Abstract
Background:
Long noncoding RNAs (lncRNAs) play pivotal roles in various malignant tumors. Epidermal growth factor receptor (EGFR) signaling is associated with the pathogenesis of cutaneous squamous cell carcinoma (cSCC). This study aimed to explore the role of LINC00520 in the development of cSCC via EGFR and phosphoinositide 3-kinase-protein kinase B (PI3K/Akt) signaling pathways.
Methods:
A microarray analysis was applied to screen differentially expressed lncRNAs in cSCC samples. The A431 cSCC cell line was transfected and assigned different groups. The expression patterns of LINC00520, EGFR, and intermediates in the PI3K/Akt pathway were characterized using reverse transcription quantitative polymerase chain reaction (RT-qPCR) and Western blotting analysis. Cell proliferation, migration, and invasion were detected using the MTT assay, scratch test, and Transwell assay, respectively. Cell-based experiments and a tumorigenicity assay were conducted to assess the effect of LINC00520 on cSCC progression. This study was ended in September 2017. Comparisons between two groups were analyzed with t-test and comparisons among multiple groups were analyzed using one-way analysis of variance. The nonparametric Wilcoxon rank sum test was used to analyze skewed data. The enumerated data were analyzed using the chi-square test or Fisher exact test.
Results:
Data from chip GSE66359 revealed depletion of LINC00520 in cSCC. Cells transfected with LINC00520 vector and LINC00520 vector + si-EGFR showed elevated LINC00520 level but decreased levels of the EGFR, PI3K, AKT, VEGF, MMP-2 and MMP-9 mRNAs and proteins, and inhibition of the growth, migration and adhesion of cSCC cells, while the si-LINC00520 group showed opposite trends (all P < 0.05). Compared with the LINC00520 vector group, the LINC00520 vector + si-EGFR group showed decreased levels of the EGFR, PI3K, AKT, VEGF, MMP-2 and MMP-9 mRNAs and proteins, and inhibition of the growth, migration and adhesion of cSCC cells, while the LINC00520 vector + EGFR vector group showed opposite results (all P < 0.05).
Conclusion:
Based on our results, LINC00520-targeted EGFR inhibition might result in the inactivation of the PI3K/Akt pathway, thus inhibiting cSCC development.
Keywords: LINC00520, EGFR, PI3K/Akt signaling pathway, Cutaneous squamous cell carcinoma, Lymphatic vessel invasion, Invasion, Metastasis
Introduction
Cutaneous squamous cell carcinoma (cSCC) is the second most frequently diagnosed type of nonmelanoma skin cancer, with a high annual mortality rate, and the disease often occurs in keratinocytes of the epidermis due to sun exposure.[1,2] Nonmelanoma skin cancer is caused by numerous risk factors, such as exposure to ultraviolet (UV) radiation, older age, male sex, chronic skin ulcers and burns scars, and immunosuppression.[3] cSCC is also reported to have a high metastatic risk, usually for the lymph nodes.[4] cSCC is sensitive to radiotherapy, which has been administered to patients who underwent incomplete unresectable excision or adjuvant treatment after complete lymph node resection.[5] Nevertheless, patients with regional lymphatic metastasis or distant metastases have a less than 20% 10-year survival rate, revealing the substantial challenge in treating advanced and metastatic cSCC.[6] Very little is currently known about the genetic mutations driving aggressive cSCC.[7] Therefore, an understanding of mutations in cSCC is urgently needed to develop an effective targeted approach for the treatment of cSCC.
Long noncoding RNAs (lncRNAs) play pivotal roles in various malignant tumors by regulating tumor cell proliferation, differentiation, invasion and metastasis.[8] Based on recent evidence, lncRNAs are involved in maintaining keratinocyte stem cells and their differentiation toward keratinocytes in the epidermis.[9] Interestingly, long intergenic nonprotein coding RNA 520 (LINC00520) is reported to play an important role in cell migration, invasion, and metastasis in breast cancer.[10] The expression of LINC00520, which is transcriptionally enriched in immortalized mammary epithelial cells, is induced by Src, an activator of transcription 3 (Stat3), and PI3K.[10] The PI3K/Akt signaling pathway regulates the development and progression of cSCC.[11] Additionally, epidermal growth factor receptor (EGFR) and transforming growth factor-beta (TGF-beta) are implicated in the tumorigenesis of SCC.[12]EGFR functions as a potential target for the treatment of cSCC.[13] Of crucial importance, both EGFR and the PI3K/Akt signaling pathway are activated in cSCC, subsequently increasing Stat3 activity to enhance cell proliferation and survival.[14] Based on the aforementioned literature, a hypothesis has been proposed that EGFR and the PI3K/Akt signaling pathway might be involved in the underlying mechanism of LINC00520 in cSCC. In this study, we aimed to identify the roles of LINC00520 in the invasion and metastasis of cSCC and characterize the potential mechanism.
Methods
Ethics statement
All experimental procedures (This study was ended in September 2017) adhered to the International Convention on Experimental Animal Ethics and conformed to the relevant regulations of the nation. All efforts were made to minimize animals’ suffering.
Differentially expressed lncRNA screen and target gene prediction
The GEO database (http://www.ncbi.nlm.nih.gov/geo) was used for the bioinformatics predictions. Chip data (GSE66359) and annotated probe files of cSCC were acquired. The Affy installation package of R software was used for background correction and normalization of data from each chip.[15] The Linear Models and Empirical Bayes Methods of Limma installation package were applied with a combination of traditional t-tests to perform a nonspecific filtration process for gene expression profiling. Then, the differentially expressed lncRNAs and messenger RNAs (mRNAs) were screened.[16] The Multi Experiment Matrix (MEM, http://biit.cs.ut.ee/mem/) website was used to predict the targets genes of differentially expressed lncRNAs. A KEGG analysis of target genes was performed using the DAVID database (https://david.ncifcrf.gov/) to identify the genes related to the occurrence and development of cSCC.
Plasmid construction, cell grouping and transfection
Blank, LINC00520 NC, LINC00520 overexpression, LINC00520 siRNA (si-LINC00520), EGFR siRNA (si-EGFR) and EGFR overexpression plasmids were constructed by Shanghai Sangon Biotech Company (Shanghai, China) using the LINC00520 and EGFR sequences in the National Center for Biotechnology Information (NCBI) database. The human cSCC cell line A431 was obtained from the Shanghai Institute of Cell Biology of Chinese Academy of Sciences (Shanghai, China), and cultured with RPMI 1640 culture medium (Gibco, Carlsbad, CA, USA) containing 100 g/L fetal bovine serum (FBS) (Jiangsu Ke Te Biological Co., Ltd., Jiangsu, China)[17] at 37°C in a 5% CO2 atmosphere. When the confluence reached 80% to 90%, cSCC cells were treated with 0.25 g/L trypsin (Shanghai Ru Ji Biotechnology Co., Ltd., Shanghai, China) and subcultured.
