Abstract
A variety of organisms, such as bacteria, fungi, and plants, produce secondary metabolites, also known as natural products. Natural products have been a prolific source and an inspiration for numerous medical agents with widely divergent chemical structures and biological activities, including antimicrobial, immunosuppressive, anticancer, and anti-inflammatory activities, many of which have been developed as treatments and have potential therapeutic applications for human diseases. Aside from natural products, the recent development of recombinant DNA technology has sparked the development of a wide array of biopharmaceutical products, such as recombinant proteins, offering significant advances in treating a broad spectrum of medical illnesses and conditions. Herein, we will introduce the structures and diverse biological activities of natural products and recombinant proteins that have been exploited as valuable molecules in medicine, agriculture and insect control. In addition, we will explore past and ongoing efforts along with achievements in the development of robust and promising microorganisms as cell factories to produce biologically active molecules. Furthermore, we will review multi-disciplinary and comprehensive engineering approaches directed at improving yields of microbial production of natural products and proteins and generating novel molecules. Throughout this article, we will suggest ways in which microbial-derived biologically active molecular entities and their analogs could continue to inspire the development of new therapeutic agents in academia and industry.
Keywords: natural products, biologics, biological activity, microbial cell factories, genetic engineering, combinatorial biosynthesis, production improvement
Introduction
Natural products originate as secondary metabolites from a myriad of sources, including terrestrial plants, animals, marine organisms, microorganisms, terrestrial vertebrates and invertebrates (Chin et al., 2006). These structurally and chemically diverse molecules act as a remarkable class of therapeutics to heal various ailments. The earliest documentation of the application of natural products to improve human health dates back to the ancient Mesopotamia’s sophisticated medicinal system from 2900 to 2600 BCE (Borchardt, 2002; Siddiqui et al., 2014). By the early 1900’s, approximately 80% of all medicines were obtained from plant sources (Sneader, 1997; Siddiqui et al., 2014). The discovery of penicillin from Penicillium notatum by Alexander Fleming in 1928 marked a significant shift from plants to microorganisms as a source of natural products (Fleming, 1944). Since then, microorganism-derived compounds have been utilized in medicine, agriculture, food industry and scientific research (Sanchez et al., 2012). The early years of antibiotic research discovered streptomycin from Streptomyces griseus (Waksman et al., 1946), chloramphenicol from Streptomyces venezuelae (Duggar, 1948), chlortetracycline from Streptomyces aureofaciens (Ehrlich et al., 1947), cephalosporin C from Cephalosporium acremonium (Newton and Abraham, 1955), erythromycin from Saccharopolyspora erythraea and vancomycin from Amycolatopsis orientalis (Geraci et al., 1956). Given these historical successes, large pharmaceutical companies have continued to invest in this traditional domain (Dias et al., 2012). Currently, approximately 60% of approved small molecule medicines are related to natural products, and 69% of all antibacterial agents originate from natural products (Patridge et al., 2016; Matsumura et al., 2018). However, many natural compounds with potential as novel drug candidates occur in low concentrations in nature, often making drug discovery and development burdensome and economically impractical. Therefore, an emerging alternative solution is to express biosynthetic genes from the original producers in microbial hosts, notably bacteria and fungi (Song et al., 2014). Engineered microbes can produce appreciable amounts of scarce natural compounds, thereby facilitating the synthesis of the target novel compound and potent derivatives, as well as the validation of their activities (Matsumura et al., 2018).
The natural product sector is not the only area that has undergone substantial growth or utilizes therapeutic products generated in/from living organisms. Prokaryotic and eukaryotic microbial cells, in combination with the advancement of recombinant DNA techniques, have been responsible for an explosion of biologics. Biologics are a set of molecules whose active pharmaceutical ingredients are derived from living organisms such as animals, plants, microorganisms, human blood products, and tissue transplants that are too complex to be produced through organic synthesis (Revers and Furczon, 2010). They can be categorized into five main classes: (1) monoclonal antibodies, like trastuzumab (Herceptin®) and rituximab (Rituxan®); (2) blood factor derivatives, like coagulation factor VIIa (NovoSeven RT®) and epoiten alfa (Epogen®); (3) vaccines; (4) enzymes; and (5) recombinant proteins, such as immunomodulatory cytokines, and thrombolytic agents (Lacana et al., 2007). Since the approval of recombinant human insulin and recombinant human growth hormone as some of the first modern biopharmaceuticals, large numbers of additional biopharmaceuticals have been developed, approved, and marketed using different microbial expression systems; many more are currently in the development pipeline (Graumann and Premstaller, 2006). After the successful production of the recombinant human insulin Humulin®, Escherichia coli quickly became the prevalent expression platform in the 1980s when the biopharmaceutical sector emerged and was followed by yeast Saccharomyces cerevisiae (Sanchez-Garcia et al., 2016). Microbial cells constitute the majority of hosts employed in the production of currently approved recombinant pharmaceuticals for human treatment, mainly because of their lack of unconventional post-translational modifications, proteolytic instability, poor solubility and activation of cell stress responses (Graumann and Premstaller, 2006). This demonstrates that microbial hosts represent convenient and robust platforms for the efficient production of recombinant proteins despite some bottlenecks and obstacles.
Herein, we will summarize the biological activities and applications of a variety of natural products and biologics and review the microbial systems used to produce these pharmaceutical compounds. We will also cover past and current attempts at improving the microbial production of these biological molecules and generating new molecules using diverse engineering approaches. In addition, we will discuss the challenges of the production of natural products and biologics in microbial systems and advances that can help overcome them for drug discovery and development. Future prospects for cutting-edge developments and technological advances in microbial production of bioactive natural products and recombinant proteins as the most valuable sources of therapeutics are also discussed.
Biological Activities of Natural Products and Biologics
Natural products have diverse biological activities relevant to human health, including antibiotic, antifungal, anticancer, immunosuppressive, anti-inflammatory, biofilm inhibitory activities, etc. In this section, we will focus on the biological activities of natural products, which can be grouped into several categories. The biological activities of microbial recombinant proteins will be also reviewed.
Antibiotics
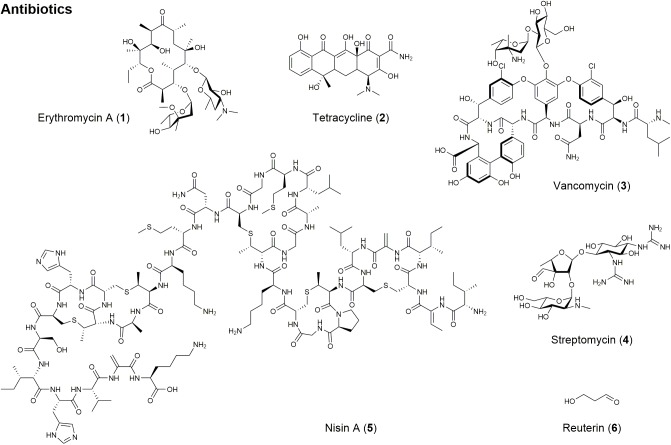
Natural products are a rich source for antibiotic drug development, but the most clinically useful of these scaffolds can be classified as polyketides, non-ribosomal peptides, and aminoglycosides (Wright, 2014). Polyketides, assembled by polyketide synthases (PKS), make up one of the largest classes of chemically diverse natural products and are among the most important secondary metabolites for their applications in medicine, agriculture, and industry (Tae et al., 2007). For example, pikromycin was the first known polyketide antibiotic produced from S. venezuelae in 1950 (Vazquez, 1967; Jung et al., 2006). It has been reported that pikromycin is very potent against multi-drug resistant respiratory pathogens (Woo et al., 2014). Another remarkable polyketide antibiotic with significant clinical applications is erythromycin A (1; Figure 1 and Table 1), which was first discovered in 1952 as a broad-spectrum antibiotic produced by S. erythraea (McGuire et al., 1952). This antibiotic is prescribed to a treat wide variety of bacterial infections, such as respiratory and gastrointestinal infections, whooping cough, syphilis, and acne, especially in patients who have adverse reactions against penicillin (Cobb et al., 2013). While many natural antibiotics fail to inhibit Gram-negative organisms, tetracyclines (2; Figure 1 and Table 1) are active against both Gram-positive and Gram-negative bacteria (Chopra and Roberts, 2001; Demain, 2009).
FIGURE 1.
Structures of natural products with antibiotic activity.
Table 1.
Biological activities of microbial-derived natural products and biologics.
| Name | Origin | Biological activity | References |
|---|---|---|---|
| Antibiotic | |||
| Erythromycin A (1) | Saccharopolyspora erythraea | Antibacterial | McGuire et al., 1952; Zhang et al., 2010; Cobb et al., 2013 |
| Tetracycline (2) | Streptomyces rimosus | Antibacterial | Chopra and Roberts, 2001; Demain, 2009 |
| Vancomycin (3) | Amycolatopsis orientalis | Antibacterial | Geraci et al., 1956; Dasgupta, 2012 |
| Streptomycin (4) | Streptomyces griseus | Antibacterial | Schatz et al., 1944; Waksman et al., 1946 |
| Nisin A (5) | Lactococcus lactis | Antimicrobial | Li and Vederas, 2009; Gyawali and Ibrahim, 2014 |
| Reuterin (6) | Lactobacillus reuteri | Antimicrobial | Talarico et al., 1988; Gyawali and Ibrahim, 2014 |
| Antifungal Agents | |||
| Amphotericin B (7) | Streptomyces nodosus | Antifungal | Abu-Salah, 1996; Tevyashova et al., 2013 |
| Ieodoglucomide C (8) | Bacillus licheniformis | Antifungal | Tareq et al., 2015 |
| Anticancer and Antitumor | |||
| Bleomycin (9) | Streptoalloteichus hindustanus, Streptomyces verticillus | Squamous cell carcinomas, Hodgkin’s lymphomas and testis tumors | Einhorn and Donohue, 2002; Demain and Vaishnav, 2011 |
| Ddaunorubicin (10) | Streptomyces peucetius and various related strains | Acute lymphoblastic or myeloblastic lymphoma | Di Marco et al., 1981; Giddings and Newman, 2013 |
| Immunosuppressant/Anti-inflammatory Agents | |||
| Rapamycin (11) | Streptomyces rapamycinicus (formerly, Streptomyces hygroscopicus ATCC 29253), Streptomyces iranensis, and Actinoplanes sp. N902-109 | Immunosuppressive, antifungal, antitumor, neuroprotective, neuroregenerative, and lifespan extension activities, growth inhibitory activity against several fungi | Vezina et al., 1975; Mann, 2001; Law, 2005; Pan et al., 2008; Anisimov et al., 2011; Song et al., 2015; Yoo et al., 2017 |
| FK506 (12) | Streptomyces tsukubaensis and several Streptomyces species | Immunosuppressive, antifungal, anti-inflammatory, neuroprotective and neuroregenerative activities, rheumatoid arthritis treatment | Tanaka et al., 1987; Mann, 2001; Migita and Eguchi, 2003; Demain, 2014; Ban et al., 2016; Yoo et al., 2017 |
| Biofilm-Inhibitory Agents | |||
| Cahuitamycins (13) | Streptomyces gandocaensis | Inhibitors of Acinetobacter baumannii biofilms | Park et al., 2016 |
| Others | |||
| Avermectins (14) | Streptomyces avermitilis | Onchocerciasis and lymphatic filariasis | Shen, 2015 |
| Mollemycin A 20 (15) | Streptomyces sp. (CMB-M0244) | Gram-positive and Gram-negative bacteria, antimalarial activity | Blunt et al., 2016 |
| Lipstatin (16) | Streptomyces toxytricini | Pancreatic lipase inhibitor for obesity and diabetes | Weibel et al., 1987; Sanchez et al., 2012 |
As previously mentioned, penicillin is a well-known antibiotic secondary metabolite from P. notatum and is effective against Gram-positive bacteria, which are responsible for diseases such as scarlet fever, pneumonia, gonorrhea, meningitis, and diphtheria (Fleming, 2001; Tan and Tatsumura, 2015). Penicillin belongs to non-ribosomal peptide antibiotics along with vancomycin (Fischbach and Walsh, 2006). Non-ribosomal peptides, assembled by non-ribosomal peptide synthetase (NRPS), possess bioactivity that can be exploited for therapeutic applications and are amongst the most widespread and structurally diverse secondary metabolites. Vancomycin (3; Figure 1 and Table 1) is another potent non-ribosomal peptide against pathogenic bacteria, including Clostridium difficile, Listeria monocytogenes, Streptococcus pneumoniae, Staphylococcus epidermidis, and methicillin-resistant Staphylococcus aureus (MRSA) (Dasgupta, 2012).
Aminoglycosides are another class of antibiotics that act by binding to the rRNA subunit of the 30S bacterial ribosome and inhibiting protein synthesis (Moazed and Noller, 1987). Streptomycin (4; Figure 1 and Table 1) produced by S. griseus is the first aminoglycoside discovered in 1944 and effective against pulmonary tuberculosis (Schatz et al., 1944). Since the discovery of streptomycin, aminoglycoside antibiotics such as kanamycin, gentamicin, sisomicin, and lividomycin have been discovered and widely used to treat infectious organisms that have developed resistance against streptomycin after prolonged use (Park et al., 2013). Despite their excellent antibacterial activity, aminoglycosides have met with resistant organisms. In order to combat antibiotic resistance to aminoglycoside antibiotics, semi-synthetic aminoglycoside antibiotics were specifically tailored to shield against these enzymes (Van Bambeke et al., 2017). Semi-synthetic aminoglycoside antibiotics such as amikacin, netilmicin, dibekacin, and isepamicin are developed as a result of semi-synthetic derivatives of the natural product (Miller et al., 1976; Leggett, 2015).
Natural antimicrobials have also been important to the food industry in terms of food safety against foodborne pathogens. Microbes such as lactic acid bacteria, produce a wide range of chemicals that have been shown to inhibit the growth and development of other microbial species. Nisin A (5; Figure 1 and Table 1), a bacteriocin produced by Lactococcus lactis, is approved to preserve food in over 50 countries and is very active against Gram-positive bacteria resistant to conventional antibiotics (Li and Vederas, 2009; Gyawali and Ibrahim, 2014). Reuterin (6; Figure 1 and Table 1) from Lactobacillus reuteri has been shown to have antimicrobial activities against foodborne pathogens and spoilage organisms when evaluated in milk, dairy, and meat products (Talarico et al., 1988; Gyawali and Ibrahim, 2014).
Antifungal Agents
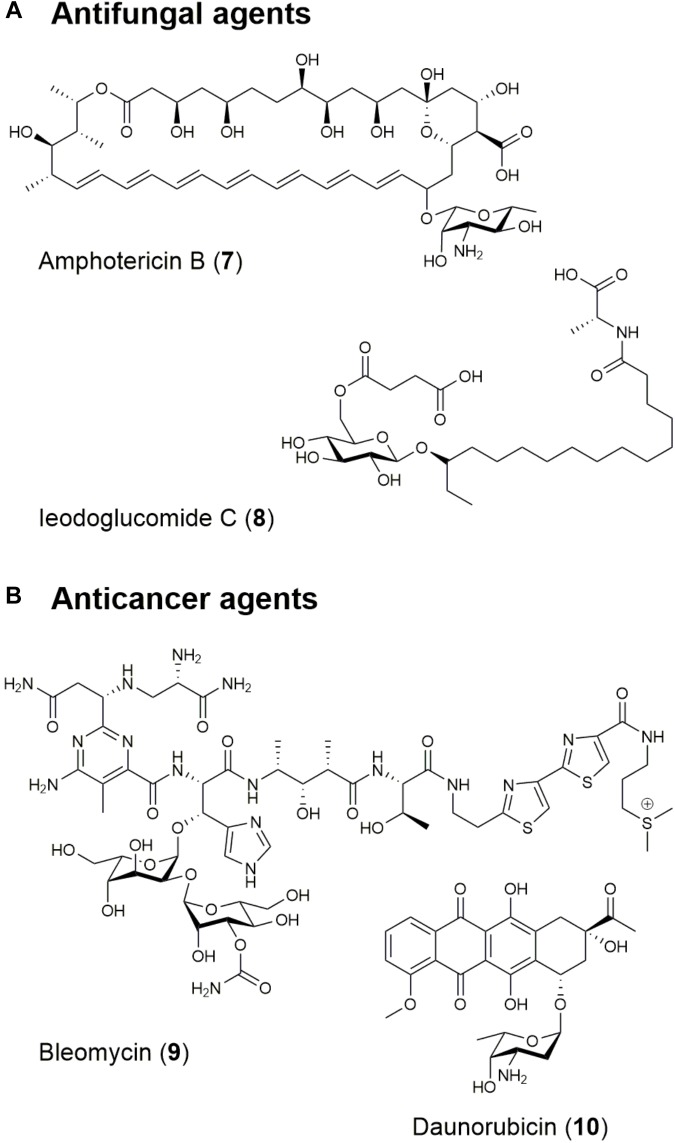
Nystatin, one of the first effective polyene antifungal agent, was obtained from Streptomyces noursei in 1950 and effective against Aspergillus species (Stanley and English, 1965). Clinically, nystatin plays a significant role as a topical antifungal agent in treating oral, gastro-intestinal, and genital candidosis (Fjærvik and Zotchev, 2005). Amphotericin B (7; Figure 2 and Table 1) is also a traditional polyene antifungal product of Streptomyces nodosus utilized against life-threatening fungal infections caused by Aspergillus species, and especially effective in patients who have undergone organ transplantation, received aggressive chemotherapy or with acquired immunodeficiency syndrome (Abu-Salah, 1996; Tevyashova et al., 2013).
FIGURE 2.
Structures of natural products with (A) antifungal and (B) anticancer/antitumor activities.
Recently, in a review of natural products with anti-Candida albicans activity, 71 substances of the 142 evaluated were determined to have antifungal activity under the criteria of having minimum inhibitory concentration (MIC) values below 8 mg/mL (Zida et al., 2017). The glycolipids ieodoglucomide C (8; Figure 2 and Table 1) and ieodoglycolipid were isolated from the marine bacterium Bacillus licheniformis and exhibited antifungal activities with a 21 μg/L MIC against Aspergillus niger, Rhizoctonia solani, Botrytis cinerea, Colletotrichum acutatum, and C. albicans (Tareq et al., 2015). Both ieodoglucomide C and ieodoglycolipid also exhibit good antibiotic properties against S. aureus, Bacillus subtilis, Bacillus cereus, Salmonella typhi, E. coli and Pseudomonas aeruginosa with MICs ranging from 0.01 to 0.05 μM, establishing these compounds as strong potential candidates for the development of new fungicides (Tareq et al., 2015).
Anticancer Agents
There are many microbe-derived anticancer agents that have been evaluated through clinical trials. For instance, the polyketide actinomycin was isolated from Streptomyces parvulus in 1940 and was the first antibiotic shown to have anticancer activity (Waksman and Woodruff, 1940; Hollstein, 1974). In particular, actinomycin D, also known as dactinomycin, is approved by FDA and has been widely used in clinical practice as an anticancer drug for treating many tumors, such as Wilms’ tumor, childhood rhabdomyosarcoma, Ewing’s sarcoma, and metastatic, non-seminomatous testicular cancer.
Another notable example is the therapeutic combination of the microbial product bleomycin (9; Figure 2 and Table 1), the plant compound etoposide, and the synthetic agent cisplatin, which has played a significant role in increasing the cure rate for disseminated testicular cancer from 5% in 1974 to 90% in 2011 (Einhorn and Donohue, 2002; Demain and Vaishnav, 2011). Bleomycin is a glycopeptide produced by Streptoalloteichus hindustanus and has been used for squamous cell carcinomas, melanomas, sarcomas, testicular, and ovarian cancer, Hodgkin’s and non-Hodgkin’s lymphomas, and testis tumors as an anticancer agent (Demain and Vaishnav, 2011). Its derivative, blenoxane is also used clinically with other compounds against lymphomas, skin carcinomas, and tumors of the head, neck, and testicles (Demain and Vaishnav, 2011). The anthracyclines are also an important family of polyketides produced by Streptomyces species by iterative PKS pathways and include daunorubicin (10; Figure 2 and Table 1) (Di Marco et al., 1981), epirubicin (Cersosimo and Hong, 1986), and doxorubicin (Metsä-Ketelä et al., 2008). The FDA approved the use of daunorubicin and doxorubicin for cancer therapy in the 1960s. Daunorubicin is used in the treatment of acute lymphoblastic or myeloblastic lymphoma. Meanwhile, doxorubicin is used in the treatment of breast cancer, solid tumors in children, soft tissue sarcomas, and aggressive lymphomas (Giddings and Newman, 2013).
