Abstract
Toll-like receptor 4 (TLR4) is a proinflammatory cascade initiator in poststroke inflammation. In this study, miR-1906, a novel regulator of TLR4, was identified via in silico analysis and microRNA profiling in male adult mice and its expression was then quantitated in the ischemic hemisphere. We found miR-1906 to be significantly brain enriched in the ischemic hemisphere and even more drastically enriched in the peri-infarct regions. Furthermore, in vitro experiments demonstrated that, during oxygen–glucose deprivation, miR-1906 expression was increased in glial cells but decreased in neurons. Surprisingly, despite the augmentation of intracellular abundance, miR-1906 expression in extracellular vesicles was decreased in astrocyte cell culture supernatants, suggesting reduced sources of miR-1906 from glia to neurons. When exogenous miR-1906 was administered, decreased TLR4 protein expression was observed both in vitro and in vivo. Using Cy3 labeling, exogenous miR-1906 uptake by astrocytes, microglia, and neurons was visualized directly in vivo. Reduced infarct volumes and improved functional outcomes were observed in middle cerebral artery occlusion mice receiving miR-1906. However, the protective effects of miR-1906 disappeared with the genetic knock-out of TLR4, suggesting that TLR4 is a major target of miR-1906 through which the microRNA exerts its therapeutic effects.
SIGNIFICANCE STATEMENT The current study identified miR-1906 as a novel specific regulator of Toll-like receptor 4 (TLR4) and depicted its distinct expression patterns in different cerebral regions and cell types during ischemic attack. Therefore, the therapeutic supplementation of miR-1906 can be beneficial in the modulation of poststroke inflammation. Using Cy3 labeling, exogenous miR-1906 expression was visualized and shown to enter astrocytes, microglia, and neurons successfully in vivo. Supplemental therapeutic miR-1906 resulted in reduced TLR4 expression and improved outcomes after middle cerebral artery occlusion in a mouse model, but its neuroprotective function was TLR4 dependent, suggesting that TLR4 is a major target of miR-1906.
Keywords: cerebrovascular disease, inflammation, microRNA, stroke, Toll like receptor 4
Introduction
Stroke is one of the most devastating and complicated neurological disorders in the world (Moskowitz et al., 2010), accounting for 11.8% of total deaths (American Heart Association, 2016). Although 87% of all strokes are ischemic (American Heart Association, 2016), the current available therapeutic options against acute ischemic stroke (AIS) are limited. The compelling disease burden and paucity of therapeutic options underlies the urgency to understand the mechanism of stroke (Eltzschig and Eckle, 2011) and to find novel potential therapeutic targets (Moskowitz et al., 2010; Macrez et al., 2011).
Recent studies have revealed the critical involvement of immune response in AIS (Iadecola and Anrather, 2011). Although the CNS and immune system have been regarded as two distinct entities for decades (Engelhardt and Ransohoff, 2005), accumulating evidence has unveiled a mutual interaction between these two systems (Chamorro et al., 2012; Fu et al., 2015). Inflammation plays a paradoxical role in AIS in that it is beneficial during the recovery phase but deleterious during the acute phase (Engelhardt and Ransohoff, 2005; Eltzschig and Eckle, 2011; Iadecola and Anrather, 2011; Macrez et al., 2011; Chamorro et al., 2012; Xu and Jiang, 2014). Therefore, immune modulation during the acute phase is a promising therapeutic approach against aggravating ischemic injuries (Fu et al., 2015).
Toll-like receptors (TLRs), a conserved family of pattern recognition receptors (PRRs), are first line sensors that trigger inflammatory cascades (Janssens and Beyaert, 2003; Molteni et al., 2016). Immediately after blood flow cessation, dying and dead cells in the ischemic territory release various danger signals that are identified as danger-associated molecular patterns (DAMPs) to initiate a subsequent inflammatory cascade via PRRs expressed in resident brain cells (Chen and Nuñez, 2010), which leads to the upregulation of proinflammatory molecules that further amplify the inflammatory response, ultimately releasing more DAMPs (Shi et al., 2003; Kono and Rock, 2008). TLR4 is an essential sensor of endogenous DAMPs resulting from ischemia (Arumugam et al., 2009) and is involved in the pathology of AIS (Hakimizadeh et al., 2016). In genomic studies, the association between TLR4 polymorphisms and the progression of atherogenesis in AIS is well established (Lin et al., 2005; Weinstein et al., 2014). Preclinical studies further illustrated that genetic TLR4 knock-out (KO) was associated with improved outcomes after AIS (Caso et al., 2007; Hyakkoku et al., 2010). TAK-242, a TLR4 antagonist, exhibited protective effects against AIS in a mouse model (Hua et al., 2015). These findings suggest that TLR4 is warranted as a potential therapeutic target in AIS.
MicroRNAs regulate ∼30% of genes in the neuronal network (Ouyang et al., 2013; Samanta et al., 2016) and the abrupt cessation of cerebral blood flow dramatically alters microRNA expression within and beyond the brain, which has been demonstrated in both preclinical studies (Jeyaseelan et al., 2008; Dharap et al., 2009; Liu et al., 2010; Lusardi et al., 2014) and clinical observations (Tan et al., 2009; Sørensen et al., 2014). Experimental microRNA studies have ranged from their use as biomarkers to functional regulators in the pathology of AIS (Liu et al., 2015; Ni et al., 2015) and the long-term recovery process (Caballero-Garrido et al., 2015; Li et al., 2015; Liu et al., 2017). Furthermore, the role of microRNAs in poststroke immunomodulation was also reported in recent studies (Li et al., 2017; Voloboueva et al., 2017). However, the specific regulation of TLR4 by microRNAs after cerebral ischemia is not yet fully understood. Based on the current knowledge of poststroke inflammation and microRNA-based posttranscriptional regulation, the current study focused on the regulation of TLR4 in the brain at the microRNA level.
Materials and Methods
Animals.
Male adult C57BL/10 mice weighing 23–25 g (8–10 weeks old), female mice 16–18 d pregnant, pups within 24 h of birth, and male TLR4 KO mice (TLR4lps-del, C57BL/10 background, J003752) weighing 23–25 g (8–10 weeks old) were purchased from the Model Animal Research Institute of Nanjing University (Nanjing, Jiangsu, China). All experimental protocols were approved by the animal subject review board of Jinling Hospital and were conducted in accordance with recommendations from the National Institutes of Health's Guide for the Care and Use of Laboratory Animals (NIH Publications No. 8023, revised 2011). Animals were allowed ad libitum access to food and water and were housed in a 12 h light/dark cycle at ∼25°C and a relative humidity of 65%. All efforts were made to minimize the number of animals killed and their suffering in the study.
Transient middle cerebral artery occlusion (MCAO).
The MCAO procedure was performed as demonstrated previously (Wu et al., 2014). In brief, mice were anesthetized with 2–3% isoflurane (RWD Life Science, Shenzhen, China) and their body temperatures were maintained at 37.0 ± 0.5°C with a heating pad. Their right middle cerebral arteries were occluded with silicone-coated 6–0 nylon monofilaments. Blood flow reduced >70% of that at the baseline, as monitored by a laser Doppler flowmeter (Perimed PF5000), was regarded as a successful occlusion. Reperfusion was achieved by withdrawing the suture 90 min after occlusion.
Microarray analysis.
Peri-infarct cortex tissues derived from the MCAO and control groups (n = 3) were used for microarray analysis. The integrity of each RNA sample was determined with an Agilent Technologies 2100 Bioanalyzer as recommended. MicroRNA profiling was performed with the SurePrint G3 Mouse Unrestricted miRNA 8 × 60 K Microarray (G4872A-046065; Agilent Technologies) as instructed by the service provider. The microarray chip was designed based on Sanger miRBase (release 19.0) and contained 1247 mouse microRNAs. The acquired microarray images were analyzed with Feature Extraction software version 10.7.1.1; GeneSpring GX software version 12.0 was used for subsequent data analyses (both Agilent Technologies). Comparisons between groups were performed with two-tailed Student's t tests and p < 0.01 and fold changes >2 were considered significant.
In situ hybridization.
In situ hybridization was performed with a microRNA in situ hybridization kit (Boster) following the manufacturer's recommendations. In brief, frozen brain tissue sections mounted on slides were fixed in 4% paraformaldehyde (PFA), pH 7.2–7.6, containing 1% diethyl pyrocarbonate and air-dried after washing. After being immersed in methanol with 0.6% H2O2 for 30 min and prehybridized with a prehybridization solution at 42°C for 4 h, the slides were incubated with a microRNA probe overnight. After washing and blocking on the second day, the slides were incubated with a biotin-labeled secondary antibody at 37°C for 1 h. Signals were visualized sequentially by a streptavidin–biotin complex, a biotin-peroxidase complex, and diaminobenzidinetetrahydrochloride (DAB) with an ImmPACT DAB peroxidase substrate kit (Vector Laboratories). Finally, nuclei were stained with hematoxylin (Sigma-Aldrich) and the slides were mounted with neutral balsam-mounting medium after gradient ethanol and xylene washes.
Cell culture.
Cortical neurons were isolated from fetal mice of embryonic days 16–18, whereas astrocytes and microglia were isolated from neonatal pups within 24 h of birth. In brief, cortical tissues were digested with 0.125% trypsin (Invitrogen), homogenized by pipetting, and filtered with a 100 μm cell strainer (Biologix). The cells were then centrifuged and resuspended in DMEM (Invitrogen) with 10% FBS (Invitrogen) and penicillin–streptomycin (Invitrogen). For neurons, cell suspensions were seeded into six-well plates (106 cells/ml) and their culture medium was replaced after 2 h with Neurobasal medium (Invitrogen) containing 1% l-glutamate and 2% B27. Astrocyte and microglial cells were seeded in flasks (Costar) coated with poly-l-lysine (Sigma-Aldrich) to obtain mixed glial cultures. When the glial cultures reached confluency for 3 d, the flasks were shaken overnight at 200 rpm. Adherent cells were reseeded in cell culture dishes (Costar) to obtain astrocyte cultures (Jang et al., 2013; Kim et al., 2016) and floating cells were collected and seeded in six-well plates to obtain microglia (Frei et al., 1986; Giulian and Baker, 1986; Ganter et al., 1992). Human embryonic kidney 293T cells were purchased from Shanghai Institutes for Biological Sciences (Shanghai, China) and cultured with OPTI-MEM (Invitrogen) before, during, and after transfection.
Oxygen-glucose deprivation (OGD).
OGD was performed according to a previously established protocol (Yang et al., 2015; Xu et al., 2016). Briefly, the cells were transferred into a sealed chamber flushed with 95% N2 and 5% CO2, and the culture medium was then replaced with glucose-free DMEM (Invitrogen). After 2 h for neurons and 8 h for glial cells (or other specifically indicated time periods), the cells were returned to their normal incubators and initial culture media, thus allowing 24 h reperfusion.
Real-time PCR.
RNA extracted using TRIzol reagent (Sigma-Aldrich) was reverse transcribed into cDNA with the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific), with 200 ng of mRNA used per sample. Specific primers were designed for each microRNA for microRNA reverse transcription (Table 1), whereas oligo (dT) 18 was used for total mRNA reverse transcription. PCR was performed with UltraSYBR Mixture (CWBio) using the Mx3000P Real-Time PCR System (Agilent Technologies). The forward microRNA primers are listed in Table 2 and the other primers are as follows: TLR4, 5′-AGTCAGAATGAGGACTGGGTGAG-3′ (forward), 5′-GTAGTGAAGGCAGAGGTGAAAGC-3′ (reverse); GAPDH, 5′-CATGTTCGTCATGGGTGTGAAC-3′ (forward), 5′-CAGTCTTCTGGGTGGCAGTGAT-3′ (reverse); U6, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ (forward), 5′-CGCTTCACGAATTTGCGTGTCAT-3′ (reverse); and the common reverse primer shared by all microRNAs, 5′-CCAGTGCAGGGTCCGAGGT-3′.
Table 1.
MicroRNA-specific primers for reverse transcription
| MicroRNAs | Primers |
|---|---|
| miR-1906 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGCCCT |
| miR-125a-3p | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGGCTCCCA |
| miR-710 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGGCTCCCA |
| miR-494-3p | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGAGGTTTC |
| miR-290-5p | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAGTGCC |
| miR-2861 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCCGCCCGC |
| miR-7a-5p | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACAACAAA |
| miR-337-3p | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAGGCAT |
| miR-148b-3p | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACAAAGTT |
| miR-380-3p | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAGATGTG |
| miR-1198a-5p | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCCAAGCCA |
| miR-199a-3p | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTAACCAAT |
| U6 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAATA |
Table 2.
