Abstract
Purpose:
The four kallikrein panel, commercially available as the 4kScore, is a statistical model that has been shown to predict accurately both Gleason grade group ≥2 (high-grade) cancer on biopsy and long-term risk of distant prostate cancer metastases. The panel includes two novel markers, intact prostate specific antigen (PSA) and human kallikrein-related peptidase 2 (hK2). It has been questionned whether these two additional markers add discrimination to the clinical predictors (age, digital rectal exam, prior biopsy) and established molecular markers (total and free PSA).
Materials and methods:
We conducted an individual patient data meta-analysis of published studies where the four kallikrein panel was measured in men undergoing prostate biopsy. We assess the improvement in discrimination associated with including intact PSA and hK2 along with total and free PSA in the statistical model.
Results:
We included 14,580 men from 10 different studies. The fixed-effects meta-analytic estimate for the discrimination of the model without intact PSA and hK2 was 0.742 (95% CI 0.727, 0.756) compared to 0.813 (95% CI 0.801, 0.825) for the full kallikrein model. The 95% C.I. do not overlap and the difference in discrimination is highly statistically significant (0.069; 95% CI 0.057, 0.080; p<0.0001). Both intact PSA (increase in discrimination 0.059; 95% CI 0.050, 0.069; p<0.0001) and hK2 (increase in discrimination 0.024; 95% CI 0.020, 0.029; p<0.0001) added independently to the model.
Conclusions:
The clinical value of the panel cannot be replicated using data readily available to urologists without measuring intact PSA and hK2.
Introduction
It is widely recognized that prostate specific antigen (PSA) has a low positive predictive value for clinicaly significant prostate cancer. A number of markers are now available to aid prostate cancer decision-making for the man with a moderately elevated PSA.1 The four kallikrein panel, commercially available as the 4Kscore, is a statistical model incorporating age, total PSA, free PSA, intact PSA, human kallikrein-related peptidase 2 (hK2), digital rectal exam (DRE) and prior biopsy. The panel has been shown to accurately predict both the risk of Gleason grade group ≥2 (high-grade) cancer on prostate biopsy and long-term risk of prostate cancer metastasis2. Two markers distinguish the kallikrein panel from other models, intact PSA and hK2. It is therefore obvious to ask whether these two additional markers add discrimination to the clinical predictors (age, DRE, prior biopsy) and established molecular markers (total and free PSA).
This question has been directly addressed in some studies. For instance, in the prospective US validation study of the 4Kscore3, including 1012 men undergoing biopsy at one of 26 urology clinics, the area-under-the-curve (AUC) of 0.821 was reduced to 0.751 if intact PSA and hK2 were excluded from the model. However, analysis of the marginal benefit of the novel markers has not been reported in all cohorts and results have not been combined across studies. We conducted an individual patient data meta-analysis to assess the improvement in discrimination by adding intact PSA and hK2 to a pre-specified model including age, total PSA, and free PSA.
Materials and Methods
Data were received from the following biopsy cohorts as for a prior individual patient data meta-analysis4: ProtecT, Rotterdam ERSPC, Rotterdam Repeat Biopsy, UPCA, Tarn ERSPC, Göteborg ERSPC, OPKO. One additional study on a Veterans Affairs (VA) cohort, published after the prior meta-analysis, was also included.5 The IMPACT study was not included as there were only 7 high grade cancers in this cohort.6 The authors of one study7 did not give us permission to use their data in this meta-analysis.
Three previously published models were used to generate predictions of the risk of high grade disease (Gleason grade group ≥ 2, Gleason score ≥7) based on the kallikrein panel. The first model, the Goteborg model8, was replaced by the Rotterdam model9 when changes were made to the kallikrein assays. To reflect changes in both biopsy practice (use of extended biopsy schemes) and grading (some Gleason 3 pattern cancers being redefined as pattern 4), the ProtecT model10 was developed for contemproary cohorts. Models were applied to each cohort as per the original publication: the Goteborg model8 was applied to the Goteborg ERSPC cohorts, the Rotterdam model9 was applied to the Rotterdam and Tarn ERSPC cohorts, and the ProtecT model10 was applied to the contemporary cohorts (ProtecT, UPCA, Opko and VA). The AUC of the four kallikrein model was compared to a model of age, total PSA and free PSA but setting the value of all hK2 and intact PSA levels to zero. As a sensitivity analysis, the AUC of the kallikrein models was compared to the performance of comparator models built on the same cohorts but which excluded intact PSA, hK2 or both markers. All models also included DRE result if this information was available.
Predictions from the 4Kscore are based only on total PSA when PSA ≥ 25 ng/ml, therefore, patients who had PSA levels higher than 25 ng/ml were excluded from all analyses. With the exception of the Rotterdam Repeat Biopsy cohort, where all patients had a prior negative biopsy, patients with a history of prior negative biopsy were excluded.
