Abstract
The proper formation of synapses—specialized unitary structures formed between two neurons—is critical to mediating information flow in the brain. Synaptic cell adhesion molecules (CAMs) are thought to participate in the initiation of the synapse formation process. However, in vivo functional analysis demonstrates that most well known synaptic CAMs regulate synaptic maturation and plasticity rather than synapse formation, suggesting that either CAMs work synergistically in the process of forming synapses or more CAMs remain to be found. By screening for unknown CAMs using a co-culture system, we revealed that protein tyrosine phosphatase receptor type O (PTPRO) is a potent CAM that induces the formation of artificial synapse clusters in co-cultures of human embryonic kidney 293 cells and hippocampal neurons cultured from newborn mice regardless of gender. PTPRO was enriched in the mouse brain and localized to postsynaptic sites at excitatory synapses. The overexpression of PTPRO in cultured hippocampal neurons increased the number of synapses and the frequency of miniature EPSCs (mEPSCs). The knock-down (KD) of PTPRO expression in cultured neurons by short hairpin RNA (shRNA) reduced the number of synapses and the frequencies of the mEPSCs. The effects of shRNA KD were rescued by expressing either full-length PTPRO or a truncated PTPRO lacking the cytoplasmic domain. Consistent with these results, the N-terminal extracellular domain of PTPRO was required for its synaptogenic activity in the co-culture assay. Our data show that PTPRO is a synaptic CAM that serves as a potent initiator of the formation of excitatory synapses.
SIGNIFICANCE STATEMENT The formation of synapses is critical for the brain to execute its function and synaptic cell adhesion molecules (CAMs) play essential roles in initiating the formation of synapses. By screening for unknown CAMs using a co-culture system, we revealed that protein tyrosine phosphatase receptor type O (PTPRO) is a potent CAM that induces the formation of artificial synapse clusters. Using loss-of-function and gain-of-function approaches, we show that PTPRO promotes the formation of excitatory synapses. The N-terminal extracellular domain of PTPRO was required for its synaptogenic activity in cultured hippocampal neurons and the co-culture assay. Together, our data show that PTPRO is a synaptic CAM that serves as a potent initiator of synapse formation.
Keywords: co-culture, electrophysiology, morphologic, PTPRO, synapse formation, synaptic cell adhesion molecules
Introduction
A synapse is an elementary structure that allows a neuron to communicate with other neurons (or target cells) through the release of neurotransmitters. The proper formation of a synapse is essential to the construction of neural circuits and cognitive functions and alterations in this process lead to many neurological disorders such as autism spectrum disorders (ASDs) and mental retardation (McAllister, 2007; Südhof, 2008; Zhang et al., 2009; Boda et al., 2010). Major efforts have been made to define the molecular composition of synapses (Südhof, 2004; Südhof and Malenka, 2008; Harris and Weinberg, 2012; Pereda, Alberto, 2014; Sando et al., 2017) and now our understanding of how synapses function in presynaptic terminals and postsynaptic spines has greatly expanded. However, much less is known about the molecular machinery that determines the targeted initiation of synapse formation, partly due to the enormous variety of synapses in the brain. Synaptic cell adhesion molecules (CAMs) were originally assumed to enable mechanical cell–cell recognition and play an important role in initiating the formation of synapses through trans-synaptic interactions (Sanes and Yamagata, 2009; Missler et al., 2012; Yang et al., 2014). Moreover, CAMs are responsible for assembling neurotransmitter receptors and the cytoskeleton (Biederer and Südhof, 2001; Missler et al., 2003; Zhang et al., 2010). For instance, deletion of presynaptic neurexins impairs the function of the postsynaptic NMDA or AMPA receptor in an isoform-specific manner (Kattenstroth et al., 2004; Aoto et al., 2013). Furthermore, presynaptic neurexins and postsynaptic neuroligins (NLs) form tight trans-synaptic interactions controlled by various splicing sites on both of these proteins (Ullrich et al., 1995), providing an intriguing hypothesis for the synaptic specificity that requires further investigation. SynCAMs are also considered likely candidates for initiating the formation of synapses because they bind to themselves in a trans-synaptic and calcium-independent manner (Biederer et al., 2002; Robbins et al., 2010). This calcium independence appears to be important because the removal of calcium does not prevent the formation of immature synapses or morphologically disrupt existing synapses in cultured neurons (Pfenninger, 1971; Cotman and Taylor, 1972).
However, evidence from analyses of most known CAM-knock-out (KO) mice indicates that these CAMs mediate synapse maturation and synaptic plasticity rather than the initiation of synapse formation. For example, knocking out neurexins 1, 2, and 3 impairs Ca2+-triggered neurotransmitter release, but has little effect on the number of synapses formed (Missler et al., 2003). Similarly, the deletion of NL1 reduces the NMDA/AMPA receptor ratio in hippocampal CA3-CA1 synapses (Chubykin et al., 2007). Although NL1/2/3 triple-KO mice exhibit a postnatal lethal phenotype and impaired synaptic transmission at the GABAergic/glycinergic and glutamatergic synapses, the number of synapses is normal in the brain of NL1/2/3 triple-KO mice, suggesting that NLs are not essential for initiating synapse formation (Varoqueaux et al., 2006). Moreover, SynCAM1-KO mice exhibit a modest (10 ± 3%) decrease in the number of excitatory synapses, but not inhibitory synapses, in the hippocampal CA1 stratum radiatum (Robbins et al., 2010). Furthermore, the number of presynaptic and postsynaptic terminals is normal in the hippocampal CA1 and dentate gyrus areas in EphB2-KO mice (Henderson et al., 2001), leucine-rich transmembrane 1 (LRRTM1)-KO mice (Linhoff et al., 2009), and N-cadherin cKO mice (Bozdagi et al., 2010). Though many known CAMs regulate the strength of the synaptic transmission, neuronal activity does not seem to be required for the formation of synapses (Verhage et al., 2000; Sando et al., 2017). Therefore, the complete molecule machinery for synapse formation remains elusive.
In this study, we performed an unbiased screen using an artificial synapse formation (ASF) assay that uses co-cultured non-neuronal cells and neurons together to identify the activity of synaptogenesis. The ASF assay has been shown to be a powerful system for screening CAMs and several families of known CAMs have tested positive in ASF assays, including neurexins and NLs, SynCAMs (Biederer et al., 2002; Robbins et al., 2010), ephrinBs and EphBs (Kayser et al., 2006, 2008; Aoto et al., 2007), NGLs/LRRC4s (Wang et al., 2003; Kim et al., 2006; Woo et al., 2009; DeNardo et al., 2012), and LRRTMs (Ko et al., 2009, 2011; Linhoff et al., 2009; Ko, 2012). By screening an unbiased cDNA collection containing 286 full-length open reading frames (ORFs) with sizes exceeding 3 kb in the present study, we identified PTPRO as a potent mediator of synapse formation. Interestingly, three intronic single nucleotide polymorphisms (SNPs) in the gene encoding PTPRO (also called glomerular epithelial protein 1, or GLEPP1) were reported recently to be strongly correlated to learning and memory function in patients with schizophrenia and bipolar disorder (LeBlanc et al., 2012; Hendriks and Pulido, 2013). We revealed that the synaptogenic function of PTPRO is dependent on its extracellular region. Further analysis of PTPRO function using immunocytochemistry and electrophysiology in cultured hippocampal neurons suggested that PTPRO promotes synapse formation and increases synaptic strength in neurons.
Materials and Methods
ATTL/ATTR site (LR).
ORFs with sizes exceeding 3 kb in the hORFeome V8.1 library were selected. Gateway@ LR cloning reactions were performed according to the manufacturer's instructions (Thermo Fisher Scientific). Briefly, the entry clone (75 ng/μl and 75 ng per reaction) insert was cloned in the pcDNA3.2-v5-DEST (75 ng/μl and 75 ng per reaction) expression vector using LR reactions at pH 8.0 TE buffer (10 mm Tris-HCl, pH 8.0, 1 mm EDTA; 2 μl/reaction). LR Clonase enzyme mix (0.5 μl/reaction) was added to each reaction and the reactions were incubated at 25°C for 2 h. Proteinase K solution (0.25 μl/reaction) was added to each reaction and the reactions were incubated for 10 min at 37°C to digest the LR Clonase. pENTR-gus (50 ng/μl) was used as a positive control. Four microliters of each LR reaction were transformed using 50 μl of competent Escherichia coli cells using the heat-shock method, and the E. coli were then plated on ampicillin plates. The clones were screened by digesting the colony DNA with the BglII restriction enzyme and the selected colonies were sent to a sequencing facility at Peking University for verification. The screening library was composed of all full-length ORFs with sizes exceeding 3 kb from the 2 public genome-wide cDNA libraries: the hORFeome V8.1 library and the hORFeome V8.1 lenti collection.
Experimental constructs.
The coding sequence of V5 (GKPIPNPLLGLDST)-tagged PTPRO was PCR amplified from the original plasmid (pcDNA3.2 PTPRO-v5-DEST) and subcloned into the NheI and BamHI site of a pFUGW expression vector (Jiang et al., 2015) with PCR. Isoform 2 of PTPRO was cloned into the NheI and XhoI site of PCAG using overlapping PCR. Isoforms 1, 3, and 4 of PTPRO were cloned into the NheI and XhoI site of the PCAG vector using PCR. PTPROCTD encoded the extracellular sequences, the transmembrane region, and a short cytoplasmic tail. PTPRONTD coded for the signal peptide, the transmembrane region, and the full cytoplasmic tail. Mutations in PTPRO were made in the NheI and XhoI site of the PCAG vector using PCR. PCR was performed with the following protocol on a MyCycler Thermal Cycler (Bio-Rad): 98°C for 90 s, 98°C for 30 s, 55°C for 30 s, 72°C for 2 min (30 cycles), 72°C for 5 min, and a final hold at 4°C. Primers are described in Table 1.
Table 1.
Summary of constructs and oligos used in this study
| Name | Forward oligo | Reverse oligo |
|---|---|---|
| Fugw-PTPRO-V5 | gcCCATTGTGTCTGTGGTGTCGCTGACCTGCCAGA | GCGGATCCCTCATTACTAACCGGTACG |
| PCAG-PTPRO isoform 1 | CTAGCTAGCGCCACCATGGGGCACCTG | CCGCTCGAGCTACAAGGACTTGCTAACATTCTCGT |
| PCAG-PTPRO isoform 2 | CTAGCTAGCGCCACCATGGGGCACCTG | CCGCTCGAGCTACAAGGACTTGCTAACATTCTCGT |
| PCAG-PTPRO isoform 3 | CTAGCTAGCGCCACCATGGTTACAGAGATGAATCCCAAT | CCGCTCGAGCTACAAGGACTTGCTAACATTCTCGT |
| PCAG-PTPRO isoform 4 | CTAGCTAGCGCCACCATGGTTACAGAGATGAATCCCAAT | CCGCTCGAGCTACAAGGACTTGCTAACATTCTCGT |
| PCAG-PTPRO CTD | CTAGCTAGCGCCACCATGGGGCACCTG | CCGCTCGAGCTATTTATAGTCAGAGTCTTTGGCCATAT |
| PCAG-PTPRO NTD | CTAGCTAGCGCCACCATGGGGCACCTG | CCGCTCGAGCTACAAGGACTTGCTAACATTCTCGT |
Human embryonic kidney (HEK) 293T cell culture and transfection.
HEK 293T cells were grown at 37°C supplied with 5% CO2 in an incubator with a humidified atmosphere (Jiang et al., 2015). The cells were grown in DMEM containing 10% fetal bovine serum. The cells were washed once using PBS and digested with 0.05% TE buffer (Invitrogen) at 37°C for routine passage of the cells. All of the HEK 293T cell transfections were performed using the polyethylenimine (PEI) method. The PEI (1 mg/ml in ddH2O):DNA ratio was 3:1. The PEI/DNA mixture was incubated for 30 min at room temperature before the mixture was added to the HEK 293T cell cultures dropwise. For screening, 1.5 μg of each plasmid was transfected in 1 well of a 24-well plate together with 0.5 μg of pFUGW-GFP. For the isoforms of and mutations in PTPRO, 3 μg of plasmid was transfected in 1 well of a 6-well plate together with 1 μg of pFUGW-GFP.