MicroRNA-specific forward primers for PCR
| MicroRNAs | Primers |
|---|---|
| miR-1906 | TGCAGCAGCCTGAGGC |
| miR-125a-3p | ACACTCCAGCTGGGACAGGTGAGGTTCTTG |
| miR-710 | ACACTCCAGCTGGGCCAAGTCTTGGGGAGA |
| miR-494-3p | ACACTCCAGCTGGGUGAAACAUACACGGGA |
| miR-290-5p | ACACTCCAGCTGGGACTCAAACTATGGGGG |
| miR-2861 | ACACTCCAGCTGGGGGGGCCTGGCGGC |
| miR-7a-5p | ACACTCCAGCTGGGUGGAAGACUAGUGAUUU |
| miR-337-3p | ACACTCCAGCTGGGUCAGCUCCUAUAUGAU |
| miR-148b-3p | ACACTCCAGCTGGGUCAGUGCAUCACAGAA |
| miR-380-3p | ACACTCCAGCTGGGTATGTAGTATGGTCCA |
| miR-1198a-5p | ACACTCCAGCTGGGTATGTGTTCCTGGCTG |
| miR-199a-3p | ACACTCCAGCTGGGACAGTAGTCTGCACAT |
Dual-luciferase reporter gene assay.
Briefly, 293T cells were plated in 12-well plates (Costar) and cultured to 90–95% confluence. Each well was then cotransfected with 1 μg of a firefly control vector (Promega) and 1 μg of a Renilla-tagged pGL3 promoter vector (Promega) containing wild-type (WT) or mutant TLR4 3′-UTR with Lipofectamine 2000 (Invitrogen). The miR-1906 agomir or the mismatch agomir was added at a concentration of 100 nm. Cells were harvested after 48 h with Passive Lysis Buffer (Promega) and luminescence was detected using a Dual-Luciferase Reporter Assay System (catalog #E1910; Promega) with the GloMax-Multi+ Detection System (Promega). The results are presented as the relative Renilla/firefly value.
RNA extraction from extracellular vesicles (EVs).
Total RNA, including miRNAs in exosomes and other EVs, was isolated from cell culture supernatants with the exoRNeasy Serum/Plasma Maxi Kit (catalog #77064; QIAGEN) according to the manufacturer's instructions. Briefly, 40 ml of serum-free cell culture medium was collected from astrocytes with or without OGD. After prefiltration with syringe filters (catalog #SLAA033SB; Millipore) and enrichment with Ultra-15 centrifugal filter concentrators (catalog #UFC900324; Millipore), the supernatants were mixed with buffer XBP at a 1:1 ratio and loaded onto exoEasy spin columns. After washing with XWP, the columns were transferred into new collection tubes, and 700 μl of QIAzol was added to collect the lysates by centrifugation. Then, 90 μl of chloroform was mixed with each lysate sample and the samples were centrifuged at 12,000 × g for 15 min at 4°C. After centrifugation, the samples were separated into three phases and the upper phase was collected and mixed with two volumes of 100% ethanol. The mixtures were then added to RNeasyMinElute spin columns and washed with RWT and RPE. Finally, columns were transferred into new collection tubes and 14 μl of RNase-free water was added to collect the RNA.
Immunofluorescence staining.
Cultured cells and 20-μm-thick frozen sections were fixed with 4% PFA, pH 7.0, and sequentially immersed in 0.3% Triton X-100 (Sigma-Aldrich) and 0.5% skim milk before being incubated in primary antibody overnight at 4°C. On the second day, the slides were incubated in the dark with a secondary antibody for 2 h and DAPI (Sigma-Aldrich) for 15 min. Images were acquired with a FluoView FV10i confocal laser scanning microscope (Olympus) and the following antibodies were used: anti-TLR4 (1:200, catalog #ab13556; Abcam), anti-GFAP (1:200, catalog #ab4648; Abcam), anti-β III-tubulin (1:200, catalog #ab78078; Abcam), anti-Iba-1 (1:200, catalog #ab15690; Abcam), anti-rabbit Alex Fluor 647-conjugated secondary antibody (1:200, Cat#ab150063; Abcam), and anti-mouse Alex Fluor 488-conjugated secondary antibody (1:200, catalog #ab150109; Abcam).
Western blotting.
Western blotting (WB) was performed as described previously (Xie et al., 2015). In brief, brain tissues or cultured cells were lysed in RIPA lysis buffer (Cell Signaling Technology) with 1 mm phenylmethanesulfonyl fluoride and protein concentrations were determined using the BCA protein assay kit (Thermo Scientific). Equal amounts of protein were loaded onto 7.5% SDS-PAGE gels for separation and then transferred to polyvinylidenedifluoride membranes (Millipore). After being blocked in 5% skim milk (Generay Biotech), the membranes were incubated in primary antibodies (TLR4: 1:500, catalog #ab13556; Abcam; β-actin: 1:1000, catalog #4970S; Cell Signaling Technology) at 4°C overnight. An HRP-conjugated anti-rabbit secondary antibody (1:2000, catalog #7074S; Cell Signaling Technology) and enhanced chemiluminescence (Merck) were used to detect protein bands on the second day.
Intraventricular injection of the mismatch or miR-1906 agomirs.
Under isoflurane anesthesia, mice were placed in a stereotaxic frame (model 900; David Kopf Instruments) and the ventricle of each mouse was injected with 5 μl of the mismatch or miR-1906 agomirs (100 nm; bregma: 0.8 mm, lateral: 1.5 mm, dorsoventral: 3.5 mm) at a speed of 1 μl every 5 min. The needle remained in place for 5 min after each injection.
Behavioral tests.
After 24 h of reperfusion, the neurological functions of the mice were assessed according to modified Neurological Severity Scores (mNSS) (Chen et al., 2001). The mNSS were a composite scoring system consisting of five neurological function sub-items: motor functions (including tail raising test and gait evaluation), sensory functions (including placing and proprioceptive tests), balancing functions (balance beam test), reflex functions (including pinna, corneal and startle reflexes), and abnormal movements such as seizure. The total scores ranged from 0 (no deficits) to 18 (maximal deficits) and higher scores indicated worse neurological functions. Investigators blinded to the animal groupings evaluated each animal and recorded their sub-item and total scores.
Fluoro-Jade C staining (FJC).
Following the manufacturer's instructions (Millipore), frozen sections were immersed sequentially in a 1% NaOH/80% ethanol solution, 70% ethanol, and a 0.06% potassium permanganate solution. The sections were then incubated with a 0.0001% FJC working solution (Histo-Chem) for 15 min, followed by immersion in xylene and mounting with neutral balsam-mounting medium.
Terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL).
TUNEL staining was performed with the In Situ Cell Death Detection Kit (Roche) according to the manufacturer's instructions. Slides were fixed with 4% PFA and immersed in 15% glacial acetic acid for 2 min. They were then incubated sequentially with a reaction solution (Roche) and converter (Roche). The signals were visualized using a BCIP/NBT solution (Beyotime Biotechnology).
H&E staining.
Frozen sections were fixed in methanol and immersed in hematoxylin (Sigma-Aldrich) for 10 min. After sufficient washing, the slides were immersed in 1% hydrochloric acid in ethanol for 10 s, immediately followed by sufficient washing and immersion in 0.1% ammonia for 1 h. The slides were then stained with an eosin staining solution (Beyotime) for 1 min, gradient washed with ethanol and xylene, and mounted with neutral balsam-mounting medium.
MRI.
Under isoflurane-induced anesthesia, mice underwent MRI examinations using a 7.0 T MRI scanner (Bruker). Their heart and respiration rates were monitored and their body temperatures were kept at 37.0 ± 0.5°C with a heating pad during scanning. T2-weighted images (T2-WIs) were acquired using a turboRARE-T2 sequence with the following parameters: repetition time/echo time = 2500/36.0 ms, field of view = 2.00/1.80 cm, MTX 384 matrix size = 256 × 256, and slice thickness = 0.5 mm (20 slices per animal). Parameters for diffusion weight imaging (DWI) were as follows: repetition time/echo time = 5000/22.0 ms, field of view = 2.00/1.80 cm, MTX 128 matrix size = 256 × 256, and slice thickness = 0.5 mm (20 slices per animal, matching T2-WI). The infarcts were identified by high signals acquired from T2-WI and further verified by DWI.
2,3,5-triphenyltetrazolium chloride (TTC) staining.
After the mice were killed, their brains were immediately sectioned into consecutive coronal slices of 1 mm thickness. The slices were incubated in 2% TTC (Sigma-Aldrich) for 15 min at 37°C and then fixed in 4% paraformaldehyde.
Infarct volume calculation.
In both TTC staining and MRI, the relative infarct volume was expressed as the percentage of noninfarcted volume in the ischemic hemisphere relative to that in the contralateral hemisphere (Caballero-Garrido et al., 2015). To calculate the noninfarcted volume in the lesioned hemisphere (VL), intact areas in each slide were summed and then multiplied by the distance between slides (1 mm for TTC and 0.5 mm for MRI) and the volume of the contralateral hemisphere (VC) was identified similarly. Therefore, the relative infarct volume was calculated as 100% × (VC − VL)/VC.
Statistics.
Statistical analyses were performed with Prism software version 6.0c (GraphPad) and SPSS version 24 (IBM), and all data are expressed as the mean ± SE. Differences between groups were determined with two-tailed Student's t tests for comparisons between two groups or by one-way ANOVA followed by Fisher's least significant difference post hoc tests for comparisons of more than two groups. Statistical significance was determined as p < 0.05 unless otherwise stated.
Results
Identifying a novel TLR4 regulator
TLR4 exacerbates cerebral ischemic injury
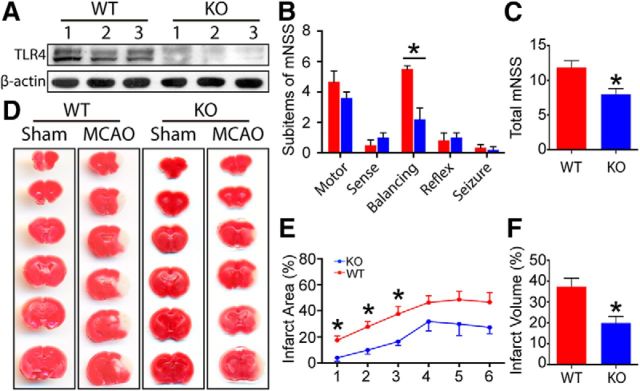
Abolishment of TLR4 protein expression was verified in TLR4 KO mice by WB analysis (Fig. 1A). Both KO and WT mice were subjected to 90 min of MCAO, followed by 24 h of reperfusion. Compared with WT mice, the mNSS (Fig. 1B,C) and infarct volumes of the TLR4 KO mice were reduced significantly (Fig. 1D–F).
Figure 1.
TLR4 KO reduced infarct volume and neurological deficits after MCAO. A, Successful ablation of TLR4 protein expression was confirmed by WB (n = 3). B, C, TLR4 KO led to better neurological function after MCAO. Among the sub-items of the modified neurological severity score system, TLR4 KO mainly contributed to improved performance in the balance beam test (p = 0.0012). The KO mice also showed lower total scores (p = 0.0175). D, Representative images of coronal brain sections from sham and MCAO mice in the WT and KO groups stained with TTC. E, Significantly decreased infarct areas were detected in the first, second, and third sections (p = 0.0228, 0.0146, and 0.0352, respectively). F, Infarct volume of total brain was also reduced in the KO group (p = 0.0188). *p < 0.05 by Student's t test, n = 4–8.
microRNA profiling and verification: miR-1906 suppresses TLR4
To clarify TLR4 regulation at the microRNA level, microarray profiling and in silico analysis were performed collaboratively (Fig. 2A). The microarray results were cross-compared with potential TLR4 microRNAs in the TargetScan (Lewis et al., 2005) (http://www.targetscan.org) and miRanda (John et al., 2004) (http://www.microrna.org) databases (Fig. 2B,C). After examining potential microRNAs by PCR, miR-1906 was identified as the most likely regulator (Fig. 2D).
Figure 2.
