Abstract
Comprehensive phosphoproteomic analysis of small populations of cells remains a daunting task due primarily to the insufficient MS signal intensity from low concentrations of enriched phosphopeptides. Isobaric labeling has a unique multiplexing feature where the “total” peptide signal from all channels (or samples) triggers MS/MS fragmentation for peptide identification, while the reporter ions provide quantitative information. In light of this feature, we tested the concept of using a “boosting” sample (e.g., a biological sample mimicking the study samples but available in a much larger quantity) in multiplexed analysis to enable sensitive and comprehensive quantitative phosphoproteomic measurements with <100 000 cells. This simple boosting to amplify signal with isobaric labeling (BASIL) strategy increased the overall number of quantifiable phosphorylation sites more than 4-fold. Good reproducibility in quantification was demonstrated with a median CV of 15.3% and Pearson correlation coefficient of 0.95 from biological replicates. A proof-of-concept experiment demonstrated the ability of BASIL to distinguish acute myeloid leukemia cells based on the phosphoproteome data. Moreover, in a pilot application, this strategy enabled quantitative analysis of over 20 000 phosphorylation sites from human pancreatic islets treated with interleukin-1β and interferon-γ. Together, this signal boosting strategy provides an attractive solution for comprehensive and quantitative phosphoproteome profiling of relatively small populations of cells where traditional phosphoproteomic workflows lack sufficient sensitivity.
Graphical Abstract
Phosphorylation is one of the key post-translational modifications (PTMs) regulating cellular processes, such as cell signaling, gene expression, cell survival, apoptosis, and mitosis.1,2 However, the dynamic and substoichiometric nature of phosphorylation as well as the relatively low ionization efficiency of phosphorylated peptides present significant analytical challenges for mass spectrometry (MS)-based phosphoproteomics. Over the past decade, the recovery and selectivity of the phosphopeptide enrichment have been greatly improved using titanium dioxide (TiO2) or immobilized metal ion affinity chromatography (IMAC) using Fe3+, Ga3+, or Ti4+.3–8 Combining these efficient front-end enrichment methods and state-of-the-art LC-MS instrumentation, current phosphoproteomic workflows allow for identification of ~10 000 phosphorylation sites (phosphosites) in a single LC-MS/MS analysis9,10 and more than 30 000 phosphosites in deep phosphoproteome profiling using 2D-LC-MS/MS approaches.11–13 Nevertheless, comprehensive phosphoproteomics still requires a relatively large amount of starting material (e.g., 300 μg to 1 mg of extracted proteins per sample), which hampers its application with small-sized samples, such as clinical specimens often providing <10 μg of extracted peptides (equivalent to 100 000 or fewer cells).
Insufficient MS signal intensity from the lower abundance peptides (especially phosphopeptides) is the main reason for low proteome coverage in LC-MS/MS analysis of small-sized samples. This is due to the inability to produce MS/MS spectra with sufficient quality for confident peptide identifications. Several data analysis and instrumental acquisition strategies may help alleviate this issue. One strategy is to align m/z and retention time measurements of the unidentified low-abundant peptides to those in a library and/or reference LC MS/MS run14 (match between runs (MBR))15,16 to yield additional peptide identification.15,17 By combining MBR with a new MS acquisition method, termed BoxCar, which boosts the MS1 signal and uses a comprehensive peptide library, more than 10 000 proteins can be identified in a 100 min LC-MS/MS analysis.18 In addition, isobaric labeling has been increasingly used for multiplexed analysis of small-sized samples.19 In isobaric labeling, the combined signal intensity from all multiplexed samples (i.e., differently labeled samples from all channels are combined) triggers MS/MS fragmentation, leading to peptide identification instead of relying on signal intensities of an individual sample. Thus, the MS/MS spectra quality is dependent on the combined peptide signal from all channels. This feature inspired us to test the concept of using a “boosting” sample to enhance the detection of phosphopeptides from small-sized samples. In this boosting to amplify signal with isobaric labeling (BASIL) strategy (Figure 1A), the combined signal intensity of a given peptide is significantly increased (e.g., by 30-fold) by including a large amount of boosting sample (i.e., a related or nonrelated sample with similar phosphoproteome content to the study samples) in one of the isobaric tandem mass tag (TMT) channels. This design enhances the detectability of the MS1 signal and identifiability of MS/MS spectra of the same peptide. Our rationale is that if a peptide can be confidently identified from the MS/MS spectrum (enhanced by the boosting samples), then reporter ion intensities from individual study sample channels will be sufficient for robust quantification. Several advantages in the BASIL strategy should be noted for comprehensive phosphoproteomic analysis of small-sized clinical samples. First, the large contribution from the boosting sample would allow for the use of fractionation in order to mitigate sample loss to achieve comprehensive phosphoproteome coverage. Second, convenient relevant cell-line samples can be used as boosting samples to mimic the phosphorylation events in clinical samples.
