Abstract
Regulatory RNAs play versatile roles in bacteria in the coordination of gene expression during various physiological processes, especially during stress adaptation. Photosynthetic bacteria use sunlight as their major energy source. Therefore, they are particularly vulnerable to the damaging effects of excess light or UV irradiation. In addition, like all bacteria, photosynthetic bacteria must adapt to limiting nutrient concentrations and abiotic and biotic stress factors. Transcriptome analyses have identified hundreds of potential regulatory small RNAs (sRNAs) in model cyanobacteria such as Synechocystis sp. PCC 6803 or Anabaena sp. PCC 7120, and in environmentally relevant genera such as Trichodesmium, Synechococcus and Prochlorococcus. Some sRNAs have been shown to actually contain μORFs and encode short proteins. Examples include the 40-amino-acid product of the sml0013 gene, which encodes the NdhP subunit of the NDH1 complex. In contrast, the functional characterization of the non-coding sRNA PsrR1 revealed that the 131 nt long sRNA controls photosynthetic functions by targeting multiple mRNAs, providing a paradigm for sRNA functions in photosynthetic bacteria. We suggest that actuatons comprise a new class of genetic elements in which an sRNA gene is inserted upstream of a coding region to modify or enable transcription of that region.
Keywords: comparative transcriptome analysis, cyanobacteria, differential RNA-seq, gene expression regulation, photosynthesis, sRNAs
Hundreds of potentially regulatory small RNAs (sRNAs) have been identified in model cyanobacteria and, despite recent significant progress, their functional characterization is substantial work and continues to provide surprising insights of general interest.
INTRODUCTION
All bacteria have developed extensive regulatory systems to adapt to limiting nutrient concentrations and abiotic and biotic stress factors. Non-coding RNAs are an essential component of bacterial regulatory systems, acting primarily at the post-transcriptional level (Storz, Vogel and Wassarman 2011). Cyanobacteria and other photosynthetic bacteria use sunlight as their major energy source; therefore, they are exposed to a particular set of additional regulatory challenges distinct from other bacteria.
In addition to the interest in oxygenic photosynthesis, cyanobacteria are studied as prokaryotic hosts for sustainable biofuel production and for their ecological role as many species are important primary producers. Cyanobacteria occupy very diverse ecological niches. Many are free-living species that thrive in marine and freshwater, others belong to terrestrial ecosystems but there are also obligate symbionts (Nowack, Melkonian and Glockner 2008; Ran et al.2010; Thompson et al.2012; Hilton et al.2013) and endolithic forms (Gaylarde, Gaylarde and Neilan 2012). Accordingly, members of the phylum exhibit vastly different morphological and metabolic adaptations, including filamentous and multicellular forms. Their physiological capabilities include oxygenic photosynthesis, photosynthetic carbon fixation and also dinitrogen fixation in several species. Some of them even live in extreme environments like deserts, arctic regions or thermal waters. Recently, cyanobacteria have been discovered that lack photosystem II (Thompson et al.2012), whereas members of the genus Gloeobacter (Nakamura et al.2003; Saw et al.2013) possess a fully competent photosynthetic apparatus but lack thylakoids, the intracellular membrane systems otherwise considered indispensable for oxygenic photosynthesis in cyanobacteria, algae and plants. This diversity is reflected also at the genomic level. Genome sizes within the cyanobacterial phylum vary from 1.44 Mbp in Candidatus Atelocyanobacterium thalassa (UCYN-A) (Thompson et al.2012; Bombar et al.2014) to 12.1 Mbp in Scytonema hofmanni PCC 7110 (Dagan et al.2013). Therefore, great heterogeneity exists in the regulatory systems of different cyanobacteria, which is in accordance with their genomic, metabolic, physiological and morphological diversity. Regulatory RNA is likely to be an integral part of this regulatory complexity and diversity.
Indeed, during recent years, comprehensive transcriptome analyses have identified hundreds of regulatory RNA candidates in model cyanobacteria, such as Synechocystis sp. PCC 6803 (Mitschke et al.2011a; Billis et al.2014; Kopf et al.2014b), Synechocystis sp. PCC 6714 (Kopf et al.2015a), Anabaena sp. PCC 7120 (Flaherty et al.2011; Mitschke et al.2011b) and Synechococcus PCC 7942 (Vijayan, Jain and O'Shea 2011; Billis et al.2014), as well as in environmentally relevant genera, such as Trichodesmium (Pfreundt et al.2014), Nodularia (Voss et al.2013; Kopf et al.2015b), marine Synechococcus (Gierga, Voss and Hess 2012) and Prochlorococcus (Steglich et al.2008; Thompson et al.2011; Waldbauer et al.2012; Voigt et al.2014). Most of these regulatory RNA candidates are non-coding small RNAs (sRNAs), while others are several kb long. Still others turned out to be small mRNAs rather than non-coding transcripts or were identified as the crRNAs of the CRISPR-Cas systems (Hein et al.2013; Scholz et al.2013). These crRNAs act as guide RNAs within the prokaryotic RNA-based defense system against invading DNA or RNA and are reviewed within another article of this issue (Plagens et al.2015).
In this review, we provide an overview of these potential regulatory RNA molecules with a focus on recent reports from cyanobacteria. Regulatory RNAs in non-oxygenic photosynthetic bacteria and the functions of the RNA-binding chaperone Hfq in anoxygenic and oxygenic bacteria have been recently reviewed (Hess et al.2014) and will not be covered here. We pay special attention to new concepts based on recent findings in photosynthetic cyanobacteria, including small proteins as an emerging field of research and the identification of actuatons as a novel class of genetic elements.
Identification of regulatory RNA candidates in cyanobacteria
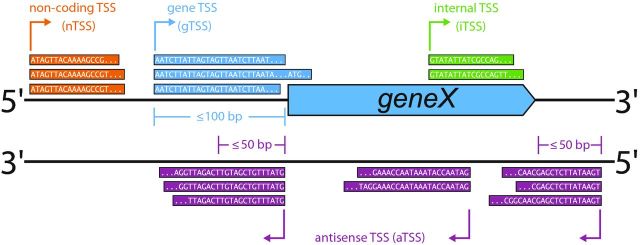
Regulatory RNA candidates in cyanobacteria were identified by computational prediction and subsequent experimental verification (Axmann et al.2005; Voss et al.2009; Ionescu et al.2010), microarray-based approaches (Steglich et al.2008; Georg et al.2009; Gierga, Voss and Hess 2012) and RNA sequencing (RNA-Seq; Mitschke et al.2011a, b; Waldbauer et al.2012; Voss et al.2013; Billis et al.2014; Kopf et al.2014b; Pfreundt et al.2014; Voigt et al.2014). Among these methods, an RNA-Seq variant called differential RNA-Seq (dRNA-Seq; Sharma et al.2010) proved as the most prolific approach for sRNA identification. dRNA-Seq allows for the precise identification of all active transcription start sites (TSS) at single nucleotide resolution (Fig. 1), including those that give rise to putative sRNAs (nTSS, non-coding RNA transcriptional start site).
Figure 1.
TSS classification based on fixed-length thresholds. Based on fixed-length thresholds, a TSS can be annotated as gTSS, aTSS, iTSS or nTSS. In this example, a TSS is classified as gTSS (gene TSS, TSS and exemplary sequencing reads are shown in blue) if it is at most 100 bp upstream of a protein-coding gene. An aTSS (antisense TSS, shown in purple) must be within 50 bp of a protein coding gene or directly antisense to it, and an iTSS (internal TSS, shown in green) must be located in sense orientation within a coding sequence. A TSS that is located in IGRs and thereby does not match any of the previous criteria is classified as nTSS (non-coding TSS, shown in orange) that gives rise to sRNA candidates (another common nomenclature for this class is orphan TSS or oTSS; Sharma et al.2010; Dugar et al.2013).
An overview of the dRNA-Seq-based transcriptome analyses in cyanobacteria is presented in Table 1. The comparison of dRNA-Seq data from seven different cyanobacteria consistently shows that only 25–33% of all TSS drive the transcription of annotated genes. The majority of the transcriptional output is likely non-coding, but with a surprisingly broad percentage of nTSS among all TSSs, ranging from 5.1% in Prochlorococcus MED4 to 26% in Trichodesmium erythraeum sp. IMS101 (Table 1). When compared to the non-coding RNA fraction of other bacteria, in the Campylobacter jejuni SuperGenome a conserved share of 1.8% nTSS was reported (Dugar et al.2013), 3.4% nTSS was reported in Escherichia coli MG1655 (Thomason et al.2015), 3.5% in Listeria pneumophila strain Paris (Sahr et al.2012), 5.4% in Helicobacter pylori (Sharma et al.2010), 6% in Salmonella enterica serovar Typhimurium (Kröger et al.2012), 6.4% in L. monocytogenes (Wurtzel et al.2012) and 12.2% in Bacillus subtilis (Irnov et al.2010). The different percentages of nTSS predicted by different published dRNA-seq data sets need to be considered with caution. There are differences in the RNA isolation and biochemical protocols, in sequencing depths, in the numbers of studied growth conditions as well as in the definition of transcript types. Nevertheless, the percentage of reported nTSS and of sRNAs among all transcriptional units is higher in cyanobacteria than in many other bacteria.
Table 1.
Numbers and types of putative TSSs mapped for different cyanobacteria by dRNA-Seq. TSS classification into four types from which the transcription of protein-coding genes (gTSS), antisense and intragenic transcripts (aTSS and iTSS) and non-coding RNAs (nTSS) is given according to the mentioned references (see Fig. 1 for classification of different TSS types). The respective percentage of nTSS from all TSS is indicated. Ana, Anabaena sp. PCC 7120; Nspu, N. spumigena CCY9414; MED4 and MIT9313, Prochlorococcus sp. MED4 and MIT9313; S6714 and S6803, Synechocystis sp. PCC 6714 and 6803; IMS101, T. erythraeum sp. IMS101.
| No. | Ana | Nspu | MED4 | MIT9313 | Sy6803 | Sy6714 | IMS101 |
|---|---|---|---|---|---|---|---|
| gTSS | 4186 | 1472 | 1059 | 1284 | 2245 | 1924 | 1858 |
| aTSS | 4172 | 1460 | 658 | 2256 | 2327 | 1976 | 855 |
| iTSS | 3933 | 1476 | 1566 | 3231 | 1734 | 862 | 1746 |
| nTSS | 1414 | 621 | 176 | 639 | 371 | 306 | 1621 |
| % nTSS | 10.3 | 12.3 | 5.1 | 8.6 | 5.5 | 6 | 26.7 |
| All TSS | 13 705 | 5029 | 3459 | 7410 | 6677 | 5068 | 6080 |
| Reference | Mitschke et al. (2011b) | Voss et al. (2013); Kopf et al. (2015a) | Voigt et al. (2014) | Voigt et al. (2014) | Mitschke et al. (2011b); Kopf et al. (2014a) | Kopf et al. (2015a) | Pfreundt et al. (2014) |
In Prochlorococcus MED4 and MIT9313, 176 and 639 nTSS, respectively, were reported to drive putative non-coding RNA transcription from intergenic regions (IGRs). These numbers correspond to 5.1 and 8.6% of all TSS, respectively. Prochlorococcus is an ecologically important primary marine producer that numerically dominates the phytoplankton of the oligotrophic open oceans with up to 105 cells per ml, and it typically thrives within the euphotic layer of the tropical and subtropical regions (reviewed by Partensky, Hess and Vaulot 1999). Prochlorococcus occurs in several distinct ecotypes; the most important of these have been defined according to their ability to adapt to high light (e.g. the laboratory model strain MED4) or low-light regimes (e.g. the MIT9313 strain) (Moore, Rocap and Chisholm 1998). Therefore, these two strains were analyzed. Prochlorococcus typically contains a compact and streamlined genome of 1.6–2.4 Mbp (Rocap et al.2003), with only few annotated genes encoding protein regulators, which raised the question whether the regulatory network is highly dependent on regulatory RNA (Steglich et al.2008).
