Abstract
Microglia are the resident immune cells of the CNS, and their response to infection, injury and disease is well documented. More recently, microglia have been shown to play a role in normal CNS development, with the fractalkine-Cx3cr1 signaling pathway of particular importance. This work describes the interaction between the light-sensitive photoreceptors and microglia during eye opening, a time of postnatal photoreceptor maturation. Genetic removal of Cx3cr1 (Cx3cr1GFP/GFP) led to an early retinal dysfunction soon after eye opening [postnatal day 17 (P17)] and cone photoreceptor loss (P30 onward) in mice of either sex. This dysfunction occurred at a time when fractalkine expression was predominantly outer retinal, when there was an increased microglial presence near the photoreceptor layer and increased microglial–cone photoreceptor contacts. Photoreceptor maturation and outer segment elongation was coincident with increased opsin photopigment expression in wild-type retina, while this was aberrant in the Cx3cr1GFP/GFP retina and outer segment length was reduced. A beadchip array highlighted Cx3cr1 regulation of genes involved in the photoreceptor cilium, a key structure that is important for outer segment elongation. This was confirmed with quantitative PCR with specific cilium-related genes, Rpgr and Rpgrip1, downregulated at eye opening (P14). While the overall cilium structure was unaffected, expression of Rpgr, Rpgrip1, and centrin were restricted to more proximal regions of the transitional zone. This study highlighted a novel role for microglia in postnatal neuronal development within the retina, with loss of fractalkine–Cx3cr1 signaling leading to an altered distribution of cilium proteins, failure of outer segment elongation and ultimately cone photoreceptor loss.
SIGNIFICANCE STATEMENT Microglia are involved in CNS development and disease. This work highlights the role of microglia in postnatal development of the light-detecting photoreceptor neurons within the mouse retina. Loss of the microglial Cx3cr1 signaling pathway resulted in specific alterations in the cilium, a key structure in photoreceptor outer segment elongation. The distribution of key components of the cilium transitional zone, Rpgr, Rpgrip1, and centrin, were altered in retinae lacking Cx3cr1 with reduced outer segment length and cone photoreceptor death observed at later postnatal ages. This work identifies a novel role for microglia in the postnatal maturation of retinal photoreceptors.
Keywords: cilium, Cx3cr1, fractalkine, microglia, photoreceptor, retina
Introduction
Microglia are the resident immune cells of the CNS, forming part of the innate immune system (Waisman et al., 2015). Microglia arise from myeloid progenitor cells in the yolk sac and colonize the brain and retina early during embryonic development (Santos et al., 2008; Ginhoux et al., 2010). Once established, they survey the local environment, extending and contracting their processes, contacting neurons, blood vessels, and other glia. As modulators of the immune environment, their response to injury and infection has been well studied and is characterized by the presence of an amoeboid phenotype and the production of inflammatory cytokines and/or macrophage-like phagocytic activity (Colton, 2009; Kettenmann et al., 2011). Given the importance of inflammation in CNS pathology, the role of microglia in neurodegeneration within the brain, spinal cord, and retina has been extensively explored (Combadière et al., 2007; Kigerl et al., 2009; Fuhrmann et al., 2010).
More recently, the role of microglia in establishing tissue architecture and maintaining homeostasis has received some attention. During CNS development, neuronal number is tightly regulated, with a subset of cells targeted for programmed cell death. Microglia actively regulate key aspects of this process, in addition to removing dead or dying cells (Frost and Schafer, 2016). Studies performed in the brain, retina, and spinal cord have shown that the depletion of microglia reduces the number of dead or dying cells, thereby increasing neuronal cell number (Frade and Barde, 1998; Sedel et al., 2004; Hose et al., 2005; Cunningham et al., 2013). While the exact mechanism remains to be fully elucidated, microglial-derived factors such as nerve growth factor, insulin-like growth factor, and interleukin-1b have been identified as key trophic factors (Frade and Barde, 1998; Ueno et al., 2013; Shigemoto-Mogami et al., 2014). In addition to modulating neuronal fate, microglia are known to make transient, activity-dependent contact with synapses, thereby regulating neuronal circuitry (Wake et al., 2009; Tremblay et al., 2010). Through this interaction, microglia eliminate immature, less active synapses via the complement system (Schafer et al., 2012; Bialas and Stevens, 2013). A recent study also suggests that adult retinal synapses are dependent upon microglial involvement (Wang et al., 2016). Because of this modulation, it is perhaps not surprising that several reports suggest that microglia affect overall brain function and behavior (Bilbo et al., 2005; Parkhurst et al., 2013).
Microglial–neuronal interactions are regulated by several signaling pathways, including fractalkine-Cx3cr1. Fractalkine (Cx3cl1) can exist in transmembrane and soluble forms and is mostly expressed by neurons, while its sole receptor, Cx3cr1, is predominantly found on microglia within the CNS. Work undertaken in the retina has indicated that the dynamics of microglial processes are modulated by fractalkine/Cx3cr1 signaling (Liang et al., 2009), while microglial migration is impaired in the somatosensory barrel cortex of Cx3cr1-deficient mice (Hoshiko et al., 2012; Arnoux and Audinat, 2015). These data suggest that fractalkine/Cx3cr1 signaling has a key role in regulating microglia–neuronal interaction. This is supported by work showing that loss of Cx3cr1 signaling leads to altered synapse formation in the brain and retina, impaired trophic support, and impaired or delayed synaptic pruning within the hippocampus and barrel cortex (Paolicelli et al., 2011; Hoshiko et al., 2012; Ueno et al., 2013; Wang et al., 2016).
In the present study, we explored the role of microglia in postnatal maturation of the retina. Using the Cx3cr1GFP/GFP mouse, we investigated the functional and structural effects of Cx3cr1 loss at periods before, during, and after eye opening, which are key time points in the maturation of the light-sensitive photoreceptors. Our work shows that early after eye opening there is a defect in the photoreceptor cilium, a structure that is critical to normal protein transport and maturation of the outer segment (OS). Loss of Cx3cr1 results in reduced outer segment length, an inhibition of the coordinated increase in photopigment (opsin) expression and ultimately a reduction in cone photoreceptor number. These Cx3cr1-mediated effects occur at a time when fractalkine expression is predominantly outer retinal, and there is altered photoreceptor–microglial interaction. These data identify an additional role for microglial Cx3cr1 signaling in the maturation of neurons within the CNS.
Materials and Methods
Animal models and antibodies.
Cx3cr1GFP/GFP transgenic mice on a C57BL/6J background were used to study the effect of Cx3cr1 signaling on postnatal photoreceptor development. This model is based upon the line produced by Jung et al. (2000) in which the first 390 bp of the Cx3cr1 gene was replaced by the gene for enhanced green fluorescent protein (EGFP). It was provided by Professor P. McMenamin (Monash University, Clayton, VIC, Australia) and backcrossed in our local animal facility for at least 10 generations onto the C57BL/6J line obtained from a single colony maintained at the Animal Resources Centre (Canning Vale, WA, Australia). Sequencing showed the Cx3cr1GFP/GFP and wild-type (WT; C57BL/6J) controls to be free from previously described murine retinal degenerations (Table 1; Baehr and Frederick, 2009). For select experiments, Cx3cr1GFP/+ heterozygote animals were generated from a Cx3cr1GFP/GFP × WT cross. Fractalkine expression was investigated using the BAC Cx3cl1cherry reporter mouse line (Kim et al., 2011; B6.Cg.Tg(Cx3cl1/mCherry)1Jung/ J; stock 025525, The Jackson Laboratory), hemizygous and WT (C57BL/6J) crosses were performed, and animals were genotyped as per supplied protocols (https://www2.jax.org/protocolsdb/f?p=116:5:0::NO:5:P5_MASTER_PROTOCOL_ID,P5_JRS_CODE:12797,025525). Cilium gene expression was also examined in the Pro23His (line 2) model of retinal degeneration (LaVail et al., 2018). All animal work was approved by the University of Melbourne Animal Ethics Committee (approval number #1313032) and adhered to the Association for Research in Vision and Ophthalmology statement for the use of animals in ophthalmic and visual research. Animals of either sex were used for this study.
Table 1.
Genotyping Cx3cr1GFP/GFP for known retinal degenerations
Antibodies to glutamine synthetase (1:1000; Merck Millipore) and glial fibrillary acidic protein (GFAP; 1:20,000; DAKO) were used to immunolabel gliotic Müller cells. Peanut agglutinin (PNA; Vector Laboratories; rhodamine conjugated, 1:250; fluorescein conjugated, 1:500) and transducin (1:1000; Gαt2; Santa Cruz Biotechnology) were used to label cone photoreceptors, while terminals were colabeled with an antibody to C-terminal binding protein-2 (Ribeye; 1:1000; BD Biosciences). A rabbit polyclonal antibody to mCherry was used to intensify labeling (1:200; Abcam), while commercial antibodies to SDCCAG8 (1:200; ProteinTech), centrin (1:1000; 20H5, Merck Millipore), and acetylated α-tubulin (Sigma-Aldrich; cryosections, 1:200, paraffin, 1:3000) were used to label photoreceptor cilium. Additional antibodies to the photoreceptor cilium were donated by Associate Professor Paulo Ferreira from Duke University Medical Center (Durham, NC; Rpgr no2, 5 μg/ml) and Dr. Tiansen Li from the National Eye Institute [Bethesda, MD; retinitis pigmentosa GTPase regulator (Rpgr) interacting protein 1 (Rpgrip1), 1:1000]. Human and wild-type mouse retinae were labeled with the monocyte marker ionized calcium-binding adapter molecule 1 (Iba1; 1:1500; Wako Pure Chemical Industries), PNA, and calbindin (1:2000; Swant). Fluorescently conjugated anti-rabbit and anti-mouse secondary antibodies were used (1:500; Alexa Fluor 488 and 594 nm; Invitrogen), and cell nuclei were counterstained with DAPI (Life Technologies) or bisbenzimide (Sigma-Aldrich).
Histology and immunohistochemistry.
Cx3cr1GFP/GFP and C57BL/6J (wild-type) animals were terminally anesthetized (ketamine/xylazine 67:13 mg/kg followed by pentabarbitone phosphate 120 mg/kg), their eyes enucleated, and the anterior segment removed. For retinal histology, posterior eyecups were processed into paraffin or resin as previously reported (Vessey et al., 2014). For immunohistochemistry, eyecups and retinal wholemounts were fixed in 4% paraformaldehyde (in 0.1 m phosphate buffer, pH 7.4) for 30 min and processed as previously reported (Vessey et al., 2012; Jobling et al., 2015). For photoreceptor cilium staining, retinal samples were used unfixed, snap frozen in Tissue-Tek O.C.T. compound (Sakura Finetek USA), and then processed as described above. All retinal sections and wholemounts were imaged using a confocal microscope (Meta, Pascal LSM-5 and LSM800, Zen Software, Zeiss) with either a 40× or 60× objective. For rendering of the photoreceptor outer segments and cone–microglial interactions, z-stack images were obtained at 1.2 μm increments. Müller cell gliosis was quantified in the S1 sublaminar of the inner plexiform layer, and retinal sections were equally divided into three sections to quantify central and peripheral retinal regions (Ly et al., 2011). For fractalkine expression, fluorescent intensity was calculated for each retinal layer and then expressed as a proportion of total retinal fluorescence. Quantification of immunofluorescent labeling was performed on at least three representative sections from one individual sample, with at least three independent samples quantified, and all imaging parameters were held constant for each outcome measure. Microglial cell and process number were quantified in the outer retina, which incorporated the outer plexiform and outer nuclear layers (ONLs), in addition to the subretinal space. Cx3cr1GFP/GFP and wild-type microglia were imaged using EGFP and Iba1, respectively. Validation experiments showed absolute colocalization between the two microglial markers (see Fig. 3B, inset). For quantification of cilium immunolabeling length, images were thresholded to exclude labeled areas <0.04 and >0.70 μm2. This ensured that only single fully resolved cilia were included in the analysis. Images were analyzed using MetaMorph Imaging Software (Molecular Devices) or ImageJ (NIH), while 3D renderings were produced via Imaris Image Software (Biplane).
Figure 3.
