Abstract
The hippocampus is a key brain area to encoding and storing memories. Hippocampal place cells encode the position of an animal in space by firing when the subject is at a specific location in the environment. Therefore, place cells are considered essential to spatial memory and navigation. It has recently been revealed that place-cell activity is not constant even in a familiar environment, but changes dynamically over time. However, the mechanism behind these changes in activity is not yet fully understood. In this study, the activity of hippocampal CA1 neurons of male mice was tracked during repeated performances of a spatial task in a virtual reality environment. By comparing place-cell ensemble representations among repeated performance of the task, the overlap rate of the active place-cell population was found to be time dependent but independent of the number of tasks within a fixed time. These findings suggest that place codes change automatically and at a constant speed. Furthermore, the dynamics of place-cell activity were found to be suppressed by an NMDA receptor antagonist. In summary, the spontaneously dynamic nature of place-cell activity is at least in part regulated by NMDA receptors, and the dynamics may encode temporal information of episodes.
SIGNIFICANCE STATEMENT Place-cell activity in the hippocampal CA1 area is not stable even in a familiar environment, but changes dynamically over time. However, the mechanism behind these changes is unknown. Using in vivo calcium imaging, activity of CA1 neurons were tracked during multiple sessions with variable intervals. The overlap rate of the active place-cell population was constant regardless of the number of tasks within a fixed time. Furthermore, the dynamics were suppressed by an NMDA receptor antagonist. This NMDA receptor-dependent, continuous change in the place-cell activity may encode temporal information of episodes.
Keywords: calcium imaging, hippocampus, place cell, virtual reality
Introduction
Memories are critical to animal life. However, the method by which memories are formed and maintained in neural networks is poorly understood. Neural activity in individual cells has been recorded over days, weeks, and even months in an attempt to understand the mechanism underlying memory storage. Many studies have focused on the hippocampus, which has been shown to play an important role in episodic memory formation (Scoville and Milner, 1957; Tulving and Markowitsch, 1998; Rolls, 2010; Allen and Fortin, 2013). Some hippocampal neurons called place cells are active at distinct locations within a given space, thus suggesting that these cells provide the spatial information for episodic memory (O'Keefe and Dostrovsky, 1971; O'Keefe and Nadel, 1978; Moser et al., 2015). The prominent feature of these cells is that they allow a quantitative assessment of neural activity dynamics in the hippocampus. Classical electrophysiological recordings have reported that some place cells in the hippocampal CA1 area have place fields that are stable for >100 d (Thompson and Best, 1990). By contrast, recent electrophysiological recordings have revealed that the activity of place cells varies with time during repeated exposure to the same environment (Manns et al., 2007; Mankin et al., 2012). Because of difficulties in tracking the same neurons over a long period of time by using electrophysiological recordings, the comprehensive characterization of the neural dynamics of hippocampal cells has not been achieved using this method. However, recent advances in calcium imaging techniques provide the ability to track large numbers of neurons over days and weeks (Huber et al., 2012; Ziv et al., 2013). Tracking the activity of thousands of hippocampal CA1 cells by using calcium imaging has revealed that the activity of place cells changes dynamically over time, but the locations of their place fields generally remain stable (Ziv et al., 2013). Such kinds of dynamic neural representation have also been found in other brain areas, such as the parietal cortex (Driscoll et al., 2017). Theoretical work suggests that unstable neural codes can be useful to learning because this instability prevents the settling of learning in the local minima (Rokni et al., 2007). Neural dynamics in the hippocampus may also be important in encoding the temporal information of episodes, i.e., events occurring at different times in the same place are assigned different neural codes in the hippocampus (Manns et al., 2007; Mankin et al., 2012; Ziv et al., 2013; Rubin et al., 2015). However, the mechanism responsible for the temporal dynamics of hippocampal activity is unknown. In the present study, the detailed dynamics of activity in the populations of place cells and the mechanism underlying these dynamics were investigated. One possibility is that a new place code is assigned for each new episode (Fig. 1A, left). In this case, the rate of change would depend on the frequency of the experience. Alternatively, place codes may change automatically and at a constant speed (Fig. 1A, right). To distinguish between these hypotheses, the calcium activity of mouse hippocampal CA1 neurons was imaged during the repeated performance of a spatial memory task, and the rates of change in the activity of place-cell populations over multiple sessions were measured. The contribution of NMDA receptors to the dynamics of place-cell activity was also tested.
Figure 1.
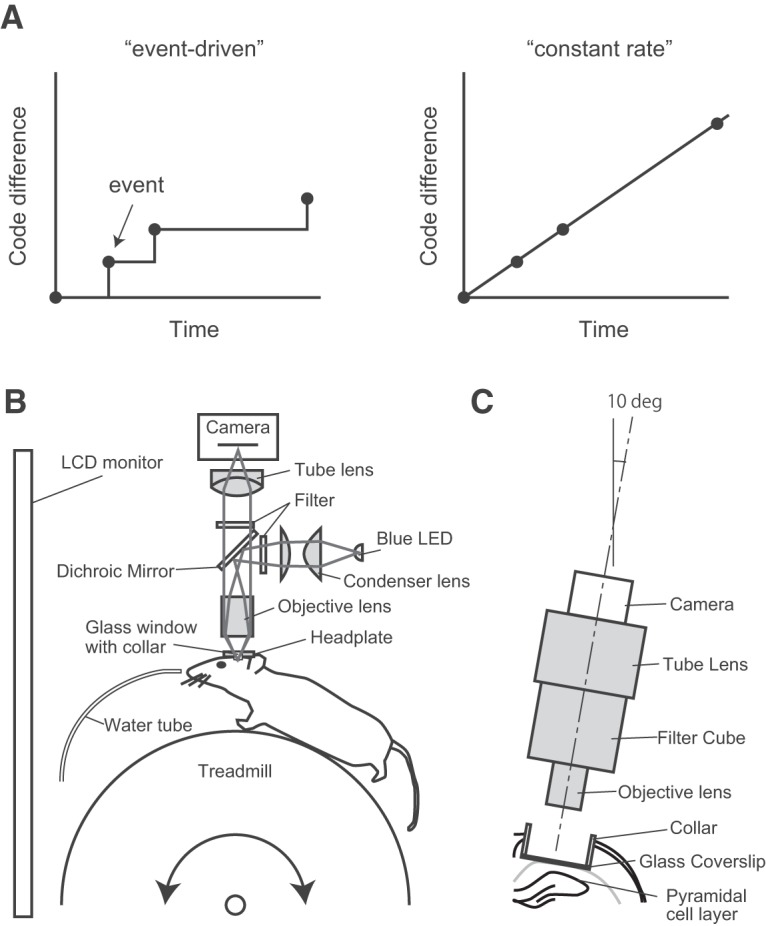
Two models for the dynamics of place-cell activity and schematic view of the experimental apparatus. A, Left, A new place code is assigned for each new episode. In this case, the rate of change will depend on the frequency of the experience. Right, Place codes change automatically and at constant speed. In this case, the rate will always be constant. B, Simplified schematic representation of the imaging system. The blue line indicates the illumination pathway, and the green line indicates the light collection pathway. Illumination light from a blue LED was collected with a condenser lens, passed through an excitation filter, reflected off a dichroic mirror, and irradiated through an objective lens. The fluorescence image was focused on a CMOS camera. C, Schematic representation of the experimental setup showing the chronic window implant above CA1. The optical axis of the fluorescence microscope was angled at 10°.
Materials and Methods
Calcium imaging with a custom wide-field microscope.
A custom wide-field microscope for in vivo calcium imaging has previously been described (Hayashi et al., 2017; Fig. 1B). Excitation light was emitted from a blue LED (LXK2-PB14-P00, Lumileds). The light passed through an excitation filter (FF480/40-25, Semrock) and was reflected by a dichroic mirror (FF506-Di02-25×36, Semrock) onto the tissue through an objective lens (LUCPlan FLN20/0.45, Olympus). The fluorescent emissions collected by the objective lens were passed through the dichroic mirror and an emission filter (FF535/50–25, Semrock). The fluorescence image was focused by a tube lens (Nikkor 50 mm f/1.8D, Nikon) and was captured by a CMOS camera (FL3-U3–13S2M-CS, FLIR Systems). The optical axis of the microscope was angled at 10° to be perpendicular to the pyramidal cell layer of the CA1 hippocampal region (Fig. 1C).