In the unicellular Synechocystis strains PCC 6803 and PCC 6714, dRNA-Seq identified 371 and 306 nTSS, corresponding to 5.5 and 6% of all TSS, respectively (Table 1) (Mitschke et al.2011a; Kopf et al.2014b, 2015a). Synechocystis sp. PCC 6803 is a popular cyanobacterial model organism as it is amenable to straightforward genetic manipulation and was the first phototrophic organism and the third organism ever for which a complete genome sequence was determined (Kaneko et al.1996). Synechocystis sp. PCC 6714 is closely related to Synechocystis sp. PCC 6803 as they share 99.4% 16S rDNA identity (Kopf et al.2014a) and 2854 protein-coding genes, leaving 829 unique genes in Synechocystis sp. PCC 6803 and 916 in Synechocystis sp. PCC 6714 (Kopf et al.2014a,c).
High numbers of putative sRNAs have also been reported for filamentous and dinitrogen-fixing cyanobacteria (Table 1). Anabaena sp. PCC 7120 (also known as Nostoc sp. PCC 7120) is a filamentous cyanobacterium capable of nitrogen assimilation by dinitrogen fixation and is used as a model for the developmental biology of heterocyst differentiation (for reviews, see Flores and Herrero 2010; Muro-Pastor and Hess 2012). The primary transcriptome of wild-type Anabaena sp. PCC 7120 and of a strain with mutated hetR gene, the central regulator of heterocyst differentiation, was studied under nitrogen-replete conditions and 8 h after nitrogen step-down. This analysis identified 1414 nTSS, corresponding to 10.3% of all TSS (Mitschke et al.2011b).
Nodularia spumigena CCY9414 frequently dominates the annual late summer cyanobacterial blooms in brackish water ecosystems such as the Baltic Sea. Its genome is smaller than that of Anabaena sp. PCC 7120 (Voss et al.2013). Accordingly, its transcriptome is less complex, yet 621 nTSS corresponding to 12.3% of all TSS were identified in the dRNA-Seq-based transcriptome analysis under three different conditions (Voss et al.2013; Kopf et al.2015b). Finally, the highest percentage of putative nTSS was identified in T. erythraeum sp. IMS101, as discussed below (Pfreundt et al.2014).
A high percentage of putative sRNAs in Trichodesmium
From all cyanobacteria studied thus far, the T. erythraeum sp. IMS101 transcriptome stands out as 26.7% putative nTSS were identified from all TSS (1621 of 6080 TSS) (Table 1). Many of these nTSS give rise to sRNAs that were independently verified by Northern analyses (Pfreundt et al.2014). When the TSS were ranked according to the absolute numbers of associated sequencing reads, 18 of the 20 strongest TSS were nTSS. Among them were the TSS of one of the two rRNA operons and the RNA component of RNase P, a ribozyme involved in tRNA maturation. Closer inspection revealed that several of these highly abundant sRNAs were associated with two different series of sequence repeats. The most highly expressed sRNAs originated from a >6000 bp long tandem repeat array consisting of 7 repeats, each being 736–973 bp long (Pfreundt et al.2014). The transcribed portion of repeats 2–6 was ∼250 nt long and almost identical. The function of these sRNAs is still unclear but their high abundance suggests that they are functionally relevant. Another sRNA that accumulated as an abundant 265 nt transcript was the template repeat RNA of a diversity generating retroelement (DGR). Such elements were also found in other cyanobacteria, for example, N. spumigena sp. CCY 9414 (Voss et al.2013). DGRs introduce sequence diversity into a short, defined section of a protein-coding gene without interrupting it. The mechanism is based on the hypermutation of a variable sequence element within the 3′ region of protein-coding genes through recombination with a mutated cDNA copy generated by the element-encoded reverse transcriptase from the template repeat sRNA (Doulatov et al.2004; Guo et al.2008; Schillinger and Zingler 2012). The identification of the template repeat sRNA allows for the identification of its putative target genes due to sequence similarity. The previously documented DGRs have one or two such targets in a bacterial genome that harbors such an element. Unexpectedly, the DGR of T. erythraeum sp. IMS101 diversifies residues of at least 12 different proteins (Pfreundt et al.2014).
Another interesting finding was the identification of mRNAs for several genes that had not been annotated during T. erythraeum sp. IMS101 genome annotation. Genes encoding small proteins (<50 amino acids) are often not modeled by automatic genome annotation due to the high background of theoretically possible reading frames. Therefore, such cryptic protein-coding genes are often initially misclassified as sRNAs in transcriptome analyses. However, it is possible to evaluate all sRNA candidates for their coding potential using the program RNAcode (Washietl et al.2011) by comparing them against possible homologs. In the case of T. erythraeum sp. IMS101, this approach led to the identification of 13 genes for small proteins, including three encoding photosynthetic proteins (petN, psaM and psbM), emphasizing the validity of this approach (Pfreundt et al.2014). These three genes encode small subunits of three different photosynthetic complexes. They are functionally important and principally well conserved. Their missing annotation in the Trichodesmium genome sequence underscores the problems in the identification of μORFS also in bacteria. Otherwise, these are very conserved proteins: petN encodes the 28 amino acids cytochrome b6-f complex subunit PetN and exhibits 85% sequence identity with the Synechocystis sp. PCC 6803 homolog. The gene psaM encodes the 31 amino acids subunit XII of the photosystem I reaction center (77% identity with the Synechocystis homolog) and psbM encodes the 39 amino acids photosystem II reaction center protein M (60% identity with the Synechocystis homolog). Almost all of the 10 remaining small protein-coding genes have homologs in other cyanobacteria, qualifying them as candidates for more detailed analysis. Whereas the identification of such small protein-coding genes is a useful by-product of transcriptome sequencing, this strategy also reinforced the view that the majority of initially identified nTSS indeed give rise to sRNAs. The high incidence of non-coding transcripts in T. erythraeum sp. IMS101 matches the high percentage of nTSS in its genome sequence. Only 60% of its genome encodes proteins compared to ∼85% in other sequenced cyanobacterial genomes (Larsson, Nylander and Bergman 2011). The recent comparison to other Trichodesmium draft and metagenome sequences suggests that the high non-coding genome share is a conserved characteristic of this genus (Walworth et al.2015). However, it is difficult to make functional assignments for these sRNAs in this cyanobacterium. Although recently developed bioinformatics approaches (Wright et al.2013, 2014) can help, the definite identification of sRNA functions requires their genetic manipulation, which is not possible due to the lack of a genetic system for Trichodesmium.
From sRNA identification to function: regulatory RNAs in Synechocystis sp. PCC 6803
In any given bacterium, the total number of existing sRNAs should be estimated and the most interesting candidates identified, before focusing on individual sRNAs and their functions. Criteria for further analysis then include the abundance, regulation of expression, conservation of individual sRNAs in other species and the proximity of the genes to other genes of interest.
In Synechocystis sp. PCC 6803, one of the most well-studied cyanobacterial models, substantial sRNA transcription, intragenic transcripts and antisense transcripts have been reported. Approximately, 64% of all TSSs give rise to these transcript types in a genome that is otherwise 87% protein coding (Mitschke et al.2011a). Therefore, it is interesting to determine the total number of sRNAs in Synechocystis sp. PCC 6803. A weakness of most existing dRNA-Seq-based studies is that a fixed-length threshold was used to assign the identified TSS to mRNA, sRNA or cis-encoded antisense RNA (asRNA) (Fig. 1). For example, in the initial analysis of Synechocystis sp. PCC 6803, a TSS was classified as a gene TSS (gTSS) if it was located 100 nt upstream of an annotated gene, in the study of C. jejuni, 300 nt was used, and in the study of Anabaena sp. PCC 7120, 200 nt was used (Mitschke et al.2011b; Dugar et al.2013). These arbitrary values are required for automatic annotation but can be biologically incorrect because the actual lengths of the untranslated regions (UTRs) can differ greatly. For example, the transcription factor HetR in Anabaena sp. PCC 7120 is transcribed from four different gTSS, yielding 5′ UTRs of 696 nt and 728 nt for the two most distal TSSs (Buikema and Haselkorn 2001; Muro-Pastor et al.2002; Ehira and Ohmori 2006; Rajagopalan and Callahan 2010). Even when additional information was considered, for example from primer extension experiments, only a minority of TSS was re-classified. Therefore, it is necessary to consider genome-wide biological information when assigning transcript lengths and coverage.
In the comparative analysis of the Synechocystis sp. PCC 6803 primary transcriptome, the information from a classical RNA-Seq dataset was included to define transcriptional units (Kopf et al.2014b) according to a newly developed protocol (Bischler, Kopf and Voss 2014). This strategy directly links the TSS, operon and UTR information and lowers the potential for false positive TSS predictions as a TSS must be followed by a region covered by reads from the classical RNA-Seq library. Hence, genome-wide maps of active TSSs under 10 different conditions were linked to the respective transcriptional units. From the 4091 transcriptional units identified, only 191 were true non-coding transcripts (Kopf et al.2014b), compared to 429 nTSS defined in the first Synechocystis sp. PCC 6803 genome-wide TSS map (Mitschke et al.2011a). In contrast, the number of transcriptional units encompassing protein-coding genes was determined to be 2012 (Kopf et al.2014b) compared to the previously identified 1165 gTSS under standard growth conditions (Mitschke et al.2011a). These differences primarily result from the fact that many TSSs that are antisense, intragenic or intergenic to annotated genes also give rise to transcriptional units that include protein-coding genes. In contrast, some transcripts that clearly accumulate in the sRNA form belong to longer transcriptional units because downstream genes are transcribed by read-through over the sRNA's terminator of transcription or because the sRNA is post-transcriptionally processed from the longer transcript. Examples in Synechocystis sp. PCC 6803 include the SyR9 sRNA/sll0208 transcript (Klähn et al.2014), NsiR4 and Ncr0700 (Kopf et al.2014b).