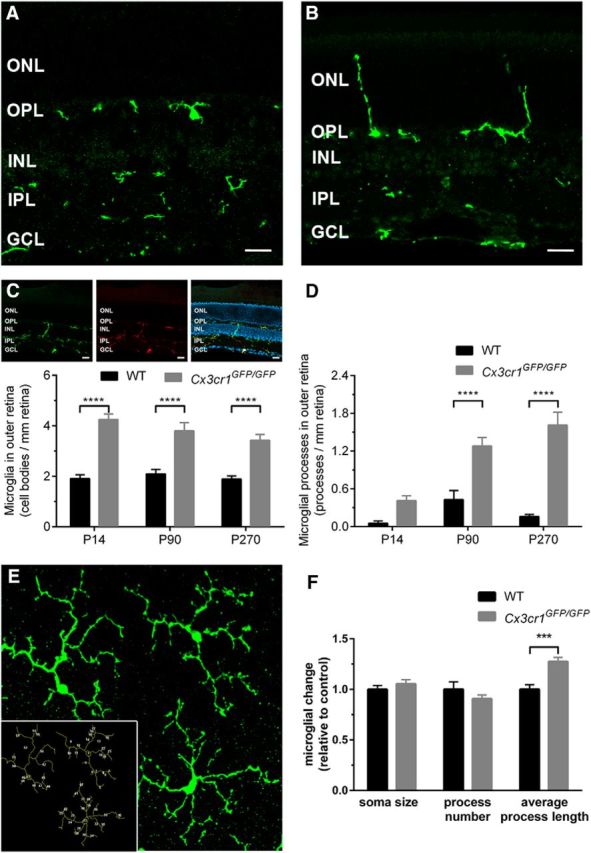
Retinal microglia are altered early in the Cx3cr1GFP/GFP retina, yet are not classically activated. A, B, Retinal microglia were imaged in wild-type (A; Iba1, green) and Cx3cr1GFP/GFP (B; EGFP) retinae, and cell bodies were restricted to the outer and inner plexiform layers in both genotypes. C, Inset, To validate morphological assessment, all GFP-expressing microglia were observed to colabel with Iba1. C, D, When quantified, an increased number of microglia and their processes were observed in the outer nuclear layer of Cx3cr1GFP/GFP retinae. E, F, Microglial morphology was assessed in ImageJ at the level of the photoreceptor terminals (E, inset), and there was no reduction in soma size or process number, while increased process length was observed in the Cx3cr1GFP/GFP retina (F). Data are presented as the mean ± SEM; n = 5 (C, D). ***p < 0.001, ****p < 0.0001, two-way ANOVA. Scale bar, 20 μm. INL, Inner nuclear layer.
Retinal function.
Retinal function was assessed using the electroretinogram (ERG) as previously described. Animals at ages postnatal day 17 (P17), P30, P90, and P270 (n > 7 for each group) were dark adapted overnight, anesthetized (ketamine/xylazine, 67:13 mg/kg), received additional corneal anesthetic (0.5%; Alcaine, Alcon Laboratories), and had their pupils dilated (atropine 0.5%; Mydriacyl, Alcon Laboratories). Recordings were made using custom AgCl recording/reference electrodes after a full field flash of 2.1 log cd/s/m2 (SB-900 Speedlight Flash, Nikon). Two flashes were delivered with a 0.8 s interstimulus interval to elicit responses from the rod and cone pathways (mixed) and the cone pathway alone (Scope version 3.6.9, ADInstruments). Data were filtered for 60 Hz noise, amplified, and digitized at 10 kHz over a 200 ms epoch (gain × 5000; −3 dB at 1 Hz and 1 kHz; ADInstruments). The cone response was digitally subtracted from the mixed response to generate the rod response.(Lyubarsky et al., 1996; Lyubarsky and Pugh, 1996), and ERG component analysis was completed as previously described (Weymouth and Vingrys, 2008; Jobling et al., 2013). Rod photoreceptor responses (rod a-wave) were analyzed using a modified PIII model, while the cone postreceptoral (cone b-wave) response was analyzed as a result of the small number of cone photoreceptors in the mouse.
Array and gene expression.
Retinae from P14 C57BL/6J (wild-type) and Cx3cr1GFP/GFP animals were isolated and snap frozen, and total RNA were isolated using silica spin column technology (Qiagen). Total RNA from three retinae were pooled, and an expression beadchip array (mouse WG-6 version 2.0, Illumina) was performed on three independent samples per group (Australian Genome Research Facility, Melbourne, VIC, Australia). The dataset was uploaded and analyzed using the Stemformatics portal (https://www.stemformatics.org; Wells et al., 2013). Based on our data showing cone outer segment alteration, we performed hierarchical clustering on cilium genes taken from the European project SysCilia site (http://www.syscilia.org/goldstandard.shtml). To validate select cilium-related genes (Rpgr, Rpgrip1, Ift140, Kif3b), quantitative gene expression analysis was performed as described previously (Jobling et al., 2013, 2015). Briefly, total RNA was isolated from dissected retinae, as described above; reverse transcribed (Tetro, Bioline); and amplified (Sensifast SYBR, Bioline) using the Rotor-Gene 3000 (Qiagen). Gene expression was quantified relative to gene-specific standards and two housekeeping genes (Hprt and Gapdh) and was expressed as a copy of gene of interest/copy housekeeper. The gene-specific primers are listed in Table 2.
Table 2.
Gene-specific oligonucleotide primer sequences
| Gene | Sequence | Forward primer | Reverse primer | Product size (bp) |
|---|---|---|---|---|
| Opn1mw | NM_008106.2 | GCCCAGACGTGTTCAGCG | GACCATCACCACCACCAT | 212 |
| Opn1sw | NM_007538.3 | CAGCCTTCATGGGATTTG | GTGCATGCTTGGAGTTGA | 339 |
| Rho | NM_145383.1 | AGCAGCAGGAGTCAGCCACC | CCGAAGTTGGAGCCCTGGTG | 145 |
| Rpgr | NM_001177950.1 | CTGTGGAAGGAACCACACCT | TTCAGAATTGTCACCCCACA | 229 |
| Rpgrip1 | NM_023879.3 | GGAGCATCCCTCACATCCTA | TCATCGCTGTCGGTAGTCTG | 102 |
| Kif3b | NM_008444.4 | TCTGTGAAGAACCCCAAAGG | TTCCCAGTTCCTGTTTGTCC | 182 |
| Ift140 | NM_134126.3 | TTGAAAAGGCTGTGGAGCTT | CTTGGTGGCCAGGTGATAGT | 215 |
| Hprt | NM_013556.2 | CCTAAGATGAGCGCAAGTTGAA | CCACAGGACTAGAACACCTGCTAA | 86 |
| Gapdh | NM_008084.2 | TGTGTCCGTCGTGGATCTGA | TTGCTGTTGAAGTCGCAGGAG | 150 |
Mouse-specific primers were designed relative to the respective published sequences. Primer sequences are shown 5′ to 3′, and the product sizes are included. For the external standards, the primers contained a T7 promoter sequence at the 5′ end of the forward primers, while a poly T15 sequence was included at the 5′ end of the reverse primers.
Experimental design and statistical analysis.
All quantitative analysis was performed on age-matched C57BL/6J (wild-type) and Cx3cr1GFP/GFP animals. For immunohistochemical analysis, at least three sections were imaged per sample and data were derived from at least n > 3 independent experiments (see individual figure legends for exact numbers). For gene expression analysis, n = 9 animals per genotype were used. All data were analyzed using a two-way ANOVA with Tukey's or Bonferroni's post hoc analysis (Prism, GraphPad Software), with variables being age and genotype or retinal eccentricity and genotype. Sex differences were assessed for each outcome measure, with no significant change observed (p > 0.05).
Results
Loss of Cx3cr1 signaling produces early retinal change
Within the retina, previous studies have identified a role for the Cx3cr1 signaling pathway in age-related macular degeneration (AMD; Tuo et al., 2004; Chen et al., 2007; Raoul et al., 2010), with the Cx3cr1GFP/GFP mouse model exhibiting fundus lesions and retinal degeneration at 18 months of age (Combadière et al., 2007; Raoul et al., 2008). To validate the Cx3cr1GFP/GFP model in our hands, we sought to confirm this late-stage pathology. Fundus images taken from 3-month-old Cx3cr1GFP/GFP animals appeared normal (Fig. 1A), while those from 16-month-old Cx3cr1GFP/GFP animals showed numerous white/yellow lesions (Fig. 1B). This age-dependent fundus pathology was reflected in the retinal histology, with normal structure observed in a 3-month-old Cx3cr1GFP/GFP retina (Fig. 1C), while cell loss was evident in the ONL of a 16-month-old retina (Fig. 1D). The Cx3cr1GFP/GFP mouse was also found to be free from known murine retinal degenerations (Table 1; Baehr and Frederick, 2009). These data show similar retinal pathology in aged mice compare with that previously reported (Combadière et al., 2007; Raoul et al., 2008).
Figure 1.
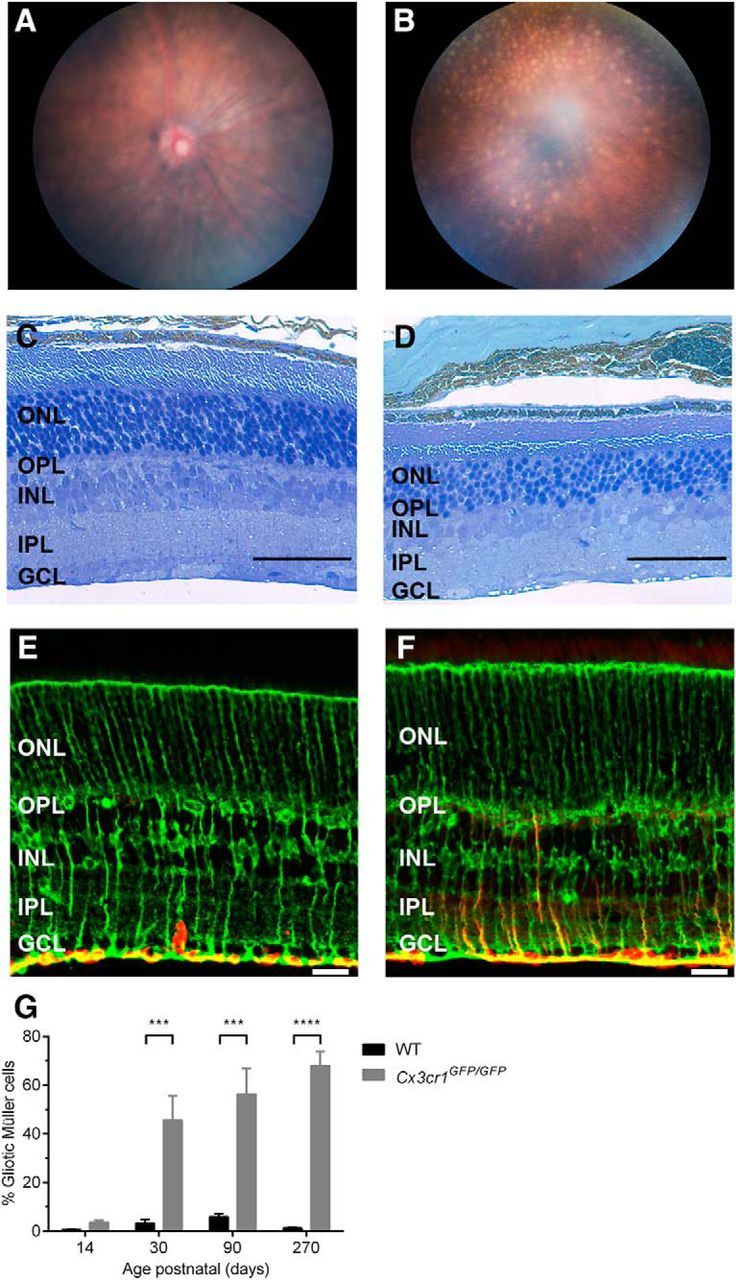
Cx3cr1GFP/GFP animals show early pathology before late-stage retinal degeneration. In vivo fundus imaging and retinal histology were used to validate late-stage retinal degeneration in Cx3cr1GFP/GFP animals. A, B, Micron III fundus images from 3-month-old (A) and 16-month-old (B) animals show white/yellow lesions are only found in older animals. C, D, Vertical sections of 3-month-old (C) and 16-month-old (D) retinae were stained with toluidine blue, with the ONL in the 16-month-old retina showing a reduced cell number, indicative of retinal degeneration. E, F, Age-matched WT (C57BL/6J) and 3-month-old Cx3cr1GFP/GFP retinae were immunolabeled with glutamine synthetase (green; E) and GFAP (red; F) to assess Müller cell gliosis. G, Gliosis was quantified in WT and Cx3cr1GFP/GFP retinae at various ages from P14 to P270, with increased gliosis evident from P30. Data are presented as the mean ± SEM; n > 3. ***p < 0.001, ****p < 0.0001, two-way ANOVA. Scale bars: E, F, 20 μm; C, D, 50 μm. INL, Inner nuclear layer.