Animals.
All animal care and use was performed in accordance with the protocols approved by Hokkaido University Institutional Animal Care and Use Committee (Project License 16-0042). Adult male C57BL/6J mice (10–16 weeks old) were maintained on a 12 h light/dark schedule, with lights off at 8:00 A.M. Behavioral tasks and recording sessions occurred in the dark phase (9:00 A.M. to 3:00 P.M.).
Surgery.
Cranial window preparation over the hippocampal CA1 area followed a former description (Dombeck et al., 2010). The animals were anesthetized using isoflurane and then injected subcutaneously with 0.1 mg/kg buprenorphine. The skull was exposed, and a small hole (<0.5 mm) was made over the right hemisphere (1.5 mm lateral to the midline, 2.3 mm posterior to the bregma). AAV1-syn-GCaMP6f-WPRE (University of Pennsylvania Vector Core) was diluted to 5 × 1012 particles/ml in PBS, and 150 nl was injected into CA1 (1.2 mm ventral from the brain surface). One week after the viral injection, the animals were anesthetized using isoflurane, and a 2.8-mm-diameter craniotomy was performed. The dura was removed, and the underlying cortex was aspirated. A stainless-steel cannula (2.76 mm outer diameter, 2.40 mm inner diameter, 1.5 mm height) covered by a glass coverslip (0.12 mm thick) was inserted over the dorsal CA1. Dental cement (Shofu) was used to glue a titanium head plate (25 × 10 mm, 1 mm thick) to the skull.
Virtual reality system.
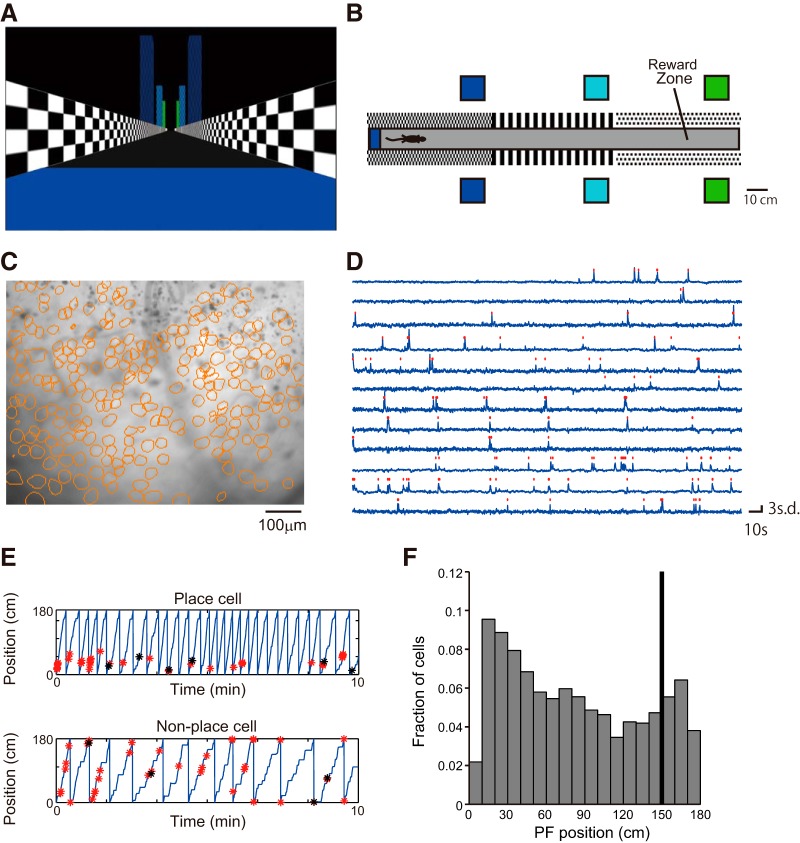
The virtual reality (VR) system was previously described (Hayashi et al., 2017; Figs. 1B, 2A,B). Mice ran along a virtual linear track (1.8 m long) and received a small sugar water reward (4 μl) in an unmarked goal zone (1.5 m from the start point of the track; Fig. 2B). The reward was delivered immediately after the animal reached the goal zone. Therefore, the animal did not need to memorize the position of the goal and needed only to run through it to get the reward. Once the mice reached the end of the track, they were transported back to the start of the track.
Figure 2.
Hippocampal activity during the VR task. A, View from the start point of the virtual linear track. B, Top view of the track. C, Sample image of mean fluorescence during an imaging session. The contours of cells extracted from the fluorescent imaging movie are shown in orange circles (236 cells). D, Relative fluorescence changes (dF/F) for 12 cells. Identified calcium transients are shown as red dots. E, Representative CA1 cell activity. Top, Place cell (mutual information = 1.7, Monte Carlo p value = 0.00); bottom, nonspecific cell (mutual information = 0.13, Monte Carlo p value = 0.48). The position of the mouse along the virtual track is shown by the blue line. Identified calcium activities during running are shown with red asterisks, and those during rest are shown with black asterisks. F, Histograms of place-field position along the linear track. The black bar indicates the reward position.
Behavioral training.
At least 2 weeks after the cranial window implantation, the mice underwent water restriction (2 ml per day) and began training on the virtual linear track (10 or 15 min sessions per day, 5 d per week). After 3–4 weeks of training, mice that achieved high performance levels (>10 rewards per single 10 min session) were used for the recording session.
Pharmacology.
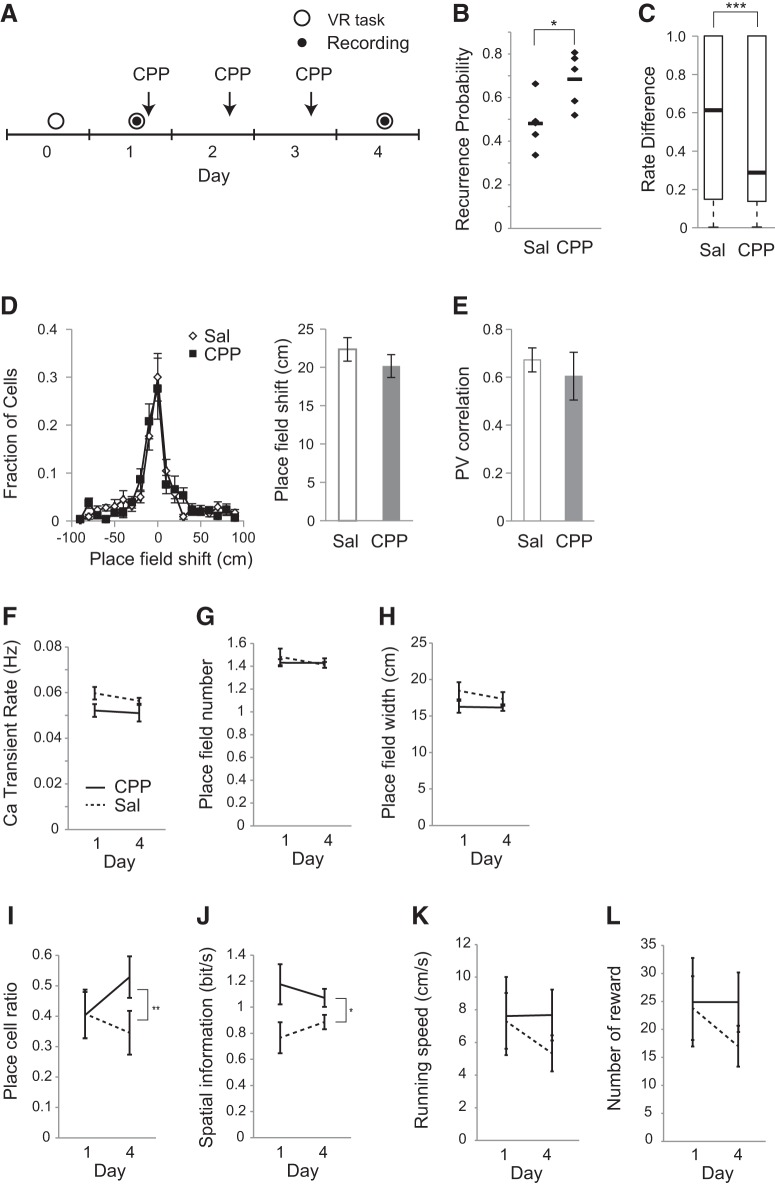
Mice received intraperitoneal injections of either saline or 10 mg/kg 3-[(R)-2-carboxypiperazin-4-yl)-propyl-1-phosphonic acid (CPP); Tocris Bioscience] after the first recording session and daily thereafter for 2 d (see Fig. 6A).