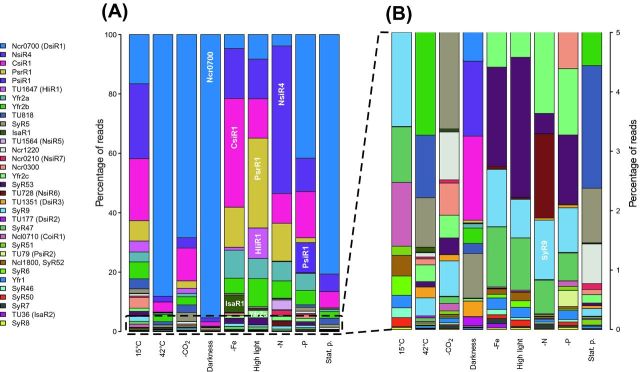
Many sRNAs are also highly regulated. Bacterial sRNAs are frequently only conditionally expressed, and several sRNAs were not expressed in Synechocystis sp. PCC 6803 under standard laboratory growth conditions (Mitschke et al.2011a), whereas they were strongly and specifically upregulated under certain stress conditions. This was studied in cultures exposed to 10 different conditions, including darkness, high light, cold and heat stress, depletion of iron, phosphate, nitrogen or inorganic carbon, exponential and stationary growth phase (Kopf et al.2014b). Here, we have summarized these findings by plotting the relative read numbers for 33 abundant sRNAs in Synechocystis sp. PCC 6803 (Fig. 2). For further detailed information, we have provided the exact sRNA sequences (Supplemental file 1). The accumulating sRNA pool is dominated by the very abundant sRNA Ncr0700, which accumulates in the dark, stationary phase and under heat stress. Its accumulation as a separate sRNA has been demonstrated, despite its association with the ssr2227 gene as a chimeric transcriptional unit (Mitschke et al.2011a; Kopf et al.2014b). Interestingly, Ncr0700 accumulation peaks in the night phase of rapidly growing cultures with a diurnal cycle (Beck et al.2014).
Figure 2.
Accumulation of 33 abundant sRNAs in Synechocystis sp. PCC 6803 under nine different growth conditions. (A and B) The 10 most abundant sRNAs and an additional 23 abundant sRNAs with interesting expression patterns were chosen. The relative abundance of sRNAs was estimated by the number of associated reads in dRNA-seq analysis (Kopf et al.2014a). Selected sRNAs with condition-dependent high accumulation were independently verified. These include the high abundance of PsrR1 in high light conditions (Mitschke et al.2011b; Kopf et al.2014a), IsaR1 in the -Fe condition (Hernandez-Prieto et al.2012; Kopf et al.2014a), NsiR4, CsiR1 and PsiR1 during nitrogen, carbon or phosphate depletion (Kopf et al.2014a) and the accumulation of SyR9 as part of the two-gene locus for alkane biosynthesis (Klähn et al.2014). High PsrR1 expression under high light is functionally relevant (Georg et al.2014). By analogy, other stress-inducible sRNAs are top candidates as regulatory molecules under each respective condition. Some identified sRNAs may encode short peptides. The phosphate stress-inducible sRNA PsiR1 contains two short reading frames (see also Fig. 6), and the high light-inducible sRNA HliR1 contains a short reading frame with 33 of its 37 residues conserved in the gene product D082_13860 in the sister strain Synechocystis sp. PCC 6714 (Kopf et al.2014b). (B) Enlargement of the 23 less abundant sRNAs from panel (A) for better resolution. Note that the sequences of the 33 transcripts were inferred from transcriptome analysis (Mitschke et al.2011b; Kopf et al.2014a) and are available for download (Supplemental file 1).
Taking these results into consideration, the total number of sRNAs in Synechocystis sp. PCC 6803 is 371 (Table 1).
Altogether, the Synechocystis sp. PCC 6803 transcriptome includes more than 4000 transcriptional units, of which approximately half represent non-coding RNAs. Most of these are antisense transcripts (asRNAs) (Kopf et al.2014b). Among these antisense RNAs are at least four important photosynthetic gene expression regulators, IsrR, As1-Flv4 and PsbA2R and PsbA3R (Dühring et al.2006; Eisenhut et al.2012; Sakurai et al.2012). Interestingly, these asRNAs appear to have repressive (IsrR and As1-Flv4) and activating (PsbA2R and PsbA3R) effects on gene expression.
Synechocystis sp. PCC 6803 PsbA2R and PsbA3R originate from the 5′ UTR of the psbA2 and psbA3 genes, just upstream and on the complementary strand of the ribosome binding site (Sakurai et al.2012). These genes encode the D1 reaction center protein of photosystem II and are highly conserved from cyanobacteria to higher plants (Cardona, Murray and Rutherford 2015). When cells are exposed to excess light, the D1 protein becomes rapidly damaged and must be continuously replaced (Järvi, Suorsa and Aro 2015). Accordingly, several different mechanisms exist to sustain maximum psbA gene expression, particularly under high light. One mechanism is gene amplification—most cyanobacteria have three to six copies of the psbA gene (Cardona, Murray and Rutherford 2015). An extreme case is Leptolyngbya sp. Heron Island J with eight copies (Paul et al.2014). Other mechanisms include strong promoters, and codon usage poised for efficient translation. The regulation of psbA gene expression occurs primarily at the transcriptional level in cyanobacteria (Golden, Brusslan and Haselkorn 1986; Mulo, Sakurai and Aro 2012). Intriguingly, the PsbA2R and PsbA3R asRNAs are transcribed from aTSS located just 19 nt upstream of the respective start codons, leading to a 30 and 69 nt overlap with the 5′ UTR of the psbA2 and psbA3 mRNAs, respectively (Sakurai et al.2012). This particular location enables PsbA2R and PsbA3R to specifically protect their cognate mRNAs from a particular form of endonucleolytic attack. The RNA endonuclease RNase E is well known in many bacteria as a key player in determining transcript stability and mediating post-transcriptional control, often together with trans-encoded sRNAs (Saramago et al.2014). The psbA2 and psbA3 mRNAs possess an RNase E-sensitive site in their 5′ UTRs, the ‘AU box’, which is located very close to the ribosome binding site (Horie et al.2007). The psbA mRNAs are cleaved at these AU boxes under darkness when the transcript is not required (Agrawal et al.2001; Horie et al.2007), but they are not recognized by RNAse E under light. The overlap with PsbA2R and PsbA3R is just long enough to shield these sites from RNase E cleavage, in concert with the initiation of translation at the ribosome binding sites (Sakurai et al.2012). Therefore, PsbA2R and PsbA3R are positively coregulated with their mRNA targets upon the transfer of cultures to higher light intensities, but jointly disappear when cells are shifted to darkness. This protective effect has physiological relevance (Sakurai et al.2012). Consequently, this example illustrates that asRNAs can act as positive post-transcriptional gene expression regulators.
In contrast, IsrR and As1-Flv4 possess negative regulatory functions by controlling the concentrations of their respective mRNAs in a codegradation mechanism (Dühring et al.2006; Eisenhut et al.2012). Codegradation between an mRNA and its frequently inversely regulated asRNA has been observed in many bacterial systems and has been reviewed in great detail separately (Georg and Hess 2011).
Whereas the targets of potential asRNA:mRNA interactions are obvious, it is very different to assess the possible regulatory functions of the plethora of sRNA candidates. However, it is very interesting to note that a similar percentage of putative trans-encoded sRNAs (46.4%) and mRNAs (43%) showed significantly reduced or enhanced expression when cyanobacterial cultures were exposed to three conditions considered relevant for photosynthetic growth (Mitschke et al.2011a), and the extension of this type of analysis to several more stress conditions confirmed these findings (Fig. 2). Thus, in a ‘guilty-by-association’ approach, the sRNAs with the most pronounced regulation of expression are likely to be functionally relevant in the conditions when their expression is at a maximum. Following this logic, Ncr0700 is an interesting candidate in the dark, stationary phase and under heat stress, NsiR4, PsiR1 and IsaR1 are interesting during nitrogen, phosphate or iron depletion and PsrR1 (for Photosynthesis regulatory RNA1) is interesting in the high light condition (Fig. 2). In particular, there are several sRNAs whose expression is connected to the availability of inorganic carbon: Ncr0700, CsiR1, Ncr1220 and SyR52 (Klähn et al.2015). Inorganic carbon availability is an important environmental factor for photosynthetic cyanobacteria. Therefore, these sRNAs are very interesting candidates for further study, although their functions are currently entirely unknown.
The trans-encoded sRNA PsrR1 controls oxygenic photosynthesis by targeting multiple mRNAs
The sRNA PsrR1, initially named SyR1 (for Synechocystis RNA1), was first identified by biocomputational prediction and experimental validation in Synechocystis sp. PCC 6803 and two other cyanobacteria species (Voss et al.2009). Transcriptome pyrosequencing and the inclusion of PsrR1 probes in custom microarrays revealed its high expression at higher light intensities and its rapid downregulation when cultures were transferred to darkness (Georg et al.2009; Mitschke et al.2011a). During the diurnal cycle, PsrR1 expression peaks early in the morning (Beck et al.2014). In Synechocystis sp. PCC 6803, PsrR1 is 131 nt long and originates from the IGR between the fabF (slr1332) and hoxH (sll1226) genes (Voss et al.2009). The two neighboring genes encode a 3-oxoacyl-(acyl carrier protein) synthase II (fabF) and the hydrogenase large subunit (hoxH), which do not provide insight into the role of PsrR1. However, when PsrR1 was overexpressed from an inducible promoter under non-stress conditions, a bleaching phenotype with considerably reduced amounts of photosynthetic pigments rapidly developed (Mitschke et al.2011a). This result suggested a function of PsrR1 in the control of pigment biosynthesis, photoprotection or photosynthetic protein synthesis. Moreover, psrR1 gene homologs can be predicted in cyanobacteria genomes that belong to morphologically and phylogenetically distant groups (Fig. 3). Based on different morphotypes, five cyanobacteria subsections have been defined (Rippka et al.1979), and genome sequences are available for representatives from all five subtypes (Shih et al.2013). By sequence similarity, PsrR1 homologs can be predicted in cyanobacteria from all five subsections, including unicellular species such as Synechocystis, Cyanothece, Microcystis and filamentous cyanobacteria capable of cell differentiation such as the Nostoc and Anabaena species. Homologs are absent or undetectable in some section V (ramified) cyanobacteria, in Thermosynechococcus sp. BP1 (Voss et al.2009), T. erythreaum and in α-cyanobacteria such as the marine picocyanobacteria Prochlorococcus spp. and Synechococcus spp. (Fig. 3). Moreover, actual PsrR1 transcription has been observed in the transcriptome datasets available for Synechococcus sp. PCC 7002 (Ludwig and Bryant 2012), N. spumigena sp. CCY9414 (Voss et al.2013; Kopf et al.2015b), Anabaena sp. PCC 7120 (Mitschke et al.2011b) and Synechocystis sp. PCC 6714 (Kopf et al.2015a). Its regulation in Synechocystis sp. PCC 6803 and its broad occurrence suggest a widely conserved and important function for PsrR1.
