Abstract
The serotonin-1A (5-HT1A) receptor is a key regulator of serotonergic activity and is implicated in mood and emotion. However, its post-transcriptional regulation has never been studied in humans. In the present study, we show that the “intronless” human 5-HT1A gene (HTR1A) is alternatively spliced in its 3′-UTR, yielding two novel splice variants. These variants lack a ∼1.6 kb intron, which contains an microRNA-135 (miR135) target site. Unlike the human HTR1A, the mouse HTR1A lacks the splice donor/accepter sites. Thus, in the mouse HTR1A, splicing was not detected. The two spliced mRNAs are extremely stable, are resistant to miR135-induced downregulation, and have greater translational output than the unspliced variant. Moreover, alternative HTR1A RNA splicing is oppositely regulated by the splice factors PTBP1 and nSR100, which inhibit or enhance its splicing, respectively. In postmortem human brain tissue from both sexes, HTR1A mRNA splicing was prevalent and region-specific. Unspliced HTR1A was expressed more strongly in the hippocampus and midbrain versus the prefrontal cortex (PFC), and correlated with reduced levels of nSR100. Importantly, HTR1A RNA splicing and nSR100 levels were reduced in the PFC of individuals with major depression compared with controls. Our unexpected findings uncover a novel mechanism to regulate HTR1A gene expression through alternative splicing of microRNA sites. Altered levels of splice factors could contribute to changes in regional and depression-related gene expression through alternative splicing.
SIGNIFICANCE STATEMENT Alternative splicing, which is prevalent in brain tissue, increases gene diversity. The serotonin-1A receptor gene (HTR1A) is a regulator of serotonin, which is implicated in mood and emotion. Here we show that human HTR1A RNA is alternately spliced. Splicing removes a microRNA site to generate ultrastable RNA and increase HTR1A expression. This splicing varies in different brain regions and is reduced in major depression. We also identify specific splice factors for HTR1A RNA, showing they are also reduced in depression. Thus, we describe a novel mechanism to regulate gene expression through splicing. Altered levels of splice factors could contribute to depression by changing gene expression.
Keywords: 5-HT1A receptor, alternative splicing, major depression, RNA splice factors, RNA stability, serotonin
Introduction
The 5-HT1A receptor is widely expressed throughout the brain, both as a somatodendritic autoreceptor that negatively regulates the activity of midbrain 5-HT neurons and as a key postsynaptic heteroreceptor that mediates downstream 5-HT actions (Albert and Lemonde, 2004). Mouse models have demonstrated that the expression of presynaptic and postsynaptic 5-HT1A receptors dictate depression-like and anxiety-like behavior, as well as response to antidepressants (Santarelli et al., 2003; Richardson-Jones et al., 2010, 2011; Samuels et al., 2015; Vahid-Ansari et al., 2017). Suppression of 5-HT1A autoreceptors increases 5-HT neuronal activity and enhances stress resilience/antidepressant response, while their overexpression has the opposite effect (Richardson-Jones et al., 2010; Vahid-Ansari et al., 2017). Postsynaptic 5-HT1A heteroreceptors in the hippocampus are critical for antidepressant response, while those in the prefrontal cortex (PFC) provide protection from anxiety and depression phenotypes (Albert et al., 2014; Samuels et al., 2015). In humans, reductions in postsynaptic 5-HT1A receptors, especially in the PFC, are associated with depression and anxiety (Savitz et al., 2009; Szewczyk et al., 2009). Conversely, an increase in presynaptic 5-HT1A receptors is seen in depression and depressed suicides and correlates with resistance to treatment with antidepressant drugs (Hesselgrave and Parsey, 2013). However, the molecular basis for these dynamic changes in 5-HT1A receptor expression remains elusive.
At the level of the human 5-HT1A gene (HTR1A), a functional C(-1019)G promoter polymorphism (rs6295) associated with depression, mood disorders, suicide risk, and antidepressant resistance, alters transcription-factor binding to affect HTR1A expression in vivo (Lemonde et al., 2003, 2004; Le François et al., 2008; Donaldson et al., 2016). However, although meta-analyses have confirmed these associations, they are not seen in all cohorts, suggesting that additional regulatory mechanisms control 5-HT1A gene expression. While the HTR1A promoter and coding sequences have been intensively studied (Albert et al., 2014), the gene's full mRNA sequence, including the 3′-untranslated regions (3′-UTR), remains unknown. However, within the HTR1A 3′-UTR, a site for microRNA-135 (miR135) has been identified and miR135 has been shown to regulate 5-HT1A autoreceptor levels, 5-HT system activity, and antidepressant action (Issler et al., 2014).
Alternative splicing is a prevalent post-transcriptional modification, especially in human neuronal genes (Kang et al., 2011), resulting in a greater diversity of RNA transcripts (Darnell, 2013; Raj and Blencowe, 2015). Alternative splicing is regulated by RNA binding proteins that dictate exon inclusion or skipping (Lee and Rio, 2015). These include Nova1 and Nova2, the ubiquitous poly-pyrimidine tract binding protein 1 (PTBP1) and its neuronal paralog PTBP2 (nPTB), or the RNA splicing domain protein nSR100, which positively regulates a vast splicing regulatory network critical for neuronal differentiation and function (Calarco et al., 2009; Irimia et al., 2014). In the present study, we are the first to demonstrate that the human 5-HT1A mRNA 3′-UTR is alternatively spliced to yield three mRNA isoforms that differ drastically in their half-lives and translational output. Interestingly, expression levels of the 5-HT1A mRNA variants vary across brain regions in humans and correlate with nSR100 levels, indicating that this molecular event is dynamically regulated. We show that this novel splicing event is regulated by PTBP1 and nSR100 in an antagonistic fashion. Also, the 5-HT1A RNA splicing ratio is decreased in the PFC of depressed individuals, correlating with a decrease in the expression of nSR100. These studies highlight the importance of alternative splicing and splice factors in the regulation of gene expression in the brain, and show how regulation of global splicing may be altered in depression.
Materials and Methods
Plasmids, reporter constructs, and 3′RACE.
The 3056 bp human 5-HT1A 3′-UTR was amplified by PCR from HEK293 genomic DNA using primers 5′-CAGTGATGACGGAGGAGTAGCC and 5′-CCAGAGACTGTAGTTTTCCTCCAT with NheI/SalI sites and cloned in XbaI/SalI-cut pGL3P (Promega, catalog #E1761), replacing the SV40 polyadenylation site, generating pGL3P-HTR1A 3′-UTR [wild type (WT)]. Our constructs correspond to the WT human 5-HT1A genotype (Rs6449693-T, Rs878567-C, Rs749099-A). For 3′RACE and transfections, RNA was extracted 48 h post-transfection, DNase treated, and cDNA synthesized using (dT)20 primer (Integrated DNA Technologies, catalog #51-01-15-01). The 5-HT1A 3′UTR was amplified by PCR using the primers 5′-GACGAAGTACCGAAAGGT and 5′-CACTGGAGTTGTCCCAATTC. Products were cloned in pGEM-TEasy (Promega, catalog #A1360) and sequenced. The pGL3P-unspliceable 5-HT1A-3′-UTR construct was obtained by mutating the 3′-splice acceptor using primers fwd 5′-CCATTTATTTGTGTCTTTTACTCAATTGTATGACAATG and rev 5′-CATTGTCATACAATTGAGTAAAAGACACAAATAAATGG (mutation in boldface). The pGL3P-spliced 5-HT1A 3′-UTR construct was obtained by subcloning BamHI/BpmI-digested 5-HT1A 3′-UTR [long spliced (LS)] 814 bp fragment into pGL3P 5-HT1A 3′UTR. Nova1, Nova2, nPTB, and nSR100 cDNAs were amplified by PCR from mouse brain cDNA and cloned in SmaI/HindIII-digested pTRIEX4 (Millipore-Sigma, catalog #70824-3).
Cell lines, transfections, and luciferase assays.
HEK293, 293T, and SKN-SH cells were maintained in DMEM supplemented with 10% FBS in 5% CO2 at 37°C. For RNA/protein extraction, cells were transfected with plasmids (1.5 μg) using polyethylenimine and harvested after 36–48 h. For luciferase assays, cells were cotransfected with 750 ng of luciferase constructs and 750 ng of pCMV β-galactosidase using Lipofectamine 2000 (Life Technologies) and harvested after 36–48 h in reporter lysis buffer (Promega). Similarly, 50 nm of either miR135b or control mimic (Dharmacon) were cotransfected with 1 μg of the indicated reporter constructs in SKN-SH and HEK293 cells using Lipofectamine 2000 (Thermo Fisher Scientific, catalog #11668019) and harvested 36–48 h later in reporter lysis buffer. Luciferase activity was normalized to β-galactosidase activity measured using chlorophenol red-β-D-galactoside (Santa Cruz Biotechnology, catalog #sc-257242).
