Abstract
Myelin sheath formed by oligodendrocytes (OLs) is essential for the rapid propagation of action potentials in the vertebrate CNS. Myelin regulatory factor (MYRF) is one of the critical factors that control OL differentiation and myelin maintenance. Previous studies showed that MYRF is a membrane-bound transcription factor associated with the endoplasmic reticulum (ER). After self-cleavage, the N-fragment of MYRF is released from the ER and translocated into the nucleus where it functions as a transcription factor to activate myelin gene expression. At present, it remains unknown whether MYRF self-cleavage and functional activation can be regulated during OL differentiation. Here, we report that TMEM98, an ER-associated transmembrane protein, is capable of binding to the C-terminal of MYRF and inhibiting its self-cleavage and N-fragment nuclear translocation. In the developing CNS, TMEM98 is selectively expressed in early maturing OLs in mouse pups of either sex. Forced expression of TMEM98 in embryonic chicken spinal cord of either sex suppresses endogenous OL differentiation and MYRF-induced ectopic expression of myelin genes. These results suggest that TMEM98, through inhibiting the self-cleavage of MYRF, functions as a negative feedback regulator of MYRF in oligodendrocyte differentiation and myelination.
SIGNIFICANCE STATEMENT MYRF protein is initially synthesized as an ER-associated membrane protein that undergoes autoproteolytic cleavage to release the N-fragment, which is then transported into the nucleus and activates the transcription of myelin genes. To date, the molecular mechanisms that regulate the self-cleavage and function of MYRF in regulating oligodendrocyte differentiation have remained unknown. In this study, we present the molecular and functional evidence that TMEM98 membrane protein physically interacts with MYRF in the ER and subsequently blocks its self-cleavage, N-terminal nuclear translocation, and functional activation of myelin gene expression. To our knowledge, this is the first report on the regulation of MYRF self-proteolytic activity and function by an interacting protein, providing new insights into the molecular regulation of OL differentiation and myelinogenesis.
Keywords: MYRF, oligodendrocyte differentiation, self-cleavage, TMEM98
Introduction
Compact myelin sheaths are assembled by oligodendrocytes (OLs) in the CNS and provide electrical insulation and trophic support for the axons (Baumann and Pham-Dinh, 2001; Simons and Nave, 2015). During embryonic development, oligodendrocyte progenitor cells (OPCs) are initially generated from ventricular neuroepithelial cells and later differentiate into myelinating oligodendrocytes (Richardson et al., 2006). Deficits of myelin are associated with several neurological diseases, such as multiple sclerosis (MS; Franklin, 2002; Cui et al., 2013; Dulamea, 2017) and leukodystrophies (Aubourg, 1993; Barañano, 2016). In spinal injuries and acute MS lesions, OPCs still exist but fail to differentiate effectively into myelinating oligodendrocytes (Raine and Wu, 1993; Wolswijk, 1998; Chang et al., 2002; Jennings and Carroll, 2010). Thus, OL differentiation is a rate-limiting step of axonal remyelination in pathological conditions (Gallo and Deneen, 2014), and understanding the molecular regulation of OL differentiation is critical for generating therapeutics to enhance myelin repair in demyelinating disorders.
Recent studies have identified a cohort of transcriptional factors (e.g., Sox10, Nkx2.2, Olig1, and Myrf) that plays a pivotal role in the regulation of oligodendrocyte differentiation. Among these, Sox10 promotes the terminal differentiation of OL lineage, and the Sox10 null mutation results in an almost complete inhibition of myelin gene expression (Stolt et al., 2002; Liu et al., 2007). Nkx2.2 and Olig1 regulate the timing of oligodendrocyte differentiation in the CNS (Huang et al., 2013), and disruption of both Olig1 and Nkx2.2 results in a significant delay of OL differentiation (Qi et al., 2001; Paes de Faria et al., 2014; Zhu et al., 2014). Unlike other transcriptional factors, myelin regulatory factor (MYRF) is a membrane-bound transcription factor that is required for OL differentiation, myelin formation, and maintenance (Emery et al., 2009; Koenning et al., 2012). MYRF is initially synthesized as a transmembrane protein in the endoplasmic reticulum (ER) but later undergoes autoproteolytic cleavage mediated by the intramolecular chaperone auto-cleavage (ICA) domain (Li et al., 2013). The freed N-fragment (N-MYRF) is then translocated into the nucleus and activates target-gene expression, whereas the C-fragment (C-MYRF) remains tethered to the ER membrane (Bujalka et al., 2013). Recent studies showed that MYRF first assembles into a homotrimeric complex preceding self-cleavage, and the homo-trimerization defines the DNA-binding specificity of the N-fragment (Muth et al., 2016; Kim et al., 2017). MYRF family proteins are highly conserved in protein sequence and domain structure among metazoans (Li and Richardson, 2016); furthermore, Dictyostelium (slime mold) MYRFA and Caenorhabditis elegans (nematode) MYRF-1/MYRF-2 exhibit ER localization and cleavage-dependent nuclear translocation (Senoo et al., 2012; Meng et al., 2017), suggesting the conserved molecular regulation of MYRF protein activity.
It has been demonstrated that many membrane-bound transcription factors (e.g., Notch, SREBPs, and ATP6) often undergo ligand- or stress-regulated intramembrane proteolysis to release nuclear-targeted products for regulating gene transcription (Brown and Goldstein, 1997; De Strooper et al., 1999; Haze et al., 1999). However, it is not known whether MYRF also undergoes a similar regulated proteolytic cleavage process.
In this study, we identified TMEM98 as a binding protein of MYRF in the ER and a regulator of MYRF activity. TMEM98 is an ER-associated transmembrane protein and highly conserved during evolution (Ng et al., 2014). The expression and function of TMEM98 are mostly unknown, although it has been implicated in T Helper 1 cell differentiation (Fu et al., 2015) and human nanophthalmos diseases (Awadalla et al., 2014). The C. elegans ortholog TMEM98 (K10C3.4) is identified as a direct-target let-7 miRNA and involved in vulval development (Andachi, 2008). Here, we demonstrate that Tmem98 is selectively expressed in postmitotic oligodendrocytes in the developing CNS with temporal and spatial patterns of expression similar to that of Myrf. Molecular and functional analyses have identified TMEM98 as a negative regulator of MYRF activity during OL differentiation.
Materials and Methods
Animals and cell culture.
All animal experiments were performed in accordance with the institutional guidelines drafted by the Laboratory Animal Center at Hangzhou Normal University (Hangzhou, China) and were approved by the Animal Ethics Committee of Hangzhou Normal University. Mouse lines for Olig1-Cre, Myrfflox, and Nkx2.2flox mice were described previously (Lu et al., 2002; Emery et al., 2009; Mastracci et al., 2013). Animals of either sex were used for analyses. HEK293T cells were cultured in DMEM (10569044, Invitrogen) supplemented with 10% fetal bovine serum (04-001-1A, Biological Industries). CG4 cells were cultured in DMEM/F-12 medium with N2, B27, and 10 ng/ml PDGF-AA (100–13A, PeproTech). After they were transfected with viruses, CG4 cells were cultured in PDGF-AA-containing medium for 2 d to allow gene expression. Cells were then cultured in growth factor-free medium for 3 d and in a differentiation medium containing 40 ng/ml 3,3′,5-Triiodo-L-thyronine (T-2752, Sigma-Aldrich) for another 2 d. All reagents used for CG4 cells were described previously (Chen et al., 2007). Cells were maintained at 37°C in a humidified incubator with 5% CO2. HEK293T cells were transfected with DNA constructs using MegaTran 1.0 (OriGene Technologies) as per the manufacturer's instructions.