Figure 3.
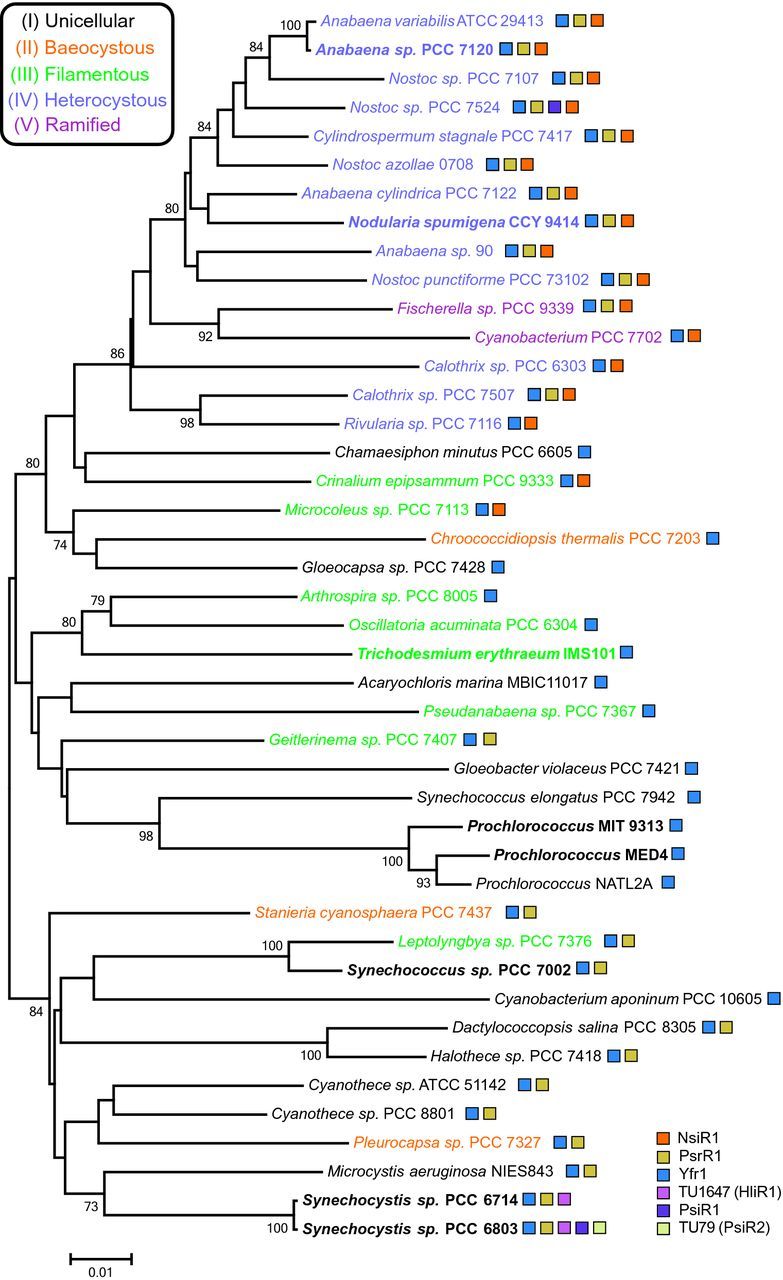
Unrooted cyanobacterial species tree and the distribution of selected sRNAs. The tree is based on an alignment of 16S rRNA genes from selected cyanobacteria. Major morphological characteristics are color coded according to the designations by Shih et al. (2013). Bootstrap values ≥70% are given at the respective nodes. The distribution of four non-coding sRNAs and two putative coding small mRNAs (PsiR1 and HliR2) is indicated by colored rectangles. The non-coding sRNA NsiR1 was first described in Anabaena sp. PCC 7120 and verified to exist in at least 19 other cyanobacteria that share the capability for nitrogen fixation and heterocyst differentiation (Ionescu et al.2010). NsiR1 is transcribed from a tandem array of direct repeats upstream of hetF (Ionescu et al.2010), a known regulator of heterocyst differentiation (Wong and Meeks 2001). NsiR1 expression is controlled by NtcA and HetR, two transcription factors critical for N2 fixation and heterocyst development and is restricted to developing heterocysts (Ionescu et al.2010; Muro-Pastor 2014). The non-coding sRNA Yfr1 (cyanobacterial functional RNA1) was initially identified in marine picocyanobacteria (Axmann et al.2005) and later found to be widely distributed throughout the cyanobacterial phylum (Voss et al.2007). Yfr1 has been suggested to control sbtA expression, which encodes the sodium-dependent bicarbonate transporter SbtA in Synechococcus elongatus PCC 6301 (Nakamura et al.2007) and outer membrane proteins (soms or porins) in Prochlorocococus sp. MED4 (Richter et al.2010). PsrR1 was initially predicted by comparative genome analysis (Georg et al.2009) and has recently been functionally characterized (Fig. 4 and (Georg et al.2014).
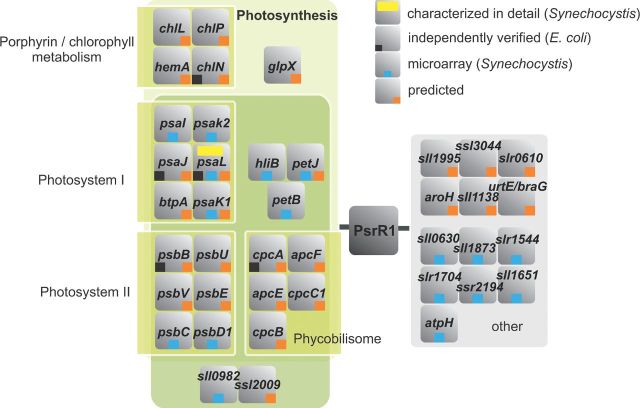
Therefore, PsrR1 was chosen for detailed functional analysis that included the detailed characterization of the phenotypic effects of knock-out and overexpression mutations and the molecular analysis of possible target genes. In addition to the effects on photosynthetic pigments, PsrR1 overexpression led to a considerable decrease in the PSI trimer-to-monomer ratio (Georg et al.2014). The combination of microarray analysis upon pulse PsrR1 overexpression with recently developed advanced computational target prediction (Wright et al.2013, 2014) yielded 26 possible target mRNAs. The majority of these target candidates may be functionally linked to photosynthesis or thylakoid membrane function (Fig. 4). Among them were the mRNAs for α- and β-phycocyanin subunits (genes cpcA and cpcB), cpcC1, apcE and apcF encoding phycobilisome linker proteins, btpA, psaK1, psaJ and psaL encoding several photosystem I or photosystem I-related proteins, petJ, encoding cytochrome c553 and chlN encoding the subunit N of the light-independent protochlorophyllide reductase (Georg et al.2014). Seven targets were tested at the molecular level by fusing their respective 5′ UTR sequences to a sequence coding for the superfolder green fluorescence protein (Corcoran et al.2012) and transforming them into an E. coli strain overexpressing PsrR1. The selected target mRNAs were cpcA, psaL, psaK1, hemA, chlN, psbB and psaJ, which were ranked on positions 1, 3, 6, 8, 34, 38 and 41 of the CopraRNA prediction (Georg et al.2014). The results of this work demonstrated the putative function of PsrR1 as a post-transcriptional repressor of psaL, psaJ, chlN, psbB and cpcA, whereas none or only minor effects were detected for the hemA and psaK1 5′ UTR fusions (Georg et al.2014). To scrutinize these results further, microarray experiments were performed with an inducible PsrR1 overproducer strain. Altogether, the levels of 16 different mRNAs were identified as significantly affected by PsrR1 overexpression. The microarray results revealed significant overlap with the computational predictions and the results of the E. coli reporter assay. Among them psaK1, petJ as well as psaL and psaI, which form a dicistronic operon. But also the other affected genes pointed strongly toward the regulation of photosynthesis (Fig. 4). Hence, these results supported the function of PsrR1 in the regulation of photosynthesis-related gene expression when cells are exposed to high light intensities. But the results also extended the computational predictions as several of the genes identified in the microarray experiment had not been predicted as putative PsrR1 targets.
Figure 4.
Regulon controlled by the sRNA PsrR1. Based on the functional enrichment analysis of its computationally predicted targets, PsrR1 is predicted to control genes (orange squares) that encode proteins involved in photosynthesis and tetrapyrrole metabolism (Georg et al.2014). Those genes that were verified experimentally in a heterologous reporter assay as PsrR1 targets are labeled by a black square, and targets suggested by microarray analysis (Georg et al.2014) by a blue square. The computational predictions were done using CopraRNA (Wright et al.2013, 2014). All top 15 CopraRNA target predictions are shown plus selected genes from the top 85 predicted candidates that were functionally enriched. Reprinted from the publication (Georg et al.2014) in modified form and with the courtesy of ASPB and the authors.
In case of the psaL mRNA encoding the photosystem I reaction center protein subunit XI, the interaction with PsrR1 inhibits ribosome binding (Georg et al.2014). In addition, a single cleavage site for the RNA endonuclease E becomes exposed within the third codon, which during active translation would be protected by ribosomes. Hence, there are two cooperating processes, first the inhibition of initiation of translation and then the endonuclease-mediated destabilization of the mRNA that lead to the post-transcriptional repression of psaL (Georg et al.2014). With these data, PsrR1 has been established as a novel regulator of the high light response in cyanobacteria. In addition, this work demonstrated the multiple-target regulation by PsrR1. Furthermore, not only psaL, psaJ, chlN, psbB and cpcA, but also several other photosynthesis-related gene products are likely controlled by PsrR1. The elucidation of the exact molecular mechanisms is an interesting topic of further research. In analogy to PsrR1, other stress-inducible sRNAs such as Ncr0700, NsiR4, CsiR1, PsiR1 and IsaR1 appear as top candidates for regulatory factors in response to darkness, or to the depletion of nitrogen, inorganic carbon, phosphate or iron.
