Abstract
GABA is the key inhibitory neurotransmitter in the cortex but regulation of its synthesis during forebrain development is poorly understood. In the telencephalon, members of the distal-less (Dlx) homeobox gene family are expressed in, and regulate the development of, the basal ganglia primodia from which many GABAergic neurons originate and migrate to other forebrain regions. The Dlx1/Dlx2 double knock-out mice die at birth with abnormal cortical development, including loss of tangential migration of GABAergic inhibitory interneurons to the neocortex (Anderson et al., 1997a). We have discovered that specific promoter regulatory elements of glutamic acid decarboxylase isoforms (Gad1 and Gad2), which regulate GABA synthesis from the excitatory neurotransmitter glutamate, are direct transcriptional targets of both DLX1 and DLX2 homeoproteins in vivo. Further gain- and loss-of-function studies in vitro and in vivo demonstrated that both DLX1 and DLX2 are necessary and sufficient for Gad gene expression. DLX1 and/or DLX2 activated the transcription of both Gad genes, and defects in Dlx function disrupted the differentiation of GABAergic interneurons with global reduction in GABA levels in the forebrains of the Dlx1/Dlx2 double knock-out mouse in vivo. Identification of Gad genes as direct Dlx transcriptional targets is significant; it extends our understanding of Dlx gene function in the developing forebrain beyond the regulation of tangential interneuron migration to the differentiation of GABAergic interneurons arising from the basal telencephalon, and may help to unravel the pathogenesis of several developmental brain disorders.
SIGNIFICANCE STATEMENT GABA is the major inhibitory neurotransmitter in the brain. We show that Dlx1/Dlx2 homeobox genes regulate GABA synthesis during forebrain development through direct activation of glutamic acid decarboxylase enzyme isoforms that convert glutamate to GABA. This discovery helps explain how Dlx mutations result in abnormal forebrain development, due to defective differentiation, in addition to the loss of tangential migration of GABAergic inhibitory interneurons to the neocortex. Reduced numbers or function of cortical GABAergic neurons may lead to hyperactivity states such as seizures (Cobos et al., 2005) or contribute to the pathogenesis of some autism spectrum disorders. GABAergic dysfunction in the basal ganglia could disrupt the learning and development of complex motor and cognitive behaviors (Rubenstein and Merzenich, 2003).
Keywords: forebrain development, GABA, glutamic acid decarboxylase, homeobox, interneuron
Introduction
Rapid progress is being made in describing the genetic programs that regulate regional specification, morphogenesis, cell-type specification, neuronal migration, and connectivity in the developing mammalian forebrain. Yet, few molecular pathways have been fully elucidated. When examining the relative contribution of interneuronal development to that of the telencephalon overall, distal-less (Dlx)1 and Dlx2 homeobox genes are essential for interneuronal differentiation and migration in the developing forebrain.
Most cortical GABA-producing (GABAergic) interneurons are born in the subpallial telencephalon and migrate tangentially to the neocortex in several migratory streams (Marin et al., 2000; Anderson et al., 2002; Wang et al., 2011). These neurons use GABA as the major inhibitory neurotransmitter which has several roles, including the regulation of proliferation, migration, differentiation, and synapse formation during embryonic development (Barker et al., 1998; Luján et al., 2005; Kwakowsky et al., 2007). Recently, investigators have focused on the lineage and temporal-spatial distribution of clonally-related GABAergic interneuronal progenitor populations derived from the basal forebrain that will eventually populate the striatum, hippocampus, and cortex (Harwell et al., 2015; Mayer et al., 2015, 2016; Sultan et al., 2016; Turrero García et al., 2016). However, there has been less emphasis placed on understanding the developmental regulation of GABA synthesis from the excitatory neurotransmitter glutamate.
GABA is mainly synthesized from glutamate by the enzyme glutamic acid decarboxylase (Gad), which has two molecular isoforms, Gad1 (GAD67) and Gad2 (GAD65; Erlander et al., 1991; Martin et al., 2000). Other GABA synthetic pathways require conversion of putrescine to GABA and pyrrolidinone; the latter compound can be converted to GABA via hydrolysis (Petroff, 2002). Gene expression analysis of the embryonic basal telencephalon reveals nearly overlapping patterns of expression of Dlx1 and Dlx2 with Gad1 and Gad2 in the mouse and zebrafish, primarily in regionally restricted domains including the ventricular zone (VZ) and subventricular zone (SVZ; Liu et al., 1997; Eisenstat et al., 1999; MacDonald et al., 2010). Similar expression data in the human fetal neocortex was recently described (Al-Jaberi et al., 2015). Previous studies showed that in mice lacking both Dlx1 and Dlx2 gene function, known as the Dlx1/2 double knock-out (DKO) mouse (which dies at birth), there is significant and almost complete reduction of tangential interneuronal migration from the subcortical telencephalon to the neocortex, resulting in few GABA-expressing cells residing in the mouse neocortex (Anderson et al., 1997a,b), as well as in zebrafish (MacDonald et al., 2013). Ectopic expression of the Dlx genes through gain-of-function assays in slice and primary cell cultures of the embryonic mouse forebrain can induce cortical cells to express the Gad genes in vitro (Stühmer et al., 2002a; Li et al., 2012). However, these prior studies have not explained the underlying molecular mechanism of Dlx genes with respect to differentiation of GABAergic interneurons during mouse forebrain development.
Using chromatin immunoprecipitation (ChIP) of embryonic forebrain, electrophoretic mobility shift assays, reporter gene experiments, gene expression analysis in wild-type and Dlx1/2 DKO mice, as well as confirmatory gain- and loss-of function assays, we have discovered that Dlx1 and Dlx2 regulate GABA expression through direct transcriptional activation of Gad1 and Gad2 isoforms by binding to specific canonical homeodomain DNA binding tetranucleotide TAAT/ATTA motifs located in their respective proximal gene promoters. By combining genetic and molecular approaches to identify these specific Dlx-dependent downstream target genes in vitro and in vivo, we further understand the role of Dlx genes as critical effectors of both GABAergic interneuron differentiation as well as tangential migration during vertebrate forebrain development.
Materials and Methods
Animal and tissue preparation
Dlx1/2 DKO mice (a kind gift from Dr. John Rubenstein, University of California, San Francisco) were maintained in a CD1 background. The null mutants were genotyped using published protocols (Qiu et al., 1995; Anderson et al., 1997b). For comparative studies, all mutants were paired with wild-type littermate controls; mutants and controls were of either sex. Embryonic age was determined by the day of appearance of the vaginal plug (E0.5) and dissected tissues [neocortex, ganglionic eminences (GE), hindbrain] were processed as previously described (de Melo et al., 2003). All animal protocols were conducted in accordance with guidelines set by the Canadian Council on Animal Care and were approved by the University of Manitoba and University of Alberta animal care committees.
Chromatin immunoprecipitation
ChIP assays were performed as described previously (Zhou et al., 2004; Le et al., 2007). The medial and lateral GEs (express Dlx1/Dlx2) and hindbrain tissue (used as a negative control since this tissue does not express any Dlx family members) were dissected from E13.5 CD1 mice. Specific rabbit polyclonal high affinity DLX1 and DLX2 antibodies were previously characterized (Anderson et al., 1997a; Eisenstat et al., 1999) and used to immunoprecipitate genomic DNA targets cross-linked to DLX1 or DLX2 homeoproteins. Genomic DNA from an E13.5 mouse embryo was used as a positive control. DNA derived from E13.5 hindbrain was used as a negative control.
Thermal cycling/PCR
Oligonucleotide primer pairs were designed to amplify distinct regions of the Gad promoters (Table 1; Fig. 1). The PCR was performed using specific primer pairs with the isolated E13.5 ChIP DNA, with genomic DNA as positive control, and with hindbrain DNA as negative control. PCR products were separated by gel electrophoresis, purified, and then ligated into the pCR2.1 TOPO vector using a TOPO TA cloning kit (Invitrogen). Recombinant plasmid DNA was extracted using a Plasmid MiniPrep Kit (Qiagen) and the M13 reverse universal primer was used for sequence confirmation.
Table 1.
Primers designed for downstream targeted Gad promoter regions of DLX proteins in ChIP assay
| DLX target | Target GenBank no. | Target region | Primers for specific regions |
|---|---|---|---|
| Gad1 | z49978 | GAD67i | Sense primer: 5′-CGCCCTCTGTGGGAAATTTT-3′ |
| Antisense primer: 5′-CCTGGAGAGGGGTAAAAGAA-3′ | |||
| GAD67ii | Sense primer: 5′-GATACGGGATGGAGGGCTAA-3′ | ||
| Antisense primer: 5′-GACTGCCTCTGGAGCTTTGT-3′ | |||
| Gad2 | AB032757 | GAD65i | Sense primer: 5′-TTTCTGGGTGGCTCACAGT-3′ |
| Antisense primer: 5′-TCCGGGTTGTTGATAACAAA-3′ | |||
| GAD65ii | Sense primer: 5′-AAAAGGGAAACAGAAAGGA-3′ | ||
| Antisense primer: 5′-AGAAAGGCTGCTGATTGAA-3′ |
Figure 1.
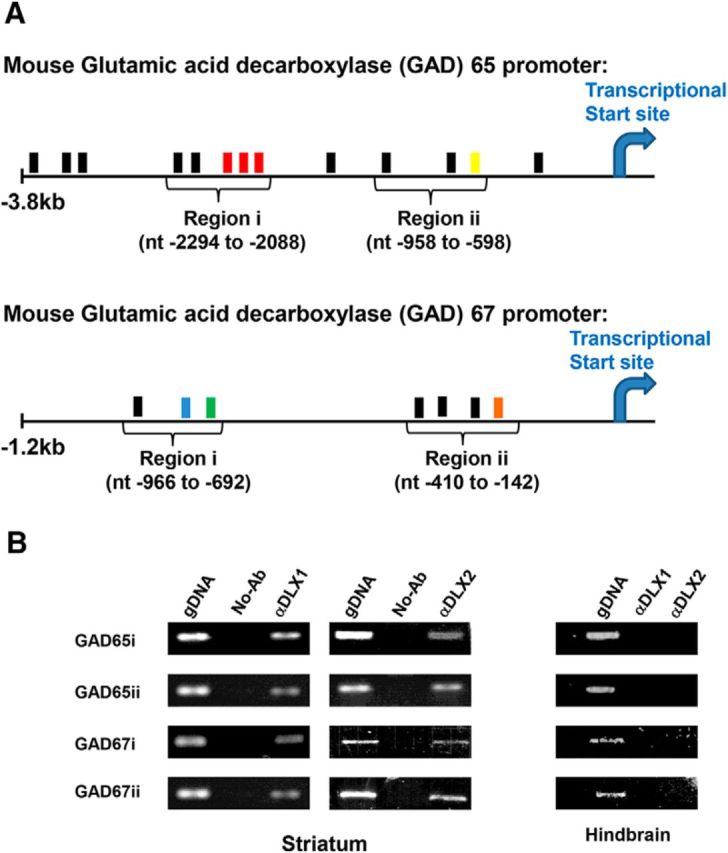
Candidate DLX target sequences within Gad1 (GAD67) and Gad2 (GAD65) promoter regions were identified through the presence of canonical homeodomain DNA binding motifs within the Gad promoters and confirmed by chromatin immunoprecipitation (ChIP). A, The specific regions within the regulatory elements of the mouse GAD65/Gad2 (GenBank AB032757; Gad2 NM_008078.2) and GAD67/Gad1 (GenBank Z49978; Gad1 NM_008077.5) promoters, designated GAD65i/ii and GAD67i/ii, contain putative homeodomain DNA-binding sites (TAAT/ATTA), shown in filled boxes. DLX proteins occupy colored boxes and do not occupy black boxes, respectively, based on subsequent experiments. Nucleotide positions are indicated with respect to transcriptional start sites. B, DLX1 and DLX2 occupy the Gad1 and Gad2 promoters in E13.5 GE tissues using ChIP. ChIP assays of GAD65/Gad2 and GAD67/Gad1 promoter regions showed that DLX1 and DLX2 occupy both regions (region i and region ii) of each Gad gene promoter using E13 GE tissues and specific DLX1 and DLX2 antibodies. Negative controls used no addition of antibodies and/or E13.5 hindbrain tissues. The positive controls used E13.5 genomic DNA. PCR bands were subcloned and sequenced for confirmation.