Constructs.
For double-immunostaining assays in HEK293T cells, CopGFP in pCDH-CMV-MCS-EF1-CopGFP vectors (CD511B-1, System Biosciences) was replaced by the ORF of Tmem98 (NM_029537.1) or Myrf (XM_006526929.3). For immunoprecipitation assays, ORFs of Tmem98 and full-length/truncated Myrf as diagrammed in the figures were cloned into pCDH-CMV-MCS-EF1-copGFP vectors or pRK5 vector (a gift from Dr. Ying Shen, Zhejiang University School of Medicine, Hangzhou, China). For chicken in ovo electroporation, ORFs of Tmem98, Tmem98-ΔGCIP (Δ55–152 aa), and Myrf were cloned into RCASBP(B) vectors (Morgan and Fekete, 1996). For virus production, ORFs of Tmem98 and Myrf were inserted after the CMV promoter or EF1 promoter in pCDH vectors. Tags fused with TMEM98 and full-length/truncated MYRF were as diagrammed in the figures.
In ovo chicken electroporation and gene delivery into CG4 cells.
For in ovo electroporation, ∼5 μl of RCASBP(B) plasmids (4 μg/μl) was mixed with 1 μl of pCAGGS-EGFP plasmids (6 μg/μl) and 1 μl of 1% Fast Green. The mixed constructs were injected into the chick spinal cord at Hamburger and Hamilton stages HH13-HH15 (chicken embryonic day 2, cE2), and the injected embryos were then subjected to five short pulses of electrical shock (25 V, 50 ms for each pulse, 950 ms interval; Momose et al., 1999) with a Electro Square Porator (ECM 830, BTX). Embryos of either sex were then allowed to develop for various time periods before they were fixed in 4% PFA for gene expression analysis. For gene delivery into CG4 cells, pCDH vectors with/without target genes were cotransfected into HEK293T cells with psPAX2 (12260, Addgene) and pMD2.G (12259, Addgene) to produce viruses. Viruses were collected every 24 h three times and centrifuged for 90 min at 20,000 × g. Condensed viruses were used to infect CG4 cells in a 6 cm dish for 6 h. For MYRF and TMEM98 coexpression, cells were infected with viruses containing pCDH-CMV-EF1-Tmem98 for 6 h before adding pCDH-CMV-Myrf-EF1-CopGFP viruses.
In situ hybridization.
The procedure of in situ hybridization (ISH) has been described previously (Huang et al., 2017). For mouse Tmem98 riboprobes, an ∼1 kb fragment corresponding to Tmem98 (200–1200 nt of NM_029537.1) was cloned into pBluescript II KS(−) vectors. To generate riboprobes for detecting gene expression in chicken spinal cords, the DNA fragments corresponding to cMBP (270–1100 nt of NM_205280.1), cPLP (100–970 nt of NM_205277.1), cENPP6 (3161–4160 nt of XM_420512.6), and cPDGFRA (600–2400 nt of NM_204749.2) were cloned into pBluescript II KS(−) vectors. The linearized plasmids were used as templates for in vitro transcription with T3 RNA polymerase (P4024, Promega) according to the manufacturer's instructions.
Immunostaining and immunohistochemical staining.
Tissues were fixed in 4% PFA at 4°C overnight and transferred into 25% sucrose in PBS overnight, embedded in OCT medium, and sectioned (14 μm thickness) on a cryostat. Cells were fixed with 4% PFA for 30 min at room temperature before immunostaining. Tissue sections/cells were then rinsed three times in PBS, blocked with 5% goat serum in PBS with 0.2% Triton X-100 for 30 min, and immediately incubated with a primary antibody in a blocking solution at 4°C overnight. Sections/cells were then washed three times in PBS and incubated with secondary antibodies (Alexa Fluor 488/594, Thermo Fisher Scientific) for 1 h at room temperature. After being rinsed three times in PBS, sections/cells were mounted in Mowiol mounting medium with DAPI. For double-labeling experiments, tissue sections were first subjected to ISH with the Tmem98 riboprobe, followed by immunohistochemical staining with the Vectastain ABC kit (catalog #PK-4001; Vector Laboratories; RRID:AB_2336810). The primary antibodies used were as follows: anti-FLAG (catalog #F1804, 1:1000, Sigma-Aldrich; RRID:AB_262044), anti-HA (catalog #ab9110, 1:1000, Abcam; RRID:AB_307019), anti-MYC (catalog #2276, 1:1000, Cell Signaling Technology; RRID:AB_331783), anti-MYC (catalog #9E 10, 1:100, DSHB; RRID:AB_2266850), anti-OLIG2 (catalog #AB9610, 1:1000, Millipore; RRID:AB_570666), anti-MAG (catalog #MAB1567, 1:500, Millipore; RRID:AB_2137847), anti-CC1 (catalog #ab16794, 1:500, Abcam; RRID:AB_443473), anti-PDGFRA (sc-338, 1:200, Santa Cruz Biotechnology), and anti-V5 (catalog #V8137, 1:1000, Sigma-Aldrich; RRID:AB_261889). Anti-MYRF (1:500) and anti-SOX10 (1:1000) are gifts from Dr. Michael Wegner (Universitaet Erlangen-uernberg, Erlangen, Germany).
Immunoprecipitation.
Forty-eight hours after transfection, HEK293T cells in each 10 cm dish were washed with PBS and lysed in 1.5 ml of lysis buffer containing 50 mm Tris, pH 7.4, 150 mm NaCl, 1% Triton X-100, 1 mm EDTA, and protease inhibitor cocktail (Sangon Biotech) at 4°C for 20 min. Cell lysate was spun down at 13,000 × g for 10 min at 4°C. A total of 0.2 ml of lysate was used as the input sample. The left was added into 2.5 ml of lysis buffer and filtered with a 0.22 μm strainer. For MYC-tagged proteins, cleared lysate was mixed with anti-MYC primary antibodies (catalog #2276, Cell Signaling Technology; RRID:AB_331783) overnight at 4°C. Then, G-protein-coupled Sepharoses (GE17-0886-01, Sigma) were added, and the mixture was incubated for another 4 h at 4°C. For FLAG-tagged proteins, cleared lysate was mixed with 40 μl of anti-FLAG magnetic beads (catalog #M8823, Sigma-Aldrich; RRID:AB_2637089) and incubated overnight at 4°C. Sepharoses and beads were used as per the manufacturer's instructions. After that, Sepharoses and beads were washed with lysis buffer three times. Proteins bound to the anti-FLAG beads were eluted with 300 μl of TBS containing 20 μg of 3xFLAG peptides at room temperature for 30 min, and Sepharoses were heated in 200 μl of 1% SDS buffer at 95°C for 5 min to release the bounded proteins. A total of 20 μl of input/immunoprecipitation (IP) samples were used for Western blot.