Among numerous recent examples, rapamycin, wortmannin, and geldanamycin have been found to have antiproliferative activities during clinical use as novel chemotherapeutic agents (da Rocha et al., 2001). Rapamycin, a natural product derived from Streptomyces rapamycinicus has anticancer activity in addition to its immunosuppressive, anti-inflammatory, and antifungal activities (Kim et al., 2014). It performs antitumor activity on a tumor cell by hindering its proliferation, triggering apoptosis, and inhibiting angiogenesis (Law, 2005). Wortmannin is a fungal furanosteroid derivative of Penicillium funiculosum (Davidson et al., 2013). It has shown as an effective selective inhibitor of phosphoinositide 3-kinases (PI3Ks) and PI3K- related enzymes which are play a key role in intracellular signaling pathways (Sieber et al., 2010). A study on the proliferation and apoptosis of human breast MCF-7 cells treated with wortmannin uncovered that wortmannin shows antitumor activity by triggering apoptosis and impeding proliferation of cancer cells by suppressing PI3K/Akt signaling and NF-κB protein expression (Yun et al., 2012). Geldanamycin is a benzoquinone ansamycin antitumor compound derived from Streptomyces hygroscopicus var. geldanus (Singh et al., 2010). Geldanamycin prevents ATPase activity by binding to the heat shock protein and hindering the stability and function of oncogenic protein kinases involved in signal amplification cascade that controls proliferation and apoptosis (Singh et al., 2010). Geldanamycin and its analogs play a key role as anticancer agent in multiple myeloma, breast, and prostate cancer (Gorska et al., 2012). Another example is epothilone, an anticancer agent produced from mycobacterium Sorangium cellulosum. It obstructs microtubule depolymerization thereby causing G2-M interphase arrest of the cell cycle (Molnar et al., 2000). There are also marine microbial natural products that have anticancer activities, such as dolastatin, which is originated from cyanobacteria of the genera Symploca and Lyngbya (Simmons et al., 2008).
Immunosuppressive Agents
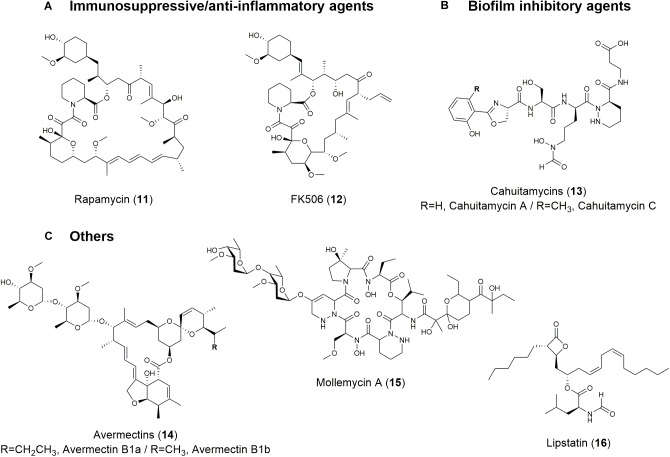
Rapamycin (also known as sirolimus) (11; Figure 3 and Table 1) and FK506 (tacrolimus) (12; Figure 3 and Table 1) are microbial natural products with immunosuppressive properties. Rapamycin blocks the proliferation of most cell types in response to activation by IL-2, IL-3, platelet-derived growth factor, epidermal growth factor, and insulin (Vezina et al., 1975). Rapamycin also exhibits synergism with other immunosuppressants, such as cyclosporin, to significantly reduce kidney toxicity and acute renal allograft rejection (Yoo et al., 2017). This compound has been developed to coat coronary stents and prevent organ transplant rejection and lymphangioleiomyomatosis; it was approved by the FDA for wider use in 1999 (Mann, 2001). In addition to its immunosuppressive activity, rapamycin possesses several other biological activities, including antitumor, neuroprotective/neuroregenerative, antineoplastic, and lifespan extension activities (Law, 2005; Pan et al., 2008; Yoo et al., 2017).
FIGURE 3.
Structures of natural products with (A) immunosuppressive/anti-inflammatory, (B) biofilm-inhibitory, and (C) other activities.
FK506 is also an immunosuppressive drug and was first discovered in soil samples containing Streptomyces tsukubaensis and several other Streptomyces species (Tanaka et al., 1987). FK506 is used to minimize organ rejection and to induce immunosuppression via calcineurin inhibition and interruption of T cell activation pathway (Migita and Eguchi, 2003). It has been demonstrated to be more effective than cyclosporin and non-toxic in low doses (Demain, 2014). The discovery of its immunosuppressive activity led to its use in heart, liver, and kidney transplants with overwhelming success (Demain, 2014). Like rapamycin, FK506 possesses various biological activities, including antifungal, anti-inflammatory, neuroprotective, and neuroregenerative activities (Ban et al., 2016).
Anti-inflammatory Agents
Some natural products also have anti-inflammatory activities. FK506 has shown efficacy in the treatment of refractory rheumatoid arthritis, a chronic inflammatory disease (Migita and Eguchi, 2003). Rapamycin also inhibits the inflammatory response after spinal cord injury by diminishing the activation and proliferation of inflammatory cells and the expression of inflammatory cytokines, thereby reducing secondary injury in the spinal cord and providing a neuroprotective effect (Song et al., 2015). Recently, strepsesquitriol, isolated from Streptomyces sp. SCSIO 10355, has been found to have anti-inflammatory activity through the inhibition of tumor necrosis factor-α production in lipopolysaccharide-activated macrophages (Yang et al., 2013). Salinamides A and B from marine Streptomyces sp. CNB-091 also displayed potent topical anti-inflammatory activity through a phorbol ester-induced mouse ear edema assay (Trischman et al., 1994). One study evaluated 7 peptides found in the Faecalibacterium prausnitzii supernatant, all belonging to a protein named microbial anti-inflammatory molecule (Breyner et al., 2017). These peptides were able to inhibit the NF-κB pathway in vitro and showed anti-inflammatory properties in vivo in a dinitrobenzene sulfate-induced colitis model (Breyner et al., 2017).
Biofilm Inhibitory Agents
Parasitic microorganisms adhere to solid surfaces and form layers of a complex polysaccharide matrix called a biofilm that confers resistance against antibiotics as wells as inflicts significant chronic bacterial infections (Singh et al., 2017). Analogs of 5-benzylidene-4-oxazolidinones are small molecules derived from marine natural products. These molecules inhibit 89% of biofilm formed by MRSA at 0.78 μM and disperses pre-formed biofilms at 4.7 μM (Edwards et al., 2017). A synthetic library of 2-aminoimidazole triazoles was able to successfully inhibit 94% of biofilm formation in Acinetobacter baumannii and MRSA at 100 μM (Rabin et al., 2015). Another recent example is cahuitamycins A-C (13; Figure 3 and Table 1) derived from the marine bacterium Streptomyces gandocaensis. Cahuitamycins have been evaluated as inhibitors of A. baumannii biofilms and it has been found that cahuitamycin C shows half maximal inhibitory concentration (IC50) at 14.5 μM. Modifications of cahuitamycins through selective mutasynthesis have yielded cahuitamycins D and E with an increased the potency of antibiofilm activity against A. baumannii (Park et al., 2016). The FDA-approved antitumor agent actinomycin D has also significant biofilm inhibitory activity against methicillin resistant- and sensitive-strains of S. aureus (Fracchia et al., 2010; Lee et al., 2016). In addition to bacterial biofilm, fungal biofilm associated with Candida pathogens is responsible for serious C. albicans infections linked to biofilm formation on medical devices. One study showed that Lactobacillus biosurfactants displayed high anti-adhesive biofilm formation properties against C. albicans and also prevented biofilm formation of L. monocytogenes, Salmonella arizonae, E. coli, and S. aureus (Fracchia et al., 2010).
Others
Natural products can also act as antiparasitic agents. The avermectins (14; Figure 3 and Table 1) and the derivative ivermectin have shown antiparasitic activity by significantly lowering the incidence of onchocerciasis and lymphatic filariasis (Shen, 2015). Spinosad and milbemycin also have insecticidal activity. Spinosad is a combination of spinosyn A and D, which are both produced by Saccharopolyspora spinosa and have insecticidal activity against livestock external parasites via the disruption of nicotinic acetylcholine receptors (Sanchez et al., 2012). Milbemycin is an isolated fermentation product of S. hygroscopicus subsp. aureolacrimosus that acts as an insecticide and acaricide with GABAergic activity on the post-synaptic membranes of the inhibitory motor neurons of mites and arthropods through hyperpolarization and impeding neuronal signal transduction mechanisms (Copping and Duke, 2007). Mollemycin A 20 (15; Figure 3 and Table 1) is a first-in-class glycol-hexadepsipeptide-polyketide from a Streptomyces sp. and has antibacterial properties against certain Gram-positive and Gram-negative bacteria, as well as extremely potent antimalarial activity against drug sensitive and MDR Plasmodium falciparum clones (Blunt et al., 2016). Microbial natural products also function as enzyme inhibitors. Lipstatin (16; Figure 3 and Table 1) is a pancreatic lipase inhibitor produced by Streptomyces toxytricini that is used to combat obesity and diabetes by interfering with the gastrointestinal absorption of fat (Weibel et al., 1987). Lipstatin contains a beta-lactone structure that is likely responsible for irreversibly binding to the active site of lipase (Sanchez et al., 2012).
Biological Activity of Microbial Biologics
Since Humulin®(Figure 4A) became the first recombinant biopharmaceutical as a treatment for diabetes (Johnson, 1983), additional FDA-approved microbial biologics have been produced by E. coli. Somatrem (Protropin®) and somatropin (Humatrope®) are used to treat children with growth hormone deficiency (Baeshen et al., 2015; Sanchez-Garcia et al., 2016). Another biopharmaceutical produced from E. coli is pegloticase (Krystexxa®) for the treatment of chronic gout and interferon (IFN) α-2b (Intron®A; Figure 4B) for the treatment of certain types of genital warts, malignant melanoma, hairy cell leukemia, follicular lymphoma, Kaposi’s sarcoma, and chronic Hepatitis B or C (Baeshen et al., 2015; Sanchez-Garcia et al., 2016). Top selling biopharmaceuticals of 2015 from microorganisms include insulin glargine (Lantus®) derived from E. coli, which functions as an insulin analog, and the pneumococcal vaccines (Prevnar®family) derived from S. pneumoniae and Corynebacterium diphtheriae (Jozala et al., 2016; Sanchez-Garcia et al., 2016). Biopharmaceuticals are also utilized for their antitumoral properties, such as the cytokines filgrastim (Neupogen®) and granulocyte colony stimulating factor pegfilgrastim (Neupeg®; Figure 4C), which are both derived from E. coli. Flgrastim stimulates hematopoiesis for bone marrow transplantation and cancer chemotherapy-induced neutropenia, whereas pegfilgrastim stimulates the differentiation, proliferation and activation of neutrophilic granulocytes for cancer chemotherapy-induced neutropenia (Sanchez-Garcia et al., 2016). Recombinant human interleukin-3 (hIL-3; Figure 4D) protein is a cytokine that regulates the differentiation and proliferation of the various cells of the immune system (Hercus et al., 2013). The hIL-3 protein is derived from B. subtilis, B. licheniformis, and E. coli and has utility as a laboratory reagent in hematology for cell cultures, differentiation studies and functional assays. It has shown that hIL-3 has potential in treating bone marrow failure, hematological malignancies, and can support engraftment after bone marrow transplantation (Westers et al., 2006). In addition, recombinant Pfs48/45 is a disulfide-rich malaria transmission-blocking vaccine produced by L. lactis that provides immunization against malaria from P. falciparum (Song et al., 2017).
FIGURE 4.
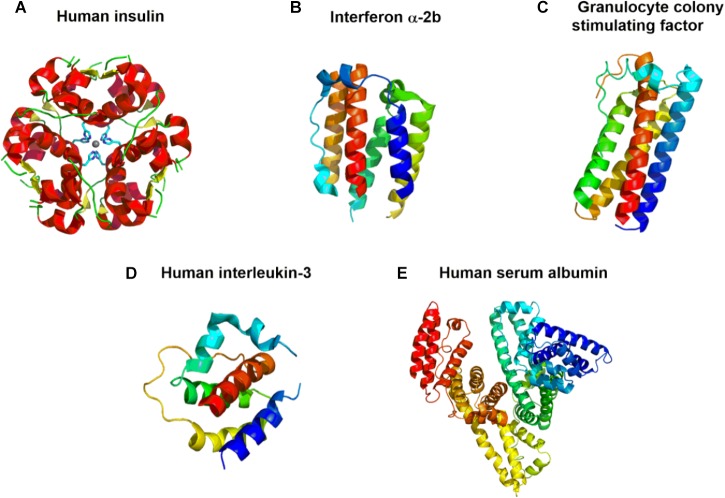
Crystal structures of (A) recombinant human insulin (Humulin®) (PDB 4F0N) (Favero-Retto et al., 2013); (B) interferon (IFN) α-2b (PDB 3SE3) (Thomas et al., 2011); (C) granulocyte colony growth factor pegfilgrastim (Neupeg®) (PDB 1HRG) (Hill et al., 1993); (D) human interleukin-3 (PDB 5UV8) (Broughton et al., 2018); and (E) human serum albumin (Recombumin®and Albucult®) (PDB 1AO6) (Sugio et al., 1999). The models are colored according to the sequence by a rainbow color from the N-terminus (blue) to the C-terminus (red).
Recombivax is produced by S. cerevisiae and can prevent infection of all known subtypes of the Hepatitis B virus (Sanchez-Garcia et al., 2016). Some examples of currently approved protein therapeutics derived from yeast include human serum albumin (Recombumin®and Albucult®; Figure 4E), human insulin (Actrapid®) and primary immunization for infants born of Hepatitis B virus (HBV) surface antigen (Pediarix®), all of which are obtained exclusively from S. cerevisiae (McAleer et al., 1984; Ballance, 1999; Nielsen, 2013; Nandy and Srivastava, 2018). Recombinant human serum albumin is utilized to increase the shelf life of protein drugs by preventing physical and chemical degradation. Actrapid®is used to treat diabetes, and subcutaneous injections of Pediarix is designed for immunization against diphtheria, tetanus, pertussis, poliomyelitis, and infection caused by all known subtypes of HBV (Nandy and Srivastava, 2018). Ecallantide (Kalbitor®) is an FDA-approved recombinant peptide produced by Pichia pastoris for the treatment of hereditary angioedema (Sheffer et al., 2011). Additionally, anakinra (Kineret®) was approved in 2001 in the United States for rheumatoid arthritis (Baeshen et al., 2015). Anakinra is expressed in E. coli and functions as an IL-1 receptor antagonist that is effective and safe for patients with systemic-onset juvenile idiopathic arthritis, adult-onset Still’s disease, hereditary autoinflammatory syndromes, and Schnitzler’s syndrome (Kalliolias and Liossis, 2008; Jozala et al., 2016).
Microbial Cell Factories
Selecting a suitable host strain is one of the most important aspects in the design of natural product and recombinant protein bioprocesses. We will review the characteristics of the microbial strains used to produce natural products and biologics in this section. We will also present the tools and strategies that facilitate engineering of the hosts as microbial cell factories for the production of biopharmaceutical compounds (Table 2).
Table 2.
Comparison between different microbial host systems for production of recombinant proteins and natural products.
| Microbial hosts | Advantages | Disadvantages | Compounds | References |
|---|---|---|---|---|
|
Gram-negative Escherichia coli |
||||
|
|
|
Johnson, 1983; Abdin et al., 2003; Chang et al., 2007; Zhang et al., 2010; Ferrer-Miralles and Villaverde, 2013; Baeshen et al., 2015; Jozala et al., 2016; Sanchez-Garcia et al., 2016 | |
|
Gram-positive Lactococcus lactis |
||||
|
|
|
Steidler et al., 1998; Drouault et al., 2000; Martínez et al., 2000; Le Loir et al., 2005; Mierau and Kleerebezem, 2005; Glenting et al., 2007; Morello et al., 2008; Li and Vederas, 2009; Linares et al., 2010; Gyawali and Ibrahim, 2014; Bermúdez-Humarán et al., 2015; Li et al., 2015; Song et al., 2017 | |
| Streptomyces sp. |
|
|
|
Stanley and English, 1965; Kaslow et al., 1994; Trischman et al., 1994; Mann, 2001; Jung et al., 2006; Copping and Duke, 2007; Park et al., 2008, 2016; Vrancken and Anne, 2009; Fracchia et al., 2010; Anné et al., 2012; De Lima Procópio et al., 2012; Sanchez et al., 2012; Yang et al., 2013; Kim et al., 2015; Blunt et al., 2016; Jozala et al., 2016; Gao et al., 2017 |
| Bacillus sp. |
|
|
|
Palva et al., 1983; Peypoux et al., 1984; Bellini et al., 1991; Kim et al., 2001; Westers et al., 2006; Deleu et al., 2008; Chang et al., 2011; Van Dijl and Hecker, 2013; Wang T. et al., 2015; El-Hossary et al., 2017 |
|
Fungi/yeast Saccharomyces cerevisiae |
||||
|
|
|
McAleer et al., 1984; Guisez et al., 1991; Kaslow et al., 1994; Ballance, 1999; Ferrer-Miralles et al., 2009; Nielsen, 2013; Paddon et al., 2013; Baeshen et al., 2014; Ding et al., 2014; Meehl and Stadheim, 2014; Moses et al., 2014; Kung et al., 2018; Nandy and Srivastava, 2018 | |
| Aspergillus sp. |
|
|
|
Gaffar and Shethna, 1977; Carrez et al., 1990; Hiort et al., 2004; Papagianni, 2004; Ward et al., 2004; Grimm et al., 2005; Maheshwari, 2006; Pel et al., 2007; Maiya et al., 2009; Meyer et al., 2011; Cragg and Newman, 2013 |
| Hansenula polymorpha |
|
|
|
Janowicz et al., 1991; Gellissen et al., 1992; Hollenberg and Gellissen, 1997; Cox et al., 2000; Heijtink et al., 2002; Müller et al., 2002; Böer et al., 2007; Kunze et al., 2009; Celik and Calik, 2012 |
Gram-Negative Bacteria
Escherichia coli
Escherichia coli has been seen as one of the optimal systems for the production of natural products because it is easily manipulated, highly productive, there is an availability of genetic tools to use with it and there is a deep knowledge of its physiology. Artemisinin, a sesquiterpene lactone endoperoxide from Artemisia annua L. plants, has strong antimalarial activity against the multi-drug resistant parasite P. falciparum (Abdin et al., 2003). Yet the synthesis of artemisinin is costly and low yields are isolated from the natural plant source. Researchers reported the production of approximately 24 mg/L of amorpha-4,11-diene (amorphadiene), an artemisinin precursor, by the expression of a codon-optimized synthetic amorphadiene synthase gene and the mevalonate pathway from S. cerevisiae in E. coli. Additionally, after further processing modifications and optimal conditions, they were able to produce 105 mg/L of artemisinic acid (Chang et al., 2007). However, there are some obstacles and limitations with E. coli as a dominant host in natural product biosynthesis. E. coli requires extensive genetic manipulation and lacks native natural product biosynthetic machinery and/or precursors. An example is phosphopantetheinyl transferase, which is responsible for the activation of the carrier protein domains of the PKSs and NRPSs. This enzyme must be introduced into E. coli to support of natural product biosynthesis (Zhang M.M. et al., 2016). There have been efforts to overcome these hurdles, such as the production of erythromycin A and its derivatives in the engineered E. coli strain (Zhang et al., 2010). The study generated two analogs through directed manipulation of polyketide biosynthesis in which variations were made to the deoxyerythronolide B synthase (DEBS) 1 and DEBS3 enzymes in order to utilize the multi-catalytic capability of the modular polyketide synthase (Zhang et al., 2010).
Escherichia coli has also been the pioneering host for recombinant protein production. To date, E. coli continues to be the first-choice microorganism for manufacturing recombinant proteins at laboratory and industrial scales. Its success is mostly due to its fast growth, simple culture procedures, cost-effectiveness, unusually high versatility, and the associated systems that make it adaptable to varying production demands (Ferrer-Miralles and Villaverde, 2013; Sanchez-Garcia et al., 2016). From 2004 to 2013, 24% of the biopharmaceuticals approved by the FDA and the European Medicines Agency were derived from E. coli (Baeshen et al., 2014). Currently, biopharmaceuticals produced from E. coli are used in the treatment of diabetes, growth hormone-deficiency in children, leukemia, gout, and many other therapeutic indications as previously discussed in Section “Biological Activity of Microbial Biologics” (Baeshen et al., 2015). A major concern when using E. coli as a production platform is the lack of post-translational modifications (PTMs) present in most eukaryotic proteins; lacking PTMs can lead to protein products being insoluble, unstable, or inactive (Ferrer-Miralles et al., 2009). However, it is possible to add synthetic PTMs to generate versions of the protein that are more stable than the original naked product (Ferrer-Miralles et al., 2009). Examples of this include pegylated products, like human growth hormone, stimulating factor, IFNs α-2a and α-2b, (Ferrer-Miralles et al., 2009). Additionally, there is a risk of translational errors due to the presence of a large number of rare codons that appear in human genes that are different from those occurring in E. coli genes. Even at low levels, these errors may cause an impact on the tertiary structure, premature termination of protein synthesis or amino acid misincorporation which results in low protein expression (Gustafsson et al., 2004). One strategy to bypass the issue with codon bias is to synthesize the whole human gene based on codon usage in E. coli through site-directed mutagenesis, which is currently a preferred method; however, it is limited by the high cost of production and time consumption (Sørensen and Mortensen, 2005). An alternative method that is less time consuming utilizes the co-transformation of E. coli strains with a plasmid(s) containing a gene encoding the tRNA cognate to the rare codons (Dieci et al., 2000). Increasing the copy number allows for E. coli to be manipulated to match the codon usage frequency in heterologous genes (Dieci et al., 2000). Currently, there are numerous commercial E. coli strains available that harbor plasmids containing gene sequences encoding the tRNA for rare codons, such as BL21(DE3) CodonPlus-RIL, BL21(DE3) CodonPlus-RP and Rosetta (DE3) (Baeshen et al., 2015). Another common problem associated with recombinant protein expression in E. coli involve inclusion body formation, which refers to insoluble and inactive protein aggregates (Hartley and Kane, 1988). E. coli producing recombinant proteins have the ability to assemble in cells and form conglomerates of inclusion bodies as well as result in erroneous protein folding which hinder the extraction of proteins directly from the cell leading to costly purification of proteins (Zweers et al., 2008). Inclusion bodies formed from lack of proportion between protein solubilization and aggregation can be resolved by combining the desired protein with a solubility enhancer fusion partner acting as an intrinsic chaperone in order to ensure the production of soluble recombinant proteins (Rosano and Ceccarelli, 2014). The fusion of maltose-binding protein to polypeptides such as human tissue inhibitor of metalloproteinase and p16 improved their solubility significantly in E. coli (Kapust and Waugh, 1999).
