Abstract
Microglia rapidly respond to changes in neural activity and inflammation to regulate synaptic connectivity. The extracellular signals, particularly neuron-derived molecules, that drive these microglial functions at synapses remains a key open question. Here, whisker lesioning, known to dampen cortical activity, induces microglia-mediated synapse elimination. We show that this synapse elimination is dependent on the microglial fractalkine receptor, CX3CR1, but not complement receptor 3, signaling. Further, mice deficient in the CX3CR1 ligand (CX3CL1) also have profound defects in synapse elimination. Single-cell RNAseq then revealed that Cx3cl1 is cortical neuron-derived and Adam10, a metalloprotease that cleaves CX3CL1 into a secreted form, is upregulated specifically in layer IV neurons and microglia following whisker lesioning. Finally, inhibition of Adam10 phenocopies Cx3cr1−/− and Cx3cl1−/− synapse elimination defects. Together, these results identify novel neuron-to-microglia signaling necessary for cortical synaptic remodeling and reveal context-dependent immune mechanisms are utilized to remodel synapses in the mammalian brain.
INTRODUCTION
Microglia are resident CNS macrophages that are becoming increasingly appreciated as dynamic regulators of synaptic connectivity. This includes developmental synaptic pruning, whereby microglia are ‘listening’ to neural activity and engulfing synapses from less active neurons1,2. Mechanisms regulating this process of activity-dependent, microglial synapse elimination have largely focused on surface receptors expressed by microglia. Whether there are activity-dependent neuronal cues that instruct microglia to eliminate synapses remains an open question. The importance of elucidating these mechanisms is further emphasized in a large array of neurological disorders, including neurodegenerative diseases, where dysregulated microglia-mediated synapse elimination has now been implicated3.
Two of the major molecular pathways identified to modulate microglia function at synapses are phagocytic signaling through complement receptor 3 (CR3) and chemokine signaling through the fractalkine receptor (CX3CR1). In the developing mouse visual thalamus, complement proteins C3 and C1q localize to synapses and microglia engulf synapses via CR3 expressed by microglia2,4. Blocking this synaptic engulfment in C3, C1q, or CR3-deficient mice results in sustained synaptic pruning defects. A similar molecular mechanism also appears to regulate early synapse loss in mouse models of neurodegeneration5–7. CX3CR1 is a G-protein coupled chemokine receptor highly enriched in microglia8. While CR3-dependent phagocytic signaling regulates synaptic pruning in the developing visual system, studies have demonstrated that these effects are independent of CX3CR19,10. Instead, in the developing hippocampus and barrel cortex, CX3CR1-deficient mice exhibit a transient delay in microglial recruitment to synapse-dense brain regions and a concomitant delay in functional maturation of synapses11,12. Long term, CX3CR1-deficient mice demonstrate defects in social interactions and functional synaptic connectivity13. It is less clear how CX3CR1 is exerting these effects and the relative involvement of the canonical CX3CR1 ligand fractalkine (CX3CL1) is unknown.
Here, we used the mouse barrel cortex system to identify activity-dependent mechanisms by which neurons communicate with microglia to regulate synapse remodeling. Sensory endings from trigeminal neurons transmit sensory information from the whisker follicles on the snout to the brain stem, then to the ventral posteromedial (VPM) nucleus of the thalamus. VPM neurons then project and form thalamocortical (TC) synapses largely within layer IV of the barrel cortex. These TC synapses form a highly precise topographic map in which each individual whisker is represented in the barrel cortex by a discrete bundle of TC synapses (i.e. barrels) separated by septa14. This is a particularly powerful system for studying synapse remodeling as TC synapses are highly sensitive to whisker manipulation, and removal of the whiskers results in dampened activity in the barrel cortex and elimination of TC synapses15–21. Despite a clear role for neural activity, the mechanism(s) by which changes in activity elicit TC synapse remodeling is an open question.
We used whisker cauterization and trimming in postnatal mice, paradigms known to reduce activity in the corresponding barrel cortex15–20. We identify synapse elimination within 1 week of whisker removal and robust microglia-mediated synaptic engulfment. Unlike the developing visual system2, synapse elimination in the barrel cortex is CR3-independent. Instead, we identify profound defects in TC synapse elimination in mice deficient in either CX3CR1 enriched in microglia or its ligand CX3CL1. Using single-cell RNAseq, we further uncover that Cx3cl1 is enriched in cortical neurons and Adam10, a metalloprotease known to cleave CX3CL1 into a secreted form, is increased specifically in layer IV neurons and microglia following sensory lesioning. Strikingly, pharmacological inhibition of ADAM10 phenocopies synapse elimination defects observed in CX3CL1 and CX3CR1-deficient mice. Together, these data provide new insight into activity-dependent molecular mechanisms by which neurons communicate with microglia. Our single-cell RNAseq further provides an unbiased approach to identify novel mechanisms underlying intercellular communication in response to sensory perturbations.
RESULTS
Whisker lesioning induces rapid and robust elimination of thalamocortical inputs in the barrel cortex
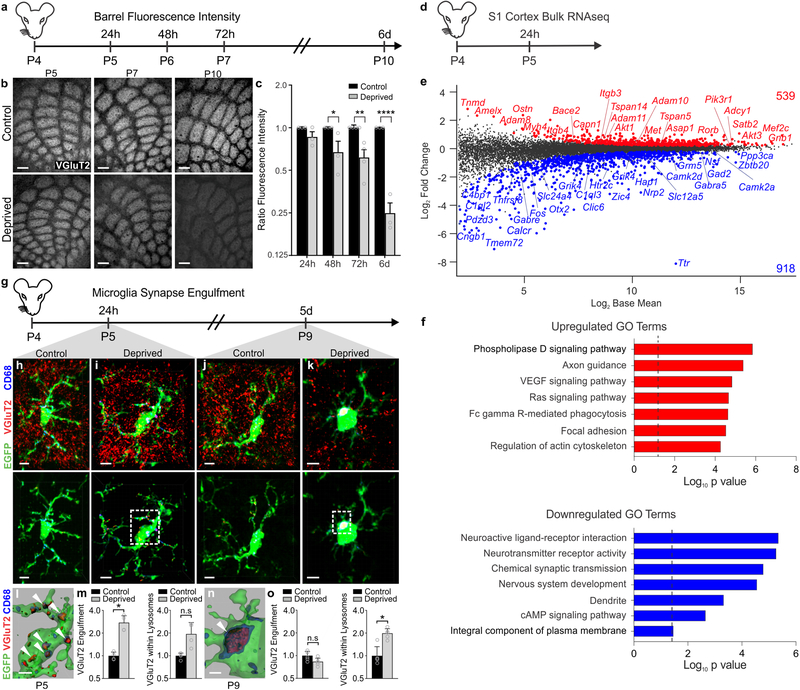
To interrogate neuron-microglia signaling in regulating synapse remodeling, we performed two different manipulations in separate cohorts of postnatal day 4 (P4) mice: unilateral whisker trimming and unilateral whisker lesioning by cauterization (Fig. 1 and Supplemental Fig.1a–b). These paradigms are known to reduce activity in the barrel cortex and maximize TC remodeling in the neonate while avoiding the critical window (P0–P3) where sensory loss disrupts initial TC input wiring and can induce neuronal apoptosis22. Also, each model has its own internal control as the whiskers are left intact on the other side of the snout. Using anti-vesicular glutamate transporter 2 (VGluT2) immunostaining, we observed a decrease in TC presynaptic terminals within layer IV of the barrel cortex by 17 days-post the beginning of whisker trimming (Supplemental Fig.1a–b). These results are consistent with previous work showing decreased TC inputs within the barrel cortex following whisker trimming in neonates and adults18,19,21. This decrease in presynaptic terminals was accelerated in the whisker lesioning model. Within 6 days-post lesioning, VGluT2 immunoreactivity was decreased by ~75% in the deprived compared to the control barrel cortex within the same animal (Fig. 1b–c). Demonstrating that this effect was not due to a downregulation of VGluT2, we observed similar effects in mice that express a fluorescent reporter in thalamic neurons and their cortical projections (SERT-Cre; Rosa26LSL.TdTomato/+) (Supplementary Fig. 1c–d). To note, presynaptic terminals and axons are labeled with tdTomato as well as other cortical projections in SERT-Cre;Rosa26LSL.TdTomato/+ mice, which results in less robust detection of TC input elimination.
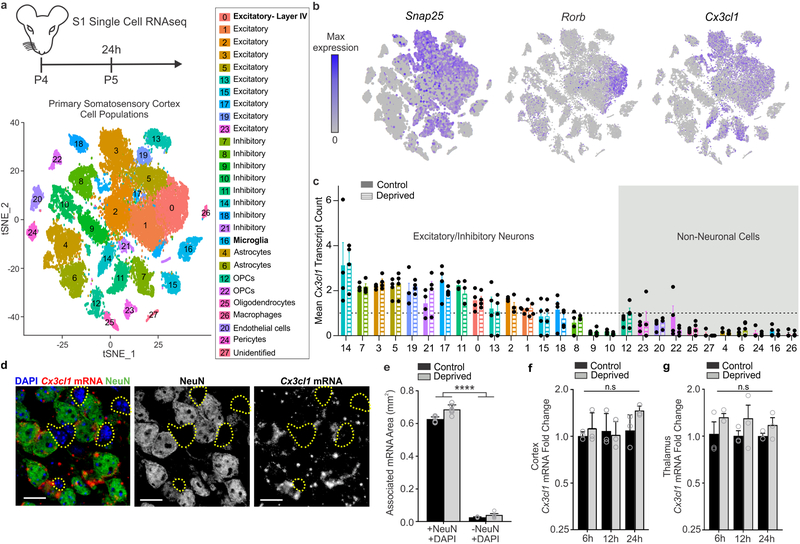
Figure 1. Whisker lesioning induces microglial engulfment and elimination of TC inputs within the barrel cortex.
a, Timeline for analysis of TC input elimination following whisker lesioning at P4. b, Tangential sections of layer IV contralateral control (top panel) and deprived (bottom panel) barrel cortices immunolabeled for anti-VGluT2 show a decrease in TC inputs by P10. Scale bar, 150 μm. c, Quantification of fluorescence intensity of VGluT2-positive TC inputs in the barrel cortex in the deprived (gray bars) compared to the control barrel cortex (black bars) at each time point post-whisker removal. Data normalized to the control, non-deprived hemisphere within each animal. (Two-way ANOVA with Sidak’s post hoc; control vs deprived 24h, n = 3 animals, P = 0.5323, t = 1.419, df = 18; control vs deprived 48h, n = 3 animals, P = 0.0142, t = 3.349, df = 18; control vs deprived 72h, n = 4 animals, P = 0.0011, t = 4.516, df = 18; control vs deprived 6d, n = 3 animals, P <0.0001, t = 7.631, df = 18). d, Timeline for bulk RNAseq of the barrel cortex 24 hours after whisker lesioning. e-f, MA plot (representing log-ratio (M) on the y axis and mean average (A) on the x axis) shows gene expression changes (red, upregulated; blue, downregulated) in the deprived somatosensory cortex at P4 (DESeq2, n = 5 mice, P5). Horizontal bar graphs show selected gene ontology (GO) annotations (Enrichr) enriched for up- (red) or downregulated (blue) genes; dotted lines, P = 0.05. g, Timeline for analysis of TC input engulfment by microglia. h,i,j,k, Fluorescent images of microglia (green) within layer IV of the control (h,j) and deprived (i,k) barrel cortices 24 h (h,i) and 5 d (j,k) after unilateral whisker removal. Microglial lysosomes are labeled with anti-CD68 (blue). Raw fluorescent images, top panel; VGlut2 signal internalized within microglia, bottom panel. Scale bar, 5 μm. l,n, 3D surface-rendered inset of i,k (bottom panel). Arrows depict VGlut2 (red) internalized within microglia (green) and within lysosomes (blue). Scale bar, 2 μm. m,o, Quantification of VGlut2 engulfment within microglia (left) and VGlut2 engulfment within lysosomes (right) 24 h (m; within microglia, Two-sided Student’s t-test, n = 4 animals, P = 0.0305, t = 2.642 df =6; within lysosomes, Two-sided Student’s t-test, n = 4 animals, P = 0.2955, t = 1.146, df =6) and 5 d (o; within microglia, Two-sided Student’s t-test, n = 5 animals, P = 0.3319, t =1.033, df =8; within lysosomes, Two-sided Student’s t-test, n = 5 animals, P = 0.0272, t =2.251, df =8) after whisker removal in Cx3cr1EGFP/+ microglia reveals increased VGluT2 within microglia at 24 h (m) and increased VGluT2 within microglial lysosomes at 5 d-post whisker removal (o). Data normalized to engulfment in microglia in the control hemisphere within each animal. All data presented as mean ± SEM.