While gross retinal structure was normal in 3-month-old Cx3cr1GFP/GFP, there was evidence of early retinal change. Unlike the wild-type retina in which expression of the intermediate filament protein GFAP (red) was limited to astrocytes in the ganglion cell layer (GCL; Fig. 1E), 3-month-old Cx3cr1GFP/GFP retinae showed extensive GFAP expression in the Müller cell processes (Fig. 1E, glutamine synthetase, green), particularly within the inner plexiform layer (IPL; Fig. 1F). In fact, when quantified (Fig. 1G), the Cx3cr1GFP/GFP retina showed increased Müller cell gliosis from around the eye opening (at P14; C57BL/6J, 0.5 ± 0.2%; Cx3cr1GFP/GFP, 3.6 ± 0.8%), becoming significant from 1 month of age (3.3 ± 1.6% vs 45.6 ± 10%, p = 0.0002). As Müller cell gliosis is used as a marker for retinal stress, such data suggest that loss of Cx3cr1 signaling produces early (≤P30) retinal change, well before the development of the late-stage retinal degeneration.
Loss of Cx3cr1 signaling produces early cone photoreceptor dysfunction and loss
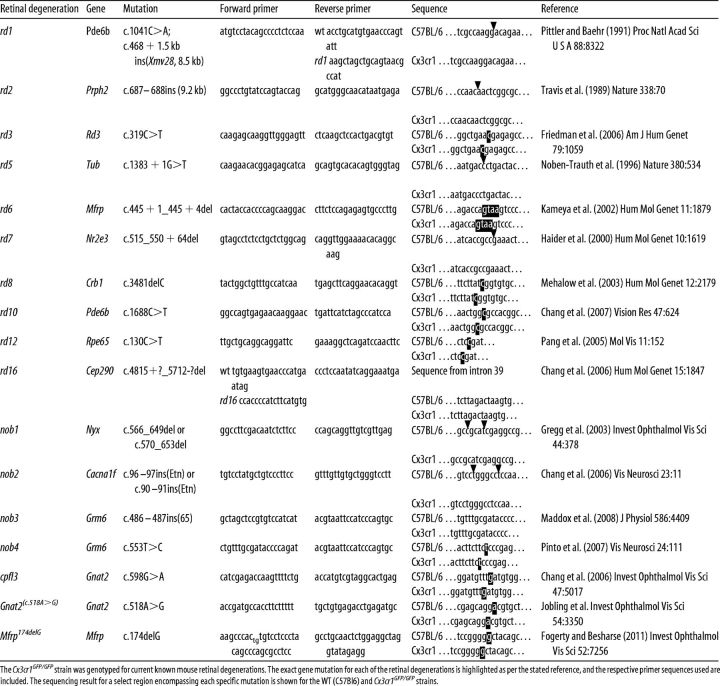
To explore this early change, retinal function was probed using the ERG at ages ranging from soon after eye opening (P17) to 9 months of age (P270). Representative waveforms for the rod photoreceptor pathway (Fig. 2A) were modeled, with the rod photoreceptor response amplitude (a-wave) in wild-type animals increasing from P17 to P30, after which time there was an age-dependent decrease in response (Fig. 2B). This initial increase in ERG response has been linked to postnatal maturation (outer segment elongation) of the photoreceptor (Gibson et al., 2013), while the age-related decrease in ERG response has also been reported previously (Li et al., 2001). The Cx3cr1GFP/GFP response was similar to that of the age-matched controls, and there was no effect of the loss of Cx3cr1 signaling on rod photoreceptor function until 9 months of age (P270), when there was a 22% decrease in amplitude (Fig. 2B; C57BL/6J, 347 ± 17 μV; Cx3cr1GFP/GFP, 272 ± 11 μV; p = 0.014). This functional decrease likely reflects the late onset of retinal degeneration observed in Figure 1 and described in previous reports (Combadière et al., 2007; Raoul et al., 2008).
Figure 2.
Loss of microglial Cx3cr1 signaling causes a cone photoreceptor loss. The electroretinogram was used to assess the effect of Cx3cr1 loss on retinal function. A–D, A twin flash protocol was used to isolate rod (A) and cone (C) waveforms, which were modeled to gain the photoreceptor amplitude across age (B and D, respectively). Rod photoreceptor function (a-wave) in Cx3cr1GFP/GFP animals was similar to wild-type counterparts until 9 months of age when a decrease in amplitude became evident (B). Cone-related function was decreased in Cx3cr1GFP/GFP animals from soon after eye opening (P17; D). E–H, Cone photoreceptor number (E and F; cone transducin, green; PNA, red) was quantified in wild-type, Cx3cr1GFP/GFP, and Cx3cr1GFP/+ retinae and showed reduced cone number from P30 in the periphery (G), while central retina (H) was affected at P90. Outer nuclear layer thickness was also quantified as an indirect measure of rod photoreceptor number and showed no alteration in either the peripheral or central retina (G, H, inset). Data are presented as the mean ± SEM; n > 4 (G, H), n > 6 (B, D, I). *p < 0.05, ****p < 0.0001, two-way ANOVA. Scale bar, 20 μm. INL, Inner nuclear layer.
In contrast, the cone pathway response was altered from an early stage in the Cx3cr1GFP/GFP retina (Fig. 2C). Cone-related function in the Cx3cr1GFP/GFP retina was reduced from P17 compared with the age-matched controls (Fig. 2D; C57BL/6J, 153 ± 13 μV; Cx3cr1GFP/GFP, 88 ± 8 μV; p = 0.014) and showed no increase in response coincident with photoreceptor maturation. This functional deficit remained at P30 (C57BL/6J, 236 ± 29 μV; Cx3cr1GFP/GFP 129 ± 14 μV; p < 0.0001) and P90 (C57BL/6J, 181 ± 14 μV; Cx3cr1GFP/GFP 73 ± 10 μV; p < 0.0001), while there was no significant difference between genotypes at P270, likely due to the age-related reduction in amplitude in the control animals at this later time point (C57BL/6J, 101 ± 7 μV; Cx3cr1GFP/GFP 80 ± 4 μV; p = 0.46).
Retinal sections were labeled with the cone markers cone transducin and PNA to explore whether this early functional deficit was a result of cone photoreceptor loss. As can be observed in Figure 2E, 3-month-old wild-type retinae show cone-specific immunostaining in the OSs (α-transducin in green and PNA in red) and outer plexiform layer (OPL; PNA in red). Cx3cr1GFP/GFP retinae showed reduced labeling (Fig. 2F), and when this was quantified, reduced cone photoreceptor numbers were evident in the peripheral retina from P30 onward compared with the wild-type control (Fig. 2G; C57BL/6J, 155 ± 1 cones/mm retina; Cx3cr1GFP/GFP, 111 ± 6 cones/mm retina; p = 0.025), while the central retina only became involved from P90 (Fig. 2H; C57BL/6J, 136 ± 4 cones/mm retina; Cx3cr1GFP/GFP, 99 ± 6 cones/mm retina; p < 0.0002), with the P270 data showing a trend for decreased cone number (C57BL/6J, 140 ± 8 cones/mm retina; Cx3cr1GFP/GFP, 117 ± 3 cones/mm retina; p = 0.08). Similar differences were observed when Cx3cr1GFP/GFP retinae were compared with those isolated from the heterozygous line (peripheral retinae: Cx3cr1GFP/+, 139 ± 4 cones/mm retina; central retinae: Cx3cr1GFP/+, 125 ± 4 cones/mm retina). To determine whether the loss of microglial Cx3cr1 also resulted in rod photoreceptor loss, ONL thickness was quantified since this layer is mostly composed of rod photoreceptor nuclei in the mouse. Neither central nor peripheral ONL thickness was altered in the Cx3cr1GFP/GFP retina (Fig. 2G, insert, H, insert, P30; two-way ANOVA, genotype, p = 0.998 and 0.929). These data suggest that loss of Cx3cr1 receptor signaling in the murine retina results in reduced cone pathway function soon after eye opening and the abolition of the increased functional response during photoreceptor maturation. This dysfunction precedes cone photoreceptor loss.
Loss of Cx3cr1 signaling alters microglia–photoreceptor interactions during postnatal maturation
As microglia are the predominant cell type that express Cx3cr1 within the healthy retina, we sought to determine whether loss of signaling through this receptor and associated cone photoreceptor dysfunction/loss was correlated with altered retinal microglia. Wild-type retinae showed characteristic microglial tiling, with cell bodies generally populating the IPL and OPL, while their processes extended into the ganglion cell inner nuclear and outer nuclear layers (Fig. 3A). The Cx3cr1GFP/GFP retina exhibited a similar expression profile with microglial somata commonly found in the plexiform layers. However, microglial processes were often observed to extend deep into the outer nuclear layer where the photoreceptors reside (Fig. 3B, ONL). To validate the quantification of microglial number/morphology in the wild-type and Cx3cr1GFP/GFP retinae, EGFP-expressing microglia were colabeled with Iba-1 and showed 100% colocalization (Fig. 3C, inset). When quantified, there was an increase in microglia numbers in the ONL of Cx3cr1GFP/GFP animals from P14 (Fig. 3C; C57BL/6J, 1.9 ± 0.15 microglia/mm retina; Cx3cr1GFP/GFP, 4.2 ± 0.22 microglia/mm retina; p < 0.0001) and a trend for increased microglial process number from P14 (C57BL/6J, 0.05 ± 0.04 microglial processes/mm retina; Cx3cr1GFP/GFP, 0.4 ± 0.08 microglial processes/mm retina; p = 0.06), becoming significant at P90 (Fig. 3D; C57BL/6J, 0.43 ± 0.14 microglial processes/mm retina; Cx3cr1GFP/GFP, 1.3 ± 0.31 microglial processes/mm retina; p < 0.0001). When expressed relative to control, the increased microglial numbers in the Cx3cr1GFP/GFP were independent of age (P14, 222 ± 12%; P90, 181 ± 15%; and P270, 181 ± 12%; p < 0.0001); however, the presence of microglial processes in the outer retina was increased at P14 compared with P90 (P14, 787 ± 146%; P90, 286 ± 33%; p = 0.0013), while levels further increased at P270 (956 ± 130%), which is likely due to the beginning of retinal degeneration (Fig. 2B).
To determine whether these microglia exhibited an activated (inflammatory) phenotype, microglial morphology was assessed at the level of the photoreceptor terminals (Fig. 3E, inset). When quantified, their soma size (Fig. 3F; C57BL/6J, 1.0 ± 0.04; Cx3cr1GFP/GFP, 1.05 ± 0.04; p = 0.79) and process number (Fig. 3F; C57BL/6J, 1.0 ± 0.07; Cx3cr1GFP/GFP, 0.91 ± 0.04; p = 0.42) remained unchanged relative to the wild-type retina, while process length increased in the Cx3cr1GFP/GFP retina (Fig. 3F; C57BL/6J, 1.0 ± 0.05; Cx3cr1GFP/GFP, 1.28 ± 0.04; p = 0.0002). These characteristics are unlike those described for activated microglia, which typically exhibit increased soma size, and reduced process number and length.
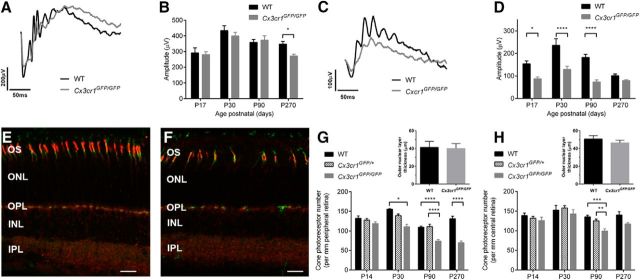
While Cx3cr1 is expressed on microglia within the retina, its ligand fractalkine is generally expressed in inner retinal neurons (Silverman et al., 2003; Zhang et al., 2012; Zieger et al., 2014), well away from the photoreceptor layer. However, studies investigating fractalkine expression in the retina have been performed in the adult eye, and there is no information on early postnatal expression, particularly during photoreceptor maturation (P14 to P30) when the Cx3cr1GFP/GFP cone-related change is evident. Using the Cx3cl1/mCherry reporter mouse, fractalkine expression was predominantly found in the outer plexiform layer at developmental times before and including eye opening (≤P14; Fig. 4A). Interestingly, at defined periods pre-eye opening, specifically at P8 to P10, fractalkine expression was also evident at the tips of the developing photoreceptor outer segments (Fig. 4A, inset). To determine whether microglia made contact with the tips of the developing outer segments at P8 to P10, Cx3cr1GFP/+ retinae were labeled with the cone marker (Fig. 4B and inset PNA, red). Microglial processes were observed to extend through the outer nuclear layer to the developing cone outer segments (Fig. 4B, inset). By contrast to the early postnatal retina, fractalkine expression in the adult retina (≥P30) was limited to cell bodies and processes within the ganglion cell and inner nuclear layers (Fig. 4C), as previously reported (Silverman et al., 2003; Zhang et al., 2012; Zieger et al., 2014). When fractalkine expression was mapped across the retina as a function of age (P8 to P90), outer retinal expression predominated before P21, while from P30 onward expression was mainly limited to the inner retina (Fig. 4D, central retina; Fig. 4E, peripheral retina). While fractalkine expression profile was spatially regulated within the retina during postnatal development, Cx3cl1 gene expression was not altered in Cx3cr1GFP/GFP retinae compared with their wild-type counterparts (Fig. 4F).