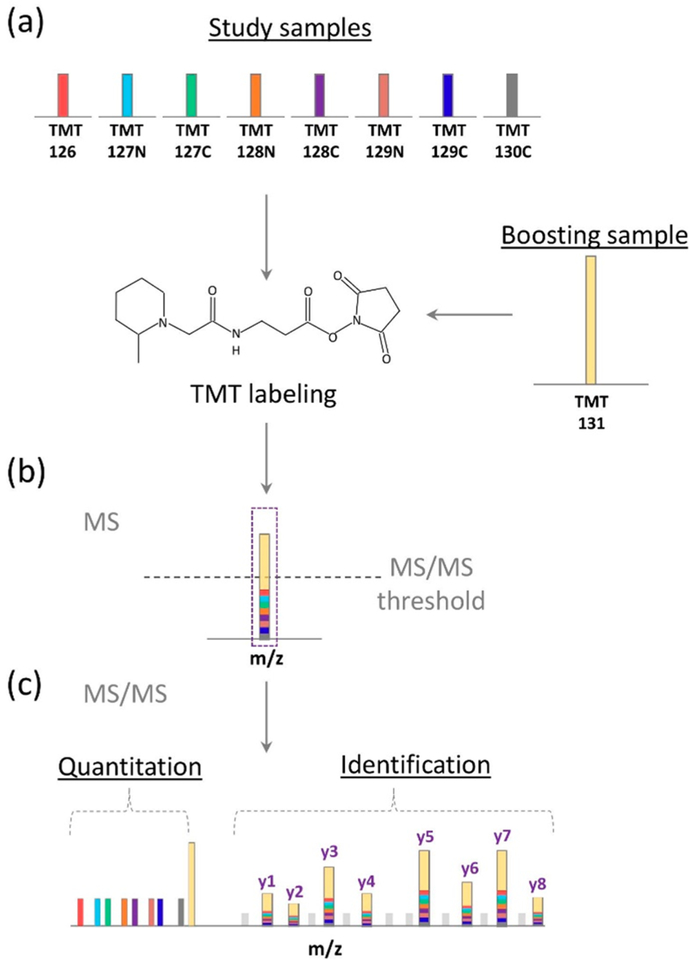
Figure 1.
BASIL quantitative strategy. (a) The small amount of study samples and the much larger amount of boosting sample were individually labeled with different TMT tags. (b) The differentially labeled peptides appear as a single peak (identical m/z at MS1 level), which represents the sum of intensities from both the study and boosting samples. (c) Sequence information can be obtained after MS/MS fragmentation of the peptide backbone and quantification information on the study samples can be obtained from the intensities of the individual TMT reporter ions.
To demonstrate the feasibility of this strategy, we evaluated the potential interference from the boosting channel and the reproducibility of quantification in other study sample channels using a study sample to boosting sample ratio of 1:30. As a proof of concept, BASIL was initially applied to differentiate acute myeloid leukemia (AML) cell lines based on their phosphoproteome profiles. We further demonstrated its application in a pilot study for quantifying phosphorylation changes in human pancreatic islets from eight individual donors treated with interleukin-1β (IL-1β) and interferon-γ (IFN-γ). Our results demonstrated that this strategy not only provided deep phosphoproteome coverage but also precisely recapitulated the dynamic changes in protein phosphorylation in the different types of small-sized samples; suggesting its potential broad applications in biological and biomedical research.
EXPERIMENTAL SECTION
Reagents.
Urea, dithiothreitol (DTT), iodoacetamide, iron chloride, ammonium bicarbonate, trifluoroacetic acid (TFA), ethylenediaminetetraacetic acid (EDTA), acetonitrile (ACN), and formic acid (FA) were obtained from Sigma (St. Louis, MO). The Ni-NTA agarose beads were obtained from Qiagen (Valencia, CA), and Empore C18 extraction disks were obtained from 3 M (St. Paul, MN). The TMT-10 labeling kit was purchased from Thermo Fisher Scientific, Inc. (Waltham, MA).
Cell Culture and Treatment.
The breast cancer cell line MCF-7 was obtained from the American Type Culture Collection and was grown as previously described.20 For the AML cells, MOLM-14 and K562 cells were maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS); CMK cells were maintained in RPMI-1640 medium supplemented with 20% FBS. Human islets from eight nondiabetic cadaveric donors were obtained from the Integrative Islet Distribution Program (IIDP). About 150 islets per condition were cultured in 2 mL of Standard Islet Medium (Prodo) supplemented with human AB serum (Prodo), ciprofloxacin (Fisher), and glutamine and glutathione (Prodo) at 37 °C under 100% humidity and 5% CO2. Islet cultures were allowed to acclimate overnight and then were either treated with cytokines IL-1β and IFN-γ by adding fresh medium containing 50 and 1000 U/mL of IL-1β and IFN-γ, respectively, or left untreated by adding fresh medium without cytokines for 24 h. EndoC-βH2 cells, a conditionally immortalized human β-cell line,21 were cultured in Matrigelfibronectin-coated (100 and 2 μg/mL, respectively)22 1 50 mm culture dishes in Dulbecco’s modified Eagle’s medium. An indicated number of cells were washed with PBS, pelleted, and stored at −80 °C for phosphoproteomic analyses. Further details of cell culture are described in the SI Methods.
Protein Extraction and Digestion.
For protein extraction, cell pellets were resuspended in cell lysis buffer (100 mM NH4HCO3, pH 8.0, 8 M urea, 1% phosphatase inhibitor, pH 8.0) and sonicated in an ice-bath for 3 min. Cell lysates were centrifuged, and the protein concentrations were measured with a Pierce BCA protein assay (Thermo Fisher Scientific). For bulk digestion of MCF-7 and EndoC-βH2 cell samples, proteins were reduced with 5 mM DTT for 1 h at 37 °C and alkylated with 20 mM iodoacetamide in the dark for 1 h at room temperature. The resulting samples were diluted 8-fold with 100 mM NH4HCO3, pH 8.0, and digested by sequencing grade-modified trypsin (Promega Corporation, Madison, WI) with a 1:50 trypsin: protein ratio (w/w) overnight at 37 °C on an Eppendorf Thermomixer. Samples were acidified to ~pH 3 with 10% TFA. The supernatant of each sample was desalted by C18 SPE extraction and concentrated for BCA assay. Then, 10 μg or 300 μg peptides were aliquoted and dried down for further TMT labeling. For AML cell-line samples (MOLM14, K562, and CMK), 10 μg cell-line protein, based on BCA assay, was added to a tube precoated with 10 μg of BSA protein and then digested by trypsin with a 1:10 trypsin:protein ratio. No further BCA assay was performed at the peptide level. For human islet samples, each sample tube was coated with 10 μg of BSA protein prior to protein extraction, and no BCA assay was performed to minimize protein loss. Then, 4 μg of trypsin was used to digest the protein in each islet-containing tube. After digestion, 1/8 of each islet sample was combined in a new tube and then aliquoted equally into 2 tubes for use as a reference channel in two sets of TMT labeling experiments.