Then, cells were transferred into a 24-well plate and treated with trypsin. Monolayer cells were then grouped and transfected: blank group (cSCC cells transfected with the blank plasmid), NC group (cSCC cells transfected with the LINC00520 NC nonsense sequence), LINC00520 vector group (cSCC cells transfected with the LINC00520 overexpression plasmid), si-LINC00520 group (cSCC cells transfected with the si-LINC00520 plasmid), si-EGFR group (cSCC cells transfected with the si-EGFR plasmid), LINC00520 vector + si-EGFR group (cSCC cells co-transfected with the LINC00520 vector plasmid and si-EGFR plasmid) and LINC00520 vector + EGFR vector group (cSCC cells co-transfected with LINC00520 overexpression and EGFR overexpression plasmids). In detail, cells were inoculated into a 6-well Plate 24 hours before transfection. When the density reached 30% to 50%, cells were transfected using Lipofectamine 2000 (11668–019, Invitrogen). Specifically, 100 pmol/L plasmid was diluted with 250 μL of serum-free medium Opti-MEM (51985042, Gibco, Gaithersburg, MD, USA) to a final concentration of 50 nmol/L, followed by a 5-minute incubation at room temperature. Meanwhile, 5 μL of Lipofectamine 2000 was diluted with 250 μL Opti-MEM, followed by a 5-minute incubation at room temperature. After the incubation, the two dilutions described above were mixed well and incubated for 20 minutes at room temperature. Then, cells were treated with the mixed solution and cultured for 6 to 8 hours at 37°C with 5% CO2. After the incubation, cells were transferred to complete medium and cultured for 24 to 48 hours.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
The RNA extraction kit (10296010, Invitrogen Company, Shanghai, China) was used to extract total RNA from tissues and cells. After confirming the purity and integrity of the RNA, a PrimeScript RT Kit (RR014A, Takara Biomedical Technology Co., Ltd. Beijing, China) was used for the reverse transcription of RNA to cDNA, with 10 μL of the reverse transcription system. The following reaction conditions were employed: reverse transcription for 15 minutes at 37°C, followed by inactivation for 5 seconds at 85°C. The primers for LINC00520, EGFR, PI3K, Akt, VEGF, MMP-2, MMP-9, internal reference U6 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were designed and synthesized by the Takara Company (Dalian, Liaoning, China) [Table 1]. RT-qPCR was conducted with a PCR kit (KR011A1, Beijing Tian Gen Biochemical Technology Co., Ltd., Beijing, China). The reaction conditions were: pre-denaturation at 95°C for 5 minutes; 30 cycles of denaturation at 95°C for 40 seconds, annealing at 57°C for 40 seconds and extension at 72°C for 40 seconds; extension at 72°C for 10 minutes and an incubation at 4°C for 5 minutes. The reaction system included 10 μL of SYBR Premix Ex TaqTM II, 0.4 μL of PCR Forward Primer (10 μmol/L), 0.4 μl of PCR Reverse Primer (10 μmol/L), 2 μL of DNA template, and 7.2 μL of sterile distilled water. The expression of genes in each group was detected and calculated using the 2−ΔΔCt method. Formula: ΔΔCT = ΔCtexperimental group − ΔCtcontrol group. ΔCt = Cttarget gene − Ctinternal reference.
Table 1.
RT-qPCR primer sequences.

Western blotting analysis
Total proteins were extracted from tissues and cells using a radioimmunoprecipitation assay (RIPA) kit (R0010, Beijing Solarbio Science & Technology Co. Ltd., Beijing, China) and the protein concentrations were measured using the bicinchoninic acid (BCA) method. Then, proteins were separated by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) and wet transferred to a nitrocellulose (NC) membrane that was then blocked with 5% BSA for 1 hour at room temperature. After blocking, the membrane was incubated with the following rabbit polyclonal primary antibodies at 4°C overnight: anti-EGFR (1:1000, ab5644), anti-PI3K (1:2000, ab1678), anti-p-AKT (1:500, ab8805), anti-VEGF (1:1000, ab11939), anti-MMP-2 (1:1000, ab37150), anti-MMP-9 (1:900, ab73734) and anti-GAPDH (1:2500, ab9485). The antibodies were purchased from Abcam Inc. (Cambridge, MA, USA). After 5 washes with PBS for 5 minutes each, the membrane was incubated with a horseradish peroxidase (HRP)-labeled goat anti-rat IgG antibody (1:5000; Beijing Zhongshan Biotechnology Co., Ltd., Beijing, China) for 1 hour at room temperature. After 3 washes with TBST for 5 minutes each, the membrane was incubated with the enhanced chemiluminescence (ECL) solution for development, followed by X-film exposure and photographing. The intensities of protein bands were analyzed using a gel imaging analysis system. Protein expression was calculated as the average intensity of the target band relative to the internal reference band.
Dual luciferase reporter gene assay
Luciferase reporter vectors carrying the wild-type EGFR sequence (EGFR-Wt, containing the binding site) and mutant forms (EGFR-Mut) were constructed. These plasmids were co-transfected into cSCC cells with the LINC00520 vector or negative control (NC), respectively. At 48 h post-transfection, the medium was discarded and cSCC cells were washed with PBS twice. Lysed cells were collected. Then, the luciferase activity was detected using a dual luciferase reporter gene detection system (Dual-Luciferase® Reporter Assay System, E1910, Promega, Madison, WI, USA). In this system, firefly luciferase and renilla luciferase are used to assess the relative luciferase activity. Specifically, 10 μL of cell suspension was mixed with 50 μL of the firefly luciferase working solution to detect the activity of firefly luciferase. Another 50 μL of renilla luciferase working solution was added to detect renilla luciferase activity.
RNA fluorescence in situ hybridization (RNA-FISH)
The subcellular localization of LINC00520 in A431 cells was identified with bioinformatics tools (http://lncatlas.crg.eu/) and then confirmed using RiboTM lncRNA FISH probe Mix(Red) (Ribo Biological Technology Co., Ltd., Guangzhou, China). In detail, A431 cells were inoculated on a coverslip that was placed in a 6-well culture plate and cultured for 1 day until the confluence reached 80%. Then, the coverslip was removed and washed with PBS. After washing, cells were fixed with 1 mL of 4% polyformaldehyde at room temperature and treated with protease K (2 μg/mL), glycine and phthalide reagents. Then, cells were mixed with 250 μL of the prehybridization solution and incubated for 1 hour at 42°C. After removing the prehybridization solution, cells were mixed with 250 μL of the hybridization solution containing probes (300 ng/mL) and incubated at 42°C overnight. After washes with phosphate-buffered saline containing Tween-20 (PBST), cells were incubated with 4′,6-diamidino-2-phenylindole (DAPI) (diluted in PBST, 1:800) for 5 minutes in a 24-well culture plate to label the nuclei. After three washes with PBST for 3 minutes each, the anti-fluorescent quencher was added to the coverslip. The slides were then observed under a fluorescence microscope (Olympus Optical Co., Ltd., Tokyo, Japan) and 5 randomly selected fields from each slide were analyzed.
Immunofluorescence staining
After routine detachment and transfection, cells were counted and cultured in an immunofluorescence chamber at a density of 2 × 105 cells/well. When the confluence reached approximately 90%, the cells were washed with PBS three times (this process was performed on ice). Then, cells were fixed with 1 mL of 4% paraformaldehyde for 15 minutes; cells were washed with PBS 3 times and permeabilized with 0.3% Triton. Ten minutes later, the cells were washed with PBS three times, blocked with goat serum for 30 minutes, incubated overnight at 4°C with the PBS-diluted primary antibodies against CD133 (1:1000, ab19898, Abcam Inc., Cambridge, MA, USA) and p-STAT3 (1:500, ab32143, Abcam Inc., Cambridge, MA, USA) and washed with PBS 3 times. Cells were incubated with the secondary antibody for 1 hour at room temperature in the dark. Next, the cells were washed with PBS 3 times and incubated with DAPI for 15 minutes in the dark. Cells were washed with PBS 3 times, a fluorescence quenching agent was used for mounting and cells were photographed under a fluorescence microscope.
3-(4,5-Dimethylthiazol-2-Yl)-2,5-diphenyltetrazolium bromide (MTT) assay
At 48 h post-transfection, cells in good condition were inoculated into a 96-well culture plate at a final density of 1 × 104 cells/well, and cultured with 200 μL of culture medium per well. Twenty-four hours after cell plating and adhesion, cells were subcultured at 37°C in a 5% CO2 atmosphere. Three-time intervals were established: 24 h, 48 h, and 72 h. Twenty microliters of the MTT solution (5 mg/mL; Sci-Meds Biomart, Wuhan, Hubei, China) were added to each well and incubated for 4 hours at 37°C. After removing the supernatant, 150 μL of dimethyl sulfoxide (DMSO) was added to each well and incubated with gentle shaking for 10 minutes. Subsequently, a microplate reader (Multiskan FC, Thermo Technology Co., Ltd., New York, USA) was used to measure the optical density (A) of each well at a wavelength of 450 nm.