RT-qPCR, BrdU staining, and TUNEL assay.
RNA was extracted from mouse spinal cord using RNAiso Plus (9109, Takara), reverse transcribed using the cDNA Synthesis kit (6210A, Takara), and used to analyze expression levels by quantitative PCR on a Bio-Rad CFX96 Real Time PCR System. The following primer pairs were used: 5′-CCTCTTTGCTGCTGGTTACCAAGT-3′ and 5′-GGCTCGACTTTGGAGATTGAAGC-3′ for Myrf; 5′-TCATTTGCTGCTTTGGTGGTGGTC-3′ and 5′-TCCACTCTGGGGCTAATCCGTTTG-3′ for Tmem98. Transcript levels were normalized to Gapdh. For BrdU staining, 0.5 ml of 10 mg/ml BrdU solution was added to every chicken embryo, and tissues were harvested after 6 h. Sections were subjected to immunostaining with anti-BrdU (catalog #G3G4, 1:50, DSHB; RRID:AB_2618097) after being treated with 2 mol/L HCl for 20 min. TUNEL assay was performed according to the manufacturer's instructions (G3250, Promega).
Western blot.
Samples with 1× loading buffer were heated at 95°C for 5 min. Twenty-microliter samples were loaded on SDS-PAGE gels. Proteins in gels were then transferred to PVDF membranes (ISEQ00010, Millpore). Membranes were blocked with 5% milk at room temperature for 1 h and incubated with a primary antibody at 4°C overnight. After being rinsed three times with TBST, membranes were probed with horseradish peroxidase-conjugated secondary antibodies (Promega) and rinsed three times in TBST. Proteins were detected using the enhanced chemiluminescence detection system (catalog #32109, Thermo Fisher Scientific) and x-ray films. Primary antibodies were used as follows: anti-FLAG (catalog #F1804, 1:10,000, Sigma-Aldrich; RRID:AB_262044), anti-HA (catalog #ab9110, 1:10,000, Abcam; RRID:AB_307019), anti-MYC (catalog #2276, 1:10000, Cell Signaling Technology; RRID:AB_331783).
Experimental design and statistical analyses.
For each analysis, results from independent transfections or animals were treated as biological replicates (n ≥ 3). The two-tailed Student's t test was performed to determine whether differences in cell numbers and transcript levels were statistically significant. Error bars represented the SDs. Statistical significance was considered to be at *p ≤ 0.05, **p ≤ 0.01, and ***p ≤ 0.001; ns, no significant difference. The exact p values are as mentioned in Results.
Results
TMEM98 is a MYRF-interacting protein specifically expressed in differentiated oligodendrocytes in the developing CNS
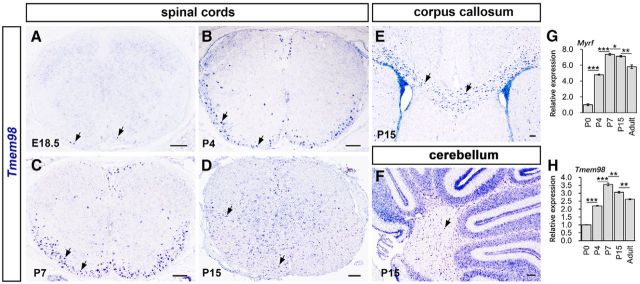
As the first step to identify MYRF-interacting proteins, we generated a C. elegans strain carrying a transgene that expresses MYRF-1 fused with GFP (Meng et al., 2017) and performed affinity purification using an anti-GFP antibody followed by mass spectrometry analyses. One of the interacting proteins identified by this strategy is TMEM98, a novel and evolutionarily conserved single-pass transmembrane protein located in the endoplasmic reticulum (Ng et al., 2014) with an unknown function. To explore their functional relationship in oligodendrocyte development, we first examined the expression of mouse Tmem98 in the developing CNS by RNA ISH. In the spinal cord region, Tmem98 expression was initially detected in the ventral white matter at embryonic day 18.5 (E18.5; Fig. 1A), upregulated rapidly afterward, and peaked at early postnatal stages from P4 to P7 (Fig. 1B,C). At P15, Tmem98 expression started to be detected in the gray matter of spinal cord but gradually disappeared thereafter (Fig. 1D), which was further confirmed by RT-qPCR (Fig. 1H). In addition, Tmem98 expression was also observed in corpus callosum (Fig. 1E) and the white matter of cerebellum (Fig. 1F) at P15 stages. The pattern of Tmem98 expression in the white matter is similar to that of mature OL markers such as Myrf (Fig. 1G,H). Collectively, these results suggest that Tmem98 is selectively expressed in newly differentiated OLs in the CNS.
Figure 1.
Developmental expression pattern of Tmem98 in the CNS. A–D, In situ hybridization for Tmem98 was performed on the sections of spinal cord from E18.5 (A), P4 (B), P7 (C), and P15 (D) wild-type mice. E, F, Sections from P15 corpus callosum (E) and cerebellum (F) of wild-type mice were subjected to ISH with Tmem98 riboprobes. G, H, Transcriptional level of Tmem98 and Myrf mRNA at different developmental stages was quantified by RT-qPCR. Representative Tmem98-positive cells are indicated by arrows. *p < 0.05, **p < 0.01, ***p < 0.001. Scale bars, 100 μm.
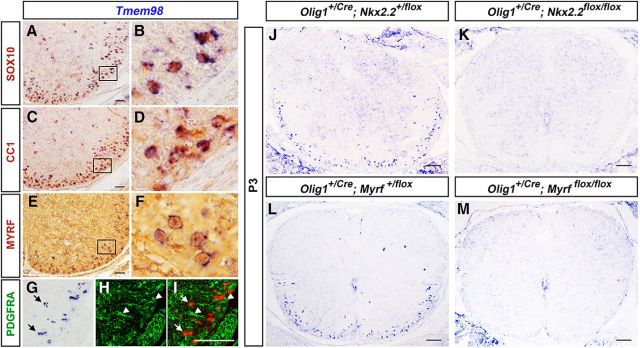
To confirm the specific expression of Tmem98 in differentiated oligodendrocytes, we performed Tmem98 ISH followed by immunohistochemical staining with oligodendrocyte lineage-specific markers (PDGFRA, SOX10, CC1, and MYRF). Double-labeling experiments showed that Tmem98 is mostly coexpressed with the SOX10 transcription factor (Fig. 2A,B), which labels both OPCs and mature OLs (Stolt et al., 2002); CC1 (Fig. 2C,D), which marks young differentiated OLs (Redwine and Evans, 2002); and MYRF (Fig. 2E,F), which is specifically expressed in postmitotic myelinating OLs (Emery et al., 2009). In additional support, Tmem98 is not expressed in PDGFRA+ immature OPCs (Fig. 2G–I). For further verification, we next examined Tmem98 expression in Myrf and Nkx2.2 conditional knock-out (CKO) mice. The disruption of the Nkx2.2 and Myrf genes results in retardation of oligodendrocyte differentiation in the spinal cord (Emery et al., 2009; Zhu et al., 2014). As expected, the number of Tmem98-positive cells was severely decreased in both Nkx2.2 (Fig. 2J,K) and Myrf CKO mice (Fig. 2L,M). Together, these findings indicate that Tmem98 is specifically expressed in differentiated oligodendrocytes in early postnatal CNS tissues.