TMT Labeling.
The digested peptides were dissolved in 200 mM HEPES (pH = 8.5) and were mixed with TMT-10 reagents which were dissolved in 100% ACN for the 1 h reaction. The labeling reactions were stopped by adding 5% hydroxylamine for 15 min and then acidified with TFA. All of the peptides labeled with different TMT tags were mixed into the same tube, and the concentration of ACN was adjusted to below 5% (v/v), and the samples were desalted by C18 SPE. For highly fractionated samples (a TMT set of MOLM14, K562, and CMK cell-line samples and two TMT sets of human islet samples), 1.3 mg of digested BSA peptides was added as “carrier peptides” to mitigate sample loss in fractionation and IMAC enrichment.
Peptide Fractionation by Basic Reversed-Phase Liquid Chromatography (bRPLC).
The peptides were fractionated using a reversed-phase Waters XBridge C18 column (250 mm × 4.6 mm column packed with 3.5-μm particles) on Aan gilent 1200 HPLC System with details described in the SI Methods.
IMAC Enrichment.
For IMAC, peptides were reconstituted at 1 μg/μL of 80% ACN/0.1%TFA prior to enrichment. The Fe3+-NTA agarose beads were prepared by replacing the Ni2+ ion on the Ni-NTA beads with Fe3+ through buffer exchange. Phosphopeptide enrichment was performed as previously described.20 Details are described in the SI Methods.
LC-MS/MS Analysis.
Lyophilized phosphopeptides were reconstituted in 12 μL of 0.1%FA with 2% ACN, and a 5 μL sample was injected directly into a nanoACQUITY UPLC system (Waters Corp., Milford, MA). Details of LC-MS/MS analysis are described in the SI Methods.
Data Analysis.
The raw MS/MS data were processed with MaxQuant.23,24 The type of LC-MS run was set to “Reporter ion MS2” with “10plex TMT” as the isobaric label. A peptide search was performed with full tryptic digestion and allowed a maximum of two missed cleavages against a human UniProt database (version May 20, 2015). The acetylation (protein N-term), oxidation (M), and phospho (STY) were set as variable modifications, and the carbamidomethyl (C) was set as a fixed modification. The false discovery rate (FDR) was set to 1% at the level of proteins, peptides, and modifications. The intensities of all ten TMT reporter ions were extracted from MaxQuant outputs, evidence files for phosphopeptides, and phospho (STY) site files for phosphorylation sites. For the data processing of human islet samples, the pooled reference sample was labeled with TMT 130N reagent, allowing for comparison of relative phosphopeptide abundances across different TMT-plexes. The relative abundances from 2 TMT-plexes were log2 transformed, and two data matrices from two TMT-plexes were combined after row-and-column centering by median values separately. For the data processing of AML cell lines, the abundances of TMT were log2 transformed and normalized by the median of the column. The normalized TMT signals were further analyzed by Perseus,25 Instant Clue,26 and MeV (http://www.tm4.org/mev.html)27 for statistical analyses. For pathway analysis, the significant phosphorylated proteins upregulated in human islet samples after cytokine treatment were used for pathway enrichment by Reactome.28 The identified phosphorylated peptides and sites are listed in Table S1.
RESULTS
The BASIL strategy (Figure 1) utilizes all study-sample channels containing inherently small amounts of tryptic peptides (e.g., from small populations of cells) used with a much larger amount (e.g., > 30-fold) of peptides in the boosting channel (TMT131) that are labeled separately with different TMT tags and combined in the same TMT set for multiplexed MS/MS analysis (Figure 1a). One of the major advantages of using isobaric tags for relative quantification is that the differentially labeled phosphopeptides from both the study and boosting samples appear as a single precursor ion at MS1 level. This leads to a significant increase in total intensity, greater than the individual sample intensities, for triggering MS/MS as well as enhancing MS/MS spectral quality and aiding phosphopeptide identification (Figure 1b). The relative quantification of phosphopeptides across study-sample channels is readily determined using reporter-ion intensities of the different TMT tags (Figure 1c).
Evaluation of Potential Channel Interference Using the Boosting Strategy.
