Abstract
Amyloid precursor protein (App) plays a crucial role in Alzheimer's disease via the production and deposition of toxic β-amyloid peptides. App is heavily expressed in neurons, the focus of the vast majority of studies investigating its function. Meanwhile, almost nothing is known about App's function in glia, where it is also expressed, and can potentially participate in the regulation of neuronal physiology. In this report, we investigated whether Appl, the Drosophila homolog of App, could influence sleep–wake regulation when its function is manipulated in glial cells. Appl inhibition in astrocyte-like and cortex glia resulted in higher sleep amounts and longer sleep bout duration during the night, while overexpression had the opposite effect. These sleep phenotypes were not the result of developmental defects, and were correlated with changes in expression in glutamine synthetase (GS) in astrocyte-like glia and in changes in the gap-junction component innexin2 in cortex glia. Downregulating both GS and innexin2, but not either one individually, resulted in higher sleep amounts, similarly to Appl inhibition. Consistent with these results, the expression of GS and innexin2 are increased following sleep deprivation, indicating that GS and innexin2 genes are dynamically linked to vigilance states. Interestingly, the reduction of GS expression and the sleep phenotype observed upon Appl inhibition could be rescued by increasing the expression of the glutamate transporter dEaat1. In contrast, reducing dEaat1 expression severely disrupted sleep. These results associate glutamate recycling, sleep, and a glial function for the App family proteins.
SIGNIFICANCE STATEMENT The amyloid precursor protein (App) has been intensively studied for its implication in Alzheimer's disease (AD). The attributed functions of App are linked to the physiology and cellular biology of neurons where the protein is predominantly expressed. Consequences on glia in AD are generally thought to be secondary effects of the pathology in neurons. Researchers still do not know whether App plays a role in glia in nonpathological conditions. We report here that glial App plays a role in physiology and in the regulation of sleep/wake, which has been shown recently to be involved in AD pathology. These results also associate glutamate recycling and sleep regulation, adding further complexity to the physiological role of App and to its implication in AD.
Keywords: amyloid precursor protein, Drosophila, glia, glutamate, sleep/wake
Introduction
Since the identification 20 years ago of App, the gene for amyloid precursor protein (App), the accumulating evidence of its involvement in Alzheimer's disease (AD), the main cause of age related dementia worldwide, has attracted much attention. The production of particular β-amyloid peptides in neuronal cells by sequential cleavage of App, and their deposition in senile plaques, has been shown to have toxic consequences in a diversity of cell culture and in vivo model systems, including Drosophila. Age, genetic, and environmental factors are known to increase the severity of the pathophysiological processes triggered by β-amyloid peptides. In recent years, the hypothesis that sleep plays a critical and potentially bidirectional role in this context has received increasing support (Ju et al., 2013). Sleep is significantly disrupted in AD patients, and the excessive production of β-amyloid peptides has been shown to disrupt sleep in animal models (Ju et al., 2014; Tabuchi et al., 2015; Gerstner et al., 2016). Conversely, the clearance of β-amyloid peptides from the interstitial fluid and CSF is facilitated during sleep (Kang et al., 2009, Xie et al., 2013). In addition, a recent report using Drosophila suggests that accumulation of β-amyloid peptides is increased following sleep deprivation and is linked to neuronal hyperactivity (Tabuchi et al., 2015). App, as well as the paralogs Aplp1 and Aplp2, are evolutionary conserved cell-surface proteins that have been attributed multiple functions, including cell adhesion, interactions with the extracellular matrix, cell signaling, and the regulation of vesicular trafficking. In mouse knock-out models as well as in Drosophila, where a single App-like protein is expressed (Appl; Luo et al., 1990; Martin-Morris and White, 1990), studies have shown that these proteins modulate intracellular signaling, gene expression, neurite outgrowth, synaptic function, and plasticity (Allebrandt et al., 2013). Inhibition of Appl expression in Drosophila mushroom body neurons, a central brain structure that plays a key role in learning/memory as well as in sleep regulation, results in long-term memory deficits (Goguel et al., 2011; Bourdet et al., 2015a), which is similar to previous findings obtained in mouse App knock-out models (Dawson et al., 1999; Phinney et al., 1999). These results suggest that the loss of App function could contribute to the memory loss observed in AD patients and may be in synergy with the accumulation of abnormal β-amyloid peptides (Bourdet et al., 2015b). Nevertheless, the physiological functions of the App family of proteins and the extent to which these functions are disrupted in AD by toxic β-amyloid peptides remain unclear (Phinney et al., 1999). In the brain, App is heavily expressed in neurons, the focus of most studies investigating App's function and the location of the majority of App's molecular partners identified so far (Martin-Morris and White, 1990; Apelt et al., 1997; Allebrandt et al., 2013). In contrast, almost nothing is known about the role of App in glia, where it is also expressed and where it can potentially participate in the regulation of neuronal physiology (Grolla et al., 2013), with consequences on behavior. In this study, we investigated the potential role of endogenous glial Appl in sleep/wake regulation, and glial gene expression changes linked to glutamate recycling and gap junctions. These results extend the functional complexity of the role played by the App family of proteins.
Materials and Methods
Fly stocks.
The following fly stocks were used: CantonS (Cs), Inx2BG02429 (RRID:BDSC_12834), NP2222-Gal4 (RRID:DGGR_112830), UAS-Appl-RNAiJF02878 (RRID:BDSC_28043), UAS-Appl-RNAi HMS01931 (RRID:BDSC_39013), repo-Gal4 (RRID:BDSC_7415), Tub-Gal80ts (RRID:DGGR_130452), UAS-dicer2, dEaat1(GMR75H11)-Gal4 (RRID:BDSC_39914), UAS-GS2-RNAiHMS02197 (Bloomington Drosophila Stock Center), UAS-Appl-RNAiKK102543 (RRID:FlyBase_FBst0480124), UAS-Appl-RNAiGD3170 (RRID:FlyBase_FBst0464704; Vienna Drosophila Stock Center), mz0709-Gal4, alrm-Gal4 (M. Freeman, University of Massachusetts), 9-137-Gal4 (U. Heberlein, Janielia Research Campus), repo-Gal80 (V. Morel, Centre National de la Recherche Scientifique, Lyon), UAS-Appl (T. Preat, Centre National de la Recherche Scientifique, Paris), UAS-inx2 (R. Bauer, University of Bonn), dEaat1-Gal4, UAS-dEaat1 (RRID:BDSC_8202), UAS-dEaat1-RNAiIRX, UAS-dEaat1-RNAiIR2 (S. Birman, Centre National de la Recherche Scientifique, Paris), UAS-GFP (B. Mollereau, École normale supérieure de Lyon), UAS-RFP:inx2, UAS-inx2:RFP (A. Brand, University of Cambridge). Stocks were outcrossed ≥3 times to wild-type flies before being used for experimentation. Unless otherwise stated, UAS-dicer2 was always associated with UAS-Appl-RNAi to increase the efficiency of the knock down. Flies were reared at 25°C, 50–60% humidity, in a 12 h light/dark cycle in a standard food containing inactivated yeast, molasses, sucrose, and agar.
Sleep monitoring.
Freshly eclosed females were collected from culture vials under CO2 anesthesia. Adult 3–5-d-old flies were transferred individually to 65 mm glass vials for continuous sleep recordings in Trikinetiks locomotor activity monitors for ≥5 d. Sleep parameters were evaluated as described previously (Shaw et al., 2000). Sleep deprivation was performed with a sleep-nullifying apparatus system as previously described (Shaw et al., 2000, 2002). Flies were sleep deprived for 25 h starting at lights on. White area in baseline sleep graphs shows the photophase; the shaded area shows the scotophase.
Immunofluorescence in whole-mount brains.
Brains were dissected in cold PBS, fixed for 20 min in a 4% paraformaldehyde PBS, washed in 0.3% Triton X-100 PBS (PBS-T) and blocked for ≥45 min in 3% goat serum in PBS-T, before overnight incubation in the corresponding primary antibody: rabbit anti-GFP (1:2000; Invitrogen, catalog #A6455; RRID:AB_221570), rabbit anti-inx2 (1:50; GenScript), mouse anti-glutamine synthetase (anti-GS; 1:250; Millipore, catalog #MAB302; RRID:AB_2110656). After primary antibody incubation, brains were washed in PBS-T and incubated with Alexa 488-conjugated and Alexa 633-conjugated secondary antibodies (Thermo Fisher Scientific, catalog A11008; RRID:AB_143165; catalog #A21050 RRID:AB_141431). Brains were mounted in hard-set Vectashield with DAPI (Vector Laboratories, catalog #H-1500; RRID:AB_2336788) and imaged using a confocal microscope (Leica, TCS-SP5 X). ImageJ (RRID:SCR_003070) was used for image processing and fluorescence quantification. Mean fluorescence of GS was measured within the calyx. Innexin2 (Inx2) levels were measured by quantifying mean fluorescence levels in the calyx and cortex region. In all cases, quantification was done averaging 10–12 Z planes of ≥3 brains for each condition. Maximal projection images are shown.
Quantitative real-time PCR.