Gram-Positive Bacteria
Lactococcus lactis
Lactococcus lactis is becoming an attractive alternative in genetic engineering for the production of various recombinant proteins. Unlike E. coli, which uses intracellular production strategies that involve expensive and often challenging purification processes, L. lactis utilizes extracellular secretion system. This is because L. lactis has a monolayer cell wall that allows direct secretion into the extracellular environment (Schneewind and Missiakas, 2012). The presence of exported proteases such as HtrA in L. lactis contributes to recombinant protein production by minimizing the destruction of heterologous proteins in the medium (Morello et al., 2008; Song et al., 2017). Additionally, L. lactis does not generate undesired glycosylation of protein, is generally recognized as safe (GRAS), does not produce endotoxins, and has probiotic properties (Singh et al., 2018). Another advantage of L. lactis includes a lack of inclusion body formation (Theisen et al., 2017). There is a diverse selection of cloning and inducible expression vectors available for use with this host that are compatible with large-scale upstream and downstream processes (Song et al., 2017).
Lactococcus lactis has been used for centuries in the fermentation of food, especially in cheese, yogurt, and sauerkraut because of its production of nisin (Song et al., 2017; Singh et al., 2018). Beyond the food industry, lactic acid is used as an emulsifier and moisturizing agent in the cosmetic industry and as an important raw material in the pharmaceutical industry (Papagianni, 2012). The L. lactis host has also been chosen after researchers had unsuccessfully attempted to obtain correct conformation using a variety of other prokaryotic and eukaryotic recombinant protein expression systems. L. lactis has multiple advantages over E. coli for expression of 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase (Pfs) recombinant proteins, including the following: (1) codon-optimization of the recombinant gene is not necessary to achieve successful expression in L. lactis; (2) the recombinant protein is secreted into the L. lactis culture supernatant, which results in easier and less expensive down-stream processing, and (3) there is no lipopolysaccharide contamination in L. lactis expression (Singh et al., 2018). L. lactis has been used in the successful production of recombinant Pfs48/45, a vaccine candidate against P. falciparum (Song et al., 2017). GMZ2, a recombinant fusion protein expressed in L. lactis, is also a malaria vaccine candidate that has been shown to elicit high levels of active IgG antibodies with inhibitory activity against a broad range of P. falciparum strains (Jepsen et al., 2013). A recent study concluded phase 2 trial of GMZ2 adjuvanted with aluminum hydroxide in a cohort of 1,849 children revealed GMZ2 as well tolerated with modest efficacy (Sirima et al., 2016). Not only is L. lactis being utilized for the production of recombinant proteins for vaccines, but the host is also being tested as a factory for antigen production, allowing the bacteria to function as live vaccines. Using L. lactis as a vaccine carrier is beneficial because it can induce both mucosal and systemic immune responses, has adjuvant properties and is free from the risks associated with the use of conventional attenuated live pathogens, such as Salmonella species and Mycobacterium species (Song et al., 2017). However, while L. lactis has been studied against an array of antigens from various pathogens, there is no current live vaccines under clinical trial which may be due to a lack of containment strategies (Bahey-El-Din, 2012). Without a plan for containment, studies on live L. lactis vectors risk the chance of proliferation and dispersion. An additional limitation of AT rich L. lactis as a cell factory is due to codon usage as well as distribution of rare codons to express heterologous genes (Mierau and Kleerebezem, 2005).
Streptomyces Species
Another major species that has shown promise as a cell factory through its wide production of natural products and biologics is Streptomyces. This Gram-positive bacterium has been studied for more than 70 years and has proven to be of great use in biotechnology due to its ability to produce natural products acting as antibiotics, anticancer agents, and immunosuppressants (Yoo et al., 2015). Some examples include tetracycline, daptomycin, and chloramphenicol (De Lima Procópio et al., 2012). There are many species of Streptomyces currently to produce various natural products and biologics. Among the Streptomyces species, Streptomyces coelicolor, Streptomyces lividans, Streptomyces albus, and S. venezuelae are favored heterologous hosts to produce specialized metabolites due to the relative ease of their genetic manipulation, the availability of their genome sequences, and the abundant supply of their natural substrates (Park et al., 2010). Streptomyces has also been used to produce a wide array of heterologous proteins of both eukaryotic and prokaryotic origin (Gilbert et al., 1995) because Streptomyces has well-developed genetic manipulation and fermentation technologies as well as efficient protein secretion systems such as the secretory (Sec) pathway and the twin-arginine-translocation (Tat) pathway (Hamed et al., 2018). The Sec-pathway catalyzes the translocation of unfolded proteins while the Tat pathway allows for the export of folded proteins across the cytoplasmic membrane (Natale et al., 2008). Tumor necrosis factor (TNF) α and human interleukin (hIL) 10 are able to be expressed in both the Sec- and Tat-pathways (Schaerlaekens et al., 2004). In particular, S. lividans could be the ideal Streptomyces host due to limited restriction-modification systems and low endogenous protease activity (Nakashima et al., 2005). Streptokinase (Pimienta et al., 2007), transforming growth factor-α (Taguchi et al., 1995), IL-2 (Bender et al., 1990) and many other proteins have been successfully expressed and secreted from S. lividans. However, aside from its efficient secretory pathways, when in culture, Streptomyces grows as mycelial networks, leading to the formation of pellets or clumps (Van Dissel et al., 2015). These pellets are unappealing from an industrial standpoint because of mass-transfer problems, slow growth, and culture heterogeneity which leads to lower product yield (Van Dissel et al., 2015).
Bacillus Species
Bacillus species are some of the most popular species used in enzyme production. It accounts for roughly 50% of enzymes market within the industrial sphere (Barros et al., 2013). Certain species, like B. subtilis, Bacillus amyloliquefaciens, and B. licheniformis are favored because of their outstanding fermentation properties, high protein production yield, and their completely toxin-free production (Van Dissel et al., 2015). The fermentation capacity of Bacillus species in acid, neutral, and alkaline pH ranges in addition to thermophiles accounts for the prolific production of enzymes that have desirable temperature, pH, and stability, which makes them appealing for specific use in various industries (Schallmey et al., 2004). Bacillus species are known for their production of iturins and fengycin which have antifungal activity as well as surfactin for its function as a surfactant (Wang T. et al., 2015).
Among these species, B. subtilis is the most widely studied due to (1) its flexibility during genetic engineering, (2) its naturally high secretory capacity, (3) its ability to produce valuable antibiotics, such as bacillomycins D-L and bacitracin, and (4) its ability to produce enzymes, such as stable alkaline cellulase, alkaline protease and alkaline α-amylase. It may also elicit better folding conditions, leading to the prevention of inclusion bodies (Peypoux et al., 1984; Van Dijl and Hecker, 2013). In addition, its ability to adapt to varying environmental conditions as well as its classification as toxic free GRAS has contributed tremendously to its success in the industrial platform (Baysal and Yıldız, 2017). B. subtilis as an endotoxin free host amplified its utilization in the production of sterile recombinant and therapeutic proteins expression as compared to E. coli which could have potential contamination due to the lipopolysaccharide endotoxins (Wang et al., 2013). For instance, B. subtilis and Bacillus megaterium were the preferred hosts over E. coli in the production of bioengineered heparin in order to diminish toxin contamination (Wang et al., 2013). Moreover, B. subtilis is able to produce high yield in enzyme as it secretes the enzymes straight into the fermentation medium due to the absence of outer membrane which allows easy recovery of purified proteins from the medium into their active form (Zweers et al., 2008). It has the capacity to secrete about 20–25 g/L of enzymes into the medium (Schallmey et al., 2004). Enzymes produced by B. subtilis has a wide variety of applications ranging from pharmaceutical, leather, detergent, food, and waste management industries (Singh et al., 2016).
Aside from enzyme production, cytokines like hIL-3 have been produced by B. subtilis and B. licheniformis. The production of hIL-3 has been tested in various host organisms, including E. coli. However, the production of IL-3 within other organisms has often exhibited problems, such as insolubility or the degradation of produced hIL-3. This led to the use of B. licheniformis and B. subtilis to minimize such complications. The production of hIL-3 in B. licheniformis was engineered to lack four C-terminal residues, resulting in a fully active hIL-3 protein. However, residual proteolytic degradation of some hIL-3 still occurred, leading to use B. subtilis to achieve complete folding and full biological activity of hIL-3 (Westers et al., 2006).
Among the Bacillus species, Bacillus thuringiensis is best known for being widely used within the agricultural industry due to its insecticidal properties through its production of parasporal crystals during the stationary phase of its growth cycle (Höfte and Whiteley, 1989; Schnepf et al., 1998). Upon ingestion, the parasporal crystals are solubilized in the midgut of insects, resulting in the release of protoxin proteins known as δ-endotoxins, leading to the formation of pores throughout the cell membrane (Höfte and Whiteley, 1989; Gill et al., 1992). Parasporal proteins also have selective cytotoxicity against liver and colon cancer cells while leaving normal cells unharmed (Ito et al., 2004).
However, the use of Bacillus has been restricted to mainly enzyme production and non-recombinant protein therapeutics, which may be due to the lack of associated expression vectors, plasmid instability and the presence of native proteases (Westers et al., 2004). Despite B. subtilis success as the industrial workhorse, it has its drawbacks in the production of heterologous proteins. Heterologous protein yield could diminish when using the Bacillus as a host due to the proteolytic destruction of foreign protein by host secreted extracellular proteases (Nijland and Kuipers, 2008). Efforts have been made to improve the production of heterologous protein by manipulating the expression of proteins involved in the post translocation phase resulting in amplified levels of heterologous protein secretion (Vitikainen et al., 2005). In contrast to E. coli, the absence of distinguished and controllable promoters in B. subtilis interferes with successful expression of heterologous genes resulting in inefficient production of heterologous proteins (Schallmey et al., 2004).
Yeast/Fungi
Saccharomyces cerevisiae
As with E. coli, S. cerevisiae has been extensively used for the production of recombinant human insulin since the early 1980s, and it currently produces half of the world’s supply of insulin (Meehl and Stadheim, 2014). S. cerevisiae is preferred because it is also cost-effective, fast growing, technically practical, and is amenable to large-scale fermentation in bioreactors. Yeast is often utilized as a cell factory when the target protein is not produced in a soluble form in prokaryotic systems or when a specific PTM cannot be produced or added to the naked product. S. cerevisiae, as with other yeast species, can perform many PTMs such as O-linked glycosylation, phosphorylation, acetylation, and acylation, which allows recombinant proteins to be expressed in a soluble, correctly folded, and functionally active form (Ferrer-Miralles et al., 2009; Baeshen et al., 2014). Some examples of currently approved protein therapeutics derived from yeast include human serum albumin, insulin, and primary immunization for infants born of HBV surface antigen, all which are obtained in S. cerevisiae (McAleer et al., 1984; Ballance, 1999; Nielsen, 2013; Nandy and Srivastava, 2018). However, the significant drawback to producing protein therapeutics from S. cerevisiae is that higher eukaryotes have a different pattern of N-linked glycosylation, which can decrease the half-life and cause hyper-immunogenicity, leading to less effective therapeutics (Ferrer-Miralles et al., 2009). In recent years, there have been some advances in limiting S. cerevisiae hypermannosylation. These yeast glycoengineering techniques involve two main stages, (1) the removal of yeast hypermannosylation and (2) the conversion to complex glycoforms containing terminal sugars, such as N-acetylglucosamine, galactose, or sialic acid. These recent reports on yeast N-glycan humanization indicate a move from the proof of concept phase to implementation (Meehl and Stadheim, 2014).
Another current area of study is the production of plant and microbe-derived secondary metabolites. Due to the structural complexity of secondary metabolites, chemical synthesis is an inefficient route for large-scale production, and fermentation is the main mode for economic commercial production of pharmaceutically useful natural products (McDaniel et al., 2001). S. cerevisiae could be an ideal candidate as a microbial host as it boasts relatively rapid growth, and it is accompanied by highly developed genetic tools and advanced fermentation science. Like E. coli, S. cerevisiae has been shown to be an outstanding production host for artemisinic acid, a precursor to the antimalarial agent artemisinin, with a high productivity (25 g/L) that led to the industrial production of semi-synthetic artemisinin beginning in 2013 (Paddon et al., 2013; Kung et al., 2018). Research has also produced the paclitaxel (Taxol®) precursor taxadiene (∼73 mg/L) by engineering the taxol biosynthetic genes in S. cerevisiae (Ding et al., 2014). Besides plant secondary metabolites, S. cerevisiae has generated a remarkable titer (1.7 g/L) of microbial polyketide 6-methylsalicylic acid in un-optimized shake-flask fermentations. In addition, S. cerevisiae has been developed as a heterologous host to express fungal cryptic BGCs and their respective metabolites. In this study, 30 ADH2-like promoters in Saccharomyces species were developed as tools for expression of 41 heterologous BGCs, 22 of which were capable of producing heterologous compounds, including novel compounds. For example, BGCs derived from basidiomycete were found to produce N-, S-bis-acylated amino acids and a leucine O-methyl ester with an additional polyketide chain amidated to the amino ester (Harvey et al., 2018). However, barriers still exist to the heterologous production of complex molecules. This includes the production of polyketides in S. cerevisiae, as the host lacks some required polyketide precursor pathways, its codon usage is biased toward A + T (most microbial polyketide producers have high G + C genome content) and it lacks the appropriate endogenous phosphopantetheinyl transferase capable of the necessary PTMs (Mutka et al., 2006).
Aspergillus Species
Multicellular filamentous fungi, such as A. niger and Aspergillus oryzae, can also offer great potential in the production of a desired substance by fermentation due to the following reasons: (1) they are well-characterized GRAS organisms, (2) are amenable to scaled-up fermentation, (3) can be genetically engineered, (4) they are capable of secreting a high level of proteins and (5) can withstand adjustable cultivation conditions, including temperature (5–45°), pH (2–11), salinity (as much as 34%), water activity (0.92–0.98), and both nutrient rich and poor environments (Maheshwari, 2006; Meyer et al., 2011).
Aspergillus niger has been predominantly used for industrial-level production of citric acid through anaerobic fermentation process. As a weak acid, citric acid can serve as a natural preservative, flavoring agent in food and beverages, antioxidant, acidulant, pH-regulator, chelating agent or vegetable rinse, as well as comparable applications in the pharmaceutical and cosmetics industries (Cairns et al., 2018). Due to its wide variety applications, its ease of production, and economical potential of citric acid, the market of citric acid has become one of the fastest-growing regions of the food additives market due to the rising demand: according to estimations, in 2007, the market value of citric acid exceeded $2 billion in 2014 and is predicted to rise to $3.6 billion by 2020 (Show et al., 2015; Cairns et al., 2018). Phytase is another example that have produced by A. niger fermentation (Papagianni et al., 1999). The significance of phytase enzymes lie in its ability to interact with the nutrient rich compounds known as phytate. Phytate, or phytic acid, is a common energy source found in oilseeds, cereals, and legumes, which are used in providing nutrition to animal feeds (Schlemmer et al., 2009). Combining citric acid with phytase has also been shown to enhance phytase activity on phytate, producing greater nutrient outcomes in tested animals (Boling et al., 2000). In addition, two different humanized immunoglobulin G1(κ) antibodies and an Fab′ fragment were produced by A. niger, and the antibodies were successfully secreted into the culture supernatant (Ward et al., 2004). Aspergillus strains have been also used to produce the human iron-binding glycoprotein lactoferrin and hIL-2 with the yields of 25 and 150 mg/L, respectively (Maheshwari, 2006). Bicoumanigrin with cytotoxic activity against human cancer cell lines and aspernigrin B with neuroprotective effects have both been isolated from A. niger (Hiort et al., 2004). The product spectrum of Aspergillus species is not restricted to biologic molecules. As an example, a novel cyclic peptide compound, KK-1, with potent antifungal activity was produced in A. oryzae by introducing gene clusters spanning approximately 40 kb from the plant-pathogenic fungus Curvularia clavata into the genome of A. oryzae. Although the amount of KK-1 produced by the host was lower than that produced by the original producer C. clavata, this result indicated that a gene twice as large as the largest native gene in A. oryzae could be successfully expressed (Yoshimi et al., 2018). Furthermore, when the industrial fungus A. niger was engineered as a heterologous host, it produced high titers (up to 4,500 mg/L) of enniatin belonging to non-ribosomal peptides with antibacterial, antiviral, and anticancer activities (Richter et al., 2014).
Hansenula polymorpha
Another industrially important yeast species that has shown promise in the production of peptides is Hansenula polymorpha (Gellissen et al., 1992; Boer et al., 2007). H. polymorpha is a methylotrophic yeast species with the ability to use and grow on methanol, glucose, or glycerol as its primary carbon source (Gellissen et al., 1992). Like S. cerevisiae and Aspergillus species, H. polymorpha, classified as GRAS organism, does not harbor pyrogens, toxins, pathogens, or viral inclusions (Ubiyvovk et al., 2011). It is distinguished by very high cell densities in bioreactors and characterized by simple cultivation mode in inexpensive growth media. For example, H. polymorpha has allowed for cost-effective production of phytase through cheap carbon sources (Mayer et al., 1999). It possesses well-established genetic tools such as strong regulatory and constitutive promoters, which consequently give high product yield (Van Dijk et al., 2000). It also has thermotolerance properties, making H. polymorpha successful in crystallographic studies and in the production of recombinant proteins like IFN-2α, IL-6, recombinant human serum albumin, glucose oxidase, and catalase (Kunze et al., 2009; Celik and Calik, 2012). A notable feature of H. polymorpha is the significant growth of peroxisomes when grown on methanol which allows for high storage capacity of soluble proteins. The lack of protein modifying enzymes in the matrix of peroxisomes also provides an advantage for the development of heterologous proteins that are susceptible to proteolytic degradation (Van Dijk et al., 2000). Furthermore, the host has been used to produce L antigens found on the HBV viral envelope in attempt to produce the HBV vaccine. The L protein produced by H. polymorpha has increased stability in comparison to other yeast species, such as S. cerevisiae and P. pastoris (Janowicz et al., 1991). In addition to its use in vaccine production, H. polymorpha is also used in the production of human hemoglobin through the use of a single expression vector (Hollenberg and Gellissen, 1997). However, hyperglycosylation has been observed as a main drawback of H. polymorpha to produce heterologous products (Müller et al., 1998).
Efforts in Product Improvements and Generation of New Analogs
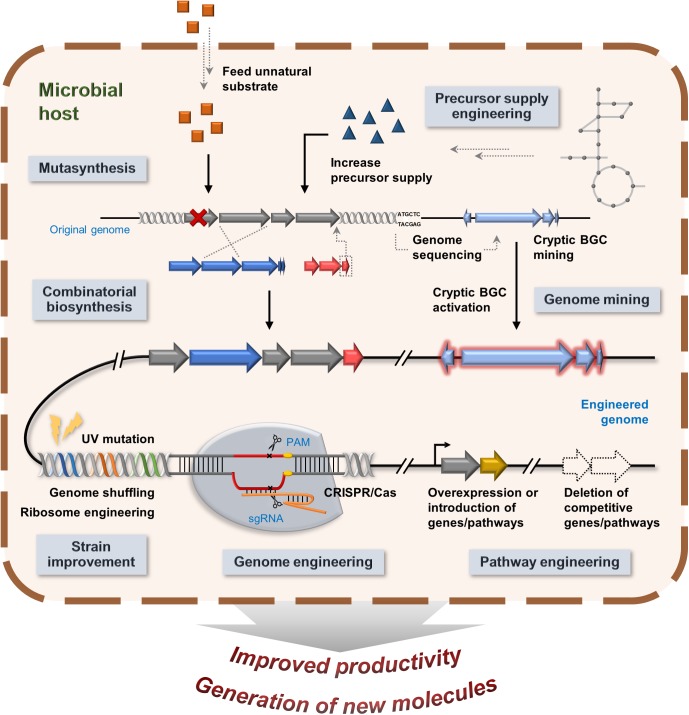
There are multiple approaches that have been taken to advance product improvement for microbial natural products and biologics. This section will discuss efforts to combat the challenges of production of natural products and its analogs, including strain improvement, increasing precursor supply, pathway engineering, combinatorial biosynthesis, and genome mining (Figure 5).