To obtain a standard error for the improvement in the AUC when including the additional markers, the DeLong, DeLong, Clarke-Pearson method was used11. For the Goteborg round 1, Rotterdam round 1 and ProtecT cohorts, where models where built and assessed on the same cohort, the Delong method is invalid for the difference in nested models, and we used bootstrap resampling to estimate the standard error around the difference in AUC. We then meta-analyzed the difference in AUC and the standard error of this difference to test whether the addition of intact PSA and hK2 significantly improved discrimination. As a sensitivity analysis, we repeated these analyses including only the four contemporary biopsy cohorts (Opko, ProtecT, UPCA and VA). All analyses were performed using Stata 13 (StataCorp, College Station, TX).
Results
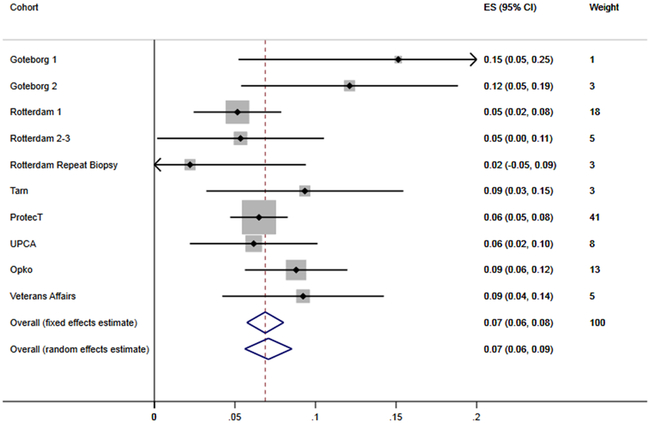
A total of 14,510 men from 10 cohorts were included in this individual patient data meta-analysis. Patient and disease characteristics are reported in Table 1. Results from individual studies and meta-analytic estimates are shown in table 2 and figure 1. Note that the results reported may not match that reported in the original publications due to the exclusion of some patients with prior negative biopsy and those with total PSA >25 ng/ml. The fixed-effects meta-analytic estimate of the AUC for the model without intact PSA and hk2 was 0.742 (95% CI 0.727, 0.756) compared to 0.813 (95% CI 0.801, 0.825) for the full kallikrein model. The 95% C.I. do not overlap and the difference in discrimination is highly statistically significant: 0.069 (95% CI 0.057, 0.080, p<0.0001).
Table 1.
Disease characteristics, N=14,510. Data are presented as median (quartiles) or frequency (%).
| Age | 64 (60, 68) |
| Total PSA measurement | 4.5 (3.5, 6.3) |
| DRE result (N=9,073) | |
| Normal | 7112 (78%) |
| Abnormal | 1961 (22%) |
| Biopsy result | |
| Negative | 9965 (69%) |
| Low Grade (Gleason grade group 1) | 3063 (21%) |
| High Grade (Gleason grade group ≥2) | 1482 (10%) |
| Clinical T stage (N=6,818) | |
| T1 | 4686 (69%) |
| T2 | 1779 (26%) |
| T3 | 346 (5.1%) |
| T4 | 7 (0.1%) |
Table 2.
Meta-analytic results comparing a model of total PSA, free PSA, age and DRE (available) with and without intact PSA and hK2.
| Model without intact PSA and hK2 |
Model including intact PSA and hK2 |
Difference | |
|---|---|---|---|
| Cohort | AUC (95% CI) | AUC (95% CI) | Increase in AUC (95% CI) |
| Goteborg 1st Round | 0.749 (0.636, 0.861) | 0.900 (0.837, 0.964) | 0.151 (0.052, 0.251) |
| Goteborg 2nd Round | 0.675 (0.590, 0.760) | 0.796 (0.730, 0.862) | 0.121 (0.054, 0.188) |
| Rotterdam 1st Round | 0.807 (0.772, 0.842) | 0.859 (0.828, 0.889) | 0.052 (0.024, 0.079) |
| Rotterdam 2-3rd Round | 0.744 (0.682, 0.805) | 0.797 (0.748, 0.847) | 0.053 (0.002, 0.105) |
| Rotterdam Repeat Biopsy | 0.861 (0.759, 0.963) | 0.883 (0.803, 0.963) | 0.022 (−0.050, 0.094) |
| Tarn | 0.763 (0.683, 0.844) | 0.857 (0.798, 0.916) | 0.093 (0.032, 0.155) |
| ProtecT | 0.734 (0.713, 0.756) | 0.799 (0.781, 0.818) | 0.065 (0.047, 0.083) |
| UPCA | 0.703 (0.652, 0.755) | 0.765 (0.718, 0.812) | 0.062 (0.022, 0.101) |
| Opko | 0.717 (0.675, 0.760) | 0.805 (0.771, 0.840) | 0.088 (0.056, 0.120) |
| Veterans Affairs | 0.659 (0.591, 0.726) | 0.751 (0.693, 0.810) | 0.092 (0.042, 0.142) |
| Overall (fixed effects estimate) | 0.742 (0.727, 0.756) | 0.813 (0.801, 0.825) | 0.069 (0.057, 0.080) |
| Overall (random effects estimate) | 0.739 (0.707, 0.770) | 0.818 (0.792, 0.844) | 0.071 (0.056, 0.085) |
Figure 1.