Figure 2.
Selective expression of Tmem98 in differentiated oligodendrocytes. A–I, Spinal cord sections from P7 wild-type mice were subject to Tmem98 ISH, followed by immunohistochemical staining with anti-SOX10 (A, B), anti-CC1 (C, D), anti-MYRF (E, F), or anti-PDGFRA (G–I). B, D, and F are higher magnifications of the outlined areas in A, C, and E, respectively. J–M, Tmem98 expression in Nkx2.2-CKO (J, K) and Myrf-CKO (L, M) mutant spinal cords is dramatically reduced at P3 stages. Scale bars, 100 μm.
TMEM98 is a transmembrane protein associated with the endoplasmic reticulum
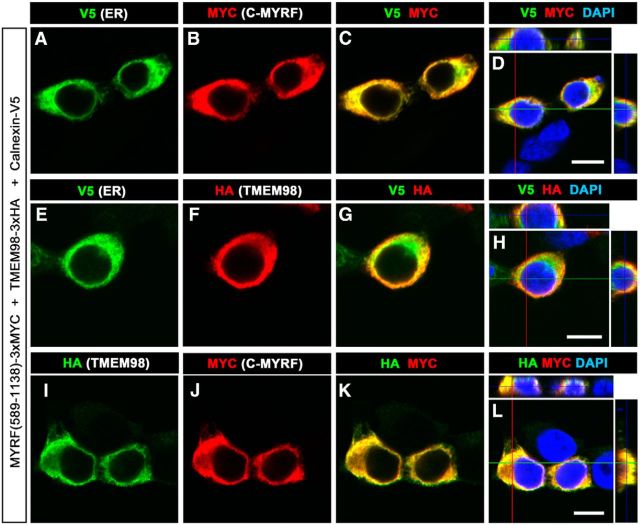
Because MYRF is an ER-associated membrane-bound transcription factor (Bujalka et al., 2013; Li et al., 2013), the physical interaction between TMEM98 and MYRF suggests that TMEM98 is an ER membrane protein as well. To examine this possibility, we expressed mouse TMEM98-3xHA (HA fused at the C terminal), MYRF (589–1138)-3xMYC, and V5-tagged ER transmembrane protein Calnexin in HEK293T cells. Whereas C-terminal MYRF (589–1138) was localized on the ER (Bujalka et al., 2013; Li et al., 2013), anti-MYC signals were cytoplasmic and colocalized with the ER marker Calnexin-V5 (Fig. 3A–D). Moreover, anti-HA signals were colocalized with Calnexin-V5 as well (Fig. 3E–H). Since TMEM98 and MYRF are similarly localized on the ER, we also tested whether they are colocalized in transfected cells. Double immunostaining revealed that TMEM98 was indeed colocalized with the C-fragment of MYRF (Fig. 3I–L), in support of their physical interaction on the ER.
Figure 3.
Colocalization of TMEM98 and MYRF C-fragments in ER. A–H, Both C-fragments of MYRF (3xMYC-tagged; A–D) and TMEM98 (E–H) were colabeled with V5-tagged Calnexin, an ER marker. I–L, C-fragments of MYRF were colabeled with TMEM98 in HEK293T cells. Scale bars, 10 μm.
TMEM98 binds to the C-fragment of MYRF protein
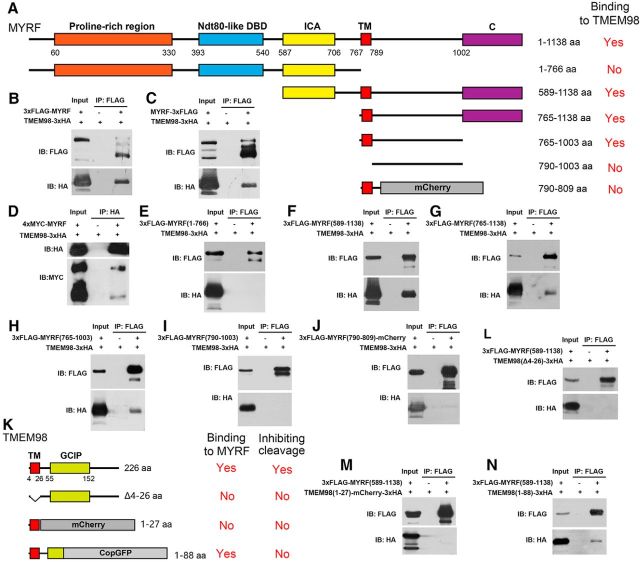
To determine whether TMEM98 can directly bind to the MYRF protein in mammalian cells, we coexpressed full-length TMEM98 and MYRF proteins in HEK293T cells and found that MYRF can be coimmunoprecipitated with TMEM98 (Fig. 4B–D). To test which part of MYRF confers binding with TMEM98, we generated a series of MYRF truncate mutants (Fig. 4A) and performed co-IP analysis with TMEM98. It was found that the N-terminal fragments (1–766) did not bind to TMEM98 (Fig. 4E), whereas the C-terminal fragments of MYRF (589–1138, 765–1138, and 765–1003) displayed strong binding to TMEM98 (Fig. 4F–H). Moreover, deletion of the transmembrane (TM) domain or the adjacent region C terminal to TM abolished its interaction with TMEM98 (Fig. 4I,J). In addition, we also mapped the TMEM98 region that interacts with MYRF and found that the majority of the C terminal of TMEM98, including the grap2 and cyclin D interacting protein (GCIP) domain, is not required for interaction, but deletion of the TM domain and the immediate C-region of TMEM98 eliminated its binding to MYRF (Fig. 4K–N). Together, these analyses indicate that the TM domains of MYRF and TMEM98 and the immediate adjacent regions are indispensable for their efficient interaction.
Figure 4.
Physical interactions between TMEM98 and MYRF proteins. A, Schematic of various truncated MYRF proteins. B–D, TMEM98 proteins physically bind to full-length MYRF proteins. E, TMEM98 does not bind to the cytoplasmic fragments (1–766) of MYRF. F–H, TMEM98 protein binds to the C-fragments of MYRF (589–1138, 765–1138, and 765–1003). I, Deletion of the TM domain in MYRF abolishes the interaction. J, Deletion of the region C terminal to the TM domain in MYRF abolishes the efficient binding between MYRF and TMEM98. K–N, The N-terminal region of TMEM98, including the TM domain, is involved in the MYRF–TMEM98 interaction, whereas the C terminal of TMEM98 is required for inhibiting MYRF cleavage. DBD, DNA-binding domain; C, unknown conserved domain; IB, immunoblot.