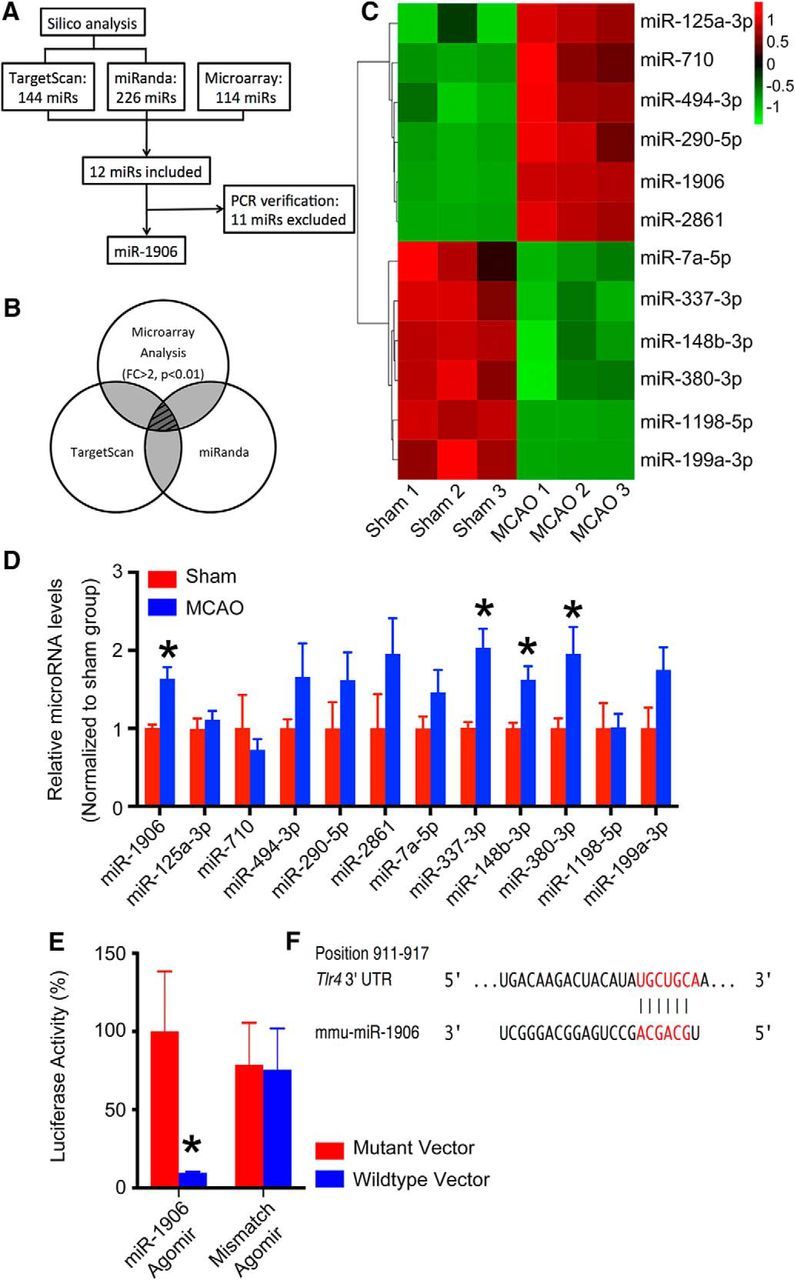
Systematic screening and verification of TLR4-targeting microRNAs. A, Flowchart of the systematic screening process. B, Combination of in silico and microarray analyses was included in subsequent experiments. From the TargetScan and miRanda databases, 144 and 226 microRNAs were retrieved as predicted regulators of TLR4 in mice, respectively. Significant alterations (fold change >2, p < 0.01) in expression were identified in 114 microRNAs by microarray analysis after MCAO. The in silico and microarray analyses included 12 microRNAs. C, Among those 12, 6 were increased and 6 were decreased after MCAO (n = 3). D, Expression levels of the 12 microRNAs as determined by PCR (n = 6). Four microRNAs (miR-1906, miR-337-3p, miR-148b-3p, and miR-380-3p) showed significantly increased expression, although the latter 3 (miR-337-3p, miR-148b-3p, and miR-380-3p) were excluded because their PCR results conflicted with the microarray results. Therefore, only miR-1906 (p = 0.0156) was included in the subsequent studies. E, Repressed luciferase activity was associated with the miR-1906 agomir, but not the mismatch agomir, in cells transfected with the TLR4 WT 3′-UTR mRNA sequence in the dual-luciferase reporter gene assay. No difference was observed in cells transfected with plasmids carrying the mutant TLR4 3′-UTR mRNA sequence (n = 4, assayed in triplicate). F, miR-1906 seed sequence and the corresponding binding target in the 3′-UTR of TLR4. *p < 0.05 by Student's t test, n = 4–8.
To determine whether miR-1906 could specifically recognize the 3′-UTR of TLR4 mRNA, the dual-luciferase reporter gene assay was performed. Vectors containing the Renilla luciferase reporter gene and either WT or mutant 3′-UTR TLR4 mRNA sequences were constructed and cotransfected with vectors carrying the firefly luciferase control reporter and the miR-1906 or mismatch control agomirs into 273T cells. Administration of the miR-1906 agomir decreased the luciferase activity of cells transfected with WT TLR4 3′-UTR but not mutant 3′-UTR TLR4 (Fig. 2E,F).
miR-1906 expression profile
Distributions of miR-1906 and TLR4
As shown in Figure 3, miR-1906 was enriched in the brain and colocalized with TLR4 in all regions of healthy mouse brain tissue. To characterize the temporal and regional alterations of TLR4 and miR-1906 after cerebral ischemia, the ischemic hemispheres of MCAO mouse models were collected and divided into the peri-infarct area and ischemic core after 2, 4, 6, 12, and 24 h of reperfusion. The expression levels of miR-1906 (Fig. 4A,B) and the TLR4 protein (Fig. 4E–H) were increased within a few hours after MCAO. TLR4 protein expression peaked after the second hours of reperfusion before returning to its normal level at the fourth hour despite the constant increase of TLR4 mRNA (Fig. 4E–H). In contrast, the miR-1906 levels began to elevate during the second hour of reperfusion, peaked after 4 h, and then declined gradually, but remained significantly higher than those of the sham group (Fig. 4A,B). The fact that TLR4 protein expression failed to correlate with the augmentation of TLR4 mRNA expression indicated the probable involvement of posttranscriptional regulation and a possible role of miR-1906 as a regulator of TLR4 via translational repression.
Figure 3.
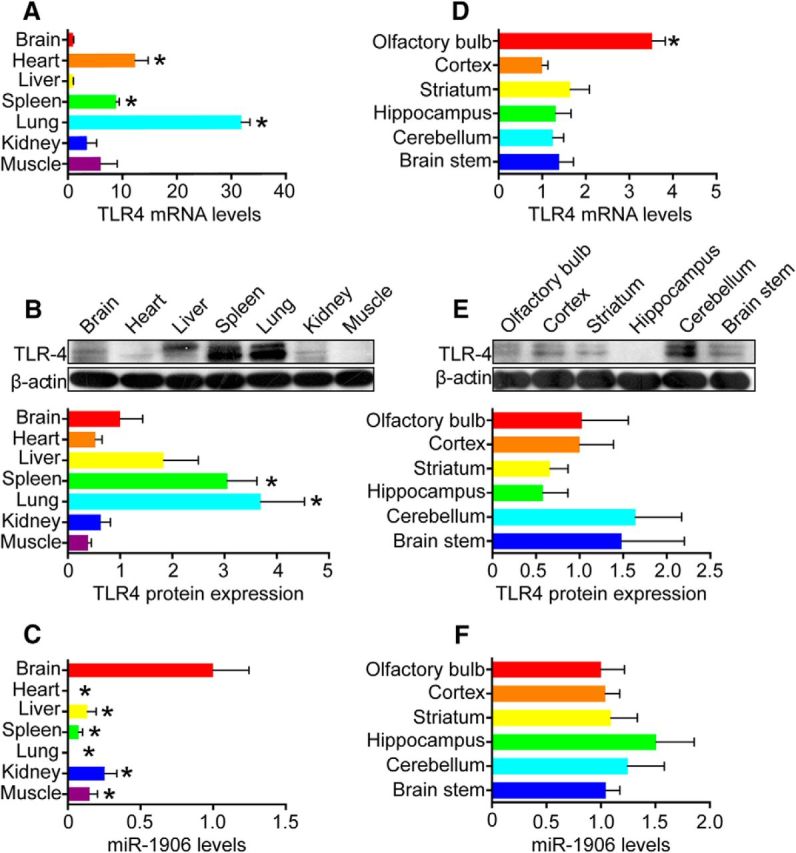
TLR4 and miR-1906 expression in major organs and the brain. A, B, TLR4 mRNA and protein levels in major organs relative to those in the brain as verified by PCR and WB. The TLR4 mRNA levels were higher in heart (p = 0.0003), spleen (p < 0.0001), and lung (p < 0.0001) tissue. The TLR4 protein levels in spleen (p = 0.0280) and lung (p = 0.0291) tisuse were higher than those in the brain (Student's t test, n = 6 for PCR, n = 4 for WB). C, miR-1906 levels in major organs relative to those in the brain. Compared with heart (p = 0.0024), liver (p = 0.0067), spleen (p = 0.0039), lung (p = 0.0024), kidney (p = 0.0169), and muscle (p = 0.0072) tissue, the brain showed significant miR-1906 enrichment (Student's t test, n = 6). D, E, TLR4 mRNA and protein levels in different regions of the brain relative to the cortex. F, miR-1906 levels in different regions of the brain. The TLR4 mRNA levels were significantly higher in the olfactory bulb than in the cortex (p < 0.0001). No significant difference was detected between the cortex and other cerebral regions (Student's t test, n = 6). Expression of the TLR4 protein (Student's t test, n = 4) and miR-1906 (Student's t test, n = 6) was statistically equivalent in each cerebral region compared with those in the cortex. *p < 0.05 by Student's t test, n as indicated.
Figure 4.
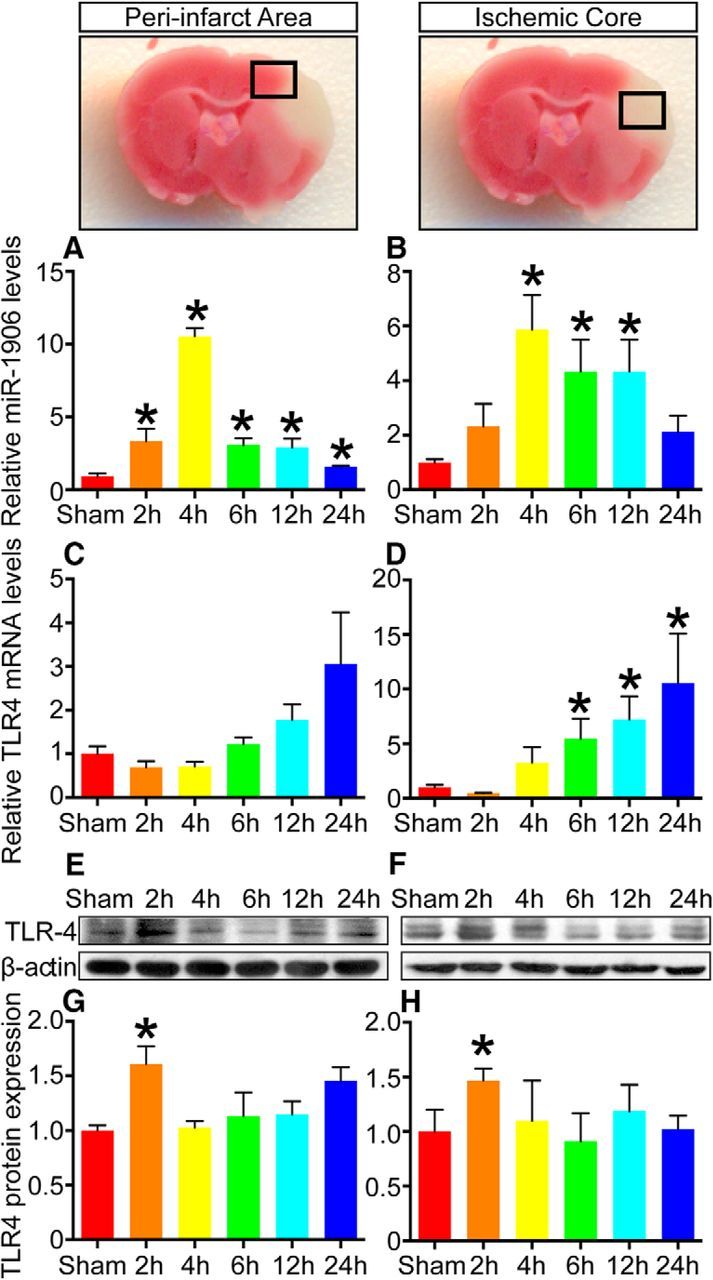
Spatiotemporal expression profile of miR-1906 and TLR4 after MCAO. A, miR-1906 levels in the peri-infarct area were increased significantly (3.34-fold, p = 0.0246) as early as 2 h after reperfusion, peaked in the fourth hour (10.54-fold, p < 0.0001), and remained high through 24 h of reperfusion. B, In the ischemic core, miR-1906 expression began to increase 4 h after reperfusion (5.87-fold, p = 0.0032) and the elevation remained significant through 12 h of reperfusion. C, mRNA levels of TLR4 failed to exhibit statistically significant increases in the peri-infarct area. D, In the ischemic core, the increase in TLR4 mRNA abundance reached significance at 6 h of reperfusion (5.457-fold, p = 0.0257) and remained elevated up to 24 h (10.52-fold, p = 0.0456). E, G, TLR4 protein levels were increased in the second hour of reperfusion in both the peri-infarct area (1.61-fold, p = 0.0279) and F, H, the ischemic core (1.47-fold, p = 0.0331), but returned to normal levels 4–24 h after reperfusion. *p < 0.05 by Student's t test, each sample was compared with the sham group, n = 6.