FIGURE 5.
An overview of multiple strategies of product improvements and generation of new analogs.
Strain Improvement
Whole-genome shuffling is a process that utilizes the advantages of the multi-parental crossing allowed by DNA shuffling with the genome recombination normally associated with conventional breeding (Zhang et al., 2002). Genome shuffling has been successfully improved the titers of variety of microorganisms. For example, two strains of Streptomyces fradiae generated from two rounds of genome shuffling were able to produce up to a ninefold increase in antibacterial tylosin production in comparison to the initial strain (Zhang et al., 2002). Using genome shuffling in a combination of protoplast fusion, mutant strain of S. cellulosum GSUV3-205 generated a 130-fold increase (104 mg/L) in production of epothilone when compared to starting strain S. cellulosum So0157-2 (0.8 mg/L) (Gong et al., 2007). Ribosome engineering is also a method useful in increasing secondary metabolite production titer and productivity (Sun and Alper, 2015). Studies demonstrate that rpoB mutations are effective in activating silent and poorly expressed secondary metabolite biosynthetic gene clusters (BGCs) at the transcriptional level in S. griseus, S. coelicolor, and S. erythraea (Ochi and Hosaka, 2013). For example, the H437R mutant of rpoB from S. erythraea was screened for drug resistance and was found to have an increased production of erythromycin (Tanaka et al., 2013; Ochi et al., 2014). Another study found a 37-fold increased production of avilamycin in a recombinant Streptomyces viridochromogenes strain due to a mutation in ribosome protein S12 (rps12) acquired through a combination of gene shuffling and ribosome engineering (Lv et al., 2013).
Engineering Precursor Supply
Precursor supply is defined as the enhancement of the availability of primary metabolites or molecules derived from primary metabolism involved in the biosynthesis of natural products (Shiba et al., 2007). Precursor supply engineering can be achieved by manipulating either the pathways or enzymes involved with the precursor supply. Malonyl-CoA and methylmalonyl-CoA are the most commonly used and metabolically available precursors for the biosynthesis of polyketides. One study found that supplying methyl oleate enhanced the internal concentration of methylmalonyl-CoA, which is a biosynthetic precursor for FK506, and led to a 2.5-fold increase in FK506 production in Streptomyces clavuligerus CKD1119 (Mo et al., 2009). In another study, propionyl-CoA carboxylase with supplementation of propionate was found to effectively increase methylmalonyl-CoA and rapamycin titers in the mutant strain S. rapamycinicus UV2-2 induced by ultraviolet mutagenesis in comparison to wild-type strain (7 ∼ 8 mg/L) (Jung et al., 2011). The mutant strain was found to have a 3.2-fold improvement (23.6 mg/L) in comparison to wild-type strain S. rapamycinicus ATCC 29253 (7 ∼ 8 mg/L) (Jung et al., 2011). Further, Méndez and coworkers improved precursor metabolite pools for the production of the antitumor polyketide mithramycin in Streptomyces argillaceus by increasing the precursor supply of malonyl-CoA and glucose-1-phosphate (Zabala et al., 2013). Several classes of natural products utilize aromatic amino acids or other metabolites derived from the shikimate pathway as precursors, including flavonoids, alkaloids, polyketides, and non-ribosomal peptides (Knaggs, 2001). The production of the vancomycin analog balhimycin was increased 2.5-fold in Amycolatopsis sp. Y-89,21022. This was achieved by increasing the non-ribosomal peptide precursor 3-deoxy-D-arabino-heptulosonate7-phosphate synthase, the first enzyme in the shikimate pathway (Thykaer et al., 2010). In addition, manipulating key enzymes that direct carbon flux through core biochemical pathways involved in glucose, fatty acid, and amino acid metabolism can increase biosynthetic precursor pools. A study on the modulation of carbon flux between the pentose phosphate pathway and the glycolysis pathway found that a deletion of phosphofructokinase isoenzymes led to the enhanced production of antibiotics actinorhodin and undecylprodigiosin in S. coelicolor by increasing carbon flux through the pentose phosphate pathway (Borodina et al., 2008).
Precursor supply engineering has been successfully used to produce most of the major classes of natural products, with application to heterologous producing strains as well as native producers. When the native host is slow-growing or cannot be easily manipulated genetically, this process could be performed effectively in an appropriate heterologous host. A study bypassed the native deoxyxylulose 5-phosphate pathway and instead introduced the mevalonate pathway from S. cerevisiae to E. coli, which allowed for an increased production of amorpha-4,11-diene which is a precursor to antimalarial artemisinin (Martin et al., 2003). Combined approach of exogenous supplementation and engineering of intracellular pathway responsible for precursors can be also performed in a heterologous host. The biosynthetic process of the hybrid non-ribosomal peptide-polyketide yersiniabactin was known to rely on the supply of salicylate, L-cysteine, S-adenosyl-L-methionine, malonyl-CoA, and NADPH. When exogenous cysteine was fed to the culture of E. coli harboring yersiniabactin BGC and an additional set of genes (hmwp1-2) responsible for yersiniabactin precursor biosynthesis was introduced, the yersiniabactin production in E. coli was boosted to approximately 175 mg/L (Ahmadi and Pfeifer, 2016).
Precursor engineering strategy can be also employed to increase recombinant protein production by reducing unwanted by-products. One of the primary obstacles observed in high cell density cultivations of E. coli for the production of recombinant proteins is the formation of acetate, which is a by-product caused by an excess influx of carbon during aerobic fermentation. This acetate accumulation hampers cell growth and recombinant protein formation, even at low concentrations (Waegeman and Soetaert, 2011). A number of engineering approaches have focused on minimizing acetate formation in order to enhance recombinant protein production in E. coli. When a heterologous anaplerotic pyruvate carboxylase from Rhizobium etli is overexpressed in E. coli, the resulting strain had a 57% reduction in acetate formation and a 68% increase in β-galactosidase production (March et al., 2002). It is also possible to combine different strategies to reduce the formation of undesired by-products, including acetate. For example, one study found that a mutant E. coli strain containing a defective acetate pathway and an overexpressed phosphoenolpyruvate carboxylase-encoding ppC reduced acetate and other by-product formation and produced five time more β-galactosidase activity when compared the wild type strain (De Mey et al., 2010).
Pathway Engineering
Metabolic pathway engineering can be performed in the native host through repetitive gene expression, gene deletion, and introduction of new genes to enhance production of natural products (Pickens et al., 2011). For example, overexpression of the 4–12 tandem copies of the actinorhodin cluster resulted in a 20-fold increase in actinorhodin production in S. coelicolor (Murakami et al., 2011). Additionally, a S. hygroscopicus strain with 3–5 tandem copies of the 40 kb validomycin A cluster showed a 34% increase in production and a maximum titer of approximately 20 g/L (Zhou et al., 2014). Deletion of genes may be useful to eliminate competing pathways that may siphon off important precursors or intermediates, or simply contribute to an unnecessary use of cellular resources which result to improve yields of products of interest. During in vivo bioconversion of lovastatin intermediate monacolin J to simvastatin using E. coli expressing heterologous acyltransferase LovD, it was found that E. coli could unexpectedly hydrolyze the synthetic thioester substrate. The responsible hydrolase BioH was knocked out to improve simvastatin production (Xie et al., 2007). The regulatory component of the pathway can be manipulated to enhance production of the resulting natural product. Negative regulation by pathway specific repressors can help regulate secondary metabolite pathways. For example, one study improved the titer by 100-fold of antibiotics platensimycin (323 mg/L) and platencin (255 mg/L) through the inactivation of a gene encoding protein PtmR1 belonging to GntR family of transcriptional repressors (Smanski et al., 2009). On the other side of the spectrum, Streptomyces antibiotic regulatory protein (SARP) is a positive regulator of antibiotic production (Chen et al., 2010). Overexpression of SARPs and/or increasing SARP gene dosage using multi-copy plasmids has been demonstrated to increase production titers. Overexpression of mgsA or chxA, SARP family members that are positive regulators for the iso-migrastatin and cycloheximide biosynthetic machinery, respectively, in Streptomyces amphibiosporus ATCC 53964 led to a fivefold increased production of antibiotic lactimidomycin (Zhang et al., 2016). Members of the large ATP-binding regulators of the LuxR (LAL) family also generally function as transcriptional activators, and constitutive overexpression of these LAL-type activators was found to increase production of rapamycin in S. rapamycinicus and FK506 in S. tsukubaensis (Kušèer et al., 2007; Mo et al., 2012).
Competing pathways can also be deleted to ensure the production of important precursors or intermediates and to save useful cellular resources. When deleting pathways, the idea is to create a host with a minimized genome to ensure the efficient production of necessary secondary metabolites. Deleting non-essential genes and directing cellular resources toward pathways that are essential for the survival and product biosynthesis can improve cellular efficiency and streamline biochemical production. For example, the genome of Streptomyces avermitilis was effectively minimized to 83% of its original size. When heterologous streptomycin gene cluster was introduced into the genome-minimized S. avermitilis, the resulting strain produced a higher titer of streptomycin than both the parent S. avermitilis carrying the same heterologous gene cluster and the native streptomycin producer S. griseus (Komatsu et al., 2010). However, large scale deletions may result in unintended effects as the complete workings of the cell are not yet entirely understood.
Similar approaches have been employed to improve the secretion capability and productivity of biologics. Engineering the protein trafficking pathway represents one successful approach to improve the secretion of heterologous recombinant proteins. For example, the secretion of the heterologous proteins human insulin precursor and α-amylase from A. oryzae in S. cerevisiae was improved by the over-expression of Sec1/Munc18 proteins, which are involved in the protein secretory pathway (Hou et al., 2012). Increasing the copy number of genes that are associated with protein secretion can also enhance protein secretion. This is seen with the Necator americanus secretory protein (Na-ASP1), which shows potential as a vaccine protein for hookworm infections. Increasing the Na-ASP1 gene copy number caused saturation of secretory capacity in P. pastoris, a species of methylotrophic yeast, led to a decreased amount of secreted protein. This was remedied by the overexpression of the protein disulfide isomerase, which allowed for the increased secretion of Na-ASP1 protein in high copy clones (Inan et al., 2006). Another study showed that deletion of obstructive protease genes involved in fission could lead to the enhanced secretion of protease-sensitive human growth hormones (hGH) in Schizosaccharomyces pombe. The production of hGH was hampered by the intracellular retention of secretory hGH, and it was determined that the multi-protease deletant strain plays a role in hGH retention. Deletion of vps10, which encodes a carboxypeptidase Y sorting receptor and is involved in the traffic between the late-Golgi and prevacuolar compartments, resulted in an approximate twofold increase in hGH secretion (Idiris et al., 2006).
Combinatorial Biosynthesis
Combinatorial biosynthesis is one genetic engineering application that can modify biosynthetic pathways in order to yield new and altered natural product structures (Hopwood et al., 1985). This approach exploits indiscriminate substrates and uses engineered enzymes and pathways for the production of new natural product analogs.
Modular megasynthases, such as PKS and NRPS enzymes, constitute a class of multifunctional proteins that govern complex enzymatic mechanisms and catalyze multiple reactions useful for combinatorial biosynthesis. Type 1 PKSs consist of multiple modules which are responsible for incorporating acyl-CoAs into a polyketide backbone for elongation. Meanwhile, NRPSs are composed of a modular set of repeating enzyme domains for the activation and incorporation of amino acids (Park and Yoon, 2015). The modular NRPSs typically consist of a condensation domain, adenylation domain, and a thiolation domain, while type I PKSs generally contain a ketosynthase domain, acyltransferase domain, and an acyl carrier protein (Komaki et al., 2015; Skiba et al., 2018). Natural product structures can be modified by mixing and matching the megasynthases at the subunit, module, and domain levels. Genetic manipulation of PKS and NRPS encoding genes can result in predictable changes in structure that is difficult to achieve with standard chemical derivatization or total synthesis methods (Park et al., 2010). This approach to manipulating substrate incorporation and biosynthetic PKS and NRPS machinery has allowed for the generation of a great number of natural product analogs. Examples include erythromycin (McDaniel et al., 1999), pikromycin from type I modular PKS (Yoon et al., 2002) and daptomycin from NRPS (Robbel and Marahiel, 2010).
Post-assembly modifications, such as glycosylation, oxidation, and halogenation are performed by diverse enzymes and can lead to structurally and biologically diverse natural compounds (Park et al., 2010). Sugar moieties attached to the core structure of polyketides or non-ribosomal peptides by glycosyltransferases can also contribute to an extension of combinatorial biosynthesis. Since several glycosyltransferases have been known to be flexible toward sugar donors and sugar accepters, arrays of analogs differing in glycosylation patterns via tailoring enzymes can also be generated by combinatorial engineering of glycosyltransferases from different pathways. For example, one study found that A. orientalis-derived glycosyltransferases accepted the unnatural sugar 4-epi-vancosamine in the presence of vancomycin pseudoaglycone or the glucosylated teicoplanin scaffold to generate novel hybrid glycopeptide compounds such as 4-epi-vancosaminyl form of vancomycin (Losey et al., 2001). Besides sugar biosynthesis, combinatorial biosynthesis can be applied for other modifications such as oxidation and halogenation. Oxidase genes from polyketide pathways have been used to induce structural alterations of important functional groups that are essential for biological activities. It has been reported that 5-O-desosaminyl erythronolide A, a potent precursor of ketolides and the latest generation of antibiotic compounds derived from erythromycin A, was produced by expressing the monooxidase gene pikC from the pikromycin pathway in a mutant strain of S. erythraea lacking of a EryBV glycosyltransferase (Basnet et al., 2008). In addition, a recent study obtained nine analogs of the antitumor antibiotic xantholipin through the individual in-frame mutagenesis of five tailoring enzymes (Zhang et al., 2012). In another study, fluorosalinosporamide, a derivative of the potent anticancer agent salinosporamide A, was produced by replacing the chlorinase gene salL from Salinispora tropica with the fluorinase gene flA from Salinispora cattleya (Eustaquio et al., 2010).
However, a common concern with this approach regards limited tolerance of downstream enzymes or domains to the new substance introduced by combinatorial biosynthesis and metabolic engineering. Rational design or directed evolution is one solution to this concern. Rational design is the strategy of creating new molecules with a certain functionality based on predicting how the molecule’s structure will affect its behavior, while directed evolution refers to methods to alter enzyme function using mutagenesis and selection (Nannemann et al., 2011). In a recent study, the reactivity of PikC was modified through protein engineering driven by molecular dynamics and quantum mechanical calculations. The computation-driven PikC engineering yielded a PikCD50N mutant that showed improved catalytic efficiency compared to the wild-type PikC (Narayan et al., 2015). This study demonstrated that a rationally designed protein using a crystal structure of protein and/or a computational analysis can develop a predictive model for substrate scope and selectivity of natural product biosynthesis-mediated reactions. Directed evolution is also a powerful tool to modify the activity of key enzymes responsible for the biosynthesis of natural products and can lead a higher diversity of natural products by generating novel and more potent analogs (Williams et al., 2013). As an example, a few rounds of directed evolution restored and enhanced the activity of an impaired chimerical enterobactin NRPS that has been swapped with a non-cognate aryl-carrier protein (Zhou et al., 2007). In order to reduce the risk of limited tolerance and reduce concerns of efficiency, directed evolution requires a large, high-quality library and an efficient screening strategy. The swapping of functional domains often results in non-functional or heavily impaired chimerical enzymes, and this remains an existing problem when manipulating modular PKS and NRPS systems.
Mutasynthesis
Novel natural product analogs can also be generated through gene disruption and mutasynthesis. Disruption of a gene, such as a tailoring enzyme acting downstream in a pathway, can serve to introduce a structural change. Two FK506 analogs, 9-deoxo-31-O-demethylFK506 and 31-O-demethylFK506, were produced by targeting gene disruption in Streptomyces sp. MA6548 (Shafiee et al., 1997; Ban et al., 2013). These two recombinant mutants were genetically engineered via disruption of fkbD and fkbM genes that code for 31-O-demethylFK506 methyltransferase and 9-deoxo-31-demethylFK506 hydroxylase/oxidase (Shafiee et al., 1997; Ban et al., 2013). Inactivation of individual domains within the multidomain modular PKSs and NRPSs serves as an alternative to the deletion of a whole gene. Mutasynthesis involves the coupling of a gene inactivation strategy with precursor feeding to generate new structural analogs. Precursor feeding is useful due to the substrate-promiscuity of the biosynthetic enzyme. Precursor feeding may lead to the acceptance of similar substrates or mutasynthons, a natural substrate substitute that can replace the natural substrate of a disrupted gene after being added to the growth medium, to ultimately generate new analogs. Mutasynthesis can generate new analogs for many classes of compounds. For example, the analog cahuitamycin D was produced through mutasynthetic generation with twofold-enhanced biofilm inhibitory activity in comparison to its natural product counterpart (Park et al., 2016). Recently, this approach was applied to generate nonbenzoquinone analogs of the Hsp90 inhibitor geldanamycin, which has anti-proliferative activity on tumor cells (Shin et al., 2008; Wu et al., 2011). By removing the biosynthetic genes for the 3-amino-5-hydroxybenzoic acid starter unit and feeding the culture with various 3-aminobenzoic acids and related heterocycles, a chloro-substituted nonbenzoquinone analog with significantly improved therapeutic properties was produced along with other geldanamycin analogs (Kim et al., 2007, 2009). This has been also seen in the generation of new analogs of rapamycin (Khaw et al., 1998), balhimycin (Weist et al., 2002), and novobiocin/chlorobiocin (Li and Heide, 2005).
Future Prospects
An increasing number of natural products and natural product-derived compounds have been launched over the years (Butler et al., 2014). Since 2000, 77% of FDA-approved antibiotics are natural products, all of which were derived from microbes (Patridge et al., 2016). There have been extensive reviews of natural products, semi-synthetic natural products, and nature-inspired molecules currently approved by the FDA that show the continued importance of natural products for medicine and health (Sanchez et al., 2012; Newman and Cragg, 2016). Microbial biologics are expected to remain prominent in the global biologics market, which was valued at 277 billion USD in 2015 and was recently estimated to reach 400 billion USD by 2025 (Grand View Research, 2017). While many of the biological activities of microbial natural products and biologics are well known, new advances and insights continue to be discovered. Chemical diversity from microbial natural products continue to be relevant to future drug discovery, with a continuing need for novel drugs with antibiotic, anticancer, and immunosuppressant effects, along with other pharmacological activities (Sanchez et al., 2012).
At the same time, there are a multitude of challenges facing microbial production of natural products and biologics. Some challenges to natural products-based drug discovery involve low production titers, difficulty in product isolation or structural identification. Similarly, there is much room for improvement in terms of the expression of recombinant proteins in microbial platforms. Accumulation of the end product in the microbial cell can cause global stress responses that result in cell growth inhibition. Also, the formation of misfolded and biologically inactive proteins can lower the yield of recombinant proteins. In particular, membrane proteins, high-molecular weight proteins, and multi-domain proteins are often expressed in inclusion bodies. Additionally, expressing eukaryotic proteins in a prokaryotic-based heterologous system can result in a product that is not correctly modified by post-translational enzymes, which are often required for functionality (Rosano and Ceccarelli, 2014). However, a wide variety of engineering strategies can be used with the conventional recombinant DNA technologies, including genome editing, ribosome engineering, precursor engineering, mutagenesis, and overexpression of structural genes, making it possible to facilitate the efficient production of natural products and pharmaceuticals in microbial systems.
Current technologies, such as CRISPR/Cas (Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR associated protein), should also be considered as tools for genome editing for additional improvements and to increase production (Gaj et al., 2013). For example, in vitro CRISPR-Cas9 cloning with Gibson assembly has provided an alternate strategy for heterologous expression of cryptic BGCs from genetically recalcitrant actinomycetes strains (Wang J.W. et al., 2015; Jiang and Zhu, 2016). Additionally, CRISPR-Cas9 has many prospects in the analyses of biochemical pathways in Streptomyces strains due to the development of a pCRISPomyces expression system (Cobb et al., 2015). With genome editing tools, it is also possible that non-model native hosts can be engineered to be heterologous production hosts to become platforms for combinatorial biosynthesis to create synthetic natural products and natural product derivatives. Some intrinsic limitations for non-model microbial hosts can be also improved by this genetic modification. Corynebacterium glutamicum, a GRAS organism, has been used for the industrial production of various amino acids for over five decades (Becker et al., 2011). Recently, it also showed promising potential for use as a protein expression system (Kallscheuer et al., 2016). However, this bacterium has intrinsic disadvantages, including a much lower transformation efficiency and lower yields of protein production. The CRISPR/Cas9 system was successfully employed to disrupt four different genes in C. glutamicum, opening new possibilities to use non-model strains as improved cell factories for the production of recombinant proteins and natural products (Peng et al., 2017).