Electrophoretic mobility shift assay
Selected regions of Gad promoters as putative DLX targets were obtained from ChIP and screened by sequencing as potential targets (Table 2). The previously cloned DNA fragments were excised from the pCR2.1-TOPO vector (Invitrogen), then the 5′ overhangs were filled in with radiolabeled [α-P32] using the Klenow large fragment of DNA polymerase I (Promega). The binding reaction mixture with “cold competition” and “supershift” assays were performed as previously described (Zhou et al., 2004; Le et al., 2007). Within each target region, sequences of putative TAAT/ATTA homeodomain binding motifs (Zerucha et al., 2000) were used to generate individual synthetic oligonucleotides (16–32 bp). These double-stranded oligonucleotides were also subjected to electrophoretic mobility shift assay (EMSA) analysis. For the complete list of binding motifs studied, see Table 3.
Table 2.
Potential target regions of Gad promoters obtained from ChIP
Table 3.
Putative TAAT/ATTA homeodomain binding motifs within individual Gad promoter regions as potential DLX targets
| DLX target | Target region | Motif no. | Nucleotide position |
|---|---|---|---|
| Gad1 | GAD67i | 2nd | −744 to −740 |
| GAD67i | 3rd | −723 to −719 | |
| GAD67ii | 4th | −172 to −168 | |
| Gad2 | GAD65i | 3rd, 4th, 5th | −2131 to −2122 |
| GAD65ii | 3rd | −624 to −620 |
Reporter gene assays
Effector plasmids expressing the mouse Dlx1 or Dlx2 genes under control of a CMV promoter were constructed separately by inserting a PCR-amplified 790 bp Dlx1 cDNA and 1020 bp Dlx2 cDNA (gifts from Dr. John Rubenstein, University of California at San Francisco) into the pcDNA3 vector (Invitrogen). Reporter plasmids were constructed by inserting Gad1 and Gad2 targeted promoter regions, which were obtained from ChIP and PCR (Table 2), into the pGL3-Basic vector (Promega), in front of the luciferase gene. The correct orientation of all of the cloned fragments was verified by restriction digestion and sequencing. Site-directed mutagenesis of putative DLX-binding sites (TAAT/ATTA; Table 3) was performed using the Quick-Change Mutagenesis kit (Stratagene). Mutations were complete deletions of candidate TAAT/ATTA motifs and were verified by DNA sequencing.
Transient cotransfection experiments were performed in the HEK293 human embryonic kidney cell line (courtesy of Dr. S. Gibson, University of Manitoba) or the P19 murine embryonic carcinoma cell line (a gift from Dr. M. McBurney, University of Ottawa) or the C6 rat glioma cell line (American Type Culture Collection). Cells were transiently transfected using Lipofectamine 2000 reagent (Invitrogen) with luciferase reporter plasmids (1 μg), effector plasmids (1 μg), and pRSV-βgal (Promega; 0.4 μg) as an internal control for transfection efficiency (Le et al., 2007). Subsequently, cells were harvested 48 h later, and luciferase activities were measured with the Luciferase Reporter Assay System (Promega) and a standard luminometer, normalizing luciferase activity with β-gal activity.
Tissue preparation and immunofluorescence
For a complete list of secondary antibodies and tertiary molecules used, see Table 5.
Table 5.
Secondary antibodies used for immunofluorescence
| Secondary antibody | Dilution | Source |
|---|---|---|
| FITC-conjugated goat anti-rabbit | 1:100 | Sigma-Aldrich |
| Texas Red-conjugated donkey anti-rabbit | 1:200 | Jackson ImmunoResearch |
Tissues were prepared as previously described (Eisenstat et al., 1999; Zhou et al., 2004; Le et al., 2007). E13.5 tissues (whole embryos) or E16.5 and E18.5 tissues (whole brains), were fixed using 4% paraformaldehyde (PFA), and cryopreserved using sucrose gradients followed by embedding in OCT medium (Tissue-Tek). Tissue samples were then sectioned coronally at a thickness of 12–15 μm using a cryostat (ThermoShandon Cryotome). Single and double immunofluorescence (IF) experiments were performed as described previously (Eisenstat et al., 1999; de Melo et al., 2003; Zhou et al., 2004). For IF, we used the following primary antibodies (Table 4), and the following secondary antibodies (Table 5). Negative controls were performed by omitting the primary antibody.
Table 4.
Primary antibodies used for immunofluorescence
| Primary antibody | Dilution | Source | Reference |
|---|---|---|---|
| Rabbit anti-DLX1 | 1:200 | Dr. J. Rubenstein, University of California, San Francisco | Eisenstat et al., 1999 |
| Rabbit anti-DLX2 | 1:400 | Dr. J. Rubenstein, University of California, San Francisco | Eisenstat et al., 1999 |
| Rabbit anti-GABA | 1:1000 | Sigma-Aldrich | Le et al., 2007 |
| Rabbit anti-GAD65 | 1:500 | Dev. Studies Hybridoma Bank, University of Iowa | Chang and Gottlieb, 1988 |
| Rabbit anti-GAD67 | 1:1000 | Chemicon | Bickford et al., 2000 |
Tissue preparations and in situ hybridization
Nonradioactive in situ hybridization using digoxigenin (DIG)-UTP was performed as previously described (Eisenstat et al., 1999). For a complete list of antisense probes used, see Table 6. Sense probes were used as controls.
Table 6.
Antisense riboprobes used in in situ hybridization studies
| Riboprobe cDNA | Source | Reference |
|---|---|---|
| Gad65 | Dr. J. Rubenstein, University of California, San Francisco | Erlander et al., 1991 |
| Gad67 | Dr. J. Rubenstein, University of California, San Francisco | Erlander et al., 1991 |
Reverse-transcription PCR and semiquantitative real-time PCR
Total RNA of mouse E13.5 tissues (striatum and neocortex) were extracted using RNA-Bee reagent (Tel-Test). One microgram of total RNA was used as a template to synthesize the cDNA. Before RT, DNase I (Sigma-Aldrich) was added to the sample for 15 min at 37°C to digest the genomic DNA. To denature the DNase I and to linearize the RNA, the reaction was heated at 70°C for 10 min and then put on ice immediately. Reverse-transcription was performed for 5 min at 25°C, 30 min at 42°C, and later 5 min at 85°C with iScript Reverse Transcriptase (Bio-Rad). The cDNA was used as a template for both subsequent PCR and real-time PCR.
Real-time PCR was performed using the IQ SYBR Green Supermix (Bio-Rad) and 1 μl of the cDNA, 5 pmol of each primer (see list below) for each sample in a 25 μl reaction on an iCycler iQTM Multi-Color Real-Time PCR Detection System (Bio-Rad). PCR was performed at 95°C for 3 min, 95°C for 15 s, 57°C for 15 s, and 72°C for 30 s; for 40 cycles; then 95°C for 1 min, 57°C for 1 min, 57°C for 10 s; for 77 cycles in total with increasing set-point temperature after cycle 2 by 0.5°C per cycle. For real-time PCR, accumulation of the product was measured as an increase in SYBR green fluorescence and analyzed by the iCycler software. Standard curves relating initial template copy number to fluorescence and amplification cycle were generated using plasmid DNA as a template (standard), and were then used to calculate the mRNA copy number in each sample. The ratio of the intensities of the Gad to Gapdh (internal control) signals was considered to be a relative measure of the Gad mRNA level in each sample.
Gad2 Forward primer: 5′-CAAGATAAGCACTATGACCTGTCC-3′; Tm = 57°C.
Reverse primer: 5′-CTCTGCTAGCTCCAAACACTTATC-3′ .
Gad1 Forward primer: 5′-TGACACCCAGCACGTACTC-3′; Tm = 57°C.
Reverse primer: 5′-CCAGTTTTCTGGTGCATCC-3′ .
Gapdh Forward primer: 5′-TTGCCATCAATGACCCCTTCA-3′; Tm = 57°C.
Reverse primer: 5′-CGCCCCACTTGATTTTGGA-3′ .
High performance liquid chromatography
HPLC system.
The high-performance liquid chromatography (HPLC) system consisted of a Beckman model 116 solvent delivery system controlled by Beckman System Gold software. The mobile phase consisted of 0.1 m Na2HPO4, 0.13 mm Na2EDTA, 28% methanol, adjusted to pH 6.0 (Donzanti and Yamamoto, 1988). The pump flow rate was 1 ml/min. Standards and samples were separated using a 3 μm HR-80 (4.6 × 80 mm inner diameter; ESA) reverse phase column. Homoserine was used as an internal standard.
Tissue preparation.
E18.5 forebrains (excluding olfactory bulb) were individually weighed, frozen, then homogenized and extracted in the mobile phase to a final concentration of 25 mg/ml to adjust for the weight difference (Vriend et al., 1993). The supernatants were separated by centrifugation, filtered, diluted, and derivatized with o-phthaldialdehyde/2-mercaptoethanol solution (Sigma-Aldrich) before injection.
Standard solutions.
Amino acid stock solutions containing glutamate, glutamine, GABA, and homoserine were prepared by dissolving 100 μg of each powder (Sigma-Aldrich) in 1 ml of mobile phase. Working standard solutions were prepared by diluting the stock solutions to GABA (50, 100, 200, 400 ng/ml), glutamate and glutamine (250, 500, 1000, 2000 ng/ml), and homoserine (200, 400, 800, 1600 ng/ml). These standards were derivatized as described above.
Data analysis.