TMEM98 suppresses the self-cleavage and nuclear localization of MYRF
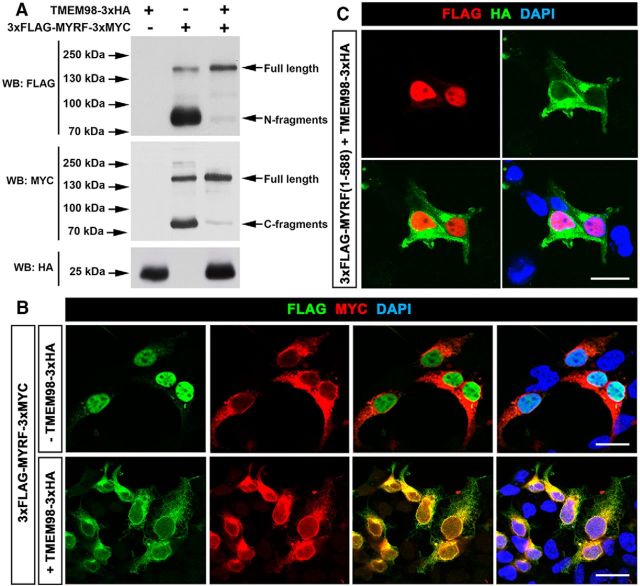
It was previously reported that MYRF undergoes autoproteolysis to produce the N-terminal and C-terminal fragments (Bujalka et al., 2013; Li et al., 2013). To examine whether TMEM98 binding can affect the auto-cleavage and subcellular localization of MYRF, we transfected HEK293T cells with tagged TMEM98 (TMEM98–3xHA), MYRF (3xFLAG-MYRF-3xMYC), or both. Consistent with the previous findings (Li et al., 2013; Kim et al., 2017), MYRF expression alone produced a band of ∼160 kDa corresponding to the full-length MYRF, an ∼80 kDa band corresponding to the N-fragment, and a ∼75 kDa band corresponding to the C-fragment (Fig. 5A). However, when TMEM98 protein (∼25 kDa) was coexpressed, the intensity of the full-length band was drastically increased, whereas those of the N-fragment and C-fragment bands were nearly undetectable (Fig. 5A). Therefore, TMEM98 coexpression strongly inhibited the self-cleavage of MYRF protein in HEK293T cells. Interestingly, in addition to the TM domain, the C terminal of TMEM98 (including the GCIP domain) is also necessary for inhibiting MYRF self-cleavage, even though it is not required for the interaction between TMEM98 and MYRF (Figs. 4K, 7I,I′).
Figure 5.
Inhibition of MYRF self-cleavage by TMEM98. A, The self-cleavage of MYRF was suppressed when TMEM98 was coexpressed. B, The nuclear translocation of MYRF N-fragments released from full-length proteins was blocked when TMEM98 was coexpressed. C, TMEM98 does not interfere with the nuclear translocation of N-fragments of MYRF (1–588). Scale bars, 20 μm. WB, Western blot.
Figure 7.
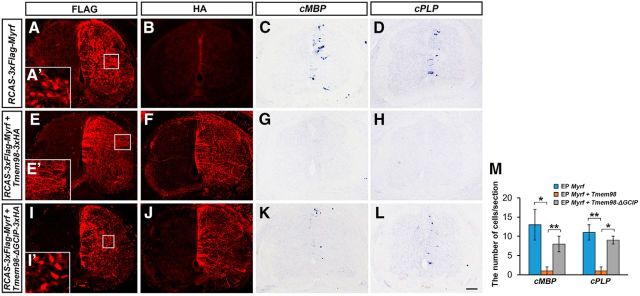
Inhibition of MYRF-induced myelin gene expression by TMEM98 in chicken spinal cords. A–D, Overexpression of MYRF induced ectopic expression of MBP and PLP in chicken spinal cords at cE7 stages. E–H, MYRF-induced MBP/PLP expression was almost completely blocked by TMEM98 coexpressoin. I–L, TMEM98-ΔGCIP was not capable of inhibiting MYRF self-cleavage and repressing MYRF functions. M, Quantitative analyses of MBP+ and PLP + cells. A′, I′, N-fused Flag (N-fragments of MYRF) was located in the nucleus when MYRF was expressed alone or with TMEM98-ΔGCIP. E′, N-fused Flag was detected in cell processes, but not in the nucleus when TMEM98 was coexpressed in chicken spinal cords. *p < 0.05, **p < 0.01. Scale bar, 100 μm. EP, electroporating.
We next examined the effect of TMEM98 expression on the nuclear translocation of the N-fragment of MYRF protein. In MYRF-transfected cells, the N-fragment tag was detected in the nucleus whereas the C-fragment tag was observed in the cytoplasm (Fig. 5B). However, in the presence of TMEM98, both the N- and C-terminal fragments were now localized in the cytoplasm (Fig. 5B), consistent with the biochemical inhibition of MYRF self-cleavage by TMEM98. To ensure that TMEM98 did not interfere with the nuclear translocation of the N-fragment itself, we expressed the N-fragment of MYRF (1–588) and TMEM98 simultaneously in HEK293T cells. As shown in Figure 5C, TMEM98 expression did not affect the nuclear translocation of the MYRF N-fragment.
TMEM98 suppresses OL differentiation in the developing spinal cord
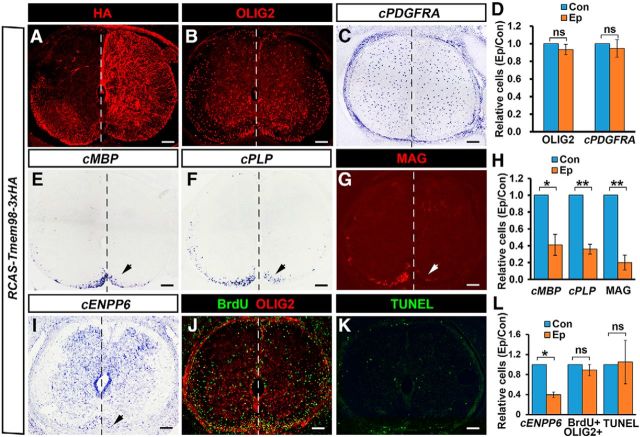
Previous studies demonstrated that the self-cleavage and N-fragment nuclear localization are required for MYRF to promote oligodendrocyte differentiation and myelin gene expression (Bujalka et al., 2013; Li et al., 2013). Our findings of TMEM98 inhibiting MYRF self-proteolysis raise the possibility that TMEM98 expression in OLs may negatively regulate the differentiation process of oligodendrocytes by interfering with MYRF activity. To test this hypothesis, we overexpressed TMEM98–3xHA protein in E2 chicken spinal cord via in ovo electroporation and analyzed its effects on myelin gene expression at the E10 stage. Exogenous expression of TMEM98 was identified by anti-HA immunostaining (Fig. 6A). It was found that overexpression of TMEM98 had no effect on the expression of OLIG2 and the OPC marker PDGFRA (p = 0.19 for OLIG2 and p = 0.47 for PDGFRA, n = 3; Fig. 6B–D) but caused >60% reduction in the number of MBP-, PLP-, and MAG-positive mature OLs and ENPP6+ newly formed OLs in the electroporated side compared with the control side (p = 0.015 for cMBP, p = 0.003 for cPLP, p = 0.004 for MAG, and p = 0.003 for cENPP6, n = 3; Fig. 6E–I). Furthermore, BrdU staining and TUNEL assay were used to detect cell proliferation and apoptosis, respectively. The results showed that the proliferation and cell death of OLIG2+ OLs were not affected by TMEM98 overexpression in the electroporated side of chicken spinal cords (Fig. 6J–L). Thus, overexpression of TMEM98 did not affect the initial specification or proliferation of oligodendrocytes in embryonic chicken spinal cord; instead, it suppressed the differentiation of oligodendrocytes.