Figure 4.
Photoreceptors express fractalkine early in postnatal maturation, and the loss of Cx3cr1 signaling alters photoreceptor–microglial interaction. The Cx3cl1/mCherry reporter mouse was used to assess retinal fractalkine expression at various postnatal ages (P8 to P90). Early postnatal (P8 to P14) fractalkine expression (red) was limited to the outer plexiform layer, including the tips of the developing photoreceptor outer segments (PNA, green; A, inset), while microglial processes were observed to extend through the outer nuclear layer to the developing tips of the outer segment (B, inset; PNA, red; Cx3cr1GFP/+ microglia, green). C, Unlike the early postnatal retina, fractalkine expression in the adult retina was predominantly inner retinal. D, E, When fractalkine expression was quantified in central (D) and peripheral (E) retina, P8 to P14 tissue exhibited an outer retinal profile, while from P21 an inner retinal expression predominated. F, Fractalkine (Cx3cl1) gene expression was unaltered with age in the Cx3cr1GFP/GFP compared with WT animals. G, H, Microglia–cone photoreceptor interaction was evident in the human (G, inset) and mouse (H, inset) retinae, and when quantified in wild-type and Cx3cr1GFP/GFP retinae (H), increased contacts were observed both centrally and peripherally. Data are presented as the mean ± SEM; n > 9 (D–F, H). **p < 0.01, ****p < 0.0001, two-way ANOVA. Scale bars, 20 μm. INL, Inner nuclear layer.
Microglia are known to sample neuronal synapses, and data from human retinae (Fig. 4G) and murine retinae (Fig. 4H, inset, rendered view of a retinal microglia [Cx3cr1GFP/GFP, green] interacting with cone photoreceptor terminals [PNA labeled, red]) show that microglia make direct contact with cone photoreceptors. Thus, given that fractalkine expression is predominantly outer retinal during photoreceptor maturation and that there is an increased microglial presence within the Cx3cr1GFP/GFP outer retina, we sought to determine whether loss of Cx3cr1 altered photoreceptor–microglial interaction. At P21, a time characterized by a cone functional loss (see Fig. 2D), microglial–cone interactions were increased in the central (Fig. 4H; C57BL/6J, 43 ± 2.5%; Cx3cr1GFP/GFP, 56 ± 1.8%; p < 0.0001) and peripheral (Fig. 4H; C57BL/6J, 49 ± 1.4%; Cx3cr1GFP/GFP, 59 ± 1.6%; p = 0.001) Cx3cr1GFP/GFP retinae. This increased association did not correlate with gross changes in cone terminal structure (data not shown) and was significantly less than the near doubling of microglia in the outer retina, suggesting that they are interacting with other structures. Overall, these data show that fractalkine expression is predominantly expressed in the outer retina (including the plexiform layer and outer segment) during photoreceptor maturation and that interrupting Cx3cr1 signaling produces altered photoreceptor–microglia interactions at a time when cone photoreceptor dysfunction becomes evident.
Photoreceptor maturation and cilium structure is abnormal due to a loss of Cx3cr1 signaling
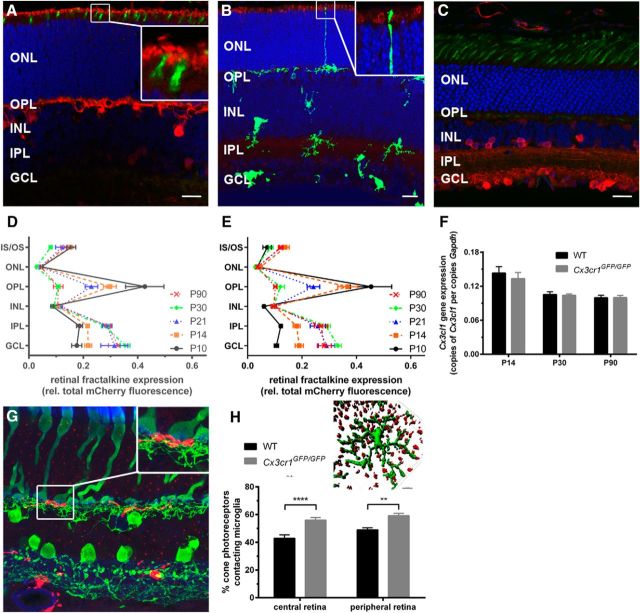
Postnatal maturation of the retinal photoreceptor is characterized by elongation of the outer segment, increased photopigment (opsin) expression and enhanced functional output (Timmers et al., 1999; Gibson et al., 2013). To determine whether the microglial Cx3cr1 pathway may have a role in coordinating this process, the expressions of middle- and short-wavelength opsin (Opn1_mw, Opn1_sw) and rhodopsin (Rho) were quantified. The expression of middle-wavelength opsin (Fig. 5A) showed an age-related decline in expression with no difference observed between wild-type and Cx3cr1GFP/GFP retinae. Similar to previously published data (Timmers et al., 1999), short-wavelength opsin (Fig. 5B) and rhodopsin (Fig. 5C) showed an increase in gene expression from P14 to P30 in wild-type retinae, reflecting the increase in outer segment length that occurs during that time. While there was no difference between the two genotypes at P14, Cx3cr1GFP/GFP retinae did not show increased Opn1_sw or Rho expression at P30, with expression remaining at P14 levels. Imaging the cone outer segments at P21 (PNA immunolabeling and 3D rendering) in wild-type (Fig. 5D) and Cx3cr1GFP/GFP retinae (Fig. 5E) showed evidence of shortened outer segments in the Cx3cr1GFP/GFP retina (Fig. 5E, highlighted yellow outer segments) and when retinal histology was used to quantify photoreceptor outer segment length at P21, photoreceptors from the Cx3cr1GFP/GFP retina consistently showed shorter outer segments, both centrally and peripherally (Fig. 5F; central C57BL/6J, 59 ± 1.4 μm; Cx3cr1GFP/GFP, 51 ± 2.0 μm; p = 0.004; peripheral C57BL/6J, 53 ± 2.8 μm; Cx3cr1GFP/GFP, 41 ± 1.4 μm; p = 0.03). Thus, the Cx3cr1GFP/GFP retina exhibits similar opsin expression to the wild-type retinae at P14 (an indirect measure of outer segment length), yet fails to show the increase coincident with outer segment elongation and exhibits shorter outer segments at P21.
Figure 5.
Microglial Cx3cr1 signaling is required for photoreceptor outer segment elongation. Photoreceptor outer segment elongation was characterized in wild-type and Cx3cr1GFP/GFP retinae. Outer segment elongation was indirectly assessed by quantifying opsin gene expression from P14 to P90. A–C, While there was no alteration in middle-wavelength opsin expression (A, Opn1_mw), short-wavelength opsin (B, Opn_sw), and rhodopsin (C, Rho), gene expression was reduced in the Cx3cr1GFP/GFP retina at P30, indicative of reduced outer segment elongation. D, E, Outer segments of cone photoreceptors were imaged (D, wild-type; E, Cx3cr1GFP/GFP; PNA, green), shorter outer segment length was evident in the Cx3cr1GFP/GFP retina (highlighted in yellow) and when photoreceptor outer segment length was quantified in P21 retina, the Cx3cr1GFP/GFP retina showed shorter outer segments both in the central and peripheral retina. Data are presented as the mean ± SEM; n = 9 (A–C), n > 5 (F). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, two-way ANOVA.
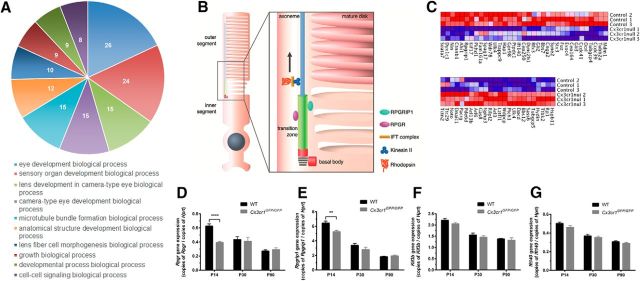
A mouse expression beadchip array (>45,200 transcripts) was performed on P14 wild-type and Cx3cr1GFP/GFP retinae to assess the gene expression changes at this period of photoreceptor maturation, before neuronal loss and outer segment change. Differential gene expression analysis showed 4857 transcripts to be significantly altered in the Cx3cr1GFP/GFP retinae, representing 4451 genes (difference due to multiple transcripts per gene). Bioinformatic analysis indicated that these genes cover several biological processes (Fig. 6A, GO enrichment), including eye development and sensory organ development (enrichment scores of 26 and 24, respectively). A table of the top 100 regulated genes obtained from the array is included as extended data and shows the genes representing crystallin heat shock proteins (cryg, crya) to be highly regulated, indicating retinal stress (Table 3).
Figure 6.
Loss of Cx3cr1 signaling alters the expression of cilium genes during photoreceptor maturation. A, Bead-chip array was performed on P14 retinal tissues from wild-type and Cx3cr1GFP/GFP retinae, and GO enrichment analysis showed the differentially expressed genes to represent ocular development processes (respective enrichment scores highlighted). B, The photoreceptor contains a modified cilium that is responsible for the transportation of proteins to the outer segment. Associated with this structure are a number of key cilium-related proteins (Rpgr and Rpgrip1). C, Analysis of cilium-related genes (http://www.syscilia.org/goldstandard.shtml) showed dysregulation of a number of genes. Quantitative PCR was used to further explore several of the key photoreceptor cilium genes (Rpgr, Rpgrip1, Kif3b, Ift140) during photoreceptor maturation (P14 to P30) and in the adult retina (P90). D–G, All genes exhibited an age-dependent decrease, particularly during outer segment elongation (P14 to P30, D–G). Loss of Cx3cr1 signaling altered the expression of select genes, with only Rpgr and Rpgrip1 gene expression decreased at P14 (D, E, respectively). Data are presented as the mean ± SEM; n = 9. **p < 0.01, ****p < 0.0001, two-way ANOVA.
Table 3.