Isolation of proteins from HEK 293T cells.
The transfected HEK 293T cells were harvested 2 d after transfection. The HEK 293T cells were washed with PBS once, kept at −80°C overnight, and thawed at 37°C for 1 min. Then, the cells were collected and centrifuged at 12,000 × g for 1 min at 4°C to obtain the cell pellet. The cell pellets were incubated at 4°C for 60 min in buffer A (20 mm HEPES-NaOH, pH 7.4, containing 1% Triton, 0.1 mm EDTA, 2 mm CaCl2, 1 mm MgCl2, and 100 mm NaCl with protease inhibitors, including 1 mm PMSF, 1 μg/ml pepstatin, 1 μg/ml leupeptin, and 2 μg/ml aprotinin). The supernatant containing the membrane fraction was collected for further analysis by removing the insoluble tissues with centrifugation at 12,000 × g for 30 min.
Fraction preparations and Western blotting.
Presynaptic and postsynaptic fractions were prepared as described previously (Yang et al., 2015). Briefly, brains were homogenized in HEPES-buffered sucrose solution (0.32 m sucrose, 4 mm HEPES, pH 7.4) and centrifuged at 1000 × g for 10 min at 4°C. The supernatant was centrifuged at 10,000 × g for 15 min. The crude synaptosomal pellet was then lysed in 4 mm HEPES, pH 7.4, and rotated for 30 min at 4°C after washing once with the HEPES-buffered sucrose solution. The lysate was centrifuged at 20,000 rpm (rotor: TLS 55) for 20 min to yield the pellet (the lysed synatosomal membrane fraction). The pellet was resuspended in buffer B (50 mm HEPES, pH 7.4, 2 mm EDTA, 0.5% Triton X-100, and proteinase inhibitors) and centrifuged at 22,000 rpm after being rotated for 15 min at 4°C. The supernatant extracts were the presynaptic fractions and the pellets were resuspended in buffer B as the postsynaptic fractions.
Protein extracts were denatured at 80°C for 10 min and separated on NuPAGE (Life Technologies) precast 10% SDS-PAGE gels at 200 V for ∼1 h. The proteins were transferred to nitrocellulose (NC) filters at 40 V for 2.5 h. The NC membrane was initially blocked with 5% nonfat milk and 2% goat serum (v/v) in Tris-buffered saline with 0.1% Tween 20 (TBS-T) at room temperature for 1 h. Monoclonal antibodies to β-actin (CW0096A; Cwbiotech), PSD95 (BD Biosciences), synaptophysin (D-4; Santa Cruz Biotechnology), the V5 tag (CW0094M; Cwbiotech), and polyclonal antibodies to PTPRO (SC-66908; Santa Cruz Biotechnology) and GAPDH (M20006; Abmart) were used for Western blot analyses as primary antibodies at 4°C overnight. After 3 washes of 5 min each with TBS-T, goat anti-rabbit or anti-mouse IgG was added at a dilution of 1:20,000 as the secondary antibody. The NC membrane was scanned with an infrared imaging system (Odyssey; LI-COR).
Animals.
C57BL/6J male wild-type mice at postnatal day 0 (P0)–P56 were used for this study. All animal studies were conducted at the Association for Assessment and Accreditation of Laboratory Animal Care-approved Animal Facility in the Laboratory Animal Center, Peking University. Experiments were undertaken in accordance with the National Institutes of Health's Guide for the Care and Use of Laboratory Animals (eighth edition). All experimental protocols were approved by the Institutional Animal Care and Use Committee of Peking University. Mice were housed separately in a temperature- and humidity-controlled room under a 12 h light/dark cycle with ad libitum access to food and water. All efforts were made to minimize animal suffering and to reduce the number of animals used.
Neuronal culture and artificial synapse formation.
Cultured neurons were obtained from C57BL/6J mouse hippocampal cells, as described previously (Lee et al., 2017; Wei et al., 2017). Briefly, mouse hippocampal cells were dissected from P0 wild-type mice, dissociated with 0.25% trypsin (Invitrogen), digested for 12 min at 37°C, plated on poly-D-lysine-coated glass coverslips (∅ 8 mm) at a density of 80,000 neurons per coverslip (μScope CellCounter Basic; C.E.T.), and maintained at 37°C in 5% CO2. ASF assays were performed using HEK 293T cells as described previously (Zhang et al., 2009). Briefly, for the expression screen, HEK 293T cells were transfected in 24-well plates with 1.5 μg of plasmid from each ORF sequence and 0.5 μg of pFUGW-GFP as a visual marker. After 24 h, the transfected HEK 293T cells were digested with 0.05% TE and seeded on the hippocampal neuron cultures at 9 d in vitro (DIV). The co-cultured mixture was maintained in an incubator for 36–48 h for immunocytochemistry. The plasmids of the positive control, NL2 (a gift from Dr. Thomas C. Südof, Stanford University) and NL3 (clone HsCD00438909 in the hORFeome V8.1 lenti collection), were included in every batch of ASF screening.
To quantify the synaptic activity of all molecules in ASF screening, a custom program was used in high-content microscopy to analyze the images. Briefly, the oval model (defined radius range: 5–30 μm) was first used to find the transfected (GFP-positive) HEK 293T cells as the target. Then, the fluorescence intensity of synapsin 1 was obtained to illustrate all of the synapses. Third, the synapsin density in a ring that included a 2 μm radius inward and outward from the edge of the target HEK 293T cells was counted to reflect the clustered synapse number (DensityZ1synapsin). After that, the synapsin density in a 4-μm-radius ring 10 μm away from the target HEK 293T cells was counted as the background synapse distribution (DensityZ2synapsin). The co-culture index (CI) was calculated with the function CI = (DensityZ1synapsin − DensityZ2synapsin)/DensityZ2synapsin). The transfected HEK 293T cells with CI ≥ 1 were counted as positive cells.
Calcium phosphate transfection.
Hippocampal neurons were transfected using the calcium phosphate transfection method after 10 d of incubation (DIV 10) and analyzed on DIV 14–15, as described previously (Zhang et al., 2010). Briefly, for each coverslip on a 48-well plate, 0.6 μg of the total plasmid was mixed with 0.99 μl of the 2 m CaCl2 solution and dH2O to reach a final volume of 8 μl and the DNA/CaCl2 solution was added slowly to 8 μl of 2× HBS (per each 500 ml: 8 g of NaCl, 0.213 g of Na2HPO4, and 6.5 g of HEPES, pH 7.00–7.05). The DNA/CaCl2/HBS solution was incubated at room temperature for 30 min and then added to the neuronal cell cultures and incubated for 30 min in an incubator. The cells were washed once with a medium containing MgCl2 and maintained in an incubator for 3–5 d before electrophysiological recordings or immunocytochemistry.
Development and validation of PTPRO shRNAs.
The shRNA plasmid (target sequence: 5′-GCT AAG AAT GTA GTT CCT AT-3′) targeting the intracellular domain of mouse PTPRO (PTPROKD) was selected from the Sigma-Aldrich MISSION shRNA Library. The knock-down (KD) effects of the PTPRO shRNAs were validated in cultured hippocampal neurons with quantitative immunostaining and RT-PCR.
RNA isolation and quantitative RT-PCR.
The samples were homogenized in a glass–Teflon homogenizer according to the protocol supplied with the TRIzol Reagent (Life Technologies). The concentration of RNA was measured with spectrophotometry. The reaction volume consisted of 2 μg of total RNA, 5× buffer (Takara), Rt enzyme mix (Takara), oligo (dT) (Takara), Random6 primer (Takara), and RNase-free H2O (to a final volume of 20 μl). The amplification program was as follows: 37°C for 15 min, 85°C for 5 s, and a final hold at 4°C. Quantitative PCR was performed in an MX 3000PTM (Agilent Technologies) RT-PCR system with 2× SYBR Green qPCR Mix (Aidlab PC3302) using the designed primers. Relative expression levels were calculated using the 2−ΔΔCT method. To quantify the KD efficiency, the cultured neurons were infected with a virus that expressed shRNA against PTPRO at DIV 4; the mRNA was collected at DIV 14 and quantified using the RT-PCR method described above.
Immunocytochemistry and culture imaging.
For screening, the co-cultures were fixed for 12 min with 4% paraformaldehyde and 4% sucrose in PBS, pH 7.4, followed by permeabilization with 0.2% Triton X-100 (v/v) in PBS. An initial blocking step was performed with PBS-MILK/NGS (PBS containing 5% milk and 3% normal goat serum) for 30 min at room temperature. Co-cultures for overexpression or KD were incubated overnight with anti-synapsin 1 (1:20,000; Synaptic Systems), anti-vGLUT1 (1:2000; Sigma-Aldrich), anti-GAD-65 (1:5000; Sigma-Aldrich), anti-vGAT (1:500; Synaptic Systems), or anti-PTPRO (1:50; Santa Cruz Biotechnology) antibodies diluted in PBS-MILK/NGS. After washing with PBS, co-cultures or cultures for overexpression or KD were incubated with an Alexa Fluor 546-conjugated goat anti-rabbit (1:500; Invitrogen) or Fluor 546-conjugated goat anti-mouse (1:500; Invitrogen) antibody to detect synapsin 1, vGLUT1, GAD-65, vGAT, or PTPRO. After washing in PBS, the samples were mounted with a mounting medium (Southern Biotech).
For the co-cultures, images were acquired with a high-content microscope (Molecular Devices) with a 40× objective lens. For co-localization, images were acquired with a confocal microscope (Olympus FV1000) and a superresolution microscope (Leica TCS SP8 STED). The co-localization ratio was measured based on the overlap of PTPRO/VGlut1 or PTPRO/GAD65. For overexpression or KD, transfected neurons were chosen randomly and images were acquired using a confocal microscope with a 60× objective lens; all image settings were maintained for all samples. Z-stacked confocal images were converted to maximum projections and analyzed with respect to the size and density of the presynaptic terminals using ImageJ software. To quantify the KD efficiency using immunostaining, the fluorescent intensity of the transfected neurons was normalized with the neighboring untransfected neurons. The laser intensity for each batch of cultures was set up carefully so that there was no saturation in the fluorescent intensity to avoid the ceiling effect. All imaging experiments and image processing were performed with the operators blinded.
Electrophysiological recordings.
Electrophysiological recordings were performed as described previously (Maximov et al., 2007; Wei et al., 2016). Whole-cell voltage-clamp recordings were obtained from hippocampal neurons with a MultiClamp 700A amplifier (Molecular Devices). Patch pipettes were pulled from borosilicate glass capillary tubes (World Precision Instruments) using a pipette puller. The resistance of the pipettes filled with the intracellular solution varied between 3 and 5 MΩ. The bath solution contained the following (in mm): 150 NaCl, 4 KCl, 1 MgCl2, 2 CaCl2, 10 HEPES, and 10 glucose, pH 7.4, adjusted with NaOH. The pipette solution contained the following (in mm): 145 KCl, 5 NaCl, 10 HEPES, 5 EGTA, 0.3 Na2GTP, and 4 MgATP, pH 7.2, adjusted with KOH. In all recordings, pyramidal neurons were voltage clamped at −60 mV. The data were digitized at 10 kHz with a 2 kHz low-pass filter. The miniature EPSCs (mEPSCs) were monitored in the presence of 1 μm tetrodotoxin (TTX) and 100 μm picrotoxin (PTX). The miniature IPSCs (mIPSCs) were recorded in the presence of 1 μm TTX and 10 μm CNQX. Series resistance was compensated to 60–70% and recordings with series resistances of >;20 MΩ were rejected. The data were analyzed using Clampfit 9.02 (Molecular Devices), Igor 4.0 (WaveMetrics), and Prism 5 (GraphPad) software. The frequency and amplitude of the mEPSCs and the mIPSCs were measured using a built-in template-search method in Clampfit software.