Figure 6.
Suppression of OL differentiation by TMEM98 overexpression. A, Detection of TMEM98 expression at the electroporated side (right side) of chicken spinal cords at cE10 stages by anti-HA staining. B–D, The expression of OLIG2 and OPC marker PDGFRA were not impacted at the electroporated side. E–I, As indicated by arrows, the expression of MBP, PLP, MAG, and ENPP6 at the electroporated side was dramatically reduced. J–L, Cell proliferation and apoptosis were not affected by TMEM98 overexpression. *p < 0.05; **p < 0.01; ns, no significant difference; n = 3. Scale bars, 100 μm. Con, control side (left side). Ep, electroporated side.
We next examined whether TMEM98 directly acts to inhibit MYRF's activity in regulating myelin gene transcription. When full-length FLAG-MYRF was overexpressed in embryonic chicken spinal cords via electroporation, anti-FLAG signals were localized in the nucleus (Fig. 7A,A′), confirming the nuclear localization of its N-terminal fragment. As reported previously (Emery et al., 2009), overexpression of full-length FLAG-MYRF protein induced ectopic expression of myelin genes MBP and PLP (Fig. 7C,D). However, when TMEM98 was coexpressed in embryonic chicken spinal cord, the nuclear translocation of anti-FLAG signals was blocked (Fig. 7E,E′,F). More importantly, coexpression of TMEM98 almost completely inhibited the MYRF-induced ectopic expression of the MBP and PLP genes in electroporated tissues (Fig. 7G,H). To confirm that TMEM98 blocks myelin gene expression through disturbing MYRF self-cleavage, truncated TMEM98 without the GCIP domain was coexpressed with MYRF. As expected, TMEM98-ΔGCIP was not capable of inhibiting MYRF cleavage (Fig. 7I′) and did not suppress MYRF-induced myelin gene expression (Fig. 7I–M).
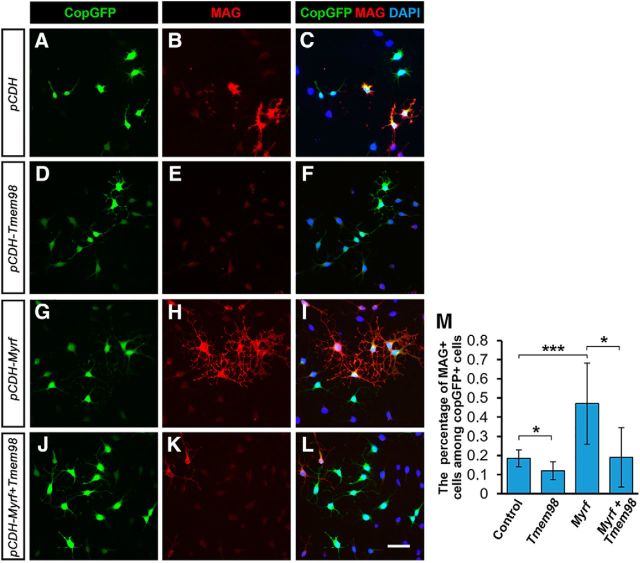
Furthermore, we tested TMEM98 function in CG4 cells, which are capable of differentiating into OLs in a serum-free medium (Louis et al., 1992). Consistent with the results in chicken spinal cords, TMEM98 overexpression inhibited CG4 cell differentiation and suppressed MYRF-induced MAG expression (Fig. 8). Together, these data strongly suggested that Tmem98 negatively regulates OL differentiation through inhibiting MYRF activity.
Figure 8.
Inhibition of MYRF-induced myelin gene expression by TMEM98 in CG4 cells. A–L, TMEM98 overexpression reduces spontaneous OL differentiation (A–F) and inhibits MYRF-induced MAG expression in CG4 cells (G–L). M, Quantitative analyses of MAG+ cells. *p < 0.05, ***p < 0.001. Scale bar, 25 μm.
Discussion
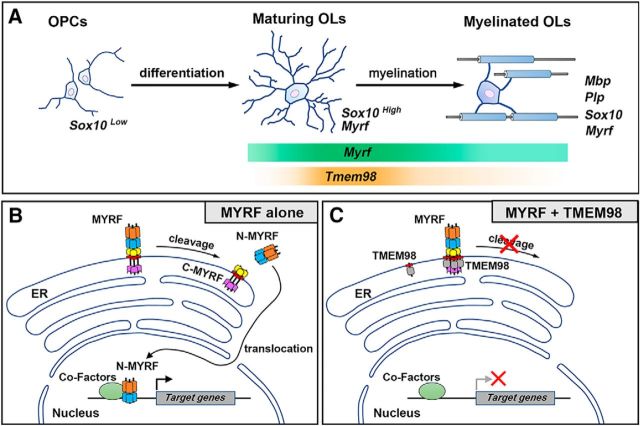
MYRF was first identified as an essential transcription factor for oligodendrocyte differentiation and myelin gene expression (Emery et al., 2009). Unexpectedly, full-length MYRF is initially produced as an ER membrane protein, and subsequent self-cleavage generates the nucleus-targeted N-terminal fragment conferring sequence-specific transcriptional activity. However, it remains unknown how MYRF's activity on the ER is regulated during oligodendrocyte differentiation. In this study, we identify TMEM98 as an interactor of MYRF on the ER membrane and provide the molecular and biochemical evidence that TMEM98 specifically binds MYRF on its C terminus and inhibits its self-cleavage, subcellular localization, and function (Figs. 4–7). Since Tmem98 is transcriptionally downstream of MYRF (Fig. 2L,M), it is possible that TMEM98 provides a negative feedback for MYRF-mediated OL differentiation. Together, these data suggest that the cleavage of MYRF on the ER is a regulated process and provide new insights into the mechanisms how MYRF plays the essential role in OL differentiation.
The nuclear fragment of MYRF is cleaved from the N end of ICA domain of MYRF. Downstream to the ICA domain is a single transmembrane domain that allows MYRF to be anchored on the ER membrane. Thus, both the N-MYRF and ICA are cytoplasmic, whereas the C-terminal MYRF (790–1138) faces to the ER lumen (Fig. 9; Li et al., 2013). Protein sequence analyses reveal that the transmembrane domain of TMEM98 is located at its very N-terminal end; thus, almost all TMEM98 protein is in the ER lumen (Fig. 9). This predicts that TMEM98 and MYRF interact through their ER-luminal motifs. Our protein interaction analyses strongly support this membrane-configuration model, as the TMEM98–MYRF interaction occurs on the transmembrane and the immediate adjacent regions. However, considering the requirement of the ER location for the interaction, we cannot rule out the possibility that some other regions are involved in the interaction as well. In support, the C-fragment of TMEM98 is required for inhibiting MYRF self-cleavage, even though the interaction is mediated by the N terminal of TMEM98 protein. At this stage, the molecular mechanism by which TMEM98 regulates MYRF cleavage is not known. Previous studies indicated that the self-proteolytic function of MYRF depends on its catalytic ICA domain and the trimerization of ICA domain (Kim et al., 2017). Based on our biochemical interaction analysis, TMEM98 binds to the C-MYRF excluding the catalytic ICA domain. Therefore, it is unlikely that the binding of TMEM98 protein directly interferes with its intramolecular cleavage activity. There are a number of possibilities for how TMEM98 may inhibit the cleavage of MYRF on the ER. One possibility is that TMEM98 binding prevents the oligomerization of MYRF, consequently affecting cleavage of MYRF. Another possibility is that the cleavage of MYRF may need a specific configuration on the ER and the bound TMEM98 may prevent the configuration process.