Lentiviruses and 5-HT1A minigenes.
WT 5-HT1A minigene containing 4316-bp 5-HT1A ORF and 3′UTR was obtained by PCR amplification of HEK293 gDNA using primers fwd 5′-ATGGATGTGCTCAGCCCT and rev 5′-CCAGAGACTGTAGTTTTCCTCCAT, containing 5′-Flag epitope and XhoI site and 3-SnaBI site, and cloned in XhoI/SnaBI-cut pWPXLd (Addgene, catalog #12258). The unspliceable and spliced minigenes were obtained by replacing the WT 3′-UTR with the respective BamHI/SbfI fragments of pGL3-P constructs. The 1269 bp 5-HT1A ORF was amplified from HEK293 genomic DNA using primers fwd 5′-ATGGATGTGCTCAGCCCT and rev 5′-TCACTGGCGGCAGAACTT, with Flag-epitope/PmeI and EcoRI sites, and cloned in PmeI/EcoRI-cut pWPXLd.
For virus production, 293T cells were transfected with pWPXLd or pGIPZ shRNA clones with pMDG2 and psPAX2 using polyethylenimine. Supernatants were harvested 48 h post-transfection, concentrated using Lenti-X concentrator (Clontech, catalog #631231), and titered using a p24 ELISA kit (Clontech, catalog #632200). Cells were infected in the presence of 10 μg/ml Polybrene (Santa Cruz Biotechnology, catalog #sc-134220). For PTBP1 and PTBP2 knockdown, cells stably expressing the shRNAs (GE Healthcare, pGIPZ clones V3LHS_645699 and V3LHS_635160 respectively) were selected with 2 μg/μl puromycin (Thermo Fisher Scientific, catalog #A1113802) for ≥1 week.
Western blot.
Cells were lyzed in 1% SDS supplemented with protease inhibitors and sonicated for 20–30 s. Protein concentration was determined with the BCA assay (Thermo Fisher Scientific, catalog #23225). Then, 30–80 μg of total protein was resolved on a 10% SDS PAGE gel and transferred to a 0.45 μm nitrocellulose or PVDF membrane. Following blocking with 5% milk in PBST, membranes were incubated overnight with antibodies against FLAG epitope (1:1000; Sigma-Aldrich, catalog #F1804, RRID:AB_262044), S-Tag (1:1000; Cell Signaling Technology, catalog #8476, RRID:AB_10949895), PTBP1 (1:1000; Santa Cruz Biotechnology, catalog #sc-16547, RRID:AB_2253470), PTBP2 (1:2000; Abcam, catalog #ab57619, RRID:AB_2284865), and α-tubulin (1:5000; Sigma-Aldrich, catalog #T6074, RRID:AB_477582). After five washes in PBST, membranes were incubated 1 h with anti-goat HRP (1:2500; Santa Cruz Biotechnology, catalog #sc-2020, RRID:AB_631728), anti-rabbit HRP (1:2500; Santa Cruz Biotechnology, catalog #sc-2004, RRID:AB_631746), or anti-mouse-HRP (1:2500; Santa Cruz Biotechnology, catalog #sc-2371, RRID:AB_634824) secondary antibodies, washed five more times in PBST, and then developed using the ECL Prime detection reagent (GE Healthcare, catalog #RPN2232). Densitometry analysis was performed using National Institutes of Health ImageJ software.
Human brain samples.
Postmortem brain tissues from subjects of both sexes (Table 1) were collected at autopsy at the Cuyahoga County Medical Examiner's Office, Cleveland, Ohio. The Institutional Review Boards of the University of Mississippi Medical Center and University Hospitals Case Medical Center approved the protocol for recruitment, tissue collection, and interviews of the next-of-kin. Written informed consent was secured from legally defined next-of-kin. For the study of the PFC, 15 depressed subjects met criteria for major depressive disorder and one subject met criteria for adjustment disorder with depressed mood. The structured clinical interview for Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (DSM-IV) Axis I disorders (First et al., 2002) was administered to knowledgeable informants for all subjects and diagnoses were made according to the DSM-IV (American Psychiatric Association, 1994), as described previously (Mahajan et al., 2018). All the depressed subjects were experiencing depressed mood in the last month of life, and none were diagnosed with a comorbid psychoactive substance-use disorder. Tissues were also sampled from psychiatrically normal control subjects matched for age and postmortem interval (Table 1). For PFC tissues, toxicology screen of the depressed cohort revealed the following: carbon monoxide (two), diazepam (one), and fluoxetine, nortriptyline, or sertraline (one each). Gray matter was dissected with a scalpel from frozen blocks of the PFC (areas 8 and 9), placed in an RNase-free and DNase-free cold centrifuge tube on dry ice, and then stored at −80°C. Frozen sections (60 μm) were collected from the midbrain and hippocampus. A 2 mm punch was used to isolate the dorsal raphe nucleus in the midbrain sections, and the hippocampal sections included the dentate gyrus and cornu ammonis subfields. The RNA integrity number of 90% of the PFC samples was between 4 and 8, assessed using RNA6000 picochip on 2100 bioanalyzer (Agilent).
Table 1.
Demographics of the human brain samples
| PFC |
Hippocampus | Midbrain | ||
|---|---|---|---|---|
| Control | Depressed | |||
| Age (years) | 43.84 (±4.77) | 49.47 (±4.47) | 47.67 (±4.47) | 49.11 (±5.73) |
| Sex | 11 males, 8 females | 11 males, 4 females | 4 males, 8 females | 4 males, 5 females |
| Postmortem interval (hours) | 20.22 (±1.85) | 20.4 (±1.77) | 22.78 (±2.50) | 22.18 (±3.32) |
| RNA integrity number | 5.35 (±0.24) | 5.18 (±0.33) | 5.31 (±0.36) | 5.88 (±0.22) |
Age, post-mortem interval, RNA quality, and sex distribution of the subjects whose brain samples were used to quantify 5-HT1A alternative splicing across the human brain regions from control and depressed subjects. Hippocampus and midbrain were a from subset of the same controls as for PFC. For each region data is presented as mean ± SEM.
RNA extraction, cDNA synthesis, RT-PCR, and real-time PCR.
RNA was extracted from cells and tissue using TRI reagent (Thermo Fisher Scientific, catalog #AM9738), quantified, and treated with TURBO DNase (Thermo Fisher Scientific, catalog #AM2238). cDNA was synthesized and analyzed by RT-PCR and real-time PCR. Unspliced and spliced 5-HT1A 3′-UTR PCR amplifications were performed with TaqDNA polymerase in ThermoPol buffer (New England Biolabs, catalog #M0267) at 95°C for 30 s; 35 cycles of 95°C for 15 s; 50°C for 30 s, and 72°C for 20 s using the following primers: unspliced fwd 5′-GAACTTTGGGAATAGTTTGTC; unspliced rev 5′-GGCTTTACTTAGATTATGTGAGC; spliced fwd 5′-CCTATTTCCTTTGTTTCC; spliced rev 5′-GTTGGATGTTCCTTCTCC. PCR products were resolved on a 3% agarose gel. Real-time PCR was performed using PrimeTime FAM-labeled probes (Integrated DNA Technologies; LS probe 5′-AGGTATCTTTTAGAGTGGACTTAATTGT; total-spliced probe 5′-TGACTTTTGGACATTTGTTCTTTCGG; unspliced probe 5′-GCTGAGAAGGGTCTGCAGTG). Real-time PCR quantifications of total 5-HT1A, PTBP1/2, nSR100, and GAPDH RNA were performed using TaqMan Gene expression assay kits (Thermo Fisher Scientific) for human 5-HT1A (Hs00265014_s1), PTBP1 (Hs00243060p_m1), PTBP2 (Hs00221842_m1), nSR100 (Hs00916552_m1), and GAPDH (Hs02758991_g1) in a 20 μl reaction. Relative quantification was performed using the ΔΔCt method (Livak and Schmittgen, 2001), using GAPDH as a reference gene.
Statistical analysis.
All analyses were done using the Statistical Package for the Social Sciences (GraphPad Prism version 6.00 for Windows, GraphPad Software, www.graphpad.com). Data are expressed as mean ± SEM. For analysis of splicing in brain samples, Rout test of spliced/unspliced ratios was used to exclude two outliers in the hippocampal spliced dataset; no other data were excluded. Data comparing depressed versus control were analyzed using an unpaired t test. One-way ANOVA followed by Tukey's post hoc test was performed for comparing all other data.