Top 100 genes regulated in the Cx3cr1GFP/GFP retinae at P14
| Gene name | Transcript | Regulation (Cx3cr1GFP/GFP vs WT) | p value |
|---|---|---|---|
| Crygd | ILMN_240632 | 17.42 | 0.0000247 |
| Crygb | ILMN_222948 | 15.92 | 0.0000459 |
| Erdr1 | ILMN_221931 | 14.81 | 0.0018341 |
| Cryba1 | ILMN_214639 | 12.67 | 0.0003662 |
| Crygs | ILMN_218946 | 9.56 | 0.0012495 |
| Cryba2 | ILMN_212719 | 8.40 | 0.0018360 |
| Crygc | ILMN_247742 | 7.51 | 0.0000159 |
| Cryba4 | ILMN_218620 | 7.01 | 0.0009379 |
| Crybb2 | ILMN_216365 | 5.49 | 0.0031111 |
| Crybb3 | ILMN_220283 | 5.12 | 0.0007055 |
| Cryaa | ILMN_213225 | 4.09 | 0.0056395 |
| LOC665032 | ILMN_233071 | 3.87 | 0.0000047 |
| Crybb1 | ILMN_218933 | 3.63 | 0.0003985 |
| Cryga | ILMN_219207 | 3.31 | 0.0001126 |
| Xist | ILMN_187626 | 2.95 | 0.0255923 |
| Abhd14b | ILMN_246267 | 2.69 | 0.0000498 |
| Wdfy1 | ILMN_184827 | 2.57 | 0.0000172 |
| 1810013B01Rik | ILMN_224073 | 2.17 | 0.0004003 |
| BC032265 | ILMN_224441 | 2.04 | 0.0001126 |
| 1700047I17Rik1 | ILMN_221070 | 1.86 | 0.0008434 |
| Cdk5rap1 | ILMN_210158 | 1.79 | 0.0074371 |
| Cryab | ILMN_217805 | 1.64 | 0.0285807 |
| Nicn1 | ILMN_209291 | 1.64 | 0.0000678 |
| Mtap1b | ILMN_220448 | 1.58 | 0.0036510 |
| EG627624 | ILMN_247810 | 1.57 | 0.0000687 |
| LOC668038 | ILMN_316510 | 1.56 | 0.0060444 |
| Guca1a | ILMN_220524 | 1.55 | 0.0300676 |
| Kcne2 | ILMN_213229 | 1.55 | 0.0146115 |
| LOC100048530 | ILMN_311555 | 1.50 | 0.0266064 |
| Rn18s | ILMN_241508 | 1.48 | 0.0072960 |
| Slc16a6 | ILMN_233199 | 1.48 | 0.0023188 |
| Rai1 | ILMN_222328 | −1.49 | 0.0007107 |
| D030035F05Rik | ILMN_205484 | −1.49 | 0.0435819 |
| Tuft1 | ILMN_184505 | −1.50 | 0.0071321 |
| Pdhb | ILMN_206331 | −1.51 | 0.0028265 |
| scl000260.1_64 | ILMN_186860 | −1.51 | 0.0188281 |
| LOC384382 | ILMN_200605 | −1.51 | 0.0118750 |
| Raf1 | ILMN_203721 | −1.51 | 0.0004298 |
| Nrxn3 | ILMN_186459 | −1.51 | 0.0045190 |
| C230070D10Rik | ILMN_206449 | −1.52 | 0.0030692 |
| BC030499 | ILMN_250820 | −1.52 | 0.0006435 |
| Srr | ILMN_206790 | −1.52 | 0.0001989 |
| Otx1 | ILMN_217984 | −1.53 | 0.0023874 |
| Msi1 h | ILMN_206457 | −1.54 | 0.0014083 |
| LOC100046129 | ILMN_319878 | −1.54 | 0.0007598 |
| LOC382163 | ILMN_201443 | −1.54 | 0.0253172 |
| Prdm2 | ILMN_230695 | −1.55 | 0.0007373 |
| Unc13b | ILMN_248534 | −1.55 | 0.0028161 |
| F730023C13Rik | ILMN_207147 | −1.56 | 0.0049047 |
| LOC385274 | ILMN_201506 | −1.56 | 0.0212427 |
| Snca | ILMN_218187 | −1.57 | 0.0223110 |
| Gja1 | ILMN_217762 | −1.58 | 0.0023868 |
| E530004K11Rik | ILMN_206656 | −1.58 | 0.0087823 |
| Kcnma1 | ILMN_205056 | −1.58 | 0.0054532 |
| Slc13a4 | ILMN_214987 | −1.58 | 0.0011619 |
| A130070G01Rik | ILMN_204057 | −1.59 | 0.0017209 |
| A430106G13Rik | ILMN_195915 | −1.59 | 0.0006800 |
| D230046H12Rik | ILMN_206396 | −1.59 | 0.0000227 |
| Mtap7 | ILMN_201776 | −1.60 | 0.0001934 |
| 1110004P21Rik | ILMN_191964 | −1.62 | 0.0000673 |
| LOC385256 | ILMN_201494 | −1.62 | 0.0153742 |
| Zmiz1 | ILMN_258289 | −1.62 | 0.0326173 |
| Trim3 | ILMN_184744 | −1.62 | 0.0001600 |
| Prf1 | ILMN_215956 | −1.63 | 0.0043147 |
| Zxda | ILMN_257105 | −1.65 | 0.0005543 |
| (Table continues) | |||
| A230057G18Rik | ILMN_186103 | −1.69 | 0.0002916 |
| Fos | ILMN_222500 | −1.69 | 0.0091080 |
| 4933439C20Rik | ILMN_201669 | −1.71 | 0.0015183 |
| Chka | ILMN_252861 | −1.71 | 0.0007560 |
| Clasp1 | ILMN_237018 | −1.72 | 0.0008889 |
| LOC236604 | ILMN_325715 | −1.72 | 0.0048868 |
| A330068P14Rik | ILMN_191871 | −1.75 | 0.0152988 |
| F830002E14Rik | ILMN_207172 | −1.76 | 0.0122756 |
| Tmem181 | ILMN_248539 | −1.76 | 0.0003768 |
| 2810423O19Rik | ILMN_224094 | −1.79 | 0.0056417 |
| scl0002069.1_48 | ILMN_187328 | −1.84 | 0.0027987 |
| A830055I09Rik | ILMN_205168 | −1.86 | 0.0017236 |
| BC030476 | ILMN_209600 | −1.87 | 0.0004203 |
| Lrrc2 | ILMN_239373 | −1.90 | 0.0001355 |
| 2900060B14Rik | ILMN_192396 | −1.91 | 0.0136437 |
| Dusp7 | ILMN_218093 | −1.94 | 0.0005954 |
| Insig2 | ILMN_217996 | −1.97 | 0.0037118 |
| Entpd4 | ILMN_240362 | −2.02 | 0.0001330 |
| LOC381946 | ILMN_201000 | −2.07 | 0.0037097 |
| Lars2 | ILMN_221567 | −2.25 | 0.0000001 |
| H3f3b | ILMN_240518 | −2.36 | 0.0008136 |
| Gsto1 | ILMN_213260 | −2.37 | 0.0109962 |
| Stfa1 | ILMN_245739 | −2.41 | 0.0239512 |
| Emp1 | ILMN_213976 | −2.63 | 0.0261338 |
| 6430571L13Rik | ILMN_214086 | −2.77 | 0.0006210 |
| Trf | ILMN_192134 | −3.05 | 0.0000235 |
| Dorz1 | ILMN_224020 | −3.39 | 0.0000202 |
| Wdr82 | ILMN_329373 | −3.51 | 0.0000019 |
| Atp2c1 | ILMN_211584 | −3.72 | 0.0000028 |
| Egr1 | ILMN_215729 | −3.89 | 0.0002716 |
| Rpl29 | ILMN_221019 | −4.29 | 0.0000007 |
| 2810409M01Rik | ILMN_201789 | −5.03 | 0.0000038 |
| Epm2aip1 | ILMN_218771 | −8.95 | 0.0000289 |
| LOC383308 | ILMN_200537 | −11.87 | 0.0000003 |
Wild-type and Cx3cr1GFP/GFP animals were taken at P14 and subjected to an Illumina beadchip array (WG-6 version 2). The top 100 differentially regulated genes are shown below, including the extent of the regulation and the estimated p value.
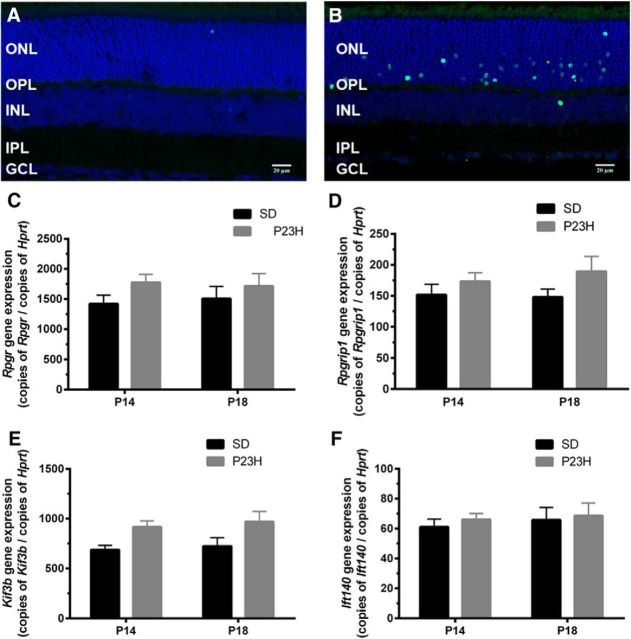
Based on the previous data indicating photoreceptor outer segment change and the fact that photoreceptor outer segment elongation is dependent upon a modified primary cilium that connects the inner and outer segments, providing for the movement of proteins (Fig. 6B; Steinberg et al., 1980; Besharse et al., 1985), cilium gene change was assessed (Fig. 6C; 272 genes collated in http://www.syscilia.org/goldstandard.shtml). Hierarchical clustering of Cx3cr1GFP/GFP samples showed specific positive (Fig. 6C, red in heatmap) and negative (Fig. 6C, blue in heatmap) regulation of several cilium genes. While transcripts within the one gene showed similar regulation, on occasion alternate regulation was observed (Ulk4). To validate these cilium gene changes, quantitative PCR was used to quantify the expression of several key cilium genes involved in cilium structure/protein packaging (Rpgr, Rpgrip1) and protein transportation (Kif3b, Ift140) from P14 onward. As observed in Figure 6, D and E, Rpgr and Rpgrip1 expression in the wild-type retinae showed an age-dependent decrease in expression from P14, while levels in the Cx3cr1GFP/GFP retinae were significantly lower at P14 and unaffected at P30 and P90 (P14: Rpgr, 0.63 ± 0.02 and 0.39 ± 0.03 copies/copy Hprt, p < 0.0001; Rpgrip1, 0.63 ± 0.02 0.39 ± 0.03 copies/copy Hprt, p = 0.001). While the transportation genes, including Ift140, showed a similar age-dependent decrease in expression from P14, there were no differences in expression between wild-type and Cx3cr1GFP/GFP retinae at any age (Fig. 6F,G). To determine whether this cilium gene dysregulation was indicative of a general response to retinal degeneration, the same genes were quantified at P14 and P18 in the Pro23His model of retinal degeneration (Fig. 7). No change in Rpgr, Rpgrip1, Ift140, or Kif3b expression was observed either before or after photoreceptor loss, indicating the changes described here for Cx3cr1GFP/GFP are not common to other models of retinal degeneration.
Figure 7.
Key cilium genes show no regulation during photoreceptor death in the Pro23His model of retinal degeneration. A, B, Retinal tissue from the Pro23His model of degeneration was isolated at P14 and P18. TUNEL was used to identify dying retinal neurons and showed limited staining at P14, while numerous photoreceptors were undergoing cell death at P18 (A and B, respectively). C–F, Quantitative PCR was used to further explore Rpgr, Rpgrip1, Kif3b, and Ift140 gene expression at these times and showed no alteration in expression. This response is unlike that observed for the Cx3cr1GFP/GFP retina. Data are presented as the mean ± SEM; n = 9. Scale bar, 20 μm. INL, Inner nuclear layer.
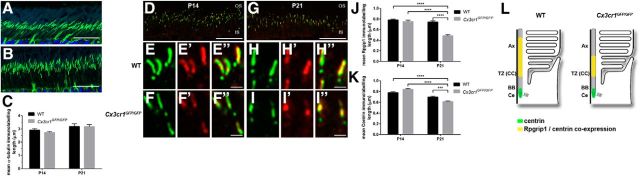
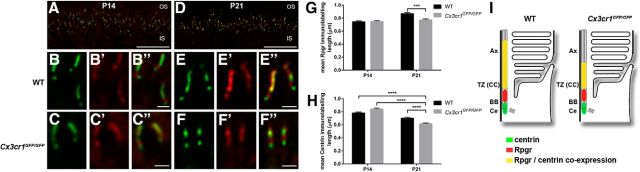
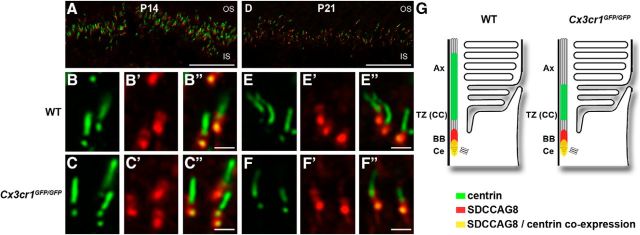
The above gene expression data suggest select alterations within the cilium rather than extensive dysregulation. To confirm this and to determine the specific Cx3cr1-mediated effects, the profile of protein expression within the photoreceptor cilium was explored. The cilium structure was investigated using acetylated α-tubulin, a biomarker for the axoneme (Fig. 8A,B; Arikawa and Williams, 1993). When the length of the immunolabeling was quantified at P14 and P21, no alteration was observed between wild-type and Cx3cr1GFP/GFP retinae, suggesting that the loss of Cx3cr1 signaling does not alter overall axoneme length (Fig. 8C). However, when the expression of select ciliary proteins was localized along the axoneme, specific alterations in expression profile were observed. Retinitis pigmentosa GTPase regulator interacting protein 1 (Rpgrip1), a key protein in the transitional zone and centrin that is expressed in the basal body and transitional zone, was found in a distinct band between the inner and outer segments at P14 and P21 (Fig. 8D,G). Closer inspection showed significant areas of coexpression along the photoreceptor cilium for both genotypes (Fig. 8E–F″, H–I″), yet the extent of the immunolabeling appeared restricted in the Cx3cr1GFP/GFP retina at P21 (Figs. 8, compare H–H″, I–I″). When the extent of immunolabeling was quantified along the axoneme, no alteration was found at P14, while the Cx3cr1GFP/GFP retina showed significantly reduced expression along the cilium at P21 for Rpgrip1 (Fig. 8J; 0.74 ± 0.03 vs 0.48 ± 0.02 μm, p < 0.0001) and centrin (Fig. 8K; 0.70 ± 0.02 vs 0.61 ± 0.01 μm, p = 0.0004). The effect of Cx3cr1 loss on the profile of cilium-specific protein expression in the photoreceptor at P21 is summarized in Figure 8L. A similar alteration was observed at P21 for the expression of Rpgr (Fig. 9; 0.87 ± 0.02 vs 0.77 ± 0.02 μm, p = 0.0005), while expression of the basal body protein SDCCAG8 was unaltered (Fig. 10). Thus, the lack of microglial Cx3cr1 expression results in restricted expression of key proteins within the transitional zone of the photoreceptor cilium, while other regions of the cilium remain unaffected.