To exclude the possibility that TC input elimination induced by whisker lesioning was due to neuronal damage and loss, we also assessed cell death, injury-induced axon degeneration, and cell stress. We observed no significant increase in cleaved Caspase3+ neurons or amyloid precursor protein (APP) accumulation in axons within the barrel cortex circuit following whisker removal (Supplementary Figure 2d–f). However, an increase in the stress marker ATF3 was observed in trigeminal neurons (Supplementary Fig. 2a–b), but not in cortical or VPM neurons (Supplementary Fig. 2c). To further characterize molecular changes, we performed bulk RNAseq of the barrel cortex following whisker lesioning. Consistent with decreased activity in the barrel cortex, immediate early genes such as Fos and genes related to neurotransmitter signaling were decreased in the deprived barrel cortex with no change in genes related to cell stress or death 24 h-post lesioning (Fig. 1d–f; Fig. 7j–k). Instead, there was an increase in genes related to axon growth and phagocytic signaling. These data are most consistent with synapse loss, and possibly axonal remodeling, resulting from decreased neural activity. Given the robust presynaptic terminal loss elicited in the absence of significant neuronal cell death or degeneration, we used whisker cauterization-induced sensory lesioning in P4 mice as a model to dampen cortical activity for the remainder of the study.
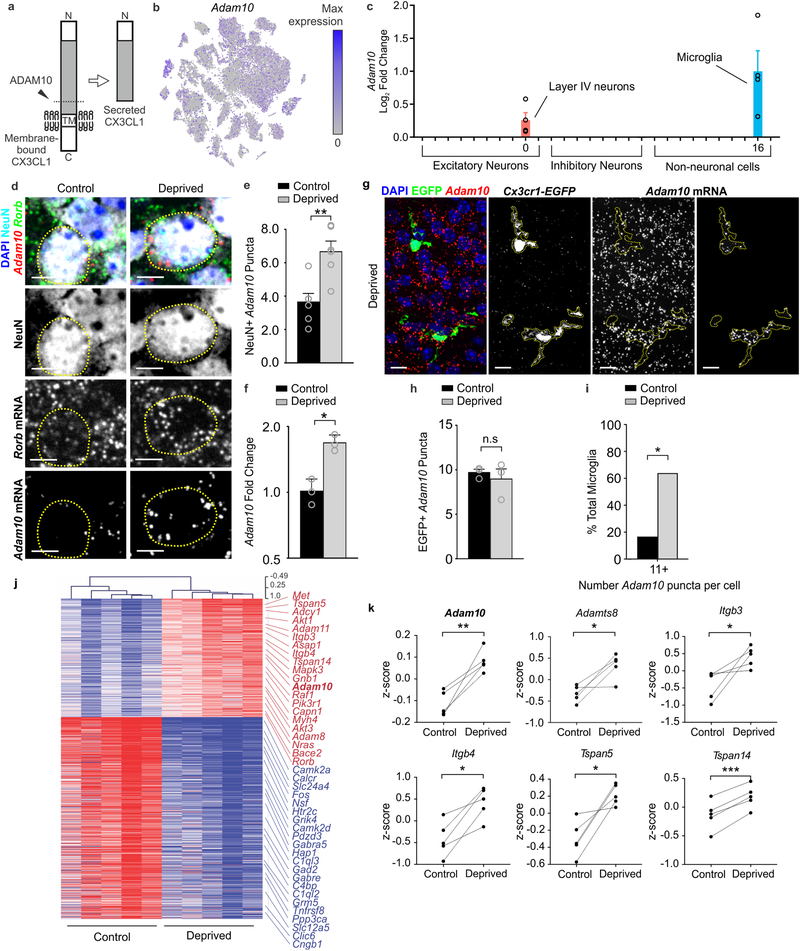
Figure 7. Adam10, a metalloprotease that cleaves CX3CL1, is increased in neurons within the barrel cortex following whisker lesioning.
a, Adam10 cleaves Cx3CL1 at the membrane (dotted line) to produce a secreted form. b, tSNE plot for Adam10 reveals broad expression across many neuronal and non-neuronal cell types. (Single cell RNAseq performed on n = 4 Cx3cr1+/− and n = 4 Cx3cr1−/− at P5, 24h after whisker lesioning). c, Fold change of Adam10 expression by single-cell RNAseq reveals significant (FDR <0.10, Monocle2) upregulation specifically in layer IV Rorb+ neurons and microglia after whisker lesioning. Each data point is the mean fold change for Adam10 within each individual Cx3cr1+/− deprived hemisphere. Data presented as mean ± SEM. d, In situ hybridization for Adam10 in the control (top panels) and deprived (bottom panels) barrel cortices. Adam10 is increased in the majority of Rorb+ layer IV excitatory neurons (NeuN+, Rorb+) assessed 24 h-post whisker lesioning compared to neurons in the control barrel cortex. Scale bar, 5 μm.e, Quantification of in situ for Adam10 puncta co-localized with layer IV neurons. (Two-tailed Student’s t-test, n = 6 Cx3cr1+/+ animals, P = 0.003, t = 0.3889, df =10). Data presented as mean ± SEM. f, qPCR for Adam10 24 h-post whisker lesioning in the control (black bars) and deprived (grey bars) whole barrel cortices reveals a significant increase in Adam 10 24 h-post whisker lesioning. (Two-tailed Student’s t-test, n = 3 Cx3cr1+/+animals, P = 0.0241, t = 3.538, df =4). Data presented as mean ± SEM. g, In situ hybridization for Adam10 within Cx3cr1EGFP/+ microglia (yellow dotted lines) in the deprived cortex 24 h-post whisker lesioning reveals increased Adam10 expression in a subset of microglia after lesioning. Representative images taken Cx3cr1+/+ animals across 1 independent experiment. h, Quantification of the average Adam10 in situ puncta per microglia averaged across all microglial cells assessed in the barrel cortex shows no significant difference in expression between the control and deprived conditions. (Two-tailed Student’s t-test, n = 3 Cx3cr1+/− animals, P = 0.5547, t = 0.6439, df =4). Data presented as mean ± SEM. i, Further quantification of Adam10 mRNA puncta within microglia reveals a significant increase in a subset of microglia expressing high levels (≥11 puncta) of Adam10 in the deprived vs. control barrel cortex (n = 36 deprived microglia, 36 control microglia from 3 Cx3cr1+/+ animals). One-tailed Chi-square test, P = 0.0306, χ2 = 3.503, df = 1, z = 1.872). Data represented as whole number percentage of the total cell population. j, Heatmap with hierarchical clustering distances shows the variation in the expression levels (z-scored log2(RPKM)) of 539 up- and 918 downregulated genes upon whisker deprivation at P4 identified by bulk RNAseq from the primary barrel cortex of whisker lesioned mice (DESeq2 software, n = 5 mice, P5, related to Figure1e). Adam10 is bolded. k, Line graphs show for individual mice z-scored log2(RPKM) changes of Adam10 and other selected genes encoding known regulators of Adam10 expression or activity. (Two-tailed Student’s t-test: Adam10, P = 0.0060, t = 5.32; Adamts8, P = 0.0205, t =3.72; Itgb3, P = 0.0454, t = 2.87; Itgb4, P = 0.0106, t = 4.53; Tspan5, P = 0.0151, t = 4.08; Tspan14, P = 0.0006, t = 9.80; n = 5 animals).
Whisker lesioning induces microglial engulfment of TC inputs within the barrel cortex independent of complement receptor 3 (CR3)
In the visual system, microglia engulf and remove synapses in response to dampened neural activity2,23,24. Therefore, we next explored whether microglia similarly engulf and eliminate TC synaptic inputs in the barrel cortex following whisker lesioning. Microglia were labeled using a transgenic mouse that expresses EGFP under the control of the fractalkine receptor (Cx3cr1EGFP/+) and TC presynaptic terminals were labeled using anti-VGluT2. Using fluorescent confocal microscopy and structured illumination microscopy, we detected an ~2-fold increase in the volume of engulfed TC inputs within microglia in the deprived barrel cortex within 24 h of removing the whiskers (Fig. 1g–o and Supplementary Fig. 3a). These inputs were largely localized within the microglia, but not yet significantly associated with lysosomes (Fig. 1g–i,l,m). At 5 days-post whisker removal, microglia had a more phagocytic morphology, with a more rounded and enlarged soma, and most engulfed TC inputs were completely localized within microglial lysosomes (Fig. 1k,n,o). In contrast, minimal TC inputs were detected within microglia in the control barrel cortex of the same animal at any time point assessed (Fig. 1h,j). In addition, the percentage of highly phagocytic microglia (phagocytic index >1% at 24 h and >2% at 5d) was increased in the deprived cortex (Supplementary Fig. 3), which may reflect microglial heterogeneity or the limitations of using static imaging to capture a dynamic process. This increase in phagocytic activity was further reflected in our RNAseq data (Fig. 1d–f). These data provide the first evidence that microglia engulf and eliminate TC inputs in the cortex several synapses away from a peripheral sensory lesion known to reduce activity in the cortex.
Complement-dependent phagocytic signaling is one of the best characterized mechanisms by which microglia engulf and remodel synapses. During synaptic pruning in the developing mouse visual system, complement proteins C1q and C3 localize to synapses4. Microglia subsequently engulf and eliminate these synapses via the microglial phagocytic receptor complement receptor 3 (CR3/CD11b)2. To determine whether this pathway also regulates microglia-mediated synapse elimination in the barrel cortex following whisker lesioning, we assessed microglial engulfment and elimination of TC inputs in CR3-deficient (CR3-KO) mice. Surprisingly, similar to wild type mice, TC inputs were eliminated by 6 days-post whisker removal in CR3-KO mice (Supplementary Fig. 4a–c) and engulfed TC inputs were still detected within CR3-KO microglia in the deprived barrel cortex (Supplementary Fig. 4d–f). Together, these data demonstrate that CR3 is dispensable for sensory lesion-induced microglial engulfment and elimination of TC synapses in the barrel cortex.
CX3CR1-deficient mice have profound defects in structural and functional synapse remodeling following whisker lesioning
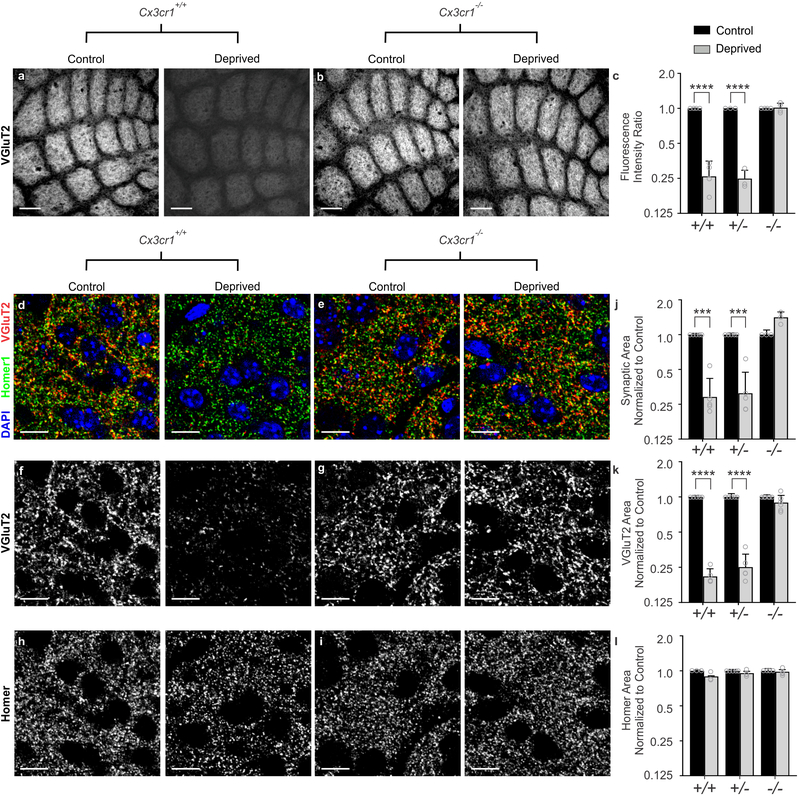
Besides CR3, the fractalkine receptor (CX3CR1), a G-protein coupled chemokine receptor highly enriched in microglia, has also been implicated in regulating microglial function at developing synapses11–13. We therefore sought to assess whisker lesion-induced TC synapse remodeling in Cx3cr1−/− mice. Remarkably, unlike CR3-KO mice (Supplementary Fig. 4), elimination of VGluT2-positive TC presynaptic inputs and structural synapses (co-localized presynaptic VGluT2 and postsynaptic Homer puncta) 6 days-post whisker removal was completely blocked in Cx3cr1−/− mice compared to Cx3cr1+/− or Cx3cr1+/+ littermates (Fig. 2). This was largely a presynaptic effect as the density of postsynaptic Homer was unaffected, even in wild-type animals (Fig. 2h–i,l). This is consistent with previous work demonstrating that microglia preferentially engulf presynaptic inputs2,25. We further confirmed that this effect was through microglial CX3CR1 and demonstrate CX3CR1 is highly enriched in microglia in control and deprived barrel cortices versus other neural or glial cell types (Supplementary Figure 5i–l). In addition, while peripheral CD45+ monocytes and F4/80+ macrophages express CX3CR1 in the whisker pad, this expression pattern does not change following whisker lesioning (Supplemental Figure 5c and h). We further measured macrophage recruitment to the whisker follicles following whisker lesioning as well as immunostaining for ATF3 in Cx3cr1−/− mice (Supplementary Fig.5). All these responses were comparable to Cx3cr1+/− controls (Supplementary Fig. 2). Last, we assessed TC input elimination following whisker trimming and found that TC input elimination elicited in this paradigm was also CX3CR1-dependent (Supplementary Fig. 5f–g). Together, these results suggest that synapse elimination defects in Cx3cr1−/− mice were not secondary to changes in injury, wound healing, or neuronal stress responses and are most consistent with microglial CX3CR1 mediating synapse elimination in the barrel cortex.