Results
Human HTR1A alternative splicing enhances its RNA stability and translation
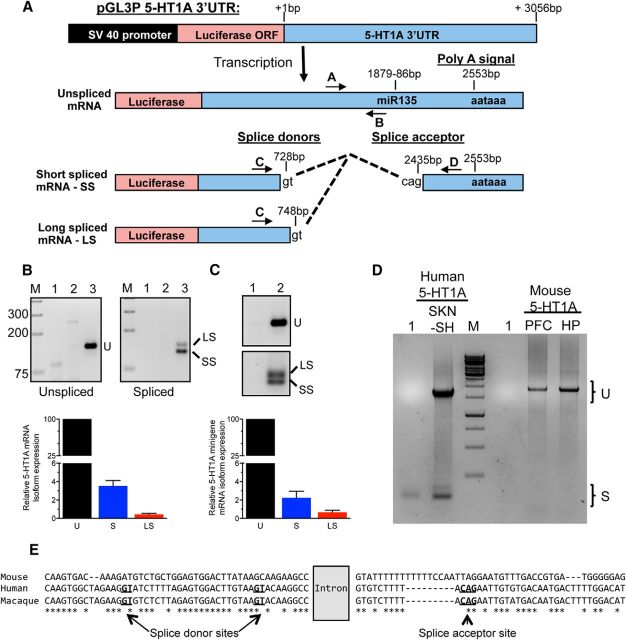
Although cloned 30 years ago (Fargin et al., 1988), the full sequence of the human 5-HT1A 3′-UTR remains unknown. Because of its potential role in RNA stability, the 3056 bp human HTR1A gene 3′-UTR was cloned downstream of the luciferase reporter (Fig. 1A) and transfected in human HEK293 cells. The RNA produced was characterized by 3′-RACE. An 885-bp-long 5-HT1A-specific RNA was identified containing the consensus polyadenylation signal located at 2553 bp but lacking a 1.6 kb intron (Fig. 1A). Since the 5-HT1A gene is considered intronless (Fargin et al., 1988; Albert et al., 1990), further RT-PCR analyses were performed. These showed that the 5-HT1A 3′-UTR is alternatively spliced to generate an unspliced isoform (2573 bp) and two spliced isoforms (885 bp LS and 865 bp SS), which use the same splice acceptor site but different splice donor sites located 20 bp apart (Fig. 1A,B). To assess the extent of HTR1A splicing, we quantified spliced and unspliced 5-HT1A RNA levels in transfected HEK293 cells using real-time PCR and found that spliced variants represented 5% of total 5-HT1A RNA (Fig. 1B). Similar results were obtained in SKN-SH human neuroblastoma cells (data not shown). To confirm whether splicing occurs in the presence of the 5-HT1A coding sequence, a lentiviral 5-HT1A minigene containing the full 5-HT1A coding and 3′-UTR sequences was stably integrated in SKN-SH cells. Under these conditions, splicing also accounted for 4% of total 5-HT1A mRNA levels (Fig. 1C). In contrast to the human HTR1A RNA, a single unspliced mRNA species in mouse PFC and hippocampus was obtained by RT-PCR (Fig. 1D). The lack of splicing is consistent with nonconserved splice donor/acceptor sites in the mouse compared with primate 3′-UTR sequences (Fig. 1E). Thus, the human, but not mouse, HTR1A gene is alternatively spliced to generate three transcripts.
Figure 1.
The 5-HT1A mRNA 3′-UTR is alternatively spliced in human cells to give rise to several mRNA isoforms. A, Schematic representation of the human 5-HT1A 3′-UTR construct used for 3′-RACE experiments, and of primers A–D used to quantify the resulting mRNA variants upon its transfection in human cells (unspliced, LS, and SS). Dashed lines represent the intron removed following splicing of the 3′-UTR. The relative location of the poly-adenylation site (polyA signal), miR135 site, and splice donor (gt dinucleotides) and acceptor (cag) sites are indicated. B, Top, RT-PCR analysis of the 5-HT1A 3′-UTR splice variants in HEK293 cells. RNA from HEK293 cells transfected with luciferase vector pGL3P or pGL3P 5-HT1A-3′-UTR was assessed by RT-PCR for the expression of unspliced (U, top, primers A/B) and spliced 3′UTR isoforms, short (SS) and long (LS) (bottom, primers C/D). M, DNA size markers; 1, negative control; 2, pGL3P; 3, pGL3P 5-HT1A-3′-UTR. Bottom, Real-time PCR quantification of unspliced (100%) and spliced HTR1A mRNA in HEK293 cells transfected with pGL3P 5-HT1A-3′-UTR using probes specific for unspliced, total spliced (S), or LS isoform 5-HT1A mRNA. C, Splicing of human 5-HT1A minigene in stably transfected SKN-SH cells. Top, RT-PCR: cDNA from uninfected cells (Lane 1) or cells infected with the 5-HT1A minigene (Lane 2) were analyzed for the unspliced 3′-UTR (U) or spliced isoforms (SS, LS) using primers shown in A. Below, real-time PCR: relative mRNA levels were normalized to GAPDH using the ΔΔCt method. Data are presented as mean ± SEM of the %unspliced from three independent experiments. D, Undetectable HTR1A RNA splicing in mouse brain. Full-length cDNA from human SKN-SH cells expressing the human 5-HT1A minigene or mouse PFC and hippocampus (HP) tissue was analyzed using two human primers C and D in A or the corresponding mouse 3′-UTR primers (521–2444 bp downstream of the stop codon). Spliced 5-HT1A mRNA isoforms (S) were only detected for the human transcript, while the unspliced form (U) was present in both. M, DNA size markers; 1, negative control. E, Sequence conservation of the mouse, human, and rhesus macaque 5-HT1A 3′-UTR sequences (701/2333 bp; 721/2462 bp; 719/2476 bp, respectively). The splice acceptor and donor sites are in bold and underlined; dashes indicate gaps; * indicates conservation among all three species. Alignment was performed with the multiple sequence comparison by log-expectation [Multiple Sequence Comparison by Log-Expectation (MUSCLE)] at the European Bioinformatics Institute (EMBL-EBI) website (https://www.ebi.ac.uk/Tools/msa/muscle/).
Because splicing of the 3′-UTR typically generates unstable mRNAs that can be targeted by the nonsense-mediated decay (NMD) pathway (Chang et al., 2007), we analyzed the relative stability of the three HTR1A mRNA variants. SKN-SH cells were transduced with the WT 5-HT1A minigene construct and treated with transcription blocker actinomycin D. Then levels of HTR1A RNA were determined by RT-PCR. Unexpectedly, the unspliced mRNA variant had a very short half-life (<30 min), while the spliced mRNA variants were extremely stable, with half-lives of >12 h (Fig. 2A). To assess the importance of splicing on 5-HT1A expression, we compared the expression of 5-HT1A minigenes or reporter constructs containing WT, unspliceable, or constitutively spliced (LS variant) 3′-UTR sequences, leading to specific expression of each RNA species (Fig. 2B,C). SKN-SH (Fig. 2B) and HEK293 cells (data not shown) were transfected with these reporter constructs. The constitutive-spliced 3′-UTR displayed a 2.5-fold increase in luciferase activity compared with the WT or unspliceable constructs (F(3,14) = 67.01, p < 0.0001, ANOVA), which is consistent with the increased RNA stability of the spliced variant (Fig. 2A). Similarly, infection of SKN-SH cells with the constitutively spliced 5-HT1A minigene led to a significant increase in the expression of 5-HT1A receptor protein compared with the WT or unspliceable minigenes (F(2,6) = 93.41, p < 0.0001, ANOVA; Fig. 2D,E), which is similar to levels achieved with the highly stable SV40 3′-UTR in the ORF construct. These data demonstrate that the spliced 5-HT1A mRNA variants are more stable than the full-length mRNA and lead to increased receptor expression in human cells.
Figure 2.