Fly heads were collected (four independent samples of 20 heads for each group) and frozen immediately in dry ice until further processing. Total RNA was extracted using RNeasy Minikit with DNase treatment (Qiagen). Reverse transcription was performed using the iScript cDNA Synthesis Kit (Bio-Rad Laboratories). Finally, quantitative real-time PCR was performed on a 7900HT Fast Real-Time PCR System using FAST SYBR Green Master Mix (Applied Biosystems). RP49 transcript levels were used to normalize results between groups. For each gene, the relative expression ratio was determined using the 2ΔΔCt calculation method. The following primers were used: RP49 primers: 5′-acgacaatctccttgcgcttct-3′ and 5′-atcggttacggatcgaacaa-3′; dEaat1 primers: 5′-tcggcattcagcctatccac-3′ and 5′-tcgaatggctactggatcgc-3′.
Statistical analysis.
Distribution and homogeneity as well as statistical group comparisons were tested using Microsoft Excel plugin software Statel. In all cases averages with SEM are plotted. The following corresponding statistical and post hoc test values are reported in the figure legends: Mann–Whitney (see Z values); ANOVA with Tukey's post hoc (see F values); or Kruskal–Wallis (see K-W values) with a posteriori test; one-tailed t tests were used to evaluate transcript levels by quantitative real-time PCR (*p < 0.05; **p < 0.001). N number is listed for each group in the following order: Gal4 control, UAS control, and experimental group.
Results
Modulation of Appl expression in cortex and astrocyte-like glia disrupts sleep
To determine whether glial Appl can affect sleep/wake regulation, we used the Gal4/UAS system to reduce or increase Appl expression in different glial subtypes of the adult brain (Fig. 1). First, we tested specific glial Gal4 drivers combined with Appl-RNAiJF02878, an Appl-RNAi construct targeting the terminal region of the Appl transcript common to all Appl isoforms reported (Ni et al., 2009; Flybase; Fig. 2A). All Gal4 constructs driving expression in perineurial, subperineurial, cortex, unsheathing, and astrocyte-like glia, or a combination of subtypes, lead to viable adults, with no obvious morphological or behavioral defects. Two Gal4 constructs that share expression in cortex glia resulted in significantly higher sleep amounts upon driving expression of Appl-RNAi (Fig. 2C–H). The most severe phenotype was observed with the dEaat1-Gal4 driver (Fig. 2C,I), which is expressed in both astrocyte-like and cortex glia (Fig. 3A). However, no effect on sleep was observed upon expression of Appl-RNAi in astrocyte-like glia (alrm-Gal4 driver; Fig. 2I), suggesting that Appl is required in cortex glia for sleep/wake regulation. Combining the dEaat1-Gal4 driver with pan-glial expression of the Gal4 inhibitor Gal80 restored a control-like sleep/wake phenotype, demonstrating that the effect on sleep/wake is not due to driving Appl-RNAi in nonglial cells (Fig. 2E,I). To further confirm this set of results and exclude the possibility that it could originate from off-targets of the Appl-RNAiJF02878 construct, we replicated the experiment using another Appl-RNAi construct that targets the first, untranslated exons of the Appl transcript (Appl-RNAiHMS01931), as well as another Gal4 driver expressed in cortex glia, Inx2BG02429-Gal4, and obtained similar results (Fig. 2F–I). Expressing two other different Appl-RNAi constructs under the control of Inx2BG02429-Gal4 also resulted in higher sleep amounts (Table 1). In almost all experimental genotypes tested, high daily sleep amounts were due to larger amounts of night sleep. Elevated night sleep was associated with longer average night-bout duration (Table 1). These sleep/wake phenotypes were not due to the use of UAS-dicer2 to enhance the efficiency of RNAi constructs (see Materials and Methods), since sleep/wake was unaffected when dicer2 alone was overexpressed (total sleep: dEaat-Gal4, 715 ± 30 min; dEaat1-Gal4 > UAS-dicer2, 718 ± 26 min).
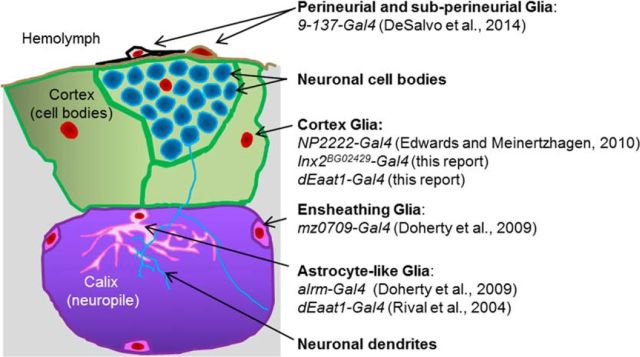
Figure 1.
The different glial subtypes of the adult Drosophila brain. The schematic represents the area surrounding the calix neuropile, which includes cell bodies and neurites of the mushroom bodies' neurons (Kenyon cells). Each glial cell type can be targeted with the listed Gal4 drivers.
Figure 2.
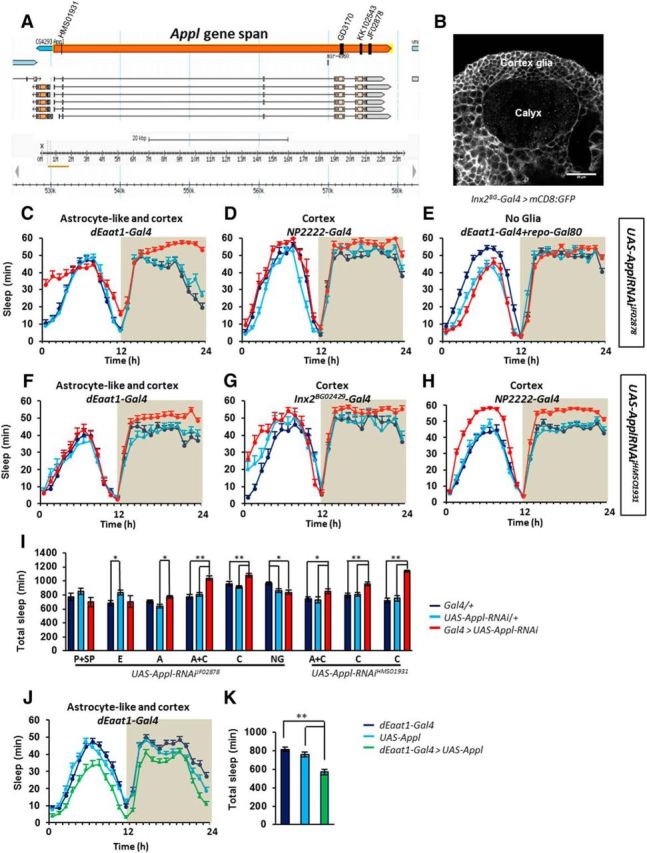
Manipulation of Appl expression in cortex and astrocyte-like glia modifies sleep. Targeting different segments of the Appl transcript with independent RNAi constructs results in higher sleep amounts during the night. A, Graphical representation of the Appl gene and localizations of the areas targeted by the different Appl RNAi constructs. Each targeted region is shared by all Appl transcripts (adapted from Flybase). B, mCD8-GFP expression driven by Inx2BG02429-Gal4 highlighting cortex glia around the calyx neuropile. Baseline sleep (minutes of sleep per hour) upon expression of UAS-Appl-RNAiJF02878 driven in (C) astrocyte-like and cortex (dEaat1-Gal4) K-W = 29.85, p < 0.00001; N = 39, 39, 35, (D) cortex (NP2222-Gal4) K-W = 14.58, p < 0.00068; N = 21, 24, 8, (E) no glia (repo-Gal80, dEaat1-Gal4) F = 6.606, p < 0.0019; N = 43, 44, 43. F–H, UAS-Appl-RNAiHMS01931 replicated the phenotype when driven in (F) cortex and astrocyte-like glia with dEaat1-Gal4 (K-W, 6.689; p < 0.035; N = 40, 41, 41) and in (G) cortex glia with Inx2BG02429-Gal4 (F = 10.9; p < 0.000035; N = 56, 56, 60) or (H) with NP2222-Gal4 (NP2222-Gal4; K-W, 50.78; p < 0.00001; N = 41, 43, 43). I, Total daily sleep for each genotype. Perineurial and subperineurial (P+SP; 9–137-Gal4); ensheathing (E; mz0709-Gal4); astrocyte-like (A; alrm-Gal4); astrocyte-like and cortex (A+C; dEaat1-Gal4); cortex [C; NP2222-Gal4 with UAS-Appl-RNAi JF02878, NP2222-Gal4 (left) and Inx2BG02429-Gal4 (right) with UAS-Appl-RNAiHMS01931)] and no glia (NG; repo-Gal80, dEaat1-Gal4). Dark blue, Glia Gal4/+ controls; light blue, UAS-Appl-RNAi/+ controls; red, corresponding glial driver controlling UAS-Appl-RNAi. J, Overexpression of Appl in cortex and astrocyte-like glia (dEaat1-Gal4>UAS-Appl) reduces sleep levels. K, Comparison of total daily sleep for each genotype (F = 24.4 p < 0.00001; N = 63, 61, 62).
Figure 3.