Amino acid content was determined by comparing the peak areas of sample chromatograms to a standard curve prepared from the peak areas of control chromatograms. To account for sample loss during the extraction procedure, an internal control homoserine was used in tissue content for balancing the experimental analysis. In addition, each sample was measured for total protein concentration (Bradford method; Bradford, 1976) and amino acid contents were recalculated accordingly. Amino acid tissue contents were expressed as nanogram/injection (average ± SD).
Generation of primary embryonic striatal or neocortical cell cultures
E16.5 and E18.5 CD1 embryos were used to derive primary cultures. On day 1, neocortex or striatum were dissected and collected in HBSS (Invitrogen). The tissue was incubated for 10 min at room temperature with 0.05 mg/ml trypsin (Invitrogen). The cells were then pelleted by centrifugation (1200 rpm, 5 min), resuspended, and gently triturated to single cell suspension in HBSS containing 100 μg/ml DNase I (Sigma-Aldrich). The cell suspension was transferred to a tube containing Neurobasal medium with B-27 (Invitrogen) and penicillin-streptomycin-fungizone (100 U/ml; Invitrogen). Cells were counted and 3 × 107 cells were plated per well using poly-d-lysine-coated 24-well plates (Fisher Scientific) and cultured at 37°C with 5% CO2 overnight.
DLX2 knockdown with transfection of primary cultures using small interfering RNA (siRNA)
The small interfering RNAs (siRNAs; Invitrogen) were targeted to the exon 2 coding sequence of Dlx2 (ensemble sequence ENSMUST00000024159). Two different siRNA duplexes and one nonsilencing control siRNA were used for the experiments as follows:
M-DLX2-R1-sense: 5′-(Fluo)GGAAGACCUUGAGCCUGAAdTdT;
M-DLX2-R1-antisense: 5′-UUCAGGCUCAAGGUCUUCCdTdT;
M-DLX2-R2-sense: 5′-(Fluo)UCUGGUUCCAGAACCGCCGdTdT;
M-DLX2-R2-antisense: 5′-CGGCGGUUCUGGAACCAGAdTdT;
CONTROL (Scramble)-sense: 5′-(Fluo)UUCUCCGAACGUGUCACGUdTdT;
CONTROL (Scramble)-antisense: 5′-ACGUGACACGUUCGGAGAAdTdT.
Transient transfection using Lipofectamine 2000 (Invitrogen) was performed on culture day 2 with 40 nm siRNAs in neurobasal medium with B-27 (Invitrogen). Forty-eight hours after transfection, cells were washed twice with cold PBS, and subjected to the immunofluorescence protocol as described previously. For primary culture, a minimum of three wild-type litters from timed-pregnant CD1 mice at E16.5 and E18.5 were used for each siRNA experiment. Neocortical or striatal tissues of each entire litter were combined to generate sufficient primary cells for tissue culture. After transfection, a minimum of three different areas of knock-down primary cultures were examined, and all cells in the selected areas were counted (n = 4).
Cell counting and statistical analysis
For DLX2 knock-down experiments using siRNA, labeled cells were counted as proportions of immunoreactive cells over total DAPI+ cells per section. All numerical results were expressed as means ± SEM in text and figures. Statistical analyses were performed using the unpaired Student's two-tailed t test for two datasets. For all statistical tests, p < 0.05 was used as the criterion for statistical significance.
Image acquisition and processing
Images were acquired using an Olympus IX81 inverted microscope with Fluoview FV500 confocal laser scanning system, or an Olympus BX51 fluorescent microscope with a SPOT 1.3.0 digital camera for photography. (Diagnostic Instruments). Images were processed using Adobe Photoshop 7.0 and/or CS5.
Results
DLX1 and DLX2 homeodomain proteins occupy the Gad1 and Gad2 promoters in vivo
We used an optimized ChIP method (Zhou et al., 2004) to identify the occupancy of DLX1 and/or DLX2 to the Gad1 (formerly Gad67) and Gad2 (formerly Gad65) promoters in embryonic GEs in vivo. We then used PCR to amplify two candidate homeodomain-binding regions in each of the Gad1 and Gad2 promoter loci. These regions were chosen based on the presence of several TAAT/ATTA canonical homeodomain tetranucleotide DNA binding motifs, and were designated GAD65 regions i and ii, and GAD67 regions i and ii (Tables 2, 3; Fig. 1A). Of interest, the ChIP assay showed that both DLX1 and DLX2 were localized to regions i and ii of the Gad65 and Gad67 promoters in situ (Fig. 1B). Mouse genomic DNA was used as a positive control for PCR. Omission of the specific antibody served as a negative control for immunoprecipitation, whereas embryonic hindbrain tissue that does not express Dlx genes served as a negative tissue control for the ChIP assay. The resulting ChIP-PCR products of the identified promoter regions were subcloned and sequenced to verify Gad1 and Gad2 promoters as DLX targets for subsequent biochemical analyses.
DLX1 and DLX2 specifically bind to the Gad promoters in vitro and in situ
To determine the specificity of binding of DLX1 and DLX2 to both regions i and ii of each of the Gad1(Gad67) and Gad2(Gad65) promoters in vitro, we used both recombinant DLX1 and DLX2 proteins or E13.5 GE nucleoprotein extracts coupled with radioactive α32P-dATP-labeled Gad65 or Gad67 promoter oligonucleotides for all regions isolated from the ChIP assay in situ.
For Gad65 (Gad2), EMSAs demonstrated specific binding of both recombinant DLX1 and DLX2 to the Gad65 promoter for both regions (region i: Fig. 2A, lanes 2, 6; region ii: B, lanes 2, 6) that can be competitively inhibited by unlabeled Gad65 probes (region i: Fig. 2A, lanes 3, 7; region ii: B, lanes 3, 7). Moreover, the addition of specific anti-DLX1 or anti-DLX2 antibodies to the protein–DNA complexes resulted in significant supershifts (Fig. 2A,B, lanes 4, 8), whereas a nonspecific polyclonal antibody failed to produce such a supershift (Fig. 2A,B, lanes 5, 9). Within the Gad65 promoter regions i and ii, we identified the homeodomain binding motifs that are necessary for the binding of DLX proteins to these regions (Table 3; Fig. 1A, bold and italicized TAAT/ATTA motifs). Using both DLX1 and DLX2 recombinant proteins (data not shown) and E13.5 GE nucleoprotein extracts, we found that DLX proteins were able to specifically bind to oligonucleotides containing 3 consecutive TAAT/ATTA motifs of the Gad65 region i (Table 3; Fig. 2C, lane 2), and to oligonucleotides containing the third TAAT/ATTA motif of Gad65 region ii (Table 3; Fig. 2D, lane 2). Unlabeled probes competitively inhibited the binding (Fig. 2C,D, lane 3), and specific DLX antibodies resulted in supershifts (Fig. 2C,D, lanes 4, 5), whereas a nonspecific polyclonal antibody failed to produce such a supershift (Fig. 2C,D, lane 6). Other candidate homeodomain binding motifs in the Gad65 promoter were not bound by DLX1 or DLX2 protein (data not shown).
Figure 2.
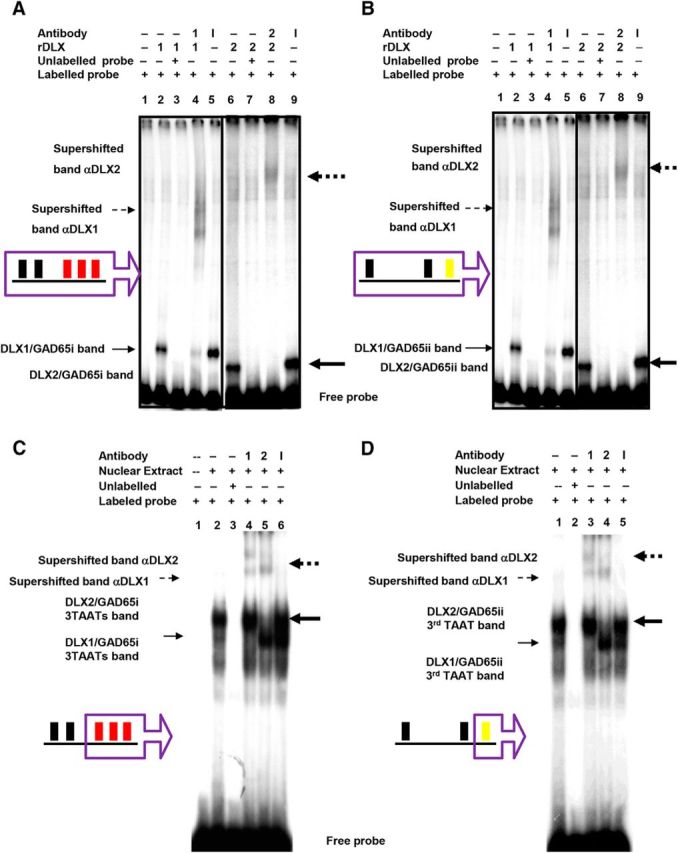
DLX1 and DLX2 proteins specifically bind to Gad65/Gad2 promoter regions i and ii in situ. EMSA showed recombinant DLX1 or DLX2 binding to α-P32-labeled (A) Gad65 promoter region i, and (B) Gad65 promoter region ii oligonucleotides containing homeodomain binding sites (Fig. 1A, regions in purple boxes). GE nuclear-extracted proteins also bound to α-P32-labeled oligonucleotides containing specific TAAT/ATTA binding motif for (C) Gad65 region i 3 TAAT motifs, and for (D) Gad65 region ii third TAAT motif (Fig. 1A, motifs in purple boxes). A, B, Radioactive oligonucleotide probes were incubated alone (lane 1), with DLX1 recombinant proteins (lanes 2–5), with DLX2 recombinant proteins (lanes 6–9), with unlabeled competitive probes (lane 3, 7), with specific DLX1 or 2 antibodies (lanes 4, 8), and with nonspecific antibodies (lanes 5, 9). C, D, Radioactive oligonucleotide probes were incubated alone (lane 1), with GE nuclear extract (lanes 2–6), with unlabeled competitive probes (lane 3), with specific DLX1 antibody (lane 4), with specific DLX2 antibody (lane 5), with nonspecific antibodies (lane 6). A–D, Binding of DLX proteins to a specific oligonucleotide sequence results in a gel shifted band, indicated by solid arrows. Binding of DLX protein to a specific oligonucleotide and to a specific DLX antibody resulted in a gel supershifted band, indicated by broken arrows. (DLX1: unbold arrow, DLX2: bold arrow). r, Recombinant; 1, DLX1 protein or anti-DLX1 antibody; 2, DLX2 protein or anti-DLX2 antibody; I, irrelevant/nonspecific antibody.
