein I, Saragovi A, et al. (2019) Long noncoding RNA MALAT1 regulates cancer glucose metabolism by enhancing mTOR-mediated translation of TCF7L2. Cancer Research 79(10): 2480. [DOI] [PubMed] [Google Scholar]
- 38. Ye G, Guo L, Xing Y, et al. (2019) Identification of prognostic biomarkers of prostate cancer with long non-coding RNA-mediated competitive endogenous RNA network. Experimental and Therapeutic Medicine 17(4): 3035–3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Arun G, Spector DL. (2019) MALAT1 long non-coding RNA and breast cancer. RNA Biology 16: 860–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yuan J, Xu XJ, Lin Y, et al. (2019) LncRNA MALAT1 expression inhibition suppresses tongue squamous cell carcinoma proliferation, migration and invasion by inactivating PI3K/Akt pathway and downregulating MMP-9 expression. European Review for Medical and Pharmacological Sciences 23(1): 198–206. [DOI] [PubMed] [Google Scholar]
- 41. Cui Y, Li G, Zhang X, et al. (2018) Increased MALAT1 expression contributes to cisplatin resistance in non-small cell lung cancer. Oncology Letters 16(4): 4821–4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Han Y, Wu Z, Wu T, et al. (2016) Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by downregulation of MMP2 and inactivation of ERK/MAPK signaling. Cell Death and Disease 7: e2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhuang M, Zhao S, Jiang Z, et al. (2019) MALAT1 sponges miR-106b-5p to promote the invasion and metastasis of colorectal cancer via SLAIN2 enhanced microtubules mobility. EBioMedicine 41: 286–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tao F, Tian X, Ruan S, et al. (2018) miR-211 sponges lncRNA MALAT1 to suppress tumor growth and progression through inhibiting PHF19 in ovarian carcinoma. FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology. Epub ahead of print 6 June DOI: 10.1096/fj.201800495RR. [DOI] [PubMed] [Google Scholar]
- 45. Xiao X, Zhou T, Guo S, et al. (2017) LncRNA MALAT1 sponges miR-204 to promote osteoblast differentiation of human aortic valve interstitial cells through up-regulating Smad4. International Journal of Cardiology 243: 404–412. [DOI] [PubMed] [Google Scholar]
- 46. Fu J, Shrivastava A, Shrivastava SK, et al. (2019) Triacetyl resveratrol upregulates miRNA-200 and suppresses the Shh pathway in pancreatic cancer: A potential therapeutic agent. International Journal of Oncology 54: 1306–1316. [DOI] [PubMed] [Google Scholar]
- 47. Liu B, Du R, Zhou L, et al. (2018) miR-200c/141 regulates breast cancer stem cell heterogeneity via targeting HIPK1/β-Catenin axis. Theranostics 8(21): 5801–5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu DM, Wang S, Wen X, et al. (2018) LncRNA SNHG15 acts as a ceRNA to regulate YAP1-Hippo signaling pathway by sponging miR-200a-3p in papillary thyroid carcinoma. Cell Death and Disease 9(10): 947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang X, Jiang F, Song H, et al. (2016) MicroRNA-200a-3p suppresses tumor proliferation and induces apoptosis by targeting SPAG9 in renal cell carcinoma. Biochemical and Biophysical Research Communications 470(3): 620–626. [DOI] [PubMed] [Google Scholar]
- 50. Imai D, Yoshizumi T, Okano S, et al. (2017) The prognostic impact of programmed cell death ligand 1 and human leukocyte antigen class I in pancreatic cancer. Cancer Medicine 6: 1614–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ahn BC, Pyo KH, Xin CF, et al. (2019) Comprehensive analysis of the characteristics and treatment outcomes of patients with non-small cell lung cancer treated with anti-PD-1 therapy in real-world practice. Journal of Cancer Research and Clinical Oncology 145: 1613–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplementary_Materials for LncRNA MALAT1 contributes to non-small cell lung cancer progression via modulating miR-200a-3p/programmed death-ligand 1 axis by Shuquan Wei, Kangwei Wang, Xiaomei Huang, Zhuxiang Zhao and Ziwen Zhao in International Journal of Immunopathology and Pharmacology