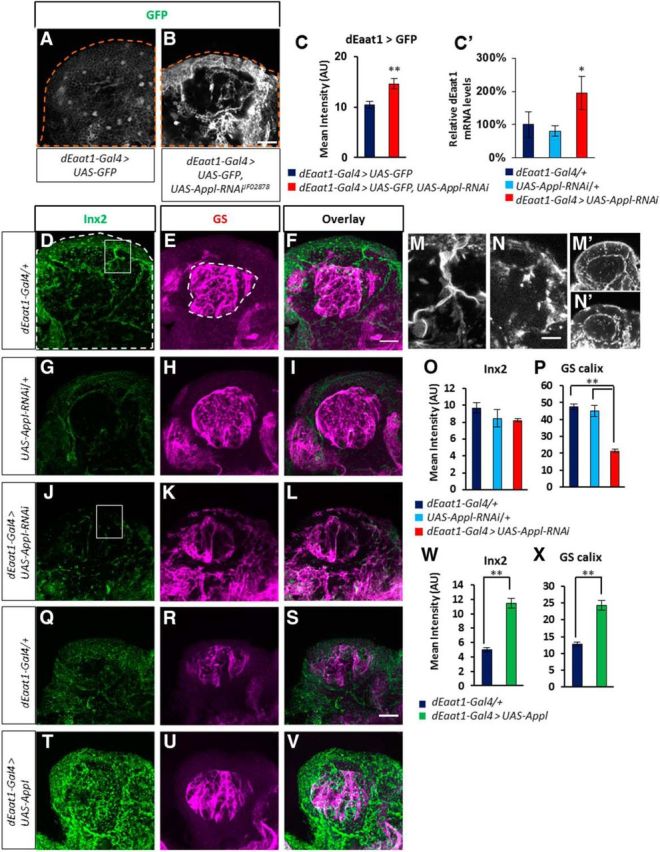
Appl knock-down is associated with changes in genes linked to glutamate recycling. A, B, Confocal images of whole-mount brains expressing GFP (green, immunolabeling) driven by dEaat1-Gal4 in cortex and astrocyte-like glia: (A) control dEaat1-Gal4 > UAS-GFP and (B) glial Appl knock-down dEaat1-Gal4 > UAS-GFP, UAS-Appl-RNAiJF02878. C, Quantification of GFP immunofluorescence reflecting dEaat1 expression levels shows increased expression in Appl mutants. GFP is not transported efficiently to the neuropiles (Rival et al., 2004) and was quantified in the cortex area (Z = 3.53, p < 0.00041, N = 4 each group). C', Quantitative PCR evaluation of dEaat1 transcripts in whole heads (p < 0.05, 1-tailed t test). D–L, Immunolabeling of Inx2 (D, F, G, I, J, L; green) and GS (E, F, H, I, K, L; magenta) in dEaat1-Gal4/+ (D–F) and in UAS-Appl-RNAiJF02878/+ (G–I) controls, and in dEaat1-Gal4 > UAS-Appl-RNAiJF02878 mutants (J–L). M, N, Close-up corresponding to the insets in D and J, showing Inx2 signal in cortex glia in (M) dEaat1-Gal4/+ control and abnormal distribution of the protein in (N) dEaat1-Gal4 > UAS-Appl-RNAiJF02878 mutant. M', N′, Inx2 signal in single-plane confocal images of the whole calyx area shown in D and J. dEaat1-Gal4/+ control (M') shows lines of Inx2 expression at the boundaries of the cortex glial cells area, while the dEaat1-Gal4 > UAS-Appl-RNAiJF02878 mutant (N′) shows more clumpy distribution. O, P, Quantification of Inx2 immunofluorescence (O; K-W, 17.33; p < 0.00017; n = 5 each group) and of GS immunofluorescence in the calyx (P; K-W, 75.53; p < 0.00017; n = 5 each group). GS immunofluorescence in the cortex area surrounding the calyx was significantly enhanced in dEaat1-Gal4 > UAS-Appl-RNAiJF02878 flies compared with controls (dEaat1-Gal4, 23.06 ± 0.41; UAS-Appl-RNAiJF02878, 22.29 ± 0.72; dEaat1-Gal4 > UAS-Appl-RNAiJF02878, 29.16 ± 0.63; K-W, 21.83; p < 0.000018; N = 5 each group). Dark-blue bar, Gal4/+ controls; light-blue bars, UAS/+ controls; red bars, Gal4 > UAS group. Q–V, Immunolabeling of Inx2 (Q, T) and GS (R, U) in brains of flies overexpressing glial Appl. W, X, Quantification of immunofluorescence shows higher Inx2 levels (W; Z = 6.98; p < 0.00001; n = 3 controls and n = 4 experimental group) and higher GS expression in calyx (X; Z = 5.96; p < 0.00001; n = 3 controls and n = 4 experimental group) but not in cortex glia surrounding the calyx (Z = 1.039; p = 0.3; N = 5 each group), upon Appl overexpression. Dark-blue bars, Gal4/+ controls; green bars, the experimental group overexpressing Appl. Scale bars: (in B, F, S) A–L, Q–V, 20 μm; (in N) M, N, 10 μm.
Table 1.
Sleep parameters evaluated in all Appl misexpression experiments
| Genotype | Glia | UAS | Total sleep | Night Sleep | Night boutd | Maximal night boutb | Night bout #a | Day sleep | Day boutd | Maximal day boutb | Day bout #a | Latencyc |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UAS-dicer2/+; repo-Gal80, Eaat1-Gal4/+; UAS-Appl-RNAiJF02878/+ | No glia | JF02878 | U | U | U | U | U | U | U | U | U | U |
| UAS-dicer2/+;9-137-Gal4/+;UAS-Appl-RNAiJF02878/+ | Perineurial and subperineurial | JF02878 | U | U | U | U | U | U | U | U | U | U |
| UAS-dicer2/+;;mz0709-Gal4/UAS-Appl-RNAiJF02878 | Ensheathing | JF02878 | U | U | U | U | U | U | U | U | U | U |
| UAS-dicer2/+;Alrm-Gal4/+;UAS-Appl-RNAiJF02878/+ | Astrocyte-like | JF02878 | U | U | U | U | U | U | U | U | U | U |
| UAS-dicer2/+;dEaat1-Gal4/+;UAS-Appl-RNAiJF02878/+ | Astrocyte-like and cortex | JF02878 | H,ss | H,ss | H,ss | H,ss | L,s | H,ss | L,s | U | H,ss | L,ss |
| UAS-dicer2/+;NP2222-Gal4/+;UAS-Appl-RNAiJF02878/+ | Cortex | JF02878 | H,s | H,ss | H,ss | H,ss | L,s | L,s | U | H,ss | U | U |
| Inx2BG02429/UAS-dicer2;Tub-Gal80ts/+;UAS-Appl-RNAiJF02878/+ | Cortex (acute) | JF02878 | H,s | H,ss | H,ss | H,ss | U | L,ss | L,s | L,s | L,s | U |
| UAS-dicer2/+;dEaat1-Gal4/UAS-Appl-RNAiHMS01931; | Astrocyte-like and cortex | HMS01931 | H,ss | H,ss | H,ss | H,s | L,s | U | U | U | U | U |
| Inx2BG02429/UAS-dicer2;UAS-Appl-RNAiHMS01931/+; | Cortex | HMS01931 | H,ss | H,ss | H,s | H,ss | L,s | U | H,s | U | L,s | L,s |
| UAS-dicer2/+;UAS-ApplRNAiHMS01931/NP2222-Gal4;; | Cortex | HMS01931 | H,ss | H,ss | H,ss | H,ss | L,ss | H,ss | H,ss | H,ss | L,s | U |
| UAS-dicer2/+;dEaat1-Gal4/+;UAS-Appl-RNAiGD3170/+ | Astrocyte-like and cortex | GD3170 | U | U | H,s | H,s | L,s | U | H,s | H,s | L,ss | H,ss |
| Inx2BG02429/UAS-dicer2;;UAS-Appl-RNAiGD3170/+ | Cortex | GD3170 | H,ss | H,ss | U | H,ss | U | H,s | U | U | U | L,ss |
| UAS-dicer2/+;dEaat1-Gal4/UAS-Appl-RNAiKK102543; | Astrocyte-like and cortex | KK102543 | H,s | U | U | U | U | H,s | H,ss | H,ss | L,ss | U |
| Inx2BG02429/UAS-dicer2;UAS-Appl-RNAiKK102543/+; | Cortex | KK102543 | H,ss | H,ss | H,s | U | L,s | H,ss | U | U | H,ss | L,s |
| ;dEaat1-Gal4/UAS-Appl; | Astrocyte-like and cortex | Appl | L,ss | L,ss | L,s | L,s | U | L,ss | U | L,ss | L,ss | H,ss |
aDay bout # and Night bout # indicate the number of sleep bouts in photophase and scotophase respectively.
bMaximal day bout and Maximal night bout indicate the longest sleep bout for each corresponding light phase.
cTime between lights off and the first sleep event.
dRefers to average sleep bout duration during the considered period.
UUnchanged values compared to both parental controls, p > 0.1.
HHigher values in the mutant compared to the controls.
LLower values in the mutant compared to the controls.
ssp < 0.05.
s0.05 < p < 0.1.