For Gad67 (Gad1), EMSA experiments also demonstrated that DLX1 and DLX2 specifically bind to Gad67 promoter regions i and ii (Fig. 3A,B, lanes 2, 6) and that this binding can be competitively inhibited by unlabeled Gad67 probes (Fig. 3A,B, lanes 3, 7). Moreover, the addition of specific anti-DLX1 or anti-DLX2 antibodies to the protein–DNA complexes resulted in significant band mobility shifts (Fig. 3A,B, lanes 4, 8), whereas a nonspecific polyclonal antibody failed to produce such a supershift (Fig. 3A,B, lanes 5,9). Within the Gad67 regions i and ii, there are homeodomain binding motifs necessary for the binding of DLX proteins to the Gad67 promoter (Table 3; Fig. 1A, bold and italicized TAAT/ATTA motifs). Recombinant DLX1 and DLX2 proteins (data not shown) and E13.5 GE nuclear extracts enriched for these homeodomain proteins both bind to oligonucleotides containing the second and third TAAT/ATTA motifs of region I (Fig. 3C and data not shown), and to oligonucleotides containing the fourth TAAT/ATTA motif of region ii (Fig. 3D). Unlabeled probes competitively inhibited the binding (Fig. 3C,D, lane 3), and specific DLX antibodies resulted in supershifts (Fig. 3C,D, lanes 4, 5), whereas a nonspecific polyclonal antibody failed to produce such a supershift (Fig. 3C,D, lane 6). Other homeodomain binding motifs in the Gad67 promoter were not significantly bound to DLX1 and DLX2 proteins (data not shown).
Figure 3.
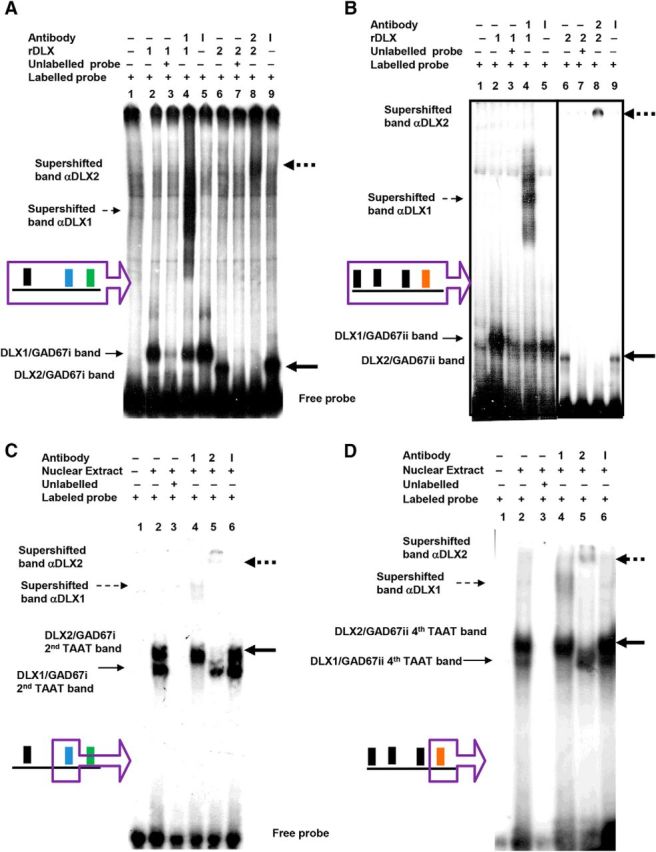
DLX1 and DLX2 proteins specifically bind to Gad67/Gad1 promoter regions i and ii in situ. EMSA showed recombinant DLX1 or DLX2 binding to α-P32-labeled (A) Gad67 promoter region i, and (B) Gad67 promoter region ii oligonucleotides containing candidate homeodomain binding sites (Fig. 1A, regions in purple boxes). GE nuclear-extracted proteins also bound to α-P32-labeled oligonucleotides containing specific TAAT/ATTA binding motif for (C) the Gad67 region i second TAAT motif, and for (D) the Gad67 region ii fourth TAAT motif (Fig. 1A, motifs in purple boxes). A, B, Radioactive oligonucleotide probes were incubated alone (lane 1), with recombinant DLX1 proteins (lanes 2–5), with recombinant DLX2 proteins (lanes 6–9), with unlabeled competitive probes (lanes 3, 7), with specific DLX1 or 2 antibodies (lanes 4, 8), and with nonspecific antibodies (lanes 5, 9). C, D, radioactive oligonucleotide probes were incubated alone (lane 1), with GE nuclear extract (lanes 2–6), with unlabeled competitive probes (lane 3), with specific DLX1 antibody (lane 4), with specific DLX2 antibody (lane 5), with nonspecific antibodies (lane 6). A–D, Binding of DLX proteins to a specific oligonucleotide sequence results in a gel shifted band, indicated by solid arrows. Binding of DLX protein to a specific oligonucleotide and to a specific DLX antibody results in a gel supershifted band, indicated by broken arrows. (DLX1: unbold arrow, DLX2: bold arrow). r, Recombinant; 1, DLX1 protein or anti-DLX1 antibody; 2, DLX2 protein or anti-DLX2 antibody; I, irrelevant/nonspecific antibody.
DLX1 and DLX2 activate transcription of Gad promoters in vitro
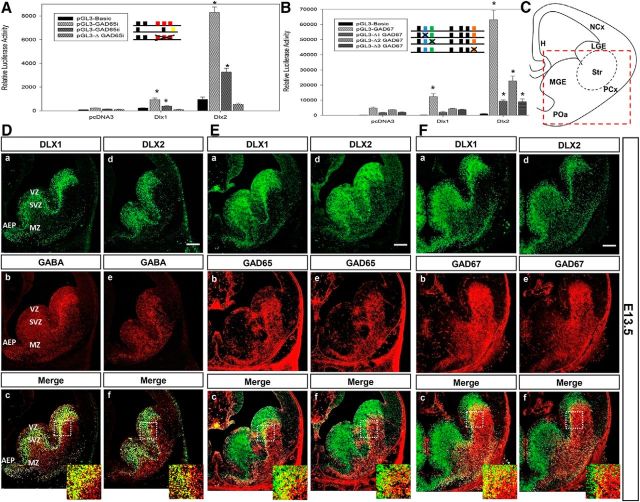
The functional significance of DLX1 and DLX2 binding to the Gad promoters was assessed using luciferase reporter gene assays in vitro. We cotransfected C6 rat glioma cells (or P19 embryonal carcinoma cells, data not shown) with an expression vector encoding DLX1 or DLX2 and a luciferase vector into which the Gad65 promoter regions i or ii (Table 2) was subcloned. For maximal activity of the Gad67 promoter, we cloned the full-length ∼1.3 kb promoter using a standard PCR protocol with flanking primers designed according to the mouse Gad67 promoter sequence and containing both regions i and ii (Table 2). Cotransfection with either wild-type Dlx1 and Dlx2 expression constructs resulted in significant increase of luciferase activity for the Gad65 reporter constructs (region i: ∼7-fold for DLX1 and ∼21-fold for DLX2; region ii: ∼2-fold for DLX1 and ∼11-fold for DLX2; Fig. 4A; p < 0.001). Wild-type DLX1 and DLX 2 proteins also activated Gad67 reporter gene expression (∼3-fold for DLX1 and ∼13-fold for DLX2; Fig. 4B; p < 0.001) in the same manner as was observed for Gad65. The increase in reporter gene activity driven by coexpression of either DLX1 or DLX2 proteins support that both DLX homeodomain proteins act as transcriptional activators of Gad1 and Gad2 promoter expression in vitro.
Figure 4.
A, B, Dlx1 and Dlx2 activate transcription of Gad1 (Gad67) and Gad2 (Gad65) reporter constructs in vitro. Transient transfection assays in C6 glioma cells with (A) Gad65 promoter regions i or ii, and (B) Gad67 1.3 kb promoter constructs (contains regions i and ii; Fig. 1A), containing homeodomain binding sites cloned into a pGL3-luciferase reporter construct, were performed in the absence or presence of DLX1 or DLX2 coexpression. DLX1 and DLX2 activate transcription of the reporter genes using the Gad65 and Gad67 promoters, with DLX2 as a more robust activator. Mutations of specific TAAT/ATTA binding motifs within (A) Gad65 or (B) Gad67 promoter sequences lead to a significant reduction of transcriptional activation of these reporter gene constructs. All luciferase activities were relative and normalized to the activity level of the internal control β-gal. Average ± SEM of at least triplicate experiments. *p < 0.05. ΔGAD65i, Mutation of 3 TAATs of Gad65 region I; Δ1GAD67, mutation of the second TAAT of Gad67 region I; Δ2GAD67, mutation of the third TAAT of Gad67 region I; Δ3GAD67, mutation of the fourth TAAT of Gad67 region ii. C–F, Coexpression of DLX homeodomain proteins and the GABA neurotransmitter in wild-type E13.5 basal telencephalon. C, Schematic diagram of coronal section of the E13.5 forebrain, showing basal telencephalon in red dashed box. Sections were double-labeled with specific antibodies against DLX1 (Da, Ea, Fa), DLX2 (Dd, Ed, Fd), GABA or GAD65 or GAD67 (Db, Eb, Fb, De, Ee, Fe) of E13.5 ganglionic eminences. Da, Ea, Fa, Dd, Ed, Fd, DLX1- or DLX2-positive cells (green) in the VZ and SVZ of the LGE, MGE, and AEP. Db, Eb, Fb, De, Ed, Fe, GABA or GAD65 or GAD67-labeled cells (red) in the same tissue sections throughout the basal telencephalon, predominantly in SVZ and MZ of the LGE and AEP. Bottom, The overlay of the two images with GABA/GAD65/GAD67 coexpressed with DLX proteins in most SVZ interneurons (yellow). Scale bar, 200 μm. Insets, Representative of a ∼10× enlargement of the dashed inset box to better demonstrate colabeled cells. H, Hippocampus; LGE, lateral ganglionic eminence; NCx, neocortex; PCx, paleocortex; POa, anterior preoptic area; Str, striatum.
To establish the functional significance of specific TAAT/ATTA DNA binding motifs for Dlx regulation of Gad isoform transcription, we performed site-directed mutagenesis of the Gad65 and Gad67 promoters to eliminate these binding motifs, then cotransfected the mutant constructs with expression vector encoding wild-type DLX1 or DLX2 into C6 glioma cells (or P19 embryonal carcinoma cells, data not shown). A Gad65 promoter region i with three TAAT/ATTA motifs mutated was not activated by DLX1 or DLX2 coexpression compared with that of controls (Fig. 4A), indicating that these three motifs localized within Gad65 region i might constitute an important binding site for DLX1 and/or DLX2. Our rationale for the simultaneous mutation of these three motifs within the Gad65 promoter was because these candidate binding sites are in very close proximity, rendering individual mutations very challenging to generate and corroborative DNA-binding assays uninterpretable. Cotransfection of the Gad67 promoter with the second or third TAAT/ATTA motif of region i mutated, or with the fourth motif of region ii mutated, resulted in a significant reduction of transcriptional activation mediated by DLX1, indicating that for DLX1, these binding sites are necessary for transcriptional activation of the Gad67 promoter (Fig. 4B). However, DLX2 coexpression still activated these mutated reporter constructs, although with significant decrease in luciferase activity compared with wild-type controls. Of note, mutation of the third TAAT/ATTA motif of region i resulted in the least reduction of luciferase activity compared with either mutation of the second motif of region i or the fourth motif of region ii of the Gad67 promoter (Fig. 4B).