Figure 6.
NMDA receptor antagonism reduced the change of place-cell activity. A, Experimental timeline. After the pretraining period, two recording sessions were conducted on Days 1 and 4. CPP was intraperitoneally administrated after the first recording session and daily thereafter for 2 d. B, Recurrence probability for the two sessions under saline and CPP conditions. Values are shown in black dots, and the mean values are shown in black bars. C, Rate difference for the two sessions under saline and CPP conditions. The box plots show the median and quartiles, and the whisker caps of the box plots show the minimum and maximum values. D, Centroid shifts of place fields between the two sessions under saline and CPP conditions. Left, Distributions of centroid shifts. Right, Average centroid shifts. E, PV correlation between the two session under saline and CPP conditions. F, Calcium transient rate. Significance was evaluated by two-way repeated-measures ANOVA on aligned rank-transformed data; main effect of drug: F(1,8) = 2.8, p = 0.13; time: F(1,8) = 1.7, p = 0.23; interaction: F(1,8) = 0.79, p = 0.40; n = 5 animals for saline, and n = 5 animals for CPP. G, Number of place fields for each cell. Significance was evaluated by two-way repeated-measures ANOVA on aligned rank-transformed data; main effect of drug: F(1,8) = 0.12, p = 0.74; time: F(1,8) = 0.53, p = 0.49; interaction: F(1,8) = 0.50, p = 0.50; n = 5 animals for saline, and n = 5 animals for CPP. H, Place-field size for each cell. Significance was evaluated by two-way repeated-measures ANOVA on aligned rank-transformed data; main effect of drug: F(1,8) = 3.2, p = 0.11; time: F(1,8) = 0.65, p = 0.44; interaction: F(1,8) = 0.14, p = 0.72; n = 5 animals for saline, and n = 5 animals for CPP. I, Fraction of cells with significant place fields. Significance was evaluated by two-way repeated-measures ANOVA on aligned rank-transformed data; main effect of drug: F(1,8) = 1.0, p = 0.34; time: F(1,8) = 0.54, p = 0.48; interaction: F(1,8) = 16, p = 0.0040; n = 5 animals for saline, and n = 5 animals for CPP. J, Spatial information content of place cells per unit time. Significance was evaluated by two-way repeated-measures ANOVA on aligned rank-transformed data; main effect of drug: F(1,8) = 5.3, p = 0.050; time: F(1,8) = 2.1, p = 0.19; and interaction: F(1,8) = 8.3, p = 0.02; n = 5 animals for saline, and n = 5 animals for CPP. K, Running speed at each recording session. Significance was evaluated by two-way repeated-measures ANOVA on aligned rank-transformed data; main effect of drug: F(1,8) = 0.23, p = 0.65; time: F(1,8) = 0.17, p = 0.69; and interaction: F(1,8) = 2.8, p = 0.13; n = 5 for saline, n = 5 for CPP). L, Number of rewards for each session. Significance was evaluated by two-way repeated-measures ANOVA on aligned rank-transformed data; main effect of drug: F(1,8) = 0.18, p = 0.68; time: F(1,8) = 0.15, p = 0.71; and interaction: F(1,8) = 2.7, p = 0.14; n = 5 for saline, n = 5 for CPP). Data are shown as mean ± SEM, unless otherwise indicated. Asterisks indicate significant differences: *p < 0.05, **p < 0.01, ***p < 0.001; otherwise, it is not significant.
Imaging sessions.
The excitation light intensity was ∼0.04 mW/mm2. FlyCapture2 software (FLIR Systems) was used to capture images at 10 Hz for 10 min. The position of the blood vessels and other biological landmarks were used to determine the imaging plane (Fig. 2C).
Image processing.
ImageJ 1.49q (National Institutes of Health) and MATLAB 2013a (MathWorks) were used for all analyses. Fluorescent images were downsampled by a factor of four. The lateral displacements of the brain were then corrected by image translation using TurboReg (Thévenaz et al., 1998). An averaged image of the entire frame was used as a reference for image alignment. Spatial filters corresponding to individual cells were identified using a principal component analysis- and independent component analysis-based algorithm (Cellsort 1.0; Mukamel et al., 2009). The contour of half-maximum intensity was calculated to select spatial filters that followed a typical cell structure, and filters with an area of contour <20 or >300 pixels or where the circularity of contour was smaller than 0.7 were discarded.
Detection of calcium transients.
Calcium activity was determined by applying the spatial filters to the time-lapse image. The calcium transients were identified by searching each trace for the local maxima that had a peak amplitude of more than three times the SD from the baseline (defined as the average of the traces calculated over the entire session; Fig. 2D).
Registration of cells across sessions.
The threshold spatial filters for all cells from each session were mapped onto a single image. Subsequently, the other filters were aligned to the first day's image via affine transform using TurboReg. Following the alignment across all sessions, cell pairs with a centroid distance <7.8 μm were registered as the same neuron.
Place fields.
To analyze the place fields, calcium events that occurred when the animal's velocity was <1 cm/s were filtered out to eliminate nonspecific activity at rest. The number of calcium transients in each bin was divided by the total occupancy time of the mouse in the bin, and a Gaussian smoothing filter was applied (σ = 2 cm). The number of bins in which a place field had a value >50% of its maximum calcium event rate was used to determine its width. The position of the place field was defined as its centroid.
For each place field of the cells, the mutual information between calcium transients and the mouse's location was calculated (Skaggs et al., 1993). To assess the significance of spatial selectivity, Monte Carlo p values were calculated. A total of 1000 distinct shuffles of the calcium transient times were performed, and the mutual information for each shuffle was calculated. The p value was defined as the fraction of shuffles that exceeded the mutual information of the cell. Cells with p < 0.05 were considered place cells.
Rate difference.
The rate difference in the place field was determined by calculating the unsigned rate difference between the peak calcium transient rates in the two recording sessions and by dividing the difference by the sum of the two rates. If a cell did not have a statistically significant place field in the second session, the rate was zero.
Population vector correlation.
To compute the population vector (PV) correlation between two sessions, the firing rate maps of all simultaneously recorded place cells were stacked into a 2D matrix (position × cell identity). At each spatial bin, the firing rate vector of all cells represents the PV for that spatial bin. To compare two recording sessions, the Pearson correlation coefficient was calculated between each pair of PVs at the corresponding locations, and the correlation coefficients of all spatial bins were averaged (Leutgeb et al., 2005).
Experimental design and statistical analysis.
Calcium activity was recorded from 19 imaging sites in 16 mice for the long-term imaging experiment or from 13 imaging sites in 13 mice for the CPP experiment. Recordings with low image quality (blurry images or inconsistent focal plane throughout the experiment) were discarded. Statistical analysis was performed using GraphPad Prism 5 (GraphPad Software), MATLAB 2013a, and R v3.4.3. Data were first tested for normality by using a Shapiro–Wilk test and for the homogeneity of variance with Bartlett's test or the F test. Normally distributed data with equal variance were compared with a paired/unpaired Student's t test or one-way repeated-measures ANOVA followed by Tukey's post hoc comparisons, as stated in the figure legends. If the data were not normally distributed, nonparametric tests (Mann–Whitney U test, Wilcoxon signed rank test, Kruskal–Wallis test, Friedman test, or two-way repeated-measures ANOVA on aligned rank transformed data) were used. Statistical significance was set at p < 0.05 for all statistical analyses.