Results
Screening for synaptogenic molecules with a custom cDNA library identified PTPRO as a candidate molecule
To search for more potential synaptogenic molecules, we performed an unbiased screen using an ASF assay, which co-cultured non-neuronal cells and neurons together to identify the activity of synaptogenesis. Previous screening for synaptogenic molecules using the ASF assay showed enrichment for small molecules, which was partly due to the construction strategy used to generate the cDNA library and partly due to the pooling strategy used for screening in the cultured cells. The exogenous expression of proteins in non-neuronal cells, such as HEK 293T cells, is known to be proportional to the size of the cDNA insert; therefore, although the pool method enables higher throughput analysis, it is less effective at screening large molecules. To overcome this shortcoming, we screened an unbiased cDNA collection containing 286 full-length ORFs with sizes exceeding 3 kb individually.
We constructed a ready-for-expression cDNA library using LR cloning methods to transfer all 217 ORFs larger than 3 kb from the hORFeome V8.1 library to the pcDNA3.2 v5-DEST backbone. Furthermore, we selected 69 additional and nonoverlapping cDNA plasmids from the hORFeome V8.1 lenti collection using the same criteria and combined these two collections to screen for CAMs (Table 2). The average size of the cDNA included in the combined library was 3.59 ± 0.03 kb and the sizes ranged from 3.0 to 7.7 kb (Fig. 1A,B). We expressed individual cDNAs in HEK 293T cells for 24 h and then co-cultured the HEK 293T cells with hippocampal neurons at DIV 9 for an additional 48 h (Fig. 1C). Synapse staining with an antibody against synapsin 1 (a presynaptic marker) was performed and a custom program (see Materials and Methods) was executed to identify positive candidates semiautomatically based on the CI measurement (Fig. 1D). NL2 (in the pCMV5 backbone) and NL3 (in the pLX304 backbone) were included as positive controls in ASF assay and an empty pCMV5 vector was used as the negative control. The screen revealed that the overexpression of PTPRO in the HEK 293T cells recruited synapsin-positive puncta from co-cultured neurons (Fig. 1E). Quantitative analysis showed that the synaptogenic activity of PTPRO was comparable to that of NL3 in the ASF assay (Fig. 1F), although the number of synapse-clustered positive HEK 293T cells with overexpression of PTPRO was lower than that of NL3.
Table 2.
Summary of the ORFs >;3 kb from the hORFeome V8.1 library and the hORFeome V8.1 lenti collection
| hORFeome v8.1 library | ||||
|---|---|---|---|---|
| ORF_ID: | ORF_SIZE: | GENE_ID: | Gene_symbol | |
| 81020@A01 | 5060 | 3006 | 9810 | RNF40 |
| 81131@D06 | 72141 | 3036 | 80031 | SEMA6D |
| 81090@E01 | 10067 | 3048 | 11234 | HPS5 |
| 81020@D09 | 10026 | 3072 | 9754 | STARD8 |
| 81020@G01 | 2449 | 3084 | 23550 | PSD4 |
| 81131@E07 | 72121 | 3111 | 153090 | DAB2IP |
| 81020@F02 | 2357 | 3006 | 23228 | PLCL2 |
| 81132@D04 | 56205 | 3072 | 54972 | TMEM132A |
| 81043@A09 | 10725 | 3084 | 64799 | IQCH |
| 81020@G03 | 56198 | 3114 | 199713 | NLRP7 |
| 81020@G02 | 2537 | 3006 | 10919 | EHMT2 |
| 81020@C01 | 4969 | 3039 | 7318 | UBA7 |
| 81020@D03 | 55889 | 3060 | 3416 | IDE |
| 81128@E01 | 11831 | 3075 | 23133 | PHF8 |
| 81020@F07 | 14246 | 3087 | 4641 | MYO1C |
| 81020@H03 | 56215 | 3114 | 3092 | HIP1 |
| 81131@F06 | 72024 | 3039 | 51230 | PHF20 |
| 81020@E03 | 55892 | 3060 | 57835 | SLC4A5 |
| 81131@B07 | 71235 | 3075 | 114791 | TUBGCP5 |
| 81043@D09 | 10424 | 3189 | 55345 | ZGRF1 |
| 81132@B04 | 70416 | 3009 | 79789 | CLMN |
| 81020@B09 | 6246 | 3042 | 478 | ATP1A3 |
| 81020@H05 | 7471 | 3060 | 23367 | LARP1 |
| 81131@C07 | 72028 | 3075 | 29953 | TRHDE |
| 81131@D07 | 72022 | 3090 | 84626 | KRBA1 |
| 81020@A04 | 56263 | 3123 | 57575 | PCDH10 |
| 81020@D07 | 14322 | 3030 | 133584 | EGFLAM |
| 81131@G06 | 11440 | 3042 | 57659 | ZBTB4 |
| 81132@E04 | 70730 | 3075 | 51191 | HERC5 |
| 81020@C12 | 53115 | 3093 | 4659 | PPP1R12A |
| 81020@F05 | 56914 | 3123 | 6900 | CNTN2 |
| 81020@E07 | 14243 | 3030 | 3980 | LIG3 |
| 81020@D12 | 52852 | 3045 | 10529 | NEBL |
| 81131@A07 | 12227 | 3063 | 3993 | LLGL2 |
| 81020@E09 | 9701 | 3078 | 55226 | NAT10 |
| 81020@H07 | 53155 | 3096 | 10198 | MPHOSPH9 |
| 81131@F07 | 11038 | 3123 | 9937 | DCLRE1A |
| 81020@A09 | 1182 | 3033 | 55753 | OGDHL |
| 81020@C09 | 9398 | 3048 | 23360 | FNBP4 |
| 81131@H06 | 71949 | 3063 | 2731 | GLDC |
| 81020@F03 | 55895 | 3078 | 64135 | IFIH1 |
| 81020@F09 | 10125 | 3102 | 9871 | SEC24D |
| 81112@D05 | 14495 | 3168 | 29761 | USP25 |
| 81128@H01 | 13706 | 4734 | 26509 | MYOF |
| 81131@B10 | 71900 | 3627 | 10752 | CHL1 |
| 81131@D10 | 72134 | 3786 | 23242 | COBL |
| 81131@F10 | 72023 | 3804 | 9895 | TECPR2 |
| 81131@H08 | 71305 | 3357 | 23196 | FAM120A |
| 81112@E04 | 10827 | 3561 | 23287 | AGTPBP1 |
| 81131@A08 | 71113 | 3201 | 49854 | ZBTB21 |
| 81131@B11 | 72146 | 4011 | 23162 | MAPK8IP3 |
| 81131@D11 | 71032 | 4026 | 387680 | FAM21A |
| 81131@F11 | 72039 | 4302 | 10076 | PTPRU |
| 81131@H09 | 1428 | 3600 | 9654 | TTLL4 |
| 81112@F04 | 10880 | 3666 | 170692 | ADAMTS18 |
| 81131@A09 | 12393 | 3453 | 5523 | PPP2R3A |
| 81131@C08 | 54625 | 3240 | 23506 | GLTSCR1L |
| 81131@E08 | 71200 | 3318 | 55729 | ATF7IP |
| 81131@G07 | 71068 | 3129 | 23517 | SKIV2L2 |
| 81131@H10 | 72173 | 3858 | 4646 | MYO6 |
| 81112@G04 | 11939 | 3729 | 3667 | IRS1 |
| 81131@A10 | 72217 | 3612 | 57530 | CGN |
| 81131@C09 | 72036 | 3519 | 7058 | THBS2 |
| 81131@E09 | 72048 | 3546 | 10207 | PATJ |
| 81131@G08 | 54264 | 3351 | 79820 | CATSPERB |
| 81131@H11 | 10256 | 4425 | 2 | A2M |
| 81127@A11 | 13519 | 3495 | 29109 | FHOD1 |
| 81131@A11 | 71706 | 3909 | 84952 | CGNL1 |
| 81131@C10 | 70975 | 3675 | 55717 | WDR11 |
| 81131@E10 | 72159 | 3801 | 23158 | TBC1D9 |
| 81131@G09 | 11113 | 3564 | 51294 | PCDH12 |
| 81127@G01 | 71355 | 1869 | 57613 | FAM234B |
| 81131@A12 | 72165 | 5010 | 50618 | ITSN2 |
| 81131@C11 | 72170 | 4017 | 5198 | PFAS |
| 81131@E11 | 10392 | 4143 | 1362 | CPD |
| 81131@G10 | 71055 | 3828 | 63967 | CLSPN |
| 81132@A05 | 70819 | 3321 | 23022 | PALLD |
| 81128@F01 | 10155 | 3390 | 3717 | JAK2 |
| 81131@B08 | 14564 | 3213 | 26018 | LRIG1 |
| 81131@D08 | 72191 | 3300 | 55035 | NOL8 |
| 81131@F08 | 71313 | 3318 | 26953 | RANBP6 |
| 81131@G11 | 9982 | 4389 | 9742 | IFT140 |
| 81132@A06 | 70236 | 3624 | 79937 | CNTNAP3 |
| 81128@G01 | 14896 | 4599 | 178 | AGL |
| 81131@B09 | 72211 | 3462 | 84441 | MAML2 |
| 81131@D09 | 72057 | 3531 | 51520 | LARS |
| 81131@F09 | 71834 | 3564 | 5784 | PTPN14 |
| 81131@H07 | 72222 | 3198 | 57585 | CRAMP1 |
| 81132@A07 | 10432 | 4059 | 23189 | KANK1 |
| 81020@H04 | 56238 | 3516 | 9889 | ZBED4 |
| 81021@B01 | 53920 | 3846 | 2903 | GRIN2A |
| 81021@F01 | 55164 | 4293 | 1612 | DAPK1 |
| 81043@C09 | 10319 | 3150 | 26091 | HERC4 |
| 81067@B10 | 14236 | 3225 | 5337 | PLD1 |
| 81090@G01 | 8467 | 3546 | 5923 | RASGRF1 |
| 81020@H06 | 3304 | 3696 | 10735 | STAG2 |
| 81021@B02 | 56347 | 4896 | 128239 | IQGAP3 |
| 81021@F02 | 56876 | 4077 | 30849 | PIK3R4 |
| 81043@E03 | 3538 | 3228 | 4775 | NFATC3 |
| 81067@C10 | 14413 | 3420 | 1639 | DCTN1 |
| 81101@E01 | 2914 | 3393 | 6601 | SMARCC2 |
| 81020@H08 | 55128 | 3750 | 83990 | BRIP1 |
| 81021@B03 | 56354 | 4263 | 8202 | NCOA3 |
| 81021@G01 | 55217 | 4308 | 11073 | TOPBP1 |
| 81043@E09 | 10765 | 3258 | 80216 | ALPK1 |
| 81067@D10 | 14242 | 3432 | 667 | DST |
| 81020@H09 | 10091 | 3150 | 51284 | TLR7 |
| 81021@C01 | 53498 | 3855 | 23090 | ZNF423 |
| 81021@G02 | 55703 | 4065 | 672 | BRCA1 |
| 81043@F03 | 5380 | 3396 | 55764 | IFT122 |
| 81067@E10 | 14264 | 3627 | 9666 | DZIP3 |
| 81101@G02 | 11638 | 3153 | 84079 | ANKRD27 |
| 81020@H10 | 4203 | 3219 | 5979 | RET |
| 81021@D01 | 55187 | 3999 | 6655 | SOS2 |
| 81021@H01 | 55210 | 5130 | 1105 | CHD1 |
| 81043@F09 | 10546 | 3492 | 9699 | RIMS2 |
| 81067@F08 | 12443 | 3264 | 6829 | SUPT5H |
| 81101@G03 | 14398 | 3399 | 9659 | PDE4DIP |
| 81020@H11 | 53071 | 3201 | 9354 | UBE4A |
| 81021@D02 | 56879 | 4146 | 22878 | TRAPPC8 |
| 81021@H02 | 56882 | 3960 | 57579 | FAM135A |