DLX1 or DLX2 are coexpressed with GAD65, GAD67, or GABA in the developing forebrain
Expression of DLX1 and DLX2 was closely examined and compared with the expression of GABA, GAD65, and GAD67 in the subpallial telencephalon (Fig. 4C). DLX1 and DLX2 expression becomes well established at E13.5 in the medial ganglionic eminence (MGE) and lateral ganglionic eminence (LGE), as well as the anterior entopeduncular (AEP) area (Fig. 4Da,Dd,Ea,Ed,Fa,Fd). These DLX homeoproteins are expressed predominantly in the VZ and SVZ of these regions with lower levels of expression in the mantle zone (MZ) of the basal forebrain. GABA expression is mainly in the SVZ and MZ of the LGE and AEP (Fig. 4Db,De). GABA expression partially overlaps with the expression of its synthetic enzymes: GAD65 (Fig. 4 compare Db,De with Eb,Ee) and GAD67 (Fig. 4Fb,Fe). Incomplete overlap of GABA with GAD65/67 may be due to a temporal lag between the onset of expression of Gad65/Gad67 RNA and protein and the onset of expression of GABA. Furthermore, concurrent radial and tangential migration of GABAergic interneurons in the basal telencephalon could contribute to only partial spatial overlap between GABA, GAD65, and GAD67. More importantly, GABA, GAD65, or GAD67 expression overlaps with DLX1 or DLX2 expression in the SVZ, especially the LGE and AEP but to a much lesser extent in the VZ or MZ of the basal telencephalon (Fig. 4Dc,Df,Fc,Ff, and insets). Coexpression studies of DLX1 or DLX2 with GABA, GAD65, or GAD67 at later embryonic stages, E14.5 to E16.5, consistently demonstrated overlapping patterns of expression of DLX1 or DLX2 with GABA or GAD65/67, especially in the SVZ of the basal telencephalon (data not shown). The absence of complete overlap of DLX1/DLX2 with GAD65/67 and/or GABA support the role for other transcription factors in the regulation of GABAergic interneuron differentiation, especially in the mantle zone (both GE) and outer SVZ (LGE > MGE, GABA > GAD65/GAD67).
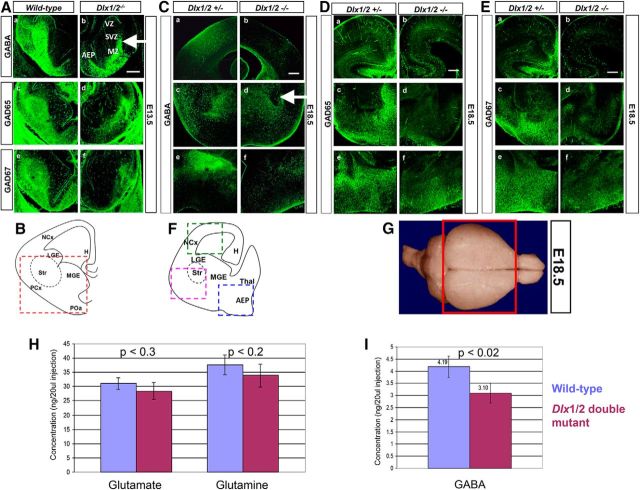
GAD65 and GAD67 expression is reduced concomitantly with decreased GABA neurotransmitter expression in the basal forebrain of Dlx1/2 double-null mice
In the wild-type E13.5 basal telencephalon (Fig. 5B), GAD65 and GAD67 are highly expressed in the SVZ and MZ of the LGE and AEP (Fig. 5Ac,Ae), overlapping with expression of GABA (Fig. 5Aa). In the absence of Dlx1 and Dlx2 function, GAD65 and GAD67 expression levels are abolished in the SVZ of the LGE and embryonic striatum, with significant reduction in the MZ (Fig. 5Ad,Af), as well as GABA reduction (Fig. 5Ab). Of significance, GAD65 and GAD67 expression are absent as well in the AEP of the Dlx1/2 DKO mutant (Fig. 5A). Concomitantly, GABA expression is also reduced at the pallial/subpallial boundary in the double-mutant compared with wild-type (Fig. 5Ab, white arrow). As development proceeds, expression of GAD65, GAD67, or GABA is strongly maintained throughout the basal telencephalon in the wild-type (Fig. 5F), with more established expression in the neocortex, striatum, and AEP at E16.5 (data not shown) and at E18.5 (Fig. 5Ca,Cc,Ce,Ea,Ec,Ee). Expression of GAD65, GAD67 and GABA are significantly and uniformly reduced throughout all regions of basal telencephalon and neocortex at E16.5 (data not shown) and at E18.5 (Fig. 5Cb,Cd,Cf,Eb,Ed,Ef). Furthermore, remaining GABA-labeled cells appear to be accumulating ectopically in the embryonic striatum (Fig. 5Ab,Cd, white arrows). These observed patterns of expression are consistent with the role of DLX proteins as important transcriptional regulators of Gad1 and Gad2 gene and GABA expression in the developing mouse forebrain, especially at later stages of development of the basal telencephalon.
Figure 5.
A–F, GABA expression in the developing forebrain of Dlx1/2 wild-type compared with Dlx1/2 DKO mice. A, In the E13.5 wild-type, GABA (Aa), GAD65 (Ac), and GAD67 (Ae) expression is predominantly localized to the SVZ and MZ of the AEP and LGE compared with that in the absence of Dlx1 and Dlx2 function (Ab, Ad, Af). B, Schematic diagram of coronal section of the E13.5 forebrain, showing basal telencephalon in red dashed box. In the E18.5 wild-type, (C) GABA, (D) GAD65, (E) GAD67 expression is shown in neocortex (Ba), striatum (Bc), and AEP (Be) compared with the Dlx1/2 double mutant (Bb, Bd, Bf). F, Schematic diagram of coronal section of the E18.5 forebrain, showing neocortex (green dashed box), striatum (pink dshed box), and AEP (blue dashed box). Scale bars, 200 μm. H, Hippocampus; LGE, lateral ganglionic eminence; NCx, neocortex; PCx, paleocortex; POa, anterior preoptic area; Str, striatum. G–I, GABA levels are reduced in the Dlx1/2-null forebrains using HPLC. G, Forebrains of wild-type and Dlx1/2 double-mutant were dissected as depicted in the red box of the diagram. H, In the E18.5 forebrain, glutamate and glutamine (precursors of GABA) levels do not change when comparing wild-type and Dlx1/2 knock-out forebrain (excluding the olfactory bulbs). I, GABA neurotransmitter level decreases by ∼26% in the Dlx1/2 knock-out compared with wild-type forebrain. Homoserine was used as internal control. Sample concentrations were measured from standard curves and expressed as nanogram per 20 μl injection into the HPLC apparatus. Average ± SEM with associated p values.
Although reduction of GABA expression in the neocortex is evident along with decreased GAD65 and GAD67 levels, it is essential to functionally quantify and compare GABA concentrations, i.e., the neurotransmitter level, in the forebrains of Dlx1/2 DKO mice compared with wild-type. There is the confounding accumulation of GABA-expressing interneurons as ectopias in the basal telencephalon from E13.5 to birth of the DKO mice (Fig. 5A,C). Abnormal accumulation of GABAergic interneurons in the embryonic basal ganglia, concurrent with the defect in tangential migration of these interneurons to the neocortex, may have partially masked the underlying reduction of GABA levels in the basal telencephalon and underestimated the important role of Dlx1 and Dlx2 genes in the terminal differentiation of these interneurons. Hence, to determine the direct role of Dlx1/2 genes in the production of GABA in the entire forebrain (excluding the olfactory bulbs), we used HPLC to measure GABA levels in whole-forebrain extracts obtained from wild-type and Dlx1/2 DKO mice (Fig. 5G). At E13.5 and E16.5, GABA levels are comparable between wild-type and knock-out littermates (data not shown). However, at E18.5 we detected a ∼26% reduction of GABA level in the entire forebrain of the DKO compared with the wild-type mice (Fig. 5I; p < 0.02, n = 6). Concentrations of the GABA precursors glutamate and glutamine were unchanged between wild-type and DKO mice (Fig. 5H; p < 0.3 and p < 0.2, respectively, n = 6). The amino acid concentrations in extracts of whole forebrain were adjusted to the frozen weight, and to the total protein concentration of individual forebrain samples, and to the concentration of an internal control, homoserine.
Gad65 and Gad67 transcription is reduced in the basal telencephalon of Dlx1/2-null mice
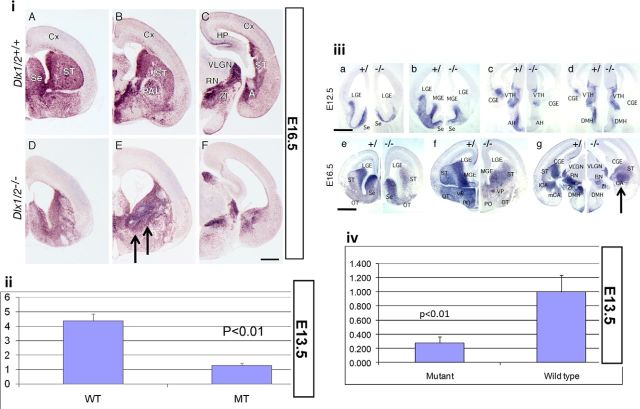
In wild-type mice, Gad65 and Gad67 RNAs are highly expressed throughout the embryonic forebrain from rostral to caudal regions at different embryonic ages, as early as E12.5 (Fig. 6i,iii), based on in situ hybridization experiments. Patterns of RNA expression were consistent with the corresponding protein expression domains in the SVZ and MZ of the basal telencephalon, including the AEP for Gad65 (Fig. 6iB), and for Gad67 (Fig. 6iiib,iiie), with overlapping pattern with DLX1 and DLX2 expression in the SVZ (Fig. 4E,F). In the absence of Dlx1 and Dlx2 function, Gad65 (Fig. 6iD–iF) and Gad67 (Fig. 6iiia–iiig) RNA expression levels were significantly lower in the basal telencephalon and diencephalon. In addition, interneurons that still expressed Gad mRNAs accumulated as ectopias in the embryonic striatum (Fig. 6i,iii, black arrows).
Figure 6.