Figure 2. Cx3cr1 expression is necessary for TC input elimination after whisker lesioning.
a-b, VGluT2 immunolabeled TC inputs within tangential sections of control and deprived barrel cortices in Cx3cr1+/+ (a) and Cx3cr1−/− mice (b) show TC inputs remain 6 d-post deprivation in mice lacking Cx3cr1. Scale bar, 150 μm. c, Quantification of fluorescence intensity of VGluT2-positive TC input immunoreactivity 6 d after deprivation in Cx3cr1+/+, Cx3cr1+/− and Cx3cr1−/− littermates demonstrates a significant decrease in VGluT2 immunoreactivity in Cx3cr1+/+, Cx3cr1+/− mice following deprivation but this is blocked in Cx3cr1−/− littermates. (Data normalized to the control, non-deprived hemisphere within each animal; Two-Way ANOVA with Sidak’s post hoc; control vs deprived Cx3cr1+/+ n = 4 animals, P < 0.0001, t = 8.967, df =16; control vs deprived Cx3cr1+/− n = 3 animals, P < 0.0001, t = 7.882, df = 16; control vs deprived Cx3cr1−/− n = 4 animals, P = 0.9976, t = 0.1722, df = 16). d-i, High magnification (63X) confocal images of TC synapses within layer IV of the control and deprived barrel cortices immunolabeled with presynaptic anti-VGluT2 (red) and postsynaptic anti-Homer (green) 6 d-post deprivation in Cx3cr1+/+ (d,f,h) and Cx3cr1−/− (e,g,i) mice. Merged channels are shown in panels d-e. The presynaptic VGluT2 channel alone is shown in panels f-g. The postsynaptic Homer channel alone is shown in panels h-i. Scale bars, 10 μm. j-l, Quantification of d-i reveals a significant decrease in structural synapses (j; colocalized VGluT2 and Homer, Two-Way ANOVA with Sidak’s post hoc, control vs deprived Cx3cr1+/+ n = 5 animals, P = 0.0004, t = 4.765, df =20; control vs deprived Cx3cr1+/− n = 5 animals, P = 0.0005, t = 4.617, df = 20; control vs deprived Cx3cr1−/− n = 3 animals, P = 0.1290, t = 2.139, df = 20) and VGluT2-positive TC presynaptic terminal density (k; VGlut2 Area, Two-Way ANOVA with Sidak’s post hoc, control vs deprived Cx3cr1+/+ n = 5 animals, P < 0.0001, t = 6.919, df =26; control vs deprived Cx3cr1+/− n = 5 animals, P <0.0001, t = 6.552, df = 26; control vs deprived Cx3cr1−/− n = 6 animals, P = 0.6907, t = 1.006, df = 26) in Cx3cr1+/+ and Cx3cr1+/− mice, which was blocked in Cx3cr1−/− littermates. There was no significant change in postsynaptic Homer density (l; Homer Area, Two-way ANOVA with Sidak’s post hoc, control vs deprived Cx3cr1+/+ n = 3 animals, P = 0.2386, t = 1.811, df =18; control vs deprived Cx3cr1+/− n = 5 animals, P =0.7852, t = 0.9918, df = 18; control vs deprived Cx3cr1−/− n = 4 animals, P = 0.9731, t = 0.3908, df = 18) in any genotype. Data normalized to the control, non-deprived cortex within each animal. All data presented as mean ± SEM.
To further determine if CX3CR1-dependent defects in TC input elimination are long-lasting, we lesioned whiskers at P4 and then assessed structural and functional remodeling at ≥6 weeks of age (Fig. 3). Similar to 6 days post-whisker removal, we observed a significant decrease in structural TC inputs in the deprived adult (P90) Cx3cr1+/+ barrel cortex (Fig. 3a), which was blocked in Cx3cr1−/− mice (Fig.3b). To assess functional connectivity, we also performed electrophysiological recordings in ≥6 week Cx3cr1−/− and Cx3cr1+/+ mice. We observed a significant decrease in spontaneous excitatory postsynaptic current (sEPSC) frequency and amplitude in layer IV stellate neurons within the deprived cortex in Cx3cr1+/+ mice (Fig. 3c–d), which is consistent with a decrease in the number and strength of functional synapses in the deprived cortex. When we assessed the same parameters in Cx3cr1−/− littermates (Fig. 3c,e), there was no longer a significant decrease in sEPSC frequency or amplitude in the deprived cortex compared to the control cortex. Interestingly, there was evidence of a decrease in baseline sEPSC frequency and amplitude in Cx3cr1−/− mice compared to Cx3cr1+/+ littermates, suggesting that CX3CR1 signaling may regulate these aspects of functional synapse maturation within the barrel cortex. These data establish that microglial CX3CR1 signaling is critical for long-term remodeling of structural and functional synapses in the barrel cortex several synapses away from a peripheral sensory lesion.
Figure 3. Microglial Cx3cr1 deficiency blocks structural and functional synaptic remodeling long-term.
a,b, VGluT2 immunolabeling of TC inputs in tangential sections of the control and deprived barrel cortex in P90 Cx3cr1+/+ (a) and Cx3cr1−/− (b) littermates. TC inputs remain in Cx3cr1−/− mice after sustained whisker removal (b, right panel). Scale bar, 150 μm. Representative images taken from 2 independent experiments/animals. c, Representative sEPSC traces from layer IV stellate neurons for the control and deprived barrel cortices of P42-P56 Cx3cr1+/+ and Cx3cr1−/− mice. d,e, Quantification of stellate neuron sEPSC frequency and amplitude in Cx3cr1+/+ (d; sEPSC Frequency: n = 17 control and 16 deprived cells from 3 Cx3cr1+/+ littermates, Two-tailed Student’s t-test, P = 0.0484, t =2.054, df =31; sEPSC Amplitude, Two-tailed Student’s t-test, P = 0.0105, t =2.723, df =31) and Cx3cr1−/− (e; sEPSC Frequency: n = 10 control and 13 deprived cells from 3 Cx3cr1−/− littermates, Two-tailed Student’s t-test, P = 0.1286, t =1.582, df =21; sEPSC Amplitude, Two-tailed Student’s t-test, P = 0.9432, t =0.07205, df =21) mice in the deprived (grey bars) compared to the contralateral control (black bars) barrel cortex. Cx3cr1−/− mice show no significant decrease in stellate neuron sEPSC frequency or amplitude. All data presented as mean ± SEM.
CX3CR1 and CX3CL1 regulate microglia-mediated engulfment of TC synapses
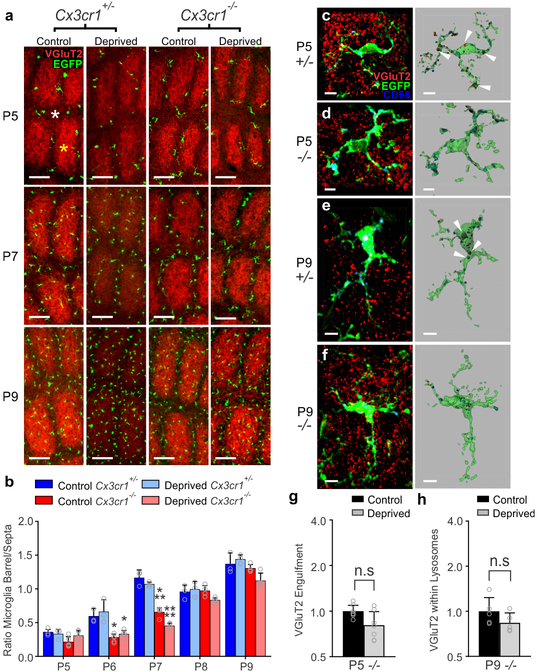
While microglial CX3CR1 signaling has previously been implicated to play an important role in regulating synapse development11–13, the mechanism by which CX3CR1 exerts these effects is largely unknown. It has been suggested that CX3CR1, a chemokine receptor, is necessary for microglial recruitment to synapses. Similar to previously published work in the barrel cortex12, we found that microglia in neonatal Cx3cr1+/− mice were concentrated in barrel septa and infiltrated the barrel centers at P6–7 and this was delayed to P8 in Cx3cr1−/− mice (Fig. 4a–b and Supplementary Fig. 6). There was no difference in the total numbers of microglia in the Cx3cr1−/− vs. Cx3cr1+/− barrel cortices (Supplementary Fig. 6). We then identified that recruitment and overall density of microglia were largely unaffected in Cx3cr1+/− and Cx3cr1−/− littermates following whisker lesioning (Fig. 4a–b, Supplementary Fig. 6). There was still a transient delay in recruitment of microglia to barrel centers in Cx3cr1−/− barrel cortex in both control and deprived barrel cortices; however, numbers of microglia within barrel centers were indistinguishable from Cx3cr1+/− mice by P8. Given that the bulk of TC synapse elimination occurs after P8 in the current paradigm (Fig. 1a–c), the delay in recruitment of microglia to the barrel centers in <P8 Cx3cr1−/− mice likely does not explain the sustained defect in synapse elimination in Cx3cr1−/− mice. To more closely assess microglial-synapse interactions in Cx3cr1−/− mice in response to whisker removal, we next analyzed microglia-mediated TC input engulfment. Unlike CR3-KO mice (Supplementary Fig. 4), loss of CX3CR1 blocked TC input engulfment by microglia in the deprived barrel cortex at P5 and P9 (Fig. 4c–h). Together, these data demonstrate that CX3CR1 signaling modulates microglia-mediated synaptic engulfment and blockade of this engulfment results in sustained defects in structural and functional synapse elimination following a peripheral sensory lesion.