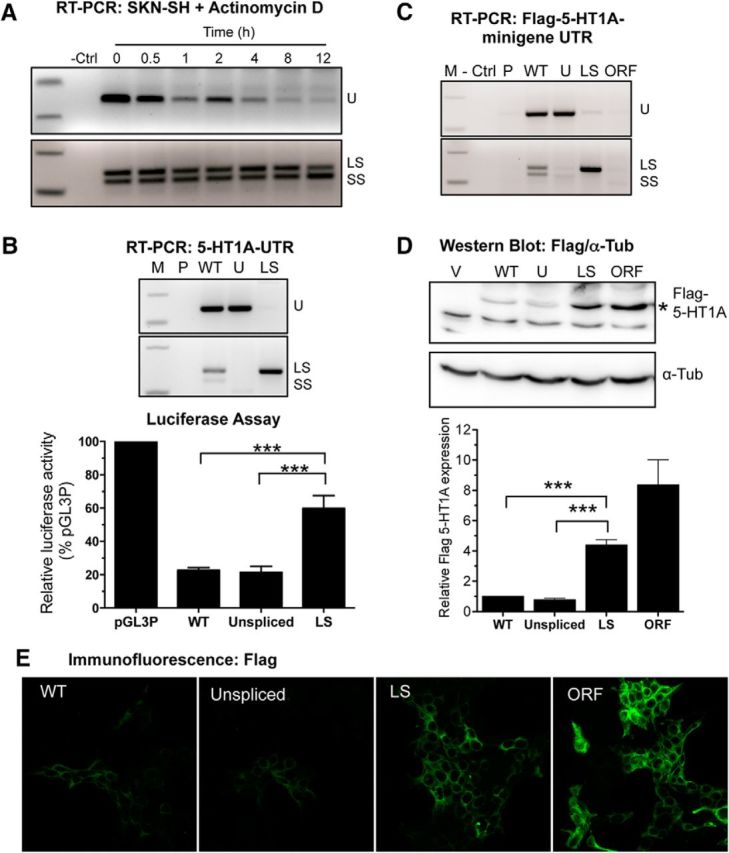
The 5-HT1A spliced mRNA variants display increased stability and lead to greater protein expression. A, Relative stability of the unspliced (U) and spliced (LS, SS) mRNA isoforms detected by RT-PCR (Fig. 1B) in SKN-SH cells infected with the 5-HT1A WT minigene lentivirus and treated with 10 μg/ml actinomycin D to inhibit de novo transcription. B, Increased luciferase activity from spliced 5-HT1A 3′-UTR. Luciferase constructs containing either the vector (P, pGL3P), WT, unspliceable (U, Unspliced) or constitutively spliced (LS, Spliced) human 5-HT1A 3′-UTR were analyzed (M, DNA size markers). Top, RT-PCR analysis shows equivalent expression of the appropriate 5-HT1A 3′-UTR isoform in SKN-SH cells transfected with the indicated construct. Bottom, SKN-SH cells cotransfected with the indicated construct and pCMV β-galactosidase were assayed for relative luciferase activity, normalized to pGL3P (100%). Data shown as mean ± SEM of 3–5 experiments. ***p < 0.001 one-way ANOVA with Tukey's post hoc test. C–E, Relative expression of Flag-5-HT1A 3′-UTR minigene isoforms. RT-PCR analysis shows equivalent expression of the appropriate 5-HT1A 3′-UTR isoform in SKN-SH cells infected at multiplicity of infection 20 for 72 h with the indicated construct (C). D, Top, Western blot for Flag-5-HT1A fusion protein (*) or tubulin (α-Tub, below) in SKN-SH cells infected with vector (V), WT, U, or LS 3′-UTR minigenes or 5-HT1A coding sequence with SV40 3′-UTR (ORF). D, Bottom, Densitometry analysis of Flag-5-HT1A protein levels relative to tubulin. E, Immunofluorescence for Flag-tag. For densitometry, mean ± SEM from three independent experiments is shown. ***p < 0.001. One-way ANOVA, Tukey's post hoc test.
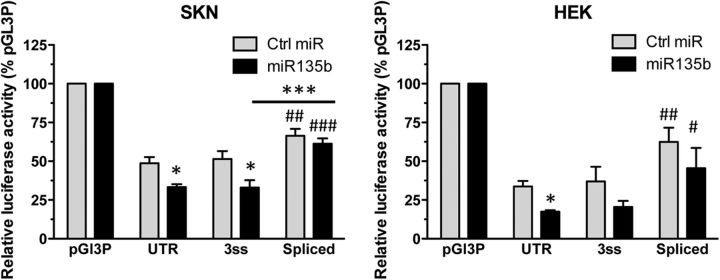
The splicing-induced increase in RNA stability could be mediated by an excision of miR sites, since miRs are known to destabilize RNA transcripts. In a screen for 5-HT neuron-specific miRs, miR135 was identified and shown to repress the HTR1A 3′-UTR at a target site located from 1879 to 1886 nt (Issler et al., 2014). Since this site is located within the intron excised in the HTR1A spliced variants (Fig. 1A), we tested whether miR135-induced repression was affected by splicing (Fig. 3). In both SKN-SH and HEK293 cells, miR135 suppressed the WT and unspliceable HTR1A 3′-UTR, but not the constitutively spliced version. Also, in both cell lines, the luciferase activity of the spliced HTR1A was greater than the WT UTR, with or without miR135 treatment. These data indicate that loss of the miR135 binding site following splicing contributes to the increased stability of the spliced HTR1A RNA.
Figure 3.
Resistance of spliced 5-HT1A 3′-UTR to miR135. SKN-SH or HEK-293 cells were transfected with luciferase constructs: vector (pGL3P), WT, unspliceable (U, Unspliced), or constitutively spliced (LS, Spliced) human 5-HT1A 3′-UTR, treated with control miR (Ctrl) or miR135, and analyzed for relative luciferase activity normalized to pGL3P (100%). Data are shown as mean ± SEM of three experiments. *p < 0.05, ***p < 0.001 versus Ctrl or as shown; ##p < 0.01, ###p < 0.001 vs WT; one-way ANOVA, Tukey's post hoc test.
HTR1A mRNA splicing is oppositely regulated by PTBP1 and nSR100
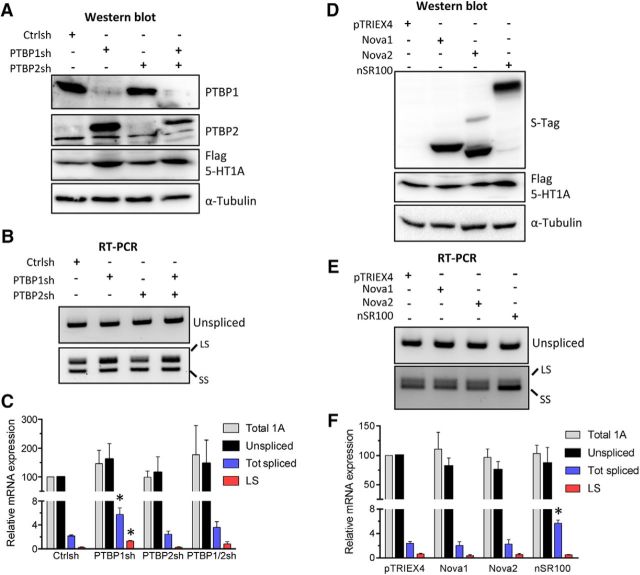
To better understand the mechanisms regulating 5-HT1A mRNA splicing, we examined the roles of specific RNA binding proteins, including the ubiquitous poly-pyrimidine-tract binding proteins (PTBP1, PTBP2), which usually repress neuronal-specific splicing (Wagner and Garcia-Blanco, 2001; Boutz et al., 2007). In SKN-SH cells expressing the WT 5-HT1A minigene, shRNA-mediated knockdown of PTBP1, but not PTBP2, induced a twofold increase in 5-HT1A receptor protein levels (Fig. 4A). Knockdown of PTBP1 also increased PTBP2 protein (Fig. 4A), which is consistent with previous findings (Boutz et al., 2007). Knockdown of PTBP1, but not PTBP2, led to a significant increase in total spliced and LS variant mRNA (F(3,12) = 4.325, p = 0.0276, ANOVA; Fig. 4B,C), which is consistent with the increase in 5-HT1A protein (Fig. 4A). Double knockdown of PTBP1 and PTBP2 did not further increase splicing or 5-HT1A receptor levels (Fig. 4A–C). This indicates that PTBP1, but not PTBP2, represses 5-HT1A mRNA splicing in SKN-SH cells.
Figure 4.
Opposite regulation of 5-HT1A mRNA splicing and protein levels by splicing factors PTBP1 and nSR100. A–C, Effect of PTBP1 and PTBP2 knockdown on 5-HT1A RNA splicing. SKN-SH cells were infected with lentiviruses expressing shRNAs against PTBP1, PTBP2, or a nontargeting control (Ctrl) shRNA at multiplicity of infection of 10 and selected using puromycin. Puromycin-resistant cells were infected with the WT 5-HT1A minigene at multiplicity of infection of 20 for 72 h and RNA and protein were extracted and analyzed. Relative expression of PTBP1, PTBP2, and Flag-5-HT1A proteins was determined by Western blot (A), while the expression of the various 5-HT1A mRNA variants was assessed by RT-PCR (B) and quantified by real-time PCR (C) using probes specific for the spliced mRNA isoforms (total spliced (Tot spliced) or LS isoform), unspliced mRNA isoforms, or total 5-HT1A mRNA (Total 1A). Relative mRNA levels were normalized to GAPDH using the ΔΔCt method. D–F, Effect of Nova1, Nova2, and nSR100 overexpression on 5-HT1A RNA splicing. SKN-SH cells were infected with the WT 5-HT1A minigene at multiplicity of infection of 20 for 72 h, and then transfected with vector (pTRIEX) or plasmids encoding the S-tagged Nova1, Nova2, and nSR100 neuronal splice regulators. The relative expression of the S-tagged proteins (S-tag), Flag-5-HT1A receptor and alpha-tubulin was determined by Western blot (D), while the expression of the various 5-HT1A mRNA isoforms was assessed by RT-PCR (E) and quantified by real-time PCR (F). Data presented as the mean and SEM of 3–4 independent experiments (C, F). *p < 0.05, one-way ANOVA, Tukey's post hoc test.