Genome mining is another alternative process to discover secondary metabolites and is done by extracting information from genome sequencing (Bachmann et al., 2014; Zhongyue et al., 2018). Evaluating silent cryptic BGCs through genome mining has provided valuable avenues to generate novel molecules. For example, a genome mining strategy combined with bioinformatics predictions was used to isolate the novel natural product orfamide A by feeding a predicted precursor to a culture of Pseudomonas fluorescens (Zerikly and Challis, 2009). In a recent study, genome mining-based combinatorial biosynthesis approach also led to the discovery of new members of the leinamycin family of natural products. Leinamycin has been considered a promising anticancer drug lead due to its potent anticancer activities, unique molecular architecture and interesting modes of action. However, no leinamycin analog had been isolated in the past three decades until this study (Pan et al., 2017).
A combination of approaches can also lead to improvements in the field of microbial natural products, such as gene shuffling and ribosome engineering for increased secondary metabolite production. Additionally, the integration of ‘omics’ information has great potential in natural product drug discovery, such as with metabolomics to accurately quantify biochemical changes and metabolic pathways. Advancements in metagenomics has allowed for further understanding of diverse and complex microbial sources, including lakes, rivers, marine environments and extreme conditions, such as sub-seafloor sites and ice cores (Chandra Mohana et al., 2018).
In terms of structural characterization, X-ray crystallography and cryo-electron microscopy are advanced techniques that can allow for structure solving with high precision. Cryo-electron microscopy has been a leading method for evaluating macromolecular structures at near-atom resolution (Shoemaker and Ando, 2018). For example, single particle electron cryo-microscopy has been used to visualize pikromycin PKS module 5 from S. venezuelae, which allowed for 3D map construction with resolutions of 7.3–9.5 Å to reveal secondary structures (Dutta et al., 2014). These techniques are just a few among many that should be considered for structural studies on natural product biosynthesis. Advancements in computational strategies have led to the identification of BGCs in genome sequences and predictions of product chemical structures. Sequencing campaigns for natural product discovery should be directed toward samples likely to yield novel natural products along with well-characterized clades, such as actinomycetes, as they are still a resource for natural product discovery and have yet to be fully exhausted. The development of algorithms to mine the ever-increasing amounts of metagenomic data will allow for the potential of genome mining to be realized (Medema and Fischbach, 2015). Finally, developing additional host platforms for high-throughput refactoring and functional expression of pathways has the potential to overcome current limitations in precursor supply, product toxicity, the ability to express very large gene clusters and more (Schmidt-Dannert, 2015).
Overall, microbial natural products and biologics will continue to broaden their diverse and integral role in human life. The potential for recombinant drugs is expanding through the utilization of new protein production platforms and efforts in product improvement. Microbial cells will remain as potent protein factories because of their versatility and cost-effectiveness. Engineering strategies and recombinant DNA technologies will also allow for the increased production of microbial natural products and recombinant proteins despite the many challenges faced. Continued efforts in natural product analog development will provide an avenue for the discovery of compounds with improved biological activities in comparison to their natural counterparts. Current advanced technologies can be utilized to further advance the field of microbial natural products, which remain a steadfast resource for novel compounds in drug discovery.
Author Contributions
SP and YY designed, directed, and coordinated this project. JP, MY, AF, MM, NM, JW, EK, HC, JR, MCS, SP, and YY made substantial contributions in providing critical feedback and drafting the manuscript. SP and JP reviewed the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by a National Research Foundation of Korea (NRF) grant (2019R1A2B5B03069338) funded by the Ministry of Science and ICT (MSIT), the Bio & Medical Technology Development Program of the NRF funded by the MSIT (2018M3A9F3079662), the Collaborative Genome Program of the Korea Institute of Marine Science and Technology Promotion (KIMST) funded by the Ministry of Oceans and Fisheries (MOF) (No. 20180430), and Cooperative Research Program for Agriculture Science & Technology Development (PJ01316001) funded by the Rural Development Administration, South Korea. MCS was supported by the RP-Grant 2019 funded by the Ewha Womans University.
References
- Abdin M. Z., Israr M., Rehman R. U., Jain S. K. (2003). Artemisinin, a novel antimalarial drug: biochemical and molecular approaches for enhanced production. Planta Med. 69 289–299. 10.1055/s-2003-38871 [DOI] [PubMed] [Google Scholar]
- Abu-Salah K. M. (1996). Amphotericin B: an update. Br. J. Biomed. Sci. 53 122–133. [PubMed] [Google Scholar]
- Ahmadi M. K., Pfeifer B. A. (2016). Improved heterologous production of the nonribosomal peptide-polyketide siderophore yersiniabactin through metabolic engineering and induction optimization. Biotechnol. Prog. 32 1412–1417. 10.1002/btpr.2369 [DOI] [PubMed] [Google Scholar]
- Anisimov V. N., Zabezhinski M. A., Popovich I. G., Piskunova T. S., Semenchenko A. V., Tyndyk M. L., et al. (2011). Rapamycin increases lifespan and inhibits spontaneous tumorigenesis in inbred female mice. Cell Cycle 10 4230–4236. 10.4161/cc.10.24.18486 [DOI] [PubMed] [Google Scholar]
- Anné J., Maldonado B., Van Impe J., Van Mellaert L., Bernaerts K. (2012). Recombinant protein production and streptomycetes. J. Biotechnol. 158 159–167. 10.1016/j.jbiotec.2011.06.028 [DOI] [PubMed] [Google Scholar]
- Bachmann B. O., Van Lanen S. G., Baltz R. H. (2014). Microbial genome mining for accelerated natural products discovery: is a renaissance in the making? J. Ind. Microbiol. Biotechnol. 41 175–184. 10.1007/s10295-013-1389-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeshen M. N., Al-Hejin A. M., Bora R. S., Ahmed M. M., Ramadan H. A., Saini K. S., et al. (2015). Production of biopharmaceuticals in E. coli: current scenario and future perspectives. J. Microbiol. Biotechnol. 25 953–962. 10.4014/jmb.1412.12079 [DOI] [PubMed] [Google Scholar]
- Baeshen N. A., Baeshen M. N., Sheikh A., Bora R. S., Ahmed M. M., Ramadan H. A., et al. (2014). Cell factories for insulin production. Microb. Cell Fact. 13:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahey-El-Din M. (2012). Lactococcus lactis-based vaccines from laboratory bench to human use: an overview. Vaccine 30 685–690. 10.1016/j.vaccine.2011.11.098 [DOI] [PubMed] [Google Scholar]
- Ballance D. J. (1999). Yeast-derived recombinant human albumin (Recombumin). Ansthesiol. Intensivmed. Notfallmed. Schmerzther. 34 775–777. 10.1055/s-1999-10842-5 [DOI] [PubMed] [Google Scholar]
- Ban Y. H., Park S. R., Yoon Y. J. (2016). The biosynthetic pathway of FK506 and its engineering: from past achievements to future prospects. J. Ind. Microbiol. Biotechnol. 43 389–400. 10.1007/s10295-015-1677-7 [DOI] [PubMed] [Google Scholar]
- Ban Y. H., Shinde P. B., Hwang J.-Y., Song M.-C., Kim D. H., Lim S.-K., et al. (2013). Characterization of FK506 biosynthetic intermediates involved in post-PKS elaboration. J. Nat. Prod. 76 1091–1098. 10.1021/np4001224 [DOI] [PubMed] [Google Scholar]
- Barros F. F. C., Simiqueli A. P. R., Andrade C. J. A., Pastore G. M. (2013). Production of Enzymes from Agroindustrial Wastes by Biosurfactant-Producing Strains of Bacillus subtilis. Biotechnol. Res. Int. 2013:9. 10.1155/2013/103960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basnet D. B., Park J. W., Yoon Y. J. (2008). Combinatorial biosynthesis of 5-O-desosaminyl erythronolide A as a potent precursor of ketolide antibiotics. J. Biotechnol. 135 92–96. 10.1016/j.jbiotec.2008.03.001 [DOI] [PubMed] [Google Scholar]
- Baysal Ö, Yıldız A. (2017). Bacillus subtilis: an industrially important microbe for enzymes production. EC Microbiol. 5 148–156. [Google Scholar]
- Becker J., Zelder O., Hafner S., Schroder H., Wittmann C. (2011). From zero to hero–design-based systems metabolic engineering of Corynebacterium glutamicum for L-lysine production. Metab. Eng. 13 159–168. 10.1016/j.ymben.2011.01.003 [DOI] [PubMed] [Google Scholar]
- Bellini A. V., Galli G., Fascetti E., Frascotti G., Branduzzi P., Lucchese G., et al. (1991). Production processes of recombinant IL-1β from Bacillus subtilis: comparison between intracellular and exocellular expression. J. Biotechnol. 18 177–192. 10.1016/0168-1656(91)90246-r [DOI] [PubMed] [Google Scholar]
- Bender E., Koller K. P., Engels J. W. (1990). Secretory synthesis of human interleukin-2 by Streptomyces lividans. Gene 86 227–232. 10.1016/0378-1119(90)90283-w [DOI] [PubMed] [Google Scholar]
- Bermúdez-Humarán L. G., Motta J. P., Aubry C., Kharrat P., Rous-Martin L., Sallenave J. M., et al. (2015). Serine protease inhibitors protect better than IL-10 and TGF-β anti-inflammatory cytokines against mouse colitis when delivered by recombinant lactococci. Microb. Cell Fact. 14:26. 10.1186/s12934-015-0198-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blunt J. W., Copp B. R., Keyzers R. A., Munro M. H., Prinsep M. R. (2016). Marine natural products. Nat. Prod. Rep. 33 382–431. 10.1039/c5np00156k [DOI] [PubMed] [Google Scholar]
- Böer E., Steinborn G., Matros A., Mock H.-P., Gellissen G., Kunze G. (2007). Production of interleukin-6 in Arxula adeninivorans, Hansenula polymorpha and Saccharomyces cerevisiae by applying the wide-range yeast vector (CoMedTM) system to simultaneous comparative assessment. FEMS Yeast Res. 7 1181–1187. 10.1111/j.1567-1364.2007.00254.x [DOI] [PubMed] [Google Scholar]
- Boer E., Steinborn G., Matros A., Mock H. P., Gellissen G., Kunze G. (2007). Production of interleukin-6 in Arxula adeninivorans, Hansenula polymorpha and Saccharomyces cerevisiae by applying the wide-range yeast vector (CoMed) system to simultaneous comparative assessment. FEMS Yeast Res. 7 1181–1187. 10.1111/j.1567-1364.2007.00254.x [DOI] [PubMed] [Google Scholar]
- Boling S. D., Webel D. M., Mavromichalis I., Parsons C. M., Baker D. H. (2000). The effects of citric acid on phytate-phosphorus utilization in young chicks and pigs. J. Anim. Sci. 78 682–689. 10.2527/2000.783682x [DOI] [PubMed] [Google Scholar]
- Borchardt J. K. (2002). The Beginnings of drug therapy: ancient mesopotamian medicine. Drug News Perspect. 15 187–192. 10.1358/dnp.2002.15.3.840015 [DOI] [PubMed] [Google Scholar]
- Borodina I., Siebring J., Zhang J., Smith C. P., Van Keulen G., Dijkhuizen L., et al. (2008). Antibiotic overproduction in Streptomyces coelicolor A3 2 mediated by phosphofructokinase deletion. J. Biol. Chem. 283 25186–25199. 10.1074/jbc.M803105200 [DOI] [PubMed] [Google Scholar]
- Breyner N. M., Michon C., De Sousa C. S., Vilas Boas P. B., Chain F., Azevedo V. A., et al. (2017). Microbial anti-inflammatory molecule (MAM) from Faecalibacterium prausnitzii shows a protective effect on DNBS and DSS-induced colitis model in mice through inhibition of NF-kappaB pathway. Front. Microbiol. 8:114. 10.3389/fmicb.2017.00114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton S. E., Hercus T. R., Nero T. L., Kan W. L., Barry E. F., Dottore M., et al. (2018). A dual role for the N-terminal domain of the IL-3 receptor in cell signalling. Nat. Commun. 9:386. 10.1038/s41467-017-02633-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler M. S., Robertson A. A., Cooper M. A. (2014). Natural product and natural product derived drugs in clinical trials. Nat. Prod. Rep. 31 1612–1661. 10.1039/c4np00064a [DOI] [PubMed] [Google Scholar]
- Cairns T. C., Nai C., Meyer V. (2018). How a fungus shapes biotechnology: 100 years of Aspergillus niger research. Fungal Biol. Biotechnol. 5:13. 10.1186/s40694-018-0054-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrez D., Janssens W., Degrave P., Van Den Hondel C. A., Kinghorn J. R., Fiers W., et al. (1990). Heterologous gene expression by filamentous fungi: secretion of human interleukin-6 by Aspergillus nidulans. Gene 94 147–154. 10.1016/0378-1119(90)90381-z [DOI] [PubMed] [Google Scholar]
- Celik E., Calik P. (2012). Production of recombinant proteins by yeast cells. Biotechnol. Adv. 30 1108–1118. 10.1016/j.biotechadv.2011.09.011 [DOI] [PubMed] [Google Scholar]
- Cersosimo R. J., Hong W. K. (1986). Epirubicin: a review of the pharmacology, clinical activity, and adverse effects of an adriamycin analogue. J. Clin. Oncol. 4 425–439. 10.1200/jco.1986.4.3.425 [DOI] [PubMed] [Google Scholar]
- Chandra Mohana N., Yashavantha Rao H. C., Rakshith D., Mithun P. R., Nuthan B. R., Satish S. (2018). Omics based approach for biodiscovery of microbial natural products in antibiotic resistance era. J. Genet. Eng. Biotechnol. 16 1–8. 10.1016/j.jgeb.2018.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. C., Chen W. C., Ho T. F., Wu H. S., Wei Y. H. (2011). Development of natural anti-tumor drugs by microorganisms. J. Biosci. Bioeng. 111 501–511. 10.1016/j.jbiosc.2010.12.026 [DOI] [PubMed] [Google Scholar]
- Chang M. C. Y., Eachus R. A., Trieu W., Ro D.-K., Keasling J. D. (2007). Engineering Escherichia coli for production of functionalized terpenoids using plant P450s. Nat. Chem. Biol. 3 274–277. 10.1038/nchembio875 [DOI] [PubMed] [Google Scholar]
- Chen Y., Smanski M. J., Shen B. (2010). Improvement of secondary metabolite production in Streptomyces by manipulating pathway regulation. Appl. Microbiol. Biotechnol. 86 19–25. 10.1007/s00253-009-2428-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin Y. W., Balunas M. J., Chai H. B., Kinghorn A. D. (2006). Drug discovery from natural sources. AAPS J 8 E239–E253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra I., Roberts M. (2001). Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65 232–260. 10.1128/mmbr.65.2.232-260.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb R. E., Luo Y., Freestone T., Zhao H. (2013). “Chapter 10 – Drug discovery and development via synthetic biology,” in Synthetic Biology, ed. Zhao H. (Boston, MA: Academic Press; ), 183–206. 10.1016/b978-0-12-394430-6.00010-8 [DOI] [Google Scholar]
- Cobb R. E., Wang Y., Zhao H. (2015). High-efficiency multiplex genome editing of Streptomyces species using an engineered CRISPR/Cas system. ACS Synth. Biol. 4 723–728. 10.1021/sb500351f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copping L. G., Duke S. O. (2007). Natural products that have been used commercially as crop protection agents. Pest. Manage. Sci. 63 524–554. 10.1002/ps.1378 [DOI] [PubMed] [Google Scholar]
- Cox H., Mead D., Sudbery P., Eland R. M., Mannazzu I., Evans L. (2000). Constitutive expression of recombinant proteins in the methylotrophic yeast Hansenula polymorpha using the PMA1 promoter. Yeast 16 1191–1203. [DOI] [PubMed] [Google Scholar]
- Cragg G. M., Newman D. J. (2013). Natural products: a continuing source of novel drug leads. Biochim. Biophys. Acta 1830 3670–3695. 10.1016/j.bbagen.2013.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Rocha A. B., Lopes R. M., Schwartsmann G. (2001). Natural products in anticancer therapy. Curr. Opin. Pharmacol. 1 364–369. 10.1016/s1471-4892(01)00063-7 [DOI] [PubMed] [Google Scholar]
- Dasgupta A. (2012). Advances in antibiotic measurement. Adv. Clin. Chem. 56 75–104. 10.1016/b978-0-12-394317-0.00013-3 [DOI] [PubMed] [Google Scholar]
- Davidson D., Amrein L., Panasci L., Aloyz R. (2013). Small molecules, inhibitors of DNA-PK, targeting DNA repair, and beyond. Front. Pharmacol. 4:5. 10.3389/fphar.2013.00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lima Procópio R. E., Da Silva I. R., Martins M. K., De Azevedo J. L., De Araújo J. M. (2012). Antibiotics produced by Streptomyces. Braz. J. Infect. Dis. 16 466–471. 10.1016/j.bjid.2012.08.014 [DOI] [PubMed] [Google Scholar]
- De Mey M., Lequeux G. J., Beauprez J. J., Maertens J., Waegeman H. J., Van Bogaert I. N., et al. (2010). Transient metabolic modeling of Escherichia coli MG1655 and MG1655 deltaackA-pta, deltapoxB deltapppc ppc-p37 for recombinant beta-galactosidase production. J. Ind. Microbiol. Biotechnol. 37 793–803. 10.1007/s10295-010-0724-7 [DOI] [PubMed] [Google Scholar]
- Deleu M., Paquot M., Nylander T. (2008). Effect of fengycin, a lipopeptide produced by Bacillus subtilis, on model biomembranes. Biophys. J. 942667–2679. 10.1529/biophysj.107.114090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demain A. L. (2009). Antibiotics: natural products essential to human health. Med. Res. Rev. 29 821–842. 10.1002/med.20154 [DOI] [PubMed] [Google Scholar]
- Demain A. L. (2014). Importance of microbial natural products and the need to revitalize their discovery. J. Ind. Microbiol. Biotechnol. 41 185–201. 10.1007/s10295-013-1325-z [DOI] [PubMed] [Google Scholar]
- Demain A. L., Vaishnav P. (2011). Natural products for cancer chemotherapy. Microb. Biotechnol. 4 687–699. 10.1111/j.1751-7915.2010.00221.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Marco A., Cassinelli G., Arcamone F. (1981). The discovery of daunorubicin. Cancer Treat. Rep. 65(Suppl. 4), 3–8. [PubMed] [Google Scholar]
- Dias D. A., Urban S., Roessner U. (2012). A historical overview of natural products in drug discovery. Metabolites 2 303–336. 10.3390/metabo2020303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieci G., Bottarelli L., Ballabeni A., Ottonello S. (2000). tRNA-assisted overproduction of eukaryotic ribosomal proteins. Protein Expr. Purif. 18346–354. 10.1006/prep.2000.1203 [DOI] [PubMed] [Google Scholar]
- Ding M. Z., Yan H. F., Li L. F., Zhai F., Shang L. Q., Yin Z., et al. (2014). Biosynthesis of Taxadiene in Saccharomyces cerevisiae: selection of geranylgeranyl diphosphate synthase directed by a computer-aided docking strategy. PLoS One 9:e109348. 10.1371/journal.pone.0109348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouault S., Corthier G., Ehrlich S. D., Renault P. (2000). Expression of the Staphylococcus hyicus lipase in Lactococcus lactis. Appl. Environ. Microbiol. 66 588–598. 10.1128/aem.66.2.588-598.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggar B. M. (1948). Aureomycin; a product of the continuing search for new antibiotics. Ann. N. Y. Acad. Sci. 51 177–181. [DOI] [PubMed] [Google Scholar]