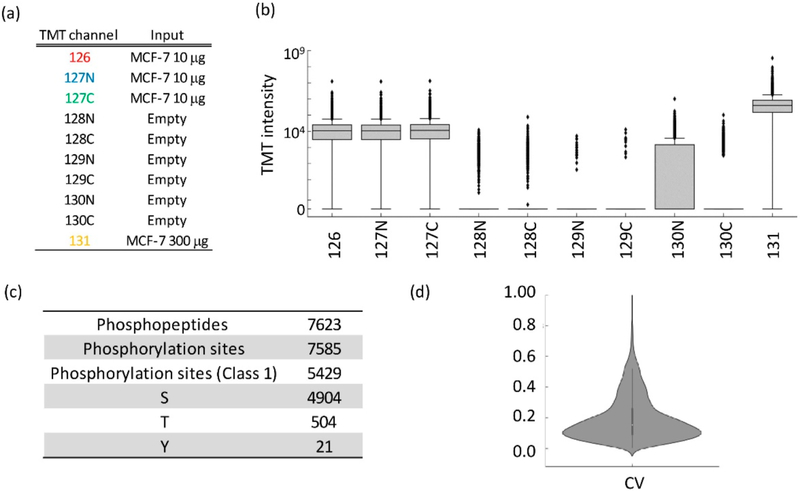
While the boosting strategy significantly increases the detectability of phosphopeptides, the much greater amount of boosting sample may affect the robustness of quantification in the other sample channels by overwhelming them through cross-channel isotopic impurity contamination. To evaluate this, tryptic peptides from MCF-7 cells were labeled with TMT-10-plex reagent, and the ratio of peptide loading in each channel was 1:1:1:0:0:0:0:0:0:30 (Figure 2a). TMT131 was selected as the boosting channel since the reporter ion in this channel has minimal isotopic “leakage” to neighboring channels based on the manufacture-provided reagent information. To mimic the small loading amount of biological samples in targeting channels, 10 μg peptides were labeled in TMT126, TMT127N, and TMT127C channels. The channels from TMT128N to TMT130C remained empty to evaluate the potential interference from the boosting channel. Enriched phosphopeptides from this TMT set were then analyzed using a single LC-MS/MS run. Figure 2b shows that the majority (>90%) of the phosphopeptides had no detectable reporter ion signal intensities in the five empty TMT channels (TMT128N, TMT128C, TMT129N, TMT129C, and TMT130C). Moreover, fewer than 10% of the peptides with reporter-ion intensities had median intensities in these channels significantly greater than 0, suggesting nearly no interference in these channels. However, a number of phosphopeptides had reporter-ion intensities in the TMT130N channel, and their intensities were much higher than the other empty channels. The median signal was 34-fold higher in TMT131 than TMT130N, which is closely matched to the impurity information on the TMT131 reagent from the manufacturer-provided reagent information. We can conclude that the signals in the TMT130N signals were due to “leakage” from the isotopic impurity of the TMT131N reagent used for the boosting channel. As a result, the TMT130N channel was excluded from use as a study-sample channel in subsequent experiments. However, the isotopic impurity is identical for the same lot of regents. Therefore, this channel can still be reserved for use as a reference channel (e.g., for normalizing across different TMT experimental sets when multiple sets are required for comparing a large number of biological samples using the same lot of TMT reagents).
Figure 2.
Evaluation of the performance of the BASIL strategy. (a) TMT-10 channel assignment. (b) Signal distribution of all TMT-10 channels. (c) Summary of identified phosphopeptides and phosphorylation sites (class 1 phosphorylation sites indicate the site localization probability is higher than 0.75). (d) CV for the TMT channels 126, 127N, and 127C.
From this single LC-MS/MS analysis, 7585 phosphorylation sites from 7623 phosphopeptides were identified (Figure 2c); among which 6020 phosphorylation sites were quantified without any missing values in the channels of TMT126, TMT127N, and TMT127C. The quantification of these phosphorylation sites was evaluated. As shown in the scatter plot of the measured TMT signals in the three replicate measurements (Figure S1), the correlation coefficient between any two of the three sample channels (TMT126, TMT127N, and TMT127C) was 0.95 (Figure S1), and the median coefficient of variation (CV) was 15.3% (Figure 2d), indicating excellent reproducibility. Taken together, these results demonstrated that quantification in the different study-sample channels (other than TMT130N) were not impacted by the signals from the boosting channel and achieved high reproducibility.
Assessment of Performance of the Boosting Strategy.
As a proof of concept, BASIL was used to test if it can effectively differentiate three AML cell lines (MOLM14, K562, and CMK) at the phosphoproteome level and at the same time achieve significantly enhanced coverage. The input amount in each channel is shown in Figure S2. Then, 300 μg of tryptic peptides pooled evenly from all three cell-line samples was used as the boosting sample in the TMT131 channel. To mimic the typical experimental setup of multiple TMT sets, TMT130N was used as a reference channel due to the presence of interference from the boosting channel, and 10 μg of the pooled tryptic peptide mixture was labeled for this channel. All the study-sample channels used 10 μg of proteins extracted from each cell-line sample. To further minimize the potential sample loss from handling the very small amount of protein input (10 μg) during sample processing, we applied the “carrier” concept29 by adding 10 μg of bovine serum albumin (BSA) to each sample tube before digestion and SPE cleanup, since the carrier content will be effectively removed during IMAC enrichment. To further increase the coverage of phosphoproteome, bRPLC was used to fractionate the TMT-10-labeled peptide mixture and was subsequently concatenated into 6 fractions before IMAC enrichment and LC-MS/MS analysis. Again, ~1.0 mg of digested BSA peptides was added as a carrier before fractionation to minimize peptide loss during fractionation and downstream IMAC enrichment for each fraction.
With this workflow, we were able to identify a total of 18 868 phosphorylation sites (~80% were class 1 phosphorylation sites,24 as defined by a localization probability of 0.75). This was a more than 4-fold increase compared to that from direct phosphoproteomic analysis of the same cell-line samples without using BASIL and fractionation (Figure S2). Even without fractionation, the use of the boosting sample led to approximately 1.9-fold more phosphopeptides being identified than by the conventional strategy without boosting (Figure S2). Note, that it is impractical to perform fractionation without the boost sample due to the limited sample amounts. However, we did notice that there was a higher percentage of the peptide spectral matches (PSMs) having one or more missing reporter-ion intensity value(s) in the sample channels with BASIL (30.4%), compared to 0.4% in the direct analysis without BASIL (Figure S3); presumably because many more identified low-abundance phosphopeptides did not provide sufficient reporter-ion signal. Nevertheless, there were still a total of 15 999 phosphopeptides that were quantifiable (no missing TMT reporter-ion signals in all study-sample channels) using BASIL. Pearson correlation showed much higher coefficients within the same cell type than that between different cell types (Figure S4). Principal component analysis (PCA) of the BASIL phosphoproteomic data also clearly classified the different AML cell lines (Figure S4). Moreover, we compared the quantification results from the BASIL strategy to those from a standard TMT-based phosphoproteomics workflow. Figure S5a shows the unsupervised clustering of phosphosites with significant differences between two AML lines using the standard workflow (~150 μg peptides for each TMT channel) and the BASIL approach. Replicates of the same AML cell-type were clustered, as expected, using both approaches. More importantly, the fold changes of significantly changing phosphosites using the two different approaches showed good correlation (Figure S5b). Taken together, these results support that comprehensive quantitative phosphoproteomics is readily achievable using BASIL.