Comparative analyses in Synechocystis sp. PCC 6714
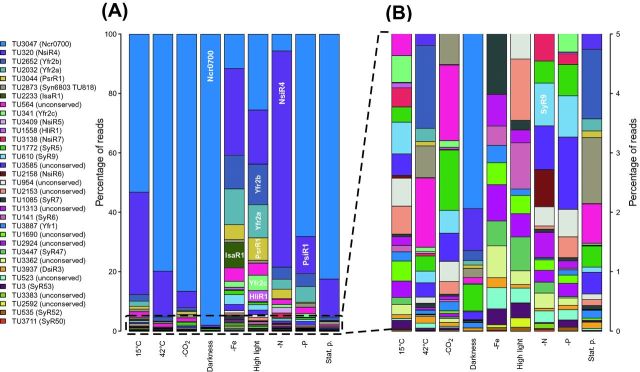
The identification of 33 abundant sRNAs in Synechocystis sp. PCC 6803 (Fig. 2) suggested that several potentially highly relevant regulators could be among them. To analyze them further, the comparison to a distinct but closely related strain can be productive. For several years, there has not been such a genome sequence available, but this has recently changed with the genome analysis of the sister strain Synechocystis sp. PCC 6714 (Kopf et al.2014a,c). Indeed, 221 of the 371 sRNAs identified in Synechocystis sp. PCC 6803 are conserved in strain 6714 (Fig. 5), and many of them show similar regulation of expression (Kopf et al.2015a). PsrR1, Ncr0700, NsiR4, IsaR1, SyR9 and Yfr1, Yfr2a, Yfr2b and Yfr2c are among the conserved sRNAs, whereas the carbon or phosphate stress-inducible sRNAs CsiR1 and PsiR1 are not conserved. Interestingly, no homologs were found in Synechocystis sp. PCC 6714 to the PsbA2R and PsbA3R asRNAs that originate from the 5′ UTR of the psbA2 and psbA3 genes in Synechocystis sp. PCC 6803. This fact is surprising, given the 97% nucleotide sequence conservation between the psbA genes of both strains. The detailed comparison revealed the existence of two nucleotide polymorphisms in the respective promoter regions. In particular, a single nucleotide polymorphism, a G-to-A transition within the –10 element of the asRNA PsbA2R promoter appears as a likely gain-of-function mutation in Synechocystis sp. PCC 6803 (or loss-of-function in Synechocystis sp. PCC 6714). Thus, a single mutation must have led to the activation or inactivation of this antisense promoter when the two strains are compared (Kopf et al.2015a). This difference is also of physiological interest at it points to a possible difference in the respective functions, here in photosystem II, between the two strains. The lack of PsbA2R and PsbA3R should make Synechocystis sp. PCC 6714 more vulnerable to high light intensities than Synechocystis sp. PCC 6803.
Figure 5.
Accumulation of 33 abundant sRNAs in Synechocystis sp. PCC 6714 under nine different growth conditions. (A and B) The 10 most abundant sRNAs and an additional 23 abundant sRNAs with interesting expression patterns were chosen. Relative abundance of sRNAs was estimated by the number of associated reads in dRNA-seq analysis (Kopf et al.2015a). If conserved in Synechocystis PCC 6803, the corresponding sRNA name or transcriptional unit is given in parentheses. With PsrR1 (high light), IsaR1 (-Fe) and NsiR4 (nitrogen depletion), selected sRNAs with condition-dependent high accumulation were independently verified for Synechocystis sp. PCC 6714 (Kopf et al.2015a). Similar to Synechocystis sp. PCC 6803, several other stress-inducible sRNAs are top regulatory candidates under each respective condition, and some sRNAs may encode short peptides. (B) Enlargement of the 23 less abundant sRNAs from panel (A) for better resolution. The sequences of these sRNAs were inferred from transcriptome analysis (Kopf et al.2015a) and are available for download (Supplemental file 2).
Emerging insight into the functions of short protein-coding transcripts
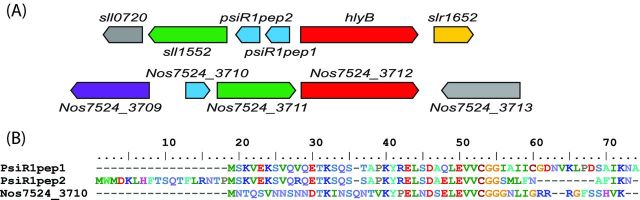
The sRNA PsiR1 is transcribed from the second most highly induced TSS under phosphate stress from a promoter containing putative pho boxes (Kopf et al.2014b), which are known to be recognized by SphR, the PhoB homolog of Synechocystis sp. PCC 6803 (Suzuki et al.2004). Although PsiR1 accumulates mainly as an sRNA of approximately 500 nt, it forms a joint transcriptional unit with the sll1552 gene encoding an uncharacterized N-acyltransferase superfamily protein. Moreover, the PsiR1 sRNA contains two possible reading frames for two small proteins, 46 and 55 codons in length, referred here as PsiR1pep1 and PsiR1pep2. These two possible proteins are very similar to each other. The only match in the database is the small protein Nos7524_3710 in the cyanobacterium Nostoc sp. PCC 7524 (Fig. 6). Its gene is directly adjacent to the Nos7524_3711 gene, which is a Sll1552 homolog (46% identical residues), suggesting that PsiR1 and sll1552 belong to a gene cassette that occurs at a low frequency within cyanobacterial genomes. However, their function within the phosphate stress regulon is unclear. Nevertheless, the similarity among the three putative short proteins is clear (Fig. 6) and suggests that some transcripts initially defined as sRNAs are in fact short mRNAs or dual-function RNAs. Information about such chimeric dual-function sRNAs is still scarce (Vanderpool, Balasubramanian and Lloyd 2011), but there are a few prominent examples for this class of regulators with the RNAIII from Staphylococcus aureus (for review, see Durand et al.2015), the SR1 regulatory sRNA from B. subtilis (Gimpel et al.2010) and the SgrS sRNA from enteric bacteria (Horler and Vanderpool 2009). Short transcripts encoding short proteins, so-called μORFs, are an emerging class of molecules with a role that has largely been underestimated (Storz, Wolf and Ramamurthi 2014). It is likely that sRNA datasets frequently contain misclassified mRNAs for such short proteins. In addition to the possible PsiR1pep1 and PsiR1pep2 peptides (Fig. 6), there are several additional candidates or confirmed examples in Synechocystis sp. PCC 6803. The HliR1 sRNA strongly induced by high light (Fig. 2) is likely an mRNA encoding a 37-amino-acid peptide. This sequence gave a single database match, to the protein D082_13860 of Synechocystis sp. PCC 6714 with 89% identity (Fig. 3) that is also induced by high light (Fig. 5). Because the transcript originates from a syntenic position upstream of the sodB gene encoding superoxide dismutase and shows the same inducibility (Kopf et al.2015a), it is likely an ortholog. Two additional examples for likely or confirmed short proteins in Synechocystis sp. PCC 6803 are the Norf1 peptide and NdhP.
Figure 6.
Putative peptides encoded by the Phosphate stress-inducible sRNA 1 (PsiR1). (A) The genome arrangement around the putative peptide-coding genes Psir1pep1 and Psir1pep2 in Synechocystis sp. PCC 6803 and Nostoc sp. PCC 7524 shows the slightly different arrangement of the conserved adjacent genes sll1552 and hlyB. Genes that are conserved in the other organism are colored and unconserved genes are shown in gray. Genes with the same color in both organisms are homologs. (B) Multiple sequence alignment of the putative PsiR1pep1 and PsiR1pep2 peptides in Synechocystis sp. PCC 6803 and of the single database match, the short protein Nos7524_3710 from Nostoc sp. PCC 7524.
Norf1 (Mitschke et al.2011a) has a 48-codon reading frame in a 378 nt long transcriptional unit in Synechocystis sp. PCC 6803 that is maximally induced under darkness, but is also induced to some extent during the stationary phase and under heat stress, with an expression pattern similar to that of Ncr0700 (Kopf et al.2014b). As it appears to be present in all β-cyanobacteria and shows the same regulation and high abundance in Synechocystis sp. PCC 6714, it is likely a relevant unknown subunit or regulatory factor. Because the transcript is more than twice as long as the coding frame, it is also possible that the norf1 transcript is a dual function RNA. The best example illustrating the functional relevance of short proteins in cyanobacteria is the sml0013 gene product. Its reading frame encodes a 40-amino-acid-long short protein that is 100% conserved in Synechocystis sp. PCC 6714, and has clear homologs in all cyanobacterial genomes and some cyanophages, with a weak similarity to plant proteins (Schwarz et al.2013). Its functional characterization revealed that sml0013 encodes a previously unknown subunit of the cyanobacterial NDH1 complex and was therefore renamed NdhP. Although it is very short, NdhP mediates coupling of the NDH1 complex to respiratory or photosynthetic electron flow (Schwarz et al.2013).
The actuaton, a new class of distinct genetic elements
In the course of sRNA identification by comparative transcriptomics, we identified a class of mRNAs that originate from read-through of an sRNA that accumulates as a discrete and abundant transcript while also serving as a 5′ UTR. We initially found that some previously known sRNAs were seemingly misclassified as gTUs because a downstream gene was in the sense orientation, lacked a specific gTSS and was apparently cotranscribed with the sRNA due to incomplete transcription termination. This arrangement exists in at least 10 cases in Synechocystis sp. PCC 6803 where abundant sRNAs belong to a chimeric precursor transcript and give rise to the respective mRNAs, which otherwise have no or only weak TSSs (Kopf et al.2015a). Examples include the CsiR1 element specific to Synechocystis sp. PCC 6803, the Ncr0700 sRNA that originates from a free-standing TU in Synechocystis sp. PCC 6714, which became part of a chimeric TU in Synechocystis sp. PCC 6803 due to rearrangement by transposition, and the sRNA SyR9 that forms a dispensable part of the mRNA encoding aldehyde deformylating oxygenase (Klähn et al.2014), the key enzyme for cyanobacterial alkane biosynthesis conserved in both Synechocystis strains discussed here (Figs 2 and 5).
Evolutionary events such as gain or loss of an sRNA within or close to a promoter may turn the respective sRNA into an actuaton that directly affects the expression of the downstream gene. The Yfr2c sRNA illustrates this effect (Fig. 7). Yfr2c belongs to a family of sRNAs that is widely distributed among cyanobacteria. The Yfr2 sRNA family genes occur in different genetic arrangements and in copy numbers from one to nine (Gierga, Voss and Hess 2012). Synechocystis sp. PCC 6803 expresses three family members, which accumulate as 80, 65 and 70 nt sRNAs (Voss et al.2009). All three are conserved in Synechocystis sp. PCC 6714 and are present in the same genomic context with yfr2c linked to the respective orthologous protein-coding gene.