- Dutta S., Whicher J. R., Hansen D. A., Hale W. A., Chemler J. A., Congdon G. R., et al. (2014). Structure of a modular polyketide synthase. Nature 510512–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards G. A., Shymanska N. V., Pierce J. G. (2017). 5-Benzylidene-4-oxazolidinones potently inhibit biofilm formation in Methicillin-resistant Staphylococcus aureus. Chem. Commun. 53 7353–7356. 10.1039/c7cc03626d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich J., Bartz Q. R., Smith R. M., Joslyn D. A., Burkholder P. R. (1947). Chloromycetin, a new antibiotic from a soil actinomycete. Science 106:417. 10.1126/science.106.2757.417 [DOI] [PubMed] [Google Scholar]
- Einhorn L. H., Donohue J. (2002). Cis-diamminedichloroplatinum, vinblastine, and bleomycin combination chemotherapy in disseminated testicular cancer. J. Urol. 168 2368–2372. 10.1097/00005392-200212000-00005 [DOI] [PubMed] [Google Scholar]
- El-Hossary E. M., Cheng C., Hamed M. M., El-Sayed Hamed A. N., Ohlsen K., Hentschel U., et al. (2017). Antifungal potential of marine natural products. Eur. J. Med. Chem. 126 631–651. [DOI] [PubMed] [Google Scholar]
- Eustaquio A. S., O’hagan D., Moore B. S. (2010). Engineering fluorometabolite production: fluorinase expression in Salinispora tropica yields fluorosalinosporamide. J. Nat. Prod. 73 378–382. 10.1021/np900719u [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favero-Retto M. P., Palmieri L. C., Souza T. A., Almeida F. C., Lima L. M. (2013). Structural meta-analysis of regular human insulin in pharmaceutical formulations. Eur. J. Pharm. Biopharm. 85 1112–1121. 10.1016/j.ejpb.2013.05.005 [DOI] [PubMed] [Google Scholar]
- Ferrer-Miralles N., Domingo-Espin J., Corchero J. L., Vazquez E., Villaverde A. (2009). Microbial factories for recombinant pharmaceuticals. Microb. Cell Fact. 8:17. 10.1186/1475-2859-8-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Miralles N., Villaverde A. (2013). Bacterial cell factories for recombinant protein production; expanding the catalogue. Microb. Cell Fact. 12:113. 10.1186/1475-2859-12-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach M. A., Walsh C. T. (2006). Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics:? logic. Mach. Mech. Chem. Rev. 106 3468–3496. 10.1021/cr0503097 [DOI] [PubMed] [Google Scholar]
- Fjærvik E., Zotchev S. B. (2005). Biosynthesis of the polyene macrolide antibiotic nystatin in Streptomyces noursei. Appl. Microbiol. Biotechnol. 67 436–443. 10.1007/s00253-004-1802-4 [DOI] [PubMed] [Google Scholar]
- Fleming A. (1944). The discovery of penicillin. Br. Med. Bull. 2 4–5. [Google Scholar]
- Fleming A. (2001). On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae. 1929. Bull. World Health Organ. 79 780–790. [PMC free article] [PubMed] [Google Scholar]
- Fracchia L., Cavallo M., Allegrone G., Martinotti M. (2010). A Lactobacillus-derived biosurfactant inhibits biofilm formation of human pathogenic Candida albicans biofilm producers. Appl. Microbiol. Biotechnol. 2 827–837. [Google Scholar]
- Gaffar S. A., Shethna Y. I. (1977). Purification and some biological properties of asparaginase from Azotobacter vinelandii. Appl. Environ. Microbiol. 33 508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T., Gersbach C. A., Barbas C. F. (2013). ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31 397–405. 10.1016/j.tibtech.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W., Wu Z., Sun J., Ni X., Xia H. (2017). Modulation of kanamycin B and kanamycin A biosynthesis in Streptomyces kanamyceticus via metabolic engineering. PLoS One 12:e0181971. 10.1371/journal.pone.0181971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellissen G., Janowicz Z. A., Weydemann U., Melber K., Strasser A. W., Hollenberg C. P. (1992). High-level expression of foreign genes in Hansenula polymorpha. Biotechnol. Adv. 10 179–189. 10.1016/0734-9750(92)90002-q [DOI] [PubMed] [Google Scholar]
- Geraci J. E., Heilman F. R., Nichols D. R., Wellman E. W., Ross G. T. (1956). Some laboratory and clinical experiences with a new antibiotic, vancomycin. Antibiot. Annu. 90–106. [PubMed] [Google Scholar]
- Giddings L. A., Newman D. J. (2013). Microbial natural products: molecular blueprints for antitumor drugs. J. Ind. Microbiol. Biotechnol. 40 1181–1210. 10.1007/s10295-013-1331-1 [DOI] [PubMed] [Google Scholar]
- Gilbert M., Morosoli R., Shareck F., Kluepfel D. (1995). Production and secretion of proteins by streptomycetes. Crit. Rev. Biotechnol. 15 13–39. 10.3109/07388559509150530 [DOI] [PubMed] [Google Scholar]
- Gill S. S., Cowles E. A., Pietrantonio P. V. (1992). The mode of action of Bacillus thuringiensis endotoxins. Annu. Rev. Entomol. 37 615–636. 10.1146/annurev.ento.37.1.615 [DOI] [PubMed] [Google Scholar]
- Glenting J., Poulsen L. K., Kato K., Madsen S. M., Frøkiær H., Wendt C., et al. (2007). Production of recombinant peanut allergen Ara h 2 using Lactococcus lactis. Microb. Cell Fact. 6:28. 10.1186/1475-2859-6-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong G.-L., Sun X., Liu X.-L., Hu W., Cao W.-R., Liu H., et al. (2007). Mutation and a high-throughput screening method for improving the production of epothilones of sorangium. J. Ind. Microbiol. Biotechnol. 34 615–623. 10.1007/s10295-007-0236-2 [DOI] [PubMed] [Google Scholar]
- Gorska M., Popowska U., Sielicka A., Kuban-Jankowska A., Sawczuk W., Knap N., et al. (2012). Geldanamycin and its derivatives as Hsp90 inhibitors. Front. Biosci. (Landmark Ed) 17 2269–2277. [DOI] [PubMed] [Google Scholar]
- Grand View Research (2017). Biologics Market Analysis by Source (Microbial, Mammalian), by Products (Monoclonal Antibodies, Vaccines, Recombinant Proteins, Antisense, RNAi), By Disease Category, by Manufacturing, & Segment Forecasts, 2018 – 2025. Biologics Market Size Forecast, Industry Growth Report, 2018-2025. San Francisco, CA: Grand View Research. [Google Scholar]
- Graumann K., Premstaller A. (2006). Manufacturing of recombinant therapeutic proteins in microbial systems. Biotechnol. J. 1 164–186. 10.1002/biot.200500051 [DOI] [PubMed] [Google Scholar]
- Grimm L. H., Kelly S., Krull R., Hempel D. C. (2005). Morphology and productivity of filamentous fungi. Appl. Microbiol. Biotechnol. 69 375–384. 10.1007/s00253-005-0213-5 [DOI] [PubMed] [Google Scholar]
- Guisez Y., Tison B., Vandekerckhove J., Demolder J., Bauw G., Haegeman G., et al. (1991). Production and purification of recombinant human interleukin-6 secreted by the yeast Saccharomyces cerevisiae. Eur. J. Biochem. 198 217–222. 10.1111/j.1432-1033.1991.tb16004.x [DOI] [PubMed] [Google Scholar]
- Gustafsson C., Govindarajan S., Minshull J. (2004). Codon bias and heterologous protein expression. Trends Biotechnol. 22 346–353. 10.1016/j.tibtech.2004.04.006 [DOI] [PubMed] [Google Scholar]
- Gyawali R., Ibrahim S. A. (2014). Natural products as antimicrobial agents. Food Control 46 412–429. 10.1016/j.foodcont.2014.05.047 [DOI] [Google Scholar]
- Hamed M. B., Anne J., Karamanou S., Economou A. (2018). Streptomyces protein secretion and its application in biotechnology. FEMS Microbiol.Lett. 365 10.1093/femsle/fny250 [DOI] [PubMed] [Google Scholar]
- Hartley D. L., Kane J. F. (1988). Properties of inclusion bodies from recombinant Escherichia coli. Biochem. Soc. Trans. 16:101. [DOI] [PubMed] [Google Scholar]
- Harvey C. J. B., Tang M., Schlecht U., Horecka J., Fischer C. R., Lin H.-C., et al. (2018). HEx: a heterologous expression platform for the discovery of fungal natural products. Sci. Adv. 4:eaar5459. 10.1126/sciadv.aar5459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijtink R. A., Bergen P. V., Melber K., Janowicz Z. A., Osterhaus A. D. (2002). Hepatitis B surface antigen (HBsAg) derived from yeast cells (Hansenula polymorpha) used to establish an influence of antigenic subtype (adw2, adr, ayw3) in measuring the immune response after vaccination. Vaccine 202191–2196. 10.1016/s0264-410x(02)00145-7 [DOI] [PubMed] [Google Scholar]
- Hercus T. R., Barry E. F., Dottore M., Mcclure B. J., Webb A. I., Lopez A. F., et al. (2013). High yield production of a soluble human interleukin-3 variant from E. coli with wild-type bioactivity and improved radiolabeling properties. PLoS One 8:e74376. 10.1371/journal.pone.0074376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C. P., Osslund T. D., Eisenberg D. (1993). The structure of granulocyte-colony-stimulating factor and its relationship to other growth factors. Proc. Natl. Acad. Sci. U.S.A. 90 5167–5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiort J., Maksimenka K., Reichert M., Peroviæ-Ottstadt S., Lin W. H., Wray V., et al. (2004). New natural products from the sponge-derived fungus Aspergillus niger. J. Nat. Prod. 67 1532–1543. 10.1021/np030551d [DOI] [PubMed] [Google Scholar]
- Höfte H., Whiteley H. R. (1989). Insecticidal crystal proteins of Bacillus thuringiensis. Microbiol. Rev. 53 242–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenberg C. P., Gellissen G. (1997). Production of recombinant proteins by methylotrophic yeasts. Curr. Opin. Biotechnol. 8 554–560. 10.1016/s0958-1669(97)80028-6 [DOI] [PubMed] [Google Scholar]
- Hollstein U. (1974). Actinomycin. Chemistry and mechanism of action. Chem. Rev. 74 625–652. 10.1021/cr60292a002 [DOI] [Google Scholar]
- Hopwood D. A., Malpartida F., Kieser H. M., Ikeda H., Duncan J., Fujii I., et al. (1985). Production of ‘hybrid’ antibiotics by genetic engineering. Nature 314:642. [DOI] [PubMed] [Google Scholar]
- Hou J., Tyo K., Liu Z., Petranovic D., Nielsen J. (2012). Engineering of vesicle trafficking improves heterologous protein secretion in Saccharomyces cerevisiae. Metab. Eng. 14 120–127. 10.1016/j.ymben.2012.01.002 [DOI] [PubMed] [Google Scholar]
- Idiris A., Tohda H., Bi K. W., Isoai A., Kumagai H., Giga-Hama Y. (2006). Enhanced productivity of protease-sensitive heterologous proteins by disruption of multiple protease genes in the fission yeast Schizosaccharomyces pombe. Appl. Microbiol. Biotechnol. 73 404–420. 10.1007/s00253-006-0489-0 [DOI] [PubMed] [Google Scholar]
- Inan M., Aryasomayajula D., Sinha J., Meagher M. M. (2006). Enhancement of protein secretion in Pichia pastoris by overexpression of protein disulfide isomerase. Biotechnol. Bioeng. 93 771–778. 10.1002/bit.20762 [DOI] [PubMed] [Google Scholar]
- Ito A., Sasaguri Y., Kitada S., Kusaka Y., Kuwano K., Masutomi K., et al. (2004). A Bacillus thuringiensis crystal protein with selective cytocidal action to human cells. J. Biol. Chem. 279 21282–21286. [DOI] [PubMed] [Google Scholar]
- Janowicz Z. A., Melber K., Merckelbach A., Jacobs E., Harford N., Comberbach M., et al. (1991). Simultaneous expression of the S and L surface antigens of hepatitis B, and formation of mixed particles in the methylotrophic yeast, Hansenula polymorpha. Yeast 7 431–443. 10.1002/yea.320070502 [DOI] [PubMed] [Google Scholar]
- Jepsen M. P., Jogdand P. S., Singh S. K., Esen M., Christiansen M., Issifou S., et al. (2013). The malaria vaccine candidate GMZ2 elicits functional antibodies in individuals from malaria endemic and non-endemic areas. J. Infect. Dis. 208 479–488. 10.1093/infdis/jit185 [DOI] [PubMed] [Google Scholar]
- Jiang W., Zhu T. F. (2016). Targeted isolation and cloning of 100-kb microbial genomic sequences by Cas9-assisted targeting of chromosome segments. Nat. Protoc. 11 960–975. 10.1038/nprot.2016.055 [DOI] [PubMed] [Google Scholar]
- Johnson I. S. (1983). Human insulin from recombinant DNA technology. Science 219 632–637. 10.1126/science.6337396 [DOI] [PubMed] [Google Scholar]
- Jozala A. F., Geraldes D. C., Tundisi L. L., Feitosa V. A., Breyer C. A., Cardoso S. L., et al. (2016). Biopharmaceuticals from microorganisms: from production to purification. Braz. J. Microbiol. 47 51–63. 10.1016/j.bjm.2016.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung W. S., Lee S. K., Hong J. S. J., Park S. R., Jeong S. J., Han A. R., et al. (2006). Heterologous expression of tylosin polyketide synthase and production of a hybrid bioactive macrolide in Streptomyces venezuelae. Appl. Microbiol. Biotechnol. 72 763–769. 10.1007/s00253-006-0318-5 [DOI] [PubMed] [Google Scholar]
- Jung W. S., Yoo Y. J., Park J. W., Park S. R., Han A. R., Ban Y. H., et al. (2011). A combined approach of classical mutagenesis and rational metabolic engineering improves rapamycin biosynthesis and provides insights into methylmalonyl-CoA precursor supply pathway in Streptomyces hygroscopicus ATCC 29253. Appl. Microbiol. Biotechnol. 91 1389–1397. 10.1007/s00253-011-3348-6 [DOI] [PubMed] [Google Scholar]
- Kalliolias G. D., Liossis S.-N. (2008). The future of the IL-1 receptor antagonist anakinra: from rheumatoid arthritis to adult-onset Still’s disease and systemic-onset juvenile idiopathic arthritis. Exp. Opin. Investig. Drugs 17 349–359. 10.1517/13543784.17.3.349 [DOI] [PubMed] [Google Scholar]
- Kallscheuer N., Vogt M., Stenzel A., Gatgens J., Bott M., Marienhagen J. (2016). Construction of a Corynebacterium glutamicum platform strain for the production of stilbenes and (2S)-flavanones. Metab. Eng. 38 47–55. 10.1016/j.ymben.2016.06.003 [DOI] [PubMed] [Google Scholar]
- Kapust R. B., Waugh D. S. (1999). Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci. 8 1668–1674. 10.1110/ps.8.8.1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaslow D. C., Bathurst I. C., Lensen T., Ponnudurai T., Barr P. J., Keister D. B. (1994). Saccharomyces cerevisiae recombinant Pfs25 adsorbed to alum elicits antibodies that block transmission of Plasmodium falciparum. Infect. Immun. 62 5576–5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaw L. E., Bohm G. A., Metcalfe S., Staunton J., Leadlay P. F. (1998). Mutational biosynthesis of novel rapamycins by a strain of Streptomyces hygroscopicus NRRL 5491 disrupted in rapL, encoding a putative lysine cyclodeaminase. J. Bacteriol. 180 809–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E. J., Yang I., Yoon Y. J. (2015). Developing Streptomyces venezuelae as a cell factory for the production of small molecules used in drug discovery. Arch. Pharm. Res. 38 1606–1616. 10.1007/s12272-015-0638-z [DOI] [PubMed] [Google Scholar]
- Kim J.-H., Wong S.-L., Kim B.-G. (2001). Optimization of staphylokinase production in Bacillus subtilis using inducible and constitutive promoters. Biotechnol. Bioprocess Eng. 6 167 10.1007/bf02932545 [DOI] [Google Scholar]
- Kim W., Lee D., Hong S. S., Na Z., Shin J. C., Roh S. H., et al. (2009). Rational biosynthetic engineering for optimization of geldanamycin analogues. Chembiochem 10 1243–1251. 10.1002/cbic.200800763 [DOI] [PubMed] [Google Scholar]
- Kim W., Lee J. S., Lee D., Cai X. F., Shin J. C., Lee K., et al. (2007). Mutasynthesis of geldanamycin by the disruption of a gene producing starter unit: generation of structural diversity at the benzoquinone ring. Chembiochem 8 1491–1494. 10.1002/cbic.200700196 [DOI] [PubMed] [Google Scholar]
- Kim Y. H., Park B. S., Bhatia S. K., Seo H. M., Jeon J. M., Kim H. J., et al. (2014). Production of rapamycin in Streptomyces hygroscopicus from glycerol-based media optimized by systemic methodology. J. Microbiol. Biotechnol. 24 1319–1326. 10.4014/jmb.1403.03024 [DOI] [PubMed] [Google Scholar]
- Knaggs A. R. (2001). The biosynthesis of shikimate metabolites. Nat. Prod. Rep. 18 334–355. 10.1039/B100399M [DOI] [PubMed] [Google Scholar]
- Komaki H., Ichikawa N., Oguchi A., Hamada M., Tamura T., Fujita N. (2015). Genome-based analysis of non-ribosomal peptide synthetase and type-I polyketide synthase gene clusters in all type strains of the genus Herbidospora. BMC Res. Notes 8:548. 10.1186/s13104-015-1526-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Uchiyama T., Omura S., Cane D. E., Ikeda H. (2010). Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc. Natl. Acad. Sci. U.S.A. 107 2646–2651. 10.1073/pnas.0914833107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung S. H., Lund S., Murarka A., Mcphee D., Paddon C. J. (2018). Approaches and recent developments for the commercial production of semi-synthetic artemisinin. Front. Plant Sci. 9:87. 10.3389/fpls.2018.00087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze G., Kang H. A., Gellissen G. (2009). “Hansenula polymorpha (Pichia angusta): biology and applications,” in Yeast Biotechnology: Diversity and Applications, eds Satyanarayana T., Kunze G. (Dordrecht: Springer; ), 47–64. 10.1007/978-1-4020-8292-4_3 [DOI] [Google Scholar]
- Kušèer E., Coates N., Challis I., Gregory M., Wilkinson B., Sheridan R., et al. (2007). Roles of rapH and rapG in positive regulation of rapamycin biosynthesis in Streptomyces hygroscopicus. J. Bacteriol. 189 4756–4763. 10.1128/jb.00129-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacana E., Amur S., Mummanneni P., Zhao H., Frueh F. (2007). The emerging role of pharmacogenomics in biologics. Clin. Pharmacol. Therap. 82 466–471. 10.1038/sj.clpt.6100334 [DOI] [PubMed] [Google Scholar]
- Law B. K. (2005). Rapamycin: an anti-cancer immunosuppressant? Crit. Rev. Oncol. Hematol. 56 47–60. 10.1016/j.critrevonc.2004.09.009 [DOI] [PubMed] [Google Scholar]
- Le Loir Y., Azevedo V., Oliveira S. C., Freitas D. A., Miyoshi A., Bermúdez-Humarán L. G., et al. (2005). Protein secretion in Lactococcus lactis: an efficient way to increase the overall heterologous protein production. Microb. Cell Fact. 4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H., Kim Y. G., Lee K., Kim C. J., Park D. J., Ju Y., et al. (2016). Streptomyces-derived actinomycin D inhibits biofilm formation by Staphylococcus aureus and its hemolytic activity. Biofouling 32 45–56. 10.1080/08927014.2015.1125888 [DOI] [PubMed] [Google Scholar]
- Leggett J. E. (2015). “25 – Aminoglycosides,” in Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases (Eighth Edition) eds Bennett J. E., Dolin R., Blaser M. J. (Philadelphia, PA: Content Repository Only!), 310.e–321.e. [Google Scholar]
- Li H. S., Piao D. C., Jiang T., Bok J. D., Cho C. S., Lee Y. S., et al. (2015). Recombinant interleukin 6 with M cell-targeting moiety produced in Lactococcus lactis IL1403 as a potent mucosal adjuvant for peroral immunization. Vaccine 33 1959–1967. 10.1016/j.vaccine.2015.02.061 [DOI] [PubMed] [Google Scholar]
- Li J. W., Vederas J. C. (2009). Drug discovery and natural products: end of an era or an endless frontier? Science 325 161–165. 10.1126/science.1168243 [DOI] [PubMed] [Google Scholar]
- Li S. M., Heide L. (2005). New aminocoumarin antibiotics from genetically engineered Streptomyces strains. Curr. Med. Chem. 12 419–427. 10.2174/0929867053363063 [DOI] [PubMed] [Google Scholar]
- Linares D. M., Kok J., Poolman B. (2010). Genome sequences of Lactococcus lactis MG1363 (Revised) and NZ9000 and comparative physiological studies. J. Bacteriol. 192 5806–5812. 10.1128/jb.00533-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losey H. C., Peczuh M. W., Chen Z., Eggert U. S., Dong S. D., Pelczer I., et al. (2001). Tandem action of glycosyltransferases in the maturation of vancomycin and teicoplanin aglycones: novel glycopeptides. Biochemistry 40 4745–4755. 10.1021/bi010050w [DOI] [PubMed] [Google Scholar]