Results
Place-cell activity in the virtual environment
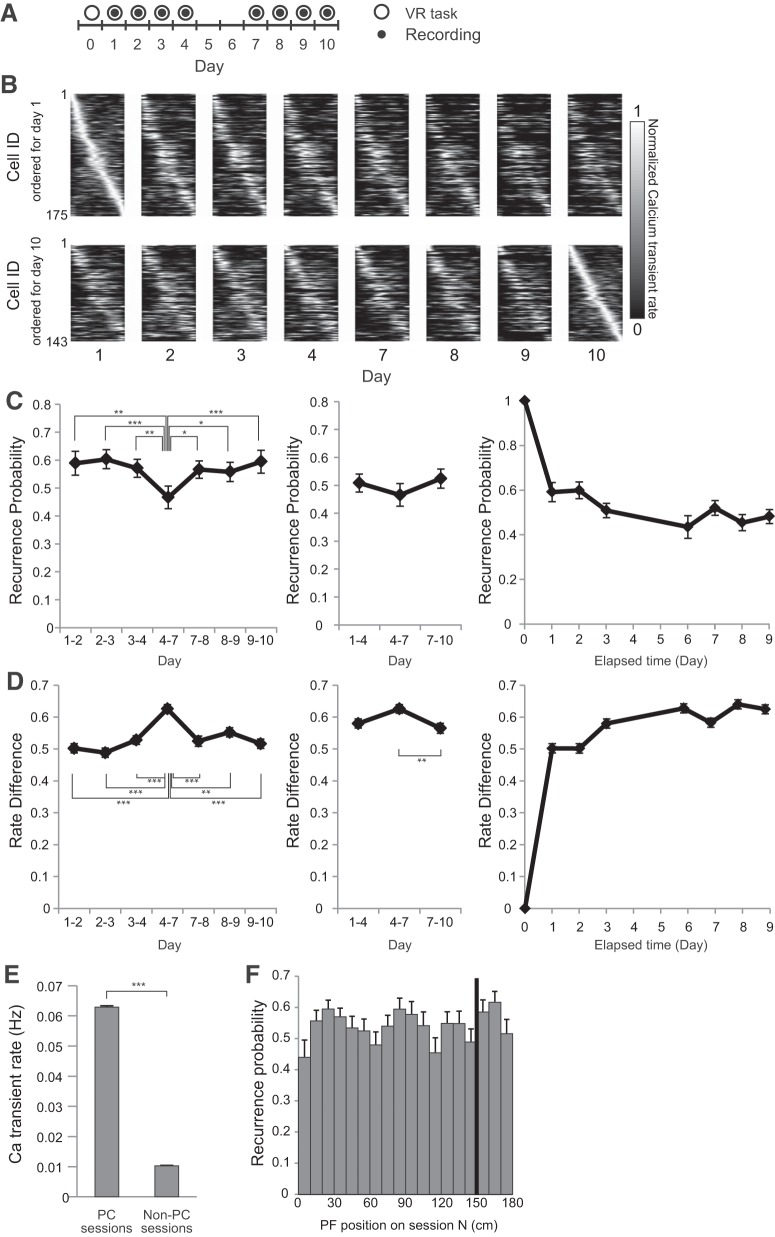
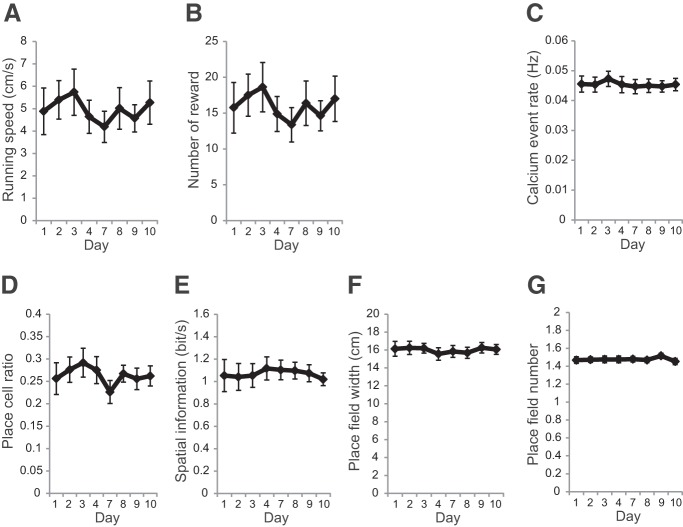
Place-cell activity in the hippocampal CA1 region was recorded using a combination of a genetically encoded calcium indicator GCaMP6f and wide-field fluorescent imaging (Figs. 1B,C, 2C). The mouse was head fixed on a spherical treadmill that allowed it to run freely (Fig. 1B). A custom VR system for rodents was used to present spatial cues (Fig. 2A,B; Hölscher et al., 2005; Dombeck et al., 2010; Youngstrom and Strowbridge, 2012; Hayashi et al., 2017). The activity patterns of the hippocampal CA1 cells were repeatedly measured in the same task with varying task intervals (Figs. 2C,D, 3A). In all recording sessions (n = 8 recording sites, 7 mice, 172–492 cells per site), the activity of the mice and their reward rate did not vary (Fig. 4A,B). Approximately one-fourth of cells exhibited location-specific activity. The basic properties of place-cell activity such as mean calcium event rate, fraction of cells having a significant place field, spatial information content, number of place fields, and place-field width did not change throughout the sessions (Fig. 4C–G). Considering that GCaMP6f was expressed under human Synapsin I promoter, which allows neuron-specific expression, both excitatory and inhibitory neurons were imaged (Nathanson et al., 2009). However, only pyramidal cells were included in the analyses because inhibitory neurons do not show clear place-specific activity (Wilson and McNaughton, 1993). Time-lapse recordings showed that the majority of place cells were only active in a subset of sessions, as described in previous studies (Fig. 3B; Ziv et al., 2013; Rubin et al., 2015).
Figure 3.
Long-term imaging of CA1 place-cell activity. A, Experimental timeline. After at least 3 weeks of pretraining, a series of four recording sessions was conducted on Days 1–4; another series of four recording sessions was conducted on Days 7–10. B, Place-field maps for the same cells on multiple days of the recording session. Maps are ordered by the place fields' centroid position on Day 1 (top) and Day 10 (bottom). The peak calcium event rate for each cell was normalized to one. C, If a cell had a statistically significant place field in one session, the odds that it had a place field in a subsequent session were displayed as recurrence probability. Left, Recurrence probability for adjacent sessions. Middle, Recurrence probability for sessions with a 3 d interval. Right, Recurrence probability between first day session and other sessions. D, Left, Changes in the peak transient rates (rate difference) for adjacent sessions. Middle, Rate difference for sessions with a 3 d interval. Right, Rate difference between first day session and other sessions. E, Calcium transient rates of the cells showing place-cell activity for at least 1 d. The rates for sessions when they exhibited place-cell activity are shown as PC sessions, whereas the rates for sessions when they did not are shown as Non-PC sessions. F, The distribution of recurrence probability as a function of place-field position. The black bar indicates the reward position. Data are shown as mean ± SEM. Asterisks indicate significant differences: *p < 0.05, **p < 0.01, ***p < 0.001; otherwise not significant.
Figure 4.