In contrast, overexpressing full-length Appl with the dEaat1-Gal4 driver resulted in the reverse phenotype: reduced sleep amounts, during both day and night (Fig. 2J,K). Compared with the controls, the latency between lights off and the first sleep bout of the night was higher (76 ± 9 vs 46 ± 5 and 50 ± 4 min for the Gal4/+ and UAS/+ controls respectively p < 0.05), suggesting difficulty to initiate sleep at night. Sleep bout duration during the night was not significantly affected, however (Table 1). Altogether, these data show that manipulating Appl expression in glia can affect sleep/wake regulation in a bidirectional way.
Inhibition of Appl in cortex glia is associated with changes in dEaat1, GS, and innexin 2 expression
Glia in mammals and in flies is known to be involved in neurotransmitter recycling functions (Rival et al., 2006; Chaturvedi et al., 2014). The reuptake and recycling of glutamate, an important neurotransmitter for learning and memory, synaptic plasticity, energy metabolism, and sleep is indeed a crucial glial function, requiring identified transporters and enzymes. These functions are impaired in AD (Shrivastava et al., 2013; Osborn et al., 2016), but it remains unclear whether this effect is a consequence of neuronal dysfunction of astrogliosis following exposure to toxic protein products, or of a failure of App function. The recycling involves the reuptake of synaptically liberated glutamate by the transporter dEaat1, and the conversion of glutamate to glutamine by GS. The genes for dEaat1 and GS are expressed in glia in Drosophila, similarly to mammals (Freeman et al., 2003; Lievens et al., 2005). To monitor glutamate transporter expression, we measured UAS-GFP expression driven by the dEaat1-Gal4 construct (dEaat1>GFP) in whole-mount brain. Inhibition of Appl expression resulted in an increase in dEaat1>GFP expression, indicating that the transcription of dEaat1 is upregulated (Fig. 3A–C). The expression is notably increased toward the periphery of the brain, where a subset of cortex glial cells is in direct contact with subperineurial glia, the Drosophila equivalent of the blood–brain barrier. Quantitative PCR evaluation of mRNA levels confirmed that dEaat1 is transcriptionally upregulated in Appl mutants (Fig. 3C'). Additionally, we evaluated GS protein using immunofluorescence in the same conditions (Fig. 3D–L) and observed a dramatic reduction of GS expression within the brain neuropiles, where the protein is normally very abundant (Fig. 3E,H,K,P). In the cortex region, GS levels were slightly increased upon inhibition of Appl expression (Fig. 3K, legend). In contrast, Appl overexpression in glia resulted in higher GS expression in the neuropiles, without any effect in the cortex region (Fig. 3R,U,X).
Gap junctions between astrocytes (Rouach et al., 2008) and between different glial cell types in Drosophila (Chaturvedi et al., 2014) have been shown to play a critical role in intercellular communications required for neurotransmitter reuptake and recycling (Giaume et al., 2010). Therefore, we used immunofluorescence to assess expression and localization of Inx2, a major glial gap junction component in Drosophila (Holcroft et al., 2013; Chaturvedi et al., 2014; Spéder and Brand, 2014). Inx2 is expressed at the boundary between astrocyte-like glia and cortex glia and between cortex glia and subperineurial glia, and along lines in the cortex region that presumably correspond to the limits of each cortex glial cell (Fig. 3D,M,M'). The expression of Inx2 within the cortex area was strongly upregulated upon Appl overexpression (Fig. 3Q,T,W). Inhibition of Appl expression using Appl-RNAiJF02878 did not result in a significant reduction of Inx2 expression (Fig. 3D,G,J,O), but changes in the cellular distribution of Inx2 were observed. In mutant flies, the continuous expression of Inx2 along cellular boundaries was lost, becoming interrupted and clumpier (Fig. 3M,M',N,N′). In addition, we further replicated these findings with the Appl-RNAiHMS01931 transgene, which targets a different segment of the Appl transcript (Fig. 2A). With this construct, Inx2 cellular distribution was not only disrupted but also significantly reduced compared with controls (dEaat1-Gal4, 23.71 ± 1.77; UAS-Appl-RNAiHMS01931, 19.19 ± 0.88; dEaat1-Gal4 > UAS-Appl-RNAiHMS01931, 10.70 ± 0.75; K-W, 46.23; p < 0.00001; N = 5 brains each group) and GS levels in calyx were similarly reduced (dEaat1-Gal4, 57.00 ± 2.90; UAS-Appl-RNAiHMS01931, 39.35 ± 1.98; dEaat1-Gal4 > UAS-Appl-RNAiHMS01931, 29.87 ± 1.94; K-W, 45.54; p < 0.00001; N = 3, 3, 4 brains). The effect GS and Inx2 expression were not due to the use of UAS-dicer2 to enhance the efficiency of RNAi constructs (see Materials and Methods), since their expressions remain unchanged when dicer2 alone was overexpressed (data not shown). The quantifications shown in Figure 3 are centered on the calyx, the input neuropile of the mushroom-body neurons (Kenyon cells), which play critical roles in behavior, learning, and memory, as well as in sleep regulation. The cell bodies of Kenyon cells are in the cortex surrounding the calyx (Fig. 1). The expression of Inx2 and GS was similarly affected in other regions of the central brain.
To exclude that potential developmental effects could explain the phenotypes observed, and to further control the time course of Appl manipulation, we used the TARGET (Temporal and Regional Gene Expression Targeting) system (McGuire et al., 2004). The Inx2BG02429-Gal4 driver, which is restricted to cortex glia (Fig. 2B) was used in these experiments. Flies were raised at 18°C (permissive temperature) until the first days of adulthood and then transferred to 30°C to allow the expression of Appl-RNAi. We observed no difference in sleep between the Inx2BG02429-Gal4 > Appl-RNAi flies and the genetic controls at 18°C, as expected in the presence of a functional Gal80 (Fig. 4A,C,D). Similarly, expression of GS was unchanged in Inx2BG02429-Gal4 > Appl-RNAi compared with controls (data not shown) while Inx2 was slightly upregulated (23.6 ± 1.6 vs 16.8 ± 0.9 for the controls, p < 0.5). In contrast, Inx2BG02429-Gal4 > Appl-RNAi flies displayed higher sleep amounts than the controls during the first day of induction at 30°C, and this change remained stable afterward. The increase in sleep occurred during the night, a period during which chronic Appl inhibition produced consistently the biggest changes in sleep (Figs. 2, 4B–D). The change in sleep was associated with a conspicuous decrease of GS immunofluorescence in the calyx neuropile, replicating the phenotype observed upon chronic inhibition of Appl, as shown in Figure 4F,I,L. Inx2 expression was also significantly downregulated but its cellular distribution was similar to that of controls, suggesting that the clumpy distribution observed in chronic Appl inactivation may be due to a longer inhibition and/or secondary effects (Fig. 4E,H,K). These results provide evidence that the consequences of Appl inhibition on sleep and GS/Inx2 expression are strongly correlated, and can occur in a relatively short time course at the adult stage. Since the expression of the Inx2BG02429-Gal4 driver used in these experiments is restricted to cortex glia, these results further indicate that Appl inhibition in one part of the glial network (cortex glia) may affect other, functionally connected compartments (astrocyte-like glia). Altogether, these observations suggest that Appl function in glial cells influences glutamate recycling. Further, it indicates that the communications between different glial compartments (i.e., astrocyte-like, ensheathing, and cortex glia) is abnormal in the mutant condition.
Figure 4.
Time-controlled manipulation of Appl expression in cortex glia increases night-time sleep and reduces GS and Inx2 protein levels. A, B, 24 h sleep amounts under permissive (A) and restrictive (B) temperatures. The temperature-sensitive Tub-Gal80ts construct driven by Inx2BG02429-Gal4 (inx2BG) was used to temporally control the expression of UAS-Appl-RNAiJF02878 (UAS-Appl-RNAi). At 30°C, sleep is reduced during the night as previously described (Ishimoto et al., 2012). C, D, No effect was observed at permissive temperature on either total sleep (C, left) or night-time sleep (D, left). Increasing temperature to 30°C significantly increased night-time sleep in Appl mutant flies (D, right; F = 3.854; p < 0.024; N = 37, 38, 39). E–J, Immunolabeling of Inx2 (E, H, G, J; green) and GS (F, I, G, J; magenta) in brains of flies exposed to restrictive temperatures (E–G) Inx2BG-Gal4/+;Tub-Gal80ts /+ and (H–J) Inx2BG-Gal4 Tub-Gal80ts>UAS-Appl-RNAi. K, L, Quantification of immunofluorescence show lower protein levels for Inx2 (K; Mann–Whitney; Z = 5.72; p < 0.00001; n = 3 each group) and for GS in the calyx upon conditional inhibition of Appl (L; Mann–Whitney, Z = 4.45; p < 0.00001; n = 3 each group). Scale bar, 20 μm.