Figure 4. Microglial engulfment of TC inputs following whisker lesioning is CX3CR1-dependent.
a, Representative images of microglia within the barrel cortex labeled by transgenic expression of EGFP under the control of CX3CR1 (green) and TC inputs labeled with anti-Vglut2 (red). b, Quantification of the ratio of microglia localized to the septa (denoted with white asterisk at P5) compared to the barrels (denoted with a yellow asterisk at Pt) +/− whisker deprivation. In both deprived and non-deprived barrel cortices, microglia begin to infiltrate the barrel centers from the septa by P6/7 in Cx3cr1+/− mice, which is delayed to P8 in Cx3cr1−/− mice. There is no significant difference by P8. (Two-way ANOVA and Tukeys post hoc test, control Cx3cr1+/− vs control Cx3cr1−/− at P6, n = 3 Cx3cr1+/− and 5 Cx3cr1−/− littermates, P = 0.0394, q = 3.887, df = 54; deprived Cx3cr1+/− vs deprived Cx3cr1−/− at P6, n = 3 Cx3cr1+/− and 5 Cx3cr1−/− littermates, P = 0.0273, q = 4.092, df = 54; Cx3cr1+/− vs control Cx3cr1−/− at P7, n = 3 Cx3cr1+/− and 4 Cx3cr1−/− littermates, P = 0.0005, q = 5.996, df = 54; deprived Cx3cr1+/− vs deprived Cx3cr1−/− at P7, n = 3 Cx3cr1+/− and 4 Cx3cr1−/− littermates, P <0.0001, q = 7.32, df = 54) c-f, Representative microglia from the deprived barrel cortex of Cx3cr1+/− and Cx3cr1−/− mice 24 hours and 5 d after whisker lesioning. Left panel shows raw fluorescent image with microglia (EGFP, green), VGluT2 (red), and lysosomes (CD-68, blue). Right panel shows 3D-rendered microglia within layer IV of the deprived barrel cortex 24 h (c,d) and 5 d (e,f) post deprivation. Arrows denote examples of engulfed TC inputs in Cx3cr1+/− barrel cortex (c,e) which are largely absent in Cx3cr1−/− mice (d,f). Scale bar, 5 μm. g,h, Quantification of engulfment in Cx3cr1−/− mice 24 h (g; Two-tailed Student’s t-test, n = 8 littermate animals, P = 0.3668, t = 0.938, df =12) and 5 d (h; Two-tailed Student’s t-test, n = 5 littermate animals, P = 0.5619, t =0.6051, df =8) post deprivation reveals no significant increase in engulfed TC inputs in the control vs deprived barrel cortex at any time point. Engulfment data normalized to the control, non-deprived hemisphere within each animal. All data presented as mean ± SEM.
We next sought to understand the signals upstream of CX3CR1 necessary for TC synapse elimination by microglia by assessing mice deficient in fractalkine (CX3CL1), the canonical and only known in vivo ligand of CX3CR126. Similar to Cx3cr1−/− mice, we observed significant defects in the elimination of TC inputs 6 days post-whisker removal in the Cx3cl1−/− barrel cortex (Fig. 5a–f). This synapse elimination defect was accompanied by a blockade of TC input engulfment by microglia (Fig. 5g–i) in Cx3cl1−/− mice, which phenocopies defects in Cx3cr1−/− mice. These data identify CX3CL1 as a novel regulator of TC synapse remodeling and microglia-mediated synapse engulfment and strongly suggests that microglia eliminate TC inputs through CX3CR1-CX3CL1 signaling.
Figure 5. CX3CL1 is necessary for TC input engulfment and elimination after sensory lesioning.
a-b, VGluT2 immunolabeled TC inputs within tangential sections of control and deprived barrel cortices in Cx3cl1+/+ (a) and Cx3cl1−/− mice (b). Scale bar, 150 μm c, Quantification of fluorescence intensity of VGluT2-positive TC input 6 d after deprivation shows a significant decrease in VGluT2 fluorescence intensity in Cx3cl1+/+ mice 6 d post-deprivation, which is blocked in Cx3cl1−/− littermates. Data normalized to the control, non-deprived hemisphere within each animal. (Two-Way ANOVA with Sidak’s post hoc, n = 4 animals per genotype, P <0.0001, t = 28.3, df = 12). d-f, Quantification of high magnification images of synaptic components in the barrel centers 6 d after whisker removal reveals a significant decrease in structural synapses (d, VGluT2 colocalized with Homer; Two-Way ANOVA with Sidak’s post hoc, n = 4 animals per genotype, Cx3cl1+/+ control vs deprived, P <0.0001, t =11.66, df = 12) and VGluT2-positive presynaptic terminals (e; Two-Way ANOVA with Sidak’s post hoc, n = 4 animals per genotype, Cx3cl1+/+ control vs deprived, P <0.0001, t = 7.418, df = 12) in Cx3cl1+/+ mice but no significant change in Cx3cl1−/− littermates (Colocalized Area, P = 0.0642, t = 2.415, df = 12; VGlut2 Area, P = 0.1071, t =2.125, df = 12). There was no significant change in density of homer immunoreactivity in Cx3cl1+/+ or Cx3cl1−/− mice following whisker deprivation (f; Two-Way ANOVA with Sidak’s post hoc, n = 4 animals per genotype, no significance). Data normalized to the control, non-deprived hemisphere within each animal. g-h, Representative microglia from the deprived barrel cortex of Cx3cl1+/+ (g) and Cx3cl1−/− (h) mice. Left panel displays raw fluorescent image with microglia (Anti-Iba1, green) VGluT2 inputs (red) and lysosomes (Anti-CD68, blue) labeled. Right panels shows 3D-surface rendering of these cells. Engulfed VGluT2 (red) immunoreactive TC inputs within microglia are visualized in Cx3cl1+/+ microglia (g, arrows)) but not Cx3cl1−/− microglia (h). Scale bars, 5 μm. i, Quantification of VGlut2 engulfment 24 h after whisker removal reveals that Cx3cl1−/− microglia fail to engulf TC inputs following sensory deprivation. Data normalized to engulfment in the control hemisphere within each animal. (Two-Way ANOVA with Sidak’s post hoc, n = 4 littermates per genotype, Cx3cl1+/+ control vs deprived P = 0.0111, t = 3.369, df = 12; Cx3cl1−/− control vs deprived P = 0.5963, t = 0.9422, df = 12). All data presented as mean ± SEM.
Single-cell RNA sequencing reveals that CX3CL1 is highly enriched in neurons in the barrel cortex but its transcription is not modulated by whisker lesioning
To further explore the mechanism by which CX3CL1 signals to CX3CR1 to regulate TC synapse elimination, we performed single-cell RNAseq (scRNAseq) in the deprived and non-deprived barrel cortices of Cx3cr1+/− and Cx3cr1−/− mice. Cx3cr1+/− mice were used as controls as these were the animals used for initial synaptic remodeling and engulfment analyses (Fig. 1). 24 h-post unilateral whisker lesioning, deprived and control barrel cortices were microdissected from each animal and scRNAseq was performed using inDrops27. Following principal component analysis, we identified 27 distinct clusters of CNS cells within the barrel cortex, which were reproducible across biological replicates with an average read depth of 8,815 reads per cell. (Fig. 6 and Supplementary Figs. 7–8). In agreement with a recent scRNAseq study in the adult visual cortex28, we observed gene expression changes in neurons and glia following manipulation of sensory experience (Supplementary Figs. 9–10). To our knowledge, this is the first analysis of sensory lesion and CX3CR1-dependent gene expression in the developing somatosensory cortex at single-cell resolution. From this data set, we identified some potentially interesting differences in glial, including microglial, gene expression changes following whisker lesioning, which were CX3CR1-dependent (Supplemental Fig. 10). To note, compared to neurons, the numbers of glial cells sequenced was relatively low for individual genotypes (Supplemental Fig. 10g). Therefore, future investigation to validate gene expression changes in glia is necessary. For the remainder of the study, we focused our analyses on whisker lesion-induced changes in Cx3cl1 and related genes.
Figure 6. Single-cell RNAseq reveals that Cx3cl1 is highly enriched in neurons in the barrel cortex but its transcription is not modulated by whisker lesioning.
a, Timeline for whisker removal and single cell sequencing analysis. P4 mice (n = 4 Cx3cr1+/−, n = 4 Cx3cr1−/−) underwent unilateral whisker lesioning and were sacrificed 24 h later. Barrel cortices were prepared for single-cell RNAseq. A tSNE plot of 27 distinct cell populations in the barrel cortex clustered by principal component analysis. (See Supplementary Fig 7). b, tSNE plots for Snap25 and Cx3cl1 across all 27 clusters. Cx3cl1 is enriched in most SNAP-25-poistive neuronal clusters. c, Mean Cx3cl1 RNA transcript counts per condition (control, solid bars; deprived, striped bars) in Cx3cr1+/− animals. Each data point is the mean expression across cells within each individual Cx3cr1+/− hemisphere (control or deprived). Data at or below the dotted line indicates 1 transcript or no expression. Cx3cl1 is enriched in neurons but its expression is unchanged following whisker lesioning across all cell types. d, In situ hybridization for Cx3cl1 (red) and immunohistochemistry for NeuN to label neurons (green) in Cx3cl1+/+ deprived barrel cortices validates that Cx3cl1 is enriched in NeuN-positive neurons compared to non-neuronal cells (NeuN negative, yellow dotted lines). Scale bar, 15 μm. e, Quantification of in situ for Cx3cl1 reveals enrichment in neuronal (+NeuN/+DAPI) vs. non-neuronal (-NeuN/+DAPI) cells and no change in expression 24 h-post whisker lesioning. (Two-Way ANOVA with Sidak’s post hoc test, Control +NeuN/+DAPI vs Control -NeuN/+DAPI, P <0.0001, t = 25.78, df = 20; Deprived +NeuN/+DAPI vs Deprived -NeuN/+DAPI, P <0.0001, t = 27.7, df = 27; Control +NeuN/+DAPI vs Deprived -NeuN/+DAPI, P <0.0001, t = 25.19, df = 20; Deprived +NeuN/+DAPI vs Control -NeuN/+DAPI, P <0.0001, t = 28.29, df = 20; n = 6 images from 3 animals (3 males)). All data presented as mean ± SEM. f-g, qPCR for Cx3cl1 expression in the barrel cortex (f) and VPM nucleus of the thalamus (g) 6, 12, 24, and 72 h after whisker lesioning in Cx3cr1+/+ mice in the control (black bars) and deprived (grey bars) barrel cortices. (Two-Way ANOVA with Sidak’s post hoc, n = 3 animals per time point, 6h barrel cortex control vs deprived; P = 0.9958, t = 0.3299, df = 16; 12hr barrel cortex control vs deprived; P = 0.9996, , t = 0.1766, df = 16; 24h barrel cortex control vs deprived; P = 0.7804, t = 1.036, df = 16; 72h barrel cortex control vs deprived; P = 0.9722, t = 0.5476, df = 16; 6h thalamus control vs deprived; P = 0.5295, t = 1.287, df = 12; 12hr thalamus control vs deprived; P = 0.5041, , t = 1.329, df = 12; 24h thalamus control vs deprived; P = 0.8261, t = 0.7954, df = 12). All data presented as mean ± SEM.
Similar to previous work26, we observed enrichment of Cx3cl1 mRNA in neurons compared to non-neuronal cells (Fig. 6b–c). However, we found no significant difference in Cx3cl1 mRNA in the deprived vs. control barrel cortex in any cell type (Fig.6c). These data were confirmed by in situ hybridization (Fig. 6d–e), RNAseq of whole barrel cortex (Fig 1e), and qPCR from the barrel cortex and VPM nucleus of the thalamus (Fig. 6f–g and Supplementary Fig. 11). These data demonstrate that CX3CL1 is neuron-derived, but its transcription in the barrel cortex and thalamus is unchanged following peripheral sensory lesioning.
ADAM10, a metalloprotease that cleaves CX3CL1, is increased in neurons and microglia within the barrel cortex following whisker lesioning
CX3CL1 can exist in a membrane or a secreted form26. The latter is produced upon cleavage of the membrane-bound form by metalloproteases29. Therefore, we hypothesized that this post-translational processing of CX3CL1, versus its transcription, may be modified upon whisker lesioning. Within our scRNAseq data set, we identified Adam10, a metalloprotease previously shown to be regulated by neuronal activity30 and known to cleave CX3CL1 (Fig. 7a)29, as specifically increased in Rorb+ layer IV neurons and microglia vs. other cell types upon whisker lesioning (Fig. 7b–c and Supplementary Fig. 9). We validated these scRNAseq results and demonstrated that Adam10 mRNA increases in Rorb+ layer IV neurons in the deprived barrel cortex relative to control by in situ hybridization (Fig.7d–e). We then assessed microglial Adam10 expression by in situ in the barrel cortex (Fig. 7g–i). Similar to analysis of Rorb+ neurons (Fig. 7e), we averaged the data across all the microglia. While this analysis revealed no significant difference in total microglial Adam10 expression following whisker lesioning (Fig. 7h), we observed a subset of microglia that had high Adam10 in the deprived cortex (Fig. 7g). Therefore, we binned the data based on Adam10 puncta per cell and found that microglia with the highest expression of Adam10 (>15 puncta/cell) were significantly increased in the deprived vs. control barrel cortex (Fig. 7i).