In silico analysis also identified consensus binding sites for the neuron-enriched splice regulators Nova1, Nova2, and nSR100 (SSRM4) near the HTR1A intron–exon boundary. To better understand the function of these regulators, they were overexpressed in SKN-SH cells stably expressing the WT 5-HT1A-3′-UTR minigene (Fig. 4D). Overexpression of nSR100 led to a twofold increase in the SS mRNA variant (F(3,8) = 9.426, p = 0.0053, ANOVA), while Nova1 and Nova2 overexpression had no detectable effect (Fig. 4E,F). Increased 5-HT1A splicing in nSR100-transfected cells was accompanied by an increase in 5-HT1A receptor levels (Fig. 4A). Collectively, these data demonstrate that 5-HT1A splicing and receptor expression is repressed by PTBP1 but enhanced by the neuron-enriched splice factor nSR100.
HTR1A mRNA splicing is dynamic in the human brain
To determine the extent of 5-HT1A RNA splicing in vivo, we measured and compared the expression of the 5-HT1A mRNA splice variants in the PFC (n = 19), hippocampus (n = 12), and midbrain (n = 9) of the same cohort of healthy subjects (Table 1). In human brain tissue the LS isoform was the only splice variant detected and splicing in brain tissues was greater than that observed in cell lines (Fig. 5A). Real-time PCR quantification showed a significant increase in unspliced mRNA levels in the midbrain and hippocampus compared with the PFC (F(2,37) = 14.46, p < 0.0001, ANOVA), with levels 3–4-fold greater (Fig. 5B). Splicing was highest in the PFC (30 ± 6%) compared with the hippocampus (14 ± 6%) and midbrain (22 ± 14%), but no significant difference in the spliced/unspliced ratio was detected due to the variability between samples (Fig. 5C).
Figure 5.
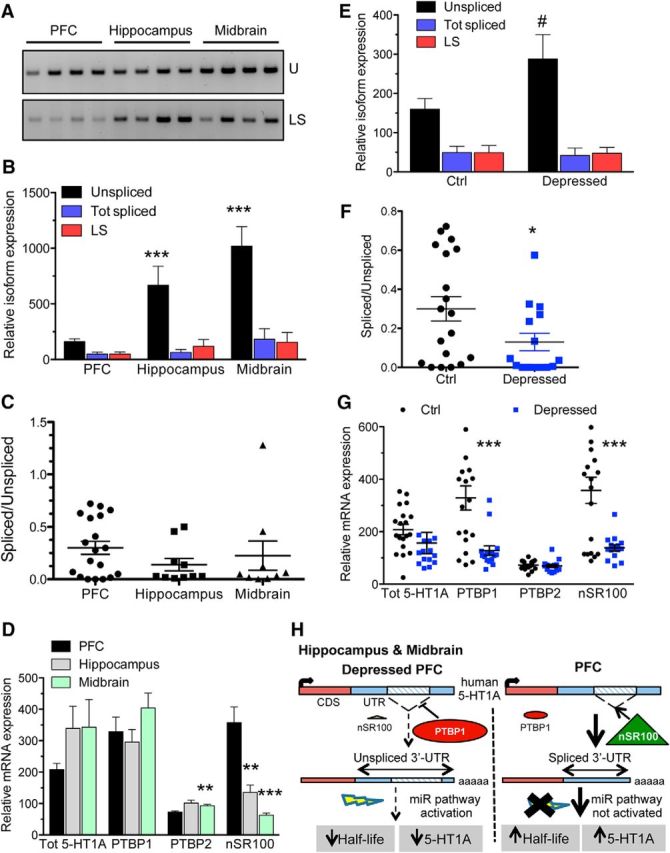
Changes in human brain regional 5-HT1A 3′-UTR splicing and splice factor RNA in depression. A–C, Regional splicing in the human brain. Relative expression of unspliced and spliced 5-HT1A mRNA isoforms was detected by RT-PCR (A) and quantified by real-time PCR (B) in human cDNA samples isolated from the PFC, hippocampus, and midbrain of control individuals. A, Unspliced and spliced 5-HT1A 3′-UTR sequences were amplified by PCR from brain cDNA samples using primers shown in Figure 1. B, The expression levels of 5-HT1A 3′-UTR unspliced, total spliced, or LS mRNA isoforms were quantified by qRT-PCR using specific probes for each mRNA variant and normalized to GAPDH RNA levels. Mean ± SEM ***p < 0.01 versus PFC unspliced, two-way ANOVA, Bonferroni post hoc test. C, Spliced/unspliced expression ratios. For each sample analyzed in B, the total spliced/unspliced expression ratio was calculated. Data presented as mean ± SEM for the PFC (n = 19), hippocampus (n = 10), and midbrain (n = 9). D, Relative expression levels of total 5-HT1A, PTBP1, PTBP2, and nSR100 mRNA in human PFC, hippocampus, and midbrain cDNA samples from controls were determined using specific Taqman probes and mRNA levels were quantified using the ΔΔCt method, with GAPDH RNA as a reference. n = 19 for the PFC, n = 10 for hippocampus, and n = 9 for midbrain for B and C. Mean ± SEM, **p < 0.01 and ***p < 0.001 versus PFC, one-way ANOVA, Tukey's post hoc test. E, F, Decreased 5-HT1A mRNA splicing in the PFC of depressed individuals. Relative expression of 5-HT1A mRNA isoforms in human PFC samples from control (Ctrl, n = 19) and depressed individuals (n = 15). E, The expression level of the unspliced, total spliced, or LS mRNA isoforms was quantified by qRT-PCR of brain cDNA samples and normalized to GAPDH levels using the ΔΔCt method. Mean ± SEM, #p = 0.0512, unpaired t test. F, For each sample analyzed in E, the total spliced/unspliced expression ratio was calculated. Mean ± SEM, *p < 0.05, unpaired t test. G, Relative expression levels of total 5-HT1A, PTBP1, PTBP2, and nSR100 mRNA in control versus depressed PFC, quantified by real-time PCR as in C. Mean ± SEM, ***p < 0.001 versus control, unpaired t tests. H, Model for the regulation of the 5-HT1A mRNA 3′-UTR alternative splicing. Left, In hippocampus, midbrain, and depressed PFC, splicing of the 5-HT1A mRNA 3′-UTR is prevented due to low levels of the neuronal splice enhancer nSR100 and by the recruitment of PTBP1, which inhibits exon inclusion. This generates the long 5-HT1A mRNA (2573 bp 3′-UTR), with a short half-life due to miR135 binding, leading to reduced 5-HT1A expression. Right, In the PFC, PTBP1 levels are low while nSR100 levels are high, increasing splicing to generate the short 5-HT1A mRNA (885 bp 3′-UTR), which is highly stable and evades the miR135 pathway, leading to higher expression of the 5-HT1A receptor.
Given the role of PTBP1 and nSR100 in regulation of 5-HT1A mRNA splicing, we measured their relative expression in the PFC, hippocampus, and midbrain. Significant differences in PTBP2 (F(2,37) = 7.046, p = 0.0026, ANOVA) and nSR100 expression (F(2,37) = 13.42, p < 0.0001, ANOVA) were observed, with PTBP2 expressed at higher levels in the hippocampus, and nSR100 expressed at higher levels in the PFC than in both other regions (Fig. 5D). No significant difference was seen in PTBP1 levels, while total 5-HT1A mRNA levels tended to be higher in the midbrain and the hippocampus, independently of splicing, likely reflecting greater transcriptional activity in these brain regions. Thus, 5-HT1A RNA splicing appears to mostly correlate with nSR100 and is robust in the PFC compared with the hippocampus and midbrain, where nSR100 expression is lower (Fig. 5D).