Inx2 and GS expression are functionally linked to sleep/wake regulation
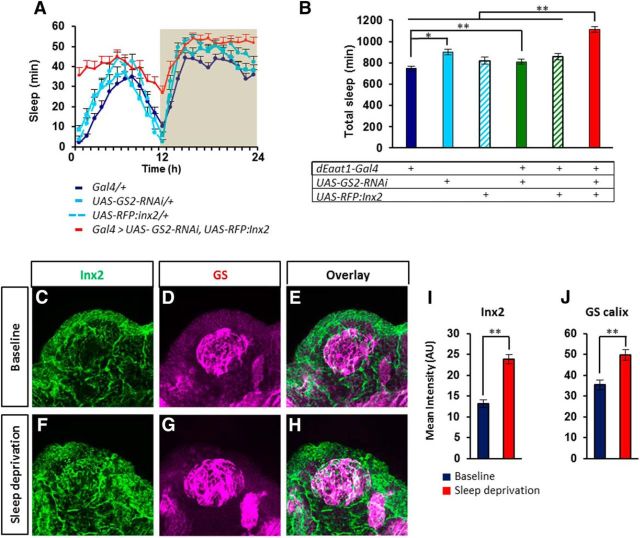
The close association between sleep and GS/Inx2 expression phenotypes observed upon Appl inhibition suggests that they could be functionally connected. To test this hypothesis, we downregulated GS expression and/or expressed a dominant-negative form of Inx2 (UAS-RFP:Inx2; Spéder and Brand, 2014) in glia. Individually, either of these two manipulations failed to affect sleep, whereas in combination they resulted in an increase in sleep similar to Appl inhibition in the same cells (Fig. 5A,B). To further assess the link between sleep/wake and GS/Inx2 expression, we evaluated their expression levels in wild-type flies under baseline condition and after a 25 h sleep deprivation. As shown in Figure 5C–J, GS and Inx2 expression levels in the sleep-deprived flies were higher than in the flies allowed to sleep, indicating that these two genes are dynamically linked to vigilance states.
Figure 5.
Inx2 and GS protein expression are linked to sleep/wake regulation. A, 24 h sleep amounts showing that the combined use of GS2-RNAi and dominant-negative UAS-RFP:Inx2 constructs to inhibit both GS and Inx2 function result in increased sleep. B, Total sleep time was only increased when both constructs where coexpressed and not upon expression of either one alone (F = 18.37; p < 0.00001; N = 32, 31, 32, 27, 32, 24). C–H, Immunolabeling of Inx2 (C, E, F, H; green) and GS (D, E, G, H; magenta) in Cs fly whole-mount brains dissected in the first hour of lights-on either under baseline condition (C–E) or after 25 h of sleep deprivation (F–H). I, J, Quantification of immunofluorescence shows increased Inx2 levels in the cortex (I; Z = 7.643; p < 0.00001; N = 8–9), and increased GS levels in the calyx (J; Z = 4.035; p < 0.000055; N = 8–9). Scale bar, 20 μm.
dEaat1 overexpression in cortex and astrocyte-like glia rescues the effect of Appl inhibition on sleep and GS expression
To confirm the implication of genes involved in glutamate recycling in the Appl-dependent regulation of sleep/wake, we overexpressed dEaat1 in cortex and astrocyte-like glia together with the Appl-RNAi transgene ApplJF02878. To sensitize the genetic background and allow the detection of functional interactions, we drove Appl-RNAiJF02878 without the UAS-dicer2 transgene, which significantly increases the efficiency of Appl-RNAi constructs. A UAS-GFP transgene was also added to the genetic background to assess a potential dilution effect of Gal4 in the presence of two UAS transgenes. In those conditions, expressing Appl-RNAiJF02878 resulted in a mild but significant increase in sleep (Fig. 6A) together with a decrease in GS expression in the calyx compared with control (Fig. 6C,D,F). These two phenotypes were rescued completely upon coexpression of UAS-dEaat1 (Fig. 6B–F), indicating that increasing the reuptake capacity of glial cells is sufficient to rescue both normal sleep/wake regulation and GS expression in the calyx. Overexpressing dEaat1 did not affect sleep/wake amounts (K-W, 8.31; p < 0.016; N = 22, 22, 23).
Figure 6.
dEaat1 overexpression in cortex and astrocyte-like glia counteracts the effect of Appl knockdown on sleep and GS expression. A, Baseline total sleep in flies coexpressing UAS-Appl-RNAiJF02878 and UAS-GFP in cortex and astrocyte-like glia under the control of dEaat1-Gal4. UAS-dicer2 was not included in the genotype. dEaat1-Gal4 > UAS-Appl-RNAiJF02878, UAS-GFP flies display significantly higher daily sleep amounts even in the absence of UAS-dicer2 (K-W, 29.24; p < 0.00001; N = 34, 35, 37). B, Coexpressing UAS-dEaat1 with UAS-Appl-RNAiJF02878 is sufficient to restore control-like sleep amounts, as confirmed by total daily sleep comparison between the genotypes (K-W, 0.72; not significant; N = 30, 31, 30). Dark blue, Glia Gal4/+ controls; light blue, UAS/+ controls; Red or green, corresponding experimental group. C–E, GS immunofluorescence quantification in calices of Eaat-Gal4/+ (C), dEaat1-Gal4 > UAS-Appl-RNAiJF02878, UAS-GFP (D), and dEaat1-Gal4 > UAS-Appl-RNAiJF02878, UAS-dEaat1 (E). F, GS immunofluorescence quantification in the calyx of the same genotypes (K-W, 43.13; p < 0.00001; N = 4 each group). Scale bar, 20 μm.
The inhibition of dEaat1 expression affects sleep/wake regulation
To further test the role of glutamate reuptake in sleep/wake regulation, we proceeded to inhibit the expression of dEaat1 in different glial-cell types and evaluated sleep–wake regulation. Higher amounts of wakefulness were observed upon inhibition of dEaat1 in perineurial and subperineurial glia, but the most severe phenotype was observed upon inhibition in astrocyte-like and cortex glia, using the dEaat1-Gal4 driver, that presumably reflects endogenous dEaat1 expression (Fig. 7A–C). Using this driver, the inhibition of dEaat1 expression led to a striking increase in wakefulness and to a disorganization of the circadian patterning of sleep. Interestingly GS levels appeared unaffected by the reduction of dEaat1 expression, suggesting that GS expression is not sensitive to the lack of glutamate uptake (Fig. 7D). These results demonstrate the importance of glutamate reuptake in sleep/wake regulation. They also indicate that the blocking of glutamate recycling at the level of the dEaat1 transporter produces a phenotype distinct from the one observed upon the inhibition of Appl expression in the same cells.
Figure 7.
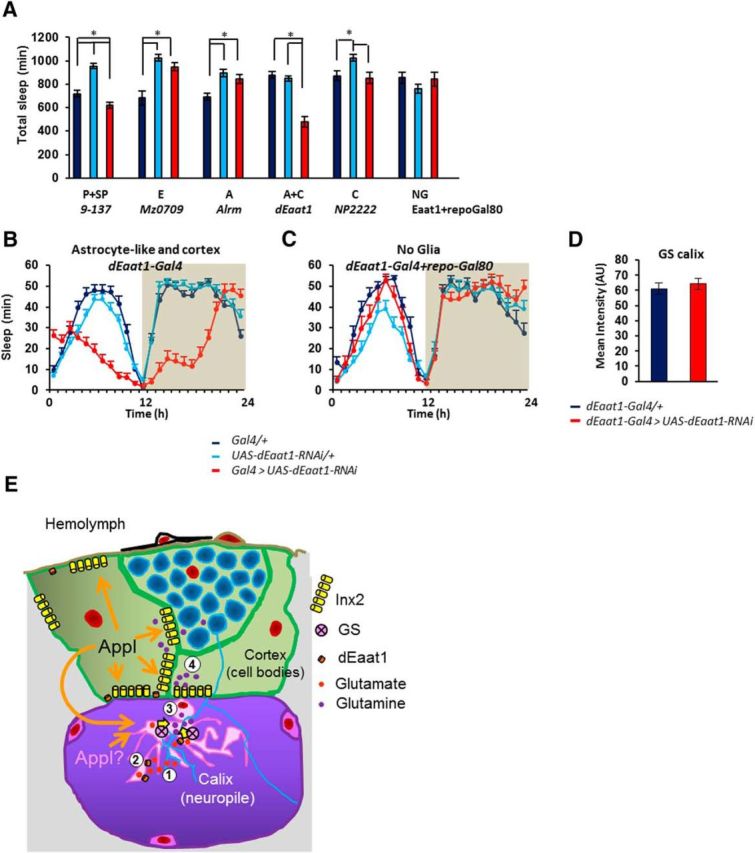
Knocking down the glutamate transporter dEaat1 specifically in cortex and astrocyte-like glia disrupts sleep. A, Total daily sleep upon expression of UAS-dEaat1-RNAiIR2 driven in different types of glia. P+SP, Perineurial and subperineurial (9–137-Gal4); F = 43.36; p < 0.00001; N = 37, 36, 38. E, Ensheathing (mz0709-Gal4); K-W, 18.56; p < 0.00093; N = 16, 16, 16. A, Astrocyte-like (Alrm-Gal4); F = 9.526; p < 0.00017; N = 32, 32, 31. A+C, Astrocyte-like and cortex (dEaat1-Gal4); K-W, 39.58; p < 0.00001; N = 36, 37, 36. C, Cortex (NP2222-Gal4); K-W, 11.66; p < 0.0024; n = 14, 16, 16. NG, No glia (repo-Gal80, dEaat1-Gal4); F = 1.019; p < 0 0.37; N = 16, 16, 16. B, Baseline sleep (minutes of sleep per hour) of dEaat1-Gal4 > UAS-dEaat1-RNAiIR2 flies: sleep is severely reduced and its circadian timing disrupted. C, Baseline sleep in repo-Gal80, dEaat1-Gal4 > UAS-dEaat1-RNAiIR2 flies is similar to that of controls. D, GS protein levels in calyx upon dEaat1 knock-down in cortex and astrocyte-like glia. Dark blue, Glia Gal4/+ controls; light blue, UAS-dEaat1-RNAiIR2/+ controls; red, corresponding glial Gal4 > UAS-dEaat1-RNAiIR2 knock-down (lines and bars). E, Model of Appl function in Drosophila glia. (1) Glutamate released at the synapses is (2) taken up by astrocyte-like glia expressing the dEaat1 transporter. Glutamate is converted to glutamine in astrocyte-like glia by GS (3) and diffuses via Inx2 gap junctions through the glial network (4). In the cortex glutamine could be eventually converted back to glutamate in neuronal cell bodies. Appl modulates both Inx2 and GS function during this process.