Scratch test
At 24 h post-transfection, cells in good condition were inoculated into a 6-well culture plate at a final density of 1 × 106 cells/well. When the confluence reached 95%, a 20 μL micropipette tip was used to create a vertical linear scratch in the monolayer on the 6-well plate. After removing the detached cells with a D-Hank's solution, cells were continuously cultured with serum-free medium for photographing. The cell migration distance was measured using Image-Pro Plus Analysis software (Media Cybernetics Company, MD, USA) at 0 and 48 hours after scratching under a light microscope.
Cell adhesion assay
At 48 hours post-transfection, cells were subjected to 24 hours of starvation in a serum-free culture medium and trypsinized. After 2 washes with PBS, cells were suspended in serum-free Opti-MEM I (31985008, Nanjing SenBeiJia Biological Technology Co. Ltd., Jiangsu, China) containing 10 g/L BSA at a final density of 3 × 104 cells/mL and transferred to a 96-well culture plate.
For the cell adhesion assay, 50 μL of Matrigel (40111ES08, Yeasen Company, Shanghai, China) was added to each well, with 5 parallel wells for each sample, followed by blocking with 2% BSA. Then, cells were incubated for 1 hour at 37°C, followed by 2 washes with PBS. After adding culture medium (100 μL/well), cells were incubated for 2 hours at 37°C. Next, the culture medium was removed and cells were washed with PBS thrice to remove non-adhesive cells. Cells were then incubated with a mixture of 100 μL of culture medium and 20 μL of MTT for 4 hours at 37°C. After the incubation, 150 μL of DMSO (Sigma-Aldrich Chemical Company, St. Louis, MO, USA) was added to each well, incubated with gentle shaking, and the A value was measured with a microplate reader at the wavelength of 490 nm. The inhibition rate of cell adhesion was then calculated. The following formula was used: inhibition rate of cell adhesion = (1−A value of adherent cells in experimental group/A value of adherent cells in si-LINC00520 group) × 100%.
Transwell assay
At 48 h post-transfection, cells were subjected to 24 h of starvation in a serum-free culture medium and then trypsinized. After 2 washes with PBS, cells were suspended in serum-free Opti-MEMI (31985008, Nanjing SenBeiJia Biological Technology Co. Ltd., Jiangsu, China) containing 10 g/L BSA at a final density of 3 × 104 cells/mL and transferred to a 96-well culture plate. Transwell chambers were placed in each well of a 24-well culture plate and Matrigel (1:8, 40111ES08, Yeasen Company, Shanghai, China) was added to the upper surface of the membrane, followed by drying at room temperature. Three wells per test group were established. Meanwhile, trypsin-digested cells were mixed with RPMI 1640 at a final density of 1 × 105 cells/mL. Then, 200 μL of the cell suspension were transferred to the upper chamber, whereas 600 μL of RPMI 1640 containing 20% FBS were loaded into the lower chamber. After 24 hours of culture, Transwell chambers were removed from the culture plate and cells that had not migrated through the membrane were removed with cotton swabs. Migrated cells were then fixed with 4% polyformaldehyde for 15 minutes, stained with a 0.5% crystal violet solution (in methanol) for 15 minutes, and washed with PBS thrice. Finally, cells were observed under an inverted light microscope (XDS-800D, Shanghai CAIKON Optical Instrument Co., Ltd., Shanghai, China). In each of the 3 parallel wells for each group, 5 randomly selected fields were analyzed and the number of invasive cells was counted.
Tumorigenicity assay
In this experiment, thymus-excised female nude mice (aged 5–6 weeks) were provided by the animal experiment center of the Fourth Military Medical University (Xi’an, China). The nude mice were housed at a constant temperature (25–27°C) and humidity (45–50%). When the confluence reached to 80% to 90%, transfected cells were detached, centrifuged, and washed with PBS; the cells were counted following resuspension, and the final density was adjusted to 1 × 107 cells/mL. Then, 20 μL of the cell suspension were subcutaneously inoculated into the axilla of nude mice. Six mice were inoculated with cell suspension from each group. After 6 weeks, nude mice were sacrificed with CO2. The size of the transplanted tumor was measured and a pathological examination of the axillary, neck and inguinal lymph nodes was performed. The volume of transplanted tumor was recorded and a growth curve was generated. The following formula was used: (a × b2)/2 (where a represents the longest diameter of the tumor and b represents the shortest diameter of the tumor). Then, tissues were fixed with formalin, embedded in paraffin, cut into slices, stained with hematoxylin and eosin (H&E) and imaged under a light microscope.
Statistical analysis
SPSS 21.0 software (IBM Corp. Armonk, NY, USA) was used to analyze the data. The measurement data collected in this study are presented as means ± standard deviations(SD). Comparisons between two groups were performed using t-tests and comparisons among multiple groups were analyzed using one-way analysis of variance (ANOVA). The nonparametric Wilcoxon rank sum test was used to analyze skewed data. The enumerated data were analyzed using the chi-square test or Fisher exact test. A value of P < 0.05 was considered statistically significant.
Results
LINC00520 targets EGFR expression in cSCC
An analysis of the data from chip GSE66359 revealed low levels of expression of the lncRNA LINC00520 in cSCC [Figure 1A]. The MEM website further predicted the first 100 target genes of LINC00520 and the KEGG website was used to conduct the enrichment analysis of the target genes. Based on our results, EGFR participated in the PI3K-Akt signaling pathway which was involved in cSCC progression [Figure 1B].
Figure 1.
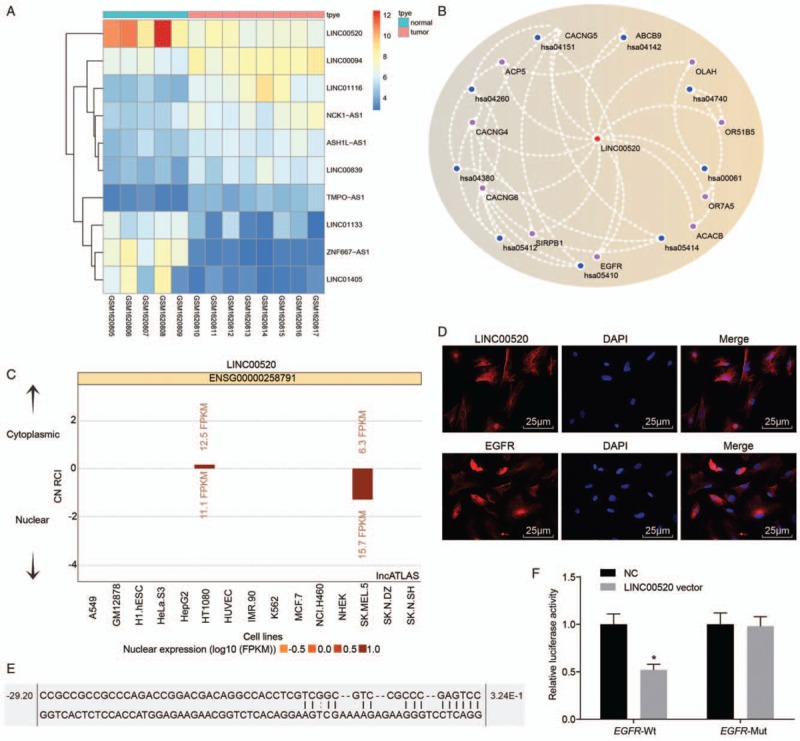
LINC00520 targets the EGFR-3’UTR in cSCC. (A) Analysis of data from chip GSE66359. (B) Predicted relationship between EGRF and LINC00520 from the MEM website (has:04151 is the PI3K-Akt signaling pathway). (C) Subcellular localization of LINC00520 predicted by the bioinformatics website. (D) FISH images and immunofluorescence staining showing the subcellular localization of LINC00520 and EGFR (scale bar = 25 μm). (E) Binding sites for LINC00520 in the EGFR gene predicted by the bioinformatics website. (F) Verification of the relationship between LINC00520 and the EGFR gene using the dual luciferase reporter gene assay. ∗P < 0.05 vs. the NC group. cSCC: Cutaneous squamous cell carcinoma; EGFR: Epidermal growth factor receptor; FISH: Fluorescence in situ hybridization; LINC00520: Long intergenic nonprotein coding RNA 520; NC: Negative control.