Figure 8.
Microglial Cx3cr1 signaling regulates Rpgrip1 and centrin expression along the transitional zone of the cilium during photoreceptor maturation. A, B, Overall photoreceptor cilium structure was assessed in the wild-type (A) and Cx3cr1GFP/GFP (B) retinae using an antibody to acetylated α-tubulin, a biomarker for the axoneme. C, Quantification of axoneme length showed no difference between the two groups at either P14 or P21. D–I″, The expression of Rpgrip1 and centrin along the cilium was also examined at P14 (D–F″) and P21 (G–I″) using immunohistochemistry (centrin, green; Rpgrip1, red). Staining was evident in a restricted band between the inner and outer segments (D, G), and there was significant colocalization independent of genotype or postnatal age (E–F″, H–I″). When the length of Rpgrip1 and centrin immunolabeling was quantified (J, K), there was no difference at P14, while the labeling in the P21 Cx3cr1GFP/GFP retina showed significant reduction for both Rpgrip1 and centrin. L, The Cx3cr1-dependent change in cilium protein expression is summarized. Data are presented as the mean ± SEM; n > 4. ***p < 0.001, ****p < 0.0001, two-way ANOVA. Scale bars: A, B, D, G, 20 μm; E–F″, H–I″, 1 μm. IS, Inner segment; Ax, axoneme; TZ, transitional zone; CC, connecting cilium; BB, basal body; Ce, centriole.
Figure 9.
Microglial Cx3cr1 signaling regulates Rpgr expression along the transitional zone of the cilium during photoreceptor maturation. A–F, The expression of Rpgr and centrin-3 along the photoreceptor cilium was examined at P14 (A–C″) and P21 (D–F″) using immunohistochemistry (centrin-3, green; Rpgr, red). Staining was evident in a restricted band between the inner and outer segments (A, D), and there was significant colocalization independent of genotype or postnatal age (B–C″, E–F″). G, H, When the length of Rpgr and centrin-3 immunolabeling was quantified (G, H), there was no difference at P14, while the labeling in the P21 Cx3cr1GFP/GFP retina showed significant reduction for both Rpgr and centrin-3 (G, H). I, The Cx3cr1-dependent change in cilium protein expression is summarized. Data are presented as the mean ± SEM; n > 4. ***p < 0.001, ****p < 0.0001, two-way ANOVA. Scale bars: A, D, 20 μm; B–C″, E–F″, 1 μm. IS, Inner segment; Ax, axoneme; TZ, transitional zone; CC, connecting cilium; BB, basal body; Ce, centriole.
Figure 10.
Microglial Cx3cr1 signaling does not alter SDCCAG8 expression in the cilium during photoreceptor maturation. A–F″, The expression of the seriologically defined colon cancer antigen-8 (SDCCAG8) and centrin-3 along the photoreceptor cilium was examined at P14 (A–C″) and P21 (D–F″) using immunohistochemistry (centrin-3, green; SDCCAG8, red). Staining was evident in a restricted band between the inner and outer segments (A, D), and there was colocalization only at the distal tip of the cilium incorporating the basal body/centriole (B″, C″, E″, and F″). There was no observable alteration in the expression profile for SDCCAG8. G, The Cx3cr1-dependent change in cilium protein expression is summarized. Scale bars: A, D, 20 μm; B–C″, E–F″, 1 μm. IS, Inner segment; Ax, axoneme; TZ, transitional zone; CC, connecting cilium; BB, basal body; Ce, centriole.
Discussion
Microglial–neuronal interaction via the Cx3cr1 signaling pathway has been extensively explored in relation to pathology, with more recent work highlighting a role in normal neuronal development. Data from the current study have identified a novel fractalkine–Cx3cr1 modulation of postnatal neuronal development in the retina. Maturation of the light-detecting photoreceptors involves elongation of the outer segment, which is coordinated with eye opening and is dependent on protein transport via the photoreceptor cilium. Our data showed that the loss of microglial Cx3cr1 signaling (Cx3cr1GFP/GFP) restricted the expression of the key cilium proteins, Rpgr, centrin, and Rpgrip1 within the transitional zone of the cilium. Subsequent to these alterations, Cx3cr1GFP/GFP retina failed to exhibit the developmental increase in opsin expression that accompanies outer segment elongation, showed shorter outer segments, and reduced retinal function within the cone pathway. These Cx3cr1-mediated changes ultimately lead to cone photoreceptor loss.
Within the healthy retina, Cx3cr1 is expressed in a stable population of endogenous microglia, which are known to constantly survey the retina. Our data show that deletion of Cx3cr1 signaling in microglia via insertion of the EGFP gene (Jung et al., 2000) resulted in the failure of outer segment elongation culminating in loss of cone photoreceptors. This microglial regulation occurred at a specific postnatal time in photoreceptor maturation when outer segment elongation was coincident with increased light levels (eye opening) and outer retinal (photoreceptor synaptic layer and developing outer segments) fractalkine expression. These data highlight a novel, as yet unidentified role of the fractalkine–Cx3cr1 signaling axis in the retina. In fact, this is the first report of Cx3cr1-mediated refinement of the normal retinal neuronal circuit. While a previous study using the Ccl2/Cx3cr1 double knock-out animal detailed early (P30) photoreceptor terminal degeneration (Zhang et al., 2013), this effect is likely due to a background mutation (rd8) rather than the absence of Ccl2/Cx3cr1 signaling pathways (Luhmann et al., 2012; Vessey et al., 2012). Work undertaken in the brain, however, has detailed several physiological roles for microglial Cx3cr1 signaling, such as providing trophic support for developing neurons (Ueno et al., 2013), controlling synaptic pruning (Paolicelli et al., 2011), and altering synaptic expression of neurotransmitter receptors (Hoshiko et al., 2012), and involvement in synaptic plasticity (Rogers et al., 2011). Thus, our work, in conjunction with the work in the brain suggests that microglial–neuronal interaction via the fractalkine–Cx3cr1 signaling pathway is important for the refinement of the neural circuits throughout the CNS.
The retinal photoreceptor represents a specialized form of a primary cilium, which is predominantly responsible for protein trafficking from the inner to the outer segment. The cilium dysregulation in the Cx3cr1GFP/GFP retina was selective, primarily affecting the transitional zone, while there was no alteration in axoneme length (α-tubulin) or localization of proteins to the basal body (centrin and SDCCAG8). Even within the transitional zone, Cx3cr1-mediated change was specific, with Rpgr and Rpgrip1 gene expression decreased, and Rpgr, Rpgrip1, and centrin protein expression restricted to the more proximal regions of the transitional zone, while genes involved in the production of motor proteins (Kif3b) and intraflagellar transport proteins (Ift140) were unaffected. Rpgr, Rpgrip1, and centrin have previously been localized to the transitional zone in mice (Mavlyutov et al., 2002; Hong et al., 2003; Giessl et al., 2004); however, their role in cilium function is not fully elucidated. The transitional zone is thought to act as a gatekeeper, controlling the movement of proteins from the inner to the outer segment of the photoreceptor, and alterations in proteins such as Rpgr, Rpgrip1, and centrin, are thought to have implications for the control of protein trafficking (Trojan et al., 2008; Won et al., 2009). In particular, Rpgrip1 is thought to act as a cilium-specific scaffold, being responsible for the recruitment of other proteins such as Rpgr, phosphodiesterase 6 delta subunit, and the Nek signaling network to the cilium (Zhao et al., 2003; Remans et al., 2014). Thus, a Cx3cr1-mediated alteration in transitional zone proteins could impact on protein recruitment within the inner segment of the photoreceptor, cilium stability, and movement of proteins between the inner and outer segments.
While several studies have reported Cx3cr1 expression on ciliated cells within airway epithelial cells and the fallopian tube (Zhang et al., 2004; Jeong et al., 2015) and fractalkine expression has been reported on the ciliated inner ear hair cells (Kaur et al., 2015), this appears to be the first report of microglial regulation of ciliary structure. Work in the olfactory system provides indirect evidence of microglial modulation of ciliated cells, with loss of galectin-3, which is expressed by activated microglia (Lalancette-Hébert et al., 2012), decreasing the number of ciliated dendritic endings in olfactory sensory neurons (Comte et al., 2011). While Cx3cr1GFP/GFP animals exhibit a photoreceptor phenotype, they do not appear to exhibit extensive change in other ciliated cells, displaying no change in respiratory tract infection rates (Hall et al., 2009), breeding rates, or hearing thresholds (Sato et al., 2010). Such a limited phenotype may suggest microglial modulation of the cilium is specific to the retinal photoreceptor. Supporting this, work by Patil et al. (2012) has shown that retinal and renal cilium structure are regulated in a cell-specific manner.
Our data in conjunction with the work of Liang et al. (2009) indicate that microglial Cx3cr1 signaling is not required for cell fate determination during fetal retinal development. However, the presence of a microglial cell population is critical for proper retinal development, with work in the developing zebrafish retina showing that a delay in microglial colonization of the retina alters neuronal fate and ultimately retinal structure (Huang et al., 2012). Similarly, depletion of retinal microglia at birth results in altered retinal vascular development (Checchin et al., 2006). Regarding the role of microglia in photoreceptor maintenance/pathology, work from Wang et al. (2016) showed that retinal microglia were required for ongoing maintenance of photoreceptor synaptic terminals in the adult mouse retina. Support for our data showing the involvement of Cx3cr1 in cone-related function comes from a study reporting increased loss of cone function and inner segment/outer segment structure in animals harboring the rd10 retinal degeneration in addition to loss of Cx3cr1 (rd10/Cx3cr1GFP/GFP) when compared with rd10 alone (Peng et al., 2014). Importantly, this study also showed increased cone degeneration and outer segment protein mislocalization in the rd10/Cx3cr1GFP/GFP, characteristic of cilium disruption.
Subsequent to the cilium change in the Cx3cr1GFP/GFP retina, cone photoreceptor number is reduced at 1 month of age. Previous work has shown that alterations in cilium structure affect photoreceptor outer segment formation and lead to neuronal death (Rachel et al., 2012). Furthermore, mutations within the RPGR gene account for 70–90% of the X-linked form of retinitis pigmentosa, a condition involving extensive photoreceptor degeneration and irreversible severe vision loss (Shu et al., 2007). Similarly, mutations in RPGRIP1 are associated with another retinal degeneration, Leber Congenital Amaurosis (Li, 2014). In addition to retinal diseases, mutations in cilium genes are known to give rise to a number of neurodegenerative diseases termed ciliopathies (i.e., Bardet–Biedl Syndrome, Joubert Syndrome). It is unclear from our data why cone photoreceptors are preferentially targeted, given that Rho gene expression also indicates a failure in rod outer segment elongation. Interestingly, 8 of the 38 currently identified genes that give rise to nonsyndromic inherited cone disorders encode proteins important in cilium-based transport (Roosing et al., 2014), while several cilium-based mutations show a predisposition toward cone photoreceptor degeneration (Li et al., 2015; Nikopoulos et al., 2016; Lessieur et al., 2017). In support of specific differences between rod and cone cilium, work by Avasthi et al. (2009) has shown that protein trafficking through the cilium is different between cone and rod photoreceptors. Given our data showing different cone opsin regulation, spectrally defined ERG and opsin immunocytochemistry will be used to determine whether there is a specific cone photoreceptor (M/L, S opsin) that is more prone to Cx3cr1-mediated outer segment pathology and cell death loss.