Figure 9.
Model for TMEM98 function in inhibiting MYRF self-cleavage and activity in the oligodendrocyte differentiation. A, Tmem98 is transiently expressed in newly differentiated oligodendrocytes. B, When expressed alone, N-fragments of MYRF are released from the ER membrane after self-cleavage and translocate into the nucleus to drive target-gene expression. C, When coexpressed with TMEM98, the release and nuclear translocation of MYRF N-fragments are suppressed, and the induction of target-gene expression by MYRF is turned off.
Although the N-terminal fragment of MYRF has been well characterized for its transcriptional function, the role of its C-terminal fragment in OL differentiation has been elusive. At the least, the C-MYRF anchors the full-length protein on the ER membrane, which may facilitate the effective oligomerization and cleavage. Moreover, as previous reports have demonstrated that certain residues on C-MYRF are glycosylated (Li et al., 2013), it is possible that additional post-translational modifications (e.g., phosphorylation, acetylation, or SUMOylation) may be required for proper nucleus translocation, DNA binding, and transcriptional activity of N-MYRF or its protein stability. TMEM98 may function to slow down MYRF self-cleavage to allow sufficient time for such post-translational modifications on the ER.
We found that Tmem98 is specifically expressed in early myelinating oligodendrocytes in the CNS, with a spatial and temporal pattern similar to that of Myrf (Fig. 1G,H). Outside the CNS, Tmem98 is expressed in a subset of non-neural tissues in midstage embryos; Moreover, Myrf and Tmem98 are expressed in tissues of similar types and positions (Huang et al., unpublished results). These observations suggest that Myrf and Tmem98 are elements of common developmental pathways. As C. elegans orthologs TMEM98 and MYRF also interact with each, we speculate that TMEM98–MYRF regulatory interaction is conserved and opted out for different developmental contexts in various species during evolution. Further investigation into TMEM98–MYRF interaction will provide more insights into the functional role of MYRF in regulating oligodendrocyte differentiation.
Footnotes
This work was supported by National Natural Science Foundation of China Grants 31572224 and 31771621.
The authors declare no competing financial interests.
References
- Andachi Y. (2008) A novel biochemical method to identify target genes of individual microRNAs: identification of a new Caenorhabditis elegans let-7 target. RNA 14:2440–2451. 10.1261/rna.1139508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubourg P. (1993) The leukodystrophies: a window to myelin. Nat Genet 5:105–106. 10.1038/ng1093-105 [DOI] [PubMed] [Google Scholar]
- Awadalla MS, Burdon KP, Souzeau E, Landers J, Hewitt AW, Sharma S, Craig JE (2014) Mutation in TMEM98 in a large white kindred with autosomal dominant nanophthalmos linked to 17p12–q12. JAMA Ophthalmol 132:970–977. 10.1001/jamaophthalmol.2014.946 [DOI] [PubMed] [Google Scholar]
- Barañano KW. (2016) Leukodystrophies. Semin Neurol 36:362–366. 10.1055/s-0036-1585455 [DOI] [PubMed] [Google Scholar]
- Baumann N, Pham-Dinh D (2001) Biology of oligodendrocyte and myelin in the mammalian central nervous system. Physiol Rev 81:871–927. 10.1152/physrev.2001.81.2.871 [DOI] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL (1997) The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89:331–340. 10.1016/S0092-8674(00)80213-5 [DOI] [PubMed] [Google Scholar]
- Bujalka H, Koenning M, Jackson S, Perreau VM, Pope B, Hay CM, Mitew S, Hill AF, Lu QR, Wegner M, Srinivasan R, Svaren J, Willingham M, Barres BA, Emery B (2013) MYRF is a membrane-associated transcription factor that autoproteolytically cleaves to directly activate myelin genes. PLoS Biol 11:e1001625. 10.1371/journal.pbio.1001625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A, Tourtellotte WW, Rudick R, Trapp BD (2002) Premyelinating oligodendrocytes in chronic lesions of multiple sclerosis. N Engl J Med 346:165–173. 10.1056/NEJMoa010994 [DOI] [PubMed] [Google Scholar]
- Chen Y, Balasubramaniyan V, Peng J, Hurlock EC, Tallquist M, Li J, Lu QR (2007) Isolation and culture of rat and mouse oligodendrocyte precursor cells. Nat Protoc 2:1044–1051. 10.1038/nprot.2007.149 [DOI] [PubMed] [Google Scholar]
- Cui QL, Kuhlmann T, Miron VE, Leong SY, Fang J, Gris P, Kennedy TE, Almazan G, Antel J (2013) Oligodendrocyte progenitor cell susceptibility to injury in multiple sclerosis. Am J Pathol 183:516–525. 10.1016/j.ajpath.2013.04.016 [DOI] [PubMed] [Google Scholar]
- De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm JS, Schroeter EH, Schrijvers V, Wolfe MS, Ray WJ, Goate A, Kopan R (1999) A presenilin-1-dependent gamma-secretase-like protease mediates release of notch intracellular domain. Nature 398:518–522. 10.1038/19083 [DOI] [PubMed] [Google Scholar]
- Dulamea AO. (2017) Role of oligodendrocyte dysfunction in demyelination, remyelination and neurodegeneration in multiple sclerosis. Adv Exp Med Biol 958:91–127. 10.1007/978-3-319-47861-6_7 [DOI] [PubMed] [Google Scholar]
- Emery B, Agalliu D, Cahoy JD, Watkins TA, Dugas JC, Mulinyawe SB, Ibrahim A, Ligon KL, Rowitch DH, Barres BA (2009) Myelin gene regulatory factor is a critical transcriptional regulator required for CNS myelination. Cell 138:172–185. 10.1016/j.cell.2009.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin RJ. (2002) Why does remyelination fail in multiple sclerosis? Nat Rev Neurosci 3:705–714. 10.1038/nrn917 [DOI] [PubMed] [Google Scholar]
- Fu W, Cheng Y, Zhang Y, Mo X, Li T, Liu Y, Wang P, Pan W, Chen Y, Xue Y, Ma D, Zhang Y, Han W (2015) The secreted form of transmembrane protein 98 promotes the differentiation of T helper 1 cells. J Interferon Cytokine Res 35:720–733. 10.1089/jir.2014.0110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo V, Deneen B (2014) Glial development: the crossroads of regeneration and repair in the CNS. Neuron 83:283–308. 10.1016/j.neuron.2014.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haze K, Yoshida H, Yanagi H, Yura T, Mori K (1999) Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell 10:3787–3799. 10.1091/mbc.10.11.3787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Zhao XF, Zheng K, Qiu M (2013) Regulation of the timing of oligodendrocyte differentiation: mechanisms and perspectives. Neurosci Bull 29:155–164. 10.1007/s12264-013-1314-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Teng P, Mei R, Yang A, Zhang Z, Zhao X, Qiu M (2017) Tmeff2 is expressed in differentiating oligodendrocytes but dispensable for their differentiation in vivo. Sci Rep 7:337. 10.1038/s41598-017-00407-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings A, Carroll W (2010) Quantification of oligodendrocyte progenitor cells in human and cat optic nerve: implications for endogenous repair in multiple sclerosis. Glia 58:1425–1436. [DOI] [PubMed] [Google Scholar]