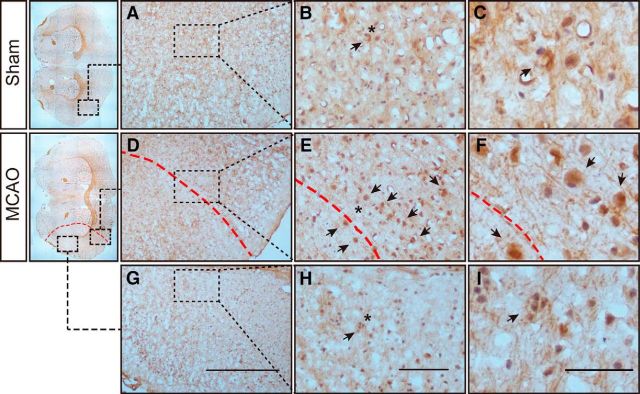
Regarding regional alteration, the variation tendencies of miR-1906 and TLR4 in the peri-infarct area (Fig. 4A,C,E,G) and ischemic core (Fig. 4B,D,F,H) were generally consistent. However, TLR4 mRNA expression increased insignificantly in the peri-infarct area (Fig. 4C) but dramatically in the ischemic core (Fig. 4D), whereas miR-1906 expression tended to be increased more significantly in the peri-infarct area (Fig. 4A) than in the core (Fig. 4B). In addition, more miR-1906-positive cells were observed surrounding the ischemic boundary (Fig. 5D–F) rather than the ischemic core (Fig. 5G–I).
Figure 5.
In situ hybridization illustrating miR-1906+ cells (arrows) around the ischemic boundary. Asterisks indicate the magnified areas on the right. A–C, Corresponding brain cortex from the sham group. D–F, Peri-infarct area in the ischemic cortex. The ischemic boundary is indicated by the red dashed line. An accumulation of miR-1906+ cells was observed around the ischemic boundary. G–I, Ischemic core of the ischemic cortex. Shrinkage of cells and nuclei was observed as a marker of cell death. Although miR-1906 was detected, it was less dense compared with miR-1906+ cells observed in the peri-infarct area. Scale bars: A, D, G, 500 μm; B, E, H, 100 μm; C, F, I, 50 μm. n = 3.
Conflicting expressional tendencies of miR-1906 across different cell types
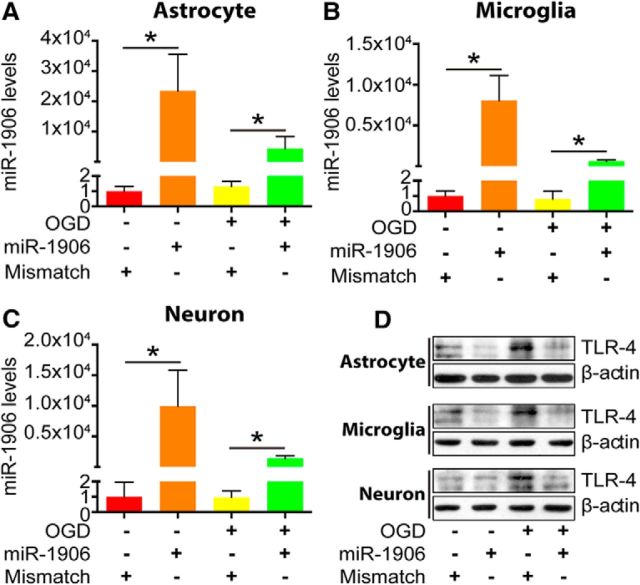
Because TLR4 is reportedly expressed in astrocytes, microglia, and neurons (Artinian et al., 2002), these cells were isolated and cultured in vitro to explore their miR-1906 expression profiles in response to OGD. miR-1906 expression was increased significantly in glial cells (Fig. 6A,C), but decreased in neurons (Fig. 6E), with OGD elongation being correlated with cell injury (Fig. 6B,D,F). In addition, after 10 h of OGD for glia and 2.5 h for neurons, the cultured cells became nonadherent (Fig. 6B,D,F), making subsequent experiments difficult. Therefore, an 8 h OGD period for glia and a 2 h period for neurons were selected for subsequent experiments. Total RNAs in exosomes and other EVs were derived from astrocyte supernatants cultured with or without OGD for 8 h to detect the levels of extracellular secreted miR-1906. Despite the increased intracellular abundance (Fig. 6A), extracellular miR-1906 secretion decreased after ischemic attack (Fig. 6G).
Figure 6.
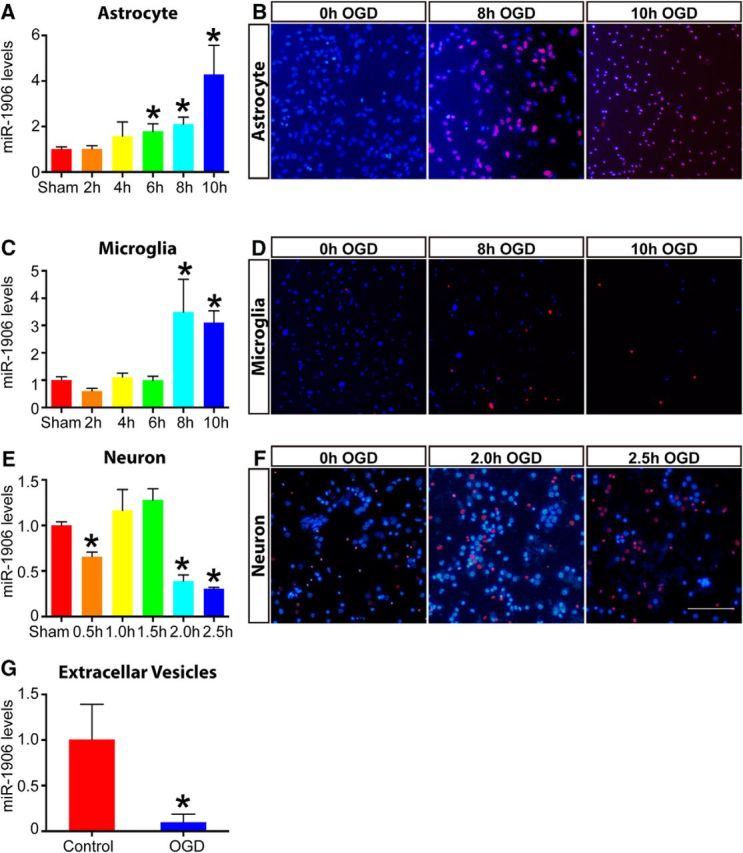
Discrepant miR-1906 expression in astrocytes, microglia, neurons, and EVs after OGD in vitro. A, In astrocytes, miR-1906 expression increased as the OGD duration extended and the increase became statistically significant after 6 h (1.78-fold, p = 0.0395), 8 h (2.10-fold, p = 0.0045), and 10 h (4.29-fold, p = 0.0116) of OGD (n = 6). B, Propidium iodide–Hochest staining of astrocytes at different time points of OGD. Because 10 hours of OGD led to excessive cell death, 8 h of OGD was used for subsequent experiments. C, In microglia, miR-1906 expression was increased significantly after 8 h (3.49-fold, p = 0.0129) and 10 h (3.11-fold, p = 0.0001) of OGD (n = 6). D, After 10 h of OGD, most microglial cells were dead and nonadherent and were thus not suitable for subsequent expreriments. Therefore, 8 h of OGD was also selected for subsequent experiments with microglial cells. E, In neurons, miR-1906 expression was decreased significantly after 0.5 h (0.66-fold, p = 0.0058), 2 h (0.39-fold, p = 0.0016), and 2.5 h (0.31-fold, p < 0.0001) of OGD (n = 3). F, For neurons, 2 h of OGD was selected because 2.5 h of OGD led to a significantly reduced number of cells. G, Abundance of miR-1906 in EVs was decreased in astrocyte cell culture supernatants after 8 h of OGD (0.01-fold, p = 0.0471, n = 5). *p < 0.05 by Student's t test, each sample was compared with the sham group, n as indicated. Scale bar, 50 μm.
Biological functions of exogenous miR-1906
miR-1906 downregulates TLR4 via translational repression: an in vitro study
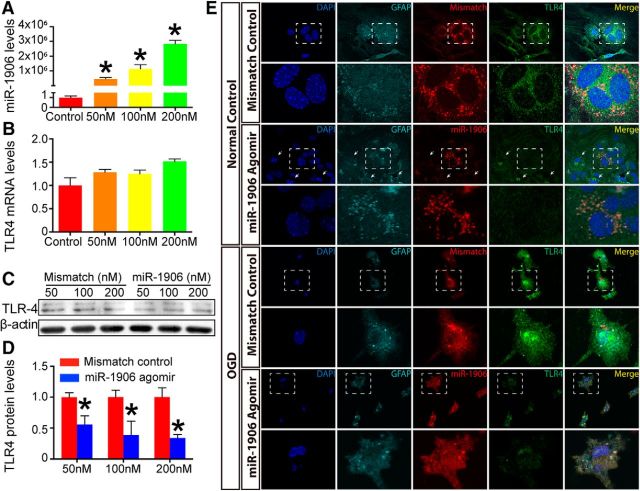
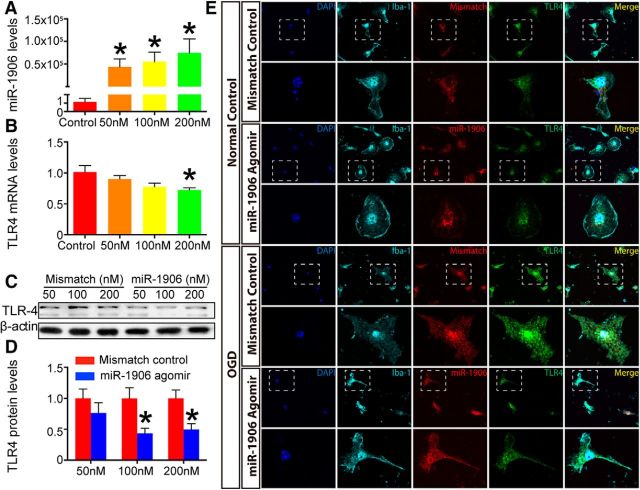
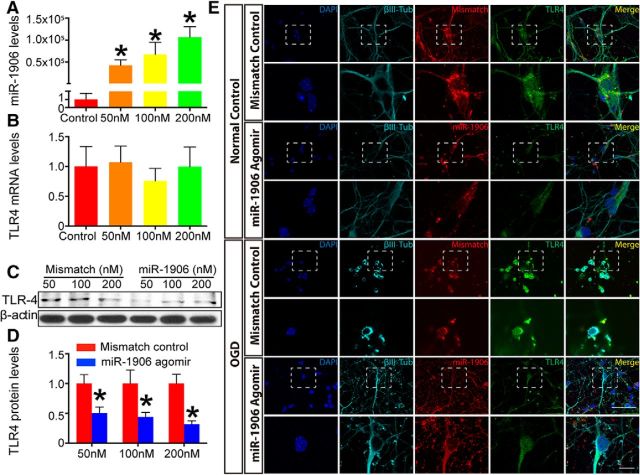
Cultured astrocytes, microglia, and neurons were administered different concentrations of the miR-1906 agomir (Figs. 7A, 8A, 9A). Among the tested dosages, 100 nm resulted in an ∼50% reduction of TLR4 protein expression in all the cell types analyzed (Figs. 7C,D, 8C,D, 9C,D). Alternatively, the mRNA levels of TLR4 in astrocytes and neurons failed to show significant decreases with increasing dosage (Figs. 7B, 9B). Although TLR4 mRNA expression decreased in microglia at the 200 nm concentration of the miR-1906 agomir (Fig. 8B), the TLR4 protein levels began to decrease with only a 100 nm concentration (Fig. 8C,D). These findings indicated that miR-1906 functions by translational repression rather than mRNA degradation.