To further validate upregulation of Adam10, we performed qPCR from whole barrel cortex and observed a similar increase in total Adam10 mRNA 24 h post-whisker lesioning (Fig. 7f). Analysis of bulk RNAseq from whole barrel cortex (Fig. 1d–f) showed a similar effect (Fig 7j–k). These RNAseq data also revealed increases in molecules known to interact with and regulate ADAM10 activity at the membrane (e.g., tetraspanins (Tspn5 and Tspn14); Fig 7k). These data demonstrate that ADAM10, a protease that cleaves CX3CL1 into a secreted form, is induced in layer IV excitatory neurons and a subset of microglia within the barrel cortex upon sensory lesioning.
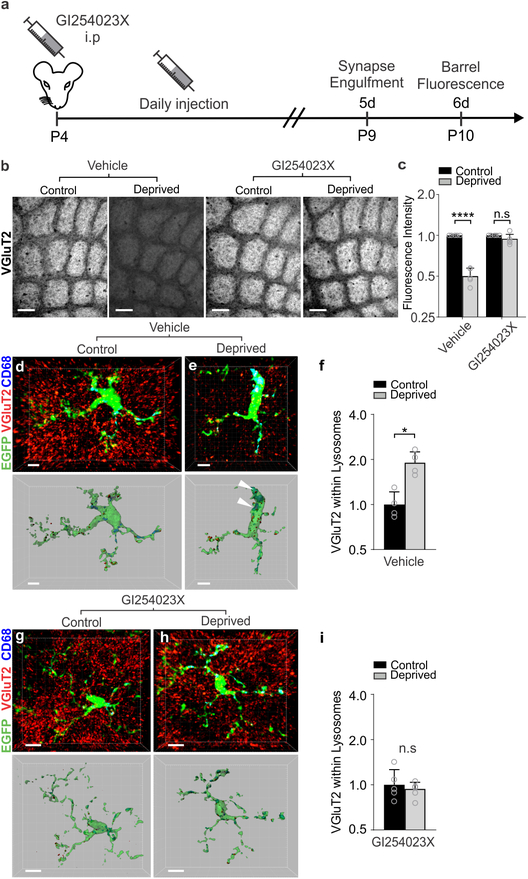
Pharmacological inactivation of ADAM10 phenocopies TC synapse elimination defects in Cx3cr1−/− and Cx3cl1−/− mice
To determine if induction of Adam10 in response to whisker lesioning translated to circuit level changes in TC synapse remodeling as observed in Cx3cr1−/− and Cx3cl1−/− mice, we pharmacologically inhibited ADAM10. Whiskers were lesioned at P4 and a pharmacological inhibitor of ADAM10 (GI254023X, 25 mg/kg), which was previously demonstrated in vivo to cross the blood brain barrier and specifically inhibit ADAM10 vs. other ADAMs31–33, was administered intraperitoneally (IP) daily starting at the time of whisker lesioning (Fig. 8a). TC synapse engulfment and elimination were subsequently assessed in the control and deprived barrel cortices. Identical to Cx3cr1−/− and Cx3cl1−/− mice, daily administration of the ADAM10 inhibitor (25mg/kg) by IP injection (P4–P10) resulted in significant disruption of TC input elimination following whisker lesioning (Fig. 8b–c) and blockade of microglia-mediated engulfment of TC inputs (Fig. 8d–i). These data suggest a mechanism by which post-translational modification of CX3CL1 by ADAM10 is increased in neurons following sensory lesioning. This secreted CX3CL1 then signals to microglia via CX3CR1 to eliminate synapses.
Figure 8. Pharmacological inactivation of ADAM10 phenocopies TC synapse elimination defects in Cx3cr1−/− and Cx3cl1−/− mice.
a, Timeline for pharmacological inhibition of ADAM10 via daily 25 mg/kg GI254023X injections intraperitoneally. b, Inhibition of ADAM10 (right panels), but not vehicle treatment (left panels), blocks TC input loss as visualized by immunostaining for VGluT2. Scale bars, 150 μm. c, Quantification of VGluT2 immunostaining intensity 5 days-post whisker lesioning and GI254023X injections (Two-Way ANOVA with Sidak’s post hoc, n = 5 Cx3cr1+/+ animals per condition, Vehicle control vs deprived, P <0.0001, t = 6.782, df = 16; GI254023X control vs deprived, P = 0.9715, t = 0.7789, df = 16). d,e, Representative microglia from the control (d) and deprived (e) cortices of vehicle treated Cx3cr1EGFP/+ mice. Top panel displays raw fluorescent image with microglia (EGFP, green) VGluT2 inputs (red) and lysosomes (Anti-CD68, blue) labeled. Bottom panels shows 3D-surface rendering of these cells. Engulfed VGluT2 (red) immunoreactive TC inputs within microglia are visualized in Cx3cr1EGFP/+ microglia (e, arrows)) in the deprived cortex but not the control cortex (d). Scale bars, 5 μm. f, Quantification of engulfed VGlut2 5 days after whisker lesioning reveals increased engulfment in the deprived cortex of vehicle treated mice. Data normalized to engulfment in the control hemisphere within each animal. (One-tailed Student’s T-test, n = 4 Cx3cr1EGFP/+ mice, control vs deprived P = 0.0455, t = 2.012, df = 6.) g,h Representative microglia from the control (d) and deprived (e) cortex of GI254023X treated Cx3cr1EGFP/+ mice 5 days after whisker lesioning. Scale bars, 5 μm. i, Quantification of engulfment 5 days-post whisker lesioning in GI254023X treated mice reveals a blockade of engulfment following ADAM10 inhibition. Data normalized to engulfment in the control hemisphere within each animal. (Two-tailed Student’s T-test, n = 4 Cx3cr1EGFP/+ mice, control vs deprived P = 0.8291, t = 0.2231, df = 6). All data presented as mean ± SEM.
DISCUSSION
We have identified a novel mechanism by which neurons signal to microglia to remodel synapses in response to sensory loss and dampened neural activity. We show that whisker lesioning and trimming induces microglial synaptic engulfment and synapse elimination via CX3CR1-CX3CL1, but not CR3, signaling. We further show by scRNAseq and other validation methods that Cx3cl1 is enriched in cortical neurons and Adam10, a gene encoding a metalloprotease known to cleave CX3CL1 into a secreted form, is specifically induced in layer IV excitatory neurons and a subset of microglia following sensory lesioning. As the first single-cell transcriptomic study in the developing barrel cortex, this is a rich dataset and likely a valuable resource for assessing gene expression in glia and neurons. Finally, to further support our scRNAseq findings, pharmacological inhibition of ADAM10 phenocopies TC synapse elimination defects observed in CX3CR1 and CX3CL1-deficient mice. Together, our data suggest a mechanism by which post-translational processing of CX3CL1 in neurons by ADAM10 is regulated by changes in neural activity following a peripheral sensory lesion. Cleaved CX3CL1 can then signal to microglia to initiate engulfment and elimination of synapses via CX3CR1. These data provide a new mechanism by which thalamocortical synapses remodel in response to peripheral sensory loss several synapses away.
Previous studies have identified that microglia contact and/or engulf synaptic elements and these interactions are activity-dependent1,2,23. However, the mechanisms underlying these activity-dependent microglia-synapse interactions and the neuron-derived signals driving this process were largely unknown. Another transcriptional profiling study has demonstrated that, under basal conditions, microglia express a number of transcripts for sensing endogenous ligands and microbes, which was termed the ‘sensome’34. Our data provide a role for one key microglial ‘sensome’ gene Cx3cr1 in synapse elimination following manipulation of circuit activity via whisker lesioning or trimming. This is consistent with work in a seizure model suggesting CX3CR1 can modulate microglia-synapse contact in response to activity35. CX3CR1 is enriched in microglia versus other CNS resident neural and glial cell types and we show that microglial synaptic engulfment is primarily affected following sensory lesioning in Cx3cr1−/− mice. Thus, our results point most directly towards microglial CX3CR1 signaling regulating engulfment and synapse elimination. Although less likely, it is important to consider the contribution of other tissue resident CX3CR1-expressing macrophages (e.g. skin, perivascular, etc.). This would require the development of new mice to specifically ablate CX3CR1 in microglia versus these other macrophage populations.
Our study raises a new question regarding how upstream and downstream CX3CR1 signaling is regulating microglial engulfment and synapse elimination. Upstream of CX3CR1, we identify a new role for neuron-derived CX3CL1 and ADAM10 in microglia-mediated synapse elimination. Going forward, it will be important to elucidate precisely how ADAM10 is modulated in neurons and microglia following sensory lesioning. This includes developing new tools to monitor CX3CL1 cleavage by ADAM10 in specific cells. Downstream of CX3CR1, our scRNAseq data provide some evidence that there is a downregulation of genes related to phagocytosis in Cx3cr1−/− microglia, even without whisker lesioning (Supplementary Fig. 12). This raises the possibility that CX3CR1 signaling could modulate basal phagocytic state through transcriptional regulation. In addition, our scRNAseq revealed that microglia, as well as other glia, modify their gene expression in response to sensory lesioning, which is largely CX3CR1-dependent. While these gene expression changes require further validation, this is an exciting first step in understanding how microglia and other glial cells respond to a changing sensory environment.
One important theme that emerges from our data is that different neural-immune signaling mechanisms are utilized by microglia to engulf and remodel synaptic connections and these mechanisms appear to be engaged in a context-dependent manner. CX3CR1 has been identified to regulate synapse maturation in the developing hippocampus and barrel cortex11,12. Work in the developing mouse retinogeniculate system has identified MHC class I molecules36,37 and microglial complement-dependent phagocytic signaling2,4 as key regulators of developmental synaptic pruning. MHC class I molecules36 and microglial P2RY1224 regulate ocular dominance plasticity in the visual cortex. Interestingly, retinogeniculate pruning and ocular dominance plasticity occur independent of CX3CR110,38. In contrast, we show that sensory lesion-induced elimination of synapses in the barrel cortex is regulated by CX3CR1-CX3CL1 signaling, but not CR3. While our TC synapse elimination defects in CX3CR1 and CX3CL1-deficient mice are robust, there may be additional pathways that are working with the CX3CR1-CX3CL1 axis that would be worthy of follow-up investigation. One intriguing pathway recently identified to regulate astrocyte-microglia phagocytic crosstalk is IL-3339. In addition, our static imaging suggests that there may be heterogeneity in levels of phagocytosis across different microglia, which may be regulated by different immune pathways. While this could be reflected in the nature of static imaging and analyzing fixed time points, it is an interesting possibility to explore.
Another important point to consider is that differences in immune signaling mechanisms may reflect differences in brain regions, types of synapses, paradigm used for eliciting synaptic changes, or activity patterns. For example, the whisker lesioning paradigm used here induces injury to the peripheral sensory endings in the snout several synapses away from the barrel cortex. Therefore, this may be an injury vs. purely an activity-dependent response. However, there are several lines of evidence arguing against this being an injury response. First, there is no change in neuronal cell death or degeneration following whisker lesioning at P4. Second, there is a similar, albeit delayed, CX3CR1-depenedent synapse elimination following whisker trimming. Last, our RNAseq data demonstrate there is a downregulation in genes related to neural activity. These data are most consistent with cortical activity modulating CX3CR1-CX3CL1 signaling via ADAM10 and microglia-mediated synapse elimination.
A broad range of neurological disorders from autism and schizophrenia to traumatic injuries causing loss of eye sight, cutaneous sensation, olfaction or hearing all result in changes in sensory inputs and TC connectivity40–42. Further, defects in CX3CR1-CX3CL1 signaling and ADAM10 have been identified to either enhance or suppress neurodegeneration in a variety of neurological disease models depending on the disease, insult, brain region, etc.43–45. This includes a recent genome-wide association meta-analysis demonstrating ADAM10 as an important late-onset Alzheimer’s disease loci46. Here, we identify that neuronal and microglial ADAM10 is modulated in the cortex by sensory lesioning and disruption of ADAM10, CX3CL1 (an ADAM10 substrate), or CX3CR1 results in profound defects in microglial synaptic engulfment and synapse elimination. This mechanism is particularly intriguing in light of recent data in mouse models of Alzheimer’s disease where changes in neural activity modulates microglial morphology, Aβ, and amyloid plaques47,48. Together, our findings have strong implications for advancing our understanding of how neural activity drives circuit remodeling via microglia and CX3CR1-CX3CL1-ADAM10 signaling. These insights are important for our basic understanding of how neurons communicate with microglia to modulate neural circuits and have significant translational potential for a variety of neurological disorders.