| 81043@G09 | 10737 | 3645 | 1659 | DHX8 |
| 81067@F10 | 14256 | 3687 | 22915 | MMRN1 |
| 81101@H01 | 8769 | 3429 | 259266 | ASPM |
| 81021@A01 | 56066 | 3807 | 5858 | PZP |
| 81021@E01 | 55150 | 4290 | 27030 | MLH3 |
| 81043@A10 | 10282 | 3750 | 7174 | TPP2 |
| 81067@A10 | 14235 | 3129 | 84640 | USP38 |
| 81067@G08 | 12412 | 3294 | 3843 | IPO5 |
| 81101@H02 | 11619 | 3444 | 6720 | SREBF1 |
| 81021@A02 | 56884 | 4938 | 23318 | ZCCHC11 |
| 81021@E02 | 55704 | 4116 | 55914 | ERBIN |
| 81043@B09 | 10760 | 3087 | 56916 | SMARCAD1 |
| 81067@A11 | 14415 | 3816 | 22828 | SCAF8 |
| 81090@F01 | 6198 | 3537 | 5091 | PC |
| 81101@H03 | 13970 | 3651 | 23236 | PLCB1 |
| 81020@A02 | 4899 | 3129 | 8449 | DHX16 |
| 81020@B04 | 55893 | 3213 | 5291 | PIK3CB |
| 81020@CO2 | 1601 | 3144 | 5921 | RASA1 |
| 81020@D04 | 56329 | 3300 | 121256 | TMEM132D |
| 81020@E05 | 56931 | 3351 | 23272 | FAM208A |
| 81020@F10 | 9540 | 3192 | 22983 | MAST1 |
| 81020@A05 | 53638 | 3540 | 3682 | ITGAE |
| 81020@B05 | 53929 | 3723 | 55160 | ARHGEF10L |
| 81020@C04 | 56249 | 3231 | 23426 | GRIP1 |
| 81020@D08 | 53660 | 3348 | 134957 | STXBP5 |
| 81020@E06 | 10044 | 3663 | 9669 | EIF5B |
| 81020@F11 | 4470 | 3306 | 47 | ACLY |
| 81020@A06 | 2462 | 3192 | 57674 | RNF213 |
| 81020@B06 | 1543 | 3465 | 118987 | PDZD8 |
| 81020@C05 | 56892 | 3660 | 26030 | PLEKHG3 |
| 81020@D06 | 4767 | 3660 | 4796 | TONSL |
| 81020@E08 | 54110 | 3426 | 5139 | PDE3A |
| 81020@G04 | 54430 | 3435 | 109 | ADCY3 |
| 81020@A07 | 2547 | 3744 | 26148 | C10orf12 |
| 81020@B07 | 5025 | 3762 | 23191 | CYFIP1 |
| 81020@C06 | 2474 | 3561 | 2073 | ERCC5 |
| 81020@D08 | 53660 | 3348 | 134957 | STXBP5 |
| 81020@E10 | 2397 | 3192 | 10013 | HDAC6 |
| 81020@G06 | 3493 | 3684 | 9785 | DHX38 |
| 81020@A08 | 53623 | 3147 | 7764 | ZNF217 |
| 81020@B08 | 56866 | 3207 | 89910 | UBE3B |
| 81020@C07 | 5228 | 3798 | 5336 | PLCG2 |
| 81020@D10 | 9930 | 3186 | 64072 | CDH23 |
| 81020@E11 | 1767 | 3279 | 10208 | USPL1 |
| 81020@G08 | 55213 | 3651 | 5800 | PTPRO |
| 81020@A10 | 7871 | 3153 | 701 | BUB1B |
| 81020@B10 | 1526 | 3186 | 10517 | FBXW10 |
| 81020@C08 | 54245 | 3312 | 1730 | DIAPH2 |
| 81020@D11 | 2395 | 3270 | 9675 | TTI1 |
| 81020@F04 | 54832 | 3414 | 23431 | AP4E1 |
| 81020@G09 | 9288 | 3138 | 479 | ATP12A |
| 81020@A11 | 8331 | 3228 | 25890 | ABI3BP |
| 81020@B11 | 5900 | 3258 | 6560 | SLC12A4 |
| 81020@C10 | 9461 | 3186 | 91662 | NLRP12 |
| 81020@E02 | 4778 | 3150 | 3678 | ITGA5 |
| 81020@F06 | 2542 | 3669 | 23262 | PPIP5K2 |
| 81020@G10 | 7715 | 3201 | 55112 | WDR60 |
| 81020@A12 | 53119 | 3162 | 57167 | SALL4 |
| 81020@B12 | 52724 | 3126 | 51311 | TLR8 |
| 81020@C11 | 2044 | 3264 | 23039 | XPO7 |
| 81020@E04 | 56064 | 3357 | 9101 | USP8 |
| 81020@F08 | 54295 | 3486 | 27127 | SMC1B |
| 81020@G11 | 3898 | 3381 | 7917 | BAG6 |
| 81132@A08 | 70210 | 5181 | 22852 | ANKRD26 |
| 81132@C05 | 55906 | 3336 | 343450 | KCNT2 |
| 81132@D08 | 70297 | 7701 | 84700 | MYO18B |
| 81132@F06 | 70209 | 3933 | 54825 | CDHR2 |
| 81132@H06 | 70293 | 4050 | 219578 | ZNF804B |
| 81132@C06 | 70415 | 3708 | 4853 | NOTCH2 |
| 81132@E01 | 70320 | 5574 | 9256 | TSPOAP1 |
| 81132@F07 | 70171 | 4515 | 51735 | RAPGEF6 |
| 81132@H07 | 70306 | 4872 | 23046 | KIF21B |
| 81132@B01 | 12011 | 4515 | 11188 | NISCH |
| 81132@C07 | 70446 | 4134 | 51199 | NIN |
| 81132@E05 | 70500 | 3135 | 5293 | PIK3CD |
| 81132@G04 | 53073 | 3276 | 108 | ADCY2 |
| 81132@B05 | 70119 | 3321 | 9732 | DOCK4 |
| 81132@C08 | 70287 | 5346 | 23094 | SIPA1L3 |
| 81132@E06 | 70377 | 3849 | 9790 | BMS1 |
| 81132@G05 | 10634 | 3555 | 2199 | FBLN2 |
| 81132@B06 | 70803 | 3678 | 10734 | STAG3 |
| 81132@D01 | 72199 | 5355 | 7249 | TSC2 |
| 81132@E07 | 70239 | 4338 | 286234 | SPATA31E1 |
| 81132@G06 | 70219 | 3951 | 84132 | USP42 |
| 81132@B07 | 70265 | 4062 | 115 | ADCY9 |
| 81132@D05 | 70259 | 3354 | 23384 | SPECC1L |
| 81132@E08 | 72181 | 5769 | 23405 | DICER1 |
| 81132@G07 | 70241 | 4584 | 8714 | ABCC3 |
| 81132@B08 | 70671 | 3222 | 3655 | ITGA6 |
| 81132@D06 | 70266 | 3714 | 5101 | PCDH9 |
| 81132@F04 | 70264 | 3273 | 23368 | PPP1R13B |
| 81132@H04 | 70807 | 3276 | 55799 | CACNA2D3 |
| 81132@C01 | 72144 | 5322 | 84624 | FNDC1 |
| 81132@D07 | 10669 | 4173 | 5797 | PTPRM |
| 81132@F05 | 70249 | 3423 | 9662 | CEP135 |
| 81132@H05 | 70697 | 3135 | 55130 | ARMC4 |
| hORFeome V8.1 Lenti collection | ||||
|---|---|---|---|---|
| lenti_clone id | ORF_ID: | ORF_SIZE: | GENE_ID: | Gene_symbol |
| ccsbBroad304_14065 | 14293 | 3000 | 11196 | SEC23IP |
| ccsbBroad304_14417 | 56286 | 5325 | 161497 | STRC |
| N/A | 6718 | 3009 | 4343 | MOV10 |
| ccsbBroad304_09184 | 2539 | 3012 | 84343 | HPS3 |
| ccsbBroad304_06534 | 53326 | 3075 | 4012 | LNPEP |
| ccsbBroad304_13940 | 9532 | 3204 | 5933 | RBL1 |
| ccsbBroad304_04224 | 10859 | 3057 | 80789 | INTS5 |
| ccsbBroad304_05864 | 13723 | 3060 | 477 | ATP1A2 |
| ccsbBroad304_08776 | 8450 | 3072 | 58484 | NLRC4 |
| ccsbBroad304_06613 | 53115 | 3090 | 4659 | PPP1R12A |
| ccsbBroad304_00972 | 11662 | 3120 | 4123 | MAN2C1 |
| ccsbBroad304_08275 | 52724 | 3123 | 51311 | TLR8 |
| ccsbBroad304_11273 | 4899 | 3123 | 8449 | DHX16 |
| ccsbBroad304_00884 | 4778 | 3147 | 3678 | ITGA5 |
| ccsbBroad304_14896 | 2586 | 3156 | 8467 | SMARCA5 |
| ccsbBroad304_09321 | 9461 | 3183 | 91662 | NLRP12 |
| ccsbBroad304_01784 | 1960 | 3213 | 7514 | XPO1 |
| ccsbBroad304_07957 | 8331 | 3225 | 25890 | ABI3BP |
| ccsbBroad304_12577 | 10772 | 3420 | 79632 | FAM184A |
| ccsbBroad304_02519 | 1972 | 3309 | 10749 | KIF1C |
| ccsbBroad304_01048 | 9491 | 3294 | 4542 | MYO1F |
| ccsbBroad304_10970 | 56210 | 3312 | 4293 | MAP3K9 |
| ccsbBroad304_06708 | 8409 | 3318 | 5159 | PDGFRB |
| ccsbBroad304_06747 | 4775 | 3321 | 5424 | POLD1 |
| ccsbBroad304_06702 | 53366 | 3336 | 5140 | PDE3B |
| ccsbBroad304_07852 | 9694 | 3651 | 23199 | KIAA0182 |
| ccsbBroad304_09069 | 14436 | 3564 | 81545 | FBXO38 |
| ccsbBroad304_06818 | 53116 | 3345 | 5794 | PTPRH |
| ccsbBroad304_14081 | 53197 | 3270 | 23368 | PPP1R13B |
| ccsbBroad304_07506 | 3877 | 3261 | 9898 | UBAP2L |
| ccsbBroad304_14233 | 53218 | 3351 | 57562 | KIAA1377 |
| ccsbBroad304_02081 | 56064 | 3354 | 9101 | USP8 |
| ccsbBroad304_02734 | 53080 | 3354 | 23196 | FAM120A |
| ccsbBroad304_07456 | 10749 | 3372 | 9645 | MICAL2 |
| ccsbBroad304_13965 | 53077 | 3372 | 6935 | ZEB1 |
| ccsbBroad304_07005 | 1880 | 3411 | 6764 | ST5 |
| ccsbBroad304_09625 | 56926 | 3459 | 146183 | OTOA |
| ccsbBroad304_12439 | 13749 | 4521 | 63977 | PRDM15 |
| ccsbBroad304_11097 | 14176 | 3444 | 5981 | RFC1 |
| ccsbBroad304_12823 | 2565 | 5502 | 84464 | SLX4 |
| ccsbBroad304_06463 | 55903 | 3456 | 3684 | ITGAM |
| ccsbBroad304_14392 | 1855 | 3447 | 144406 | WDR66 |
| ccsbBroad304_07208 | 2561 | 3474 | 8204 | NRIP1 |
| ccsbBroad304_13954 | 1643 | 3723 | 6522 | SLC4A2 |
| ccsbBroad304_02925 | 2547 | 3741 | 26148 | C10orf12 |
| ccsbBroad304_06851 | 11294 | 3741 | 5949 | RBP3 |
| ccsbBroad304_07469 | 6127 | 3558 | 9698 | PUM1 |
| ccsbBroad304_00024 | 54430 | 3432 | 109 | ADCY3 |
| ccsbBroad304_14209 | 1715 | 3963 | 55818 | KDM3A |
| ccsbBroad304_07248 | 1152 | 3606 | 8500 | PPFIA1 |
| ccsbBroad304_14026 | 8232 | 4617 | 9765 | ZFYVE16 |
| ccsbBroad304_08557 | 10357 | 3894 | 55626 | AMBRA1 |
| ccsbBroad304_07463 | 10044 | 3660 | 9669 | EIF5B |
| ccsbBroad304_02743 | 2542 | 3729 | 23262 | PPIP5K2 |
| ccsbBroad304_11346 | 7283 | 4644 | 9202 | ZMYM4 |
| ccsbBroad304_02153 | 10108 | 4275 | 9372 | ZFYVE9 |
| ccsbBroad304_07454 | 2132 | 4173 | 9631 | NUP155 |
| ccsbBroad304_14076 | 56338 | 4827 | 23240 | KIAA0922 |
| ccsbBroad304_13814 | 53302 | 4506 | 394 | ARHGAP5 |
| ccsbBroad304_02250 | 56346 | 4572 | 9816 | URB2 |
| ccsbBroad304_07265 | 10374 | 4941 | 8567 | MADD |
| ccsbBroad304_07511 | 55154 | 4944 | 9928 | KIF14 |
| ccsbBroad304_09025 | 55136 | 5100 | 80309 | SPHKAP |
| ccsbBroad304_15058 | 56063 | 3861 | 50937 | CDON |
| ccsbBroad304_07126 | 6043 | 3792 | 7407 | VARS |
| ccsbBroad304_07998 | 55496 | 3792 | 26166 | RGS22 |
| ccsbBroad304_11682 | 12685 | 4401 | 23132 | RAD54L2 |
| ccsbBroad304_08389 | 10642 | 4107 | 54505 | DHX29 |
| ccsbBroad304_02556 | 11608 | 4098 | 10908 | PNPLA6 |
Figure 1.