Phosphorylation Changes in Human Islets following Cytokine Treatment.
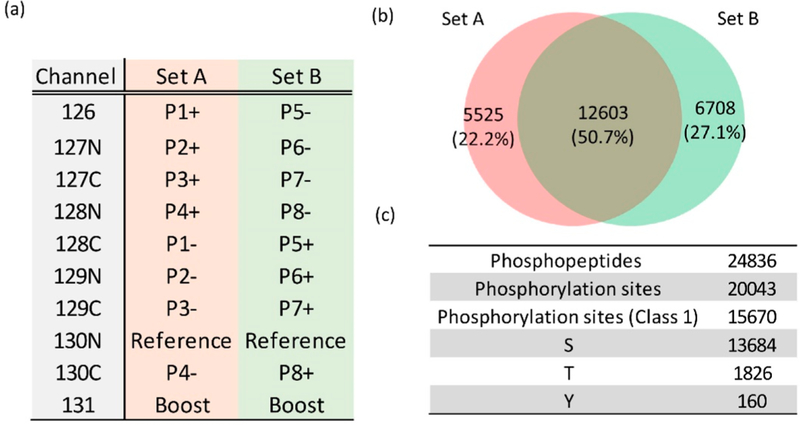
To further demonstrate an initial application using this strategy, we applied it to identify cytokine-induced phosphorylation changes in human pancreatic islets obtained from individual nondiabetic donors. Human islets typically present a significant analytical challenge for phosphoproteomics due to very limited sample/material amounts available per cadaveric donor (~2000 cells per islet and ~150 islets per condition). Moreover, the level of phosphorylated proteins from such cadaveric human islet tissue is typically much lower than that from cell lines. Here, we applied this strategy to compare a set of islet samples obtained from eight donors either with or without treatment of cytokines (IL-1β and IFN-γ) for 24 h to mimic the inflamed conditions found in islets of patients with type-1 diabetes. As shown in Figure 3a, the tryptic peptides from the islets were labeled with TMT tags in the two different TMT-10 sets (set A and set B). A pooled mixture of a small portion from all the islet samples was used in the reference channels (TMT130N), and 300 μg of tryptic peptides prepared from EndoC-βH2 cells were used in the boosting channels (TMT131). In order to achieve deep phosphoproteome coverage of the islets, the TMT-labeled peptides were further fractionated using bRPLC followed by IMAC enrichment for phosphopeptides. A total of 24 836 phosphopeptides mapped to 5162 proteins were identified from the two sets of TMT analyses. The overlap of identified phosphopeptides between TMT-10 set A and B was 50.7% (Figure 3b). About 78% of the 20 043 identified phosphorylation sites were localized with high confidence (class 1), with a median localization probability greater than 0.99. The islet phosphorylation sites (class 1) were composed of 87.3% pSer sites, 11.7% pThr sites, and 1% pTyr sites (Figure 3c). The percentages were similar to those reported from previous studies of the phosphoproteome.30
Figure 3.
Summary of quantitative phosphoproteome analysis of human islet samples. (a) The TMT-10 channel assignment for experimental sets A and B (Pn indicates the different patient; + and — signs indicate the human islets with and without cytokine treatment, respectively; Reference is the mix of all human islet samples; Boost is the EndoC-βH2 cells). (b) The overlap of identified phosphopeptides between TMT set A and B. (c) The summary of identified phosphopeptides and phosphorylation sites.
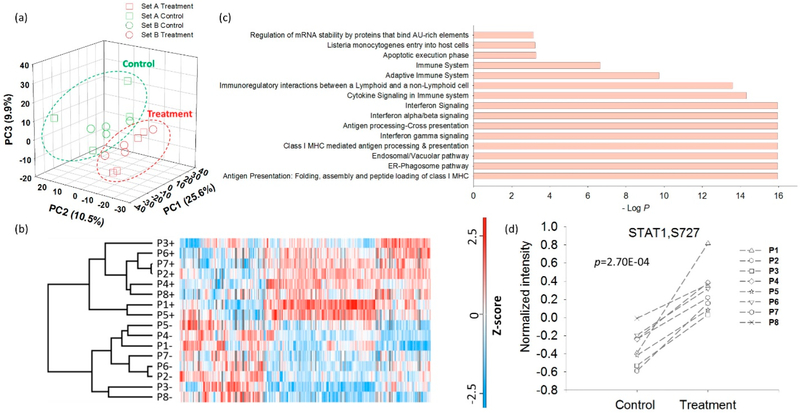
To identify phosphorylation changes in response to cytokines, the TMT-intensity data from each human islet sample channel were normalized to a reference sample in each TMT-10 set to facilitate comparison across the entire cohort. Despite the anticipated donor-to-donor variability, PCA analysis of the abundance data of all quantifiable phosphorylation sites clearly classified these samples into two groups; those with and those without cytokine treatment (Figure 4a), regardless of the TMT set in which the sample was analyzed. The overall results demonstrate the robust quantification that is achievable with BASIL even when multiple experimental TMT sets are required. However, unlike the relatively good reproducibility observed between the biological replicates of AML cell lines (Figure S4), a higher degree of donor-to-donor variability was observed, as shown in Figure 4b. To identify significant changes in phosphorylation, we applied a paired t test analysis with a relatively relaxed FDR control (<0.2) to partially address the large donor-to-donor variability observed. Overall, 429 sites displayed increased phosphorylation and 213 sites with decreased phosphorylation in response to cytokine treatment. Figure 4c shows the top 15 enriched pathways for the proteins with increased phosphorylation. Among them, several immune or inflammation-related pathways, such as antigen presentation and interferon signaling, were identified. All 8 donors demonstrated increased phosphorylation of S727 of STAT1 (Figure 4d) (a site known to be phosphorylated downstream after stimulation by interleukins and interferons), an interesting example that illustrates the data quality and the potential significance of the data set. Activation of STAT1 through cytokine induction has been previously reported as being related to the induction of apoptosis and diabetes progression in murine models of T1D;31,32 STAT1 has also been discovered as a main regulator of pancreatic β-cell apoptosis and islet inflammation.33
Figure 4.