Behavioral properties and place-cell properties in the long-term recording experiment. A, Running speed at each recording session. Significance was evaluated by Friedman test (Q = 6.7, p = 0.46, n = 8). B, Number of rewards at each session. Significance was evaluated by Friedman test (Q = 6.9, p = 0.44, n = 8). C, Calcium event rate. Significance was evaluated by one-way repeated-measures ANOVA (F(2,21) = 1.5, p = 0.19, n = 8). D, Fraction of cells with significant place cells. Significance was evaluated by Friedman test (Q = 10, p = 0.19, n = 8). E, Spatial information content of place cells per unit time. Significance was evaluated by one-way repeated-measures ANOVA (F(2,21) = 0.33, p = 0.94, n = 8). F, Place-field size for each cell. Significance was evaluated by one-way repeated-measures ANOVA (F(2,21) = 0.65, p = 0.71, n = 8). G, Number of place field for each cell. Significance was evaluated by Friedman test (Q = 3.9, p = 0.79, n = 8). Data are shown as mean ± SEM.
The spatial distribution of place fields was inhomogeneous, as reported previously (Fig. 3F; Dombeck et al., 2010; Ziv et al., 2013). Previous studies showed that the place fields of CA1 cells accumulate around the goal zone during goal-oriented tasks (Markus et al., 1995; Hollup et al., 2001; Dupret et al., 2010; Gauthier and Tank, 2018). However, in the present study, place-field density was not higher around the reward zone than around other areas (Fig. 3F), much as in the study by Duvelle et al. (2019).
Effect of task schedule on the dynamics of place-cell activity
First, the effects of interval between sessions on the recurrence probability of place-cell activity were evaluated. Recurrence probability is defined as the probability of a cell to exhibit a statistically significant place field in one session and also have a place field in a subsequent session (Ziv et al., 2013). Time-series data indicated that recurrence probability was higher between 1-d-interval sessions than 3-d-interval sessions (Fig. 4C, left; F(6,42) = 18.76, p < 0.0001, n = 8 animals, one-way repeated-measures ANOVA followed by Tukey's post hoc test). Thereafter, the effects of task frequency per fixed time on recurrence probability were tested. In the first trial block, mice performed the task every day between the recording sessions (Fig. 3A, Days 1–4). In the second block, mice did not perform the task between the recording sessions (Fig. 3A, Days 4–7). In the last block, mice performed the task every day again between the recording sessions (Fig. 3A, Days 7–10). No significant difference was observed in recurrence probability among these three trial blocks, thus indicating that task frequency had no detectable effect on the speed of change of place-cell activity (Fig. 3C, middle; F(2,14) = 1.9, p = 0.19, n = 8 animals, one-way repeated-measures ANOVA). One reason for a cell to switch from place cell to non-place cell could be a change in its activity level. To test this possibility, we tested the calcium transient rates of the cells showing place-cell activity for at least 1 d. The rates for sessions when they did and did not exhibit place-cell activity are shown as “Place cell (PC) sessions” and “Non-PC sessions”, respectively. For each cell, the calcium event rate was higher when it showed place-cell activity than when it did not (Fig. 3E; U = 9,199,000, p < 0.0001, n = 6550 sessions for PC sessions, n = 18170 sessions for Non-PC sessions, Mann–Whitney U test). The result indicates that the change of place-cell activity is mainly due to the change of mean activity rate and not the spatial specificity of firing. The change of place-cell activity between sessions was also assayed by comparing the peak calcium transient rate in the place field. For this purpose, the normalized change in calcium transient rate (rate difference: unsigned rate difference divided by rate sum) was used (Leutgeb et al., 2005). Time-series data indicated a higher rate difference between 3-d-interval sessions (Fig. 3D, left; H = 62.81, p < 0.0001, n = 895–702 cells, Kruskal–Wallis test followed by Dunn's post hoc test). These results were consistent with higher recurrence ratios in 3-d-interval sessions compared with 1-d-interval sessions. The comparison of the rate difference in different task frequencies resulted in a significant but small difference between sessions (Fig. 3D, middle; H = 9.797, p = 0.0075, n = 856–702 cells, Kruskal–Wallis test followed by Dunn's post hoc test). Although the task frequency did not have an effect on place-cell activity at the population level (Fig. 3C, middle), the result suggests that task frequency had an effect on the activity at cellular level. Comparisons between the first day and each day indicated that recurrence probability and rate difference exhibited a monotonic decrease and increase, respectively, with time (Fig. 3C, right, D, right).
To assess the effect of visual landmarks or rewards on the dynamics of place-cell activity, the relationship between place-field location and the recurrence probability was analyzed. As shown in Figure 3F, the recurrence probability of the place cells was not dependent on their place-field location (F(17,126) = 1.53, p = 0.094, n = 8 animals, one-way repeated-measures ANOVA).
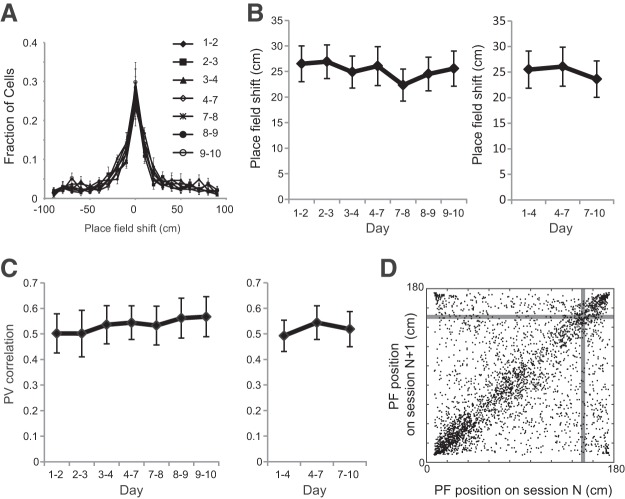
Thereafter, the effects of time and task frequency on place-field stability were evaluated. No significant difference was found in place-field shift when either the interval or task frequency was varied (Fig. 5A,B, left; F(6,42) = 1.622, p = 0.17, n = 8 animals, one-way repeated-measures ANOVA; and B, right; F(2,14) = 0.63, p = 0.55, n = 8 animals, one-way repeated-measures ANOVA). Another measure of place-field stability, namely, PV correlation, was not different between sessions with 1- and 3-d-intervals (Fig. 5C, left; Q = 3.536, p = 0.74, n = 8 animals, Friedman test). Furthermore, the task frequency did not affect the PV correlation between sessions (Fig. 5C, right; Q = 4.00, p = 0.15, n = 8 animals, Friedman test). These results are consistent with previous observations indicating that the place-field location for familiar environments is generally stable over time (Thompson and Best, 1990; Lever et al., 2002; Ziv et al., 2013). To characterize the dependence of place-field location on stability, the place-field locations for two consecutive sessions were plotted (Fig. 5D). The place fields around the start and end were more stable than those in other areas. However, the place fields around the reward zone did not show higher stability.
Figure 5.
Stability of place-field location. A, Distribution of centroid shifts in the place fields between adjacent recording sessions (1–2, 2–3, 3–4, 4–7, 7–8, 8–9, and 9–10). B, Left, Centroid shifts of the place fields between adjacent recording sessions. Right, Centroid shifts between sessions with a 3 d interval. C, Right, PV correlation of place cells between adjacent sessions. Left, PV correlation of place cells between sessions with a 3 d interval. D, The place-field location of all cells at the Nth session and N + 1th session (n = 3296 cells). Gray bars indicate the reward position. Data are shown as mean ± SEM.