Expression and level of Gad mRNA isoforms in the rostral and caudal forebrain in the wild-type and Dlx1/2 DKO demonstrated by digoxigenin in situ hybridization and quantitative real-time PCR. i, Gad65 isoform mRNA is expressed throughout the basal forebrain in a diffuse pattern in the wild-type (iA–iC, from rostral to caudal) compared with the Dlx1/2 knock-out forebrain (iD–iF, from rostral to caudal). Black arrows depicted ectopic accumulation of Gad65 mRNA in the basal ganglia. ii, Gad65 mRNA level was examined by real-time PCR in the basal ganglia and compared between wild-type (WT) and double-mutant (MT) littermates at E13.5. iii, Gad67 isoform mRNA is also expressed throughout the basal forebrain in a diffuse pattern in the wild-type (iiia–iiig, left, from rostral to caudal) and compared with Dlx1/2 DKO forebrain (iiia–iiig, right, from rostral to caudal). Black arrow depicted accumulation of Gad67 mRNA in the embryonic striatum at E16.5. iv, Gad67 mRNA levels were examined by quantitative real-time PCR in the basal ganglia and compared between wild-type and double mutant littermates at E13.5. The real-time PCR results show relative fold differences between wild-type and mutant littermates, and were normalized using the house keeping gene GAPDH as an internal control. Average ± SEM with associated p values. CGE, Caudal ganglionic eminence; LGE, lateral ganglionic eminence; MGE, medial ganglionic eminence; ST, striatum; VTH, ventral thalamus; AH, anterior hypothalamic nucleus; DMH, dorsal medial nucleus; OT, olfactory tract; SE, septum; VP, ventral pallidum; PO, preoptic area; VLGN, ventral lateral geniculate nucleus; ZI, zona incerta nucleus; RN, red nucleus. Scale bars: iA–iF, iiia–iiid, 200 μm; iiie–iiig, 400 μm .
Real-time PCR was performed on three different E13.5 litters, each of which contained at least six pairs of wild-type and mutant embryos from which embryonic ganglionic eminence tissues were dissected and total RNA was subsequently extracted. There was ∼75% reduction of Gad65 mRNA (Fig. 6ii) and a similar ∼75% reduction of Gad67 mRNA (Fig. 6iv) expression in the developing E13.5 mutant GE. The reduction of Gad65 and Gad67 mRNA expression in the Dlx1/Dlx2 DKO mice was statistically significant (p < 0.01, n = 6). These results indicate that Dlx1 and Dlx2 are necessary for the expression of Gad65 and Gad67 mRNA, to maintain GABA-producing interneurons throughout the basal telencephalon as early as E12.5.
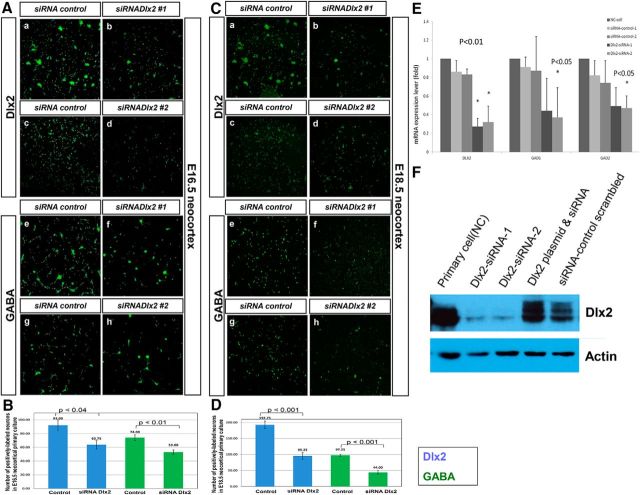
Knockdown of DLX2 expression in primary embryonic striatal and neocortical cultures results in decreased GABA expression
Our previous studies have shown that transcriptional regulation by DLX2 is more potent than DLX1, in regards to the induction of GABA expression in vitro (Fig. 4A,B), and the lack of both Dlx1 and Dlx2 function reduces GABA expression levels in the embryonic forebrain (Figs. 5H,I, 6). These results led us to investigate the potential effects of inhibition of Dlx2 expression on GABA expression in primary embryonic forebrain cultures. Reverse-transcription PCR (RT-PCR) and Western blot analysis of primary embryonic forebrain cells transfected with interfering RNA constructs demonstrated an efficient reduction of DLX2 mRNA and protein expression, respectively, indicating that our siRNA constructs were working properly (Fig. 7E,F). Moreover, knockdown with the pooled siRNAs to Dlx2 was rescued by cotransfection with a plasmid expressing Dlx2 (Fig. 7F). Furthermore, qRT-PCR assays demonstrated significant reductions in Gad1 and Gad2 expression, concomitant with reductions in Dlx2 expression using either siRNA construct but not the scrambled control (Fig. 7E). Primary cultures derived from embryonic neocortex (Fig. 7A,C) and GE (embryonic striatum; data not shown) were transfected with either scrambled control siRNA or two different siRNAs targeting DLX2 expression. When DLX2 expression was knocked down in embryonic neocortical cells (Fig. 7Ab,Ad,B for E16.5; Cb,Cd,D for E18.5), GABA expression was correspondingly reduced (Fig. 7Af,Ah,B,Cf,Ch,D), respectively. The reduction of GABA expression by the siRNA targeting Dlx2 was consistent in primary cultures derived from these neuroanatomical regions. The results were quantified as the number of DLX2- (blue column) or GABA- (green column) positive cells and compared with controls. In E16.5 neocortical cultures (Fig. 7B), there was an ∼1.4-fold reduction (p < 0.04) of DLX2-positive cells with a corresponding 1.4-fold reduction (p < 0.01) of GABA-positive cells. In the E18.5 neocortical cultures (Fig. 7D), there was an ∼2.0-fold reduction (p < 0.001) of DLX2-positive cells and 2.2-fold reduction (p < 0.001) of GABA-positive cells. This siRNA knockdown study supports a direct transcriptional role of the DLX2 homeodomain protein in the regulation of the key enzymes required for GABA synthesis, and subsequently, GABAergic interneuron differentiation. More importantly, this study has demonstrated that Dlx genes directly regulate the transcription of Gad genes, and are necessary to induce the terminal differentiation of GABAergic neurons in the developing forebrain.
Figure 7.
A–D, Knockdown of Dlx2 expression by siRNA in E16.5 and E18.5 embryonic neocortical primary cultures. Primary dissociated forebrain cultures of neocortex (A, C) at E16.5 (A) and E18.5 (C) were transfected with a duplex control scrambled siRNA or two different duplex siRNAs targeting Dlx2 coding sequences. Transfection of duplex siRNA targeting Dlx2 reduced DLX2 expression (green; Ab, Ad, Cb, Cd) and concomitantly decreased GABA expression (green; Af, Ah, Cf, Ch) in embryonic neocortical primary cultures compared with control scrambled siRNA-transfected cells (Aa, Ac, Ae, Ag, Ca, Cc, Ce, Cg). Scale bars, 200 μm. E, Using quantitative RT-PCR, different versions of siRNA (siRNA constructs 1 and 2) knocked down Dlx2 expression in primary embryonic neocortical (NC) cultures, compared with a control scrambled siRNA control (p < 0.01). Controls were performed separately for each siRNA construct to Dlx2. Embryonic neocortical cells endogenously express DLX2, and were used as a positive control. Concomitantly, transfection with either Dlx2 siRNA resulted in statistically significant decreases in Gad1 and Gad2 expression (p < 0.05). F, For testing of siRNA transfection efficiency, cotransfection of both Dlx2 siRNAs and the siRNA control, and pooled Dlx2 siRNA with pcDNA3/Dlx2 plasmid was also performed in embryonic NC cells. Western analysis with anti-DLX2 antibody showed that different versions of siRNA (siRNA constructs 1 and 2) knocked down DLX2 expression compared with endogenous DLX2 levels. Coexpression of a Dlx2 expression plasmid with Dlx2 siRNA rescued DLX2 expression. An siRNA control (scrambled siRNA) and untransfected embryonic neocortical lysate were used as controls, respectively. β-Actin was used as loading control. B, D, Quantification of DLX2- and GABA-positive cells following reduction of DLX2 expression mediated by duplex siRNA in E16.5 (B) and E18.5 (D) neocortical primary cultures (n = 4). DLX2- or GABA-positive cells were counted and compared with the total of number of immunopositive cells transfected with scrambled siRNA control. All comparisons were performed in at least three trials. Average ± SEM with associated p values.
Discussion
ChIP technology has advanced the identification of direct target genes of specific transcription factors, especially homeobox genes (Gould et al., 1990; Graba et al., 1997; Orlando et al., 1997; Kuo and Allis, 1999). Biochemical approaches, such as ChIP, rather than genetic approaches, provide several advantages. Identified target genes are directly downstream and derived from physiological homeodomain–DNA complexes obtained in vivo. DNA fragments may be from regulatory elements of known or novel genes. Cross-linking preserves naturally existing (in situ) protein-DNA interactions. Utilizing these methods, homeodomain targets have been isolated, including transcription factors, growth factors, adhesion molecules and secreted proteins (Mannervik, 1999; Komata et al., 2014; Cejas et al., 2016).
We have optimized our ChIP approach by reducing the concentration of the cross-linking reagent PFA and the incubation time to preferentially obtain homeoprotein-genomic DNA complexes from embryonic tissues in situ (Zhou et al., 2004). The polyclonal antisera to DLX1 and DLX2 have been subjected to a rigorous affinity purification process and are sensitive and highly specific (Eisenstat et al., 1999). In addition, chromatin is derived from nuclear extracts of embryonic tissues where the peak developmental expression of DLX1 and DLX2 occurs in restricted anatomical regions, hence enriching for the selection of Dlx1- and Dlx2-specific DNA target fragments. Future advances will use high-throughput sequencing platforms and ChIP-seq will become the dominant profiling approach for generation of libraries of Dlx downstream targets (Göbel et al., 2010; Reimer and Turck, 2010).
The identification of multiple Dlx target sequences in vivo will facilitate the elucidation and characterization of a consensus DLX homeodomain DNA-binding-site initially derived in vitro for Dlx3 by (Feledy et al., 1999). The consensus site identified as (A/C/G)TAATT(G/A) (C/G) was reported in zebrafish (Zerucha et al., 2000). Because the homeodomains of the Dlx family are very homologous, it is likely that DLX proteins will recognize a similar sequence. However, there still are major variations in the flanking sequences of the TAATT core motif above (Givens et al., 2005). The difference in binding capabilities between members of the Dlx family may also be due to different transcriptional cofactors of DLX proteins as well as post-translational modifications, such as phosphorylation, which have yet to be identified. Identification of the entire network of Dlx downstream targets will facilitate the elucidation and biological validation of the consensus binding sequence.