Methods
Animals
SERT-Cre mice were a generous gift from Dr. Mark Ansorage, Columbia University and provided by Dr. Sacha Nelson, Brandeis University. Cx3cl1−/− mice were provided by Dr. Sergio Lira (Ichan School of Medicine, Mount Sinai). Rosa26-TdTomato mice (Ai14; stock #007914), Cx3cr1−/− mice (Cx3cr1EGFP/EGFP; stock #005582), CR3-KO mice (stock #003991), and C57Bl6/J (stock #000664) mice were obtained from Jackson Laboratories (Bar Harbor, ME). Heterozygous breeder pairs were set up for all experiments and wild-type and heterozygote littermates were used as controls, which included males and females for each genotype. All experiments were performed in accordance with animal care and use committees and under NIH guidelines for proper animal welfare.
Whisker Removal
For sensory lesioning, whiskers were unilaterally removed at P4 with a high temperature handheld cautery kit (Bovie Medical Corporation; Clearwater, FL) applied to the right whisker pad of anesthetized pups. After whisker removal, pups were placed on a heating pad before animals were returned to their home cage. All animals within one experimental litter were cauterized at the same time blind to animal genotype. For long term whisker deprivation, whiskers were removed at P4 and assessed for whisker re-growth weekly. For whisker trimming experiments, whiskers were unilaterally trimmed using fine surgical scissors (No. 15003–08, Fine Science Tools; Foster City, CA) from the right whisker pad starting at P4. Whiskers were trimmed twice daily to minimize whisker growth up to P21.
Immunohistochemistry
Animals were perfused with 4% paraformaldehyde in 0.1M phosphate buffer (PB) prior to brain removal. In order to visualize the entire barrel field in one plane, the midbrain was dissected from each brain hemisphere and the cortex was then flattened between two slides in 4% paraformaldehyde overnight. Sections were placed in 30% sucrose in 0.1M PB for 24 hours before 50μm (P5–P11) and 40μm (>P40) tangential sections were prepared. Sections were blocked in 10% goat serum, 0.01% TritonX-100 in 0.1M PB for 1 hour before primary immunostaining antibodies were applied overnight. For analysis of synaptic engulfment within microglia, anti-CD68 (1:1000; cat# MCA1957, lot# 1708; AbD Serotec; Raleigh, NC)2 and anti-VGluT2 (1:2000; cat# AB2251, lot# 3101508; MilliporeSigma; Darmstadt, Germany)12 were used. Microglia were labeled either using transgenic expression of EGFP (CX3CR1EGFP/+) or immunostaining for anti-Iba-1 (1:1000; cat# 019–18741, lot# PTR2404; Wako Chemicals; Richmond, VA)2. For analysis of synapse density, anti-VGluT2 (1:2000; cat# AB2251, lot# 3101508; MilliporeSigma; Darmstadt, Germany)12 and anti-Homer1 (1:1000; cat#160003, lot# 1–47; Synaptic Systems; Goettingen, Germany)7,49 were used. For markers of neurodegeneration, cell death, and cell stress, anti-APP (1:1000; cat# 51–2700, lot# SA243371; ThermoFisher Scientific; Waltham, MA)50, anti-Cleaved Caspase 3 (1:200; cat# 9661; lot# 45; Cell Signaling Technology; Danvers, MA)51, and anti-ATF3 (1:500; cat# HPA001562, lot# B116285; Sigma-Aldrich; Darmstadt, Germany)52 were used. Anti-NeuN (1:1000; cat# ABN91, lot# 170621; MilliporeSigma; Darmstadt, Germany)53 was used as a marker for neuronal cell bodies. Peripheral monocytes/macrophages were labelled with anti-CD45 (Bio-Rad; Hercules, CA) and anti- F4/80 (1:1000; cat# MA-91124, lot# SJ2459832O; ThermoFisher Scientific; Waltham MA)54. For validating specificity of CX3CR1-EGFP expression before and after whisker lesioning, markers for NG2 cells (anti-NG2; 1:200; cat# AB5320, lot# 3061186; MilliporeSigma; Darmstadt, Germany)55, astrocytes (anti- ALDH1L1 clone N103/39; 1:1000; cat# MABN495, lot# 2943620; MilliporeSigma; Darmstadt, Germany)56, microglia (anti- P2RY12; 1:100; cat# 848002, lot# B244070; BioLegend; San Diego CA)57, and neurons (anti- NeuN; 1:1000; cat# ABN91, lot# 170621; MilliporeSigma; Darmstadt, Germany)53 were used. Anti-ALDH1L1 clone N103/39 required a 20 minute incubation step with L.A.B antigen retrieval solution (Polysciences, Inc; Warrington, PA) prior to blocking with 10% goat serum, 0.01% TritonX-100 in 0.1M PB for 1 hour.
Fluorescence Intensity Analysis
For fluorescence intensity, single plane 10x epifluorescence images were collected at the same exposure time with a Zeiss Observer microscope equipped with Zen Blue acquisition software (Zeiss; Oberkochen, Germany). One field of view containing all the barrels was collected per hemisphere per animal. Fluorescence intensity within each barrel was quantified similar to what has been described previously19. Briefly, each image was analyzed in ImageJ (NIH) where first all image pixel intensity thresholds were set to the full range of 16-bit images before quantification to ensure a consistent pixel range across all images. To sample fluorescence intensity, a circular region of interest (ROI) 75μm in circumference (4470.05 μm2 area) was placed within the center of 15–20 barrels per 10x field of view. A background ROI outside of the barrel field was also taken for each image. The raw integrated density of pixels within each ROI was measured and each barrel intensity value was background-corrected by subtraction of the background ROI pixel intensity. Average intensity over all barrel ROIs was quantified for each image and then normalized to the control hemisphere within each animal. All data analyses were performed blind to genotype.
Engulfment Analysis
Engulfment analysis was performed according to previously described methods2,58. Briefly, immunostained sections were imaged on a Zeiss Observer Spinning Disk Confocal microscope equipped with diode lasers (405nm, 488nm, 594nm, 647nm) and Zen acquisition software (Zeiss; Oberkochen, Germany). For each hemisphere, three to five 63x fields of view within the barrel field were acquired with 50–70 z-stack steps at 0.27 μm spacing. Images were first processed in ImageJ (NIH) and then individual images of 15–20 single cells per hemisphere per animal were processed in Imaris (Bitplane; Zurich, Switzerland) as previously described. All image files were blinded for unbiased quantification. All data was then normalized to the control, spared hemisphere within each animal. Note, Cx3cr1+/− littermates were used for comparison to Cx3cr1−/− mice as microglia within both sets of mice are labeled with EGFP and show similar changes in TC synapses (Fig 2).
Synapse Density Analysis
Synapse density analysis was performed blind to condition and genotype as described previously2,7. Briefly, immunostained sections were imaged on a Zeiss LSM700 scanning confocal microscope equipped with 405nm, 488nm, 555nm, and 639nm lasers and Zen acquisition software (Zeiss; Oberkochen, Germany). Synapse density analysis was performed on single plane confocal images in ImageJ (NIH). Three 63x fields of view per hemisphere per animal were analyzed. Sample images for each genotype and condition were manually thresholded by eye, and a consistent threshold range was determined (IsoData segmentation method, 85–255). Each channel was thresholded and the Analyze Particles function (ImageJ plugin, NIH) with set parameters for each marker (VGluT2 = 0.2-infinity; Homer1 = 0.1-infinity) was used to measure the total pre- and postsynaptic puncta area. To quantify total synaptic area, Image Calculator (ImageJ plugin, NIH) was used to visualize co-localized pre- and postsynaptic puncta and then the Analyze Particles function was used to calculate the total area of co-localized puncta. Data for each hemisphere was averaged across all three fields of view and then normalized to the control hemisphere within each animal.
Bulk RNA sequencing
Mice were killed by CO2 asphyxiation at indicated ages, and brain regions of interest were dissected. Brain tissue from one mouse was immediately homogenized with a motor-driven Teflon glass homogenizer in ice-cold polysome extraction buffer (10 mM HEPES (pH 7.3), 150 mM KCl, 5 mM MgCl2, 0.5 mM dithiothreitol (Sigma) 100 μg/mL cycloheximide (Sigma), EDTA-free protease inhibitor cocktail (Roche), 10 μL/mL RNasin (Promega) and Superasin (Applied Biosystems). Homogenates were centrifuged for 10 min at 2,000g, 4 °C, to pellet large cell debris. NP-40 (EMD Biosciences, CA) and 1,2-diheptanoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids, AL) were added to the supernatant at final concentrations of 1% and 30 mM, respectively. After incubation on ice for 5 min, the lysate was centrifuged for 10 min at 13,000g to pellet insoluble material. RNA was purified from the lysate using RNeasy Mini Kit (Qiagen) following the manufacturer’s instructions. RNA integrity was assayed using an RNA Pico chip on a Bioanalyzer 2100 (Agilent, Santa Clara, CA), and only samples with RIN > 9 were considered for subsequent analysis. Double-stranded cDNA was generated from 1–5 ng of RNA using Nugen Ovation V2 kit (NuGEN, San Carlos, CA) following the manufacturer’s instructions. Fragments of 200 bp were obtained by sonicating 500 ng of cDNA per sample using the Covaris-S2 system (duty cycle: 10%, intensity: 5.0, bursts per second: 200, duration: 120 s, mode: frequency sweeping, power: 23 W, temperature: 5.5–6 °C; Covaris Inc., Woburn, MA). Subsequently, these fragments were used to produce libraries for sequencing by TruSeq DNA Sample kit (Illumina, San Diego, CA, USA) following the manufacturer’s instructions. The quality of the libraries was assessed by 2200 TapeStation (Agilent). Multiplexed libraries were directly loaded on NextSeq 500 (Illumina) with high-output single-read sequencing for 75 cycles. Raw sequencing data was processed using Illumina bcl2fastq2 Conversion Software v2.17. Raw sequencing reads were mapped to the mouse genome (mm9) using the TopHat2 package (v2.1.0). Reads were counted using HTSeq-count (v0.6.0) against the Ensembl v67 annotation. The read alignment, read count, and quality assessment using metrics, such as total mapping rate and mitochondrial and ribosomal mapping rates, were done in parallel using an in-house workflow pipeline called SPEctRA. The raw counts were processed through a variance stabilizing transformation (VST) procedure using the DESeq2 package to obtain transformed values that are more suitable than the raw read counts for certain data mining tasks. Principal component analysis (PCA) was performed on the top 500 most-variable genes across all samples based on the VST data to visually assess whether there were any outliers. Additionally, hierarchical clustering was used to assess the outliers once again to protect against false positives/negatives from PCA, and the outliers were further justified by the aforementioned quality control metrics as well as experimental metadata. After outlier removal, all pairwise comparisons were performed on the count data of entire gene transcripts using the DESeq2 package (v1.6.3). A cutoff of adjusted P value < 0.05, and mean expression > 3.5. MA plot was made using R (v3.1.1; https://www.R-project.org). For the heatmap, expression of each gene in log2(RPKM) was normalized to the mean across all samples (z-scored). Heatmap with hierarchical clustering was made on Multiple Experiment Viewer 4.8 (v.10.2; http://www.tm4.org/) with Pearson correlation by average link clustering. Bar graphs representing RPKM of genes and line plots representing z-scored log2(RPKM) values were made on GraphPad Prism v5.01 (https://www.graphpad.com).
Single-cell transcriptomics experimental design
Non-deprived, control and deprived barrel cortices from a total of 8 mice served as the input for single-cell sequencing studies, equaling 16 samples total. The capture and barcoding of single cells was performed across four separate experiments on two different days. Per experiment, one littermate each of the genotypes Cx3cr1+/− and Cx3cr1−/− underwent unilateral whisker cauterization at P4 and their deprived and non-deprived cortices were then processed in parallel at P5, yielding 4 samples per experiment. 3,000 cells were collected from each hemisphere of each mouse, equaling a total of ~48,000 cells collected overall. All libraries were prepared in parallel then pooled and sequenced together across two sequencing runs, as described below.