Expression screening identifies PTPRO as a synaptogenic molecule. A, A total of 217 ORFs larger than 3 kb were selected from the hORFeome V8.1 gene library for expression screening. B, Representative image of an immunoblot for the V5 tag from HEK 293T cell lysates expressing a test set of expression clones. C, Schematic drawing of the screening of expression in hippocampal neurons co-cultured with HEK 293T cells expressing target DNA. D, Analysis criteria of the CI, an indication of the synaptic density. Two micrometers for the inner and outer radii of the ring were chosen for all the data presented except in this panel. Each red dot represents an individual synapse. E1, E2, Representative images of co-cultured neurons with HEK 293T cells expressing control, NL2 (positive control), NL3 (positive control), or PTPRO, stained for synapsin 1 and GFP. Scale bars: E1, 20 μm; E2, 5 μm. F, Summary graphs of the percentage of positive cells (top) and the CI of positive cells (bottom) from the control, pFUGW-PTPRO-transfected, pLX304-NL3-transfected, or pCMV5-NL2-transfected HEK 293T cells co-cultured with hippocampal neurons (control: n = 0 cell of 165 cells/3 cultures, PTPRO: n = 28 cells of 630 cells/3 cultures, NL3: n = 15 cells of 113 cells/3 cultures, and NL2: n = 32 cells of 41 cells/3 cultures). For all representative images, scale bars apply to all panels in a set. All summary graphs show the mean ± SEM; statistical comparisons were made with Student's t test (***p < 0.001; n.s., not significant).
Synaptogenic activity of PTPRO is specific to isoforms 1 and 2
Four PTPRO isoforms have been identified thus far (Beltran et al., 2003) and we next investigated whether the synaptogenic ability of PTPRO is isoform specific. Our results demonstrate that isoforms 1 and 2 specifically induced synapse formation, whereas isoforms 3 and 4 could not cluster synapses (Fig. 2A,B). Because the domain structures of isoforms 1 and 2 of PTPRO are almost identical, we focused on isoform 1 in the following study. Because isoforms 3 and 4 encode short forms of PTPRO in which most of the extracellular domain is missing, we hypothesized that this domain is required for synapse formation. To test this idea, we generated N-terminal and C-terminal deletion constructs of human PTPRO isoform 1 (hPTPRONTD and hPTPROCTD; Fig. 2A) and tested their effects in the ASF assay. We found that the extracellular domain of PTPRO is sufficient to induce the aggregation of synapsin-positive puncta and the intracellular phosphatase domain is dispensable (Fig. 2B,D). Next, to test whether the synaptogenic activity of PTPRO is specific to a particular type of synapse, we examined synapse specificity by immunostaining with antibodies against vesicular glutamate transporter 1 (vGLUT1; an excitatory presynaptic marker) and glutamic acid decarboxylase 65 (GAD-65; an inhibitory presynaptic marker). The results showed that PTPRO recruited vGLUT1- and GAD-65-positive puncta in the co-culture system (Fig. 2C,E), suggesting that PTPRO has a general role in initiating synapse formation in the ASF assay.
Figure 2.

PTPRO expressed in non-neural cells induces the formation of artificial synapses in a co-culture assay. A, Diagram of expression constructs for human PTPRO (hPTPRO) isoforms and mutations. The longest of the PTPRO isoform (isoform 1) contains a single transmembrane domain (TMD, yellow) flanked by eight fibronectin repeat III domains (gray) in the extracellular region and a phosphatase domain (green) in the cytoplasmic tail. B, Representative images of hippocampal neurons co-cultured with HEK 293T cells expressing FUGW-GFP together with hPTPRO isoforms 1–4 or hPTPROCTD or hPTPRONTD, stained for synapsin 1. Scale bars, 20 μm. C, Representative images of hippocampal neurons co-cultured with HEK 293T cells expressing either an empty vector (control) or a vector encoding hPTPRO stained for vGlut1 or GAD65. Scale bars, 20 μm. D, E, Summary graphs of the percentage of positive cells (CI values >; 1 were defined as positive cells) and CI values of positive cells from B and C; synapsin-positive cell of D, control: n = 0 cell of 169 cells/3 cultures; isoform 1: n = 11 cells of 204 cells/3 cultures; isoform 2: n = 9 cells of 183 cells/3 cultures; isoform 3: n = 0 cell of 212 cells/3 cultures; isoform 4: n = 0 cell of 225 cells/3 cultures; hPTPROCTD: n = 8 cells of 176 cells/3 cultures; hPTPRONTD: n = 0 cell of 230 cells/3 cultures; and E: vGlut1-positive cell, control: n = 0 cell of 138 cells/3 cultures; PTPRO: n = 16 cells of 88 cells/3 cultures; GAD65-positive cell, control: n = 0 cell of 87 cells/3 cultures; PTPRO: n = 14 cells of 72 cells/3 cultures. For all representative images, scale bars apply to all panels in a set. All summary graphs show the mean ± SEM; statistical comparisons were made with Student's t test (n.s., not significant).
Localization of endogenous PTPRO to synapses
PTPRO belongs to the family of type III receptor protein tyrosine phosphatases (RPTPs). The other three members of this family are vascular endothelial-protein tyrosine phosphatase (VE-PTP, also known as PTPRB), density-enriched PTP-1 (DEP-1, also known as PTPRJ), and stomach cancer-associated protein tyrosine phosphatase-1 (SAP-1, also known as PTPRH). Then, we tested whether PTPRB or PTPRH (PTPRJ is not in our cDNA library) has synaptogenic activity in the ASF assay. The results showed that PTPRB or PTPRH could not induce detectable synaptogenesis in the ASF assay (Fig. 3A). Next, to examine the tissue distribution of PTPRO, we performed RT-PCR to detect the mRNA levels of PTPRO in various tissues and to compare its distribution with that of other RPTP family members. PTPRO was found specifically in the brain and kidneys, whereas the other three RPTP family members exhibited different tissue distributions (Fig. 3B).
Figure 3.

PTPRO is enriched in the brain and localized to the synapses. A, Summary graphs of the percentage of positive cells in hippocampal neurons co-cultured with HEK 293T cells expressing FUGW-GFP together with control, type III RPTPs, type IIa RPTPs, or type IIb RPTPs stained for synapsin 1. B, RT-PCR analysis of PTPRO and type III RPTPs mRNA levels in C57BL/6J adult mice tissues, including the heart, brain, liver, kidneys, and lungs. C, RT-PCR analysis of PTPRO mRNA levels in C57BL/6J adult mice brains, including the brainstem, cortex, hippocampus, olfactory bulb, and cerebellum. D, RT-PCR quantitation of PTPRO mRNA levels in C57BL/6J mouse brains from P0 to P56. E, PTPRO expression levels in different brain regions. CBC, Cerebellar cortex; MD, mediodorsal nucleus of the thalamus; STR, striatum; AMY, amygdala; HIP, hippocampus; NCX, neocortex. The expression levels of various proteins were calculated from the public data from the Human Brain Transcriptome database. F, Western-blot detection of PTPRO (top) in postsynaptic and presynaptic fractions of the mouse brain, as well as the detection of synaptophysin (medium) and PSD95 (bottom) in these fractions as controls. G, H, Representative images of the double immunostaining of PTPRO and vGLUT1 (excitatory presynaptic marker) or the double immunostaining of PTPRO and PSD95 (excitatory postsynaptic marker) in cultured hippocampal neurons. Scale bars: G, 20 μm (top), 1 μm (bottom); H, 5 μm (top), 1 μm (bottom). For all representative images, scale bars apply to all panels in a set. All summary graphs show the mean ± SEM.
In the brain, PTPRO was expressed ubiquitously in all regions tested, including the brainstem, cortex, hippocampus, olfactory bulb, and cerebellum (Fig. 3C). Furthermore, PTPRO was highly expressed during the first week after birth and then expression declined gradually toward adulthood (Fig. 3D). This is consistent with the expression profiling of the human PTPRO protein plotted from the public data from the Human Brain Transcriptome database (http://hbatlas.org/; Fig. 3E).
Because many proteins in the RPTP family showed brain expression (Fig. 3B), brain enrichment offers PTPRO an opportunity but is not sufficient to initiate the formation of synapses. To further explore the mechanisms of PTPRO in synaptogenesis, we next examined the synaptic localization of endogenous PTPRO in cultured neurons with an antibody against the N-terminal extracellular region of PTPRO. Fractionation experiments showed that PTPRO was enriched in the PSD fraction compared with that of the presynaptic faction (Fig. 3F). Next, we examined the synaptic localization of PTPRO using immunofluorescence approaches and identified PTPRO in the soma and the synapses. Using confocal microscopy, PTPRO was found to overlap mainly with presynaptic markers for excitatory synapses (60.05 ± 6.55%; Fig. 3G) and, to a much lesser extent, inhibitory synapses (9.54 ± 1.68%). We performed double-labeling experiments for PTPRO and PSD-95 (a postsynaptic marker for excitatory synapse) and the results showed that PTPRO colocalized with PSD-95, confirming the postsynaptic localization of PTPRO in the excitatory synapses (Fig. 3H).
Overexpression of PTPRO in neurons promotes synapse formation
Because PTPRO demonstrated synaptogenic activity in the ASF assay, we next assessed whether the overexpression of PTPRO in neurons promotes the formation of neuronal synapses. To accomplish this, we transfected hippocampal neurons with expression vectors encoding PTPRO and GFP at DIV 10. Using immunocytochemistry, we quantified the number of synapses formed on the transfected (GFP-positive) neurons with an antibody against synapsin 1. The overexpression of PTPRO in the cultured hippocampal neurons significantly increased the synapse density compared with the control, as indicated by the increased number of synapsin-positive puncta along the transfected dendrites (Fig. 4A,D). To determine whether this phenomenon is restricted to a certain type of synapse, we quantified the number of vGLUT1-positive and GAD-65-positive puncta (Fig. 4B,C). The overexpression of PTPRO increased the density of vGLUT1-positive and GAD65-positive puncta (though to a less extent) compared with the control neurons (Fig. 4E,F). Overexpression of PTPRO did not change the size the of synapsin-positive, vGLUT1-positive, and GAD-65-positive puncta. Therefore, overexpression of PTPRO in the hippocampal neurons increased the density of synapses, as assessed with morphological measures.