Figure 7.
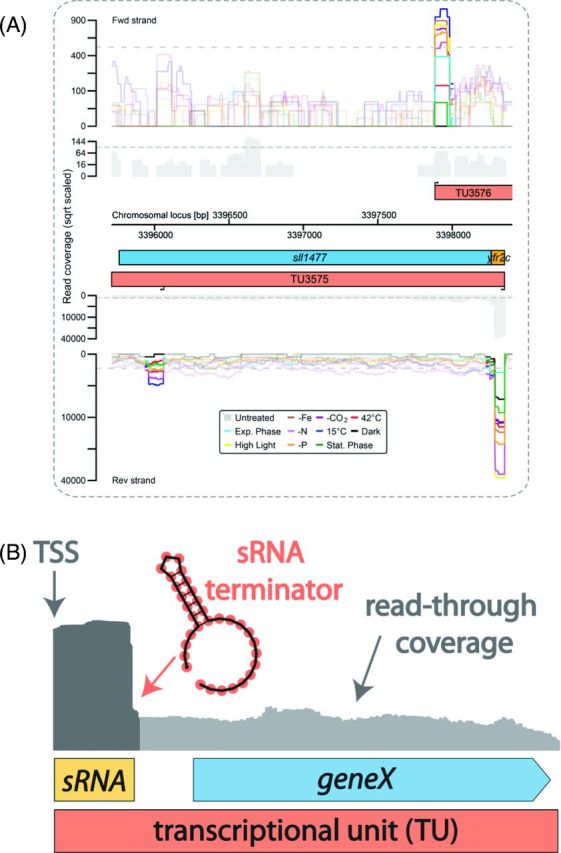
The actuaton concept. (A) The transcriptional organization of the yfr2c actuaton region in Synechocystis sp. PCC 6803. The colored graphs represent the accumulation of treated reads from a dRNA-seq analysis of Synechocystis sp. PCC 6803 under 10 different conditions (Kopf et al.2014a). All TSS positions inferred from the TEX-treated cDNA library are indicated by black arrows, and the graphs for the following 100 nt are highlighted. Transcriptional units (TUs, red) were inferred from the untreated read coverage (gray). Protein-coding genes are displayed in blue and previously predicted sRNAs are shown in yellow (Mitschke et al.2011b). Data for the forward strand is shown above and data for the reverse strand is shown below the axis that shows the chromosomal locus in bp. A gTSS upstream of sll1477 gives rise to both the sRNA Yfr2c (Voss et al.2009) and the mRNA for protein Sll1477, a protease of the abortive phage infection (abi, CAAX) family, and the respective TU3575 is therefore classified as an actuaton. Further downstream, TU3575 contains an internal start site (iTSS). On the forward strand, the TU3576 TSS gives rise to a weakly expressed asRNA to TU3575. The arrangement is well conserved in the closely related strain Synechocystis sp. PCC 6714 (Kopf et al.2015a), with the exception of the exact TSS position of the asRNA, which is slightly different. (B) Model of the actuaton concept. The expression of a downstream gene is driven by read-through (RNA-seq coverage shown in light gray) over the terminator from an sRNA that accumulates as a discrete and abundant transcript (RNA-seq coverage shown in dark gray). Because the downstream gene lacks an own TSS, it is classified in a joint transcriptional unit (TU) together with the sRNA.
Such an sRNA/mRNA structure, which we have termed an ‘actuaton’, constitutes an additional way for bacteria to remodel the output from their transcriptional network. A hallmark of these elements is that they are followed by a protein-coding gene in the sense direction that lacks a gTSS. Nevertheless, the major accumulating RNA species is an sRNA that accumulates as an abundant and discrete transcript, and therefore, constitutes a clearly separate entity. Therefore, an actuaton gives rise to an sRNA and simultaneously constitutes part of the 5′ region of a gene. Even long 5′ UTRs of well-characterized genes, such as those encoding RNase E, may belong to this class. The change in expression of the mRNA portion of an actuaton can result only from the sRNA promoter replacing the original one. In more complex scenarios, it may also result from events causing differential termination, for instance from the riboswitch-mediated attenuation of transcription. Actuatons may also be predecessors or derivatives of riboswitches in which the sRNA function is modified to serve as the metabolite-sensing entity regulating the expression of the protein-coding portion.
The actuaton concept is consistent with the increasing understanding that functional RNA elements possess plasticity (Mellin et al.2014). Other examples include a riboswitch in L. monocytogenes, which also acts as a regulatory sRNA (Loh et al.2009). Non-coding RNAs constitute the evolutionarily most flexible transcriptome component, and it is likely that actuatons are genetic elements found also in organisms outside of cyanobacteria.
CONCLUDING REMARKS
Cyanobacteria are frequently the dominant primary producers in aquatic ecosystems, but they also thrive on land as part of the microbial assemblages that form soil crusts, as well as in deserts and other arid habitats. They frequently form symbioses with organisms from very different phyla, and some species are endolithotrophs. Morphologically, many cyanobacteria are unicellular but there are also baeocystous, heterocystous and ramified morphotypes. Genome sizes vary in size from between 1.44 Mb and 1200 genes for Candidatus A. thalassa (UCYN-A) diazotrophic cyanobacteria (Tripp et al.2010; Bombar et al.2014) to 12 356 protein-coding genes in S. hofmanni PCC 7110 (Dagan et al.2013). Accordingly, great variation can be expected within the regulatory systems of different cyanobacteria, including various roles and types of regulatory RNAs. Thus, it is important to catalog the pool of potential regulatory RNAs throughout this phylum, but the analytical approaches should be as efficient as possible, involving the different types of RNA-Seq as well as tools for the definition of transcriptional units, comparative analysis and target prediction, as described in this review. Existing data show that non-coding RNAs in cyanobacteria have an important role in the control of photosynthetic functions and adaptation to abiotic stress. Functional characterization of cyanobacterial sRNAs will likely focus on the most interesting sRNA candidates in model strains that are amenable to genetic manipulation, but aspects relevant for their ecological success and possible biotechnological exploitation are equally interesting. An important aspect is the integration of regulatory RNAs with protein activities. Although it likely functions quite differently from its homolog in enteric bacteria, recent findings have shed new light on the cyanobacterial Hfq homolog (Puerta-Fernandez and Vioque 2011; Schürgers et al.2014). Other promising factors include cyanobacterial RNA helicases (Owttrim 2012) and the various, still largely uncharacterized RNA endo- and exonucleases (Matos et al.2012; Chan et al.2015; Zhang et al.2014). Finally, the use of ribosome display and related technical approaches will greatly advance the finding and characterization of dual-function sRNAs and of mRNAs encoding small proteins that were initially misclassified as non-coding transcripts but are important molecules with regulatory or modulating functions.
Supplementary Material
Acknowledgments
The authors wish to thank all their coworkers and colleagues.
FUNDING
A large part of the work discussed in this review was supported over several years by grants from the Deutsche Forschungsgemeinschaft Focus program ‘Sensory and regulatory RNAs in Prokaryotes’ SPP1258, grants HE 2544/4-1 and 4-2, from the Federal Ministry of Education and Research, grants no. 0316165 ‘RNASYS’ and GR2378/03F0640A ‘Joint German-Israeli Research Projects’, from the German-Israeli Research Foundation, project number 1133-13.8/2011 and by the EU project MaCuMBA (Marine Microorganisms: Cultivation Methods for Improving their Biotechnological Applications; grant agreement no: 311975) to W.R.H.
Conflict of interest. None declared.