Generation of single-cell suspensions
Single-cell sequencing of mouse cortex using inDrops was performed as previously described28. Mice were transcardially perfused with ice-cold choline solution containing 2.1 g NaHCO3 per liter, 2.16 g glucose per liter, 0.172 g NaH2PO4∙H2O per liter, 7.5 mM MgCl2∙6H2O, 2.5 mM KCl, 10 mM HEPES, 15.36 g choline chloride per liter, 2.3 g ascorbic acid per liter, and 0.34 g pyruvic acid per liter (all chemicals, Sigma). A caveat of sequencing techniques that involve mechanical and enzymatic dissociation is that the dissociation itself induces neural activity-dependent and injury-induced gene transcription. Therefore, because we were interested in analyzing sensory experience-dependent changes in gene expression across all cell types, a number of drugs that block neuronal activity and transcription were included in the perfusion solution. These included 1 μM TTX (Sigma), 100 μM AP-V (Thermo Fisher Scientific), 5 μg actinomycin D (Sigma) per milliliter, and 10 μM triptolide (Sigma). Following the 5 minute perfusion, deprived and non-deprived, control somatosensory cortices were microdissected and each sample was transferred to a tube with 1.65 mL of pre-incubation solution containing HBSS (Life Technologies), 10 mM HEPES, 172 mg kynurenic acid (Sigma) per liter, 0.86 g MgCl2∙6H2O per liter, and 6.3 g D-glucose (Sigma) per liter, pH 7.35, which was saturated with 95% O2 and 5% CO2 previously but was not bubbled after the sample was added. The drugs contained in the perfusion solution were also present in the pre-incubation solution at the same concentrations. After 30 minutes on ice, 1.65 mL of papain (Worthington) was added to a final concentration of 20 U per milliliter. Samples were moved to a rocker at 37° C and gently rocked for 60 minutes.
Following the papain incubation at 37° C, a series of triturations in increasingly small volumes were performed to fully dissociate the tissue. In between each round of trituration, the tissue was filtered through the corner of a 40 μM nylon cell strainer (Corning). The cells were then centrifuged at 300*g for 5 minutes and the pellet resuspended in 1 mL of trypsin inhibitor (Worthington) plus DNAse (Sigma) in preincubation solution without drugs (dissociation media, DM). The cells were washed by resuspension in DM adjusted to 0.04% BSA (Sigma) three times, then resuspended in DM containing 0.04% BSA and 15% Optiprep (Sigma) to a concentration of 100,000 cells per mL and transferred to the Single-Cell Core at Harvard Medical School for inDrops collection (see Acknowledgements).
Single-cell RNA sequencing via inDrops
For each sample, approximately 3,000 cells were encapsulated into microfluidic droplets containing polyacrylamide gels with embedded barcoded reverse transcription primers. Reverse transcription was carried out in intact droplets to generate barcoded cDNA from a single cell. Following droplet lysis, inDrops libraries were prepared as previously described27,28. All 16 libraries were indexed, pooled, and sequenced (Read 1: 54 cycles, Read 2: 21 cycles, Index 1: 8 cycles, Index 2: 8 cycles) across 2 runs on a NextSeq 500 (Illumina) with an average read depth across biological replicates of 8,815 reads per cell.
inDrops data processing
Sequenced reads were processed according to a previously published pipeline59,60. Briefly, this pipeline was used to build a custom transcriptome from Ensembl GRCm38 genome and GRCm38.84 annotation using Bowtie 1.1.1, after filtering the annotation gtf file (gencode.v17.annotation.gtf filtered for feature_type=”gene”, gene_type=“protein_coding” and gene_status=“KNOWN”). Read quality control and mapping against this transcriptome were performed. Unique molecular identifiers (UMIs) were used to link sequence reads back to individual captured molecules. All steps of the pipeline were run using default parameters unless explicitly stated.
Quality control and clustering of cells
All cells were combined into a single dataset. Nuclei with >10% mitochondrial content were excluded from the dataset. Cells with fewer than 400 UMI counts were excluded. Cells were then clustered using the Seurat R package61. The data were log normalized and scaled to 10,000 transcripts per cell. Variable genes were identified using the following parameters: x.low.cutoff = 0.0125, x.high.cutoff = 3, y.cutoff = 0.5. We limited the analysis to the top 30 principal components. Clustering resolution was set to 0.6. The expression of known marker genes was used to assign each cluster to one of the main cell types. Tubb3/Snap25 was used to identify neurons, Gad1/Gad2 was used to identify inhibitory neurons, Rorb was used to identify excitatory cortical layer IV neurons, Aldoc/Aqp4 was used to identify astrocytes, Mbp/Plp1 was used to identify mature oligodendrocytes, Pdgfra/Matn4 was used to identify immature oligodendrocytes, and P2ry12/C1qa was used to identify microglia. The analysis reported here focused on the glial population and layer IV neurons, for which the following numbers of cells per condition passed quality control filters and were included in subsequent analyses. For cell numbers see Supplemental Figure 9g.
Identification of differentially-expressed genes
Differential gene expression analyses were performed in pairwise fashion between each of the four groups using the R package Monocle262. The data were modeled using a negative binomial distribution consistent with data generated by high-throughput single-cell RNA-seq platforms such as inDrops. Unlike deep single-cell sequencing, inDrops probabilistically captures/samples the transcriptome of each cell and retrieves only a small fraction of all the present transcripts. Genes whose differential gene expression false discovery rate (FDR) was less than 0.10 (FDR < 0.10) were considered statistically significant.
Cell Counts
For microglia cell counts, single plane 10x epifluorescence images were collected within the barrel cortex at the same exposure time with a Zeiss Observer microscope equipped with Zen Blue acquisition software (Zeiss; Oberkochen, Germany). Images of entire barrel fields from 10x images were quantified blind to genotype in ImageJ (NIH). Each individual barrel per field of view was outlined and grouped as a single ROI. The same ROI was transposed to the thresholded microglia channel where the number of microglia in the barrels was quantified by counting the number of cells within the total barrel ROI. The entire perimeter of the barrel field was then outlined and the number of microglia over the entire barrel field area was analyzed. The number of microglia within the septa was quantified by subtracting the total number of microglia within the barrels from the total number of microglia within the entire S1 ROI. The level of microglia infiltration into the barrel field was quantified by calculating a ratio of the total number of microglia in the barrels divided by the total number of microglia within the septa for each time point. Note, Cx3cr1+/− littermates were used for comparison to Cx3cr1−/− mice as microglia within both sets of mice are labeled with EGFP and show similar changes in TC synapses (Fig 2). Similar results were obtained in wild-type mice labeling microglia with Iba-1 (data not shown).
For ATF3/Caspase-3/APP positive cell counts, single plane 20x (for trigeminal nerve ganglia) and single plane 10x (for thalamic VPM and primary somatosensory cortex) were taken. The number of NeuN positive and ATF3/Caspase 3/APP positive cells for each plane were counted and divided by the total number of NeuN positive cells and normalized to the total field of view area. For peripheral macrophage infiltration to the whisker follicles following cauterization, single plane 20x images were taken. The number of CX3CR1-EGFP positive and CD45 positive cells within a given whisker follicle were counted and normalized to the total area of the whisker follicle.
In situ RNA hybridization
In situ RNA hybridization was performed according to the manufacturer’s specification with slight modifications (ACDBio; Newark, CA). Briefly, mice were perfused with 4% PFA and brains were post-fixed for 24 hours. 10μm cryo-sections were prepared and stored in −80°C. Prior to in situ hybridization, cryo-sections were equilibrated to room temperature for 1 hour, then were dehydrated in a serial dilution of ethanol. Sections were incubated in hydrogen peroxide for 10 minutes and rinsed with RNase free water. Sections were treated with “Protease Plus” for 15 minutes at room temperature and rinsed with phosphate buffered saline. In situ probes were added and incubated for 2 hours at 40°C. Subsequent amplification steps were performed according to the manufacturer’s specification. Slices were immunostained following in situ hybridization. Sections were washed in 1X PBS for 10 minutes and blocked in 0.01% TritonX-100 and 2% normal goat serum for 30 minutes. Primary antibody (NeuN; Millipore) prepared in 0.01% TritonX-100 and 2% normal goat serum and slides were incubated overnight at room temperature. Secondary antibody was prepared in 0.01% TritonX-100 and 2% normal goat serum and incubated at room temperature for 2 hours. For Cx3cl1 in situ quantification, fluorescent RNA signal was localized to DAPI and NeuN positive or DAPI and NeuN negative cells with a MATLAB script (custom script made in vR2016b, available upon request). Individual channels were segmented with a set threshold for dilating the masked signal around the NeuN positive or DAPI positive channels. For Adam10 in situ quantification, fluorescent RNA signal was co-localized to NeuN positive or Cx3cr1EGFP positive cells and the number of puncta co-localized to either signal was measured with Image J (NIH).
Structured Illumination Microscopy imaging
Immunostained sections were prepared as described above. Images were acquired on a GE Healthcare DeltaVision OMX microscope for structured illumination microscopy. Images were then processed in Imaris v 8.2.1 (Bitplane; Zurich,Switzerland) in order to 3D reconstruct the cell and visualize engulfed material.
Slice Preparation and Electrophysiological Recordings
Male mice (~3 months old) were anesthetized by intraperitoneal injection of sodium pentobarbital (200 mg/kg) and then were decapitated. The brain was quickly removed and placed in an oxygenated ice-cold cutting solution containing (in mM): 2.5 KCl, 1.25 NaH2PO4•H2O, 20 HEPES, 2 Thiourea, 5 Na-ascorbate, 92 NMDG, 30 NaHCO3, 25 D-Glucose, 0.5 CaCl2•2H2O and 10 MgSO4•7H2O. Brain slices (200 μM) were made using a Leica VT1200 vibratome (Leica Biosystems Inc.). The brain slices were immediately transferred into an incubation chamber containing oxygenated cutting solution at 34°C for 20 minutes. Slices were transferred into oxygenated artificial cerebrospinal fluid (ACSF) at room temperature (24°C) for recording. ACSF solution contains (in mM): 125 NaCl, 2.5 KCl, 1.2 NaH2PO4•H2O, 1.2 MgCl2•6H2O, 2.4 CaCl2•2H2O, 26 NaHCO3, 11 D-Glucose. Slices were left in this chamber for at least 1 hour before being placed in a recording chamber at room temperature. Single slices were transferred into a recording chamber continually superfused with oxygenated ACSF (30 – 32°C) at a flow rate of ~2 ml/min for recording. Cells were visualized using infrared differential interference contrast (IR-DIC) imaging on an Olympus BX-50WI microscope. Electrophysiological recordings were recorded using an Axon Multiclamp 700B patch-clamp amplifier (Molecular Devices). Spontaneous excitatory postsynaptic current (sEPSC) was acquired in the whole cell configuration and gap-free acquisition mode in Clampex (Axon Instruments). Neurons were held at a membrane potential of −70 mV. Signals were filtered at 1 kHz using the amplifier’s four-pole, low-pass Bessel filter, digitized at 10 kHz with an Axon Digidata 1440A interface and stored on a personal computer. Pipette solution contained (in mM) 120 K gluconate; 5 KCl; 2 MgCl2•6H2O 10 HEPES; 4 ATP, 2 GTP. sEPSCs were recorded in the presence of bicuculline (20 μM) in the bath solution to block GABAA receptors.After recordings stabilized, 1 min duration of recording was taken for sEPSC analysis. sEPSC frequency and amplitude was detected using Mini Analysis Program (Synaptosoft Inc. Fort Lee, NJ). All recordings and quantification were performed blind.
Pharmacological Inhibition of ADAM10
C57Bl/6J (fluorescence intensity analysis) or Cx3cr1EGFP/+ (microglia engulfment analysis) mice were daily injected intraperitoneally with 25 mg/kg GI254023X (Millipore-Sigma, Darmstadt, Germany) following whisker cauterization at P4. The drug was prepared daily in 0.1M carbonate buffer/10% DMSO vehicle. Control littermate animals were vehicle injected following the same drug schedule for both fluorescence intensity and microglia engulfment analyses. Animals within each strain and within each litter were randomly assigned to either drug or vehicle treatment group.