Effect of NMDA receptor antagonism on the dynamics of place-cell activity
NMDA receptors have been identified as an important molecule in both synaptic plasticity and hippocampal memory (Malenka and Nicoll, 1993; Steele and Morris, 1999; Nakazawa et al., 2004). Previous studies have shown that the inhibition of NMDA receptors sustains LTP at perforant path–dentate gyrus synapses and promotes the retention of hippocampal memory (Villarreal et al., 2002; Shinohara and Hata, 2014; Sachser et al., 2016). These studies have led to the idea that the change in place-cell activity might also be an NMDA receptor-dependent process. To test this hypothesis, the NMDA receptor antagonist CPP was systemically administered straight after the first recording session and daily thereafter for 2 d (Fig. 6A). The drug administration had no effect on either running speed or reward rate (Fig. 6K,L). The recurrence probability of place-cell activity for the 3-d-interval was significantly increased in the CPP group compared with the saline group (Fig. 6B; U = 2, p = 0.032, n = 5 mice for saline, n = 5 mice for CPP, Mann–Whitney U test). The rate difference in the CPP group was lower than that in the saline group (Fig. 6C; U = 76390, p = 0.0005, n = 514 cells for saline, n = 344 cells for CPP, Mann–Whitney U test). These results indicated that NMDA receptor antagonism interfered with the change in place-cell activity. At the same time, no significant difference was observed in the amount of place-field shift between the saline and CPP groups (Fig. 6D; U = 1, p = 1.0, n = 5 mice for saline, n = 5 mice for CPP, Mann–Whitney U test). The PV correlation between the two sessions was also found to be the same between the saline and CPP groups (Fig. 6E; U = 1, p = 1.0, n = 5 mice for saline, n = 5 mice for CPP, Mann–Whitney U test). Although CPP had no effect on place-cell properties including mean calcium event rate (Fig. 6F), number of place field per cell (Fig. 6G), or place-field width (Fig. 6H), it had a significant effect on place-cell ratio (Fig. 6I) and spatial information content (Fig. 6J). These results suggest that NMDA receptor antagonism affects not only the recurrence rate of place cells but also some place-cell properties.
Discussion
The activity of hippocampal CA1 neurons was tracked during the repeated performances of a spatial task. By comparing the place-cell ensemble representations among repeated performance of the task, the overlap rate of the active place-cell population was found to be independent of the number of exposures to the environment within a fixed time but was dependent on elapsed time. The administration of an NMDA receptor antagonist suppressed this change in place-cell activity.
Effect of task frequency on the dynamics of place-cell activity
A previous long-term imaging study has shown that an active population of hippocampal CA1 place cells changes dynamically over time even when the animal is repeatedly exposed to a familiar environment (Ziv et al., 2013). In that study, place-cell activity was repeatedly measured with a constant interval. Therefore, it could not be determined whether the change of place-cell activity was triggered by the task sessions or if the activity changed automatically and continuously (Fig. 1A). To answer this question, place-cell activity in the CA1 area was recorded repeatedly at variable time intervals. The experiments determined that the rate of change of place-cell activity was constant regardless of the task frequency (Fig. 3C,D). These results support the constant rate model (Fig. 1A, right). This constant rate of change in place-cell activity provides support for the idea that place-cell dynamics work as a time code for memories of multiple events (Manns et al., 2007; Mankin et al., 2012; Ziv et al., 2013; Rubin et al., 2015; Cai et al., 2016). According to this hypothesis, the temporal proximity of any two episodes is encoded as the overlap rate of the active CA1 population. However, it cannot be denied that there are other factors modulating the rate of change in place-cell activity. One possible candidate is adult neurogenesis in the dentate gyrus of the hippocampus (Zhao et al., 2008). Although the population of newborn neuron is small in number (Kempermann et al., 1997a), they are more active than mature neurons (Danielson et al., 2016) and are known to have highly plastic synapses 4–6 weeks after generation (Ge et al., 2007). Therefore, these neurons may affect the dynamics of neural activity and hippocampal-based memory. In fact, new neurons in the dentate gyrus have been shown to play a significant role in synaptic plasticity and memory function (Clelland et al., 2009; Sahay et al., 2011; Nakashiba et al., 2012; Akers et al., 2014). Researchers have found that the rate of neurogenesis can be modulated by exercise, environmental enrichment, and stress (Gould et al., 1997; Kempermann et al., 1997b; van Praag et al., 1999). Therefore, these treatments may alter the rate of change in place-cell activity.
Does the decrease of overlap rate in place-cell population for longer intervals (Fig. 3C) affect the retention of spatial memory? The task used in this study did not require spatial memory; therefore, we could not behaviorally assess whether the interval length (1 vs 3 d) affects the memory quality. However, using in vivo calcium imaging and Bayesian decoding analysis, a previous study showed that the ensemble activity of CA1 cells retain a stable spatial representation for a month despite their activity dynamically changing over time (Ziv et al., 2013). This stable spatial representation by the CA1 cells results from their place fields having stable spatial tuning (Figs. 3B, 5A). Therefore, despite the smaller overlap in the place-cell population for longer intervals, spatial representation in the CA1 is likely preserved.
Involvement of NMDA receptors in the dynamics of place-cell activity
NMDA receptors are important in regulating synaptic plasticity and controlling memory function (Morris et al., 1986; Malenka and Nicoll, 1993). A number of studies have shown that blocking NMDA receptors impairs memory acquisition (Morris et al., 1986; Morris and Frey, 1997). In addition, the systemic administration of NMDA receptor antagonist impairs the acquisition of stable place code for novel environment (Kentros et al., 1998; Silva et al., 2015). Furthermore, several articles have reported that NMDA receptors are involved in memory retention. The antagonists of NMDA receptors block the decay of LTP in the perforant path–dentate gyrus synapses and enhance spatial memory retention (Villarreal et al., 2002; Shinohara and Hata, 2014; Sachser et al., 2016). These studies led to an idea that the rate of change of place-cell activity is also an NMDA receptor–dependent process. In the present study, the NMDA receptor blockade CPP reduced the speed of change in place-cell activity (Fig. 6B,C), thus indicating that NMDA receptors play an important role in the dynamics of CA1 neural ensembles. In the experiments presented here, rate difference in the CPP group was lower than that in the saline group (Fig. 6C), which would be the main cause of the increase of recurrence probability for the CPP group. In addition to this, the place-cell ratio for the CPP group was ∼20% higher than that for the saline group (Fig. 6I). This also partly contributed to a ∼1.4-fold increase of recurrence probability for the CPP group (Fig. 6B).
In the present study, the NMDA receptor antagonist was systemically administered; therefore, the synapses critical for the effect of the drug on the place-cell activity dynamics remain to be identified. Moreover, whether the same synapses in the brain govern the stability of the place code for both familiar and novel environments is unclear. Previous studies revealed that the hippocampal subregion-specific loss of NMDA receptor function using the Cre/loxP system had different effects on place-cell properties in the hippocampus (McHugh et al., 1996, 2007; Nakazawa et al., 2002). As well as within the hippocampus, brain regions sending input to the hippocampus such as entorhinal cortex affect the place-cell properties in the CA1 (Brun et al., 2008; Hales et al., 2014; Schlesiger et al., 2015). Similarly, within the hippocampus, the brain regions sending input to the hippocampus, such as the entorhinal cortex, affect the place-cell properties in the CA1 (Brun et al., 2008; Hales et al., 2014; Schlesiger et al., 2015). In addition, glutamatergic neurons as well as GABAergic interneurons have a significant impact on the activity of place cells in the CA1 (Royer et al., 2012). However, the sites responsible for days-to-weeks scale dynamics of place-cell activity have not still been identified. Therefore, using the brain region- or cell-type-specific manipulation of NMDA receptor function, the synapses controlling the place-cell dynamics in the CA1 remain to be identified in the future.
Dynamic changes in place-cell activity and stable place representation
The synapse is widely believed to be the site for information storage in the brain (Frey and Morris, 1997; Mayford et al., 2012). A recent imaging study revealed that the synaptic turnover rate in hippocampal CA1 cells is very high, with an estimated lifetime of ∼1–2 weeks (Attardo et al., 2015). This highly dynamic nature of synaptic turnover may be the basis for fluctuations in place-cell activity. On the contrary, although the activity of place cells changes dynamically over time, their place fields are generally stable over days and weeks (Ziv et al., 2013; Rubin et al., 2015). Moreover, the present study demonstrated that these changes in place fields are not affected by elapsed time (Fig. 5B, left), task frequency (Fig. 5B, right), or NMDA receptor blockade (Fig. 6B). This highly stable nature of the place field suggests the existence of a mechanism that stabilizes place fields against the continuous synaptic modifications that occur in the hippocampus.
Footnotes
This work was supported by JSPS KAKENHI (Grant 17K19436). I thank Enago (http://www.enago.jp) for the English language review.
The author declares no competing financial interests.