Figure 4.
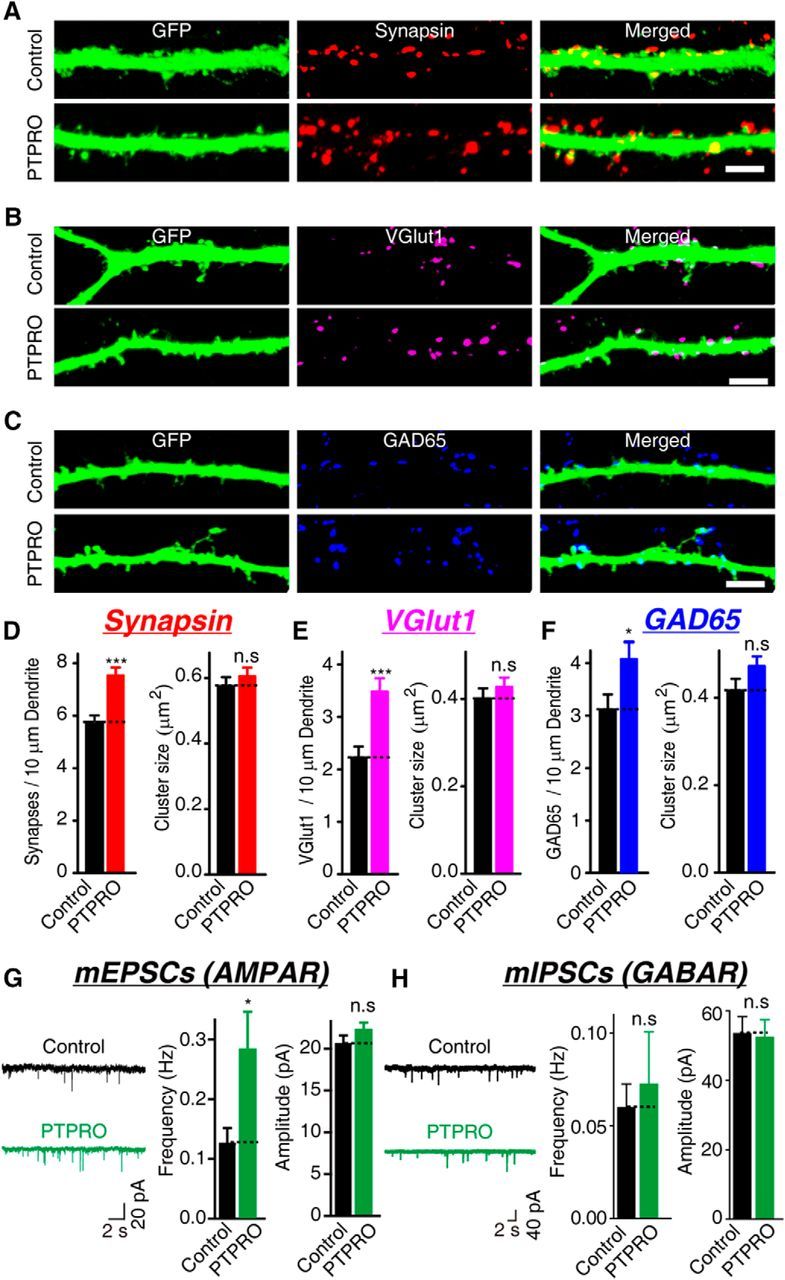
Overexpression of PTPRO increases the synapse density in cultured hippocampal neurons. A–C, Representative images of hippocampal neurons transfected with either an empty vector (control) or a vector encoding PTPRO together with pFUGW-GFP at DIV 10 and analyzed with double immunofluorescence with antibodies to GFP and synapsin 1 (A) or vGLUT1 (B) or GAD-65 (c) at DIV 14. Scale bars in A–C, 5 μm. D–F, Summary graphs of synapse density and cluster size in images in A–C. D: Synapsin: control: n = 28/3, PTPRO: n = 27/3; vGlut1: control: n = 20/3, PTPRO: n = 26/3; GAD65: control: n = 24/3, PTPRO: n = 14/3. G, H, Representative traces (left) and summary graphs of the frequencies (center) and amplitudes (right) of mEPSCs (G) recorded in 1 μm TTX and 0.1 mm PTX or mIPSCs (H) recorded in 10 μm CNQX and 1 μm TTX. G: mEPSCs: control: n = 29/3, PTPRO: n = 30/3; H: mIPSCs: control: n = 33/3, PTPRO: n = 32/3. For all representative traces and images, scale bars apply to all panels in a set. All summary graphs show the mean ± SEM; statistical comparisons were made with Student's t test (*p < 0.05; ***p < 0.001; n.s., not significant).
Next, to test whether the synapses induced by the overexpression of PTPRO are functional, we measured the mEPSCs and mIPSCs in these neurons. The frequency of mEPSCs was significantly increased in the neurons that overexpressed PTPRO compared with the control neurons transfected with a plasmid-encoding GFP (Fig. 4G). The frequency of the mIPSCs remained unchanged in the neurons that overexpressed PTPRO (Fig. 4H). The amplitudes of the mEPSCs and mIPSCs, which presumably reflected the number of postsynaptic receptors, were not changed significantly. Therefore, overexpression of PTPRO in cultured hippocampal neurons induces the formation of functional excitatory synapses.
KD of PTPRO in neurons impairs synapse formation
Because the overexpression of PTPRO resulted in synaptogenic activity in the ASF assay and in cultured neurons, we next assessed whether the loss of PTPRO in cultured hippocampal neurons would affect the formation of neuronal synapses. We transfected neurons with a plasmid expressing an shRNA targeting the intracellular domain of mouse PTPRO (PTPROKD) and then used human PTPRO as a rescue construct that was resistant to the shRNA targeting mouse PTPRO (Fig. 5A). The KD efficiency was validated with quantitative immunostaining and RT-PCR. The expression of PTPROKD in neurons suppressed the endogenous protein by 62 ± 5% and reduced mRNA by 80 ± 7% in the cultured hippocampal neurons (Fig. 5B). The total number of synapses, quantified as the number of synapsin-positive puncta, was reduced by 47.27 ± 3.42% in the PTPROKD neurons, whereas the synapse size was not significantly affected (Fig. 5C). The reduction in synapse density was fully rescued by the co-expression of a plasmid encoding full-length hPTPRO, excluding the possibility that the observed phenotypes were due to off-target effects. We also determined the number of excitatory and inhibitory synapses that formed on the PTPROKD neurons. The KD of PTPRO in the postsynaptic neurons reduced the number of vGLUT1-positive and GAD-65-positive puncta, confirming the role of PTPRO in promoting synapse formation (Fig. 5D,E).
Figure 5.

KD of PTPRO by shRNA decreases the glutamatergic and GABAergic synaptic strength. A, Sequence of an shRNA against mouse PTPRO. B, Representative images of the protein levels of PTPRO in transfected neurons (left) and quantitative graphs of relative protein levels (center) and relative mRNA levels (right). C–E, Representative images (left) and summary graphs (right) of synapsin 1 (C), vGlut1 (D), or GAD65 (E) in neurons transfected with the control plasmid, or PTPROKD construct, PTPROKD construct together with human PTPRO (synapsin: control: n = 27/3, PTPROKD: n = 24/3, rescue: n = 21/3; vGlut1: control: n = 60/3, PTPROKD: n = 70/3, rescue: n = 56/3; GAD65: control: n = 84/3, PTPROKD: n = 78/3; rescue: n = 76/3). F, G, Representative traces (left) and summary graphs of the frequencies (center) and amplitudes (right) of mEPSCs (F) and mIPSCs (G). F, Control: n = 33/3, PTPROKD: n = 14/3, and rescue: n = 17/3. G: Control: n = 23/3, PTPROKD: n = 20/3, rescue: n = 15/3. For all representative traces and images, scale bars apply to all panels in a set. All summary graphs show the mean ± SEM; statistical comparisons were made with Student's t test (*p < 0.05; **p < 0.01; ***p < 0.001; n.s., not significant).
Moreover, measurements of the synaptic neurotransmission in PTPROKD neurons revealed a significant reduction in the frequencies of the mEPSCs and mIPSCs, whereas the amplitudes remained unchanged (Fig. 5F,G). These results support the notion that the inactivation of PTPRO in neurons impairs the formation of synapses.
Because the extracellular domain of PTPRO is sufficient to induce synapse formation in an ASF assay (Fig. 2), we tested whether the extracellular domain is also sufficient to mediate synaptogenic effects in neurons. Cultured mouse hippocampal neurons transfected with hPTPROCTD at DIV 10 showed almost full restoration of the deficits in the synapse density (revealed by antibodies against synapsin 1, vGLUT1, or GAD-65) observed in the PTPROKD neurons (Fig. 6A–C). The rescue effects of the overexpressed hPTPROCTD are comparable to those of full-length hPTPRO, whereas the overexpression of hPTPRONTD failed to rescue these deficits. Measurements of the mEPSCs and mIPSCs also demonstrated that hPTPROCTD, but not hPTPRONTD, was sufficient to rescue the KD phenotype fully (Fig. 6D,E). Therefore, the extracellular domain of PTPRO is responsible for the synaptogenic activity of PTPRO in primary neurons.
Figure 6.

Expression of the extracellular region of PTPRO rescues the PTPRO KD-induced decrease in synaptic strength. A–C, Representative images (left) and summary graphs (right) of synaptic signals of synapsin (A), vGlut1 (B), or GAD65 (C) in neurons transfected with the control plasmid, PTPROKD construct, PTPROKD construct together with full-length human PTPRO, PTPROKD construct together with the hPTPROCTD construct, or PTPROKD construct together with the hPTPRONTD construct (synapsin: control: n = 16/3, PTPROKD: n = 18/3, PTPROKD + hPTPRO: n = 19/3, PTPROKD + hPTPROCTD: n = 19/3, and PTPROKD + hPTPRONTD: n = 20/3; vGlut1: control: n = 20/3, PTPROKD: n = 19/3, PTPROKD + hPTPRO: n = 17/3, PTPROKD + hPTPROCTD: n = 20/3, and PTPROKD + hPTPRONTD: n = 18/3; GAD65: control: n = 21/3, PTPROKD: n = 20/3, PTPROKD + hPTPRO: n = 20/3, PTPROKD + hPTPROCTD: n = 18/3, and PTPROKD + hPTPRONTD: n = 22/3). Scale bars in A–C, 5 μm. D, E, Representative traces (left) and summary graphs of the frequencies (center) and amplitudes (right) of mEPSCs (D) and mIPSCs (E). D: Control: n = 36/4, PTPROKD: n = 30/4, PTPROKD + hPTPRO: n = 33/4, PTPROKD + hPTPROCTD: n = 34/4, and PTPROKD + hPTPRONTD: n = 33/4. E: Control: n = 36/4; KD: n = 33/4; PTPROKD + hPTPRO: n = 24/4; PTPROKD + hPTPROCTD: n = 28/4; and PTPROKD + hPTPRONTD: n = 20/4. For all representative traces and images, scale bars apply to all panels in a set. All summary graphs show the mean ± SEM; statistical comparisons were made with Student's t test (*p < 0.05; **p < 0.01).
Discussion
Here, we report synaptogenic activity for PTPRO in heterologous cells and cultured neurons. This effect seems to be specific to PTPRO because three other type III RPTPs did not induce artificial synapse formation in an ASF assay. Consistent with previous reports (Matozaki et al., 2010), we found that PTPRO was enriched in mouse brains, where it localized to postsynaptic sites. The overexpression of PTPRO in cultured hippocampal neurons increased the density of the synapses and the frequency of the mEPSCs, whereas the KD of PTPRO expression in neurons resulted in a loss in the number of synapses and reduced the frequencies of the mEPSCs and mIPSCs. Because PTPRO is enriched in the postsynaptic site of excitatory synapses, changes in inhibitory synapses could be due to homeostatic mechanisms and whether PTPRO regulates the formation of inhibitory synapses directly remains to be tested. Furthermore, the synaptogenic effect is mediated by the cell-autonomous expression of PTPRO and the catalytic phosphatase domain of PTPRO is dispensable for this activity. Our observations clearly demonstrate that PTPRO is capable of facilitating forming synaptic connections with presynaptic neurons in heterologous cells and cultured neurons.