Because 5-HT1A receptor expression in the PFC is reduced in depression (Savitz et al., 2009) and splicing affects receptor expression, we next explored whether 5-HT1A splicing is altered in depression. Real-time quantification of HTR1A mRNA revealed an increase in the unspliced variant in depressed versus healthy control PFC tissue (t(32) = 2.025, p = 0.0512, unpaired t test; Fig. 5E) and a significant reduction in total spliced/unspliced ratio (t(32) = 2.09, p = 0.0446, unpaired t test; Fig. 5F), suggesting reduced splicing efficiency. Interestingly, both PTBP1 and nSR100 levels were significantly decreased in the PFC of depressed individuals (t(32) = 3.691, p = 0.0008; t(32) = 3.836, p = 0.0006 unpaired t tests; Fig. 5G). The reduction in nSR100 may drive reduced HTR1A splicing efficiency in depressed PFC (Fig. 5H), a change partly counteracted by reduced PTBP1 suppression and thus not sufficient to alter total 5-HT1A RNA levels. Importantly, these changes in splice factor levels could affect the expression of many other neuronal-specific mRNA variants. As a consequence, depressed subjects may have profound alterations in neural-specific splicing.
Discussion
Alternative splicing of human 5-HT1A receptor RNA alters its stability
Despite its fundamental role in serotonergic neurotransmission and mood disorders, the full 5-HT1A mRNA sequence was incompletely characterized. Although long considered an “intronless” gene (Fargin et al., 1988; Charest et al., 1993), we show that the human 5-HT1A 3′-UTR is alternatively spliced into three mRNA isoforms that differ greatly in their stability and translational output. Corroborating our results, the LS variant has been identified in RNAseq databases (National Center for Biotechnology Information Homo Sapiens annotation release 108), but never actually characterized. In rodent HTR1A genes, the splice acceptor/donor sites are poorly conserved and splicing was not detected (Fig. 1D). In contrast, sequence comparison of the rhesus macaque and human 5-HT1A 3′-UTRs shows strong homology (93% identity) and conservation of the splice sites (Fig. 1E), suggesting that 5-HT1A splicing may occur in other primates. This is consistent with genome-wide evidence that the incidence of splicing increases with evolution (Barbosa-Morais et al., 2012; Gueroussov et al., 2015), and the presence of introns in the 5-HT1A mRNA 3′-UTR confers a powerful mechanism for splice-dependent tuning of 5-HT1A receptor expression in primates. Elucidation of human-specific or primate-specific mechanisms [e.g., miRNA-dependent mechanisms (Lopez et al., 2014)] implicated in depression may provide new targets and insights that cannot be gleaned from rodent models.
The presence of 3′-UTR introns occurs in only 6% of genes (Bicknell et al., 2012) and typically yields highly unstable mRNAs degraded by NMD. However, this is not the case for the spliced 5-HT1A mRNA variants, which are extremely stable compared with the unspliced form. Hence, we addressed the role of miR135 in splicing-induced HTR1A stabilization, since miR135 acts at a highly conserved site within the HTR1A intron to negatively regulate 5-HT1A receptor RNA levels (Issler et al., 2014). In murine 5-HT neurons, miR135 is rapidly induced by antidepressants imipramine and fluoxetine, and the overexpression of miR135 in 5-HT neurons downregulates 5-HT1A receptor RNA and reduces anxiety and depression in the chronic social-defeat model (Issler et al., 2014). By contrast, knockdown of miR135 in raphe increased anxiety and revealed selective serotonin reuptake inhibitor (SSRI)-induced depression-like behavior, while preventing SSRI-induced antianxiety actions. These findings are consistent with a key role of miR135 in suppressing 5-HT1A autoreceptor expression and its behavioral consequences. Our data show that HTR1A splicing prevents miR135-mediated destabilization by removing the miRNA binding site. This finding in the HTR1A 3′-UTR provides further evidence of this novel mechanism for RNA stabilization (Mohr and Mott, 2015).
nSR100 and PTBP1 as the determinants of 5-HT1A mRNA splicing
Tissue-specific RNA binding proteins that promote or block the inclusion of alternative exons in RNA transcripts are important regulators of alternative splicing (Lee and Rio, 2015). By manipulating several brain-enriched splicing regulators and examining changes in 5-HT1A mRNA splicing, we found that 5-HT1A RNA splicing is repressed by PTBP1, but enhanced by nSR100. However, we cannot rule out the possibility that another splice factor downstream of PTBP1 or nSR100 could mediate or contribute to the regulation of 5-HT1A mRNA splicing. PTBP1, a ubiquitous repressor of splicing, is downregulated by miR124 during neuronal differentiation, enabling PTBP2 expression and activation of a neural-specific splicing program (Makeyev et al., 2007). The low basal levels of 5-HT1A mRNA splicing in cell lines appear to be mostly driven by high PTBP1 expression, since knockdown of PTBP1 increased HTR1A splicing despite an increase in PTBP2, a weaker repressor of splicing (Markovtsov et al., 2000). In contrast, nSR100 promotes inclusion of a number of neuronal-specific exons (Raj et al., 2014), and its overexpression in SKN-SH cells increased levels of the SS variant, which we did not detect in brain tissue. This suggests that additional splice regulators not present in SKN-SH cells may be required for proper 5-HT1A splicing in vivo. Opposing regulation of splicing by PTBP1 and nSR100 has been reported for other neuronal genes, including PTBP2 (Calarco et al., 2009; Raj et al., 2014). Our data suggest that in neuronal cells, lower levels of PTBP1 combined with the presence of nSR100 promote synthesis of the stable spliced 5-HT1A RNA isoform to enhance receptor levels (Fig. 5H), whereas in non-neuronal cells, PTBP1 strongly inhibits splicing and receptor expression. Importantly, regional differences in nSR100 and PTBP1 levels also appear to affect 5-HT1A splicing in vivo.
Regulation of 5-HT1A mRNA splicing in the human brain
Consistent with the prevalence of alternative splicing in the human brain (Kang et al., 2011), we found that HTR1A splicing in the postmortem human brain was greater than in cell lines and was region-specific. Region-specific HTR1A splicing appeared to be primarily driven by nSR100 expression: it was lowest in the midbrain/hippocampus, where low nSR100 and high PTBP1 expression were associated with increased unspliced HTR1A RNA, while higher levels of nSR100 in the PFC were associated with reduced unspliced mRNA and higher spliced/unspliced ratios (Fig. 5H). While nSR100 has been strongly implicated in brain developmental splicing (Calarco et al., 2009; Irimia et al., 2014; Raj et al., 2014), our results suggest it may be important in dynamic regulation of region-specific RNA expression. Moreover, reduced splicing to augment miR135-induced downregulation of 5-HT1A RNA (Issler et al., 2014) could also contribute to the preferential reduction in 5-HT1A receptors observed in the midbrain and hippocampus following chronic SSRI treatment in human depression (Gray et al., 2013) and anxiety, respectively (Spindelegger et al., 2009). The region-specificity of HTR1A splicing establishes a novel mechanism for differential regulation of postsynaptic and presynaptic 5-HT1A receptors, complementary to transcriptional mechanisms (Czesak et al., 2006, 2012).
In the PFC of depressed individuals, there was a significant reduction in the splicing ratio, consistent with a reduction in nSR100, although the reduction in PTBP1 may partially counteract the effect of reduced nSR100 levels. Both imaging and postmortem studies have reported reductions in cortical 5-HT1A receptors in depression (López-Figueroa et al., 2004; Savitz et al., 2009; Szewczyk et al., 2009), a phenotype that reduced 5-HT1A splicing could contribute to. In contrast, presynaptic 5-HT1A autoreceptors appear to be upregulated in depression (Stockmeier et al., 1998; Boldrini et al., 2008; Hesselgrave and Parsey, 2013), which is consistent with their role in inhibition of 5-HT neuronal activity. While we did not see a significant reduction in total 5-HT1A RNA in the PFC, most studies show highly localized changes in frontal cortex 5-HT1A receptors that may have been missed in the tissue we studied.
Several human HTR1A single-nucleotide polymorphisms (SNPs) are associated with depression (Kishi et al., 2013), including the functional promoter SNP rs6295 C(-1019)G (Lemonde et al., 2003), which has been associated with increased expression of 5-HT1A autoreceptors (Hesselgrave and Parsey, 2013) and a reduction in PFC 5-HT1A receptors (Kautzky et al., 2017). There are several SNPs located proximal to the spliced region, two of which are in strong linkage disequilibrium with rs6295-C/G: rs6449693 T/C and rs878567 C/T (Donaldson et al., 2016). HTR1A RNA with the rs878567-C allele is the more strongly expressed allele in a normal PFC, but is reduced in a depressed PFC (Donaldson et al., 2016). This suggests a stabilizing effect of rs878567-C, perhaps by increasing spliced HTR1A RNA. However, HTR1A RNA levels are also influenced by allele-specific transcription of the rs6295-C promoter SNP (Lemonde et al., 2003). Interestingly, the rs749099 A/G SNP sits a few nucleotides from the splice acceptor site and could affect splicing. Further studies will be needed to address the linkage between promoter and 3′-UTR SNPs and their impact on 5-HT1A splicing, receptor levels, and major depression.