Forest plot for the difference in AUC between models with and without hK2 and intact PSA
We also wanted to investigate the marginal improvement in AUC when adding intact PSA and hK2 separately. When adding intact PSA to the model including age, total PSA, free PSA and DRE, AUCs ranged from 0.708 to 0.885. AUC increased significantly by 0.059 (95% CI 0.050, 0.069, p<0.0001) to 0.802 (95% CI 0.789, 0.814, p<0.0001). The marginal improvement in discrimination when adding hK2 to the model including age, total PSA, free PSA and intact PSA was smaller but significant, 0.024 (95% CI 0.020, 0.029, p<0.0001), indicating that intact PSA and hK2 significantly increase discrimination alone as well as in combination.
There was significant heterogeneity between cohorts for the discrimination of the models (p≤0.01). This is expected as the patient groups and indications for biopsy differed between cohorts. For instance, discrimination is higher for both models in cohorts undergoing a first PSA is generally higher than those undergoing repeat screening. Critically however, there was no heterogeneity in the improvement in AUC with intact PSA and hK2 (p=0.2).
When limiting the analysis to the four contemporary cohorts, discrimination was slightly lower for all models, but increases in discrimination with the addition of intact PSA and hK2 remained significant and were consistent with those seen in the main analysis. Among these cohorts, discrimination for the model without intact PSA and hK2 was 0.723 (95% CI 0 0.706, 0.740). Discrimination when adding both intact PSA and hK2 was 0.794 (95% CI 0.779, 0.809), an increase of 0.071 (95% CI 0.057, 0.085, p<0.0001). Adding intact PSA alone increased discrimination by 0.063 (95% CI 0.052, 0.074, p<0.0001) to 0.786 (95% CI 0.771, 0.801). hK2 alone improved the discrimination by 0.026 (95% CI 0.021, 0.032, p<0.0001) to 0.753 (95% CI 0.737, 0.770).
As a sensitivity analysis requested by a reviewer, we repeated the analysis comparing the kallikrein models to the performance of comparator models built on the same cohorts which excluded intact PSA and hK2. The results were similar. The AUC for the model without intact PSA and hk2 was 0.784 (95% CI 0.771, 0.797) compared to 0.813 (95% CI 0.801, 0.825) for the full kallikrein model. The 95% C.I. do not overlap and the difference in discrimination is highly statistically significant: 0.027 (95% CI 0.021, 0.034, p<0.0001).
Discussion
Our findings clearly demonstrate that intact PSA and hK2 add importantly to discrimination of the kallikrein panel. Although the marginal value of intact PSA and hK2 has been reported in some studies, such as the US validation study3, the current meta-analysis is the first attempt to systematically estimate the value of these two markers across different studies. Hitherto, it was possible that the high discriminatory accuracy of the kallikrein panel reported across numerous studies was due to the free and total PSA components. If so, the clinical value of the panel – reducing unnecessary biopsy and overdiagnosis - could be replicated using data readily available to urologists without measuring additional markers. We have disconfirmed this hypothesis.
The value of hK2 might still be questionned on the grounds that a greater part of the value of the novel markers comes from intact PSA. However, a 0.024 increment in AUC for hK2 is far from trivial. Moreover, hK2 is of clear value in the context of multiplex testing: given that blood needs to be sent to a special laboratory for testing of intact PSA, and that total and free PSA is tested simultaneously, inclusion of hK2 has no effect on timeliness or inconvenience, only a marginal increase in cost.
In conclusion, in an individual patient data meta-analysis of intact PSA and hK2 add statistically significant predictive discrimination to the kallikrein panel, with non-overlapping confidence intervals for the estimates of discrimination of the panel with and without these two markers. The increment in discrimination is of clear clinical benefit. The clinical value of the panel cannot be replicated using data readily available to urologists without measuring intact PSA and hK2.
Acknowledgments
Monique J. Roobol (Rotterdam, Netherlands) assisted with data collection.