According to the bioinformatics analysis, LINC00520 was mainly distributed in the nucleus [Figure 1C], which was verified by FISH images, and EGFR was mainly expressed in the cytoplasm and nucleus, as detected using immunofluorescence staining [Figure 1D]. Data from the online prediction website RNA22 revealed binding sites between LINC00520 and EGFR-3′UTR [Figure 1E] and EGFR may be the target gene of LINC00520. This hypothesis was further confirmed by the dual luciferase reporter gene assay. The luciferase activity in the EGFR-Wt group was significantly decreased compared with the NC group (P < 0.05), indicating that LINC00520 specifically bound to the EGFR gene [Figure 1F]. No significant change was observed in the EGFR-Mut group (P > 0.05).
LINC00520 inactivates the PI3K/AKT signaling pathway by targeting EGFR
The expression of genes related to the PI3K/AKT signaling pathway was determined in cSCC cells in which LINC00520 was upregulated or silenced and EGFR was silenced to explore the potential mechanism of LINC00520 and EGFR in cSCC. RT-qPCR [Figure 2A] and Western blotting results [Figure 2B and 2C] showed no marked differences in LINC00520 expression, levels of the EGFR, PI3K, AKT, VEGF, MMP-2 and MMP-9 mRNAs and proteins, as well as the level of phosphorylated AKT between the blank and NC groups (P > 0.05). LINC00520 expression was significantly increased in the LINC00520 vector group, the LINC00520 vector + si-EGFR group and the LINC00520 vector + EGFR vector group compared with the blank and NC groups (P < 0.05), whereas its expression in the si-LINC00520 group was significantly decreased (P < 0.05). However, no significant differences win LINC00520 expression was observed among the blank, NC, and si-EGFR groups (P > 0.05). Compared with the blank and NC groups, levels of the EGFR, PI3K, AKT, VEGF, MMP-2 and MMP-9 mRNAs and proteins, as well as the level of phosphorylated AKT in both LINC00520 vector group and the si-EGFR group were significantly decreased (P < 0.05), but were significantly increased in the si-LINC00520 group (P < 0.05). Compared with the LINC00520 vector group, levels of the EGFR, PI3K, AKT, VEGF, MMP-2 and MMP-9 mRNAs and proteins, as well as the level of phosphorylated AKT were significantly decreased in the LINC00520 vector + si-EGFR group, but were significantly increased in the LINC00520 vector + EGFR vector group (P < 0.05). Based on these results, LINC00520 negatively regulated EGFR expression to inactivate the PI3K/AKT signaling pathway.
Figure 2.
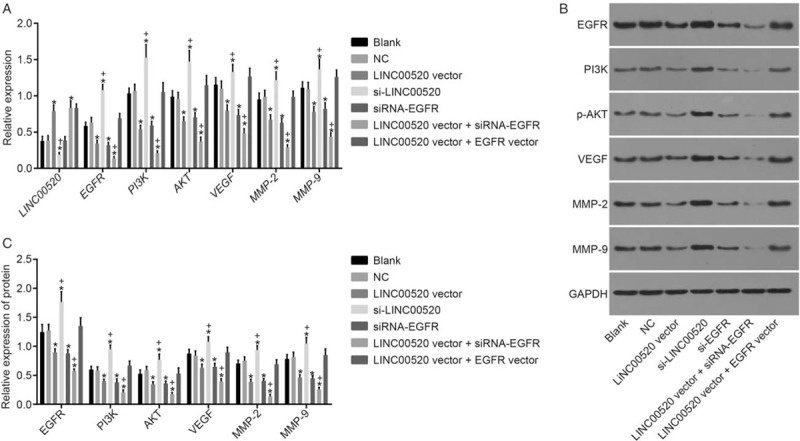
LINC00520 downregulates EGFR expression to inactivate the PI3K/AKT signaling pathway. (A) RT-qPCR detection of the expression of LINC00520 and the EGFR, PI3K, AKT, VEGF, MMP-2, and MMP-9 mRNAs. (B) Western blotting analysis showing the levels of the EGFR, PI3K, AKT, VEGF, MMP-2, MMP-9 and phosphorylated AKT proteins. (C) Bands for the EGFR, PI3K, AKT, VEGF, MMP-2, MMP-9 and phosphorylated AKT proteins; ∗P < 0.05 vs. the blank and NC groups; †P < 0.05 vs. the LINC00520 vector or si-EGFR groups. EGFR: Epidermal growth factor receptor; LINC00520: Long intergenic nonprotein coding RNA 520; MMP: Matrix metalloproteinase; NC: Negative control; PI3K: Phosphatidylinositol-3-Kinase; RT-qPCR: Reverse transcription quantitative polymerase chain reaction; VEGF: Vascular endothelial growth factor.
LINC00520 inhibits the proliferation of cSCC cells by targeting EGFR
The viability of transfected cells in which LINC00520 was upregulated or silenced and EGFR was silenced was detected using the MTT method to analyze the functions of LINC00520 and EGFR in cSCC [Figure 3]. The results showed no significant differences between the blank, NC and LINC00520 vector + EGFR vector groups (all P > 0.05). Importantly, cSCC cells in the LINC00520 vector group and si-EGFR group exhibited slower growth rates and significantly decreased A values at 48 h and 72 h compared with the blank and NC groups (P < 0.05). However, cSCC cells in the si-LINC00520 group displayed an accelerated growth rate and significantly increased A values at 48 h and 72 h than the blank and NC groups (P < 0.05). Compared with the LINC00520 vector group, cSCC cells in the LINC00520 vector + si-EGFR group exhibited a slower growth rate and significantly decreased A values at 48 h and 72 h, while the LINC00520 vector + EGFR vector group showed the opposite results (P < 0.05). Thus, LINC00520 inhibited the proliferation of cSCC cells.
Figure 3.
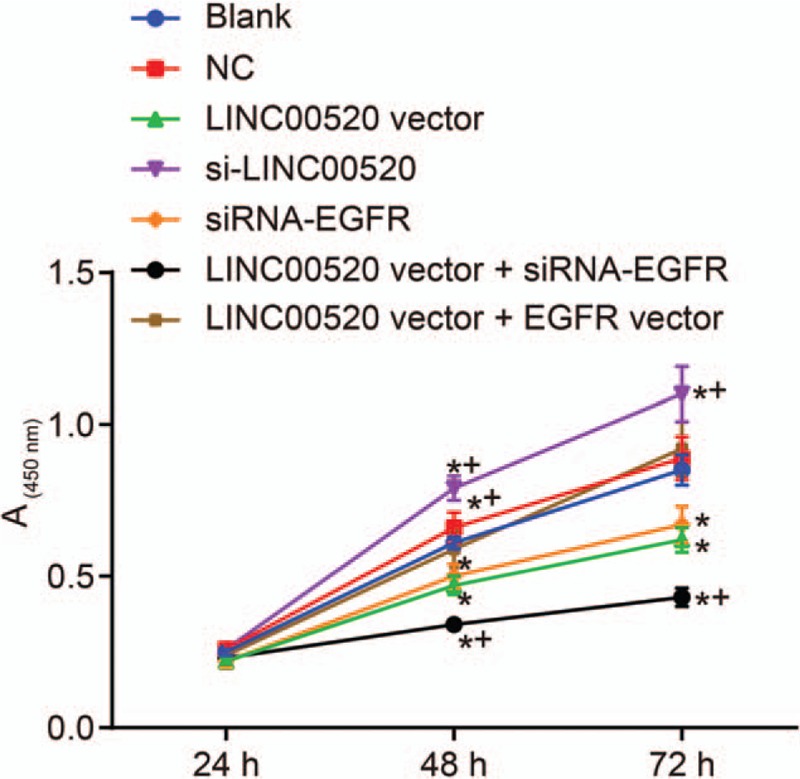
The proliferation of transfected cells is inhibited by LINC00520. ∗P < 0.05 vs. the blank and NC groups; †P < 0.05 vs. the LINC00520 vector group. EGFR: Epidermal growth factor receptor; LINC00520: Long intergenic nonprotein coding RNA 520; MTT: 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide; NC: Negative control.