Quantitative phosphoproteome analysis of human islet samples. (a) PCA readily separates human islet samples by cytokine treatment but not by the respective TMT set. (b) Unsupervised hierarchical cluster analysis of phosphoproteome of the human islet samples. (c) The enriched Reactome signaling pathways (top 15) for up-regulated phosphorylation sites after cytokine treatment. (d) Distribution of the abundance of phosphorylation sites of STAT1 (S727) in human islet samples before and after cytokine treatment.
DISCUSSION
It is appealing to take advantage of the multiplexing nature of isobaric labeling approaches (e.g., TMT) to achieve higher sensitivity and proteome coverage with limited individual sample amounts. An advantageous utilization of this concept is to use samples, with similar proteome contents to the other study samples, in a much larger quantity as a “boosting” of the sample in one of the TMT-10 channels; so that the proteins contributing to the other lower-input study samples will have a better chance of being identified and thus quantified based on reporter-ion intensities. For example, Budnik et al. used a carrier TMT channel with much higher protein input than the single-cell samples in the other channels to increase the number of detectable proteins from the single-cell samples.19 Tan et al. proposed a similar concept which used a combination of fractionation and a carrier/reference method in order to detect single-amino-acid-variant peptides in 9 PANC-1 cells.34 Russell et al. reported a TMT workflow which analyzed the tissue or cell-line-derived peptides and the bodyfluid (cerebrospinal fluid, CSF)-derived peptides in different TMT channels to increase the chance of identifying low-abundance proteins that were otherwise hard to detect from the body fluid alone due to the masking effects of high-abundance proteins in the body fluids.35 Several studies similar to these can also be carried out using highly effective “nanoscale” sample handling techniques, such as the recently developed nanodroplet processing in one pot for trace samples (nanoPOTS) platform; where ~3000 proteins can be identified from as few as 10 cells.17 However, such nanoscale sample handling techniques are largely incompatible with PTM analyses due to the additional enrichment steps that are required (e.g., IMAC). In this study, we applied the much-needed concept of BASIL for the first time to analyze the comprehensive quantitative phosphoproteome of small populations of cells and demonstrated its effectiveness and robustness in quantitative analysis of cytokine-induced phosphorylation changes in human islet samples from individual donors.
An important basis of the boosting concept is that there are zero interferences for all the sample channels from the boosting sample, except one channel (130N in this case), as defined by the isotopic purity of TMT reagents. Compared to a typical low-input TMT analysis (10 μg of protein or 100 000 cells per sample) without using the boosting sample, our BASIL strategy provides an effective protection against sample loss during processing, thereby allowing for sample fractionation. This demonstrated a >4-fold increase in phosphoproteome coverage while achieving a CV of ~15% in quantitative analysis. However, the boosting channel can only increase the MS1 signal so as to improve peptide identification but does not boost the reporter-ion signals. For some low-abundance phosphopeptides, the reporter-ion intensities in some study-sample channels may be below the detection limit (this results in missing data), even though the peptide is confidently identified from the boosting channel signals. Another caveat of BASIL analysis using TMT labeling is that the phosphopeptide identifications from TMT-labeled samples are typically lower than comparative label-free phosphopeptide samples generated from the same amount of starting material.13,36 This issue may be caused by a change in peptide fragmentation properties due to the TMT tag(s). The potential ratio compression for TMT labeling due to multiple coeluting peptides within the MS1 isolation window can still be an issue; however, the BASIL strategy is fully compatible with approaches such as the synchronous precursor selection (SPS)-based MS3 analysis37 to minimize such an effect. Nevertheless, the BASIL strategy enables broader quantification of phosphopeptides compared to typical TMT analysis for such small-sized samples. This represents a much-needed advancement in phosphoproteomics for deep phosphoproteome profiling of small-starting-amount protein samples, such as those found in clinical specimens.
There are also several areas where the current workflow could be further optimized. First, given the large difference in signal intensities between the low-input sample channels and the empty channels, we anticipate further that lowering the input down to 1 μg of peptide per study sample is possible. Second, the ratio between the study samples and boosting sample may be further optimized to achieve the best coverage and, at the same time, not sacrifice significantly on the quality of quantification. Finally, although the sample loss after the TMT labeling procedure can be minimized by the presence of a large amount of sample from the boosting sample, the experimental procedures before TMT labeling, such as cell lysis and enzymatic digestion, still need to be carried out with the least possible sample loss. In this study, we used BSA as a carrier protein for that purpose, but there is still room for optimization for small-scale sample handling.
As an application area, the in-depth phosphoproteome profiling of either human or murine pancreatic islets has been largely hindered by the limited protein amounts available from primary islet samples. For example, Li et al. used the optimized in-tip and best-ratio phosphopeptides enrichment strategy and the stable isotope labeling with amino acids in cell culture (SILAC)-based workflow for processing rat islet samples and identified 8539 phosphorylation sites mapping to 2487 proteins from each rat, which provided 20–47 μg islet proteins from each rat.38 In this study, we used cultured EndoC-βH2 cells as the boosting sample to significantly improve the detection sensitivity of phosphopeptides in human islets. Taking advantage of the boosting channel, we were able to achieve a comprehensive coverage of 24 836 phosphopeptides mapped to 5162 proteins from the human islet samples. Known pathways such as interferon signaling and cytokine signaling were enriched after cytokine treatment. Interestingly, we also found that the C-terminal phosphorylation sites of the HLA family, such as S357 and S360 of HLA-C, S353 of HLA-E, and S352 of HLA-A (Figure S6), were up-regulated.