Funding
Supported in part by funds from David H. Koch provided through the Prostate Cancer Foundation, the Sidney Kimmel Center for Prostate and Urologic Cancers, P50-CA92629 SPORE grant from the National Cancer Institute to Dr. H Scher, the P30-CA008748 NIH/NCI Cancer Center Support Grant to MSKCC, R01 CA179115 to Dr. A. Vickers and R01CA160816 to Drs. Lilja and Vickers, Swedish Cancer Society project number 14–0722 and the Swedish Research Council (VR-MH project no. 2016–02974) to Dr. Lilja, and the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre Program in UK to Dr. FC Hamdy. The ProtecT trial is funded by the UK National Institute for Health Research Health Technology Assessment Programme (projects 96/20/06, 96/20/99, and University of Oxford is the sponsor. Department of Health disclaimer: The views and opinions expressed therein are those of the authors and do not necessarily reflect those of the Department of Health. Also supported by the NIHR Collaboration for Leadership in Applied Health Research and Care West, hosted by University Hospitals Bristol NHS Foundation Trust. Swedish Cancer Society (projects no. 13–0479 and 14–0274 to Anders Bjartell, Swedish Scientific Council (projects no. K2011–67X-21861–01-6 and K2014–66X-20760–07-4 to Anders Bjartell). Further support from Ligue Nationale contre le cancer SCG 12574/2015,
Footnotes
Conflict of interest
HL hold patents on assays for free PSA, intact PSA, and HK2. AV and HL are named on a patent for a statistical method to detect prostate cancer. The method has been commercialized by OPKO Health. AV and HL receive royalties from sales of the test. HL has stock and AV has stock options in OPKO Health. MJR is a member of the advisory board of OPKO Health. SZ is a Consultant with stock options at OPKO Health.
Contributor Information
Andrew Vickers, New York, NY.
Emily A. Vertosick, New York, NY
Daniel D. Sjoberg, New York, NY
Freddie Hamdy, Oxford, United Kingdom.
David Neal, Oxford, United Kingdom.
Anders Bjartell, Lund, Sweden.
Jonas Hugosson, Göteborg, Sweden.
Jenny L. Donovan, Bristol, United Kingdom
Arnauld Villers, Lille, France.
Stephen Zappala, Andover Urology, Andover, MA.
Hans Lilja, New York, NY.
References
- 1.Lamy PJ, Allory Y, Gauchez AS et al. : Prognostic Biomarkers Used for Localised Prostate Cancer Management: A Systematic Review. Eur Urol Focus, 2017 [DOI] [PubMed] [Google Scholar]
- 2.McDonald ML, Parsons JK: 4-Kallikrein Test and Kallikrein Markers in Prostate Cancer Screening. Urol Clin North Am, 43: 39, 2016 [DOI] [PubMed] [Google Scholar]
- 3.Parekh DJ, Punnen S, Sjoberg DD et al. : A Multi-institutional Prospective Trial in the USA Confirms that the 4Kscore Accurately Identifies Men with High-grade Prostate Cancer. European Urology, 68: 464, 2015 [DOI] [PubMed] [Google Scholar]
- 4.Vickers A, Vertosick EA, Sjoberg DD et al. : Properties of the 4-Kallikrein Panel Outside the Diagnostic Gray Zone: Meta-Analysis of Patients with Positive Digital Rectal Examination or Prostate Specific Antigen 10 ng/ml and Above. J Urol, 197: 607, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Punnen S, Freedland SJ, Polascik TJ et al. : A Multi-institutional Prospective Trial in the Veterans Affairs Health System confirms the 4Kscore maintains its Predictive value among African American Men. J Urol, 2017 [DOI] [PubMed] [Google Scholar]
- 6.Bancroft EK, Page EC, Castro E et al. : Targeted prostate cancer screening in BRCA1 and BRCA2 mutation carriers: results from the initial screening round of the IMPACT study. Eur Urol, 66: 489, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nordstrom T, Vickers A, Assel M et al. : Comparison Between the Four-kallikrein Panel and Prostate Health Index for Predicting Prostate Cancer. Eur Urol, 68: 139, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vickers AJ, Cronin AM, Aus G et al. : A panel of kallikrein markers can reduce unnecessary biopsy for prostate cancer: data from the European Randomized Study of Prostate Cancer Screening in Goteborg, Sweden. BMC Med, 6: 19, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vickers A, Cronin A, Roobol M et al. : Reducing unnecessary biopsy during prostate cancer screening using a four-kallikrein panel: an independent replication. J Clin Oncol, 28: 2493, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bryant RJ, Sjoberg DD, Vickers AJ et al. : Predicting high-grade cancer at ten-core prostate biopsy using four kallikrein markers measured in blood in the ProtecT study. J Natl Cancer Inst, 107, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeLong ER, DeLong DM, Clarke-Pearson DL: Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics, 44: 837, 1988 [PubMed] [Google Scholar]