PTPRO, also known as GLEPP1, was originally cloned from rabbit kidney tissue in a search for podocyte-specific proteins using an antibody strategy (Thomas et al., 1994). PTPRO homologs have been found in multiple species, including humans, mice, rats (Tagawa et al., 1997), and chickens (Bodden and Bixby, 1996). PTPRO is a single-pass transmembrane protein with an extracellular domain containing eight fibronectin type III-like repeats and an intracellular PTPase domain (Thomas et al., 1994). PTPRO-KO mice exhibit normal birth rates and gross kidney and glomerular structures, but podocyte structures are affected and the glomerular filtration rate is reduced (Wharram et al., 2000).
PTPRO is enriched in the kidneys and the brain (Thomas et al., 1994; Beltran et al., 2003; Kotani et al., 2010) and the present data further demonstrate that PTPRO localizes to the postsynaptic sites of excitatory synapses (Fig. 3). Furthermore, the overexpression and KD experiments demonstrated the postsynaptic function of PTPRO in synapse formation, reflected as changes in the number of neuronal synapses and synaptic strength measured using the electrophysiological method (Figs. 4, 5). This complements previous observations showing that PTPRO is involved in axonogenesis in the nervous system. Axogenesis, also called axon formation, requires the precise cooperation of multiple morphogens and signaling pathways in the developing nerve system (Tessier-Lavigne and Goodman, 1996; Zou and Lyuksyutova, 2007; Onishi et al., 2014; Thakar et al., 2017). Expression of PTPRO mRNA in the brain reaches the maximum between 16 d postcoitum and 3 d postpartum (Beltran et al., 2003), temporally correlating with axonogenesis in the brain. Furthermore, mouse PTPRO mRNA was found in tropomyosin-related kinase A (TrkA)-positive and TrkC-positive neurons in dorsal root ganglion (DRG) (Beltran et al., 2003), supporting a possible role of mPTPRO in neuronal differentiation and axonogenesis. In PTPRO−/− mice, DRG neurons expressing the neuropeptide calcitonin gene-related peptide (CGRP) or TrkA receptors exhibit progressive cell loss approaching the adult period. CGRP-positive projections from the DRG to the dorsal horn exhibit abnormal patterns in PTPRO−/− mice (Gonzalez-Brito and Bixby, 2009; Tchetchelnitski et al., 2014), whereas the trigeminal ganglion neurons in these animals show enhanced axonal outgrowth and branching (Gatto et al., 2013). Therefore, PTPRO seems to participate in the different stages of synapse development, including neuronal differentiation, axonogenesis, and synapse formation, in a neuron-specific and development-specific manner. Therefore, due to the expression of PTPRO in the brain, especially the hippocampus, together with its expression as early as E16 and its role in development, it is important to determine the specific synaptic role of PTPRO after knocking it down after embryonic development.
We also demonstrated that the synaptogenic activity of PTPRO is independent of the cytoplasmic tails (Figs. 2, 6). Moreover, the PTPRO extracellular domains (ECDs) are essential in mediating the repulsive guidance signal for cultured retinal neurons. The presence of the chicken PTPRO ECD in the supporting matrix prevents neuronal adhesion and inhibits the growth of neurites in culture. Acute treatment of developing neurons in culture with PTPRO ECD collapses the growth cone in a concentration-dependent manner (Stepanek et al., 2001). However, the cytoplasmic tail of PTPRO, which harbors the phosphatase domain, seems to be important for axon growth and patterning. The cytoplasmic tail of PTPRO interacts with and dephosphorylates several neuronal proteins, including EphA4 and EphB2 receptors (Shintani et al., 2006) and neuronal pentraxin with the chromo domain (NPCD) (Chen and Bixby, 2005a), a cytoplasmic protein different from the classic neuronal pentraxins (Chen and Bixby, 2005b). Using the ephrin-A2-Fc stripe assay, Shintani et al. (2006) reported that the phosphatase activity of PTPRO determines the sensitivity of developing axons to ephrins and thus affects the pathfinding of retinal, nasal, and temporal axons. The inactivation of PTPRO in an embryonic chick spinal cord using shRNA specifically affects the outgrowth of a small subset of dorsal nerves in the limb (Stepanek et al., 2005). Therefore, PTPRO mediates synaptogenesis and axon growth using distinct domains.
In conclusion, the present data show that PTPRO promotes synaptogenesis. Overexpression of PTPRO in neurons increases the number of synapses and enhances the synaptic strength at the excitatory synapses. KD of PTPRO in postsynaptic neurons exhibits the opposite effect. The synaptogenic effect of PTPRO is solely dependent on the extracellular domains. Recently, an association between three intronic PTPRO SNPs and memory status in patients with schizophrenia and bipolar disorder has been reported (LeBlanc et al., 2012; Hendriks and Pulido, 2013). Furthermore, PTPRO was reduced in patients with Galloway–Mowat syndrome, a rare autosomal-recessive disorder characterized by early-onset nephrotic syndrome and CNS anomalies (Roos et al., 1987). This human clinical evidence suggests that variations in PTPRO might be associated with neurodevelopment deficits, which confer risk to these psychiatric illnesses. However, the synaptic function of PTPRO in the CNS remains to be established. The present findings identify PTPRO as a novel synaptic adhesion molecule and suggest a plausible link between PTPRO and neurodevelopment deficits such as schizophrenia.
Footnotes
This work was supported by the National Basic Research Program of China (Grants 2017YFA0105201, 2012YQ03026004, 2014CB942804, and 2014BAI03B01), the National Science Foundation of China (Grant 31670842 to C.Z. and Grant 31670850 to X.Y.), the Beijing Municipal Science and Technology Commission (Grants Z161100002616021 and Z161100000216154), and the National Key Research and Development Program of China(2017YFA0105201). We thank Dr. Thomas C. Südhof for beneficial discussions and critical comments on this manuscript.
The authors declare no competing financial interests.
References
- Aoto J, Ting P, Maghsoodi B, Xu N, Henkemeyer M, Chen L (2007) Postsynaptic ephrinB3 promotes shaft glutamatergic synapse formation. J Neurosci 27:7508–7519. 10.1523/JNEUROSCI.0705-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoto J, Martinelli DC, Malenka RC, Tabuchi K, Südhof TC (2013) Presynaptic neurexin-3 alternative splicing trans-synaptically controls postsynaptic AMPA receptor trafficking. Cell 154:75–88. 10.1016/j.cell.2013.05.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran PJ, Bixby JL, Masters BA (2003) Expression of PTPRO during mouse development suggests involvement in axonogenesis and differentiation of NT-3 and NGF-dependent neurons. J Comp Neurol 456:384–395. 10.1002/cne.10532 [DOI] [PubMed] [Google Scholar]
- Biederer T, Sara Y, Mozhayeva M, Atasoy D, Liu X, Kavalali ET, Südhof TC (2002) SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science 297:1525–1531. 10.1126/science.1072356 [DOI] [PubMed] [Google Scholar]
- Biederer T, Südhof TC (2001) CASK and protein 4.1 support F-actin nucleation on neurexins. J Biol Chem 276:47869–47876. 10.1074/jbc.M105287200 [DOI] [PubMed] [Google Scholar]
- Boda B, Dubos A, Muller D (2010) Signaling mechanisms regulating synapse formation and function in mental retardation. Curr Opin Neurobiol 20:519–527. 10.1016/j.conb.2010.03.012 [DOI] [PubMed] [Google Scholar]
- Bodden K, Bixby JL (1996) CRYP-2: A receptor-type tyrosine phosphatase selectively expressed by developing vertebrate neurons. J Neurobiol 31:309–324. 10.1002/ (SICI)1097-4695 (199611)31:3%3C309::AID-NEU4%3E3.0.CO%3B2-E [DOI] [PubMed] [Google Scholar]
- Bozdagi O, Wang XB, Nikitczuk JS, Anderson TR, Bloss EB, Radice GL, Zhou Q, Benson DL, Huntley GW (2010) Persistence of coordinated long-term potentiation and dendritic spine enlargement at mature hippocampal CA1 synapses requires N-cadherin. J Neurosci 30:9984–9989. 10.1523/JNEUROSCI.1223-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Bixby JL (2005a) A novel substrate of receptor tyrosine phosphatase PTPRO is required for nerve growth factor-induced process outgrowth. J Neurosci 25:880–888. 10.1523/JNEUROSCI.4365-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Bixby JL (2005b) Neuronal pentraxin with chromo domain (NPCD) is a novel class of protein expressed in multiple neuronal domains. J Comp Neurol 481:391–402. 10.1002/cne.20391 [DOI] [PubMed] [Google Scholar]
- Chubykin AA, Atasoy D, Etherton MR, Brose N, Kavalali ET, Gibson JR, Südhof TC (2007) Activity-dependent validation of excitatory versus inhibitory synapses by Neuroligin-1 versus Neuroligin-2. Neuron 54:919–931. 10.1016/j.neuron.2007.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotman CW, Taylor D (1972) Isolation and structural studies on synaptic complexes from rat brain. J Cell Biol 55:696–711. 10.1083/jcb.55.3.696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNardo LA, de Wit J, Otto-Hitt S, Ghosh A (2012) NGL-2 regulates input-specific synapse development in CA1 pyramidal neurons. Neuron 76:762–775. 10.1016/j.neuron.2012.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatto G, Dudanova I, Suetterlin P, Davies AM, Drescher U, Bixby JL, Klein R (2013) Protein tyrosine phosphatase receptor type O inhibits trigeminal axon growth and branching by repressing TrkB and ret signaling. J Neurosci 33:5399–5410. 10.1523/JNEUROSCI.4707-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Brito MR, Bixby JL (2009) Protein tyrosine phosphatase receptor type O regulates development and function of the sensory nervous system. Mol Cell Neurosci 42:458–465. 10.1016/j.mcn.2009.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KM, Weinberg RJ (2012) Ultrastructure of synapses in the mammalian brain. Cold Spring Harb Perspect Biol 4: pii: a005587. 10.1101/cshperspect.a005587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson JT, Georgiou J, Jia Z, Robertson J, Elowe S, Roder JC, Pawson T (2001) The receptor tyrosine kinase EphB2 regulates NMDA-dependent synaptic function. Neuron 32:1041–1056. 10.1016/S0896-6273(01)00553-0 [DOI] [PubMed] [Google Scholar]
- Hendriks WJ, Pulido R (2013) Protein tyrosine phosphatase variants in human hereditary disorders and disease susceptibilities. Biochim Biophys Acta 1832:1673–1696. 10.1016/j.bbadis.2013.05.022 [DOI] [PubMed] [Google Scholar]
- Jiang W, Hua R, Wei M, Li C, Qiu Z, Yang X, Zhang C (2015) An optimized method for high-titer lentivirus preparations without ultracentrifugation. Sci Rep 5:13875. 10.1038/srep13875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattenstroth G, Tantalaki E, Südhof TC, Gottmann K, Missler M (2004) Postsynaptic N-methyl-D-aspartate receptor function requires α-neurexins. Proc Natl Acad Sci U S A 101:2607–2612. 10.1073/pnas.0308626100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser MS, McClelland AC, Hughes EG, Dalva MB (2006) Intracellular and trans-synaptic regulation of glutamatergic synaptogenesis by EphB receptors. J Neurosci 26:12152–12164. 10.1523/JNEUROSCI.3072-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser MS, Nolt MJ, Dalva MB (2008) EphB receptors couple dendritic filopodia motility to synapse formation. Neuron 59:56–69. 10.1016/j.neuron.2008.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Burette A, Chung HS, Kwon SK, Woo J, Lee HW, Kim K, Kim H, Weinberg RJ, Kim E (2006) NGL family PSD-95-interacting adhesion molecules regulate excitatory synapse formation. Nat Neurosci 9:1294–1301. 10.1038/nn1763 [DOI] [PubMed] [Google Scholar]