The late-stage (>9-month-old animals) fundus pathology and retinal degeneration we observed in the Cx3cr1GFP/GFP animal is similar to results from previous reports using this model (Combadière et al., 2007; Raoul et al., 2010). This pathology, in addition to human genetic studies (Tuo et al., 2004; Chan et al., 2005), has led to the hypothesis that the loss of Cx3cr1 (Cx3cr1GFP/GFP) or reduced fractalkine binding capacity (V249I and T280M polymorphisms within the CX3CR1 gene) result in chronic retinal parainflammation, leading to AMD. Our data showing early cone photoreceptor loss in the Cx3cr1GFP/GFP retina may highlight an additional mechanism whereby individuals harboring Cx3cr1 polymorphisms may have reduced cone photoreceptor number. This lower cone number, in addition to the development of parainflammation, may predispose these individuals to a higher risk of the development of AMD. At present, there are no data showing that those with the V249I or T280M CX3CR1 polymorphisms have reduced cone photoreceptor number/function before the development of AMD.
Maturation of the light-detecting photoreceptors is a coordinated process involving eye opening, increased production of the light-detecting photopigment, and elongation of the outer segment. Our data show that microglial–neuronal interaction via the fractalkine–Cx3cr1 signaling pathway is important for outer segment maturation, with the loss of Cx3cr1 leading to altered distribution of photoreceptor cilium proteins, aberrant outer segment elongation and cone photoreceptor degeneration. This work highlights a novel role for the microglial Cx3cr1 signaling pathway in the development of the retinal neuronal circuit and may have implications for the development of age-related pathology such as age-related macular degeneration.
Footnotes
This work was supported by the National Health and Medical Research Council of Australia (Grant #1061418 to E.L.F. and A.I.J.; Grant #1061419 to E.L.F. and K.A.V.) and the Victorian Science Agenda (to E.L.F.). We thank Gene Venables for processing and imaging the acetylated α-tubulin, and Horace (Wing Hei) Chan for help with the photoreceptor cilium diagram. We also thank Associate Professor Paulo Ferreira from Duke University Medical Center (Durham, NC) and Dr. Tiansen Li from the National Eye Institute (Bethesda, MD) for the donation of antibodies to probe the photoreceptor cilium.
The authors declare no competing financial interests.
References
- Arikawa K, Williams DS (1993) Acetylated alpha-tubulin in the connecting cilium of developing rat photoreceptors. Invest Ophthalmol Vis Sci 34:2145–2149. [PubMed] [Google Scholar]
- Arnoux I, Audinat E (2015) Fractalkine signaling and microglia functions in the developing brain. Neural Plast 2015:689404. 10.1155/2015/689404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avasthi P, Watt CB, Williams DS, Le YZ, Li S, Chen CK, Marc RE, Frederick JM, Baehr W (2009) Trafficking of membrane proteins to cone but not rod outer segments is dependent on heterotrimeric kinesin-II. J Neurosci 29:14287–14298. 10.1523/JNEUROSCI.3976-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baehr W, Frederick JM (2009) Naturally occurring animal models with outer retina phenotypes. Vision Res 49:2636–2652. 10.1016/j.visres.2009.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besharse JC, Forestner DM, Defoe DM (1985) Membrane assembly in retinal photoreceptors. III. Distinct membrane domains of the connecting cilium of developing rods. J Neurosci 5:1035–1048. 10.1523/JNEUROSCI.05-04-01035.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialas AR, Stevens B (2013) TGF-beta signaling regulates neuronal C1q expression and developmental synaptic refinement. Nat Neurosci 16:1773–1782. 10.1038/nn.3560 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bilbo SD, Levkoff LH, Mahoney JH, Watkins LR, Rudy JW, Maier SF (2005) Neonatal infection induces memory impairments following an immune challenge in adulthood. Behav Neurosci 119:293–301. 10.1037/0735-7044.119.1.293 [DOI] [PubMed] [Google Scholar]
- Chan CC, Tuo J, Bojanowski CM, Csaky KG, Green WR (2005) Detection of CX3CR1 single nucleotide polymorphism and expression on archived eyes with age-related macular degeneration. Histol Histopathol 20:857–863. 10.14670/HH-20.857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checchin D, Sennlaub F, Levavasseur E, Leduc M, Chemtob S (2006) Potential role of microglia in retinal blood vessel formation. Invest Ophthalmol Vis Sci 47:3595–3602. 10.1167/iovs.05-1522 [DOI] [PubMed] [Google Scholar]
- Chen J, Connor KM, Smith LE (2007) Overstaying their welcome: defective CX3CR1 microglia eyed in macular degeneration. J Clin Invest 117:2758–2762. 10.1172/JCI33513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colton CA. (2009) Heterogeneity of microglial activation in the innate immune response in the brain. J Neuroimmune Pharmacol 4:399–418. 10.1007/s11481-009-9164-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combadière C, Feumi C, Raoul W, Keller N, Rodéro M, Pézard A, Lavalette S, Houssier M, Jonet L, Picard E, Debré P, Sirinyan M, Deterre P, Ferroukhi T, Cohen SY, Chauvaud D, Jeanny JC, Chemtob S, Behar-Cohen F, Sennlaub F (2007) CX3CR1-dependent subretinal microglia cell accumulation is associated with cardinal features of age-related macular degeneration. J Clin Invest 117:2920–2928. 10.1172/JCI31692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comte I, Kim Y, Young CC, van der Harg JM, Hockberger P, Bolam PJ, Poirier F, Szele FG (2011) Galectin-3 maintains cell motility from the subventricular zone to the olfactory bulb. J Cell Sci 124:2438–2447. 10.1242/jcs.079954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham CL, Martínez-Cerdeño V, Noctor SC (2013) Microglia regulate the number of neural precursor cells in the developing cerebral cortex. J Neurosci 33:4216–4233. 10.1523/JNEUROSCI.3441-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frade JM, Barde YA (1998) Microglia-derived nerve growth factor causes cell death in the developing retina. Neuron 20:35–41. 10.1016/S0896-6273(00)80432-8 [DOI] [PubMed] [Google Scholar]
- Frost JL, Schafer DP (2016) Microglia: architects of the developing nervous system. Trends Cell Biol 26:587–597. 10.1016/j.tcb.2016.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann M, Bittner T, Jung CK, Burgold S, Page RM, Mitteregger G, Haass C, LaFerla FM, Kretzschmar H, Herms J (2010) Microglial Cx3cr1 knockout prevents neuron loss in a mouse model of Alzheimer's disease. Nat Neurosci 13:411–413. 10.1038/nn.2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson R, Fletcher EL, Vingrys AJ, Zhu Y, Vessey KA, Kalloniatis M (2013) Functional and neurochemical development in the normal and degenerating mouse retina. J Comp Neurol 521:1251–1267. 10.1002/cne.23284 [DOI] [PubMed] [Google Scholar]
- Giessl A, Pulvermüller A, Trojan P, Park JH, Choe HW, Ernst OP, Hofmann KP, Wolfrum U (2004) Differential expression and interaction with the visual G-protein transducin of centrin isoforms in mammalian photoreceptor cells. J Biol Chem 279:51472–51481. 10.1074/jbc.M406770200 [DOI] [PubMed] [Google Scholar]
- Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, Samokhvalov IM, Merad M (2010) Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330:841–845. 10.1126/science.1194637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JD, Kurtz SL, Rigel NW, Gunn BM, Taft-Benz S, Morrison JP, Fong AM, Patel DD, Braunstein M, Kawula TH (2009) The impact of chemokine receptor CX3CR1 deficiency during respiratory infections with Mycobacterium tuberculosis or Francisella tularensis. Clin Exp Immunol 156:278–284. 10.1111/j.1365-2249.2009.03882.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong DH, Pawlyk B, Sokolov M, Strissel KJ, Yang J, Tulloch B, Wright AF, Arshavsky VY, Li T (2003) RPGR isoforms in photoreceptor connecting cilia and the transitional zone of motile cilia. Invest Ophthalmol Vis Sci 44:2413–2421. 10.1167/iovs.02-1206 [DOI] [PubMed] [Google Scholar]
- Hose S, Zigler JS Jr, Sinha D (2005) A novel rat model to study the functions of macrophages during normal development and pathophysiology of the eye. Immunol Lett 96:299–302. 10.1016/j.imlet.2004.09.017 [DOI] [PubMed] [Google Scholar]
- Hoshiko M, Arnoux I, Avignone E, Yamamoto N, Audinat E (2012) Deficiency of the microglial receptor CX3CR1 impairs postnatal functional development of thalamocortical synapses in the barrel cortex. J Neurosci 32:15106–15111. 10.1523/JNEUROSCI.1167-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T, Cui J, Li L, Hitchcock PF, Li Y (2012) The role of microglia in the neurogenesis of zebrafish retina. Biochem Biophys Res Commun 421:214–220. 10.1016/j.bbrc.2012.03.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong KI, Piepenhagen PA, Kishko M, DiNapoli JM, Groppo RP, Zhang L, Almond J, Kleanthous H, Delagrave S, Parrington M (2015) CX3CR1 is expressed in differentiated human ciliated airway cells and co-localizes with respiratory syncytial virus on cilia in a G protein-dependent manner. PLoS One 10:e0130517. 10.1371/journal.pone.0130517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobling AI, Vessey KA, Waugh M, Mills SA, Fletcher EL (2013) A naturally occurring mouse model of achromatopsia: characterization of the mutation in cone transducin and subsequent retinal phenotype. Invest Ophthalmol Vis Sci 54:3350–3359. 10.1167/iovs.13-11831 [DOI] [PubMed] [Google Scholar]
- Jobling AI, Guymer RH, Vessey KA, Greferath U, Mills SA, Brassington KH, Luu CD, Aung KZ, Trogrlic L, Plunkett M, Fletcher EL (2015) Nanosecond laser therapy reverses pathologic and molecular changes in age-related macular degeneration without retinal damage. FASEB J 29:696–710. 10.1096/fj.14-262444 [DOI] [PubMed] [Google Scholar]
- Jung S, Aliberti J, Graemmel P, Sunshine MJ, Kreutzberg GW, Sher A, Littman DR (2000) Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol 20:4106–4114. 10.1128/MCB.20.11.4106-4114.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur T, Zamani D, Tong L, Rubel EW, Ohlemiller KK, Hirose K, Warchol ME (2015) Fractalkine signaling regulates macrophage recruitment into the cochlea and promotes the survival of spiral ganglion neurons after selective hair cell lesion. J Neurosci 35:15050–15061. 10.1523/JNEUROSCI.2325-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenmann H, Hanisch UK, Noda M, Verkhratsky A (2011) Physiology of microglia. Physiol Rev 91:461–553. 10.1152/physrev.00011.2010 [DOI] [PubMed] [Google Scholar]
- Kigerl KA, Gensel JC, Ankeny DP, Alexander JK, Donnelly DJ, Popovich PG (2009) Identification of two distinct macrophage subsets with divergent effects causing either neurotoxicity or regeneration in the injured mouse spinal cord. J Neurosci 29:13435–13444. 10.1523/JNEUROSCI.3257-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KW, Vallon-Eberhard A, Zigmond E, Farache J, Shezen E, Shakhar G, Ludwig A, Lira SA, Jung S (2011) In vivo structure/function and expression analysis of the CX3C chemokine fractalkine. Blood 118:e156–e167. 10.1182/blood-2011-04-348946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalancette-Hébert M, Swarup V, Beaulieu JM, Bohacek I, Abdelhamid E, Weng YC, Sato S, Kriz J (2012) Galectin-3 is required for resident microglia activation and proliferation in response to ischemic injury. J Neurosci 32:10383–10395. 10.1523/JNEUROSCI.1498-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaVail MM, Nishikawa S, Steinberg RH, Naash MI, Duncan JL, Trautmann N, Matthes MT, Yasumura D, Lau-Villacorta C, Chen J, Peterson WM, Yang H, Flannery JG (2018) Phenotypic characterization of P23H and S334ter rhodopsin transgenic rat models of inherited retinal degeneration. Exp Eye Res 167:56–90. 10.1016/j.exer.2017.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessieur EM, Fogerty J, Gaivin RJ, Song P, Perkins BD (2017) The ciliopathy gene ahi1 is required for zebrafish cone photoreceptor outer segment morphogenesis and survival. Invest Ophthalmol Vis Sci 58:448–460. 10.1167/iovs.16-20326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Cheng M, Yang H, Peachey NS, Naash MI (2001) Age-related changes in the mouse outer retina. Optom Vis Sci 78:425–430. 10.1097/00006324-200106000-00015 [DOI] [PubMed] [Google Scholar]