Figure 7.
The miR-1906 agomir suppressed TLR4 expression in astrocytes at the protein level, but not at the mRNA level in vitro. A, Intracellular abundance of miR-1906 increased drastically as the miR-1906 agomir dosage increased to final concentrations of 50, 100, and 200 nm, respectively (compared with the control group, Student's t test, n = 3). B, No TLR4 mRNA expression reduction was observed at any of the tested concentrations (compared with the control group, Student's t test, n = 4). C, D, TLR4 protein expression was decreased in astrocyte cultures receiving the miR-1906 agomir at final concentrations of 50, 100, and 200 nm (0.56-fold, p = 0.0299; 0.39-fold, p = 0.0445; and 0.34-fold, p = 0.0160, respectively; compared with the same dose of the mismatch control, Student's t test, n = 4). E, Colocalization of the TLR4 protein and the administered agomir was visualized by immunofluorescence with a confocal laser scanning microscope. Magnified views are marked with dashed line boxes. Both the miR-1906 and mismatch agomirs could penetrate cultured astrocytes both under normal conditions and after 8 h of OGD. Under normal conditions, diminished TLR4 signals were detected in astrocytes receiving the miR-1906 agomir compared with those receiving the mismatch agomir. In addition, cells with stronger miR-1906 agomir signals showed less TLR4 signal than those with less miR-1906 agomir accumulation (arrows). After 8 h of OGD, the cells appeared injured and damaged, as marked by their shrunken and folded morphology. Consistent with those in the normal condition, both the miR-1906 and mismatch agomirs accumulated in astrocytes despite OGD and the miR-1906 agomir was associated with decreased TLR4 signals. *p < 0.05, n as indicated.
Figure 8.
miR-1906 agomir suppressed TLR4 expression in microglia in vitro. A, Intracellular accumulation of miR-1906 increased drastically with increased miR-1906 agomir administration at final concentrations of 50, 100, and 200 nm (compared with the control group, Student's t test, n = 3). B, Significantly reduced TLR4 mRNA expression was observed only when the miR-1906 agomir concentration reached 200 nm (0.72-fold, p = 0.0395, compared with the control group, Student's t test, n = 4). C, D, TLR4 protein expression decreased at concentrations of 100 and 200 nm (0.43-fold, p = 0.0242 and 0.50-fold, p = 0.0375, respectively; compared with the same dose of the mismatch control, Student's t test, n = 4). E, Confocal laser scanning microscope confirmed that the exogenous agomir entered microglia under both normal conditions and after 8 h of OGD in vitro. The overall TLR4 signal appeared stronger after OGD and the miR-1906 agomir was associated with diminished TLR4 expression with or without OGD. *p < 0.05, n as indicated.
Figure 9.
miR-1906 agomir suppressed TLR4 expression in neurons in vitro. A, Intracellular miR-1906 expression increased with increasing doses of the miR-1906 agomir (compared with the control group, Student's t test, n = 3). B, No significant differences in TLR4 mRNA expression were observed among the tested dosages (compared with the control group, Student's t test, n = 4). C, D, TLR4 protein expression was decreased in neurons receiving the miR-1906 agomir at final concentrations of 50, 100, and 200 nm (0.50-fold, p = 0.0349; 0.44-fold, p = 0.0440; and 0.32-fold, p = 0.0058, respectively; compared with the same dose of the mismatch control, Student's t test, n = 4). E, Both the miR-1906 and mismatch agomirs were able to penetrate cultured neurons in vitro under normal conditions and after OGD. Diminished TLR4 signals were observed in cell cultures receiving the miR-1906 agomir. Neurons receiving the mismatch agomir exhibited more severely damaged overall appearances after OGD than those receiving the miR-1906 agomir, mainly based on the integrity of axons and dendrites. Scale bar, 50 μm (upper scale bar), 10 μm (lower scale bar). *p < 0.05, n as indicated.
To confirm that the transfection and function of miR-1906 was not nullified after ischemia, OGD was performed after miR-1906 agomir transfection. As revealed by immunofluorescence, the Cy3-labeled miR-1906 or mismatch agomir colocalized within astrocytes (Fig. 7E), microglia (Fig. 8E), and neurons (Fig. 9E). The levels of miR-1906 were further verified by PCR (Fig. 10A–C) and elevated levels of miR-1906 were detected in cells receiving the miR-1906 agomir with or without subsequent OGD. Moreover, both WB (Fig. 10D) and immunofluorescence (Figs. 7E, 8E, 9E) confirmed that the miR-1906 agomir repressed TLR4 protein expression.
Figure 10.
Biological functions of the exogenous miR-1906 agomir in vitro. A–C, Administration of the exogenous miR-1906 agomir resulted in increased intracellular miR-1906 levels, which were sustained after OGD in astrocytes (p = 0.0418 and p = 0.0266), microglia (p = 0.0260 and p = 0.0059), and neurons (p = 0.0326 and p = 0.0273) as determined by comparing the agomir and mismatch control groups in normal and OGD conditions, respectively. Student's t test, n = 3. D, OGD resulted in increased TLR4 protein expression in astrocytes, microglia, and neurons. Reduced protein levels were observed in astrocytes, microglia, and neurons receiving the miR-1906 agomir in vivo. *p < 0.05, n as indicated.
miR-1906 agomir accesses astrocytes, microglia, and neurons in vivo
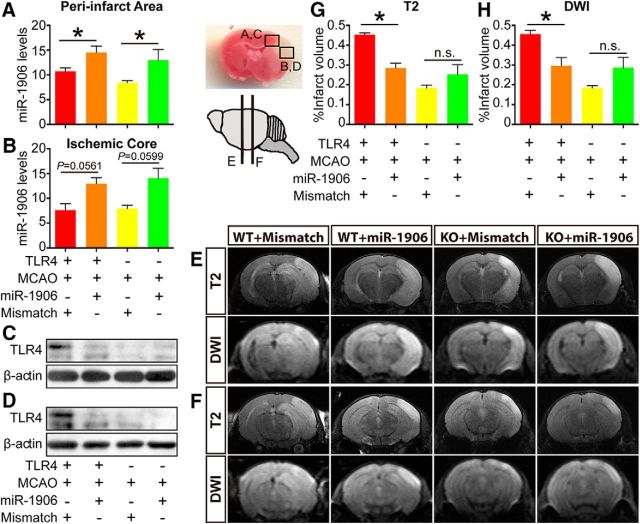
To confirm that exogenous miR-1906 or the mismatch agomir could penetrate different types of brain cells, immunofluorescence was used to visualize colocalization of the Cy3-labeled agomir and relevant cell biomarkers. Cy3 was detected in the astrocytes (Fig. 11), microglia (Fig. 12), and neurons (Fig. 13) of WT and KO mice. Compared with WT mice receiving the mismatch agomir, those receiving the miR-1906 agomir exhibited significantly increased miR-1906 expression in their peri-infarct areas (Fig. 14A) and ischemic cores (Fig. 14B). TLR4 expression was consistently decreased in the ischemic cores and peri-infarction areas of miR-1906 agomir-treated WT mice, whereas TLR4 was obviously absent in KO mice (Fig. 14C,D). Similarly, immunofluorescence illustrated repressed TLR4 fluorescence in the astrocytes (Fig. 11), microglia (Fig. 12), and neurons (Fig. 13) of WT mice receiving the miR-1906 agomir, whereas the signal was absent in the KO group.
Figure 11.
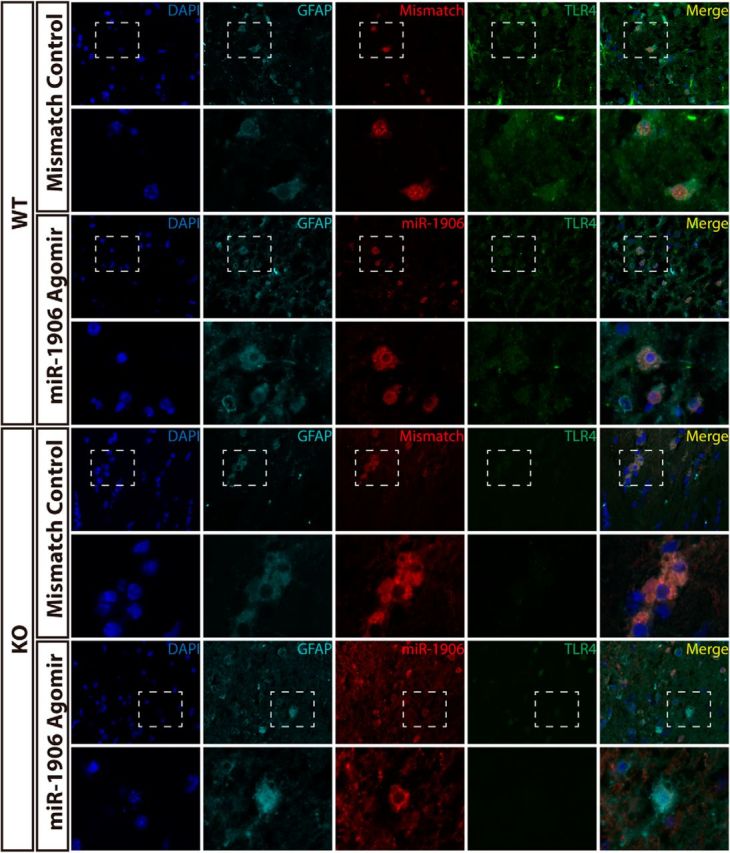
Both exogenous miR-1906 and the mismatch agomir penetrated astrocytes in WT and KO mice after MCAO, as illustrated by the colocalization of GFAP and Cy3 fluorescence. TLR4 signals were diminished in WT mice receiving the miR-1906 agomir and were barely detectable in KO mice brain sections. Magnified views are marked with dashed line boxes.
Figure 12.
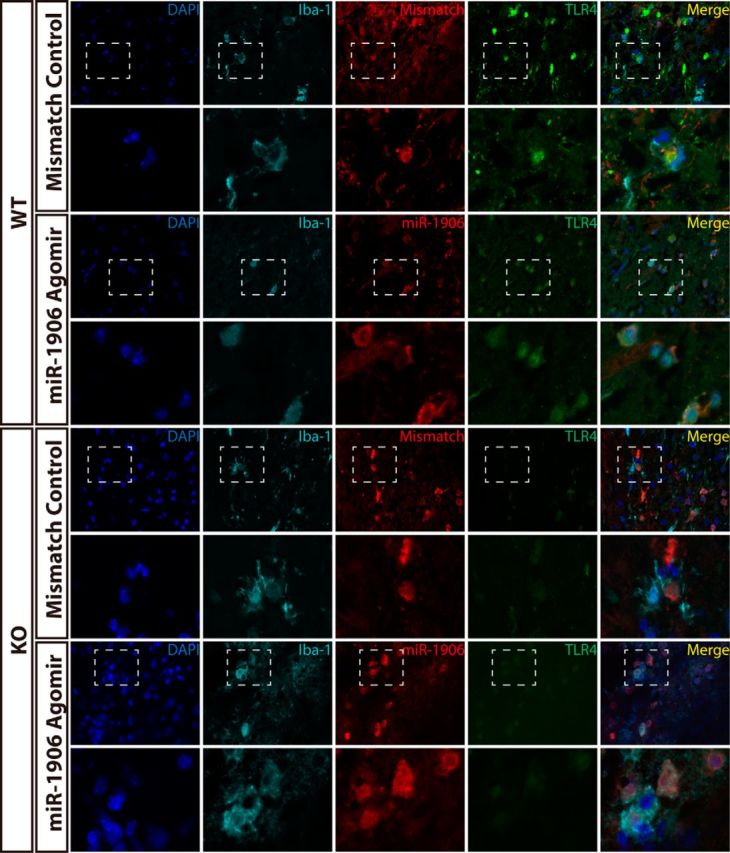
Colocalization of Iba-1 and Cy3 fluorescence demonstrated that miR-1906 and the mismatch agomir could access microglia in both WT and KO mice after MCAO. The overall intensity of the TLR4 signal was decreased in WT mice receiving the miR-1906 agomir and was barely detectable in KO mice. Magnified views are marked with dashed line boxes.