LINC00520 inhibits the invasion and migration of cSCC cells by targeting EGFR
A Transwell assay and scratch test were conducted to assess the invasion and migration of transfected cells in which LINC00520 was upregulated or silenced and EGFR was silenced, thereby analyzing the functions of LINC00520 and EGFR in cSCC. Based on the results of the Transwell assay [Figure 4], evident differences were not observed between the blank, NC and LINC00520 vector + EGFR vector groups (P > 0.05). Significantly fewer invasion cells were observed in the LINC00520 vector group and si-EGFR group than those in the blank and NC groups (P < 0.05), whereas more invasion cells were detected in the si-LINC00520 group (P < 0.05). Similar results were observed for cell migration [Figure 5]. The scratch test revealed significant decreases in the cell migration distance and ability in the LINC00520 vector group and si-EGFR group compared with the blank and NC groups (P < 0.05), whereas these parameters were increased in the si-LINC00520 group (P < 0.05). Compared with the LINC00520 vector group, the LINC00520 vector + si-EGFR group exhibited a significantly reduced number of invasion cells, cell migration distance, and cell migration ability, while the LINC00520 vector + EGFR vector group showed opposite results (P < 0.05). Based on these results, LINC00520 inhibited the invasion and migration of cSCC cells.
Figure 4.
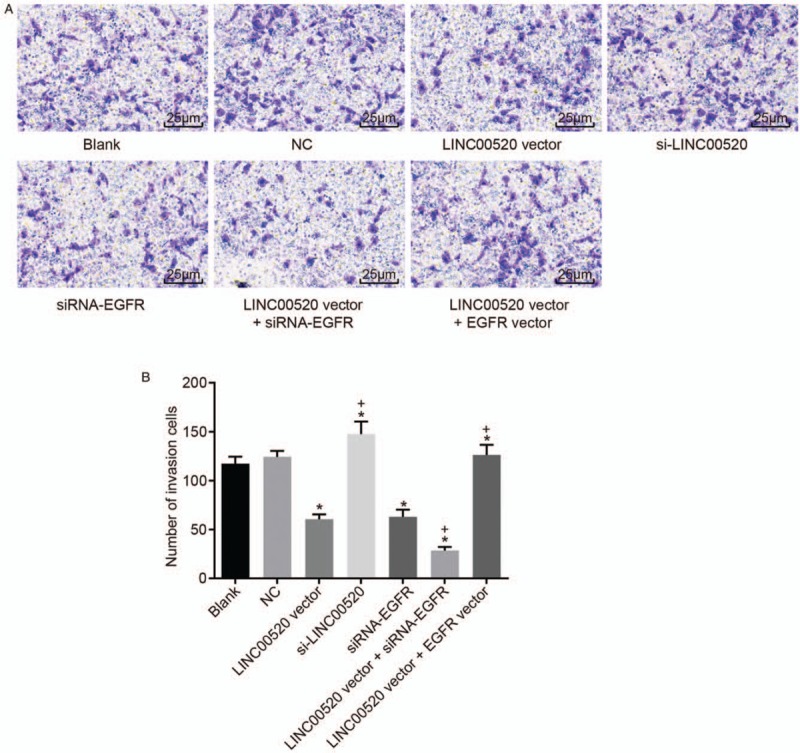
The invasion of transfected cSCC cells is attenuated by LINC00520 overexpression and EGFR silencing. (A) Representative images of invasion cells in the Transwell assay (scale bar = 25 μm). (B) The number of invasion cells among transfected cSCC cells. ∗P < 0.05 vs. the blank and NC groups; †P < 0.05 vs. the LINC00520 vector group. cSCC: cutaneous squamous cell carcinoma; EGFR: Epidermal growth factor receptor; LINC00520: Long intergenic nonprotein coding RNA 520; NC: Negative control.
Figure 5.
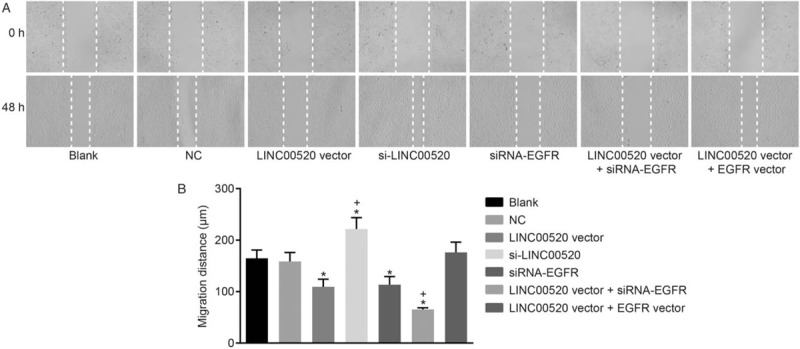
The migration of transfected cSCC cells is reduced by LINC00520 overexpression and EGFR silencing. (A) Representative images of migrating cells in the scratch test (×100). (B) Distance migrated by transfected cSCC cells. ∗P < 0.05 vs. the blank and NC groups; †P < 0.05 vs. the LINC00520 vector group. cSCC: cutaneous squamous cell carcinoma; EGFR: Epidermal growth factor receptor; LINC00520: Long intergenic nonprotein coding RNA 520; NC: Negative control.
LINC00520 inhibits the adhesion of cSCC cells by targeting EGFR
Cell adhesion assays were conducted to assess the adhesion of transfected cells in which LINC00520 was upregulated or silenced and EGFR was silenced, so as to further analyze the functions of LINC00520 and EGFR in cSCC. The results from the cell adhesion assay [Figure 6] did not identify evident differences between the blank, NC and LINC00520 vector + EGFR vector groups (P > 0.05). The inhibition rate of cell adhesion was significantly increased in the LINC00520 vector group and si-EGFR group compared with the blank and NC groups, but the cell adhesion ability was weakened (P < 0.05). However, the si-LINC00520 group exhibited the opposite trend. The inhibition rate of cell adhesion was decreased and cell adhesion ability was increased in this group (P < 0.05). Compared with the LINC00520 vector group, the inhibition rate of cell adhesion was significantly increased and the cell adhesion ability was weakened in the LINC00520 vector + si-EGFR group, while the LINC00520 vector + EGFR vector group showed the opposite trends (P < 0.05). Thus, LINC00520 inhibited the adhesion ability of cSCC cells.
Figure 6.