References
- Akers KG, Martinez-Canabal A, Restivo L, Yiu AP, De Cristofaro A, Hsiang HL, Wheeler AL, Guskjolen A, Niibori Y, Shoji H, Ohira K, Richards BA, Miyakawa T, Josselyn SA, Frankland PW (2014) Hippocampal neurogenesis regulates forgetting during adulthood and infancy. Science 344:598–602. 10.1126/science.1248903 [DOI] [PubMed] [Google Scholar]
- Allen TA, Fortin NJ (2013) The evolution of episodic memory. Proc Natl Acad Sci U S A 110:10379–10386. 10.1073/pnas.1301199110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attardo A, Fitzgerald JE, Schnitzer MJ (2015) Impermanence of dendritic spines in live adult CA1 hippocampus. Nature 523:592–596. 10.1038/nature14467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun VH, Leutgeb S, Wu HQ, Schwarcz R, Witter MP, Moser EI, Moser MB (2008) Impaired spatial representation in CA1 after lesion of direct input from entorhinal cortex. Neuron 57:290–302. 10.1016/j.neuron.2007.11.034 [DOI] [PubMed] [Google Scholar]
- Cai DJ, Aharoni D, Shuman T, Shobe J, Biane J, Song W, Wei B, Veshkini M, La-Vu M, Lou J, Flores SE, Kim I, Sano Y, Zhou M, Baumgaertel K, Lavi A, Kamata M, Tuszynski M, Mayford M, Golshani P, et al. (2016) A shared neural ensemble links distinct contextual memories encoded close in time. Nature 534:115–118. 10.1038/nature17955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clelland CD, Choi M, Romberg C, Clemenson GD Jr, Fragniere A, Tyers P, Jessberger S, Saksida LM, Barker RA, Gage FH, Bussey TJ (2009) A functional role for adult hippocampal neurogenesis in spatial pattern separation. Science 325:210–213. 10.1126/science.1173215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielson NB, Kaifosh P, Zaremba JD, Lovett-Barron M, Tsai J, Denny CA, Balough EM, Goldberg AR, Drew LJ, Hen R, Losonczy A, Kheirbek MA (2016) Distinct contribution of adult-born hippocampal granule cells to context encoding. Neuron 90:101–112. 10.1016/j.neuron.2016.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombeck DA, Harvey CD, Tian L, Looger LL, Tank DW (2010) Functional imaging of hippocampal place cells at cellular resolution during virtual navigation. Nat Neurosci 13:1433–1440. 10.1038/nn.2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll LN, Pettit NL, Minderer M, Chettih SN, Harvey CD (2017) Dynamic reorganization of neuronal activity patterns in parietal cortex. Cell 170:986–999.e16. 10.1016/j.cell.2017.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupret D, O'Neill J, Pleydell-Bouverie B, Csicsvari J (2010) The reorganization and reactivation of hippocampal maps predict spatial memory performance. Nat Neurosci 13:995–1002. 10.1038/nn.2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvelle É, Grieves RM, Hok V, Poucet B, Arleo A, Jeffery KJ, Save E (2019) Insensitivity of place cells to the value of spatial goals in a two-choice flexible navigation task. J Neurosci 39:2522–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey U, Morris RG (1997) Synaptic tagging and long-term potentiation. Nature 385:533–536. 10.1038/385533a0 [DOI] [PubMed] [Google Scholar]
- Gauthier JL, Tank DW (2018) A dedicated population for reward coding in the hippocampus. Neuron 99:179–193.e7. 10.1016/j.neuron.2018.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge S, Yang CH, Hsu KS, Ming GL, Song H (2007) A critical period for enhanced synaptic plasticity in newly generated neurons of the adult brain. Neuron 54:559–566. 10.1016/j.neuron.2007.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould E, McEwen BS, Tanapat P, Galea LA, Fuchs E (1997) Neurogenesis in the dentate gyrus of the adult tree shrew is regulated by psychosocial stress and NMDA receptor activation. J Neurosci 17:2492–2498. 10.1523/JNEUROSCI.17-07-02492.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales JB, Schlesiger MI, Leutgeb JK, Squire LR, Leutgeb S, Clark RE (2014) Medial entorhinal cortex lesions only partially disrupt hippocampal place cells and hippocampus-dependent place memory. Cell Rep 9:893–901. 10.1016/j.celrep.2014.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Yawata S, Funabiki K, Hikida T (2017) In vivo calcium imaging from dentate granule cells with wide-field fluorescence microscopy. PLoS One 12:e0180452. 10.1371/journal.pone.0180452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollup SA, Molden S, Donnett JG, Moser MB, Moser EI (2001) Accumulation of hippocampal place fields at the goal location in an annular watermaze task. J Neurosci 21:1635–1644. 10.1523/JNEUROSCI.21-05-01635.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölscher C, Schnee A, Dahmen H, Setia L, Mallot HA (2005) Rats are able to navigate in virtual environments. J Exp Biol 208:561–569. 10.1242/jeb.01371 [DOI] [PubMed] [Google Scholar]
- Huber D, Gutnisky DA, Peron S, O'Connor DH, Wiegert JS, Tian L, Oertner TG, Looger LL, Svoboda K (2012) Multiple dynamic representations in the motor cortex during sensorimotor learning. Nature 484:473–478. 10.1038/nature11039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempermann G, Kuhn HG, Gage FH (1997a) Genetic influence on neurogenesis in the dentate gyrus of adult mice. Proc Natl Acad Sci U S A 94:10409–10414. 10.1073/pnas.94.19.10409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempermann G, Kuhn HG, Gage FH (1997b) More hippocampal neurons in adult mice living in an enriched environment. Nature 386:493–495. 10.1038/386493a0 [DOI] [PubMed] [Google Scholar]
- Kentros C, Hargreaves E, Hawkins RD, Kandel ER, Shapiro M, Muller RV (1998) Abolition of long-term stability of new hippocampal place cell maps by NMDA receptor blockade. Science 280:2121–2126. 10.1126/science.280.5372.2121 [DOI] [PubMed] [Google Scholar]
- Leutgeb S, Leutgeb JK, Barnes CA, Moser EI, McNaughton BL, Moser MB (2005) Independent codes for spatial and episodic memory in hippocampal neuronal ensembles. Science 309:619–623. 10.1126/science.1114037 [DOI] [PubMed] [Google Scholar]
- Lever C, Wills T, Cacucci F, Burgess N, O'Keefe J (2002) Long-term plasticity in hippocampal place-cell representation of environmental geometry. Nature 416:90–94. 10.1038/416090a [DOI] [PubMed] [Google Scholar]
- Malenka RC, Nicoll RA (1993) NMDA-receptor-dependent synaptic plasticity: multiple forms and mechanisms. Trends Neurosci 16:521–527. 10.1016/0166-2236(93)90197-T [DOI] [PubMed] [Google Scholar]
- Mankin EA, Sparks FT, Slayyeh B, Sutherland RJ, Leutgeb S, Leutgeb JK (2012) Neuronal code for extended time in the hippocampus. Proc Natl Acad Sci U S A 109:19462–19467. 10.1073/pnas.1214107109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manns JR, Howard MW, Eichenbaum H (2007) Gradual changes in hippocampal activity support remembering the order of events. Neuron 56:530–540. 10.1016/j.neuron.2007.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus EJ, Qin YL, Leonard B, Skaggs WE, McNaughton BL, Barnes CA (1995) Interactions between location and task affect the spatial and directional firing of hippocampal neurons. J Neurosci 15:7079–7094. 10.1523/JNEUROSCI.15-11-07079.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayford M, Siegelbaum SA, Kandel ER (2012) Synapses and memory storage. Cold Spring Harb Perspect Biol 4:a005751. 10.1101/cshperspect.a005751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh TJ, Blum KI, Tsien JZ, Tonegawa S, Wilson MA (1996) Impaired hippocampal representation of space in CA1-specific NMDAR1 knockout mice. Cell 87:1339–1349. 10.1016/S0092-8674(00)81828-0 [DOI] [PubMed] [Google Scholar]