The data obtained in this study not only illustrate the effectiveness of the BASIL strategy for comprehensive quantitative phosphoproteome analysis, they also provided valuable information on the phosphorylation sites that may be useful to discover the aberrant pathways and networks to better understand islet inflammation.
CONCLUSIONS
The BASIL strategy provides a simple yet highly effective quantitative phosphoproteomic workflow suitable for multiplexed analysis of relatively small biological or clinical samples, including cells or tissues. The enabling aspect of this strategy is demonstrated well by the comprehensive quantitative profiling of the human pancreatic islet phosphoproteome obtained from individual donors, a sample type particularly challenging for phosphoproteomic analysis. While the current tested sample input level is about 10 μg of proteins or ~100 000 cells, the sample input level may be reduced further, and the ratio between the study samples and boost sample may be optimized. Overall, we anticipate this strategy should enable broad biomedical applications involving limited amounts of starting cells or tissues and should also be readily adopted for studies of different PTMs.
Supplementary Material
ACKNOWLEDGMENTS
Portions of this work were supported by NIH Grants UC4 DK104167, DP3 DK110844, and UC4 DK104197 from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK); P01 AI42288 from the National Institute of Allergy and Infectious Diseases (NIAID); U24CA210955 and U01 CA214116 from the National Cancer Institute (NCI) Clinical Proteomic Tumor Analysis Consortium (CPTAC); and P41GM103493 from the National Institutes of General Medical Sciences (NIGMS). The experimental work was performed in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the DOE and located at Pacific Northwest National Laboratory, which is operated by the Battelle Memorial Institute for the DOE under Contract DE-AC05–76RL0 1830.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.anal-chem.9b00024
Supplementary methods, figures, and tables (PDF) Identified phosphopeptides and phosphorylation sites and TMT intesities (XLSX)
Accession Codes
Raw data sets have been deposited in the Japan ProteOme STandard Repository39 (https://repository.jpostdb.org/). The accession numbers are PXD011857 for ProteomeXchange40 and JPST000522 for jPOST.
The authors declare no competing financial interest.
REFERENCES
- (1).Hunter T Cell 2000, 100 (1), 113–27. [DOI] [PubMed] [Google Scholar]
- (2).Cohen P Nat. Cell Biol 2002, 4 (5), E127–30. [DOI] [PubMed] [Google Scholar]
- (3).Tsai CF; Wang YT; Chen YR; Lai CY; Lin PY; Pan KT; Chen JY; Khoo KH; Chen YJ J. Proteome Res 2008, 7 (9), 4058–69. [DOI] [PubMed] [Google Scholar]
- (4).Larsen MR; Thingholm TE; Jensen ON; Roepstorff P; Jorgensen TJ Mol. Cell. Proteomics 2005, 4 (7), 873–86. [DOI] [PubMed] [Google Scholar]
- (5).Ruprecht B; Koch H; Medard G; Mundt M; Kuster B; Lemeer S Mol. Cell. Proteomics 2015, 14 (1), 205–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Sugiyama N; Masuda T; Shinoda K; Nakamura A; Tomita M; Ishihama Y Mol. Cell. Proteomics 2007, 6 (6), 1103–9. [DOI] [PubMed] [Google Scholar]
- (7).Tsai CF; Hsu CC; Hung JN; Wang YT; Choong WK; Zeng MY; Lin PY; Hong RW; Sung TY; Chen Y J.Anal. Chem 2014, 86 (1), 685–93. [DOI] [PubMed] [Google Scholar]
- (8).Zhou H; Low TY; Hennrich ML; van der Toorn H; Schwend T; Zou H; Mohammed S; Heck A J.Mol. Cell. Proteomics 2011, 10 (10), M110006452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Potel CM; Lin MH; Heck AJR; Lemeer S Mol. Cell. Proteomics 2018, 17 (5), 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Humphrey SJ; Azimifar SB; Mann M Nat. Biotechnol 2015, 33 (9), 990–5. [DOI] [PubMed] [Google Scholar]
- (11).Bekker-Jensen DB; Kelstrup CD; Batth TS; Larsen SC; Haldrup C; Bramsen JB; Sorensen KD; Hoyer S; Orntoft TF; Andersen CL; Nielsen ML; Olsen JV Cell Syst 2017, 4 (6), 587–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Mertins P; Tang LC; Krug K; Clark DJ; Gritsenko MA; Chen L; Clauser KR; Clauss TR; Shah P; Gillette MA; Petyuk VA; Thomas SN; Mani DR; Mundt F; Moore RJ; Hu Y; Zhao R; Schnaubelt M; Keshishian H; Monroe ME; Zhang Z; Udeshi ND; Mani D; Davies SR; Townsend RR; Chan DW; Smith RD; Zhang H; Liu T; Carr SA Nat. Protoc 2018, 13 (7), 1632–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Hogrebe A; von Stechow L; Bekker-Jensen DB; Weinert BT; Kelstrup CD; Olsen JV Nat. Commun 2018, 9 (1), 1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Tsou CC; Tsai CF; Tsui YH; Sudhir PR; Wang YT; Chen YJ; Chen JY; Sung TY; Hsu WL Mol. Cell. Proteomics 2010, 9 (1), 131–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Geyer PE; Kulak NA; Pichler G; Holdt LM; Teupser D; Mann M Cell Syst 2016, 2 (3), 185–95. [DOI] [PubMed] [Google Scholar]