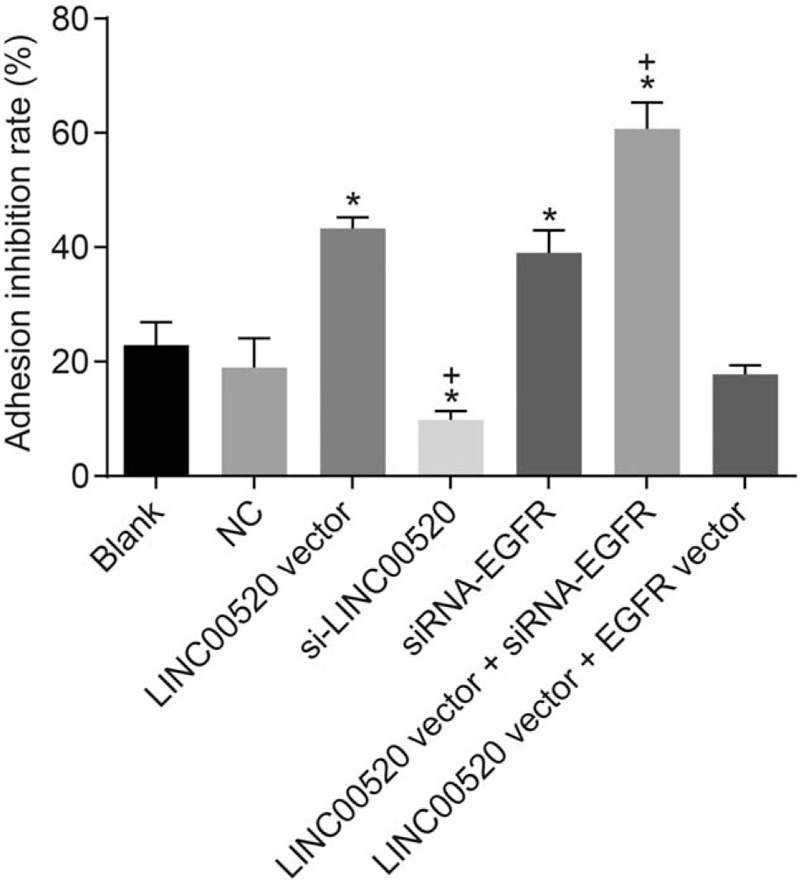
The inhibition rate of adhesion in transfected cells is increased by LINC00520 overexpression and EGFR silencing. ∗P < 0.05 vs. the blank and NC groups; †P < 0.05 vs. the LINC00520 vector group. cSCC: cutaneous squamous cell carcinoma; EGFR: Epidermal growth factor receptor; LINC00520: Long intergenic nonprotein coding RNA 520; NC: Negative control.
LINC00520 inhibits tumor growth and lymph node metastasis by targeting EGFR
Tumor formation in nude mice was evaluated to examine the effects of LINC00520 and EGFR on tumorigenesis. The results from in vivo experiments [Figure 7A and 7B] did not show remarkable differences in tumor volumes and weights between the blank and NC groups (P > 0.05). The tumor volumes and weights were decreased over time in the LINC00520 vector group and si-EGFR group but were increased in the si-LINC00520 group compared with the blank and NC groups (P < 0.05). Meanwhile, the tumor volumes and weights were decreased in the LINC00520 vector + si-EGFR group compared with the LINC00520 vector group (P < 0.05). H&E staining [Figure 7C] revealed a greater volume of metastatic tumor cells in lymph nodes than the volume of peripheral lymphocytes, and the nuclear division was observed in these cells. Few metastatic tumor cells were observed in the LINC00520 vector and si-EGFR groups. However, a large number of metastatic tumor cells were visible in the si-LINC00520 group. Rare metastatic tumor cells were detected in the LINC00520 vector + si-EGFR group, indicating that the metastasis to the lymph nodes was inhibited. Meanwhile, the number of metastatic lymph nodes was decreased in the LINC00520 vector and si-EGFR groups compared with the blank and NC groups (P > 0.05), whereas it was markedly increased in the si-LINC00520 group (P < 0.05). The number of metastatic lymph nodes was decreased in the LINC00520 vector + si-EGFR group compared with the LINC00520 vector group (P < 0.05). Therefore, overexpression of LINC00520 and silencing of the EGFR gene inhibited tumor growth and lymph node metastasis.
Figure 7.
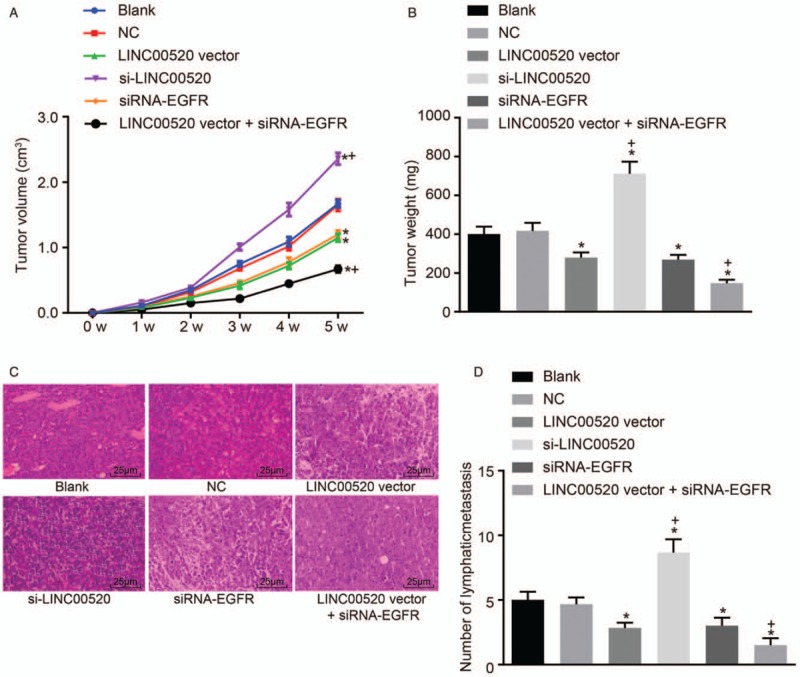
The tumor volumes and weights, as well as lymph node metastasis, in nude mice are reduced by LINC00520 overexpression and EGFR silencing in vivo. (A) Volumes of transplanted tumors in nude mice over time. (B) Tumor weights in nude mice after the implantation of transfected cells. (C) Representative images of hematoxylin and eosin (H&E)-stained lymph node sections (scale bar = 25 μm). (D) Statistical analysis of the number of metastatic lymph nodes. ∗P < 0.05 vs. the blank and NC groups; †P < 0.05 vs. the LINC00520 vector group. EGFR: Epidermal growth factor receptor; LINC00520: Long intergenic nonprotein coding RNA 520; NC: Negative control.
Discussion
As shown in the present study, LINC00520 was expressed at low levels and EGFR was expressed at high levels in cSCC. LINC00520 inhibited EGFR expression. Furthermore, LINC00520 suppressed the invasion and metastasis of cSCC by inhibiting EGFR and inactivating the PI3K-AKT signaling pathway.
cSCC is considered a common malignant tumor occurring in keratinocytes of the epidermis with dermal invasion.[18] cSCC was recently shown to be a highly metastatic tumor.[19] Abnormally expressed lncRNAs were reported to play a significant role in the development of lung SCC, providing diagnostic value for patients with this cancer.[20] An expression profile published by Sand et al[9] revealed 1516 significantly upregulated and 2586 downregulated lncRNAs in cSCC. In the present study, we confirmed that LINC00520 was expressed at a low level in cSCC, and low LINC00520 expression was correlated with cancer progression. Importantly, EGFR was verified as a target gene of LINC00520. The mRNA encoding EGFR, a transmembrane glycoprotein, was expressed at high levels in most head and neck mucosal SCC tumors.[21,22] Increased EGFR expression was also identified in tumors with poor differentiation and an advanced stage.[21] Abnormal expression of EGFR is associated with the occurrence of SCC.[23] Based on accumulating evidence, EGFR is expressed at high levels in cSCC, even in advanced stage tumors,[24,25] consistent with our results. Hence, EGFR represents a potential therapeutic target for SCC of the head and neck, lung, and skin. Meanwhile, EGFR inhibitors, including cetuximab or erlotinib, are considered targeted treatments for cSCC in patients in which mono- or polychemotherapy failed and cancer has progressed,[26] suggesting a critical role for EGFR in cSCC. In the present study, LINC00520 functionally downregulated EGFR expression, which might provide a therapeutic target for cSCC.
Additionally, EGFR tyrosine kinase mutations activate anti-apoptotic signaling pathways, such as PI3K/Akt, JAK-STAT and ERK/MAPK.[27] PI3K, a downstream signaling molecule of the EGFR gene, functions as an important factor regulating the proliferation or invasion of SCC cells, particularly the development and progression of SCC cells in the head and neck.[14,28] In another study, activation of PI3K/Akt signaling pathway was reported to promote the occurrence and metastasis of human esophageal cancer and induce the apoptosis of esophageal SCC cells.[29] Hence, the inactivation of the PI3K/Akt signaling pathway might contribute to the prevention of SCC progression. As shown in the present study, the PI3K/Akt signaling pathway was inactivated by LINC00520 overexpression and EGFR silencing in cSCC. LINC00520-targeted EGFR inhibition suppresses the activation of the PI3K/Akt signaling pathway, which might be a specific mechanism by which LINC00520 regulates cSCC.