Within the forebrain, the expression of the Dlx genes coincides with the localization of virtually all neurons that use GABA as their neurotransmitter (Cobos et al., 2005, 2007). The spatial and temporal coincidental patterns of DLX and GAD isoform expression in the SVZ of the basal telencephalon suggest that the Dlx genes may have an important role in regulating the expression of Gad genes. We have discovered that DLX1 and DLX2 bind directly to specific regions, and furthermore, to specific homeodomain binding motifs of Gad1 and Gad2 promoters in vivo and in vitro (Figs. 1–3). Reporter gene assays and site-directed mutagenesis demonstrated that transcriptional activation is mediated by DLX1 and DLX2 upon binding to the Gad1 and Gad2 promoters in vitro (Fig. 4A,B). DLX2 acted as a more potent transcriptional activator than DLX1 of both the Gad65 and Gad67 promoters, signifying a more important role for Dlx2 gene function in forebrain development. Coexpression of Dlx1/Dlx2 with various target regulatory sequences identified by ChIP in vivo linked to a reporter gene demonstrate neither synergy nor an additive effect of these two transcription factors; the limiting factor either for activation or inhibition in vitro is always expression of Dlx2 (Zhou et al., 2004) and data not shown. Indeed, mice homozygous for the Dlx1 mutation have a milder phenotype than mice homozygous for the Dlx2 mutation. Some Dlx1 mutants are viable at birth, dependent on the background mouse strain, but are small and all die within 1 month with an abnormal phenotype including a seizure disorder. Dlx2 mutants are similar to the Dlx1/2 mutants; they all die within a few hours after birth with distinctive craniofacial skeletal deformities. Dlx1, Dlx2, and Dlx1/2 heterozygotes cannot be distinguished from their wild-type littermates.
We have provided evidence that Dlx homeobox gene expression marks the majority of embryonic GABAergic cortical and striatal neurons, from the beginning of their development in the basal telencephalon (Fig. 4D and data not shown). In most of the SVZ, the Dlx genes are coexpressed with GABA, Gad1, and Gad2 genes, all of which are hallmarks of an interneuronal phenotype in the GE. The Dlx genes are essential for the development of GABAergic cortical neurons (Anderson et al., 1997a; Anderson et al., 1999; Stühmer et al., 2002a). Indeed, GABA, GAD65, and GAD67 expression are severely decreased in the SVZ of the basal telencephalon in the Dlx1/2 mutants from E13.5 to birth (Fig. 5 and data not shown). However, residual GABA and Gad isoform expression in the Dlx1/2 mutants suggests that other transcription factors can compensate for the absence of Dlx1/2 homeobox gene function. Of note, Dlx1, Dlx2, Dlx5, and Dlx6 are expressed in overlapping populations of cells in the developing forebrain, consistent with redundant functions (Bulfone et al., 1993; Liu et al., 1997; Eisenstat et al., 1999). Regulation of Gad1/2 by DLX5/DLX6 could be mediated indirectly through the activities of DLX1/2 on the expression of Dlx5/6 (Zhou et al., 2004). However, because expression of Dlx5 and Dlx6 is greatly reduced in the Dlx1/2 mutants (Zhou et al., 2004; Ghanem et al., 2008), other genes are more likely to play this role. Candidates include Ascl1 (Mash1), a bHLH transcription factor, whose expression appears to be upstream of the Dlx genes (Fode et al., 2000; Kito-Shingaki et al., 2014), and Gsh1 and Gsh2, homeobox genes whose expression is unaffected in the Dlx1/2 mutants (Yun et al., 2003). Interestingly, in the Dlx1/2 double mutants, the first wave of neurogenesis (from ∼E10–E12) appears to be undisturbed, whereas differentiation of later born neurons (E13.5 to birth) is largely affected. The primary proliferative population (located in the VZ) appears normal in the double mutants, whereas abnormalities are found in the SVZ that contains the secondary proliferative population of neuroblasts (Marin et al., 2000). These studies signify a role for Dlx genes in the differentiation of later-born interneurons, as well as provide a region-specific role of Dlx genes in the developing basal telencephalon. This concept is supported by our finding that overall GABA neurotransmitter levels do not decrease significantly in the developing forebrain until E18.5 using HPLC, substantiating the role of Dlx genes in the terminal differentiation of later-born neurons.
Although these studies have enriched our understanding of the role of Dlx1 and Dlx2 during forebrain development, important questions remain. There is limited postnatal Dlx expression in the cortical SVZ (Anderson et al., 2001; Stühmer et al., 2002b). However, it is unclear whether this expression arises from cortical progenitors, or whether these Dlx-positive cells are introduced into the cortex by tangential migration from the basal telencephalon. Through this latter mechanism, progenitor cells that are specified in the subpallial telencephalon might continue to proliferate after they reach the progenitor zone of the cortex, providing a secondary source of GABAergic interneurons. Future generation of conditional temporal, spatial, or lineage-specific knock-out mouse models could resolve the functional significance of Dlx genes after birth with respect to postnatal forebrain development. These genetic mouse models will advance our understanding of the regulation of cortical interneuron development by Dlx genes, and contribute to animal models for congenital epilepsies, autism spectrum disorders, and neuronal migration disorders.
To further assess DLX2 as a potent regulator of GABA expression, we used RNA interference technologies to acutely knock down Dlx2 expression in primary wild-type embryonic striatal and neocortical cultures (Fig. 7). However, it remains to be demonstrated what thresholds exist for functional reduction of GABA expression and whether residual GABA expression levels will have any physiological significance for the inhibitory signaling circuitry of the basal telencephalon. Neurophysiologic techniques, such as measuring inhibitory currents, may help to determine whether there is a physiologic decrease in inhibitory signal corresponding to the decrease in GABA expression in the Dlx1/2 mutant mice.
In summary, we have successfully established that DLX1 and DLX2 are necessary and sufficient for the transcriptional activation of both Gad isoforms in embryonic forebrain. Furthermore, there is a global reduction of GABA in the embryonic forebrain of Dlx1/2 DKO mice substantiating a role for these homeobox transcription factors beyond the regulation of tangential migration of GABAergic interneurons. Our study supports a critical role for Dlx genes in the differentiation of GABAergic interneurons. This work contributes toward the identification of potential targets for treating neurodevelopmental disorders whose pathogenesis may derive from an imbalance of excitation to inhibition, especially in those patients with a concomitant seizure disorder.
Footnotes
This work was supported by graduate student stipends from the Natural Sciences and Engineering Research Council of Canada and the Canadian Institutes for Health Research MD/PhD program to T.N.L, and operating grants from the Manitoba Health Research Council, the Children's Hospital Foundation of Manitoba, the Women and Children's Health Research Institute, University of Alberta, and through the Muriel and Ada Hole Kids with Cancer Society Chair in Pediatric Oncology to D.D.E.
The authors declare no competing financial interests.
References
- Al-Jaberi N, Lindsay S, Sarma S, Bayatti N, Clowry GJ (2015) The early fetal development of human neocortical GABAergic interneurons. Cereb Cortex 25:631–645. 10.1093/cercor/bht254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SA, Eisenstat DD, Shi L, Rubenstein JL (1997a) Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science 278:474–476. 10.1126/science.278.5337.474 [DOI] [PubMed] [Google Scholar]
- Anderson SA, Qiu M, Bulfone A, Eisenstat DD, Meneses J, Pedersen R, Rubenstein JL (1997b) Mutations of the homeobox genes Dlx-1 and Dlx-2 disrupt the striatal subventricular zone and differentiation of late born striatal neurons. Neuron 19:27–37. 10.1016/S0896-6273(00)80345-1 [DOI] [PubMed] [Google Scholar]
- Anderson SA, Kaznowski CE, Horn C, Rubenstein JL, McConnell SK (2002) Distinct origins of neocortical projection neurons and interneurons in vivo. Cereb Cortex 12:702–709. 10.1093/cercor/12.7.702 [DOI] [PubMed] [Google Scholar]
- Anderson SA, Marín O, Horn C, Jennings K, Rubenstein JL (2001) Distinct cortical migrations from the medial and lateral ganglionic eminences. Development 128:353–363. [DOI] [PubMed] [Google Scholar]
- Anderson S, Mione M, Yun K, Rubenstein JL (1999) Differential origins of neocortical projection and local circuit neurons: role of Dlx genes in neocortical interneuronogenesis. Cereb Cortex 9:646–654. 10.1093/cercor/9.6.646 [DOI] [PubMed] [Google Scholar]
- Barker JL, Behar T, Li YX, Liu QY, Ma W, Maric D, Maric I, Schaffner AE, Serafini R, Smith SV, Somogyi R, Vautrin JY, Wen XL, Xian H (1998) GABAergic cells and signals in CNS development. Perspect Dev Neurobiol 5:305–322. [PubMed] [Google Scholar]
- Bickford ME, Ramcharan E, Godwin DW, Erişir A, Gnadt J, Sherman SM (2000) Neurotransmitters contained in the subcortical extraretinal inputs to the monkey lateral geniculate nucleus. J Comp Neurol 424:701–717. [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Bulfone A, Puelles L, Porteus MH, Frohman MA, Martin GR, Rubenstein JL (1993) Spatially restricted expression of Dlx-1, Dlx-2 (Tes-1), Gbx-2, and Wnt-3 in the embryonic day 12.5 mouse forebrain defines potential transverse and longitudinal segmental boundaries. J Neurosci 13:3155–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejas P, Li L, O'Neill NK, Duarte M, Rao P, Bowden M, Zhou CW, Mendiola M, Burgos E, Feliu J, Moreno-Rubio J, Guadalajara H, Moreno V, García-Olmo D, Bellmunt J, Mullane S, Hirsch M, Sweeney CJ, Richardson A, Liu XS, et al. (2016) Chromatin immunoprecipitation from fixed clinical tissues reveals tumor-specific enhancer profiles. Nat Med 22:685–691. 10.1038/nm.4085 [DOI] [PubMed] [Google Scholar]
- Chang YC, Gottlieb DI (1988) Characterization of the proteins purified with monoclonal antibodies to glutamic acid decarboxylase. J Neurosci 8:2123–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobos I, Calcagnotto ME, Vilaythong AJ, Thwin MT, Noebels JL, Baraban SC, Rubenstein JL (2005) Mice lacking Dlx1 show subtype-specific loss of interneurons, reduced inhibition and epilepsy. Nat Neurosci 8:1059–1068. 10.1038/nn1499 [DOI] [PubMed] [Google Scholar]