Figure 13.
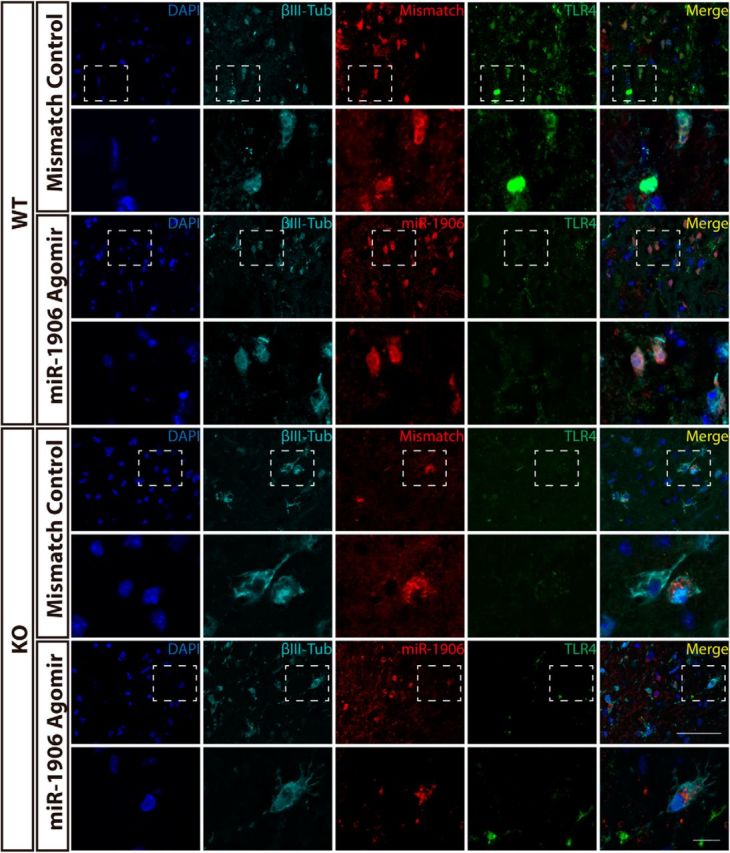
Cy3 and βIII-tubulin fluorescence were colocalized in WT and KO brain sections, indicating that both miR-1906 and the mismatch agomir were taken up by neurons in vivo. WT mice receiving the miR-1906 agomir showed less TLR4 signal fluorescence and the fluorescence was barely detectable in KO mice. Magnified views are marked with dashed line boxes. Scale bars: 50 μm (upper scale bar), 10 μm (lower scale bar).
Figure 14.
Biological functions of the miR-1906 agomir in vivo. The brains of the animals were divided into the peri-infarct region (A, C) and the ischemic core (B, D) 24 h after MCAO. In the peri-infarct region (A), miR-1906 expression was increased significantly in WT (p = 0.0209) and KO mice (p = 0.0334), whereas miR-1906 augmentation reached marginal significance in the ischemic core (WT: p = 0.0561, KO: p = 0.0599), which may have been due to severe cell death. Correspondingly, suppressed TLR4 protein expression was detected in the peri-infarct region (C) and ischemic core (D) of WT animals subjected to miR-1906 injection. MRI scans were acquired using two MR sequences: T2 and DWI. Two representative images of different coronary slices were acquired from one animal of each group, as the schematic indicates (E, F). Compared with those receiving the mismatch agomir, WT mice receiving the miR-1906 agomir showed significantly decreased infarct volume, as illustrated by T2 (E, G, p = 0.0043) and DWI (F, H, p = 0.0260), whereas no significant differences were observed in the KO animals (T2: p = 0.3049, DWI: p = 0.1630). *p < 0.05 by one-way ANOVA and Fisher's least significant difference post hoc test, n = 4.
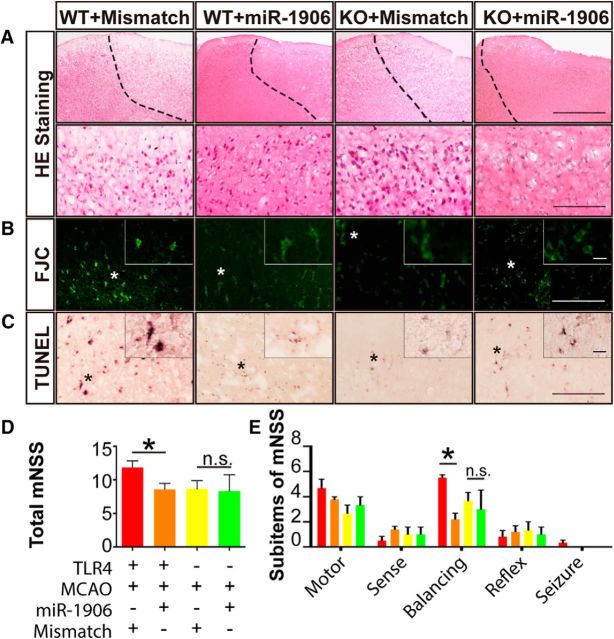
miR-1906 agomir alleviates ischemic injury in WT, but not KO, mice
miR-1906 overexpression and the corresponding TLR4 protein repression attenuated cerebral ischemic damage by reducing infarct volumes macroscopically, as determined by MRI (Fig. 14E–H). From a microscopic viewpoint, H&E staining showed alleviated brain damage in WT mice after miR-1906 agomir injection (Fig. 15A). Furthermore, the FJC and TUNEL assays also showed mitigated neuron degeneration (Fig. 15B) and cell apoptosis (Fig. 15C) in the miR-1906 agomir-treated group compared with WT mice receiving the mismatch agomir, whereas no significant morphological discrepancies were observed between the miR-1906 agomir-treated and mismatch agomir-treated KO mice. Neurological function was evaluated with an mNSS consisting of five sub-items and alleviated functional deficits were observed in WT mice receiving the miR-1906 agomir (Fig. 15D,E). In comparing the functional outcomes for each sub-item, investigators observed distinguishable differences in the balance beam test (Fig. 15E). The balance beam test required an animal to walk on a beam that was 1 inch wide, 4 feet long, and suspended 3 feet above the floor. The test evaluated sensorimotor integration and could therefore reflect complicated behavioral deficits. However, the protective effects of the miR-1906 agomir disappeared in the absence of the TLR4 protein in KO mice (Figs. 14, 15).
Figure 15.
Tissue injury and neurological function evaluation. Alleviated brain tissue damage, marked by less vacuoles within the cortex, was elucidated with H&E staining (A). The ischemic boarder line was marked with a dashed line manually based on the shrinking morphology of nuclei within the ischemic core. FJC staining (B) and the TUNEL assay (C) illustrated mitigated neuronal degeneration and apoptosis in WT mice receiving the miR-1906 agomir relative to WT animals receiving the mismatch agomir, whereas overall neuronal degeneration and apoptosis were less severe in the TLR4 KO groups regardless of which agomir was received. (D), Neurological deficit evaluations using mNSS demonstrated that the miR-1906 agomir was associated with improved functional outcomes among WT animals (p = 0.0390), but not KO animals (p = 0.973) (E). Among the sub-items, a significant difference was found in the balance beam test scores (p < 0.0001). *p < 0.05, one-way ANOVA and Fisher's least significant difference post hoc test, n = 4.
miR-1906 regulates postischemia inflammation via targeting TLR4 over time
The miR-1906 agomir and the antagomir altered TLR4 protein expression in both the peri-infarct area and the ischemic core in a time-dependent manner (Fig. 16A,B). In the mismatch control group, TLR4 protein expression was increased by the second hour of reperfusion and returned to baseline by the 24th hour. In contrast to the mismatch group and the TLR4 time course described above (Fig. 4), no significant decrease in TLR4 protein expression was observed from the second through the 24th hour of perfusion from the miR-1906 antagomir, suggesting that miR-1906 inhibition reversed decreased TLR4 protein expression. In addition, the miR-1906 agomir led to an overall inhibition of TLR4 expression at each time point. Consistent with this, expression of the inflammation markers IL-1β, TNF-α, iNOS, and MCP-1 was decreased in the peri-infarct area (Fig. 16C) and IL-1β expression was reduced in the ischemic core (Fig. 16D).
Figure 16.
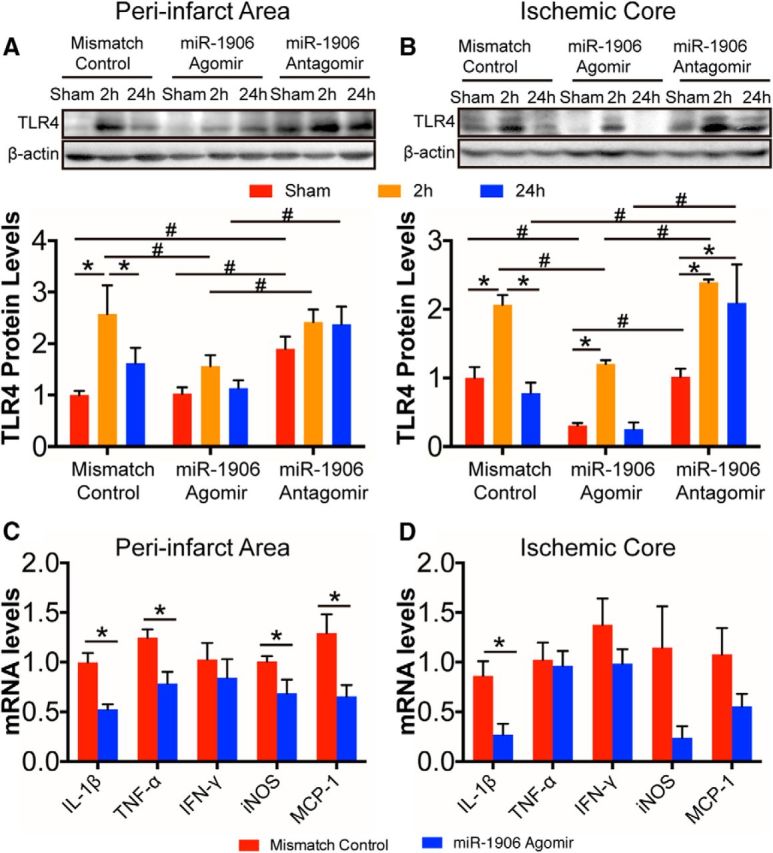
Regulation of miR-1906 affected postischemia inflammation by targeting TLR4 in a time-dependent manner after MCAO. Intracerebroventricular delivery of the miR-1906 agomir and the antagomir changed the time course of TLR4 protein expression in both the peri-infarct area (A) and the ischemic core (B). In the peri-infarct area, TLR4 expression was elevated significantly 2 h after reperfusion (p = 0.0011) and decreased significantly after 24 h (p = 0.0304) in the mismatch control group, which was not observed in the group receiving the miR-1906 agomir or the antagomir. The miR-1906 agomir inhibited TLR4 expression 2 h after reperfusion (mismatch vs agomir: p = 0.0231; agomir vs antagomor: p = 0.0149). Animals receiving the miR-1906 antagomir showed increased TLR4 expression in the sham group (mismatch vs antagomir: p = 0.0399, agomir vs antagomor: p = 0.0449) and after 24 h of reperfusion compared with those receiving the agomir (p = 0.0068). In the ischemic core, animals receiving the mismatch control demonstrated significantly increased TLR4 expression 2 h after reperfusion (mismatch control group, sham vs 2 h: p = 0.0024), which decreased after 24 h (mismatch control group, 2 h vs 24 h: p = 0.0005). A similar pattern was observed in the miR-1906 agomir group (sham vs 2 h: p = 0.0083; and marginal significance between 2 h vs 24 h: p = 0.0056), but not in the miR-1906 antagomir group (sham vs 2 h: p = 0.0002; sham vs 24 h: p = 0.0022). Conversely, the miR-1906 agomir group showed inhibited TLR4 expression in the sham group (mismatch vs agomir: p = 0.0343; agomir vs antagomir: p = 0.0310) and in the 2 h group (mismatch vs agomir: p = 0.0107; agomir vs antagomir: p = 0.0010). In addition, elevated levels of TLR4 were detected in the miR-1906 antagomir group 24 h after MCAO (mismatch vs antagomir: p = 0.0004; agomir vs antagomir: p < 0.0001). C, D, Inflammatory marker expression was suppressed significantly in the peri-infarct area and ischemic core with miR-1906 administration. Among the markers, IL-1β (p = 0.0112), TNF-α (p = 0.0373), iNOS (p = 0.0440), and MCP-1 (p = 0.0460) expression were decreased in the peri-infarct area (D), whereas IL-1β expression (p = 0.0318) was also reduced in the ischemic core (E). For A and B: *p < 0.05, comparisons among groups receiving the same treatment (mismatch control, agomir or antagomir) at different time points; #p < 0.05, comparisons among groups receiving different treatments at the same time point, one-way ANOVA and Fisher's least significant difference post hoc test, n = 3. For C and D: *p < 0.05, Student's t test, n = 4.