- Kim D, Choi JO, Fan C, Shearer RS, Sharif M, Busch P, Park Y (2017) Homo-trimerization is essential for the transcription factor function of myrf for oligodendrocyte differentiation. Nucleic Acids Res 45:5112–5125. 10.1093/nar/gkx080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenning M, Jackson S, Hay CM, Faux C, Kilpatrick TJ, Willingham M, Emery B (2012) Myelin gene regulatory factor is required for maintenance of myelin and mature oligodendrocyte identity in the adult CNS. J Neurosci 32:12528–12542. 10.1523/JNEUROSCI.1069-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Richardson WD (2016) Evolution of the CNS myelin gene regulatory program. Brain Res 1641:111–121. 10.1016/j.brainres.2015.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Park Y, Marcotte EM (2013) A bacteriophage tailspike domain promotes self-cleavage of a human membrane-bound transcription factor, the myelin regulatory factor MYRF. PLoS Biol 11:e1001624. 10.1371/journal.pbio.1001624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Hu X, Cai J, Liu B, Peng X, Wegner M, Qiu M (2007) Induction of oligodendrocyte differentiation by Olig2 and Sox10: evidence for reciprocal interactions and dosage-dependent mechanisms. Dev Biol 302:683–693. 10.1016/j.ydbio.2006.10.007 [DOI] [PubMed] [Google Scholar]
- Louis JC, Magal E, Muir D, Manthorpe M, Varon S (1992) CG-4, a new bipotential glial cell line from rat brain, is capable of differentiating in vitro into either mature oligodendrocytes or type-2 astrocytes. J Neurosci Res 31:193–204. 10.1002/jnr.490310125 [DOI] [PubMed] [Google Scholar]
- Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, Rowitch DH (2002) Common developmental requirement for olig function indicates a motor neuron/oligodendrocyte connection. Cell 109:75–86. 10.1016/S0092-8674(02)00678-5 [DOI] [PubMed] [Google Scholar]
- Mastracci TL, Lin CS, Sussel L (2013) Generation of mice encoding a conditional allele of Nkx2.2. Transgenic Res 22:965–972. 10.1007/s11248-013-9700-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng J, Ma X, Tao H, Jin X, Witvliet D, Mitchell J, Zhu M, Dong MQ, Zhen M, Jin Y, Qi YB (2017) Myrf ER-bound transcription factors drive C. elegans synaptic plasticity via cleavage-dependent nuclear translocation. Dev Cell 41:180–194.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momose T, Tonegawa A, Takeuchi J, Ogawa H, Umesono K, Yasuda K (1999) Efficient targeting of gene expression in chick embryos by microelectroporation. Dev Growth Differ 41:335–344. 10.1046/j.1440-169X.1999.413437.x [DOI] [PubMed] [Google Scholar]
- Morgan BA, Fekete DM (1996) Manipulating gene expression with replication-competent retroviruses. Methods Cell Biol 51:185–218. 10.1016/S0091-679X(08)60629-9 [DOI] [PubMed] [Google Scholar]
- Muth KN, Piefke S, Weider M, Sock E, Hermans-Borgmeyer I, Wegner M, Küspert M (2016) The dual-specificity phosphatase Dusp15 is regulated by Sox10 and myrf in myelinating oligodendrocytes. Glia 64:2120–2132. 10.1002/glia.23044 [DOI] [PubMed] [Google Scholar]
- Ng KT, Lo CM, Guo DY, Qi X, Li CX, Geng W, Liu XB, Ling CC, Ma YY, Yeung WH, Shao Y, Poon RT, Fan ST, Man K (2014) Identification of transmembrane protein 98 as a novel chemoresistance-conferring gene in hepatocellular carcinoma. Mol Cancer Ther 13:1285–1297. 10.1158/1535-7163.MCT-13-0806 [DOI] [PubMed] [Google Scholar]
- Paes de Faria J, Kessaris N, Andrew P, Richardson WD, Li H (2014) New Olig1 null mice confirm a non-essential role for Olig1 in oligodendrocyte development. BMC Neurosci 15:12. 10.1186/1471-2202-15-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Cai J, Wu Y, Wu R, Lee J, Fu H, Rao M, Sussel L, Rubenstein J, Qiu M (2001) Control of oligodendrocyte differentiation by the Nkx2.2 homeodomain transcription factor. Development 128:2723–2733. [DOI] [PubMed] [Google Scholar]
- Raine CS, Wu E (1993) Multiple sclerosis: remyelination in acute lesions. J Neuropathol Exp Neurol 52:199–204. 10.1097/00005072-199305000-00003 [DOI] [PubMed] [Google Scholar]
- Redwine JM, Evans CF (2002) Markers of central nervous system glia and neurons in vivo during normal and pathological conditions. Curr Top Microbiol Immunol 265:119–140. [DOI] [PubMed] [Google Scholar]
- Richardson WD, Kessaris N, Pringle N (2006) Oligodendrocyte wars. Nat Rev Neurosci 7:11–18. 10.1038/nrn1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoo H, Wang HY, Araki T, Williams JG, Fukuzawa M (2012) An orthologue of the myelin-gene regulatory transcription factor regulates dictyostelium prestalk differentiation. Int J Dev Biol 56:325–332. 10.1387/ijdb.120030jw [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons M, Nave KA (2015) Oligodendrocytes: myelination and axonal support. Cold Spring Harb Perspect Biol 8:a020479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Rehberg S, Ader M, Lommes P, Riethmacher D, Schachner M, Bartsch U, Wegner M (2002) Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev 16:165–170. 10.1101/gad.215802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolswijk G. (1998) Chronic stage multiple sclerosis lesions contain a relatively quiescent population of oligodendrocyte precursor cells. J Neurosci 18:601–609. 10.1523/JNEUROSCI.18-02-00601.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Zhao X, Zheng K, Li H, Huang H, Zhang Z, Mastracci T, Wegner M, Chen Y, Sussel L, Qiu M (2014) Genetic evidence that Nkx2.2 and pdgfra are major determinants of the timing of oligodendrocyte differentiation in the developing CNS. Development 141:548–555. 10.1242/dev.095323 [DOI] [PMC free article] [PubMed] [Google Scholar]