Our results also revealed that LINC00520 inhibited the invasion and metastasis of cSCC by inhibiting its target gene EGFR and the activation of the PI3K/Akt signaling pathway. Overexpression of EGFR activates intracellular tyrosine kinases that subsequently trigger various downstream phosphorylation cascades. EGFR overexpression also increases the survival, proliferation, and metastasis of SCC cells in the head and neck.[30]EGFR inhibitors and immune checkpoint blockers are regarded as an option for treating patients with SCC presenting with distant metastases.[31]EGFR inhibitors significantly reduce the tumor size of locally advanced and metastatic SCC.[32] Furthermore, high mobility group box 1 was recently shown to modulate cSCC metastasis by regulating the PI3K/Akt and MAPK signaling pathways.[33] The combination of EGFR inhibitors and inhibitors of the PI3K/AKT/mTOR signaling pathway represents a promising treatment approach for unresectable cSCC.[34] LINC00520 is involved in modulating the migration and invasion of breast cancer cells by regulating the PI3K signaling pathway.[10] Based on those findings, LINC00520-targeted EGFR inhibition confers suppressive effects on the progression and metastasis of cSCC.
LINC00520 expression regulated by oncogenic Src, PIK3CA, and Stat3 played substantial roles in the migration, invasion and metastasis of breast cancer cells.[10] Epidermal growth factor receptor (EGFR), transforming growth factor beta (TGF-beta) and the PI3K/Akt signaling pathway was also reported to participate in cSCC.[12,33]EGFR activates the PI3K/Akt signaling pathway to inhibit cancer progression.[35]EGFR was suggested to be a potential target in the treatment of cSCC.[13] Based on our own research and previous studies, LINC00520 and PI3K might exhibit negative feedback regulation. However, due to the limited funding for this research, we did not conduct further studies. In a follow-up study, we will consider investigating more specific interactions among LINC00520, EGFR and the PI3K/Akt signaling pathway.
In conclusion, LINC00520 suppresses the proliferation, invasion, and migration of cSCC cells in vitro and restrains tumor growth and metastasis in vivo. Functionally, LINC00520 inhibits its target gene EGFR, subsequently inactivating the PI3K/Akt signaling pathway [Figure 8]. Our findings might provide novel insights into the mechanism of LINC00520 in cSCC and the molecular targets for the treatment of cSCC. Further clinical experiments are needed to illustrate and verify the role of LINC00520 in regulating cSCC.
Figure 8.
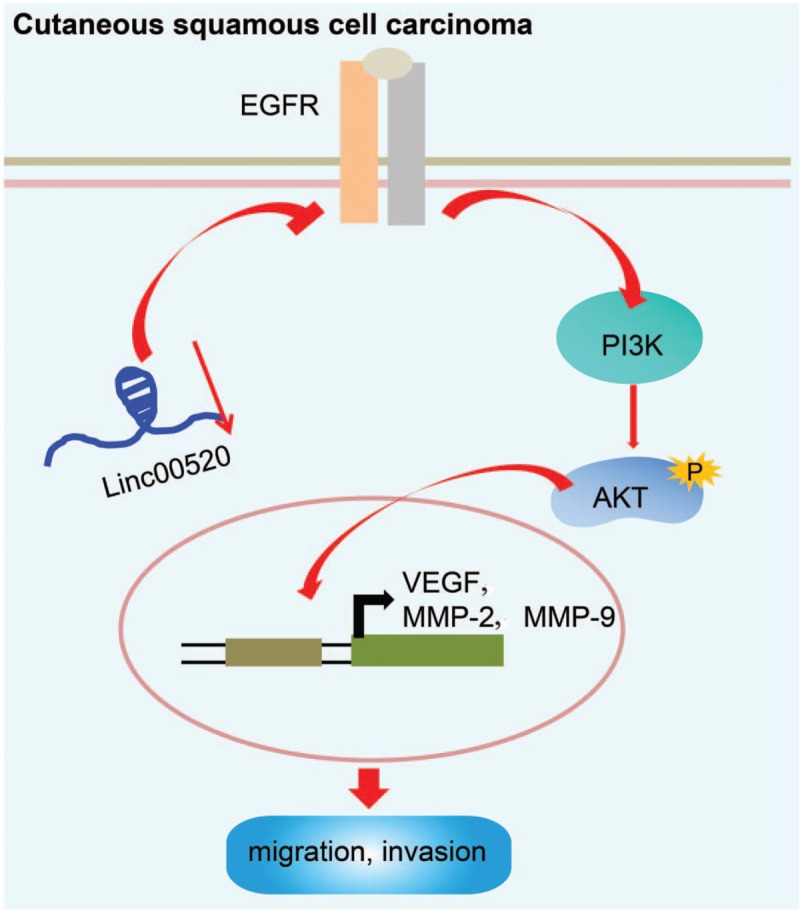
LINC00520 suppresses EGFR expression and inactivates the PI3K/Akt signaling pathway, thereby suppressing cSCC migration, invasion and metastasis. cSCC: Cutaneous squamous cell carcinoma; EGFR: Epidermal growth factor receptor; LINC00520: Long intergenic nonprotein coding RNA 520; PI3K/Akt: Phosphoinositide 3-kinase/protein kinase B.
Conflicts of interest
None.
Footnotes
How to cite this article: Mei XL, Zhong S. Long noncoding RNA LINC00520 prevents the progression of cutaneous squamous cell carcinoma through the inactivation of the PI3K/Akt signaling pathway by downregulating EGFR. Chin Med J 2019;00:00–00. doi: 10.1097/CM9.0000000000000070
References
- 1.Lansbury L, Bath-Hextall F, Perkins W, Stanton W, Leonardi-Bee J. Interventions for non-metastatic squamous cell carcinoma of the skin: systematic review and pooled analysis of observational studies. BMJ 2013; 347:f6153.doi: 10.1136/bmj.f6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu S, Chen M, Li P, Wu Y, Chang C, Qiu Y, et al. Ginsenoside rh2 inhibits cancer stem-like cells in skin squamous cell carcinoma. Cell Physiol Biochem 2015; 36:499–508. doi: 10.1159/000430115. [DOI] [PubMed] [Google Scholar]
- 3.Loeb KR, Asgari MM, Hawes SE, Feng Q, Stern JE, Jiang M, et al. Analysis of Tp53 codon 72 polymorphisms, Tp53 mutations, and HPV infection in cutaneous squamous cell carcinomas. PLoS One 2012; 7:e34422.doi: 10.1371/journal.pone.0034422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silberstein E, Sofrin E, Bogdanov-Berezovsky A, Nash M, Segal N. Lymph node metastasis in cutaneous head and neck squamous cell carcinoma. Dermatol Surg 2015; 41:1126–1129. doi: 10.1097/DSS.0000000000000488. [DOI] [PubMed] [Google Scholar]
- 5.Mateus C. Cutaneous squamous cell carcinoma. Rev Prat 2014; 64:45–52. doi: 10.1016/j.cll.2017.06.003. [PubMed] [Google Scholar]
- 6.Hillen U, Ulrich M, Alter M, Becker JC, Gutzmer R, Leiter U, et al. Cutaneous squamous cell carcinoma: a review with consideration of special patient groups. Hautarzt 2014; 65:590–599. doi: 10.1007/s00105-013-2734-7. [DOI] [PubMed] [Google Scholar]
- 7.Pickering CR, Zhou JH, Lee JJ, Drummond JA, Peng SA, Saade RE, et al. Mutational landscape of aggressive cutaneous squamous cell carcinoma. Clin Cancer Res 2014; 20:6582–6592. doi: 10.1158/1078-0432.CCR-14-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang H, Bai M, Zeng A, Si L, Yu N, Wang X, et al. LncRNA HOXD-AS1 promotes melanoma cell proliferation and invasion by suppressing RUNX3 expression. Am J Cancer Res 2017; 7:2526–2535. [PMC free article] [PubMed] [Google Scholar]
- 9.Sand M, Bechara FG, Sand D, Gambichler T, Hahn SA, Bromba M, et al. Expression profiles of long noncoding RNAs in cutaneous squamous cell carcinoma. Epigenomics 2016; 8:501–518. doi: 10.2217/epi-2015-0012. [DOI] [PubMed] [Google Scholar]
- 10.Henry WS, Hendrickson DG, Beca F, Glass B, Lindahl-Allen M, He L, et al. LINC00520 is induced by Src, STAT3, and PI3K and plays a functional role in breast cancer. Oncotarget 2016; 7:81981–81994. doi: 10.18632/oncotarget.11962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janus JM, O'Shaughnessy RFL, Harwood CA, Maffucci T. Phosphoinositide 3-Kinase-dependent signalling pathways in cutaneous squamous cell carcinomas. Cancers (Basel) 2017; 9: doi: 10.3390/cancers9070086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berger F, Geddert H, Faller G, Werner M, Dimmler A. Pattern of TGFbeta receptor 1 expression differs between kras-mutated keratoacanthomas and squamous cell carcinomas of the skin. Pathol Res Pract 2014; 210:596–602. doi: 10.1016/j.prp.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 13.Mavropoulos JC, Aldabagh B, Arron ST. Prospects for personalized targeted therapies for cutaneous squamous cell carcinoma. Semin Cutan Med Surg 2014; 33:72–75. doi: 10.12788/j.sder.0083. [DOI] [PubMed] [Google Scholar]