Discussion
In the current study, we screened systematically and verified functionally neuroprotective microRNAs that target TLR4. The major findings were as follows. First, of 1247 microRNAs screened by microarray, miR-1906 was identified as a novel regulator of TLR4 immediately after cerebral ischemia. Second, altered expression of miR-1906 and TLR4 were identified in the ischemic core and peri-infarct area. Third, miR-1906 downregulates TLR4 protein expression by repressing mRNA translation instead of mRNA degradation. Finally, the miR-1906 agomir ameliorates cerebral ischemic injury and postischemic inflammation and reduces neurological deficits after MCAO in vivo.
MicroRNAs react to cerebral ischemia immediately after ischemic attack and are actively involved in the modulation of poststroke inflammation (Ouyang et al., 2015). For example, the expression of miR-181, a crucial player in endoplasmic reticulum stress, is altered as early as 1 h after MCAO (Ouyang et al., 2012). Similarly, miR-424 is actively involved in oxidative stress after cerebral ischemia and its expression is increased 1 h after MCAO (Liu et al., 2015). In addition, Ni et al. (2015) demonstrated that miR-let-7c-5p overexpression attenuates neuroinflammation in the ischemic brain and is associated with better outcomes after experimental stroke. Consistently, the current study also revealed the therapeutic potential of microRNAs in targeting poststroke inflammation by targeting TLR4, a critical receptor of the innate immune system (Hakimizadeh et al., 2016). In the current study, increased miR-1906 expression was detected within 2 h after reperfusion and lasted for up to 24 h, which is consistent with the initiation of poststroke inflammation that occurs within minutes after ischemic onset and remains active for days (Ouyang, 2013; Hakimizadeh et al., 2016). TLR4 plays a deleterious role during the acute phase after cerebral ischemic attack. In a study performed by Hyakkoku et al. (2010), acute cerebral ischemia was induced in WT, TLR4 KO, TLR3 KO, and TLR9 KO mice from C57BL/6 and BALB/c backgrounds. Reduced infarct volume and less impaired neurological functions were observed only in TLR4 KO mice. Consistently, our current study also verified a detrimental function of TLR4 in the C57BL/10 strain and, based on this finding, a systematic screening for potential TLR4 regulators was conducted. With the combined results from microRNA profiling and in silico analysis, miR-1906 was identified as a candidate and was further verified with a dual-luciferase reporter gene assay. Furthermore, decreased TLR4 expression was observed with exogenous miR-1906 administration in vitro and in vivo, demonstrating its biological function in regulating TLR4.
Furthermore, the current study illustrated the miR-1906 expression profile with and without AIS. Under normal conditions, miR-1906 was enriched in the brain, suggesting its indispensable role in the CNS. In response to cerebral ischemic attack, miR-1906 expression increased within hours after MCAO in both the ischemic core and the peri-infarct area, especially in the latter. The ischemic core and peri-infarct area undergo distinct biological processes during focal cerebral ischemia: cells located in the core are quickly destined for death, whereas those located in the surrounding peri-infarct regions are potentially salvageable (Boisserand et al., 2017; Ginsberg, 2016) and have therefore emerged as a promising target for neuroprotective therapies (Ouyang et al., 2013; Ouyang et al., 2015). Ouyang et al. (2012) reported that miR-181a expression, a detrimental regulator in AIS, is altered in opposite directions in the ischemic core and peri-infarct areas: its expression is increased in the ischemic core in which cells are destined to die, but decreased in the potentially reversible peri-infarct tissue. Consistently, a relatively more drastic upregulation of miR-1906, a protective microRNA, was observed in the peri-infarct region in the current study, which probably also drives the fate of this region toward a favorable outcome. Moreover, delivery of this microRNA was associated with relatively more inflammatory marker suppression in the peri-infarct region, suggesting its potential therapeutic value in preserving peri-infarct tissue after ischemic attack.
In addition, the current study identified the regulation pattern of miR-1906. Two mechanisms underlie the microRNA regulation of their target mRNAs: translational repression and mRNA destabilization (Guo et al., 2010). Between these two patterns, mRNA destabilization explains >84% of all microRNA-mediated protein reductions (Guo et al., 2010), whereas translational repression acts more rapidly than mRNA destabilization (Eichhorn et al., 2014). As a combined result, during the early phase of microRNA expression alteration (during the first 5.7 h to be precise, as determined by Eichhorn et al., 2014), translational repression is dominant. Decreased protein expression recovers more rapidly after translational repression because new transcription is not necessary (Eichhorn et al., 2014). In the current study, administration of the exogenous miR-1906 agomir led to reduced TLR4 protein expression despite the TLR4 mRNA levels, suggesting that miR-1906 regulates its target via translational repression rather than mRNA destabilization. Therefore, because miR-1906 inflicts rapid inhibition and rapid recovery, it is suitable for immune modulation during the acute phase, with less negative effects on long-term recovery.
Finally, the successful delivery of miR-1906 showed protective effects against ischemic injuries. Elevated miR-1906 expression was associated with inhibited inflammation marker expression, reduced infarct volume, ameliorated tissue damage, and improved neurological function. TLR4 is widely known to contribute to brain injury after AIS, mainly via initiating downstream signaling cascades that induce the transcription of proinflammatory genes (Caso et al., 2007; Tang et al., 2007; Brea et al., 2011). However, TLR4 regulation by microRNAs and the therapeutic implication of microRNA delivery is not fully understood. In the current study, successful microRNA agomir administration was visualized readily by fluorescent labeling. Moreover, instead of using a viral-based delivery method, the current study used a cholesterol-modified microRNA agomir, which avoided safety concerns related to viral-based strategies (Maegdefessel, 2014). Because microRNAs usually have multiple targets (Ouyang et al., 2012), TLR4 KO mice were used as negative controls to determine whether TLR4 was the key target of miR-1906. miR-1906 delivery became futile in the absence of TLR4, suggesting that TLR4 is the major target of miR-1906 through which the microRNA exerts its protective effects.
The main drawback of this study was that we focused exclusively on the acute phase of experimental AIS and the effects of miR-1906 in the latter phase were undefined. However, short-term microRNA agomir administration is not expected to be preserved for long periods of time (Caballero-Garrido et al., 2015). Poststroke inflammation can be beneficial in the brain tissue recovery process (Chamorro et al., 2012) and poststroke systematic immunosuppression is also a major concern after AIS (Meisel and Meisel, 2011). Therefore, further studies regarding miR-1906 in the delayed phase are imperative for safety concerns due to the immunomodulation nature of this microRNA. In addition, although the current study readily detected extracellular miR-1906 using commercial kits (Qiagen), whether this microRNA can be transferred between different cells via exosomes or other EVs remains unknown. Because exosome- or extracellular vesicle-mediated microRNA delivery are promising drug delivery strategies (Ohno et al., 2016), further studies on extracellular microRNAs are necessary for a comprehensive understanding of intercellular communication and novel drug delivery strategies. Finally, this study was restricted to mice, which may not mimic human beings perfectly. In addition to the obvious distinction between species, the pathogenesis of AIS is remarkably different in human cases. Therefore, despite the current success of the microRNA-based treatment of hepatitis C virus (Janssen et al., 2013), clinical applications of microRNA-based strategies against AIS await further research.
Conclusions
In conclusion, our study identified miR-1906 as a novel regulator of TLR4 and proved that administration of the miR-1906 agomir to mice could ameliorate brain injury caused by experimental stroke. However, a comprehensive understanding of miR-1906 and the therapeutic implications require additional studies.
Footnotes
This work was supported by the Natural Science Foundation of China (Grants 81530038, 31200817, 81500939, and 81471182) and the Scientific Research Foundation of the Graduate School of Nanjing University (Grant 2015CL13). We thank Professor Ya Hua at the University of Michigan for assistance in revising the manuscript.
The authors declare no competing financial interests.
References
- Artinian NT, Magnan M, Sloan M, Lange MP (2002) Self-care behaviors among patients with heart failure. Heart Lung 31:161–172. 10.1067/mhl.2002.123672 [DOI] [PubMed] [Google Scholar]
- Arumugam TV, Okun E, Tang SC, Thundyil J, Taylor SM, Woodruff TM (2009) Toll-like receptors in ischemia-reperfusion injury. Shock 32:4–16. 10.1097/SHK.0b013e318193e333 [DOI] [PubMed] [Google Scholar]
- Boisserand LSB, Lemasson B, Hirschler L, Moisan A, Hubert V, Barbier EL, Rémy C, Detante O (2017) Multiparametric magnetic resonance imaging including oxygenation mapping of experimental ischaemic stroke. J Cereb Blood Flow Metab 37:2196–2207. 10.1177/0271678X16662044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brea D, Blanco M, Ramos-Cabrer P, Moldes O, Arias S, Pérez-Mato M, Leira R, Sobrino T, Castillo J (2011) Toll-like receptors 2 and 4 in ischemic stroke: outcome and therapeutic values. J Cereb Blood Flow Metab 31:1424–1431. 10.1038/jcbfm.2010.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero-Garrido E, Pena-Philippides JC, Lordkipanidze T, Bragin D, Yang Y, Erhardt EB, Roitbak T (2015) In vivo inhibition of miR-155 promotes recovery after experimental mouse stroke. J Neurosci 35:12446–12464. 10.1523/JNEUROSCI.1641-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caso JR, Pradillo JM, Hurtado O, Lorenzo P, Moro MA, Lizasoain I (2007) Toll-like receptor 4 is involved in brain damage and inflammation after experimental stroke. Circulation 115:1599–1608. 10.1161/CIRCULATIONAHA.106.603431 [DOI] [PubMed] [Google Scholar]
- Chamorro Á, Meisel A, Planas AM, Urra X, van de Beek D, Veltkamp R (2012) The immunology of acute stroke. Nat Rev Neurol 8:401–410. 10.1038/nrneurol.2012.98 [DOI] [PubMed] [Google Scholar]
- Chen GY, Nuñez G (2010) Sterile inflammation: sensing and reacting to damage. Nat Rev Immunol 10:826–837. 10.1038/nri2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li Y, Wang L, Zhang Z, Lu D, Lu M, Chopp M (2001) Therapeutic benefit of intravenous administration of bone marrow stromal cells after cerebral ischemia in rats. Stroke 32:1005–1011. 10.1161/01.STR.32.4.1005 [DOI] [PubMed] [Google Scholar]
- Dharap A, Bowen K, Place R, Li LC, Vemuganti R (2009) Transient focal ischemia induces extensive temporal changes in rat cerebral microRNAome. J Cereb Blood Flow Metab 29:675–687. 10.1038/jcbfm.2008.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichhorn SW, Guo H, McGeary SE, Rodriguez-Mias RA, Shin C, Baek D, Hsu SH, Ghoshal K, Villén J, Bartel DP (2014) mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol Cell 56:104–115. 10.1016/j.molcel.2014.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltzschig HK, Eckle T (2011) Ischemia and reperfusion–from mechanism to translation. Nat Med 17:1391–1401. 10.1038/nm.2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt B, Ransohoff RM (2005) The ins and outs of T-lymphocyte trafficking to the CNS: anatomical sites and molecular mechanisms. Trends Immunol 26:485–495. 10.1016/j.it.2005.07.004 [DOI] [PubMed] [Google Scholar]