Discussion
The function of App remains elusive in glial cells (Müller and Zheng, 2013) and in other, non-neuronal cells. We report here that Appl modulates the expression of glutamate recycling genes and regulates sleep/wake. To carry out these functions, Appl is predominantly required in cortex glia, a cell type associated with neuronal cell bodies. Interestingly, recent reports provided evidence that calcium transients in this type of glia regulate neuronal activity (Melom and Littleton, 2013; Ma et al., 2016). Importantly, our results suggest that inhibiting Appl in cortex glia affects GS expression in another glial cell type, astrocyte-like glia, which closely interacts with neurites and synapses. In connection with these findings, the expression of Inx2, a major component of glial gap junctions at the interface between cortex and astrocyte-like glia is reduced upon Appl inhibition in cortex glia (Fig. 7E).
In mammals, App and the enzymes necessary for its cleavage have been reported to be present in astrocytes (Grolla et al., 2013). Glial cells are known to be adversely impacted by AD pathology, but these defects are often thought to be consequences of neuronal dysfunction, inflammation, or other noncell autonomous processes, and therefore unrelated to App normal function in glial cells. It is noticeable however, that one of the main cellular phenotypes observed in this study (i.e., alteration of GS expression) has been reported in numerous studies in rodent AD-related models as well as in human (Robinson, 2000; Olabarria et al., 2011; Kulijewicz-Nawrot et al., 2013), suggesting that it could be linked to a disruption of App function within astrocytes. In AD patients and experimental models (Ju et al., 2014), including Drosophila (Vanderheyden et al., 2013; Tabuchi et al., 2015), the pathology is also associated with reduced and highly fragmented sleep. We find here that the inhibition of Appl expression in glial cells results in higher sleep amounts and more consolidated sleep during the night. The sleep deficits observed in AD may thus result from both neuronal and glial dysfunction and probably neuroglial-signaling abnormalities. App proteins expressed in presynapses and postsynapses have been shown to influence each other's activities and to mediate cell–cell interactions (Wang et al., 2009). Similarly, App could mediate interglial and/or neuroglial interactions, possibly through cell adhesion and/or cell signaling. Because of these mutual interactions, toxic amyloid peptides produced and released by neurons could impair App function in glia, and explain the similarity of the cellular glial phenotypes observed here with those found in AD pathology. Finally, we cannot exclude the possibility that the decrease in GS expression observed in this study may reflect an impact of Appl inhibition on glial viability. However, the observation that Appl-null mutants are viable (Luo et al., 1992) and that overexpression produces a phenotype opposite to the inhibition is more in favor of a specific regulatory role for Appl.
GS plays a major role in glutamate and ammonium recycling by catalyzing the conversion of glutamate into glutamine, impacting both neurotransmission systems, as well as brain energy metabolism. The reduction of GS expression could lead to impaired glutamate–glutamine recycling and, possibly, to an accumulation of glutamate and excitotoxicity as hypothesized for AD. How App regulates GS expression remains unclear. In AD, it has been suggested that increased oxidation triggered by the pathology modifies the conformation of GS, such that it becomes nonfunctional and undetectable to antibodies. Alternatively, GS expression could be modulated by the intracellular concentrations of glutamate or glutamine (Robinson, 2000), and could reflect ongoing neuronal activity, as seen in this report in sleep-deprived conditions (Fig. 5). Blocking glutamate reuptake through inhibition of dEaat1 expression results in a dramatic increase in wakefulness, consistent with an enhancement of neuronal excitability and with recent findings indicating that glutamate plays an important role in sleep homeostasis (Rival et al., 2004; Tomita et al., 2015; Liu et al., 2016; Robinson et al., 2016). However, inhibition of dEaat1 expression does not seem to affect GS expression, suggesting that GS levels are not sensitive to intracellular glial glutamate concentration. Furthermore, inhibiting Appl in glia results in higher sleep amounts, a phenotype opposite to the one observed upon inhibition of dEaat1. In parallel, Appl inhibition results in an upregulation of dEaat1 transcripts (Fig. 3), which appeared localized preferentially at the periphery of the brain. Increasing further dEaat1 expression can rescue the longer sleep phenotype of Appl mutants, while it has no effect in wild-type flies (Fig. 6). These observations together suggest that Appl inhibition has consequences distinct from a mere blockade of glutamate recycling, but rather elicits compensatory mechanisms as a result of abnormal glutamate recycling.
In this context, Appl appears to be mainly required in cortex glia. Cortex glia cells are associated with neuronal cell bodies and are in contact with neuropile glia (ensheathing, astrocyte-like) as well as brain-hemolymph barrier glia (perineurial and subperineurial) (Awasaki et al., 2008). Being placed at the junction between nutrient supply and synaptic activity, cortex glia cells are thus strategically located to regulate neuronal metabolism. The bulk of GS expression changes observed in this study occurred in the brain neuropiles, which receive projections of astrocyte-like glia. It may be possible, as described for histamine in the visual system, that glutamate recycling involves long-distance interactions between several glial cell types (Chaturvedi et al., 2012). In support of this idea, we find that the expression of Inx2, a major glial gap junction protein, is also modulated bidirectionally by Appl expression. Inx2 expression is prominent at the junction between astrocyte-like/ensheathing and cortex glia, as well as between cortex glia and subperineurial glia, suggesting that it plays a key role in enabling the flow of small molecules through the different glial compartments. Both glutamate and glutamine can diffuse through gap junction (Giaume et al., 1997). Since insects lack a vascular system penetrating the brain, the network properties of glial cells should play a critical role in neurotransmitter recycling and in the distribution of nutrients. In mammals, App has been shown to be upregulated in glia devoid of connexin 43, a main gap junction involved in astrocyte coupling (Iacobas et al., 2004). Interestingly, astrocyte coupling is increased in mouse overexpressing App (Peters et al., 2009). The expression of astroglial connexins is upregulated in a mouse model of AD as well as in AD patients (Mei et al., 2010; Koulakoff et al., 2012) and alterations in gap junctions may contribute to the pathophysiology of the disease (Yi et al., 2016).
We find that both Inx2 and GS levels are increased following sleep deprivation. Similarly, App transcripts have been found to increase in mouse astrocytes following sleep deprivation (Bellesi et al., 2015). In addition, paradoxical sleep deprivation in rat has been reported to increase GS protein and messenger levels throughout the brain (Sallanon-Moulin et al., 1994), and in human, a Nuclear Magnetic Resonance study in sleep-deprived healthy subjects found increased glutamate and glutamine levels in selected regions of the brain (Murck et al., 2002). Together these results suggest that altered glutamatergic recycling is an evolutionary conserved consequence of sleep deprivation and may be linked to a modulation of App function, although further work is needed to confirm this model.
Altogether, these data emphasize the role of glia in sleep/wake regulation, a topic that so far has received little attention (Frank, 2013; Petit and Magistretti, 2016). Of note, the Notch transmembrane protein, which, like Appl, depends on γ-secretase for processing, and on mint/Spoc/spen proteins for intracellular signaling, has also been implicated in the glia-dependent regulation of sleep/wake in Drosophila (Seugnet et al., 2011). Interestingly, Notch also regulates glutamate transporter as well as the expression of several innexins during development (Lechner et al., 2007; Stacey et al., 2010). In rodents, Notch signaling genes are also predominantly expressed in astrocytes (Cahoy et al., 2008). Both Notch and App interact with the epidermal growth factor receptor pathway, which can modulate the expression of the dEaat1 glutamate transporter and sleep in Drosophila and in rodents (Kramer et al., 2001; Caillé et al., 2004; Lievens et al., 2005; Foltenyi et al., 2007). These observations suggest the existence of potentially complex interactions between multiple glial signaling pathways modulating both sleep/wake and glutamate recycling. Elucidating how these pathways interact in flexible models, such as Drosophila, will be an important step in understanding how they contribute to pathologies such as AD and affect sleep/wake in this context.