In summary, our data provide the first evidence of altered HTR1A splicing in depression. Two other serotonergic genes are regulated by alternative splicing (HTR2C and TPH2), and impairment in their splicing patterns correlates with psychiatric disorders (Grohmann et al., 2010; Zhang et al., 2011; Martin et al., 2013). However, the splice factors implicated are unknown. We propose that depression-related changes in splice-factor expression or activity could potentially affect many neuronal genes subject to regulation by splicing (Calarco et al., 2009; Raj et al., 2014). Further studies like this one are needed to elucidate the mechanisms that regulate brain region-specific splicing and their impact on brain function and disease.
Footnotes
This work was supported by grants from the Canadian Institutes for Health Research (MOP-115098 and MOP-123426 to P.R.A.) and by the Postmortem Brain Core of the COBRE (Centers of Biomedical Research Excellence) Center for Psychiatric Neuroscience (P30 GM103328 to C.A.S.). We appreciate the invaluable contributions made by the families who consented to donate brain tissue and agreed to be interviewed. We also gratefully acknowledge the support of the staff of the Cuyahoga County Medical Examiner's Office, Cleveland, Ohio. We acknowledge the expert assistance of Drs. James C. Overholser, George Jurjus, and Lisa C. Konick; and of Lesa Dieter in establishing the psychiatric diagnoses, acquiring written consent, and collecting the tissues.
The authors declare no competing financial interests.
References
- Albert PR, Lemonde S (2004) 5-HT1A receptors, gene repression, and depression: guilt by association. Neuroscientist 10:575–593. 10.1177/1073858404267382 [DOI] [PubMed] [Google Scholar]
- Albert PR, Zhou QY, Van Tol HH, Bunzow JR, Civelli O (1990) Cloning, functional expression, and mRNA tissue distribution of the rat 5-hydroxytryptamine1A receptor gene. J Biol Chem 265:5825–5832. [PubMed] [Google Scholar]
- Albert PR, Vahid-Ansari F, Luckhart C (2014) Serotonin-prefrontal cortical circuitry in anxiety and depression phenotypes: pivotal role of pre- and post-synaptic 5-HT1A receptor expression. Front Behav Neurosci 8:199. 10.3389/fnbeh.2014.00199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association (1994) Diagnostic and statistical manual of mental disorders, 4th edition Washington, DC: American Psychiatric Association. [Google Scholar]
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, Kim T, Misquitta-Ali CM, Wilson MD, Kim PM, Odom DT, Frey BJ, Blencowe BJ (2012) The evolutionary landscape of alternative splicing in vertebrate species. Science 338:1587–1593. 10.1126/science.1230612 [DOI] [PubMed] [Google Scholar]
- Bicknell AA, Cenik C, Chua HN, Roth FP, Moore MJ (2012) Introns in UTRs: why we should stop ignoring them. Bioessays 34:1025–1034. 10.1002/bies.201200073 [DOI] [PubMed] [Google Scholar]
- Boldrini M, Underwood MD, Mann JJ, Arango V (2008) Serotonin-1A autoreceptor binding in the dorsal raphe nucleus of depressed suicides. J Psychiatr Res 42:433–442. 10.1016/j.jpsychires.2007.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, Shiue L, Ares M Jr, Black DL (2007) A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev 21:1636–1652. 10.1101/gad.1558107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco JA, Superina S, O'Hanlon D, Gabut M, Raj B, Pan Q, Skalska U, Clarke L, Gelinas D, van der Kooy D, Zhen M, Ciruna B, Blencowe BJ (2009) Regulation of vertebrate nervous system alternative splicing and development by an SR-related protein. Cell 138:898–910. 10.1016/j.cell.2009.06.012 [DOI] [PubMed] [Google Scholar]
- Chang YF, Imam JS, Wilkinson MF (2007) The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem 76:51–74. 10.1146/annurev.biochem.76.050106.093909 [DOI] [PubMed] [Google Scholar]
- Charest A, Wainer BH, Albert PR (1993) Cloning and differentiation-induced expression of a murine serotonin1A receptor in a septal cell line. J Neurosci 13:5164–5171. 10.1523/JNEUROSCI.13-12-05164.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czesak M, Lemonde S, Peterson EA, Rogaeva A, Albert PR (2006) Cell-specific repressor or enhancer activities of deaf-1 at a serotonin 1A receptor gene polymorphism. J Neurosci 26:1864–1871. 10.1523/JNEUROSCI.2643-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czesak M, Le François B, Millar AM, Deria M, Daigle M, Visvader JE, Anisman H, Albert PR (2012) Increased serotonin-1A (5-HT1A) autoreceptor expression and reduced raphe serotonin levels in deformed epidermal autoregulatory factor-1 (Deaf-1) gene knock-out mice. J Biol Chem 287:6615–6627. 10.1074/jbc.M111.293027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell RB. (2013) RNA protein interaction in neurons. Annu Rev Neurosci 36:243–270. 10.1146/annurev-neuro-062912-114322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson ZR, Le François B, Santos TL, Almli LM, Boldrini M, Champagne FA, Arango V, Mann JJ, Stockmeier CA, Galfalvy H, Albert PR, Ressler KJ, Hen R (2016) The functional serotonin 1a receptor promoter polymorphism, rs6295, is associated with psychiatric illness and differences in transcription. Transl Psychiatry 6:e746. 10.1038/tp.2015.226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fargin A, Raymond JR, Lohse MJ, Kobilka BK, Caron MG, Lefkowitz RJ (1988) The genomic clone G-21 which resembles a beta-adrenergic receptor sequence encodes the 5-HT1A receptor. Nature 335:358–360. 10.1038/335358a0 [DOI] [PubMed] [Google Scholar]
- First M, Gibbon M, Williams J (2002) Structured clinical interview for DSM-IV-TR axis I disorders, research version, patient edition. (SCID-I/P). Biometrics research. New York, NY: New York State Psychiatric Institute. [Google Scholar]
- Gray NA, Milak MS, DeLorenzo C, Ogden RT, Huang YY, Mann JJ, Parsey RV (2013) Antidepressant treatment reduces serotonin-1A autoreceptor binding in major depressive disorder. Biol Psychiatry 74:26–31. 10.1016/j.biopsych.2012.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohmann M, Hammer P, Walther M, Paulmann N, Büttner A, Eisenmenger W, Baghai TC, Schüle C, Rupprecht R, Bader M, Bondy B, Zill P, Priller J, Walther DJ (2010) Alternative splicing and extensive RNA editing of human TPH2 transcripts. PLoS One 5:e8956. 10.1371/journal.pone.0008956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueroussov S, Gonatopoulos-Pournatzis T, Irimia M, Raj B, Lin ZY, Gingras AC, Blencowe BJ (2015) An alternative splicing event amplifies evolutionary differences between vertebrates. Science 349:868–873. 10.1126/science.aaa8381 [DOI] [PubMed] [Google Scholar]
- Hesselgrave N, Parsey RV (2013) Imaging the serotonin 1A receptor using [11C]WAY100635 in healthy controls and major depression. Philos Trans R Soc Lond B Biol Sci 368:20120004. 10.1098/rstb.2012.0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, Weatheritt RJ, Ellis JD, Parikshak NN, Gonatopoulos-Pournatzis T, Babor M, Quesnel-Vallières M, Tapial J, Raj B, O'Hanlon D, Barrios-Rodiles M, Sternberg MJ, Cordes SP, Roth FP, Wrana JL, Geschwind DH, Blencowe BJ (2014) A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell 159:1511–1523. 10.1016/j.cell.2014.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issler O, Haramati S, Paul ED, Maeno H, Navon I, Zwang R, Gil S, Mayberg HS, Dunlop BW, Menke A, Awatramani R, Binder EB, Deneris ES, Lowry CA, Chen A (2014) MicroRNA 135 is essential for chronic stress resiliency, antidepressant efficacy, and intact serotonergic activity. Neuron 83:344–360. 10.1016/j.neuron.2014.05.042 [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, Guennel T, Shin Y, Johnson MB, Krsnik Z, Mayer S, Fertuzinhos S, Umlauf S, Lisgo SN, Vortmeyer A, Weinberger DR, Mane S, Hyde TM, Huttner A, Reimers M, Kleinman JE, Sestan N (2011) Spatio-temporal transcriptome of the human brain. Nature 478:483–489. 10.1038/nature10523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kautzky A, James GM, Philippe C, Baldinger-Melich P, Kraus C, Kranz GS, Vanicek T, Gryglewski G, Wadsak W, Mitterhauser M, Rujescu D, Kasper S, Lanzenberger R (2017) The influence of the rs6295 gene polymorphism on serotonin-1A receptor distribution investigated with PET in patients with major depression applying machine learning. Transl Psychiatry 7:e1150. 10.1038/tp.2017.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi T, Yoshimura R, Fukuo Y, Okochi T, Matsunaga S, Umene-Nakano W, Nakamura J, Serretti A, Correll CU, Kane JM, Iwata N (2013) The serotonin 1A receptor gene confer susceptibility to mood disorders: results from an extended meta-analysis of patients with major depression and bipolar disorder. Eur Arch Psychiatry Clin Neurosci 263:105–118. 10.1007/s00406-012-0337-4 [DOI] [PubMed] [Google Scholar]