- Li L, Rao KN, Zheng-Le Y, Hurd TW, Lillo C, Khanna H (2015) Loss of retinitis pigmentosa 2 (RP2) protein affects cone photoreceptor sensory cilium elongation in mice. Cytoskeleton (Hoboken) 72:447–454. 10.1002/cm.21255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T. (2014) Leber congenital amaurosis caused by mutations in RPGRIP1. Cold Spring Harb Perspect Med 5:a017384. 10.1101/cshperspect.a017384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang KJ, Lee JE, Wang YD, Ma W, Fontainhas AM, Fariss RN, Wong WT (2009) Regulation of dynamic behavior of retinal microglia by CX3CR1 signaling. Invest Ophthalmol Vis Sci 50:4444–4451. 10.1167/iovs.08-3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhmann UF, Lange CA, Robbie S, Munro PM, Cowing JA, Armer HE, Luong V, Carvalho LS, MacLaren RE, Fitzke FW, Bainbridge JW, Ali RR (2012) Differential modulation of retinal degeneration by Ccl2 and Cx3cr1 chemokine signalling. PLoS One 7:e35551. 10.1371/journal.pone.0035551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly A, Yee P, Vessey KA, Phipps JA, Jobling AI, Fletcher EL (2011) Early inner retinal astrocyte dysfunction during diabetes and development of hypoxia, retinal stress, and neuronal functional loss. Invest Ophthalmol Vis Sci 52:9316–9326. 10.1167/iovs.11-7879 [DOI] [PubMed] [Google Scholar]
- Lyubarsky AL, Pugh EN Jr (1996) Recovery phase of the murine rod photoresponse reconstructed from electroretinographic recordings. J Neurosci 16:563–571. 10.1523/JNEUROSCI.16-02-00563.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyubarsky A, Nikonov S, Pugh EN Jr (1996) The kinetics of inactivation of the rod phototransduction cascade with constant Ca2+i. J Gen Physiol 107:19–34. 10.1085/jgp.107.1.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavlyutov TA, Zhao H, Ferreira PA (2002) Species-specific subcellular localization of RPGR and RPGRIP isoforms: implications for the phenotypic variability of congenital retinopathies among species. Hum Mol Genet 11:1899–1907. 10.1093/hmg/11.16.1899 [DOI] [PubMed] [Google Scholar]
- Nikopoulos K, Farinelli P, Giangreco B, Tsika C, Royer-Bertrand B, Mbefo MK, Bedoni N, Kjellström U, El Zaoui I, Di Gioia SA, Balzano S, Cisarova K, Messina A, Decembrini S, Plainis S, Blazaki SV, Khan MI, Micheal S, Boldt K, Ueffing M, et al. (2016) Mutations in CEP78 cause cone-rod dystrophy and hearing loss associated with primary-cilia defects. Am J Hum Genet 99:770–776. 10.1016/j.ajhg.2016.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L, Ragozzino D, Gross CT (2011) Synaptic pruning by microglia is necessary for normal brain development. Science 333:1456–1458. 10.1126/science.1202529 [DOI] [PubMed] [Google Scholar]
- Parkhurst CN, Yang G, Ninan I, Savas JN, Yates JR 3rd, Lafaille JJ, Hempstead BL, Littman DR, Gan WB (2013) Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell 155:1596–1609. 10.1016/j.cell.2013.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil H, Tserentsoodol N, Saha A, Hao Y, Webb M, Ferreira PA (2012) Selective loss of RPGRIP1-dependent ciliary targeting of NPHP4, RPGR and SDCCAG8 underlies the degeneration of photoreceptor neurons. Cell Death Dis 3:e355. 10.1038/cddis.2012.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng B, Xiao J, Wang K, So KF, Tipoe GL, Lin B (2014) Suppression of microglial activation is neuroprotective in a mouse model of human retinitis pigmentosa. J Neurosci 34:8139–8150. 10.1523/JNEUROSCI.5200-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachel RA, Li T, Swaroop A (2012) Photoreceptor sensory cilia and ciliopathies: focus on CEP290, RPGR and their interacting proteins. Cilia 1:22. 10.1186/2046-2530-1-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raoul W, Feumi C, Keller N, Lavalette S, Houssier M, Behar-Cohen F, Combadière C, Sennlaub F (2008) Lipid-bloated subretinal microglial cells are at the origin of drusen appearance in CX3CR1-deficient mice. Ophthalmic Res 40:115–119. 10.1159/000119860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raoul W, Auvynet C, Camelo S, Guillonneau X, Feumi C, Combadière C, Sennlaub F (2010) CCL2/CCR2 and CX3CL1/CX3CR1 chemokine axes and their possible involvement in age-related macular degeneration. J Neuroinflammation 7:87. 10.1186/1742-2094-7-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remans K, Bürger M, Vetter IR, Wittinghofer A (2014) C2 domains as protein-protein interaction modules in the ciliary transition zone. Cell Rep 8:1–9. 10.1016/j.celrep.2014.05.049 [DOI] [PubMed] [Google Scholar]
- Rogers JT, Morganti JM, Bachstetter AD, Hudson CE, Peters MM, Grimmig BA, Weeber EJ, Bickford PC, Gemma C (2011) CX3CR1 deficiency leads to impairment of hippocampal cognitive function and synaptic plasticity. J Neurosci 31:16241–16250. 10.1523/JNEUROSCI.3667-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosing S, Thiadens AA, Hoyng CB, Klaver CC, den Hollander AI, Cremers FP (2014) Causes and consequences of inherited cone disorders. Prog Retin Eye Res 42:1–26. 10.1016/j.preteyeres.2014.05.001 [DOI] [PubMed] [Google Scholar]
- Santos AM, Calvente R, Tassi M, Carrasco MC, Martín-Oliva D, Marín-Teva JL, Navascués J, Cuadros MA (2008) Embryonic and postnatal development of microglial cells in the mouse retina. J Comp Neurol 506:224–239. 10.1002/cne.21538 [DOI] [PubMed] [Google Scholar]
- Sato E, Shick HE, Ransohoff RM, Hirose K (2010) Expression of fractalkine receptor CX3CR1 on cochlear macrophages influences survival of hair cells following ototoxic injury. J Assoc Res Otolaryngol 11:223–234. 10.1007/s10162-009-0198-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, Ransohoff RM, Greenberg ME, Barres BA, Stevens B (2012) Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 74:691–705. 10.1016/j.neuron.2012.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedel F, Béchade C, Vyas S, Triller A (2004) Macrophage-derived tumor necrosis factor alpha, an early developmental signal for motoneuron death. J Neurosci 24:2236–2246. 10.1523/JNEUROSCI.4464-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigemoto-Mogami Y, Hoshikawa K, Goldman JE, Sekino Y, Sato K (2014) Microglia enhance neurogenesis and oligodendrogenesis in the early postnatal subventricular zone. J Neurosci 34:2231–2243. 10.1523/JNEUROSCI.1619-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu X, Black GC, Rice JM, Hart-Holden N, Jones A, O'Grady A, Ramsden S, Wright AF (2007) RPGR mutation analysis and disease: an update. Hum Mutat 28:322–328. 10.1002/humu.20461 [DOI] [PubMed] [Google Scholar]
- Silverman MD, Zamora DO, Pan Y, Texeira PV, Baek SH, Planck SR, Rosenbaum JT (2003) Constitutive and inflammatory mediator-regulated fractalkine expression in human ocular tissues and cultured cells. Invest Ophthalmol Vis Sci 44:1608–1615. 10.1167/iovs.02-0233 [DOI] [PubMed] [Google Scholar]
- Steinberg RH, Fisher SK, Anderson DH (1980) Disc morphogenesis in vertebrate photoreceptors. J Comp Neurol 190:501–508. 10.1002/cne.901900307 [DOI] [PubMed] [Google Scholar]
- Timmers AM, Fox DA, He L, Hansen RM, Fulton AB (1999) Rod photoreceptor maturation does not vary with retinal eccentricity in mammalian retina. Curr Eye Res 18:393–402. 10.1076/ceyr.18.6.393.5263 [DOI] [PubMed] [Google Scholar]
- Tremblay MÈ, Lowery RL, Majewska AK (2010) Microglial interactions with synapses are modulated by visual experience. PLoS Biol 8:e1000527. 10.1371/journal.pbio.1000527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojan P, Krauss N, Choe HW, Giessl A, Pulvermüller A, Wolfrum U (2008) Centrins in retinal photoreceptor cells: regulators in the connecting cilium. Prog Retin Eye Res 27:237–259. 10.1016/j.preteyeres.2008.01.003 [DOI] [PubMed] [Google Scholar]
- Tuo J, Smith BC, Bojanowski CM, Meleth AD, Gery I, Csaky KG, Chew EY, Chan CC (2004) The involvement of sequence variation and expression of CX3CR1 in the pathogenesis of age-related macular degeneration. FASEB J 18:1297–1299. 10.1096/fj.04-1862fje [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno M, Fujita Y, Tanaka T, Nakamura Y, Kikuta J, Ishii M, Yamashita T (2013) Layer V cortical neurons require microglial support for survival during postnatal development. Nat Neurosci 16:543–551. 10.1038/nn.3358 [DOI] [PubMed] [Google Scholar]
- Vessey KA, Greferath U, Jobling AI, Phipps JA, Ho T, Waugh M, Fletcher EL (2012) Ccl2/Cx3cr1 knockout mice have inner retinal dysfunction but are not an accelerated model of AMD. Invest Ophthalmol Vis Sci 53:7833–7846. 10.1167/iovs.12-10650 [DOI] [PubMed] [Google Scholar]
- Vessey KA, Greferath U, Aplin FP, Jobling AI, Phipps JA, Ho T, De Iongh RU, Fletcher EL (2014) Adenosine triphosphate-induced photoreceptor death and retinal remodeling in rats. J Comp Neurol 522:2928–2950. 10.1002/cne.23558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waisman A, Liblau RS, Becher B (2015) Innate and adaptive immune responses in the CNS. Lancet Neurol 14:945–955. 10.1016/S1474-4422(15)00141-6 [DOI] [PubMed] [Google Scholar]
- Wake H, Moorhouse AJ, Jinno S, Kohsaka S, Nabekura J (2009) Resting microglia directly monitor the functional state of synapses in vivo and determine the fate of ischemic terminals. J Neurosci 29:3974–3980. 10.1523/JNEUROSCI.4363-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao L, Zhang J, Fariss RN, Ma W, Kretschmer F, Wang M, Qian HH, Badea TC, Diamond JS, Gan WB, Roger JE, Wong WT (2016) Requirement for microglia for the maintenance of synaptic function and integrity in the mature retina. J Neurosci 36:2827–2842. 10.1523/JNEUROSCI.3575-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells CA, Mosbergen R, Korn O, Choi J, Seidenman N, Matigian NA, Vitale AM, Shepherd J (2013) Stemformatics: visualisation and sharing of stem cell gene expression. Stem Cell Res 10:387–395. 10.1016/j.scr.2012.12.003 [DOI] [PubMed] [Google Scholar]
- Weymouth AE, Vingrys AJ (2008) Rodent electroretinography: methods for extraction and interpretation of rod and cone responses. Prog Retin Eye Res 27:1–44. 10.1016/j.preteyeres.2007.09.003 [DOI] [PubMed] [Google Scholar]
- Won J, Gifford E, Smith RS, Yi H, Ferreira PA, Hicks WL, Li T, Naggert JK, Nishina PM (2009) RPGRIP1 is essential for normal rod photoreceptor outer segment elaboration and morphogenesis. Hum Mol Genet 18:4329–4339. 10.1093/hmg/ddp385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Tuo J, Cao X, Shen D, Li W, Chan CC (2013) Early degeneration of photoreceptor synapse in Ccl2/Cx3cr1-deficient mice on Crb1(rd8) background. Synapse 67:515–531. 10.1002/syn.21674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Xu G, Liu W, Ni Y, Zhou W (2012) Role of fractalkine/CX3CR1 interaction in light-induced photoreceptor degeneration through regulating retinal microglial activation and migration. PLoS One 7:e35446. 10.1371/journal.pone.0035446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Shimoya K, Temma K, Kimura T, Tsujie T, Shioji M, Wasada K, Fukui O, Hayashi S, Kanagawa T, Kanzaki T, Koyama M, Murata Y (2004) Expression of fractalkine in the fallopian tube and of CX3CR1 in sperm. Hum Reprod 19:409–414. 10.1093/humrep/deh085 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Hong DH, Pawlyk B, Yue G, Adamian M, Grynberg M, Godzik A, Li T (2003) The retinitis pigmentosa GTPase regulator (RPGR)-interacting protein: subserving RPGR function and participating in disk morphogenesis. Proc Natl Acad Sci U S A 100:3965–3970. 10.1073/pnas.0637349100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieger M, Ahnelt PK, Uhrin P (2014) CX3CL1 (fractalkine) protein expression in normal and degenerating mouse retina: in vivo studies. PLoS One 9:e106562. 10.1371/journal.pone.0106562 [DOI] [PMC free article] [PubMed] [Google Scholar]