- 14.Bito T, Sumita N, Ashida M, Budiyanto A, Ueda M, Ichihashi M, et al. Inhibition of epidermal growth factor receptor and PI3K/Akt signaling suppresses cell proliferation and survival through regulation of Stat3 activation in human cutaneous squamous cell carcinoma. J Skin Cancer 2011; 2011:874571.doi: 10.1155/2011/874571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujita A, Sato JR, Rodrigues Lde O, Ferreira CE, Sogayar MC. Evaluating different methods of microarray data normalization. BMC Bioinformatics 2006; 7:469.doi: 10.1186/1471-2105-7-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004; 3: Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 17.Baro M, de Llobet LI, Figueras A, et al. Dasatinib worsens the effect of cetuximab in combination with fractionated radiotherapy in FaDu- and A431-derived xenografted tumours. Br J Cancer 2014; 111:1310–1318. doi: 10.1038/bjc.2014.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Green AC, McBride P. Squamous cell carcinoma of the skin (non-metastatic). BMJ Clin Evid 2014; 2014:1709. [PMC free article] [PubMed] [Google Scholar]
- 19.Brougham ND, Tan ST. The incidence and risk factors of metastasis for cutaneous squamous cell carcinoma–implications on the T-classification system. J Surg Oncol 2014; 110:876–882. doi: 10.1002/jso.23731. [DOI] [PubMed] [Google Scholar]
- 20.Chen WJ, Tang RX, He RQ, Li DY, Liang L, Zeng JH, et al. Clinical roles of the aberrantly expressed lncRNAs in lung squamous cell carcinoma: a study based on RNA-sequencing and microarray data mining. Oncotarget 2017; 8:61282–61304. doi: 10.18632/oncotarget.18058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sweeny L, Dean NR, Magnuson JS, et al. EGFR expression in advanced head and neck cutaneous squamous cell carcinoma. Head Neck 2012; 34:681–686. doi: 10.1002/hed.21802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furrukh M, Mufti T, Hamid RS, Qureshi A. Squamous cell carcinoma of external auditory canal lacking epidermal growth factor receptor protein overexpression, in an elderly Omani with oculocutaneous albinism treated with palliative radiotherapy. BMJ Case Rep 2014; 2014:bcr2013203226.doi: 10.1136/bcr-2013-203226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joseph SR, Endo-Munoz L, Gaffney DC, Saunders NA, Simpson F. Dysregulation of epidermal growth factor receptor in actinic keratosis and squamous cell carcinoma. Curr Probl Dermatol 2015; 46:20–27. doi: 10.1159/000367959. [DOI] [PubMed] [Google Scholar]
- 24.Mauerer A, Herschberger E, Dietmaier W, Landthaler M, Hafner C. Low incidence of EGFR and HRAS mutations in cutaneous squamous cell carcinomas of a German cohort. Exp Dermatol 2011; 20:848–850. doi: 10.1111/j.1600-0625.2011.01334.x. [DOI] [PubMed] [Google Scholar]
- 25.Li YY, Hanna GJ, Laga AC, Haddad RI, Lorch JH, Hammerman PS. Genomic analysis of metastatic cutaneous squamous cell carcinoma. Clin Cancer Res 2015; 21:1447–1456. doi: 10.1158/1078-0432.CCR-14-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stratigos A, Garbe C, Lebbe C, Malvehy J, del Marmol V, Pehamberger H, et al. Diagnosis and treatment of invasive squamous cell carcinoma of the skin: European consensus-based interdisciplinary guideline. Eur J Cancer 2015; 51:1989–2007. doi: 10.1016/j.ejca.2015.06.110. [DOI] [PubMed] [Google Scholar]
- 27.Uribe P, Gonzalez S. Epidermal growth factor receptor (EGFR) and squamous cell carcinoma of the skin: molecular bases for EGFR-targeted therapy. Pathol Res Pract 2011; 207:337–342. doi: 10.1016/j.prp.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 28.Simpson DR, Mell LK, Cohen EE. Targeting the PI3K/AKT/mTOR pathway in squamous cell carcinoma of the head and neck. Oral Oncol 2015; 51:291–298. doi: 10.1016/j.oraloncology.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 29.Zhang HB, Lu P, Guo QY, Zhang ZH, Meng XY. Baicalein induces apoptosis in esophageal squamous cell carcinoma cells through modulation of the PI3K/Akt pathway. Oncol Lett 2013; 5:722–728. doi: 10.3892/ol.2012.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Bryan K, Sherman W, Niedt GW, Taback B, Manolidis S, Wang A, et al. An evolving paradigm for the workup and management of high-risk cutaneous squamous cell carcinoma. J Am Acad Dermatol 2013; 69:595–602 e1.doi: 10.1016/j.jaad.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 31.Leiter U, Gutzmer R, Alter M, Ulrich C, Lonsdorf AS, Sachse MM, et al. Cutaneous squamous cell carcinoma. Hautarzt 2016; 67:857–866. doi: 10.1007/s00105-016-3875-2. [DOI] [PubMed] [Google Scholar]
- 32.Yin VT, Pfeiffer ML, Esmaeli B. Targeted therapy for orbital and periocular basal cell carcinoma and squamous cell carcinoma. Ophthalmic Plast Reconstr Surg 2013; 29:87–92. doi: 10.1097/IOP.0b013e3182831bf3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun Y, Tu Y, Ji C, Cheng BO. High mobility group box 1 regulates tumor metastasis in cutaneous squamous cell carcinoma via the PI3K/AKT and MAPK signaling pathways. Oncol Lett 2016; 11:59–62. doi: 10.3892/ol.2015.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bejar C, Maubec E. Therapy of advanced squamous cell carcinoma of the skin. Curr Treat Options Oncol 2014; 15:302–320. doi: 10.1007/s11864-014-0280-x. [DOI] [PubMed] [Google Scholar]
- 35.Pi J, Jiang J, Cai H, Yang F, Jin H, Yang P, et al. GE11 peptide conjugated selenium nanoparticles for EGFR targeted oridonin delivery to achieve enhanced anticancer efficacy by inhibiting EGFR-mediated PI3K/AKT and Ras/Raf/MEK/ERK pathways. Drug Deliv 2017; 24:1549–1564. doi: 10.1080/10717544.2017.1386729. [DOI] [PMC free article] [PubMed] [Google Scholar]