- Cobos I, Borello U, Rubenstein JL (2007) Dlx transcription factors promote migration through repression of axon and dendrite growth. Neuron 54:873–888. 10.1016/j.neuron.2007.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Melo J, Qiu X, Du G, Cristante L, Eisenstat DD (2003) Dlx1, Dlx2, Pax6, Brn3b, and Chx10 homeobox gene expression defines the retinal ganglion and inner nuclear layers of the developing and adult mouse retina. J Comp Neurol 461:187–204. 10.1002/cne.10674 [DOI] [PubMed] [Google Scholar]
- Donzanti BA, Yamamoto BK (1988) An improved and rapid HPLC-EC method for the isocratic separation of amino acid neurotransmitters from brain tissue and microdialysis perfusates. Life Sci 43:913–922. 10.1016/0024-3205(88)90267-6 [DOI] [PubMed] [Google Scholar]
- Eisenstat DD, Liu JK, Mione M, Zhong W, Yu G, Anderson SA, Ghattas I, Puelles L, Rubenstein JL (1999) DLX-1, DLX-2, and DLX-5 expression define distinct stages of basal forebrain differentiation. J Comp Neurol 414:217–237. [DOI] [PubMed] [Google Scholar]
- Erlander MG, Tillakaratne NJ, Feldblum S, Patel N, Tobin AJ (1991) Two genes encode distinct glutamate decarboxylases. Neuron 7:91–100. 10.1016/0896-6273(91)90077-D [DOI] [PubMed] [Google Scholar]
- Feledy JA, Morasso MI, Jang SI, Sargent TD (1999) Transcriptional activation by the homeodomain protein distal-less 3. Nucleic Acids Res 27:764–770. 10.1093/nar/27.3.764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fode C, Ma Q, Casarosa S, Ang SL, Anderson DJ, Guillemot F (2000) A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev 14:67–80. 10.1101/gad.14.1.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanem N, Yu M, Poitras L, Rubenstein JL, Ekker M (2008) Characterization of a distinct subpopulation of striatal projection neurons expressing the Dlx genes in the basal ganglia through the activity of the I56ii enhancer. Dev Biol 322:415–424. 10.1016/j.ydbio.2008.07.029 [DOI] [PubMed] [Google Scholar]
- Givens ML, Rave-Harel N, Goonewardena VD, Kurotani R, Berdy SE, Swan CH, Rubenstein JL, Robert B, Mellon PL (2005) Developmental regulation of gonadotropin-releasing hormone gene expression by the MSX and DLX homeodomain protein families. J Biol Chem 280:19156–19165. 10.1074/jbc.M502004200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göbel U, Reimer J, Turck F (2010) Genome-wide mapping of protein-DNA interaction by chromatin immunoprecipitation and DNA microarray hybridization (ChIP-chip): Part B. ChIP-chip data analysis. Methods Mol Biol 631:161–184. 10.1007/978-1-60761-646-7_13 [DOI] [PubMed] [Google Scholar]
- Gould AP, Brookman JJ, Strutt DI, White RA (1990) Targets of homeotic gene control in Drosophila. Nature 348:308–312. 10.1038/348308a0 [DOI] [PubMed] [Google Scholar]
- Graba Y, Aragnol D, Pradel J (1997) Drosophila Hox complex downstream targets and the function of homeotic genes. Bioessays 19:379–388. 10.1002/bies.950190505 [DOI] [PubMed] [Google Scholar]
- Harwell CC, Fuentealba LC, Gonzalez-Cerrillo A, Parker PR, Gertz CC, Mazzola E, Garcia MT, Alvarez-Buylla A, Cepko CL, Kriegstein AR (2015) Wide dispersion and diversity of clonally related inhibitory interneurons. Neuron 87:999–1007. 10.1016/j.neuron.2015.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kito-Shingaki A, Seta Y, Toyono T, Kataoka S, Kakinoki Y, Yanagawa Y, Toyoshima K (2014) Expression of GAD67 and Dlx5 in the taste buds of mice genetically lacking Mash1. Chem Senses 39:403–414. 10.1093/chemse/bju010 [DOI] [PubMed] [Google Scholar]
- Komata M, Katou Y, Tanaka H, Nakato R, Shirahige K, Bando M (2014) Chromatin immunoprecipitation protocol for mammalian cells. Methods Mol Biol 1164:33–38. 10.1007/978-1-4939-0805-9_4 [DOI] [PubMed] [Google Scholar]
- Kuo MH, Allis CD (1999) In vivo cross-linking and immunoprecipitation for studying dynamic protein: DNA associations in a chromatin environment. Methods 19:425–433. 10.1006/meth.1999.0879 [DOI] [PubMed] [Google Scholar]
- Kwakowsky A, Schwirtlich M, Zhang Q, Eisenstat DD, Erdélyi F, Baranyi M, Katarova ZD, Szabó G (2007) GAD isoforms exhibit distinct spatiotemporal expression patterns in the developing mouse lens: correlation with Dlx2 and Dlx5. Dev Dyn 236:3532–3544. 10.1002/dvdy.21361 [DOI] [PubMed] [Google Scholar]
- Le TN, Du G, Fonseca M, Zhou QP, Wigle JT, Eisenstat DD (2007) Dlx homeobox genes promote cortical interneuron migration from the basal forebrain by direct repression of the semaphorin receptor neuropilin-2. J Biol Chem 282:19071–19081. 10.1074/jbc.M607486200 [DOI] [PubMed] [Google Scholar]
- Li H, Hader AT, Han YR, Wong JA, Babiarz J, Ricupero CL, Godfrey SB, Corradi JP, Fennell M, Hart RP, Plummer MR, Grumet M (2012) Isolation of a novel rat neural progenitor clone that expresses Dlx family transcription factors and gives rise to functional GABAergic neurons in culture. Dev Neurobiol 72:805–820. 10.1002/dneu.20977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JK, Ghattas I, Liu S, Chen S, Rubenstein JL (1997) Dlx genes encode DNA-binding proteins that are expressed in an overlapping and sequential pattern during basal ganglia differentiation. Dev Dyn 210:498–512. [DOI] [PubMed] [Google Scholar]
- Luján R, Shigemoto R, López-Bendito G (2005) Glutamate and GABA receptor signalling in the developing brain. Neuroscience 130:567–580. 10.1016/j.neuroscience.2004.09.042 [DOI] [PubMed] [Google Scholar]
- MacDonald RB, Debiais-Thibaud M, Talbot JC, Ekker M (2010) The relationship between dlx and gad1 expression indicates highly conserved genetic pathways in the zebrafish forebrain. Dev Dyn 239:2298–2306. 10.1002/dvdy.22365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald RB, Pollack JN, Debiais-Thibaud M, Heude E, Talbot JC, Ekker M (2013) The ascl1a and dlx genes have a regulatory role in the development of GABAergic interneurons in the zebrafish diencephalon. Dev Biol 381:276–285. 10.1016/j.ydbio.2013.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannervik M. (1999) Target genes of homeodomain proteins. Bioessays 21:267–270. [DOI] [PubMed] [Google Scholar]
- Marin O, Anderson SA, Rubenstein JL (2000) Origin and molecular specification of striatal interneurons. J Neurosci 20:6063–6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DL, Liu H, Martin SB, Wu SJ (2000) Structural features and regulatory properties of the brain glutamate decarboxylases. Neurochem Int 37:111–119. 10.1016/S0197-0186(00)00014-0 [DOI] [PubMed] [Google Scholar]
- Mayer C, Jaglin XH, Cobbs LV, Bandler RC, Streicher C, Cepko CL, Hippenmeyer S, Fishell G (2015) Clonally related forebrain interneurons disperse broadly across both functional areas and structural boundaries. Neuron 87:989–998. 10.1016/j.neuron.2015.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Bandler RC, Fishell G (2016) Lineage is a poor predictor of interneuron positioning within the forebrain. Neuron 92:45–51. 10.1016/j.neuron.2016.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando V, Strutt H, Paro R (1997) Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods 11:205–214. 10.1006/meth.1996.0407 [DOI] [PubMed] [Google Scholar]
- Petroff OA. (2002) GABA and glutamate in the human brain. Neuroscientist 8:562–573. 10.1177/1073858402238515 [DOI] [PubMed] [Google Scholar]
- Qiu M, Bulfone A, Martinez S, Meneses JJ, Shimamura K, Pedersen RA, Rubenstein JL (1995) Null mutation of Dlx-2 results in abnormal morphogenesis of proximal first and second branchial arch derivatives and abnormal differentiation in the forebrain. Genes Dev 9:2523–2538. 10.1101/gad.9.20.2523 [DOI] [PubMed] [Google Scholar]
- Reimer JJ, Turck F (2010) Genome-wide mapping of protein-DNA interaction by chromatin immunoprecipitation and DNA microarray hybridization (ChIP-chip): Part A. ChIP-chip molecular methods. Methods Mol Biol 631:139–160. 10.1007/978-1-60761-646-7_12 [DOI] [PubMed] [Google Scholar]
- Rubenstein JL, Merzenich MM (2003) Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav 2:255–267. 10.1034/j.1601-183X.2003.00037.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stühmer T, Anderson SA, Ekker M, Rubenstein JL (2002a) Ectopic expression of the Dlx genes induces glutamic acid decarboxylase and Dlx expression. Development 129:245–252. [DOI] [PubMed] [Google Scholar]
- Stühmer T, Puelles L, Ekker M, Rubenstein JL (2002b) Expression from a Dlx gene enhancer marks adult mouse cortical GABAergic neurons. Cereb Cortex 12:75–85. 10.1093/cercor/12.1.75 [DOI] [PubMed] [Google Scholar]
- Sultan KT, Han Z, Zhang XJ, Xianyu A, Li Z, Huang K, Shi SH (2016) Clonally related GABAergic interneurons do not randomly disperse but frequently form local clusters in the forebrain. Neuron 92:31–44. 10.1016/j.neuron.2016.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrero García M, Mazzola E, Harwell CC (2016) Lineage relationships do not drive MGE/PoA-derived interneuron clustering in the brain. Neuron 92:52–58. 10.1016/j.neuron.2016.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriend J, Alexiuk NA, Green-Johnson J, Ryan E (1993) Determination of amino acids and monoamine neurotransmitters in caudate nucleus of seizure-resistant and seizure-prone BALB/c mice. J Neurochem 60:1300–1307. 10.1111/j.1471-4159.1993.tb03290.x [DOI] [PubMed] [Google Scholar]
- Wang B, Lufkin T, Rubenstein JL (2011) Dlx6 regulates molecular properties of the striatum and central nucleus of the amygdala. J Comp Neurol 519:2320–2334. 10.1002/cne.22618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun K, Garel S, Fischman S, Rubenstein JL (2003) Patterning of the lateral ganglionic eminence by the Gsh1 and Gsh2 homeobox genes regulates striatal and olfactory bulb histogenesis and the growth of axons through the basal ganglia. J Comp Neurol 461:151–165. 10.1002/cne.10685 [DOI] [PubMed] [Google Scholar]
- Zerucha T, Stühmer T, Hatch G, Park BK, Long Q, Yu G, Gambarotta A, Schultz JR, Rubenstein JL, Ekker M (2000) A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J Neurosci 20:709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou QP, Le TN, Qiu X, Spencer V, de Melo J, Du G, Plews M, Fonseca M, Sun JM, Davie JR, Eisenstat DD (2004) Identification of a direct Dlx homeodomain target in the developing mouse forebrain and retina by optimization of chromatin immunoprecipitation. Nucleic Acids Res 32:884–892. 10.1093/nar/gkh233 [DOI] [PMC free article] [PubMed] [Google Scholar]