- Lv X. A., Jin Y. Y., Li Y. D., Zhang H., Liang X. L. (2013). Genome shuffling of Streptomyces viridochromogenes for improved production of avilamycin. Appl. Microbiol. Biotechnol. 97 641–648. 10.1007/s00253-012-4322-7 [DOI] [PubMed] [Google Scholar]
- Maheshwari R. (2006). Fungi as cell factories: hype, reality and hope. Indian J. Microbiol. 46 307–324. [Google Scholar]
- Maiya S., Grundmann A., Li S. M., Turner G. (2009). Improved tryprostatin B production by heterologous gene expression in Aspergillus nidulans. Fungal Genet. Biol. 46 436–440. 10.1016/j.fgb.2009.01.003 [DOI] [PubMed] [Google Scholar]
- Mann J. (2001). Natural products as immunosuppressive agents. Nat. Prod. Rep. 18 417–430. 10.1039/b001720p [DOI] [PubMed] [Google Scholar]
- March J. C., Eiteman M. A., Altman E. (2002). Expression of an anaplerotic enzyme, pyruvate carboxylase, improves recombinant protein production in Escherichia coli. Appl. Environ. Microbiol. 68 5620–5624. 10.1128/aem.68.11.5620-5624.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin V. J., Pitera D. J., Withers S. T., Newman J. D., Keasling J. D. (2003). Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat. Biotechnol. 21 796–802. 10.1038/nbt833 [DOI] [PubMed] [Google Scholar]
- Martínez J. M., Kok J., Sanders J. W., Hernández P. E. (2000). Heterologous coproduction of Enterocin A and pediocin PA-1 by Lactococcus lactis: detection by specific peptide-directed antibodies. Appl. Environ. Microbiol. 66 3543–3549. 10.1128/aem.66.8.3543-3549.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura E., Nakagawa A., Tomabechi Y., Ikushiro S., Sakaki T., Katayama T., et al. (2018). Microbial production of novel sulphated alkaloids for drug discovery. Sci. Rep. 8:7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A. F., Hellmuth K., Schlieker H., Lopez-Ulibarri R., Oertel S., Dahlems U., et al. (1999). An expression system matures: a highly efficient and cost-effective process for phytase production by recombinant strains of Hansenula polymorpha. Biotechnol. Bioeng. 63 373–381. [DOI] [PubMed] [Google Scholar]
- McAleer W. J., Buynak E. B., Maigetter R. Z., Wampler D. E., Miller W. J., Hilleman M. R. (1984). Human hepatitis B vaccine from recombinant yeast. Nature 307 178–180. 10.1038/307178a0 [DOI] [PubMed] [Google Scholar]
- McDaniel R., Licari P., Khosla C. (2001). Process development and metabolic engineering for the overproduction of natural and unnatural polyketides. Adv. Biochem. Eng. Biotechnol. 73 31–52. 10.1007/3-540-45300-8_3 [DOI] [PubMed] [Google Scholar]
- McDaniel R., Thamchaipenet A., Gustafsson C., Fu H., Betlach M., Ashley G. (1999). Multiple genetic modifications of the erythromycin polyketide synthase to produce a library of novel ”unnatural” natural products. Proc. Natl. Acad. Sci. U.S.A. 96 1846–1851. 10.1073/pnas.96.5.1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire J. M., Bunch R. L., Anderson R. C., Boaz H. E., Flynn E. H., Powell H. M., et al. (1952). Ilotycin, a new antibiotic. Antibiot. Chemother. (Northfield) 2 281–283. [PubMed] [Google Scholar]
- Medema M. H., Fischbach M. A. (2015). Computational approaches to natural product discovery. Nat. Chem. Biol. 11 639–648. 10.1038/nchembio.1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehl M. A., Stadheim T. A. (2014). Biopharmaceutical discovery and production in yeast. Curr. Opin. Biotechnol. 30 120–127. 10.1016/j.copbio.2014.06.007 [DOI] [PubMed] [Google Scholar]
- Metsä-Ketelä M., Niemi J., Mäntsälä P., Schneider G. (2008). “Anthracycline biosynthesis: genes, enzymes and mechanisms,” in Anthracycline Chemistry and Biology I: Biological Occurence and Biosynthesis, Synthesis and Chemistry, ed. Krohn K. (Berlin: Springer; ), 101–140. 10.1007/128_2007_14 [DOI] [Google Scholar]
- Meyer V., Wu B., Ram A. F. J. (2011). Aspergillus as a multi-purpose cell factory: current status and perspectives. Biotechnol. Lett. 33 469–476. 10.1007/s10529-010-0473-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierau I., Kleerebezem M. (2005). 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl. Microbiol. Biotechnol. 68 705–717. 10.1007/s00253-005-0107-6 [DOI] [PubMed] [Google Scholar]
- Migita K., Eguchi K. (2003). FK506: anti-inflammatory properties. Curr. Med. Chem. 2:5. [Google Scholar]
- Miller G. H., Arcieri G., Weinstein M. J., Waitz J. A. (1976). Biological activity of netilmicin, a broad-spectrum semisynthetic aminoglycoside antibiotic. Antimicrob. Agents Chemother. 10 827–836. 10.1128/aac.10.5.827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo S., Ban Y. H., Park J. W., Yoo Y. J., Yoon Y. J. (2009). Enhanced FK506 production in Streptomyces clavuligerus CKD1119 by engineering the supply of methylmalonyl-CoA precursor. J. Ind. Microbiol. Biotechnol. 36 1473–1482. 10.1007/s10295-009-0635-7 [DOI] [PubMed] [Google Scholar]
- Mo S., Yoo Y. J., Ban Y. H., Lee S. K., Kim E., Suh J. W., et al. (2012). Roles of fkbN in positive regulation and tcs7 in negative regulation of FK506 biosynthesis in Streptomyces sp. strain KCTC 11604BP. Appl. Environ. Microbiol. 78 2249–2255. 10.1128/aem.06766-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. (1987). Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature 327 389–394. 10.1038/327389a0 [DOI] [PubMed] [Google Scholar]
- Molnar I., Schupp T., Ono M., Zirkle R., Milnamow M., Nowak-Thompson B., et al. (2000). The biosynthetic gene cluster for the microtubule-stabilizing agents epothilones A and B from Sorangium cellulosum So ce90. Chem. Biol. 7 97–109. 10.1016/s1074-5521(00)00075-2 [DOI] [PubMed] [Google Scholar]
- Morello E., Bermudez-Humaran L. G., Llull D., Sole V., Miraglio N., Langella P., et al. (2008). Lactococcus lactis, an efficient cell factory for recombinant protein production and secretion. J. Mol. Microbiol. Biotechnol. 14 48–58. 10.1159/000106082 [DOI] [PubMed] [Google Scholar]
- Moses T., Pollier J., Almagro L., Buyst D., Van Montagu M., Pedreño M. A., et al. (2014). Combinatorial biosynthesis of sapogenins and saponins in Saccharomyces cerevisiae using a C-16α hydroxylase from Bupleurum falcatum. Proc. Natl. Acad. Sci. U.S.A. 111 1634–1639. 10.1073/pnas.1323369111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller F., Tieke A., Waschk D., Mühle C., Müller F., Seigelchifer M., et al. (2002). Production of IFNα-2a in Hansenula polymorpha. Process Biochem. 38 15–25. 10.1016/s0032-9592(02)00037-7 [DOI] [Google Scholar]
- Müller S., Sandal T., Kamp-Hansen P., Dalbøge H. (1998). Comparison of expression systems in the yeasts Saccharomyces cerevisiae, Hansenula polymorpha, Klyveromyces lactis, Schizosaccharomyces pombe and Yarrowia lipolytica. Cloning of two novel promoters from Yarrowia lipolytica. Yeast 14 1267–1283. [DOI] [PubMed] [Google Scholar]
- Murakami T., Burian J., Yanai K., Bibb M. J., Thompson C. J. (2011). A system for the targeted amplification of bacterial gene clusters multiplies antibiotic yield in Streptomyces coelicolor. Proc. Natl. Acad. Sci. U.S.A. 108 16020–16025. 10.1073/pnas.1108124108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutka S. C., Bondi S. M., Carney J. R., Da Silva N. A., Kealey J. T. (2006). Metabolic pathway engineering for complex polyketide biosynthesis in Saccharomyces cerevisiae. FEMS Yeast Res. 6 40–47. 10.1111/j.1567-1356.2005.00001.x [DOI] [PubMed] [Google Scholar]
- Nakashima N., Mitani Y., Tamura T. (2005). Actinomycetes as host cells for production of recombinant proteins. Microb. Cell Fact. 4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandy S. K., Srivastava R. K. (2018). A review on sustainable yeast biotechnological processes and applications. Microbiol. Res. 207 83–90. 10.1016/j.micres.2017.11.013 [DOI] [PubMed] [Google Scholar]
- Nannemann D. P., Birmingham W. R., Scism R. A., Bachmann B. O. (2011). Assessing directed evolution methods for the generation of biosynthetic enzymes with potential in drug biosynthesis. Future Med. Chem. 3 809–819. 10.4155/fmc.11.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan A. R., Jimenez-Oses G., Liu P., Negretti S., Zhao W., Gilbert M. M., et al. (2015). Enzymatic hydroxylation of an unactivated methylene C-H bond guided by molecular dynamics simulations. Nat. Chem. 7 653–660. 10.1038/nchem.2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natale P., Brüser T., Driessen A. J. M. (2008). Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane—Distinct translocases and mechanisms. Biochim. Biophys. Acta (BBA) – Biomembranes 1778 1735–1756. 10.1016/j.bbamem.2007.07.015 [DOI] [PubMed] [Google Scholar]
- Newman D. J., Cragg G. M. (2016). Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 79 629–661. 10.1021/acs.jnatprod.5b01055 [DOI] [PubMed] [Google Scholar]
- Newton G. G., Abraham E. P. (1955). Cephalosporin C, a new antibiotic containing sulphur and D-alpha-aminoadipic acid. Nature 175:548. 10.1038/175548a0 [DOI] [PubMed] [Google Scholar]
- Nielsen J. (2013). Production of biopharmaceutical proteins by yeast: advances through metabolic engineering. Bioengineered 4 207–211. 10.4161/bioe.22856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijland R., Kuipers O. P. (2008). Optimization of protein secretion by Bacillus subtilis. Recent Pat. Biotechnol. 2 79–87. 10.2174/187220808784619694 [DOI] [PubMed] [Google Scholar]
- Ochi K., Hosaka T. (2013). New strategies for drug discovery: activation of silent or weakly expressed microbial gene clusters. Appl. Microbiol. Biotechnol. 97 87–98. 10.1007/s00253-012-4551-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochi K., Tanaka Y., Tojo S. (2014). Activating the expression of bacterial cryptic genes by rpoB mutations in RNA polymerase or by rare earth elements. J. Ind. Microbiol. Biotechnol. 41 403–414. 10.1007/s10295-013-1349-4 [DOI] [PubMed] [Google Scholar]
- Paddon C. J., Westfall P. J., Pitera D. J., Benjamin K., Fisher K., Mcphee D., et al. (2013). High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496 528–532. [DOI] [PubMed] [Google Scholar]
- Palva I., Lehtovaara P., Kääriäinen L., Sibakov M., Cantell K., Schein C. H., et al. (1983). Secretion of interferon by Bacillus subtilis. Gene 22 229–235. 10.1016/0378-1119(83)90107-5 [DOI] [PubMed] [Google Scholar]
- Pan G., Xu Z., Guo Z., Ma Hindra M., Yang D.et al. (2017). Discovery of the leinamycin family of natural products by mining actinobacterial genomes. Proc. Natl. Acad. Sci. U.S.A. 114 E11131–E11140. 10.1073/pnas.1716245115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T., Kondo S., Zhu W., Xie W., Jankovic J., Le W. (2008). Neuroprotection of rapamycin in lactacystin-induced neurodegeneration via autophagy enhancement. Neurobiol. Dis. 32 16–25. 10.1016/j.nbd.2008.06.003 [DOI] [PubMed] [Google Scholar]
- Papagianni M. (2004). Fungal morphology and metabolite production in submerged mycelial processes. Biotechnol. Adv. 22 189–259. 10.1016/j.biotechadv.2003.09.005 [DOI] [PubMed] [Google Scholar]
- Papagianni M. (2012). Metabolic engineering of lactic acid bacteria for the production of industrially important compounds. Comput. Struct. Biotechnol. J. 3:e201210003. 10.5936/csbj.201210003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papagianni M., Nokes S. E., Filer K. (1999). Production of phytase by Aspergillus niger in submerged and solid-state fermentation. Process Biochem. 35 397–402. 10.1016/s0032-9592(99)00088-6 24688383 [DOI] [Google Scholar]
- Park S. R., Han A. R., Ban Y. H., Yoo Y. J., Kim E. J., Yoon Y. J. (2010). Genetic engineering of macrolide biosynthesis: past advances, current state, and future prospects. Appl. Microbiol. Biotechnol. 85 1227–1239. 10.1007/s00253-009-2326-8 [DOI] [PubMed] [Google Scholar]
- Park S. R., Park J. W., Ban Y. H., Sohng J. K., Yoon Y. J. (2013). 2-Deoxystreptamine-containing aminoglycoside antibiotics: recent advances in the characterization and manipulation of their biosynthetic pathways. Nat. Prod. Rep. 30 11–20. 10.1039/c2np20092a [DOI] [PubMed] [Google Scholar]
- Park S. R., Park J. W., Jung W. S., Han A. R., Ban Y. H., Kim E. J., et al. (2008). Heterologous production of epothilones B and D in Streptomyces venezuelae. Appl. Microbiol. Biotechnol. 81 109–117. 10.1007/s00253-008-1674-0 [DOI] [PubMed] [Google Scholar]
- Park S. R., Tripathi A., Wu J., Schultz P. J., Yim I., Mcquade T. J., et al. (2016). Discovery of cahuitamycins as biofilm inhibitors derived from a convergent biosynthetic pathway. Nat. Commun. 7:10710. 10.1073/pnas.1716245115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. R., Yoon Y. J. (2015). “Antibiotics: current innovations and future trends,” in Combinatorial Biosynthesis for Antiobiotic Discovery, eds Sánchez S., Demain A. L. (Poole: Caister Academic Press; ), 294–318. [Google Scholar]
- Patridge E., Gareiss P., Kinch M. S., Hoyer D. (2016). An analysis of FDA-approved drugs: natural products and their derivatives. Drug Discov. Today 21 204–207. 10.1016/j.drudis.2015.01.009 [DOI] [PubMed] [Google Scholar]
- Pel H. J., De Winde J. H., Archer D. B., Dyer P. S., Hofmann G., Schaap P. J., et al. (2007). Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 25 221–231. [DOI] [PubMed] [Google Scholar]
- Peng F., Wang X., Sun Y., Dong G., Yang Y., Liu X., et al. (2017). Efficient gene editing in Corynebacterium glutamicum using the CRISPR/Cas9 system. Microb. Cell Fact. 16:201. 10.1186/s12934-017-0814-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peypoux F., Pommier M. T., Das B. C., Besson F., Delcambe L., Michel G. (1984). Structures of bacillomycin D and bacillomycin L peptidolipid antibiotics from Bacillus Subtilis. J. Antibiot. 37 1600–1604. 10.7164/antibiotics.37.1600 [DOI] [PubMed] [Google Scholar]
- Pickens L. B., Tang Y., Chooi Y. H. (2011). Metabolic engineering for the production of natural products. Annu. Rev. Chem. Biomol. Eng. 2 211–236. 10.1146/annurev-chembioeng-061010-114209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimienta E., Ayala J. C., Rodríguez C., Ramos A., Van Mellaert L., Vallín C., et al. (2007). Recombinant production of Streptococcus equisimilis streptokinase by Streptomyces lividans. Microb. Cell Fact. 6:20. 10.1186/1475-2859-6-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabin N., Zheng Y., Opoku-Temeng C., Du Y., Bonsu E., Sintim H. O. (2015). Agents that inhibit bacterial biofilm formation. Future Med. Chem. 7 647–671. 10.4155/fmc.15.7 [DOI] [PubMed] [Google Scholar]
- Revers L., Furczon E. (2010). An introduction to biologics and biosimilars. Part II: subsequent entry biologics: biosame or biodifferent? Can. Pharm. J. 143 184–191. 10.3821/1913-701x-143.4.184 17469473 [DOI] [Google Scholar]
- Richter L., Wanka F., Boecker S., Storm D., Kurt T., Vural Öet al. (2014). Engineering of Aspergillus niger for the production of secondary metabolites. Fungal Biol. Biotechnol. 1:4. 10.1186/s40694-014-0004-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbel L., Marahiel M. A. (2010). Daptomycin, a bacterial lipopeptide synthesized by a nonribosomal machinery. J. Biol. Chem. 285 27501–27508. 10.1074/jbc.r110.128181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosano G. L., Ceccarelli E. A. (2014). Recombinant protein expression in Escherichia coli: advances and challenges. Front. Microbiol. 5:172. 10.3389/fmicb.2014.00172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez S., Guzmán-Trampe S., Ávalos M., Ruiz B., Rodríguez-Sanoja R., Jiménez-Estrada M. (2012). “Microbial natural products,” in Natural Products in Chemical Biology, ed. Civjan N. (Hoboken, NJ: Wiley; ),65–108. [Google Scholar]
- Sanchez-Garcia L., Martin L., Mangues R., Ferrer-Miralles N., Vazquez E., Villaverde A. (2016). Recombinant pharmaceuticals from microbial cells: a 2015 update. Microb. Cell Fact. 15:33. 10.1186/s12934-016-0437-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaerlaekens K., Lammertyn E., Geukens N., De Keersmaeker S., Anne J., Van Mellaert L. (2004). Comparison of the Sec and Tat secretion pathways for heterologous protein production by Streptomyces lividans. J. Biotechnol. 112 279–288. 10.1016/j.jbiotec.2004.05.004 [DOI] [PubMed] [Google Scholar]
- Schallmey M., Singh A., Ward O. P. (2004). Developments in the use of Bacillus species for industrial production. Can. J. Microbiol. 50 1–17. 10.1139/w03-076 [DOI] [PubMed] [Google Scholar]
- Schatz A., Bugle E., Waksman S. A. (1944). Streptomycin, a substance exhibiting antibiotic activity against gram-positive and gram-negative bacteria. Proc. Soc. Exp. Biol. Med. 55 66–69. 10.3181/00379727-55-14461 [DOI] [PubMed] [Google Scholar]
- Schlemmer U., Frolich W., Prieto R. M., Grases F. (2009). Phytate in foods and significance for humans: food sources, intake, processing, bioavailability, protective role and analysis. Mol. Nutr. Food Res. 53(Suppl. 2), S330–S375. [DOI] [PubMed] [Google Scholar]
- Schmidt-Dannert C. (2015). NextGen microbial natural products discovery. Microb. Biotechnol. 8 26–28. 10.1111/1751-7915.12184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneewind O., Missiakas D. M. (2012). Protein secretion and surface display in Gram-positive bacteria. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367 1123–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnepf E., Crickmore N., Van Rie J., Lereclus D., Baum J., Feitelson J., et al. (1998). Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. MMBR 62 775–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafiee A., Motamedi H., Dumont F. J., Arison B. H., Miller R. R. (1997). Chemical and biological characterization of two FK506 analogs produced by targeted gene disruption in Streptomyces sp. MA6548. J. Antibiot. (Tokyo) 50 418–423. 10.7164/antibiotics.50.418 [DOI] [PubMed] [Google Scholar]
- Sheffer A. L., Campion M., Levy R. J., Li H. H., Horn P. T., Pullman W. E. (2011). Ecallantide (DX-88) for acute hereditary angioedema attacks: integrated analysis of 2 double-blind, phase 3 studies. J. Allergy Clin. Immunol. 128 153.e4–159.e4. 10.1016/j.jaci.2011.03.006 [DOI] [PubMed] [Google Scholar]
- Shen B. (2015). A new golden age of natural products drug discovery. Cell 163 1297–1300. 10.1016/j.cell.2015.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba Y., Paradise E. M., Kirby J., Ro D. K., Keasling J. D. (2007). Engineering of the pyruvate dehydrogenase bypass in Saccharomyces cerevisiae for high-level production of isoprenoids. Metab. Eng. 9 160–168. 10.1016/j.ymben.2006.10.005 [DOI] [PubMed] [Google Scholar]
- Shin J. C., Na Z., Lee D. H., Kim W. C., Lee K., Shen Y. M., et al. (2008). Characterization of tailoring genes involved in the modification of geldanamycin polyketide in Streptomyces hygroscopicus JCM4427. J. Microbiol. Biotechnol. 18 1101–1108. [PubMed] [Google Scholar]