- Lee Y, Rio DC (2015) Mechanisms and regulation of alternative pre-mRNA splicing. Annu Rev Biochem 84:291–323. 10.1146/annurev-biochem-060614-034316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le François B, Czesak M, Steubl D, Albert PR (2008) Transcriptional regulation at a HTR1A polymorphism associated with mental illness. Neuropharmacology 55:977–985. 10.1016/j.neuropharm.2008.06.046 [DOI] [PubMed] [Google Scholar]
- Lemonde S, Turecki G, Bakish D, Du L, Hrdina PD, Bown CD, Sequeira A, Kushwaha N, Morris SJ, Basak A, Ou XM, Albert PR (2003) Impaired repression at a 5-hydroxytryptamine 1A receptor gene polymorphism associated with major depression and suicide. J Neurosci 23:8788–8799. 10.1523/JNEUROSCI.23-25-08788.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemonde S, Du L, Bakish D, Hrdina P, Albert PR (2004) Association of the C(-1019)G 5-HT1A functional promoter polymorphism with antidepressant response. Int J Neuropsychopharmacol 7:501–506. 10.1017/S1461145704004699 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lopez JP, Lim R, Cruceanu C, Crapper L, Fasano C, Labonte B, Maussion G, Yang JP, Yerko V, Vigneault E, El Mestikawy S, Mechawar N, Pavlidis P, Turecki G (2014) miR-1202 is a primate-specific and brain-enriched microRNA involved in major depression and antidepressant treatment. Nat Med 20:764–768. 10.1038/nm.3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Figueroa AL, Norton CS, López-Figueroa MO, Armellini-Dodel D, Burke S, Akil H, López JF, Watson SJ (2004) Serotonin 5-HT1A, 5-HT1B, and 5-HT2A receptor mRNA expression in subjects with major depression, bipolar disorder, and schizophrenia. Biol Psychiatry 55:225–233. 10.1016/j.biopsych.2003.09.017 [DOI] [PubMed] [Google Scholar]
- Mahajan GJ, Vallender EJ, Garrett MR, Challagundla L, Overholser JC, Jurjus G, Dieter L, Syed M, Romero DG, Benghuzzi H, Stockmeier CA (2018) Altered neuro-inflammatory gene expression in hippocampus in major depressive disorder. Prog Neuropsychopharmacol Biol Psychiatry 82:177–186. 10.1016/j.pnpbp.2017.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T (2007) The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell 27:435–448. 10.1016/j.molcel.2007.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovtsov V, Nikolic JM, Goldman JA, Turck CW, Chou MY, Black DL (2000) Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol Cell Biol 20:7463–7479. 10.1128/MCB.20.20.7463-7479.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin CB, Ramond F, Farrington DT, Aguiar AS Jr, Chevarin C, Berthiau AS, Caussanel S, Lanfumey L, Herrick-Davis K, Hamon M, Madjar JJ, Mongeau R (2013) RNA splicing and editing modulation of 5-HT(2C) receptor function: relevance to anxiety and aggression in VGV mice. Mol Psychiatry 18:656–665. 10.1038/mp.2012.171 [DOI] [PubMed] [Google Scholar]
- Mohr AM, Mott JL (2015) Overview of microRNA biology. Semin Liver Dis 35:3–11. 10.1055/s-0034-1397344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj B, Blencowe BJ (2015) Alternative splicing in the mammalian nervous system: recent insights into mechanisms and functional roles. Neuron 87:14–27. 10.1016/j.neuron.2015.05.004 [DOI] [PubMed] [Google Scholar]
- Raj B, Irimia M, Braunschweig U, Sterne-Weiler T, O'Hanlon D, Lin ZY, Chen GI, Easton LE, Ule J, Gingras AC, Eyras E, Blencowe BJ (2014) A global regulatory mechanism for activating an exon network required for neurogenesis. Mol Cell 56:90–103. 10.1016/j.molcel.2014.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson-Jones JW, Craige CP, Guiard BP, Stephen A, Metzger KL, Kung HF, Gardier AM, Dranovsky A, David DJ, Beck SG, Hen R, Leonardo ED (2010) 5-HT(1A) autoreceptor levels determine vulnerability to stress and response to antidepressants. Neuron 65:40–52. 10.1016/j.neuron.2009.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson-Jones JW, Craige CP, Nguyen TH, Kung HF, Gardier AM, Dranovsky A, David DJ, Guiard BP, Beck SG, Hen R, Leonardo ED (2011) Serotonin-1A autoreceptors are necessary and sufficient for the normal formation of circuits underlying innate anxiety. J Neurosci 31:6008–6018. 10.1523/JNEUROSCI.5836-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels BA, Anacker C, Hu A, Levinstein MR, Pickenhagen A, Tsetsenis T, Madroñal N, Donaldson ZR, Drew LJ, Dranovsky A, Gross CT, Tanaka KF, Hen R (2015) 5-HT1A receptors on mature dentate gyrus granule cells are critical for the antidepressant response. Nat Neurosci 18:1606–1616. 10.1038/nn.4116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarelli L, Saxe M, Gross C, Surget A, Battaglia F, Dulawa S, Weisstaub N, Lee J, Duman R, Arancio O, Belzung C, Hen R (2003) Requirement of hippocampal neurogenesis for the behavioral effects of antidepressants. Science 301:805–809. 10.1126/science.1083328 [DOI] [PubMed] [Google Scholar]
- Savitz J, Lucki I, Drevets WC (2009) 5-HT(1A) receptor function in major depressive disorder. Prog Neurobiol 88:17–31. 10.1016/j.pneurobio.2009.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spindelegger C, Lanzenberger R, Wadsak W, Mien LK, Stein P, Mitterhauser M, Moser U, Holik A, Pezawas L, Kletter K, Kasper S (2009) Influence of escitalopram treatment on 5-HT(1A) receptor binding in limbic regions in patients with anxiety disorders. Mol Psychiatry 14:1040–1050. 10.1038/mp.2008.35 [DOI] [PubMed] [Google Scholar]
- Stockmeier CA, Shapiro LA, Dilley GE, Kolli TN, Friedman L, Rajkowska G (1998) Increase in serotonin-1A autoreceptors in the midbrain of suicide victims with major depression-postmortem evidence for decreased serotonin activity. J Neurosci 18:7394–7401. 10.1523/JNEUROSCI.18-18-07394.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk B, Albert PR, Burns AM, Czesak M, Overholser JC, Jurjus GJ, Meltzer HY, Konick LC, Dieter L, Herbst N, May W, Rajkowska G, Stockmeier CA, Austin MC (2009) Gender-specific decrease in NUDR and 5-HT1A receptor proteins in the prefrontal cortex of subjects with major depressive disorder. Int J Neuropsychopharmacol 12:155–168. 10.1017/S1461145708009012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahid-Ansari F, Daigle M, Manzini MC, Tanaka KF, Hen R, Geddes SD, Béïque JC, James J, Merali Z, Albert PR (2017) Abrogated freud-1/Cc2d1a repression of 5-HT1A autoreceptors induces fluoxetine-resistant anxiety/depression-like behavior. J Neurosci 37:11967–11978. 10.1523/JNEUROSCI.1668-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EJ, Garcia-Blanco MA (2001) Polypyrimidine tract binding protein antagonizes exon definition. Mol Cell Biol 21:3281–3288. 10.1128/MCB.21.10.3281-3288.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Nicholls PJ, Laje G, Sotnikova TD, Gainetdinov RR, Albert PR, Rajkowska G, Stockmeier CA, Speer MC, Steffens DC, Austin MC, McMahon FJ, Krishnan KR, Garcia-Blanco MA, Caron MG (2011) A functional alternative splicing mutation in human tryptophan hydroxylase-2. Mol Psychiatry 16:1169–1176. 10.1038/mp.2010.99 [DOI] [PMC free article] [PubMed] [Google Scholar]