- Ko J. (2012) The leucine-rich repeat superfamily of synaptic adhesion molecules: LRRTMs and slitrks. Mol Cells 34:335–340. 10.1007/s10059-012-0113-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J, Fuccillo MV, Malenka RC, Südhof TC (2009) LRRTM2 functions as a neurexin ligand in promoting excitatory synapse formation. Neuron 64:791–798. 10.1016/j.neuron.2009.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J, Soler-Llavina GJ, Fuccillo MV, Malenka RC, Südhof TC (2011) Neuroligins/LRRTMs prevent activity- and Ca2+/calmodulin-dependent synapse elimination in cultured neurons. J Cell Biol 194:323–334. 10.1083/jcb.201101072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani T, Murata Y, Ohnishi H, Mori M, Kusakari S, Saito Y, Okazawa H, Bixby JL, Matozaki T (2010) Expression of PTPRO in the interneurons of adult mouse olfactory bulb. J Comp Neurol 518:119–136. 10.1002/cne.22239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc M, Kulle B, Sundet K, Agartz I, Melle I, Djurovic S, Frigessi A, Andreassen OA (2012) Genome-wide study identifies PTPRO and WDR72 and FOXQ1-SUMO1P1 interaction associated with neurocognitive function. J Psychiatr Res 46:271–278. 10.1016/j.jpsychires.2011.11.001 [DOI] [PubMed] [Google Scholar]
- Lee SJ, Wei M, Zhang C, Maxeiner S, Pak C, Calado Botelho S, Trotter J, Sterky FH, Südhof TC (2017) Presynaptic neuronal pentraxin receptor organizes excitatory and inhibitory synapses. J Neurosci 37:1062–1080. 10.1523/JNEUROSCI.2768-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhoff MW, Laurén J, Cassidy RM, Dobie FA, Takahashi H, Nygaard HB, Airaksinen MS, Strittmatter SM, Craig AM (2009) An unbiased expression screen for synaptogenic proteins identifies the LRRTM protein family as synaptic organizers. Neuron 61:734–749. 10.1016/j.neuron.2009.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matozaki T, Murata Y, Mori M, Kotani T, Okazawa H, Ohnishi H (2010) Expression, localization, and biological function of the R3 subtype of receptor-type protein tyrosine phosphatases in mammals. Cell Signal 22:1811–1817. 10.1016/j.cellsig.2010.07.001 [DOI] [PubMed] [Google Scholar]
- Maximov A, Pang ZP, Tervo DG, Südhof TC (2007) Monitoring synaptic transmission in primary neuronal cultures using local extracellular stimulation. J Neurosci Methods 161:75–87. 10.1016/j.jneumeth.2006.10.009 [DOI] [PubMed] [Google Scholar]
- McAllister AK. (2007) Dynamic aspects of synapse formation. Annu Rev Neurosci 30:425–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missler M, Zhang W, Rohlmann A, Kattenstroth G, Hammer RE, Gottmann K, Südhof TC (2003) α-Neurexins couple Ca2+ channels to synaptic vesicle exocytosis. Nature 423:939–948. 10.1038/nature01755 [DOI] [PubMed] [Google Scholar]
- Missler M, Südhof TC, Biederer T (2012) Synaptic cell adhesion molecules. Cold Spring Harb Perspect Biol 4:a005694. 10.1101/cshperspect.a005694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi K, Hollis E, Zou Y (2014) Axon guidance and injury-lessons from Wnts and Wnt signaling. Curr Opin Neurobiol 27:232–240. 10.1016/j.conb.2014.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereda AE. (2014) Chemical synapses. Nat Rev Neurosci 15:250–263. 10.1038/nrn3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfenninger KH. (1971) The cytochemistry of synaptic densities. II. Proteinaceous components and mechanism of synaptic connectivity. J Ultrasruct Res 35:451–475. 10.1016/S0022-5320(71)80005-9 [DOI] [PubMed] [Google Scholar]
- Robbins EM, Krupp AJ, Perez de Arce K, Ghosh AK, Fogel AI, Boucard A, Südhof TC, Stein V, Biederer T (2010) SynCAM 1 adhesion dynamically regulates synapse number and impacts plasticity and learning. Neuron 68:894–906. 10.1016/j.neuron.2010.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos RA, Maaswinkel-Mooy PD, vd Loo EM, Kanhai HH (1987) Congenital microcephaly, infantile spasms, psychomotor retardation, and nephrotic syndrome in two sibs. Eur J Pediatr 146:532–536. 10.1007/BF00441612 [DOI] [PubMed] [Google Scholar]
- Sando R, Bushong E, Zhu Y, Huang M, Considine C, Phan S, Ju S, Uytiepo M, Ellisman M, Maximov A (2017) Assembly of excitatory synapses in the absence of glutamatergic neurotransmission. Neuron 94:312–321.e3. 10.1016/j.neuron.2017.03.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes JR, Yamagata M (2009) Many paths to synaptic specificity. Annu Rev Cell Dev Biol 25:161–195. 10.1146/annurev.cellbio.24.110707.175402 [DOI] [PubMed] [Google Scholar]
- Shintani T, Ihara M, Sakuta H, Takahashi H, Watakabe I, Noda M (2006) Eph receptors are negatively controlled by protein tyrosine phosphatase receptor type O. Nat Neurosci 9:761–769. 10.1038/nn1697 [DOI] [PubMed] [Google Scholar]
- Stepanek L, Stoker AW, Stoeckli E, Bixby JL (2005) Receptor tyrosine phosphatases guide vertebrate motor axons during development. J Neurosci 25:3813–3823. 10.1523/JNEUROSCI.4531-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanek L, Sun QL, Wang J, Wang C, Bixby JL (2001) CRYP-2/cPTPRO is a neurite inhibitory repulsive guidance cue for retinal neurons in vitro. J Cell Biol 154:867–878. 10.1083/jcb.200105019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof TC. (2004) The synaptic vesicle cycle. Annu Rev Neurosci 27:509–547. 10.1146/annurev.neuro.26.041002.131412 [DOI] [PubMed] [Google Scholar]
- Südhof TC. (2008) Neuroligins and neurexins link synaptic function to cognitive disease. Nature 455:903–911. 10.1038/nature07456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof TC, Malenka RC (2008) Understanding synapses: past, present, and future. Neuron 60:469–476. 10.1016/j.neuron.2008.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagawa M, Shirasawa T, Yahagi Y, Tomoda T, Kuroyanagi H, Fujimura S, Sakiyama S, Maruyama N (1997) Identification of a receptor-type protein tyrosine phosphatase expressed in postmitotic maturing neurons: its structure and expression in the central nervous system. Biochem J 321:865–871. 10.1042/bj3210865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchetchelnitski V, van den Eijnden M, Schmidt F, Stoker AW (2014) Developmental co-expression and functional redundancy of tyrosine phosphatases with neurotrophin receptors in developing sensory neurons. Int J Dev Neurosci 34:48–59. 10.1016/j.ijdevneu.2014.01.005 [DOI] [PubMed] [Google Scholar]
- Tessier-Lavigne M, Goodman CS (1996) The molecular biology of axon guidance. Science 274:1123–1133. 10.1126/science.274.5290.1123 [DOI] [PubMed] [Google Scholar]
- Thakar S, Wang L, Yu T, Ye M, Onishi K, Scott J, Qi J, Fernandes C, Han X, Yates JR 3rd, Berg DK, Zou Y (2017) Evidence for opposing roles of Celsr3 and Vangl2 in glutamatergic synapse formation. Proc Natl Acad Sci U S A 114:E610–E618. 10.1073/pnas.1612062114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PE, Wharram BL, Goyal M, Wiggins JE, Holzman LB, Wiggins RC (1994) GLEPP1, a renal glomerular epithelial cell (podocyte) membrane protein-tyrosine phosphatase: Identification, molecular cloning, and characterization in rabbit. J Biol Chem 269:19953–19962. [PubMed] [Google Scholar]
- Ullrich B, Ushkaryov YA, Südhof TC (1995) Cartography of neurexins: More than 1000 isoforms generated by alternative splicing and expressed in distinct subsets of neurons. Neuron 14:497–507. 10.1016/0896-6273(95)90306-2 [DOI] [PubMed] [Google Scholar]
- Varoqueaux F, Aramuni G, Rawson RL, Mohrmann R, Missler M, Gottmann K, Zhang W, Südhof TC, Brose N (2006) Neuroligins determine synapse maturation and function. Neuron 51:741–754. 10.1016/j.neuron.2006.09.003 [DOI] [PubMed] [Google Scholar]
- Verhage M, Maia AS, Plomp JJ, Brussaard AB, Heeroma JH, Vermeer H, Toonen RF, Hammer RE, van den Berg TK, Missler M, Geuze HJ, Südhof TC (2000) Synaptic assembly of the brain in the absence of neurotransmitter secretion. Science 287:864–869. 10.1126/science.287.5454.864 [DOI] [PubMed] [Google Scholar]
- Wang JR, Li XL, Fan SQ, Tan C, Xiang JJ, Tang K, Wang R, Li GY (2003) Expression of LRRC4 has the potential to decrease the growth rate and tumorigenesis of glioblastoma cell line U251. Ai Zheng 22:897–902. [PubMed] [Google Scholar]
- Wei M, Zhang J, Jia M, Yang C, Pan Y, Li S, Luo Y, Zheng J, Ji J, Chen J, Hu X, Xiong J, Shi Y, Zhang C (2016) α/β-Hydrolase domain-containing 6 (ABHD6) negatively regulates the surface delivery and synaptic function of AMPA receptors. Proc Natl Acad Sci U S A 113:E2695–E2704. 10.1073/pnas.1524589113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M, Jia M, Zhang J, Yu L, Zhao Y, Chen Y, Ma Y, Zhang W, Shi YS, Zhang C (2017) The inhibitory effect of α/β-hydrolase domain-containing 6 (ABHD6) on the surface targeting of GluA2- and GluA3-containing AMPA receptors. Front Mol Neurosci 10:55. 10.3389/fnmol.2017.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharram BL, Goyal M, Gillespie PJ, Wiggins JE, Kershaw DB, Holzman LB, Dysko RC, Saunders TL, Samuelson LC, Wiggins RC (2000) Altered podocyte structure in GLEPP1 (Ptpro)-deficient mice associated with hypertension and low glomerular filtration rate. J Clin Invest 106:1281–1290. 10.1172/JCI7236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo J, Kwon SK, Kim E (2009) The NGL family of leucine-rich repeat-containing synaptic adhesion molecules. Mol Cell Neurosci 42:1–10. 10.1016/j.mcn.2009.05.008 [DOI] [PubMed] [Google Scholar]
- Yang X, Hou D, Jiang W, Zhang C (2014) Intercellular protein-protein interactions at synapses. Protein Cell 5:420–444. 10.1007/s13238-014-0054-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wei M, Xiong Y, Du X, Zhu S, Yang L, Zhang C, Liu JJ (2015) Endophilin A1 regulates dendritic spine morphogenesis and stability through interaction with p140Cap. Cell Res 25:496–516. 10.1038/cr.2015.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Milunsky JM, Newton S, Ko J, Zhao G, Maher A, Tager-flusberg H, Bolliger MF, Carter AS, Boucard AA, Powell CM, Südhof TC (2009) A Neuroligin-4 missense mutation associated with autism impairs Neuroligin-4 folding and ER export clinical evaluation. J Neurosci 29:10843–10854. 10.1523/JNEUROSCI.1248-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Atasoy D, Araç D, Yang X, Fucillo MV, Robison AJ, Ko J, Brunger AT, Südhof TC (2010) Neurexins physically and functionally interact with GABAA receptors. Neuron 66:403–416. 10.1016/j.neuron.2010.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Lyuksyutova AI (2007) Morphogens as conserved axon guidance cues. Curr Opin Neurobiol 17:22–28. 10.1016/j.conb.2007.01.006 [DOI] [PubMed] [Google Scholar]