REFERENCES
- Agrawal GK, Kato H, Asayama M, et al. An AU-box motif upstream of the SD sequence of light-dependent psbA transcripts confers mRNA instability in darkness in cyanobacteria. Nucleic Acids Res. 2001;29:1835–43. doi: 10.1093/nar/29.9.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axmann IM, Kensche P, Vogel J, et al. Identification of cyanobacterial non-coding RNAs by comparative genome analysis. Genome Biol. 2005;6:R73. doi: 10.1186/gb-2005-6-9-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck C, Hertel S, Rediger A, et al. Daily expression pattern of protein-encoding genes and small noncoding RNAs in Synechocystis sp. strain PCC 6803. Appl Environ Microb. 2014;80:5195–206. doi: 10.1128/AEM.01086-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billis K, Billini M, Tripp HJ, et al. Comparative transcriptomics between Synechococcus PCC 7942 and Synechocystis PCC 6803 provide insights into mechanisms of stress acclimation. PLoS One. 2014;9:e109738. doi: 10.1371/journal.pone.0109738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischler T, Kopf M, Voss B. Transcript mapping based on dRNA-seq data. BMC Bioinformatics. 2014;15:122. doi: 10.1186/1471-2105-15-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombar D, Heller P, Sanchez-Baracaldo P, et al. Comparative genomics reveals surprising divergence of two closely related strains of uncultivated UCYN-A cyanobacteria. ISME J. 2014;8:2530–42. doi: 10.1038/ismej.2014.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buikema WJ, Haselkorn R. Expression of the Anabaena hetR gene from a copper-regulated promoter leads to heterocyst differentiation under repressing conditions. P Natl Acad Sci USA. 2001;98:2729–34. doi: 10.1073/pnas.051624898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona T, Murray JW, Rutherford AW. Origin and evolution of water oxidation before the last common ancestor of the Cyanobacteria. Mol Biol Evol. 2015 doi: 10.1093/molbev/msv024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YW, Millard A, Wheatley PJ, et al. Genomic and proteomic characterisation of two novel siphovirus infecting the sedentary facultative epibiont cyanobacterium Acaryochloris marina. Environ Microbiol. 2015 doi: 10.1111/1462-2920.12735. [DOI] [PubMed] [Google Scholar]
- Corcoran CP, Podkaminski D, Papenfort K, et al. Superfolder GFP reporters validate diverse new mRNA targets of the classic porin regulator, MicF RNA. Mol Microbiol. 2012;84:428–45. doi: 10.1111/j.1365-2958.2012.08031.x. [DOI] [PubMed] [Google Scholar]
- Dagan T, Roettger M, Stucken K, et al. Genomes of Stigonematalean cyanobacteria (subsection V) and the evolution of oxygenic photosynthesis from prokaryotes to plastids. Genome Biol Evol. 2013;5:31–44. doi: 10.1093/gbe/evs117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doulatov S, Hodes A, Dai L, et al. Tropism switching in Bordetella bacteriophage defines a family of diversity-generating retroelements. Nature. 2004;431:476–81. doi: 10.1038/nature02833. [DOI] [PubMed] [Google Scholar]
- Dugar G, Herbig A, Forstner KU, et al. High-resolution transcriptome maps reveal strain-specific regulatory features of multiple Campylobacter jejuni isolates. PLoS Genet. 2013;9:e1003495. doi: 10.1371/journal.pgen.1003495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dühring U, Axmann IM, Hess WR, et al. An internal antisense RNA regulates expression of the photosynthesis gene isiA. P Natl Acad Sci USA. 2006;103:7054–8. doi: 10.1073/pnas.0600927103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand S, Tomasini A, Braun F, et al. sRNA and mRNA biogenesis and turnover in Gram-positive bacteria. FEMS Microbiol Rev. 2015;39 doi: 10.1093/femsre/fuv007. [DOI] [PubMed] [Google Scholar]
- Ehira S, Ohmori M. NrrA directly regulates expression of hetR during heterocyst differentiation in the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 2006;188:8520–5. doi: 10.1128/JB.01314-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhut M, Georg J, Klähn S, et al. The antisense RNA As1_flv4 in the Cyanobacterium Synechocystis sp. PCC 6803 prevents premature expression of the flv4-2 operon upon shift in inorganic carbon supply. J Biol Chem. 2012;287:33153–62. doi: 10.1074/jbc.M112.391755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty BL, Van Nieuwerburgh F, Head SR, et al. Directional RNA deep sequencing sheds new light on the transcriptional response of Anabaena sp. strain PCC 7120 to combined-nitrogen deprivation. BMC Genomics. 2011;12:332. doi: 10.1186/1471-2164-12-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores E, Herrero A. Compartmentalized function through cell differentiation in filamentous cyanobacteria. Nat Rev Microbiol. 2010;8:39–50. doi: 10.1038/nrmicro2242. [DOI] [PubMed] [Google Scholar]
- Gaylarde CC, Gaylarde PM, Neilan BA. Endolithic phototrophs in built and natural stone. Curr Microbiol. 2012;65:183–8. doi: 10.1007/s00284-012-0123-6. [DOI] [PubMed] [Google Scholar]
- Georg J, Dienst D, Schürgers N, et al. The small regulatory RNA SyR1/PsrR1 controls photosynthetic functions in cyanobacteria. Plant Cell. 2014;26:3661–79. doi: 10.1105/tpc.114.129767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georg J, Hess WR. cis-antisense RNA, another level of gene regulation in bacteria. Microbiol Mol Biol R. 2011;75:286–300. doi: 10.1128/MMBR.00032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georg J, Voß B, Scholz I, et al. Evidence for a major role of antisense RNAs in cyanobacterial gene regulation. Mol Syst Biol. 2009;5:305. doi: 10.1038/msb.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierga G, Voss B, Hess WR. Non-coding RNAs in marine Synechococcus and their regulation under environmentally relevant stress conditions. ISME J. 2012;6:1544–57. doi: 10.1038/ismej.2011.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimpel M, Heidrich N, Mader U, et al. A dual-function sRNA from B. subtilis: SR1 acts as a peptide encoding mRNA on the gapA operon. Mol Microbiol. 2010;76:990–1009. doi: 10.1111/j.1365-2958.2010.07158.x. [DOI] [PubMed] [Google Scholar]
- Golden SS, Brusslan J, Haselkorn R. Expression of a family of psbA genes encoding a photosystem II polypeptide in the cyanobacterium Anacystis nidulans R2. EMBO J. 1986;5:2789–98. doi: 10.1002/j.1460-2075.1986.tb04569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Tse LV, Barbalat R, et al. Diversity-generating retroelement homing regenerates target sequences for repeated rounds of codon rewriting and protein diversification. Mol Cell. 2008;31:813–23. doi: 10.1016/j.molcel.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein S, Scholz I, Voß B, et al. Adaptation and modification of three CRISPR loci in two closely related cyanobacteria. RNA Biol. 2013;10:852–64. doi: 10.4161/rna.24160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Prieto MA, Schön V, Georg J, et al. Iron deprivation in Synechocystis: inference of pathways, non-coding RNAs, and regulatory elements from comprehensive expression profiling. G3. 2012;2:1475–95. doi: 10.1534/g3.112.003863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess WR, Berghoff BA, Wilde A, et al. Riboregulators and the role of Hfq in photosynthetic bacteria. RNA Biol. 2014;11:413–26. doi: 10.4161/rna.28035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton JA, Foster RA, Tripp HJ, et al. Genomic deletions disrupt nitrogen metabolism pathways of a cyanobacterial diatom symbiont. Nat Commun. 2013;4:1767. doi: 10.1038/ncomms2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie Y, Ito Y, Ono M, et al. Dark-induced mRNA instability involves RNase E/G-type endoribonuclease cleavage at the AU-box and SD sequences in cyanobacteria. Mol Genet Genomics. 2007;278:331–46. doi: 10.1007/s00438-007-0254-9. [DOI] [PubMed] [Google Scholar]
- Horler RS, Vanderpool CK. Homologs of the small RNA SgrS are broadly distributed in enteric bacteria but have diverged in size and sequence. Nucleic Acids Res. 2009;37:5465–76. doi: 10.1093/nar/gkp501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionescu D, Voss B, Oren A, et al. Heterocyst-specific transcription of NsiR1, a non-coding RNA encoded in a tandem array of direct repeats in cyanobacteria. J Mol Biol. 2010;398:177–88. doi: 10.1016/j.jmb.2010.03.010. [DOI] [PubMed] [Google Scholar]
- Irnov I, Sharma CM, Vogel J, et al. Identification of regulatory RNAs in Bacillus subtilis. Nucleic Acids Res. 2010;38:6637–51. doi: 10.1093/nar/gkq454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Järvi S, Suorsa M, Aro EM. Photosystem II repair in plant chloroplasts—regulation, assisting proteins and shared components with photosystem II biogenesis. Biochim Biophys Acta. 2015 doi: 10.1016/j.bbabio.2015.01.006. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Sato S, Kotani H, et al. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions (supplement) DNA Res. 1996;3:185–209. doi: 10.1093/dnares/3.3.185. [DOI] [PubMed] [Google Scholar]
- Klähn S, Baumgartner D, Pfreundt U, et al. Alkane biosynthesis genes in cyanobacteria and their transcriptional organization. Front Bioeng Biotechnol. 2014;2:24. doi: 10.3389/fbioe.2014.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klähn S, Orf I, Schwarz D, et al. Integrated transcriptomic and metabolomic characterization of the low-carbon response using an ndhR mutant of Synechocystis sp. PCC 6803. Plant Physiol. 2015 doi: 10.1104/pp.114.254045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopf M, Klähn S, Pade N, et al. Comparative genome analysis of the closely related Synechocystis strains PCC 6714 and PCC 6803. DNA Res. 2014a;21:255–66. doi: 10.1093/dnares/dst055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopf M, Klähn S, Scholz I, et al. Comparative analysis of the primary transcriptome of Synechocystis sp. PCC 6803. DNA Res. 2014b;21:527–39. doi: 10.1093/dnares/dsu018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopf M, Klähn S, Scholz I, et al. Variations in the non-coding transcriptome as a driver of inter-strain divergence and physiological adaptation in bacteria. Sci Rep. 2015a;5:9560. doi: 10.1038/srep09560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopf M, Klähn S, Voss B, et al. Finished genome sequence of the unicellular cyanobacterium Synechocystis sp. strain PCC 6714. Genome Announc. 2014c;2:e00757–14. doi: 10.1128/genomeA.00757-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopf M, Möke F, Bauwe H, et al. Expression profiling of the bloom-forming cyanobacterium Nodularia spumigena CCY9414 under high light and oxidative stress conditions. ISME J. 2015b doi: 10.1038/ismej.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kröger C, Dillon SC, Cameron ADS, et al. The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. P Natl Acad Sci USA. 2012;109:E1277–86. doi: 10.1073/pnas.1201061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson J, Nylander JA, Bergman B. Genome fluctuations in cyanobacteria reflect evolutionary, developmental and adaptive traits. BMC Evol Biol. 2011;11:187. doi: 10.1186/1471-2148-11-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh E, Dussurget O, Gripenland J, et al. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell. 2009;139:770–9. doi: 10.1016/j.cell.2009.08.046. [DOI] [PubMed] [Google Scholar]
- Ludwig M, Bryant DA. Acclimation of the global transcriptome of the cyanobacterium Synechococcus sp. strain PCC 7002 to nutrient limitations and different nitrogen sources. Front Microbiol. 2012;3:145. doi: 10.3389/fmicb.2012.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos RG, Fialho AM, Giloh M, et al. The rnb gene of Synechocystis PCC6803 encodes a RNA hydrolase displaying RNase II and not RNase R enzymatic properties. PLoS One. 2012;7:e32690. doi: 10.1371/journal.pone.0032690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellin JR, Koutero M, Dar D, et al. Riboswitches. Sequestration of a two-component response regulator by a riboswitch-regulated noncoding RNA. Science. 2014;345:940–3. doi: 10.1126/science.1255083. [DOI] [PubMed] [Google Scholar]