Footnotes
This work was supported by recurrent funding from the Institut National de la Santé et de la Recherche Médicale, Centre National de la Recherche Scientifique, Université Claude Bernard Lyon1, and by a special grant from Agence Nationale de la Recherche (AstroSleep R12123CC/RPV12027CCA). We thank T. Preat, V. Goguel, S. Birman, M. Freeman, U. Heberlein, and A. Brand for fly stocks; R. Bauer for anti-Inx2 antibodies; and the Centre de Imagerie Quantitative Lyon Est, confocal microscope facility and the ProfileXpert facility for help and technical advice. We thank S. Birman and C. Giaume for their advice during the project.
The authors declare no competing financial interests.
References
- Allebrandt KV, Amin N, Müller-Myhsok B, Esko T, Teder-Laving M, Azevedo RV, Hayward C, van Mill J, Vogelzangs N, Green EW, Melville SA, Lichtner P, Wichmann HE, Oostra BA, Janssens AC, Campbell H, Wilson JF, Hicks AA, Pramstaller PP, Dogas Z, et al. (2013) A K(ATP) channel gene effect on sleep duration: from genome-wide association studies to function in Drosophila. Molecular psychiatry 18:122–132. 10.1038/mp.2011.142 [DOI] [PubMed] [Google Scholar]
- Apelt J, Schliebs R, Beck M, Rossner S, Bigl V (1997) Expression of amyloid precursor protein mRNA isoforms in rat brain is differentially regulated during postnatal maturation and by cholinergic activity. Int J Dev Neurosci 15:95–112. 10.1016/S0736-5748(96)00073-1 [DOI] [PubMed] [Google Scholar]
- Awasaki T, Lai SL, Ito K, Lee T (2008) Organization and postembryonic development of glial cells in the adult central brain of Drosophila. J Neurosci 28:13742–13753. 10.1523/JNEUROSCI.4844-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellesi M, de Vivo L, Tononi G, Cirelli C (2015) Effects of sleep and wake on astrocytes: clues from molecular and ultrastructural studies. BMC Biol 13:66. 10.1186/s12915-015-0176-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdet I, Preat T, Goguel V (2015a) The full-length form of the Drosophila amyloid precursor protein is involved in memory formation. J Neurosci 35:1043–1051. 10.1523/JNEUROSCI.2093-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdet I, Lampin-Saint-Amaux A, Preat T, Goguel V (2015b) Amyloid-beta peptide exacerbates the memory deficit caused by amyloid precursor protein loss-of-function in Drosophila. PLoS One 10:e0135741. 10.1371/journal.pone.0135741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ, Barres BA (2008) A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci 28:264–278. 10.1523/JNEUROSCI.4178-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillé I, Allinquant B, Dupont E, Bouillot C, Langer A, Müller U, Prochiantz A (2004) Soluble form of amyloid precursor protein regulates proliferation of progenitors in the adult subventricular zone. Development 131:2173–2181. 10.1242/dev.01103 [DOI] [PubMed] [Google Scholar]
- Chaturvedi LS, Zhang P, Basson MD (2012) Effects of extracellular pressure and alcohol on the microglial response to inflammatory stimulation. Am J Surg 204:602–606. 10.1016/j.amjsurg.2012.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi R, Reddig K, Li HS (2014) Long-distance mechanism of neurotransmitter recycling mediated by glial network facilitates visual function in Drosophila. Proc Natl Acad Sci U S A 111:2812–2817. 10.1073/pnas.1323714111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson GR, Seabrook GR, Zheng H, Smith DW, Graham S, O'Dowd G, Bowery BJ, Boyce S, Trumbauer ME, Chen HY, Van der Ploeg LH, Sirinathsinghji DJ (1999) Age-related cognitive deficits, impaired long-term potentiation and reduction in synaptic marker density in mice lacking the beta-amyloid precursor protein. Neuroscience 90:1–13. 10.1016/S0306-4522(98)00410-2 [DOI] [PubMed] [Google Scholar]
- DeSalvo MK, Hindle SJ, Rusan ZM, Orng S, Eddison M, Halliwill K, Bainton RJ (2014) The Drosophila surface glia transcriptome: evolutionary conserved blood–brain barrier processes. Front Neurosci 8:346. 10.3389/fnins.2014.00346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltenyi K, Greenspan RJ, Newport JW (2007) Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nat Neurosci 10:1160–1167. 10.1038/nn1957 [DOI] [PubMed] [Google Scholar]
- Frank MG. (2013) Astroglial regulation of sleep homeostasis. Curr Opin Neurobiol 23:812–818. 10.1016/j.conb.2013.02.009 [DOI] [PubMed] [Google Scholar]
- Freeman MR, Delrow J, Kim J, Johnson E, Doe CQ (2003) Unwrapping glial biology: Gcm target genes regulating glial development, diversification, and function. Neuron 38:567–580. 10.1016/S0896-6273(03)00289-7 [DOI] [PubMed] [Google Scholar]
- Gerstner JR, Lenz O, Vanderheyden WM, Chan MT, Pfeiffenberger C, Pack AI (2016) Amyloid-β induces sleep fragmentation that is rescued by fatty acid binding proteins in Drosophila. J Neurosci Res. 10.1002/jnr.23778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaume C, Tabernero A, Medina JM (1997) Metabolic trafficking through astrocytic gap junctions. Glia 21:114–123. [DOI] [PubMed] [Google Scholar]
- Giaume C, Koulakoff A, Roux L, Holcman D, Rouach N (2010) Astroglial networks: a step further in neuroglial and gliovascular interactions. Nat Rev Neurosci 11:87–99. 10.1038/nrn2757 [DOI] [PubMed] [Google Scholar]
- Goguel V, Belair AL, Ayaz D, Lampin-Saint-Amaux A, Scaplehorn N, Hassan BA, Preat T (2011) Drosophila amyloid precursor protein-like is required for long-term memory. J Neurosci 31:1032–1037. 10.1523/JNEUROSCI.2896-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grolla AA, Fakhfouri G, Balzaretti G, Marcello E, Gardoni F, Canonico PL, DiLuca M, Genazzani AA, Lim D (2013) Aβ leads to Ca2+ signaling alterations and transcriptional changes in glial cells. Neurobiol Aging 34:511–522. 10.1016/j.neurobiolaging.2012.05.005 [DOI] [PubMed] [Google Scholar]
- Holcroft CE, Jackson WD, Lin WH, Bassiri K, Baines RA, Phelan P (2013) Innexins Ogre and Inx2 are required in glial cells for normal postembryonic development of the Drosophila central nervous system. J Cell Sci 126:3823–3834. 10.1242/jcs.117994 [DOI] [PubMed] [Google Scholar]
- Iacobas DA, Scemes E, Spray DC (2004) Gene expression alterations in connexin null mice extend beyond the gap junction. Neurochem Int 45:243–250. 10.1016/j.neuint.2003.12.008 [DOI] [PubMed] [Google Scholar]
- Ishimoto H, Lark A, Kitamoto T (2012) Factors that dfferentially affect daytime and nighttime sleep in Drosophila melanogaster. Front Neurol 3:24. 10.3389/fneur.2012.00024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju YE, Lucey BP, Holtzman DM (2014) Sleep and Alzheimer disease pathology—a bidirectional relationship. Nat Rev Neurol 10:115–119. 10.1038/nrneurol.2013.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JE, Lim MM, Bateman RJ, Lee JJ, Smyth LP, Cirrito JR, Fujiki N, Nishino S, Holtzman DM (2009) Amyloid-beta dynamics are regulated by orexin and the sleep–wake cycle. Science 326:1005–1007. 10.1126/science.1180962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koulakoff A, Mei X, Orellana JA, Sáez JC, Giaume C (2012) Glial connexin expression and function in the context of Alzheimer's disease. Biochim Biophys Acta 1818:2048–2057. 10.1016/j.bbamem.2011.10.001 [DOI] [PubMed] [Google Scholar]
- Kramer A, Yang FC, Snodgrass P, Li X, Scammell TE, Davis FC, Weitz CJ (2001) Regulation of daily locomotor activity and sleep by hypothalamic EGF receptor signaling. Science 294:2511–2515. 10.1126/science.1067716 [DOI] [PubMed] [Google Scholar]
- Kulijewicz-Nawrot M, Syková E, Chvátal A, Verkhratsky A, Rodríguez JJ (2013) Astrocytes and glutamate homoeostasis in Alzheimer's disease: a decrease in glutamine synthetase, but not in glutamate transporter-1, in the prefrontal cortex. ASN Neuro 5:273–282. 10.1042/AN20130017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner H, Josten F, Fuss B, Bauer R, Hoch M (2007) Cross regulation of intercellular gap junction communication and paracrine signaling pathways during organogenesis in Drosophila. Dev Biol 310:23–34. 10.1016/j.ydbio.2007.07.008 [DOI] [PubMed] [Google Scholar]
- Liévens JC, Rival T, Iché M, Chneiweiss H, Birman S (2005) Expanded polyglutamine peptides disrupt EGF receptor signaling and glutamate transporter expression in Drosophila. Hum Mol Genet 14:713–724. 10.1093/hmg/ddi067 [DOI] [PubMed] [Google Scholar]