- Shoemaker S. C., Ando N. (2018). X-rays in the cryo-electron microscopy era: structural biology’s dynamic future. Biochemistry 57 277–285. 10.1021/acs.biochem.7b01031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Show P. L., Oladele K. O., Siew Q. Y., Aziz Zakry F. A., Lan J. C.-W., Ling T. C. (2015). Overview of citric acid production from Aspergillus niger. Front. Life Sci. 8 271–283. 10.1080/21553769.2015.1033653 [DOI] [Google Scholar]
- Siddiqui A. A., Iram F., Siddiqui S., Sahu K. (2014). Role of natural products in drug discovery process. Int. J. Drug Dev. Res. 6 172–204. [Google Scholar]
- Sieber S. A., Böttcher T., Staub I., Orth R. (2010). “9.17 – Small molecules as versatile tools for activity-based protein profiling experiments,” in Comprehensive Natural Products II, eds Liu H.-W., Mander L. (Oxford: Elsevier; ), 629–674. 10.1016/b978-008045382-8.00159-3 [DOI] [Google Scholar]
- Simmons T. L., Coates R. C., Clark B. R., Engene N., Gonzalez D., Esquenazi E., et al. (2008). Biosynthetic origin of natural products isolated from marine microorganism-invertebrate assemblages. Proc. Natl. Acad. Sci. U.S.A. 105 4587–4594. 10.1073/pnas.0709851105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Kumar M., Mittal A., Mehta P. K. (2016). Microbial enzymes: industrial progress in 21st century. 3 Biotech 6 174–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S., Singh S. K., Chowdhury I., Singh R. (2017). Understanding the mechanism of bacterial biofilms resistance to antimicrobial agents. Open Microbiol. J. 11 53–62. 10.2174/1874285801711010053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S. B., Genilloud O., Peláez F. (2010). “2.05 – Terrestrial Microorganisms – Filamentous Bacteria,” in Comprehensive Natural Products II, eds Liu H.-W., Mander L. (Oxford: Elsevier; ), 109–140. 10.1016/b978-008045382-8.00036-8 [DOI] [Google Scholar]
- Singh S. K., Tiendrebeogo R. W., Chourasia B. K., Kana I. H., Singh S., Theisen M. (2018). Lactococcus lactis provides an efficient platform for production of disulfide-rich recombinant proteins from Plasmodium falciparum. Microb. Cell Fact. 17:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirima S. B., Mordmüller B., Milligan P., Ngoa U. A., Kironde F., Atuguba F., et al. (2016). A phase 2b randomized, controlled trial of the efficacy of the GMZ2 malaria vaccine in African children. Vaccine 34 4536–4542. [DOI] [PubMed] [Google Scholar]
- Skiba M. A., Maloney F. P., Dan Q., Fraley A. E., Aldrich C. C., Smith J. L., et al. (2018). PKS-NRPS enzymology and structural biology: considerations in protein production. Methods Enzymol. 604 45–88. 10.1016/bs.mie.2018.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smanski M. J., Peterson R. M., Rajski S. R., Shen B. (2009). Engineered Streptomyces platensis strains that overproduce antibiotics platensimycin and platencin. Antimicrob. Agents Chemother. 53 1299–1304. 10.1128/aac.01358-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneader W. (1997). Drug Prototypes and Their Exploitation Journal of the. Washington, DC: American Chemical Society, 119. [Google Scholar]
- Song A. A., In L. L. A., Lim S. H. E., Rahim R. A. (2017). A review on Lactococcus lactis: from food to factory. Microb. Cell Fact. 16:55. 10.1186/s12934-017-0669-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M. C., Kim E. J., Kim E., Rathwell K., Nam S. J., Yoon Y. J. (2014). Microbial biosynthesis of medicinally important plant secondary metabolites. Nat. Prod. Rep. 31 1497–1509. 10.1039/c4np00057a [DOI] [PubMed] [Google Scholar]
- Song Y., Xue H., Liu T. T., Liu J. M., Chen D. (2015). Rapamycin plays a neuroprotective effect after spinal cord injury via anti-inflammatory effects. J. Biochem. Mol. Toxicol. 29 29–34. 10.1002/jbt.21603 [DOI] [PubMed] [Google Scholar]
- Sørensen H. P., Mortensen K. K. (2005). Advanced genetic strategies for recombinant protein expression in Escherichia coli. J. Biotechnol. 115 113–128. 10.1016/j.jbiotec.2004.08.004 [DOI] [PubMed] [Google Scholar]
- Stanley V. C., English M. P. (1965). Some effects of nystatin on the growth of four Aspergillus species. J. Gen. Microbiol. 40 107–118. 10.1099/00221287-40-1-107 [DOI] [PubMed] [Google Scholar]
- Steidler L., Robinson K., Chamberlain L., Schofield K. M., Remaut E., Le Page R. W. F., et al. (1998). Mucosal delivery of murine interleukin-2 (IL-2) and IL-6 by recombinant strains of Lactococcus lactis coexpressing antigen and cytokine. Infect. Immun. 66 3183–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio S., Kashima A., Mochizuki S., Noda M., Kobayashi K. (1999). Crystal structure of human serum albumin at 2.5 A resolution. Protein Eng. 12 439–446. [DOI] [PubMed] [Google Scholar]
- Sun J., Alper H. S. (2015). Metabolic engineering of strains: from industrial-scale to lab-scale chemical production. J. Ind. Microbiol. Biotechnol. 42 423–436. 10.1007/s10295-014-1539-8 [DOI] [PubMed] [Google Scholar]
- Tae H., Kong E.-B., Park K. (2007). ASMPKS: an analysis system for modular polyketide synthases. BMC Bioinformat. 8:327. 10.1186/1471-2105-8-327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi S., Misawa S., Yoshida Y., Momose H. (1995). Microbial secretion of biologically active human transforming growth factor α fused to the Streptomyces protease inhibitor. Gene 159 239–243. 10.1016/0378-1119(95)00107-h [DOI] [PubMed] [Google Scholar]
- Talarico T. L., Casas I. A., Chung T. C., Dobrogosz W. J. (1988). Production and isolation of reuterin, a growth inhibitor produced by Lactobacillus reuteri. Antimicrob. Agents Chemother. 32 1854–1858. 10.1128/aac.32.12.1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S. Y., Tatsumura Y. (2015). Alexander fleming (1881–1955): discoverer of penicillin. Singapore Med. J. 56 366–367. 10.11622/smedj.2015105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H., Kuroda A., Marusawa H., Hatanaka H., Kino T., Goto T., et al. (1987). Structure of FK506, a novel immunosuppressant isolated from Streptomyces. J. Am. Chem. Soc. 109 5031–5033. 10.1021/ja00250a050 [DOI] [Google Scholar]
- Tanaka Y., Kasahara K., Hirose Y., Murakami K., Kugimiya R., Ochi K. (2013). Activation and products of the cryptic secondary metabolite biosynthetic gene clusters by rifampin resistance (rpob) mutations in actinomycetes. J. Bacteriol. 195 2959–2970. 10.1128/jb.00147-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tareq F. S., Lee H. S., Lee Y. J., Lee J. S., Shin H. J. (2015). Ieodoglucomide C and ieodoglycolipid, new glycolipids from a marine-derived bacterium Bacillus licheniformis 09IDYM23. Lipids 50 513–519. 10.1007/s11745-015-4014-z [DOI] [PubMed] [Google Scholar]
- Tevyashova A. N., Olsufyeva E. N., Solovieva S. E., Printsevskaya S. S., Reznikova M. I., Trenin A. S., et al. (2013). Structure-antifungal activity relationships of polyene antibiotics of the amphotericin B group. Antimicrob. Agents Chemother. 57 3815–3822. 10.1128/aac.00270-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theisen M., Adu B., Mordmuller B., Singh S. (2017). The GMZ2 malaria vaccine: from concept to efficacy in humans. Exp. Rev. Vaccines 16 907–917. 10.1080/14760584.2017.1355246 [DOI] [PubMed] [Google Scholar]
- Thomas C., Moraga I., Levin D., Krutzik P. O., Podoplelova Y., Trejo A., et al. (2011). Structural linkage between ligand discrimination and receptor activation by type I interferons. Cell 146 621–632. 10.1016/j.cell.2011.06.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thykaer J., Nielsen J., Wohlleben W., Weber T., Gutknecht M., Lantz A. E., et al. (2010). Increased glycopeptide production after overexpression of shikimate pathway genes being part of the balhimycin biosynthetic gene cluster. Metab. Eng. 12 455–461. 10.1016/j.ymben.2010.05.001 [DOI] [PubMed] [Google Scholar]
- Trischman J. A., Tapiolas D. M., Jensen P. R., Dwight R., Fenical W., Mckee T. C., et al. (1994). Salinamides A and B: anti-inflammatory depsipeptides from a marine streptomycete. J. Am. Chem. Soc. 116 757–758. 10.1021/ja00081a042 [DOI] [Google Scholar]
- Ubiyvovk V. M., Ananin V. M., Malyshev A. Y., Kang H. A., Sibirny A. A. (2011). Optimization of glutathione production in batch and fed-batch cultures by the wild-type and recombinant strains of the methylotrophic yeast Hansenula polymorpha DL-1. BMC Biotechnol. 11:8. 10.1186/1472-6750-11-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bambeke F., Mingeot-Leclercq M.-P., Glupczynski Y., Tulkens P. M. (2017). “137 – mechanisms of action,” in Infectious Diseases (Fourth Edition), eds Cohen J., Powderly W. G., Opal S. M. (Amsterdam: Elsevier; ), 1162.e1161–1180.e1161. [Google Scholar]
- Van Dijk R., Faber K. N., Kiel J. A., Veenhuis M., Van Der Klei I. (2000). The methylotrophic yeast Hansenula polymorpha: a versatile cell factory. Enzyme Microb. Technol. 26 793–800. 10.1016/s0141-0229(00)00173-3 [DOI] [PubMed] [Google Scholar]
- Van Dijl J. M., Hecker M. (2013). Bacillus subtilis: from soil bacterium to super-secreting cell factory. Microb. Cell Fact. 12:3. 10.1186/1475-2859-12-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dissel D., Claessen D., Roth M., Van Wezel G. P. (2015). A novel locus for mycelial aggregation forms a gateway to improved Streptomyces cell factories. Microb. Cell Fact. 14:44. 10.1186/s12934-015-0224-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez D. (1967). “Macrolide antibiotics — Spiramycin, carbomycin, angolamycin, methymycin and lancamycin,” in Antibiotics: Mechanism of Action Vol. 1 eds Gottlieb D., Shaw P. D. (Berlin: Springer; ), 366–377. 10.1007/978-3-662-38439-8_25 [DOI] [Google Scholar]
- Vezina C., Kudelski A., Sehgal S. N. (1975). Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J. Antibiot. (Tokyo) 28 721–726. 10.7164/antibiotics.28.721 [DOI] [PubMed] [Google Scholar]
- Vitikainen M., Hyyrylainen H. L., Kivimaki A., Kontinen V. P., Sarvas M. (2005). Secretion of heterologous proteins in Bacillus subtilis can be improved by engineering cell components affecting post-translocational protein folding and degradation. J. Appl. Microbiol. 99 363–375. 10.1111/j.1365-2672.2005.02572.x [DOI] [PubMed] [Google Scholar]
- Vrancken K., Anne J. (2009). Secretory production of recombinant proteins by Streptomyces. Future Microbiol. 4 181–188. 10.2217/17460913.4.2.181 [DOI] [PubMed] [Google Scholar]
- Waegeman H., Soetaert W. (2011). Increasing recombinant protein production in Escherichia coli through metabolic and genetic engineering. J. Ind. Microbiol. Biotechnol. 38 1891–1910. 10.1007/s10295-011-1034-4 [DOI] [PubMed] [Google Scholar]
- Waksman S. A., Reilly H. C., Johnstone D. B. (1946). Isolation of streptomycin-producing strains of Streptomyces griseus. J. Bacteriol. 52393–397. [DOI] [PubMed] [Google Scholar]
- Waksman S. A., Woodruff H. B. (1940). Bacteriostatic and bactericidal substances produced by a soil actinomyces. Proc. Soc. Exp. Biol. Med. 45 609–614. 10.3181/00379727-45-11768 [DOI] [Google Scholar]
- Wang J. W., Wang A., Li K., Wang B., Jin S., Reiser M., et al. (2015). CRISPR/Cas9 nuclease cleavage combined with Gibson assembly for seamless cloning. Biotechniques 58 161–170. 10.2144/000114261 [DOI] [PubMed] [Google Scholar]
- Wang T., Liang Y., Wu M., Chen Z., Lin J., Yang L. (2015). Natural products from Bacillus subtilis with antimicrobial properties. Chin. J. Chem. Eng. 23 744–754. 10.1016/j.cjche.2014.05.020 [DOI] [Google Scholar]
- Wang W., Englaender J. A., Xu P., Mehta K. K., Suwan J., Dordick J. S., et al. (2013). Expression of low endotoxin 3-O-sulfotransferase in Bacillus subtilis and Bacillus megaterium. Appl. Biochem. Biotechnol. 171 954–962. 10.1007/s12010-013-0415-8 [DOI] [PubMed] [Google Scholar]
- Ward M., Lin C., Victoria D. C., Fox B. P., Fox J. A., Wong D. L., et al. (2004). Characterization of humanized antibodies secreted by Aspergillus niger. Appl. Environ. Microbiol. 70 2567–2576. 10.1128/aem.70.5.2567-2576.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibel E. K., Hadvary P., Hochuli E., Kupfer E., Lengsfeld H. (1987). Lipstatin, an inhibitor of pancreatic lipase, produced by Streptomyces toxytricini. I. Producing organism, fermentation, isolation and biological activity. J. Antibiot. (Tokyo) 40 1081–1085. 10.7164/antibiotics.40.1081 [DOI] [PubMed] [Google Scholar]
- Weist S., Bister B., Puk O., Bischoff D., Pelzer S., Nicholson G. J., et al. (2002). Fluorobalhimycin–a new chapter in glycopeptide antibiotic research. Angew. Chem. Int. Ed. Engl. 41 3383–3385. [DOI] [PubMed] [Google Scholar]
- Westers L., Dijkstra D. S., Westers H., Van Dijl J. M., Quax W. J. (2006). Secretion of functional human interleukin-3 from Bacillus subtilis. J. Biotechnol. 123 211–224. 10.1016/j.jbiotec.2005.11.007 [DOI] [PubMed] [Google Scholar]
- Westers L., Westers H., Quax W. J. (2004). Bacillus subtilis as cell factory for pharmaceutical proteins: a biotechnological approach to optimize the host organism. Biochim. Biophys. Acta 1694 299–310. 10.1016/j.bbamcr.2004.02.011 [DOI] [PubMed] [Google Scholar]
- Williams G., Koryakina I., Mcarthur J., Draelos M., Randal S., Muddimanl D. (2013). “Reprogramming the biosynthesis of natural products by directed evolution,” in Developments in Biotechnology and Bioprocessing, eds Kantardjieff A., Coffman J. L., Asuri P., Jayapal K. (Washington, DC: American Chemical Society; ), 147–163. 10.1021/bk-2013-1125.ch009 [DOI] [Google Scholar]
- Woo M.-W., Nah H.-J., Choi S.-S., Kim E.-S. (2014). Pikromycin production stimulation through antibiotic down-regulatory gene disruption in Streptomyces venezuelae. Biotechnol. Bioprocess Eng. 19 973–977. 10.1007/s12257-014-0407-8 [DOI] [Google Scholar]
- Wright G. D. (2014). Something old, something new: revisiting natural products in antibiotic drug discovery. Can. J. Microbiol. 60 147–154. 10.1139/cjm-2014-0063 [DOI] [PubMed] [Google Scholar]
- Wu C.-Z., Moon A. N., Jang J.-H., Lee D., Kang S.-Y., Park J.-T., et al. (2011). New non-quinone geldanamycin analogs from genetically engineered Streptomyces hygroscopicus. J. Antibiot. 64 461. 10.1038/ja.2011.24 [DOI] [PubMed] [Google Scholar]
- Xie X., Wong W. W., Tang Y. (2007). Improving simvastatin bioconversion in Escherichia coli by deletion of bioH. Metab. Eng. 9 379–386. 10.1016/j.ymben.2007.05.006 [DOI] [PubMed] [Google Scholar]
- Yang X.-W., Peng K., Liu Z., Zhang G.-Y., Li J., Wang N., et al. (2013). Strepsesquitriol, a rearranged zizaane-type sesquiterpenoid from the deep-sea-derived actinomycete Streptomyces sp. SCSIO 10355. J. Nat. Prod. 76 2360–2363. 10.1021/np400923c [DOI] [PubMed] [Google Scholar]
- Yoo Y. J., Hwang J. Y., Shin H. L., Cui H., Lee J., Yoon Y. J. (2015). Characterization of negative regulatory genes for the biosynthesis of rapamycin in Streptomyces rapamycinicus and its application for improved production. J. Ind. Microbiol. Biotechnol. 42 125–135. 10.1007/s10295-014-1546-9 [DOI] [PubMed] [Google Scholar]
- Yoo Y. J., Kim H., Park S. R., Yoon Y. J. (2017). An overview of rapamycin: from discovery to future perspectives. J. Ind. Microbiol. Biotechnol. 44 537–553. 10.1007/s10295-016-1834-7 [DOI] [PubMed] [Google Scholar]
- Yoon Y. J., Beck B. J., Kim B. S., Kang H. Y., Reynolds K. A., Sherman D. H. (2002). Generation of multiple bioactive macrolides by hybrid modular polyketide synthases in Streptomyces venezuelae. Chem. Biol. 9 203–214. 10.1016/s1074-5521(02)00095-9 [DOI] [PubMed] [Google Scholar]
- Yoshimi A., Yamaguchi S., Fujioka T., Kawai K., Gomi K., Machida M., et al. (2018). Heterologous production of a novel cyclic peptide compound, KK-1, in Aspergillus oryzae. Front. Microbiol. 9:690. 10.3389/fmicb.2018.00690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun J., Lv Y. G., Yao Q., Wang L., Li Y. P., Yi J. (2012). Wortmannin inhibits proliferation and induces apoptosis of MCF-7 breast cancer cells. Eur. J. Gynaecol. Oncol. 33 367–369. [PubMed] [Google Scholar]
- Zabala D., Brana A. F., Florez A. B., Salas J. A., Mendez C. (2013). Engineering precursor metabolite pools for increasing production of antitumor mithramycins in Streptomyces argillaceus. Metab. Eng. 20 187–197. 10.1016/j.ymben.2013.10.002 [DOI] [PubMed] [Google Scholar]
- Zerikly M., Challis G. L. (2009). Strategies for the discovery of new natural products by genome mining. ChemBioChem 10 625–633. 10.1002/cbic.200800389 [DOI] [PubMed] [Google Scholar]
- Zhang B., Yang D., Yan Y., Pan G., Xiang W., Shen B. (2016). Overproduction of lactimidomycin by cross-overexpression of genes encoding Streptomyces antibiotic regulatory proteins. Appl. Microbiol. Biotechnol. 100 2267–2277. 10.1007/s00253-015-7119-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Wang Y., Wu J., Skalina K., Pfeifer B. A. (2010). Complete biosynthesis of erythromycin A and designed analogs using E. coli as a heterologous host. Chem. Biol. 17 1232–1240. 10.1016/j.chembiol.2010.09.013 [DOI] [PubMed] [Google Scholar]
- Zhang M. M., Wang Y., Ang E. L., Zhao H. (2016). Engineering microbial hosts for production of bacterial natural products. Nat. Prod. Rep. 33 963–987. 10.1039/c6np00017g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Wang L., Kong L., Wang T., Chu Y., Deng Z., et al. (2012). Unveiling the post-PKS redox tailoring steps in biosynthesis of the type II polyketide antitumor antibiotic xantholipin. Chem. Biol. 19 422–432. 10.1016/j.chembiol.2012.01.016 [DOI] [PubMed] [Google Scholar]
- Zhang Y. X., Perry K., Vinci V. A., Powell K., Stemmer W. P., Del Cardayre S. B. (2002). Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature 415 644–646. 10.1038/415644a [DOI] [PubMed] [Google Scholar]
- Zhongyue L., Deyu Z., Yuemao S. (2018). Discovery of novel bioactive natural products driven by genome mining. Drug Discov. Therap. 12 318–328. 10.5582/ddt.2018.01066 [DOI] [PubMed] [Google Scholar]
- Zhou T. C., Kim B. G., Zhong J. J. (2014). Enhanced production of validamycin A in Streptomyces hygroscopicus 5008 by engineering validamycin biosynthetic gene cluster. Appl. Microbiol. Biotechnol. 98 7911–7922. 10.1007/s00253-014-5943-9 [DOI] [PubMed] [Google Scholar]
- Zhou Z., Lai J. R., Walsh C. T. (2007). Directed evolution of aryl carrier proteins in the enterobactin synthetase. Proc. Natl. Acad. Sci. U.S.A. 104 11621–11626. 10.1073/pnas.0705122104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zida A., Bamba S., Yacouba A., Ouedraogo-Traore R., Guiguemde R. T. (2017). Anti-Candida albicans natural products, sources of new antifungal drugs: a review. J. Mycol. Med. 27 1–19. 10.1016/j.mycmed.2016.10.002 [DOI] [PubMed] [Google Scholar]
- Zweers J. C., Barák I., Becher D., Driessen A. J., Hecker M., Kontinen V. P., et al. (2008). Towards the development of Bacillus subtilis as a cell factory for membrane proteins and protein complexes. Microb. Cell Fact. 7:10. 10.1186/1475-2859-7-10 [DOI] [PMC free article] [PubMed] [Google Scholar]