- Mitschke J, Georg J, Scholz I, et al. An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. P Natl Acad Sci USA. 2011a;108:2124–9. doi: 10.1073/pnas.1015154108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitschke J, Vioque A, Haas F, et al. Dynamics of transcriptional start site selection during nitrogen stress-induced cell differentiation in Anabaena sp. PCC7120. P Natl Acad Sci USA. 2011b;108:20130–5. doi: 10.1073/pnas.1112724108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LR, Rocap G, Chisholm SW. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature. 1998;393:464–7. doi: 10.1038/30965. [DOI] [PubMed] [Google Scholar]
- Mulo P, Sakurai I, Aro EM. Strategies for psbA gene expression in cyanobacteria, green algae and higher plants: from transcription to PSII repair. Biochim Biophys Acta. 2012;1817:247–57. doi: 10.1016/j.bbabio.2011.04.011. [DOI] [PubMed] [Google Scholar]
- Muro-Pastor AM. The heterocyst-specific NsiR1 small RNA is an early marker of cell differentiation in cyanobacterial filaments. MBio. 2014;5:e01079–01014. doi: 10.1128/mBio.01079-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro-Pastor AM, Hess WR. Heterocyst differentiation: from single mutants to global approaches. Trends Microbiol. 2012;20:548–57. doi: 10.1016/j.tim.2012.07.005. [DOI] [PubMed] [Google Scholar]
- Muro-Pastor AM, Valladares A, Flores E, et al. Mutual dependence of the expression of the cell differentiation regulatory protein HetR and the global nitrogen regulator NtcA during heterocyst development. Mol Microbiol. 2002;44:1377–85. doi: 10.1046/j.1365-2958.2002.02970.x. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Naito K, Yokota N, et al. A cyanobacterial non-coding RNA, Yfr1, is required for growth under multiple stress conditions. Plant Cell Physiol. 2007;48:1309–18. doi: 10.1093/pcp/pcm098. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Kaneko T, Sato S, et al. Complete genome structure of Gloeobacter violaceus PCC 7421, a cyanobacterium that lacks thylakoids. DNA Res. 2003;10:137–45. doi: 10.1093/dnares/10.4.137. [DOI] [PubMed] [Google Scholar]
- Nowack EC, Melkonian M, Glockner G. Chromatophore genome sequence of Paulinella sheds light on acquisition of photosynthesis by eukaryotes. Curr Biol. 2008;18:410–8. doi: 10.1016/j.cub.2008.02.051. [DOI] [PubMed] [Google Scholar]
- Owttrim GW. RNA helicases in cyanobacteria: biochemical and molecular approaches. Method Enzymol. 2012;511:385–403. doi: 10.1016/B978-0-12-396546-2.00018-8. [DOI] [PubMed] [Google Scholar]
- Partensky F, Hess WR, Vaulot D. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol Mol Biol R. 1999;63:106–27. doi: 10.1128/mmbr.63.1.106-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul R, Jinkerson RE, Buss K, et al. Draft genome sequence of the filamentous cyanobacterium Leptolyngbya sp. strain heron island J, exhibiting chromatic acclimation. Genome Announc. 2014;2:e01166–13. doi: 10.1128/genomeA.01166-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfreundt U, Kopf M, Belkin N, et al. The primary transcriptome of the marine diazotroph Trichodesmium erythraeum IMS101. Sci Rep. 2014;4:6187. doi: 10.1038/srep06187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagens A, Richter H, Charpentier E, et al. DNA and RNA interference mechanisms 1 by CRISPR-Cas 2 surveillance complexes. FEMS Microbiol Rev. 2015 doi: 10.1093/femsre/fuv019. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puerta-Fernandez E, Vioque A. Hfq is required for optimal nitrate assimilation in the Cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 2011;193:3546–55. doi: 10.1128/JB.00254-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R, Callahan SM. Temporal and spatial regulation of the four transcription start sites of hetR from Anabaena sp. strain PCC 7120. J Bacteriol. 2010;192:1088–96. doi: 10.1128/JB.01297-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran L, Larsson J, Vigil-Stenman T, et al. Genome erosion in a nitrogen-fixing vertically transmitted endosymbiotic multicellular cyanobacterium. PLoS One. 2010;5:e11486. doi: 10.1371/journal.pone.0011486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter AS, Schleberger C, Backofen R, et al. Seed-based INTARNA prediction combined with GFP-reporter system identifies mRNA targets of the small RNA Yfr1. Bioinformatics. 2010;26:1–5. doi: 10.1093/bioinformatics/btp609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippka R, Deruelles J, WJ B., et al. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111:1–61. [Google Scholar]
- Rocap G, Larimer FW, Lamerdin J, et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 2003;424:1042–7. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- Sahr T, Rusniok C, Dervins-Ravault D, et al. Deep sequencing defines the transcriptional map of L. pneumophila and identifies growth phase-dependent regulated ncRNAs implicated in virulence. RNA Biol. 2012;9:503–19. doi: 10.4161/rna.20270. [DOI] [PubMed] [Google Scholar]
- Sakurai I, Stazic D, Eisenhut M, et al. Positive regulation of psbA gene expression by cis-encoded antisense RNAs in Synechocystis sp. PCC 6803. Plant Physiol. 2012;160:1000–10. doi: 10.1104/pp.112.202127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saramago M, Barria C, Dos Santos RF, et al. The role of RNases in the regulation of small RNAs. Curr Opin Microbiol. 2014;18:105–15. doi: 10.1016/j.mib.2014.02.009. [DOI] [PubMed] [Google Scholar]
- Saw JH, Schatz M, Brown MV, et al. Cultivation and complete genome sequencing of Gloeobacter kilaueensis sp. nov., from a lava cave in Kilauea Caldera, Hawai'i. PLoS One. 2013;8:e76376. doi: 10.1371/journal.pone.0076376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schillinger T, Zingler N. The low incidence of diversity-generating retroelements in sequenced genomes. Mob Genet Elements. 2012;2:287–91. doi: 10.4161/mge.23244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz I, Lange SJ, Hein S, et al. CRISPR-Cas systems in the cyanobacterium Synechocystis sp. PCC6803 exhibit distinct processing pathways involving at least two Cas6 and a Cmr2 protein. PLoS One. 2013;8:e56470.1–15. doi: 10.1371/journal.pone.0056470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürgers N, Ruppert U, Watanabe S, et al. Binding of the RNA chaperone Hfq to the type IV pilus base is crucial for its function in Synechocystis sp. PCC 6803. Mol Microbiol. 2014;92:840–52. doi: 10.1111/mmi.12595. [DOI] [PubMed] [Google Scholar]
- Schwarz D, Schubert H, Georg J, et al. The gene sml0013 of Synechocystis species strain PCC 6803 encodes for a novel subunit of the NAD(P)H oxidoreductase or complex I that is ubiquitously distributed among Cyanobacteria. Plant Physiol. 2013;163:1191–202. doi: 10.1104/pp.113.224287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma CM, Hoffmann S, Darfeuille F, et al. The primary transcriptome of the major human pathogen, Helicobacter pylori. Nature. 2010;464:250–5. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- Shih PM, Wu D, Latifi A, et al. Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. P Natl Acad Sci USA. 2013;110:1053–8. doi: 10.1073/pnas.1217107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steglich C, Futschik ME, Lindell D, et al. The challenge of regulation in a minimal photoautotroph: non-coding RNAs in Prochlorococcus. PLoS Genet. 2008;4:e1000173. doi: 10.1371/journal.pgen.1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell. 2011;43:880–91. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G, Wolf YI, Ramamurthi KS. Small proteins can no longer be ignored. Annu Rev Biochem. 2014;83:753–77. doi: 10.1146/annurev-biochem-070611-102400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Ferjani A, Suzuki I, et al. The SphS-SphR two component system is the exclusive sensor for the induction of gene expression in response to phosphate limitation in Synechocystis. J Biol Chem. 2004;279:13234–40. doi: 10.1074/jbc.M313358200. [DOI] [PubMed] [Google Scholar]
- Thomason MK, Bischler T, Eisenbart SK, et al. Global transcriptional start site mapping using differential RNA sequencing reveals novel antisense RNAs in Escherichia coli. J Bacteriol. 2015;197:18–28. doi: 10.1128/JB.02096-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AW, Foster RA, Krupke A, et al. Unicellular cyanobacterium symbiotic with a single-celled eukaryotic alga. Science. 2012;337:1546–50. doi: 10.1126/science.1222700. [DOI] [PubMed] [Google Scholar]
- Thompson AW, Huang K, Saito MA, et al. Transcriptome response of high- and low-light-adapted Prochlorococcus strains to changing iron availability. ISME J. 2011;5:1580–94. doi: 10.1038/ismej.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripp HJ, Bench SR, Turk KA, et al. Metabolic streamlining in an open-ocean nitrogen-fixing cyanobacterium. Nature. 2010;464:90–4. doi: 10.1038/nature08786. [DOI] [PubMed] [Google Scholar]
- Vanderpool CK, Balasubramanian D, Lloyd CR. Dual-function RNA regulators in bacteria. Biochimie. 2011;93:1943–9. doi: 10.1016/j.biochi.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayan V, Jain IH, O'Shea EK. A high resolution map of a cyanobacterial transcriptome. Genome Biol. 2011;12:R47. doi: 10.1186/gb-2011-12-5-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt K, Sharma CM, Mitschke J, et al. Comparative transcriptomics of two environmentally relevant cyanobacteria reveals unexpected transcriptome diversity. ISME J. 2014;8:2056–68. doi: 10.1038/ismej.2014.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss B, Bolhuis H, Fewer DP, et al. Insights into the physiology and ecology of the brackish-water-adapted Cyanobacterium Nodularia spumigena CCY9414 based on a genome-transcriptome analysis. PLoS One. 2013;8:e60224. doi: 10.1371/journal.pone.0060224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss B, Georg J, Schön V, et al. Biocomputational prediction of non-coding RNAs in model cyanobacteria. BMC Genomics. 2009;10:123. doi: 10.1186/1471-2164-10-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss B, Gierga G, Axmann IM, et al. A motif-based search in cyanobacterial genomes identifies the ortholog of the small RNA Yfr1 in all lineages of cyanobacteria. BMC Genomics. 2007;8:375. doi: 10.1186/1471-2164-8-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldbauer JR, Rodrigue S, Coleman ML, et al. Transcriptome and proteome dynamics of a light-dark synchronized bacterial cell cycle. PLoS One. 2012;7:e43432. doi: 10.1371/journal.pone.0043432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walworth N, Pfreundt U, Nelson W, et al. Trichodesmium genome maintains abundant, widespread noncoding DNA in situ, despite oligotrophic lifestyle. P Natl Acad Sci USA. 2015 doi: 10.1073/pnas.1422332112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl S, Findeiss S, Muller SA, et al. RNAcode: robust discrimination of coding and noncoding regions in comparative sequence data. RNA. 2011;17:578–94. doi: 10.1261/rna.2536111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong FC, Meeks JC. The hetF gene product is essential to heterocyst differentiation and affects HetR function in the cyanobacterium Nostoc punctiforme. J Bacteriol. 2001;183:2654–61. doi: 10.1128/JB.183.8.2654-2661.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright PR, Georg J, Mann M, et al. CopraRNA and IntaRNA: predicting small RNA targets, networks and interaction domains. Nucleic Acids Res. 2014;42:W119–23. doi: 10.1093/nar/gku359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright PR, Richter AS, Papenfort K, et al. Comparative genomics boosts target prediction for bacterial small RNAs. P Natl Acad Sci USA. 2013;110:E3487–96. doi: 10.1073/pnas.1303248110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzel O, Sesto N, Mellin JR, et al. Comparative transcriptomics of pathogenic and non-pathogenic Listeria species. Mol Sys Biol. 2012;8:583. doi: 10.1038/msb.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JY, Deng XM, Li FP, et al. RNase E forms a complex with polynucleotide phosphorylase in cyanobacteria via a cyanobacterial-specific nonapeptide in the noncatalytic region. RNA. 2014;20:568–79. doi: 10.1261/rna.043513.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.