- (16).Cox J; Hein MY; Luber CA; Paron I; Nagaraj N; Mann M Mol. Cell. Proteomics 2014, 13 (9), 2513–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Zhu Y; Piehowski PD; Zhao R; Chen J; Shen Y; Moore RJ; Shukla AK; Petyuk VA; Campbell-Thompson M; Mathews CE; Smith RD; Qian WJ; Kelly RT Nat. Commun 2018, 9 (1), 882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Meier F; Geyer PE; Virreira Winter S.; Cox J; Mann M Nat. Methods 2018, 15 (6), 440–448. [DOI] [PubMed] [Google Scholar]
- (19).Budnik B; Levy E; Harmange G; Slavov N Genome Biol 2018, 19 (1), 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Yi L; Shi T; Gritsenko MA; X’Avia Chan CY; Fillmore TL; Hess BM; Swensen AC; Liu T; Smith RD; Wiley HS; Qian W J. Anal. Chem 2018, 90 (8), 5256–5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Scharfmann R; Pechberty S; Hazhouz Y; von Bulow M; Bricout-Neveu E; Grenier-Godard M; Guez F; Rachdi L; Lohmann M; Czernichow P; Ravassard P J. Clin. Invest 2014, 124 (5), 2087–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Ravassard P; Hazhouz Y; Pechberty S; Bricout-Neveu E; Armanet M; Czernichow P; Scharfmann R J.Clin. Invest 2011, 121 (9), 3589–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Cox J; Mann M Nat. Biotechnol 2008, 26 (12), 1367–72. [DOI] [PubMed] [Google Scholar]
- (24).Tyanova S; Temu T; Cox J Nat. Protoc 2016, 11 (12), 2301–2319. [DOI] [PubMed] [Google Scholar]
- (25).Tyanova S; Temu T; Sinitcyn P; Carlson A; Hein MY; Geiger T; Mann M; Cox J Nat. Methods 2016, 13 (9), 731–40. [DOI] [PubMed] [Google Scholar]
- (26).Nolte H; MacVicar TD; Tellkamp F; Kruger M Sci. Rep 2018, 8 (1), 12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Wang YE; Kutnetsov L; Partensky A; Farid J; Quackenbush J Cancer Res 2017, 77 (21), e11–e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Croft D; Mundo AF; Haw R; Milacic M; Weiser J; Wu G; Caudy M; Garapati P; Gillespie M; Kamdar MR; Jassal B; Jupe S; Matthews L; May B; Palatnik S; Rothfels K; Shamovsky V; Song H; Williams M; Birney E; Hermjakob H; Stein L; D’Eustachio P Nucleic Acids Res 2014, 42, D472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Shi T; Gaffrey MJ; Fillmore TL; Nicora CD; Yi L; Zhang P; Shukla AK; Wiley HS; Rodland KD; Liu T; Smith RD; Qian W J. Commun. Biol 2018, 1, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Olsen JV; Blagoev B; Gnad F; Macek B; Kumar C; Mortensen P; Mann M Cell 2006, 127 (3), 635–48. [DOI] [PubMed] [Google Scholar]
- (31).Callewaert HI; Gysemans CA; Ladriere L; D’Hertog W; Hagenbrock J; Overbergh L; Eizirik DL; Mathieu C Diabetes 2007, 56 (8), 2169–73. [DOI] [PubMed] [Google Scholar]
- (32).Gysemans CA; Ladriere L; Callewaert H; Rasschaert J; Flamez D; Levy DE; Matthys P; Eizirik DL; Mathieu C Diabetes 2005, 54 (8), 2396–403. [DOI] [PubMed] [Google Scholar]
- (33).Moore F; Naamane N; Colli ML; Bouckenooghe T; Ortis F; Gurzov EN; Igoillo-Esteve M; Mathieu C; Bontempi G; Thykjaer T; Orntoft TF; Eizirik DL J. Biol. Chem 2011, 286 (2), 929–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Tan Z; Yi X; Carruthers NJ; Stemmer PM; Lubman DM J. Proteome Res 2018, 18 (1), 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Russell CL; Heslegrave A; Mitra V; Zetterberg H; Pocock JM; Ward MA; Pike I Rapid Commun. Mass Spectrom. 2017, 31(2), 153–159. [DOI] [PubMed] [Google Scholar]
- (36).Paulo JA; Navarrete-Perea J; Erickson AR; Knott J; Gygi SP J. Am. Soc. Mass Spectrom. 2018, 29 (7), 1505–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).McAlister GC; Nusinow DP; Jedrychowski MP; Wuhr M; Huttlin EL; Erickson BK; Rad R; Haas W; Gygi SP Anal. Chem 2014, 86 (14), 7150–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Li J; Li Q; Tang J; Xia F; Wu J; Zeng R J. Proteome Res 2015, 14 (11), 4635–46. [DOI] [PubMed] [Google Scholar]
- (39).Okuda S; Watanabe Y; Moriya Y; Kawano S; Yamamoto T; Matsumoto M; Takami T; Kobayashi D; Araki N; Yoshizawa AC; Tabata T; Sugiyama N; Goto S; Ishihama Y Nucleic Acids Res 2017, 45 (D1), D1107–D1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Vizcaino JA; Deutsch EW; Wang R; Csordas A; Reisinger F; Rios D; Dianes JA; Sun Z; Farrah T; Bandeira N; Binz PA; Xenarios I; Eisenacher M; Mayer G; Gatto L; Campos A; Chalkley RJ; Kraus HJ; Albar JP; Martinez-Bartolome S; Apweiler R; Omenn GS; Martens L; Jones AR; Hermjakob H Nat. Biotechnol 2014, 32 (3), 223–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.