- Frei K, Bodmer S, Schwerdel C, Fontana A (1986) Astrocyte-derived interleukin 3 as a growth factor for microglia cells and peritoneal macrophages. J Immunol 137:3521–3527. [PubMed] [Google Scholar]
- Fu Y, Liu Q, Anrather J, Shi FD (2015) Immune interventions in stroke. Nat Rev Neurol 11:524–535. 10.1038/nrneurol.2015.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganter S, Northoff H, Männel D, Gebicke-Härter PJ (1992) Growth control of cultured microglia. J Neurosci Res 33:218–230. 10.1002/jnr.490330205 [DOI] [PubMed] [Google Scholar]
- Ginsberg MD. (2016) Expanding the concept of neuroprotection for acute ischemic stroke: the pivotal roles of reperfusion and the collateral circulation. Prog Neurobiol 145–146:46–77. [DOI] [PubMed] [Google Scholar]
- Giulian D, Baker TJ (1986) Characterization of ameboid microglia isolated from developing mammalian brain. J Neurosci 6:2163–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP (2010) Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466:835–840. 10.1038/nature09267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimizadeh E, Kazemi Arababadi M, Shamsizadeh A, Roohbakhsh A, Allahtavakoli M (2016) The possible role of Toll-like receptor 4 in the pathology of stroke. Neuroimmunomodulation 23:131–136. 10.1159/000446481 [DOI] [PubMed] [Google Scholar]
- Hua F, Tang H, Wang J, Prunty MC, Hua X, Sayeed I, Stein DG (2015) TAK-242, an antagonist for Toll-like receptor 4, protects against acute cerebral ischemia/reperfusion injury in mice. J Cereb Blood Flow Metab 35:536–542. 10.1038/jcbfm.2014.240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyakkoku K, Hamanaka J, Tsuruma K, Shimazawa M, Tanaka H, Uematsu S, Akira S, Inagaki N, Nagai H, Hara H (2010) Toll-like receptor 4 (TLR4), but not TLR3 or TLR9, knock-out mice have neuroprotective effects against focal cerebral ischemia. Neuroscience 171:258–267. 10.1016/j.neuroscience.2010.08.054 [DOI] [PubMed] [Google Scholar]
- Iadecola C, Anrather J (2011) The immunology of stroke: from mechanisms to translation. Nat Med 17:796–808. 10.1038/nm.2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang E, Kim JH, Lee S, Kim JH, Seo JW, Jin M, Lee MG, Jang IS, Lee WH, Suk K (2013) Phenotypic polarization of activated astrocytes: the critical role of lipocalin-2 in the classical inflammatory activation of astrocytes. J Immunol 191:5204–5219. 10.4049/jimmunol.1301637 [DOI] [PubMed] [Google Scholar]
- Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, Persson R, King BD, Kauppinen S, Levin AA, Hodges MR (2013) Treatment of HCV infection by targeting microRNA. N Engl J Med 368:1685–1694. 10.1056/NEJMoa1209026 [DOI] [PubMed] [Google Scholar]
- Janssens S, Beyaert R (2003) Role of Toll-like receptors in pathogen recognition. Clin Microbiol Rev 16:637–646. 10.1128/CMR.16.4.637-646.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyaseelan K, Lim KY, Armugam A (2008) MicroRNA expression in the blood and brain of rats subjected to transient focal ischemia by middle cerebral artery occlusion. Stroke 39:959–966. 10.1161/STROKEAHA.107.500736 [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS (2004) Human microRNA targets. PLoS Biol 2:e363. 10.1371/journal.pbio.0020363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BW, Jeong KH, Kim JH, Jin M, Kim JH, Lee MG, Choi DK, Won SY, McLean C, Jeon MT, Lee HW, Kim SR, Suk K (2016) Pathogenic upregulation of glial lipocalin-2 in the parkinsonian dopaminergic system. J Neurosci 36:5608–5622. 10.1523/JNEUROSCI.4261-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono H, Rock KL (2008) How dying cells alert the immune system to danger. Nat Rev Immunol 8:279–289. 10.1038/nri2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15–20. 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- Li P, Shen M, Gao F, Wu J, Zhang J, Teng F, Zhang C (2017) An antagomir to microRNA-106b–5p ameliorates cerebral ischemia and reperfusion injury in rats via inhibiting apoptosis and oxidative stress. Mol Neurobiol 54:2901–2921. [DOI] [PubMed] [Google Scholar]
- Li Y, Mao L, Gao Y, Baral S, Zhou Y, Hu B (2015) MicroRNA-107 contributes to post-stroke angiogenesis by targeting Dicer-1. Sci Rep 5:13316. 10.1038/srep13316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Chang YM, Yu JM, Yen JH, Chang JG, Hu CJ (2005) Toll-like receptor 4 gene C119A but not Asp299Gly polymorphism is associated with ischemic stroke among ethnic Chinese in Taiwan. Atherosclerosis 180:305–309. 10.1016/j.atherosclerosis.2004.12.022 [DOI] [PubMed] [Google Scholar]
- Liu DZ, Tian Y, Ander BP, Xu H, Stamova BS, Zhan X, Turner RJ, Jickling G, Sharp FR (2010) Brain and blood microRNA expression profiling of ischemic stroke, intracerebral hemorrhage, and kainate seizures. J Cereb Blood Flow Metab 30:92–101. 10.1038/jcbfm.2009.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Zhao H, Wang R, Wang P, Tao Z, Gao L, Yan F, Liu X, Yu S, Ji X, Luo Y (2015) MicroRNA-424 protects against focal cerebral ischemia and reperfusion injury in mice by suppressing oxidative stress. Stroke 46:513–519. 10.1161/STROKEAHA.114.007482 [DOI] [PubMed] [Google Scholar]
- Liu XS, Chopp M, Pan WL, Wang XL, Fan BY, Zhang Y, Kassis H, Zhang RL, Zhang XM, Zhang ZG (2017) MicroRNA-146a promotes oligodendrogenesis in stroke. Mol Neurobiol 54:227–237. 10.1007/s12035-015-9655-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusardi TA, Murphy SJ, Phillips JI, Chen Y, Davis CM, Young JM, Thompson SJ, Saugstad JA (2014) MicroRNA responses to focal cerebral ischemia in male and female mouse brain. Front Mol Neurosci 7:11. 10.3389/fnmol.2014.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrez R, Ali C, Toutirais O, Le Mauff B, Defer G, Dirnagl U, Vivien D (2011) Stroke and the immune system: from pathophysiology to new therapeutic strategies. Lancet Neurol 10:471–480. 10.1016/S1474-4422(11)70066-7 [DOI] [PubMed] [Google Scholar]
- Maegdefessel L. (2014) The emerging role of microRNAs in cardiovascular disease. J Intern Med 276:633–644. 10.1111/joim.12298 [DOI] [PubMed] [Google Scholar]
- Meisel C, Meisel A (2011) Suppressing immunosuppression after stroke. N Engl J Med 365:2134–2136. 10.1056/NEJMcibr1112454 [DOI] [PubMed] [Google Scholar]
- Molteni M, Gemma S, Rossetti C (2016) The role of Toll-like receptor 4 in infectious and noninfectious inflammation. Mediators Inflamm 2016:6978936. 10.1155/2016/6978936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskowitz MA, Lo EH, Iadecola C (2010) The science of stroke: mechanisms in search of treatments. Neuron 67:181–198. 10.1016/j.neuron.2010.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Wang X, Chen S, Liu H, Wang Y, Xu X, Cheng J, Jia J, Zhen X (2015) MicroRNA let-7c–5p protects against cerebral ischemia injury via mechanisms involving the inhibition of microglia activation. Brain Behav Immun 49:75–85. 10.1016/j.bbi.2015.04.014 [DOI] [PubMed] [Google Scholar]
- Ohno S, Drummen GP, Kuroda M (2016) Focus on extracellular vesicles: development of extracellular vesicle-based therapeutic systems. Int J Mol Sci 17:172. 10.3390/ijms17020172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang YB. (2013) Inflammation and stroke. Neurosci Lett 548:1–3. 10.1016/j.neulet.2013.05.031 [DOI] [PubMed] [Google Scholar]
- Ouyang YB, Lu Y, Yue S, Xu LJ, Xiong XX, White RE, Sun X, Giffard RG (2012) miR-181 regulates GRP78 and influences outcome from cerebral ischemia in vitro and in vivo. Neurobiol Dis 45:555–563. 10.1016/j.nbd.2011.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang YB, Stary CM, Yang GY, Giffard R (2013) microRNAs: innovative targets for cerebral ischemia and stroke. Curr Drug Targets 14:90–101. 10.2174/138945013804806424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang YB, Stary CM, White RE, Giffard RG (2015) The use of microRNAs to modulate redox and immune response to stroke. Antioxid Redox Signal 22:187–202. 10.1089/ars.2013.5757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta S, Balasubramanian S, Rajasingh S, Patel U, Dhanasekaran A, Dawn B, Rajasingh J (2016) MicroRNA: A new therapeutic strategy for cardiovascular diseases. Trends Cardiovasc Med 26:407–419. 10.1016/j.tcm.2016.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Evans JE, Rock KL (2003) Molecular identification of a danger signal that alerts the immune system to dying cells. Nature 425:516–521. 10.1038/nature01991 [DOI] [PubMed] [Google Scholar]
- Sørensen SS, Nygaard AB, Nielsen MY, Jensen K, Christensen T (2014) miRNA expression profiles in cerebrospinal fluid and blood of patients with acute ischemic stroke. Transl Stroke Res 5:711–718. 10.1007/s12975-014-0364-8 [DOI] [PubMed] [Google Scholar]
- Tan KS, Armugam A, Sepramaniam S, Lim KY, Setyowati KD, Wang CW, Jeyaseelan K (2009) Expression profile of MicroRNAs in young stroke patients. PLoS One 4:e7689. 10.1371/journal.pone.0007689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang SC, Arumugam TV, Xu X, Cheng A, Mughal MR, Jo DG, Lathia JD, Siler DA, Chigurupati S, Ouyang X, Magnus T, Camandola S, Mattson MP (2007) Pivotal role for neuronal Toll-like receptors in ischemic brain injury and functional deficits. Proc Natl Acad Sci U S A 104:13798–13803. 10.1073/pnas.0702553104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voloboueva LA, Sun X, Xu L, Ouyang YB, Giffard RG (2017) Distinct effects of miR-210 reduction on neurogenesis: increased neuronal survival of inflammation but reduced proliferation associated with mitochondrial enhancement. J Neurosci 37:3072–3084. 10.1523/JNEUROSCI.1777-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein JR, Schulze J, Lee RV, Phillips H, Zierath D, Tanzi P, Shibata D, Cain KC, Becker KJ (2014) Functional polymorphisms in toll-like receptor 4 are associated with worse outcome in acute ischemic stroke patients. Neuroreport 25:580–584. 10.1097/WNR.0000000000000140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Jiang Y, Zhu J, Wen Z, Xu X, Xu X, Xie Y, Yang L, Xu L, Lan W, Xu G, Liu X (2014) Orosomucoid1: involved in vascular endothelial growth factor-induced blood-brain barrier leakage after ischemic stroke in mouse. Brain Res Bull 109:88–98. 10.1016/j.brainresbull.2014.09.007 [DOI] [PubMed] [Google Scholar]
- Writing Group Members, Mozaffarian D, Benjamin EJ, Go AS, et al.; American Heart Association Statistics Committee; Stroke Statistics Subcommittee (2016) Heart disease and stroke statistics-2016 update: a report from the American Heart Association. Circulation 133:e38–360. 10.1161/CIR.0000000000000350 [DOI] [PubMed] [Google Scholar]
- Xie Y, Liu W, Zhang X, Wang L, Xu L, Xiong Y, Yang L, Sang H, Ye R, Liu X (2015) Human albumin improves long-term behavioral sequelae after subarachnoid hemorrhage through neurovascular remodeling. Crit Care Med 43:e440–e449. 10.1097/CCM.0000000000001193 [DOI] [PubMed] [Google Scholar]
- Xu L, Wang L, Wen Z, Wu L, Jiang Y, Yang L, Xiao L, Xie Y, Ma M, Zhu W, Ye R, Liu X (2016) Caveolin-1 is a checkpoint regulator in hypoxia-induced astrocyte apoptosis via Ras/Raf/ERK pathway. Am J Physiol Cell Physiol 310:C903–C910. 10.1152/ajpcell.00309.2015 [DOI] [PubMed] [Google Scholar]
- Xu X, Jiang Y (2014) The Yin and Yang of innate immunity in stroke. Biomed Res Int 2014:807978. 10.1155/2014/807978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Jiang Y, Wen Z, Xu X, Xu X, Zhu J, Xie X, Xu L, Xie Y, Liu X, Xu G (2015) Overexpressed EGR1 may exaggerate ischemic injury after experimental stroke by decreasing BDNF expression. Neuroscience 290:509–517. 10.1016/j.neuroscience.2015.01.020 [DOI] [PubMed] [Google Scholar]