Statistics and Reproducibility
GraphPad Prism v7.01 and v5.01 (La Jolla, CA) provided the platform for all statistical and graphical analyses. All data sets were first tested and found to be normally distributed and parametric stats were subsequently run. Analyses included Students t-test when comparing 2 conditions or two-way ANOVA followed by Sidak’s, Tukey’s, or Dunnett’s post hoc analyses (indicated in figure legends). For population data (percentage of cells expressing or binned data) a two-tailed Fisher’s exact test or a Chi-square was used (indicated in figure legends). All p and n values are specified within each figure legend. No statistical tests were used to predetermine sample sizes but our sample sizes are similar to those reported in previous publications2,7,27. All images shown in figures are representative images of data quantified in corresponding graphs. For all whisker deprivation experiments, each animal served as its own internal control and relative changes were normalized to the spared, control hemisphere.
Supplementary Material
Acknowledgments
We thank M. Freeman (OHSU), V. Budnik (UMMS), E. Baehrecke (UMMS), M. Francis (UMMS), P. Greer (UMMS), and R. Bruno (Columbia University) for critical reading of the manuscript. The Cx3cl1−/− mice and SERT-Cre mice were generously provided by S. Lira (MSSM) and M. Ansorage/S. Nelson (Columbia University/Brandeis University), respectively. We thank M. Cahill (UMMS) and A. Lotun (UMMS) for assistance assessing microglia within the barrel cortex and S. Becker (UMMS) and J. Jung (UMMS) for assistance with tissue preparation and whisker trimming experiments. H. Learnard (UMMS), A. Song (UMMS), Z. Zhang (Boston Children’s Hospital), and C. Woolf (Boston Children’s Hospital) for assistance with experiments to assess ATF3. We thank D. Bergles (John’s Hopkins) for advice and discussions related to identification of OPCs by Matn4 expression in the single-cell RNAseq data set. This work was funded by NIMH-R00MH102351 (DPS), NIMH- R01MH113743 (DPS), NIMH- R21MH115353 (DPS and AS), Charles H. Hood Foundation (DPS), Brain & Behavior Research Foundation (DPS), Worcester Foundation (DPS), and the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (DPS).
Footnotes
Accession Codes
The data discussed in this publication has been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE129150 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE129150).
Declaration of Interests
Authors declare no conflict of interest.
Data availability
All data that support the findings, tools, and reagents will be shared on an unrestricted basis. All requests should be directed to the corresponding author.
References
- 1.Wu Y, Dissing-Olesen L, MacVicar BA & Stevens B Microglia: Dynamic Mediators of Synapse Development and Plasticity. Trends in immunology 36, 605–613 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schafer DP, et al. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 74, 691–705 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hong S, Dissing-Olesen L & Stevens B New insights on the role of microglia in synaptic pruning in health and disease. Current opinion in neurobiology 36, 128–134 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stevens B, et al. The classical complement cascade mediates CNS synapse elimination. Cell 131, 1164–1178 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Vasek MJ, et al. A complement-microglial axis drives synapse loss during virus-induced memory impairment. Nature 534, 538–543 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lui H, et al. Progranulin Deficiency Promotes Circuit-Specific Synaptic Pruning by Microglia via Complement Activation. Cell 165, 921–935 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hong S, et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science 352, 712–716 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolf Y, Yona S, Kim KW & Jung S Microglia, seen from the CX3CR1 angle. Frontiers in cellular neuroscience 7, 26 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schecter RW, et al. Experience-Dependent Synaptic Plasticity in V1 Occurs without Microglial CX3CR1. The Journal of neuroscience : the official journal of the Society for Neuroscience 37, 10541–10553 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lowery RL, Tremblay ME, Hopkins BE & Majewska AK The microglial fractalkine receptor is not required for activity-dependent plasticity in the mouse visual system. Glia 65, 1744–1761 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paolicelli RC, et al. Synaptic pruning by microglia is necessary for normal brain development. Science 333, 1456–1458 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Hoshiko M, Arnoux I, Avignone E, Yamamoto N & Audinat E Deficiency of the microglial receptor CX3CR1 impairs postnatal functional development of thalamocortical synapses in the barrel cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience 32, 15106–15111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhan Y, et al. Deficient neuron-microglia signaling results in impaired functional brain connectivity and social behavior. Nature neuroscience 17, 400–406 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Woolsey TA & Van der Loos H The structural organization of layer IV in the somatosensory region (SI) of mouse cerebral cortex. The description of a cortical field composed of discrete cytoarchitectonic units. Brain Res 17, 205–242 (1970). [DOI] [PubMed] [Google Scholar]
- 15.Van der Loos H & Woolsey TA Somatosensory cortex: structural alterations following early injury to sense organs. Science 179, 395–398 (1973). [DOI] [PubMed] [Google Scholar]
- 16.Fox K A critical period for experience-dependent synaptic plasticity in rat barrel cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience 12, 1826–1838 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glazewski S, McKenna M, Jacquin M & Fox K Experience-dependent depression of vibrissae responses in adolescent rat barrel cortex. The European journal of neuroscience 10, 2107–2116 (1998). [DOI] [PubMed] [Google Scholar]
- 18.Oberlaender M, Ramirez A & Bruno RM Sensory experience restructures thalamocortical axons during adulthood. Neuron 74, 648–655 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wimmer VC, Broser PJ, Kuner T & Bruno RM Experience-induced plasticity of thalamocortical axons in both juveniles and adults. The Journal of comparative neurology 518, 4629–4648 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Erzurumlu RS & Kind PC Neural activity: sculptor of ‘barrels’ in the neocortex. Trends in neurosciences 24, 589–595 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sadaka Y, Weinfeld E, Lev DL & White EL Changes in mouse barrel synapses consequent to sensory deprivation from birth. The Journal of comparative neurology 457, 75–86 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Erzurumlu RS & Gaspar P Development and critical period plasticity of the barrel cortex. The European journal of neuroscience 35, 1540–1553 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tremblay ME, Lowery RL & Majewska AK Microglial interactions with synapses are modulated by visual experience. PLoS biology 8, e1000527 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sipe GO, et al. Microglial P2Y12 is necessary for synaptic plasticity in mouse visual cortex. Nat Commun 7, 10905 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weinhard L, et al. Microglia remodel synapses by presynaptic trogocytosis and spine head filopodia induction. Nature communications 9, 1228 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim KW, et al. In vivo structure/function and expression analysis of the CX3C chemokine fractalkine. Blood 118, e156–167 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klein AM, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hrvatin S, et al. Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex. Nature neuroscience 21, 120–129 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hundhausen C, et al. The disintegrin-like metalloproteinase ADAM10 is involved in constitutive cleavage of CX3CL1 (fractalkine) and regulates CX3CL1-mediated cell-cell adhesion. Blood 102, 1186–1195 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Suzuki K, et al. Activity-dependent proteolytic cleavage of neuroligin-1. Neuron 76, 410–422 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Madoux F, et al. Discovery of an enzyme and substrate selective inhibitor of ADAM10 using an exosite-binding glycosylated substrate. Scientific reports 6, 11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shackleton B, Crawford F & Bachmeier C Inhibition of ADAM10 promotes the clearance of Abeta across the BBB by reducing LRP1 ectodomain shedding. Fluids and barriers of the CNS 13, 14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Venkatesh HS, et al. Targeting neuronal activity-regulated neuroligin-3 dependency in high-grade glioma. Nature 549, 533–537 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hickman SE, et al. The microglial sensome revealed by direct RNA sequencing. Nature neuroscience 16, 1896–1905 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eyo UB, et al. Regulation of Physical Microglia-Neuron Interactions by Fractalkine Signaling after Status Epilepticus. eNeuro 3(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Datwani A, et al. Classical MHCI molecules regulate retinogeniculate refinement and limit ocular dominance plasticity. Neuron 64, 463–470 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huh GS, et al. Functional requirement for class I MHC in CNS development and plasticity. Science 290, 2155–2159 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schecter RW, et al. Experience-Dependent Synaptic Plasticity in V1 Occurs without Microglial CX3CR1. The Journal of neuroscience : the official journal of the Society for Neuroscience 37, 10541–10553 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vainchtein ID, et al. Astrocyte-derived interleukin-33 promotes microglial synapse engulfment and neural circuit development. Science 359, 1269–1273 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woodward ND, Giraldo-Chica M, Rogers B & Cascio CJ Thalamocortical dysconnectivity in autism spectrum disorder: An analysis of the Autism Brain Imaging Data Exchange. Biological psychiatry. Cognitive neuroscience and neuroimaging 2, 76–84 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woodward ND, Karbasforoushan H & Heckers S Thalamocortical dysconnectivity in schizophrenia. The American journal of psychiatry 169, 1092–1099 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen R, Cohen LG & Hallett M Nervous system reorganization following injury. Neuroscience 111, 761–773 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Lauro C, Catalano M, Trettel F & Limatola C Fractalkine in the nervous system: neuroprotective or neurotoxic molecule? Annals of the New York Academy of Sciences 1351, 141–148 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Pruessmeyer J & Ludwig A The good, the bad and the ugly substrates for ADAM10 and ADAM17 in brain pathology, inflammation and cancer. Seminars in cell & developmental biology 20, 164–174 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Ransohoff RM & Benveniste EN Cytokines and the CNS, Second Edition, (Taylor and Francis Group, 2006). [Google Scholar]
- 46.Kunkle BW, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nature genetics 51, 414–430 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iaccarino HF, et al. Gamma frequency entrainment attenuates amyloid load and modifies microglia. Nature 540, 230–235 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martorell AJ, et al. Multi-sensory Gamma Stimulation Ameliorates Alzheimer’s-Associated Pathology and Improves Cognition. Cell 177, 256–271 e222 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chozinski TJ, Halpern AR, Okawa H, Kim H-J, Tremel GJ, Wong ROL, Vaughan JC. Expansion microscopy with conventional antibodies and fluorescent proteins. Nature Methods 11;13:485 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang C, Cai C-Z, Song J-X, Tan J-Q, Durairajan SSK, Iyaswamy A, Wu M-Y, Chen L-L, Yue Z, Li M, Lu J-H. NRBF2 is involved in the autophagic degradation process of APP-CTFs in Alzheimer disease models. Autophagy 2;13(12):2028–2040 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Manabe Y, Nagano I, Gazi MSA, Murakami T, Shiote M, Shoji M, Kitagawa H, Abe K. Glial cell line-derived neurotrophic factor protein prevents motor neuron loss of transgenic model mice for amyotrophic lateral sclerosis. Neurological Research 1;25(2):195–200 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Manue49l G, Renate W, Corinna S, T PM, Daniela S, Bernd K. Atf3 mutant mice show reduced axon regeneration and impaired regeneration-associated gene induction after peripheral nerve injury. Open Biol. Royal Society April 12;6(8):160091 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hao F, Yang C, Chen S-S, Wang Y-Y, Zhou W, Hao Q, Lu T, Hoffer B, Zhao L-R, Duan W-M, Xu Q-Y. Long-term protective effects of AAV9-mesencephalic astrocyte-derived neurotrophic factor gene transfer in parkinsonian rats. Exp Neurol. Academic Press May 1;291:120–133 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Droguett A, Krall P, Burgos ME, Valderrama G, Carpio D, Ardiles L, Rodriguez-Diez R, Kerr B, Walz K, Ruiz-Ortega M, Egido J, Mezzano S. Tubular Overexpression of Gremlin Induces Renal Damage Susceptibility in Mice. Dussaule J-C, editor. PLoS One July 18;9(7):e101879 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Viganò F, Schneider S, Cimino M, Bonfanti E, Gelosa P, Sironi L, Abbracchio MP, Dimou L. GPR17 expressing NG2-Glia: Oligodendrocyte progenitors serving as a reserve pool after injury. Glia 2016 February 1;64(2):287–299 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Kluge MG, Jones K, Kooi Ong L, Gowing EK, Nilsson M, Clarkson AN, Walker FR. Age-dependent Disturbances of Neuronal and Glial Protein Expression Profiles in Areas of Secondary Neurodegeneration Post-stroke. Neuroscience November 21;393:185–195 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Funk KE, Klein RS. CSF1R antagonism limits local restimulation of antiviral CD8+ T cells during viral encephalitis. J Neuroinflammation December 31;16(1):22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schafer DP, Lehrman EK, Heller CT & Stevens B An engulfment assay: a protocol to assess interactions between CNS phagocytes and neurons. Journal of visualized experiments : JoVE (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zilionis R et al. Single-cell barcoding and sequencing using droplet microfluidics. Nat. Protoc 12, 44–73 (2017). [DOI] [PubMed] [Google Scholar]
- 60.Langmead B, Trapnell C, Pop M & Salzberg SL Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Satija R, Farrell JA, Gennert D, Schier AF & Regev A Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trapnell C et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol 32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.