- McHugh TJ, Jones MW, Quinn JJ, Balthasar N, Coppari R, Elmquist JK, Lowell BB, Fanselow MS, Wilson MA, Tonegawa S (2007) Dentate gyrus NMDA receptors mediate rapid pattern separation in the hippocampal network. Science 317:94–99. 10.1126/science.1140263 [DOI] [PubMed] [Google Scholar]
- Morris RG, Frey U (1997) Hippocampal synaptic plasticity: role in spatial learning or the automatic recording of attended experience? Philos Trans R Soc Lond B Biol Sci 352:1489–1503. 10.1098/rstb.1997.0136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RG, Anderson E, Lynch GS, Baudry M (1986) Selective impairment of learning and blockade of long-term potentiation by an N-methyl-d-aspartate receptor antagonist, AP5. Nature 319:774–776. 10.1038/319774a0 [DOI] [PubMed] [Google Scholar]
- Moser MB, Rowland DC, Moser EI (2015) Place cells, grid cells, and memory. Cold Spring Harb Perspect Biol 7:a021808. 10.1101/cshperspect.a021808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukamel EA, Nimmerjahn A, Schnitzer MJ (2009) Automated analysis of cellular signals from large-scale calcium imaging data. Neuron 63:747–760. 10.1016/j.neuron.2009.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashiba T, Cushman JD, Pelkey KA, Renaudineau S, Buhl DL, McHugh TJ, Rodriguez Barrera V, Chittajallu R, Iwamoto KS, McBain CJ, Fanselow MS, Tonegawa S (2012) Young dentate granule cells mediate pattern separation, whereas old granule cells facilitate pattern completion. Cell 149:188–201. 10.1016/j.cell.2012.01.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa K, Quirk MC, Chitwood RA, Watanabe M, Yeckel MF, Sun LD, Kato A, Carr CA, Johnston D, Wilson MA, Tonegawa S (2002) Requirement for hippocampal CA3 NMDA receptors in associative memory recall. Science 297:211–218. 10.1126/science.1071795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa K, McHugh TJ, Wilson MA, Tonegawa S (2004) NMDA receptors, place cells and hippocampal spatial memory. Nat Rev Neurosci 5:361–372. 10.1038/nrn1385 [DOI] [PubMed] [Google Scholar]
- Nathanson JL, Yanagawa Y, Obata K, Callaway EM (2009) Preferential labeling of inhibitory and excitatory cortical neurons by endogenous tropism of adeno-associated virus and lentivirus vectors. Neuroscience 161:441–450. 10.1016/j.neuroscience.2009.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe J, Dostrovsky J (1971) The hippocampus as a spatial map: preliminary evidence from unit activity in the freely-moving rat. Brain Res 34:171–175. 10.1016/0006-8993(71)90358-1 [DOI] [PubMed] [Google Scholar]
- O'Keefe J, Nadel L (1978) The hippocampus as a cognitive map. New York: Oxford UP. [Google Scholar]
- Rokni U, Richardson AG, Bizzi E, Seung HS (2007) Motor learning with unstable neural representations. Neuron 54:653–666. 10.1016/j.neuron.2007.04.030 [DOI] [PubMed] [Google Scholar]
- Rolls ET. (2010) A computational theory of episodic memory formation in the hippocampus. Behav Brain Res 215:180–196. 10.1016/j.bbr.2010.03.027 [DOI] [PubMed] [Google Scholar]
- Royer S, Zemelman BV, Losonczy A, Kim J, Chance F, Magee JC, Buzsáki G (2012) Control of timing, rate and bursts of hippocampal place cells by dendritic and somatic inhibition. Nat Neurosci 15:769–775. 10.1038/nn.3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin A, Geva N, Sheintuch L, Ziv Y (2015) Hippocampal ensemble dynamics timestamp events in long-term memory. eLife 4:e12247. 10.7554/eLife.12247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachser RM, Santana F, Crestani AP, Lunardi P, Pedraza LK, Quillfeldt JA, Hardt O, de Oliveira Alvares L (2016) Forgetting of long-term memory requires activation of NMDA receptors, L-type voltage-dependent Ca2+ channels, and calcineurin. Sci Rep 6:22771. 10.1038/srep22771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahay A, Scobie KN, Hill AS, O'Carroll CM, Kheirbek MA, Burghardt NS, Fenton AA, Dranovsky A, Hen R (2011) Increasing adult hippocampal neurogenesis is sufficient to improve pattern separation. Nature 472:466–470. 10.1038/nature09817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesiger MI, Cannova CC, Boublil BL, Hales JB, Mankin EA, Brandon MP, Leutgeb JK, Leibold C, Leutgeb S (2015) The medial entorhinal cortex is necessary for temporal organization of hippocampal neuronal activity. Nat Neurosci 18:1123–1132. 10.1038/nn.4056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoville WB, Milner B (1957) Loss of recent memory after bilateral hippocampal lesions. J Neurol Neurosurg Psychiatry 20:11–21. 10.1136/jnnp.20.1.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara K, Hata T (2014) Post-acquisition hippocampal NMDA receptor blockade sustains retention of spatial reference memory in Morris water maze. Behav Brain Res 259:261–267. 10.1016/j.bbr.2013.11.016 [DOI] [PubMed] [Google Scholar]
- Silva D, Feng T, Foster DJ (2015) Trajectory events across hippocampal place cells require previous experience. Nat Neurosci 18:1772–1779. 10.1038/nn.4151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaggs WE, McNaughton BL, Gothard KM, Markus EJ (1993) An information-theoretic approach to deciphering the hippocampal code (Hanson SJ, Cowan JD, Giles CI, eds). San Mateo, CA: Morgan Kaufmann. [Google Scholar]
- Steele RJ, Morris RG (1999) Delay-dependent impairment of a matching-to-place task with chronic and intrahippocampal infusion of the NMDA-antagonist d-AP5. Hippocampus 9:118–136. [DOI] [PubMed] [Google Scholar]
- Thévenaz P, Ruttimann UE, Unser M (1998) A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process 7:27–41. 10.1109/83.650848 [DOI] [PubMed] [Google Scholar]
- Thompson LT, Best PJ (1990) Long-term stability of the place-field activity of single units recorded from the dorsal hippocampus of freely behaving rats. Brain Res 509:299–308. 10.1016/0006-8993(90)90555-P [DOI] [PubMed] [Google Scholar]
- Tulving E, Markowitsch HJ (1998) Episodic and declarative memory: role of the hippocampus. Hippocampus 8:198–204. [DOI] [PubMed] [Google Scholar]
- van Praag H, Kempermann G, Gage FH (1999) Running increases cell proliferation and neurogenesis in the adult mouse dentate gyrus. Nat Neurosci 2:266–270. 10.1038/6368 [DOI] [PubMed] [Google Scholar]
- Villarreal DM, Do V, Haddad E, Derrick BE (2002) NMDA receptor antagonists sustain LTP and spatial memory: active processes mediate LTP decay. Nat Neurosci 5:48–52. 10.1038/nn776 [DOI] [PubMed] [Google Scholar]
- Wilson MA, McNaughton BL (1993) Dynamics of the hippocampal ensemble code for space. Science 261:1055–1058. 10.1126/science.8351520 [DOI] [PubMed] [Google Scholar]
- Youngstrom IA, Strowbridge BW (2012) Visual landmarks facilitate rodent spatial navigation in virtual reality environments. Learn Mem 19:84–90. 10.1101/lm.023523.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Deng W, Gage FH (2008) Mechanisms and functional implications of adult neurogenesis. Cell 132:645–660. 10.1016/j.cell.2008.01.033 [DOI] [PubMed] [Google Scholar]
- Ziv Y, Burns LD, Cocker ED, Hamel EO, Ghosh KK, Kitch LJ, El Gamal A, Schnitzer MJ (2013) Long-term dynamics of CA1 hippocampal place codes. Nat Neurosci 16:264–266. 10.1038/nn.3329 [DOI] [PMC free article] [PubMed] [Google Scholar]