- Liu S, Liu Q, Tabuchi M, Wu MN (2016) Sleep drive is encoded by neural plastic changes in a dedicated circuit. Cell 165:1347–1360. 10.1016/j.cell.2016.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo LQ, Martin-Morris LE, White K (1990) Identification, secretion, and neural expression of APPL, a Drosophila protein similar to human amyloid protein precursor. J Neurosci 10:3849–3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Tully T, White K (1992) Human amyloid precursor protein ameliorates behavioral deficit of flies deleted for Appl gene. Neuron 9:595–605. 10.1016/0896-6273(92)90024-8 [DOI] [PubMed] [Google Scholar]
- Ma Z, Stork T, Bergles DE, Freeman MR (2016) Neuromodulators signal through astrocytes to alter neural circuit activity and behaviour. Nature 539:428–432. 10.1038/nature20145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Morris LE, White K (1990) The Drosophila transcript encoded by the beta-amyloid protein precursor-like gene is restricted to the nervous system. Development 110:185–195. [DOI] [PubMed] [Google Scholar]
- McGuire SE, Mao Z, Davis RL (2004) Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci STKE 2004:pl6. [DOI] [PubMed] [Google Scholar]
- Mei X, Ezan P, Giaume C, Koulakoff A (2010) Astroglial connexin immunoreactivity is specifically altered at beta-amyloid plaques in beta-amyloid precursor protein/presenilin1 mice. Neuroscience 171:92–105. 10.1016/j.neuroscience.2010.08.001 [DOI] [PubMed] [Google Scholar]
- Melom JE, Littleton JT (2013) Mutation of a NCKX eliminates glial microdomain calcium oscillations and enhances seizure susceptibility. J Neurosci 33:1169–1178. 10.1523/JNEUROSCI.3920-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller UC, Zheng H (2013) Physiological functions of APP family proteins. Cold Spring Harb Perspect Med 2:a006288. 10.1101/cshperspect.a006288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murck H, Struttmann T, Czisch M, Wetter T, Steiger A, Auer DP (2002) Increase in amino acids in the pons after sleep deprivation: a pilot study using proton magnetic resonance spectroscopy. Neuropsychobiology 45:120–123. 10.1159/000054949 [DOI] [PubMed] [Google Scholar]
- Ni JQ, Liu LP, Binari R, Hardy R, Shim HS, Cavallaro A, Booker M, Pfeiffer BD, Markstein M, Wang H, Villalta C, Laverty TR, Perkins LA, Perrimon N (2009) A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics 182:1089–1100. 10.1534/genetics.109.103630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olabarria M, Noristani HN, Verkhratsky A, Rodríguez JJ (2011) Age-dependent decrease in glutamine synthetase expression in the hippocampal astroglia of the triple transgenic Alzheimer's disease mouse model: mechanism for deficient glutamatergic transmission? Mol Neurodegener 6:55. 10.1186/1750-1326-6-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn LM, Kamphuis W, Wadman WJ, Hol EM (2016) Astrogliosis: an integral player in the pathogenesis of Alzheimer's disease. Prog Neurobiol 144:121–141. 10.1016/j.pneurobio.2016.01.001 [DOI] [PubMed] [Google Scholar]
- Peters O, Schipke CG, Philipps A, Haas B, Pannasch U, Wang LP, Benedetti B, Kingston AE, Kettenmann H (2009) Astrocyte function is modified by Alzheimer's disease-like pathology in aged mice. J Alzheimers Dis 18:177–189. 10.3233/JAD-2009-1140 [DOI] [PubMed] [Google Scholar]
- Petit JM, Magistretti PJ (2016) Regulation of neuron-astrocyte metabolic coupling across the sleep–wake cycle. Neuroscience 323:135–156. 10.1016/j.neuroscience.2015.12.007 [DOI] [PubMed] [Google Scholar]
- Phinney AL, Calhoun ME, Wolfer DP, Lipp HP, Zheng H, Jucker M (1999) No hippocampal neuron or synaptic bouton loss in learning-impaired aged beta-amyloid precursor protein-null mice. Neuroscience 90:1207–1216. 10.1016/S0306-4522(98)00645-9 [DOI] [PubMed] [Google Scholar]
- Rival T, Soustelle L, Strambi C, Besson MT, Iché M, Birman S (2004) Decreasing glutamate buffering capacity triggers oxidative stress and neuropil degeneration in the Drosophila brain. Curr Biol 14:599–605. [DOI] [PubMed] [Google Scholar]
- Rival T, Soustelle L, Cattaert D, Strambi C, Iché M, Birman S (2006) Physiological requirement for the glutamate transporter dEAAT1 at the adult Drosophila neuromuscular junction. J Neurobiol 66:1061–1074. 10.1002/neu.20270 [DOI] [PubMed] [Google Scholar]
- Robinson JE, Paluch J, Dickman DK, Joiner WJ (2016) ADAR-mediated RNA editing suppresses sleep by acting as a brake on glutamatergic synaptic plasticity. Nat Commun 7:10512. 10.1038/ncomms10512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson SR. (2000) Neuronal expression of glutamine synthetase in Alzheimer's disease indicates a profound impairment of metabolic interactions with astrocytes. Neurochem Int 36:471–482. [DOI] [PubMed] [Google Scholar]
- Rouach N, Koulakoff A, Abudara V, Willecke K, Giaume C (2008) Astroglial metabolic networks sustain hippocampal synaptic transmission. Science 322:1551–1555. 10.1126/science.1164022 [DOI] [PubMed] [Google Scholar]
- Sallanon-Moulin M, Touret M, Didier-Bazes M, Roudier V, Fages C, Tardy M, Jouvet M (1994) Glutamine synthetase modulation in the brain of rats subjected to deprivation of paradoxical sleep. Brain Res Mol Brain Res 22:113–120. [DOI] [PubMed] [Google Scholar]
- Seugnet L, Suzuki Y, Merlin G, Gottschalk L, Duntley SP, Shaw PJ (2011) Notch signaling modulates sleep homeostasis and learning after sleep deprivation in Drosophila. Curr Biol 21:835–840. 10.1016/j.cub.2011.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PJ, Cirelli C, Greenspan RJ, Tononi G (2000) Correlates of sleep and waking in Drosophila melanogaster. Science 287:1834–1837. 10.1126/science.287.5459.1834 [DOI] [PubMed] [Google Scholar]
- Shaw PJ, Tononi G, Greenspan RJ, Robinson DF (2002) Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature 417:287–291. 10.1038/417287a [DOI] [PubMed] [Google Scholar]
- Shrivastava AN, Kowalewski JM, Renner M, Bousset L, Koulakoff A, Melki R, Giaume C, Triller A (2013) β-amyloid and ATP-induced diffusional trapping of astrocyte and neuronal metabotropic glutamate type-5 receptors. Glia 61:1673–1686. 10.1002/glia.22548 [DOI] [PubMed] [Google Scholar]
- Spéder P, Brand AH (2014) Gap junction proteins in the blood–brain barrier control nutrient-dependent reactivation of Drosophila neural stem cells. Dev Cell 30:309–321. 10.1016/j.devcel.2014.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey SM, Muraro NI, Peco E, Labbé A, Thomas GB, Baines RA, van Meyel DJ (2010) Drosophila glial glutamate transporter Eaat1 is regulated by fringe-mediated notch signaling and is essential for larval locomotion. J Neurosci 30:14446–14457. 10.1523/JNEUROSCI.1021-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi M, Lone SR, Spira AP, Wu Correspondence MN, Liu S, Liu Q, Zhang J, Wu MN (2015) Sleep interacts with Aβ to modulate intrinsic neuronal excitability. Curr Biol 25:702–712. 10.1016/j.cub.2015.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita J, Ueno T, Mitsuyoshi M, Kume S, Kume K (2015) The NMDA receptor promotes sleep in the fruit fly, Drosophila melanogaster. PLoS One 10:e0128101. 10.1371/journal.pone.0128101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderheyden WM, Gerstner JR, Tanenhaus A, Yin JC, Shaw PJ (2013) ERK phosphorylation regulates sleep and plasticity in Drosophila. PLoS One 8:e81554. 10.1371/journal.pone.0081554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wang B, Yang L, Guo Q, Aithmitti N, Songyang Z, Zheng H (2009) Presynaptic and postsynaptic interaction of the amyloid precursor protein promotes peripheral and central synaptogenesis. J Neurosci 29:10788–10801. 10.1523/JNEUROSCI.2132-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Kang H, Xu Q, Chen MJ, Liao Y, Thiyagarajan M, O'Donnell J, Christensen DJ, Nicholson C, Iliff JJ, Takano T, Deane R, Nedergaard M (2013) Sleep drives metabolite clearance from the adult brain. Science 342:373–377. 10.1126/science.1241224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C, Mei X, Ezan P, Mato S, Matias I, Giaume C, Koulakoff A (2016) Astroglial connexin43 contributes to neuronal suffering in a mouse model of Alzheimer's disease. Cell Death Differ 23:1691–1701. 10.1038/cdd.2016.63 [DOI] [PMC free article] [PubMed] [Google Scholar]