Abstract
In the adult mammalian brain, neural stem cells (NSCs) generate new neurons throughout the mammal's lifetime. The balance between quiescence and active cell division among NSCs is crucial in producing appropriate numbers of neurons while maintaining the stem cell pool for a long period. The Notch signaling pathway plays a central role in both maintaining quiescent NSCs (qNSCs) and promoting cell division of active NSCs (aNSCs), although no one knows how this pathway regulates these apparently opposite functions. Notch1 has been shown to promote proliferation of aNSCs without affecting qNSCs in the adult mouse subependymal zone (SEZ). In this study, we found that Notch3 is expressed to a higher extent in qNSCs than in aNSCs while Notch1 is preferentially expressed in aNSCs and transit-amplifying progenitors in the adult mouse SEZ. Furthermore, Notch3 is selectively expressed in the lateral and ventral walls of the SEZ. Knockdown of Notch3 in the lateral wall of the adult SEZ increased the division of NSCs. Moreover, deletion of the Notch3 gene resulted in significant reduction of qNSCs specifically in the lateral and ventral walls, compared with the medial and dorsal walls, of the lateral ventricles. Notch3 deletion also reduced the number of qNSCs activated after antimitotic cytosine β-D-arabinofuranoside (Ara-C) treatment. Importantly, Notch3 deletion preferentially reduced specific subtypes of newborn neurons in the olfactory bulb derived from the lateral walls of the SEZ. These results indicate that Notch isoforms differentially control the quiescent and proliferative steps of adult SEZ NSCs in a domain-specific manner.
SIGNIFICANCE STATEMENT In the adult mammalian brain, the subependymal zone (SEZ) of the lateral ventricles is the largest neurogenic niche, where neural stem cells (NSCs) generate neurons. In this study, we found that Notch3 plays an important role in the maintenance of quiescent NSCs (qNSCs), while Notch1 has been reported to act as a regulator of actively cycling NSCs. Furthermore, we found that Notch3 is specifically expressed in qNSCs located in the lateral and ventral walls of the lateral ventricles and regulates neuronal production of NSCs in a region-specific manner. Our results indicate that Notch3, by maintaining the quiescence of a subpopulation of NSCs, confers a region-specific heterogeneity among NSCs in the adult SEZ.
Keywords: adult neurogenesis, neural stem cell, notch signaling, olfactory bulb, quiescence, stem-cell maintenance
Introduction
In the adult mammalian brain, the subependymal zone (SEZ) of the lateral ventricles is the largest neurogenic niche, where neural stem cells (NSCs) generate neurons. Most NSCs (or B1 cells) in the adult SEZ are quiescent NSCs (qNSCs). That is, they are NSCs kept in a quiescent state. After entering the cell cycle, the activated NSCs (aNSCs) undergo amplification and differentiation, sequentially becoming transit-amplifying progenitors (TAPs or C cells), neuroblasts (or A cells), and postmitotic immature neurons (Kriegstein and Alvarez-Buylla, 2009; Chaker et al., 2016). These immature neurons then migrate to the olfactory bulb (OB) and modify innate and adaptive olfactory responses (Sakamoto et al., 2011, 2014). NSCs in the SEZ are heterogeneous, and their spatial distribution correlates with distinct cellular fates. For instance, periglomerular cells (PGCs) expressing calretinin (CR+), calbindin (CalB+), and tyrosine hydroxylase (TH+) are preferentially produced by medial (septal), lateral/ventral, and dorsal walls of the SEZ, respectively (Merkle et al., 2007; Young et al., 2007; Fernández et al., 2011). It is quite likely that NSC subpopulations are differentially regulated in an area-dependent and context-dependent manner and, indeed, NSCs in the ventral SEZ are specifically regulated by Shh signaling and POMC axons (Ihrie et al., 2011; Paul et al., 2017). Even so, little is known about the molecular mechanisms involved in subpopulation-specific regulation of NSCs in the SEZ.
Regulation of NSC quiescence is crucial for determining the rate of neurogenesis. Disruption of NSC quiescence causes a short-term increase in the number of aNSCs and their descendants, and therefore results in an enhanced neurogenesis. In addition, it later leads to a gradual exhaustion of NSCs and a reduction of neurogenesis, suggesting that NSC quiescence is important for the long-term maintenance of the total NSC pool and neurogenesis (Groszer et al., 2006; Nam and Benezra, 2009; Renault et al., 2009; Imayoshi et al., 2010; Mira et al., 2010; Kawaguchi et al., 2013; Martynoga et al., 2013; Porlan et al., 2013; Furutachi et al., 2015).
The Notch signaling pathway, in addition to maintaining the undifferentiated state of NSCs throughout the CNS, plays a pivotal role in the regulation of NSC quiescence by limiting the activation of qNSCs in the adult SEZ. Deletion of RBP-J (recombination signal-binding protein for immunoglobin κJ region), a downstream mediator of Notch signaling, induces transient activation of qNSCs, which subsequently causes depletion of NSCs (Imayoshi et al., 2010). Meanwhile, deletion of Notch1, one of the four mammalian Notch receptors, does not significantly reduce qNSCs and instead results in a selective loss of aNSCs (Basak et al., 2012), suggesting that Notch1 promotes aNSC proliferation (Nyfeler et al., 2005; Androutsellis-Theotokis et al., 2006; Aguirre et al., 2010; Basak et al., 2012). It thus remains unclear which Notch receptor(s) are involved in the regulation of NSC quiescence in the adult SEZ.
Notch3, another mammalian Notch receptor, has been associated with the quiescence of adult tissue stem cells, such as satellite cells in the muscles (Kitamoto and Hanaoka, 2010), luminal progenitor cells in the mammary gland (Lafkas et al., 2013), and NSCs in the hippocampal dentate gyrus (Ehret et al., 2015). In zebrafish, a Notch3 homolog suppresses, and a Notch1 homolog promotes, the cell cycle of qNSCs and aNSCs, respectively, in the adult brain (Alunni et al., 2013). We therefore asked whether Notch3 plays any role in regulating qNSCs in the adult mouse SEZ.
In this study, we found that Notch3 is selectively expressed in qNSCs located at the lateral and ventral walls of the SEZ. Acute knockdown of Notch3 in these cells abrogated their quiescence. We also found that the numbers of qNSCs and newborn neurons were reduced in the lateral and ventral walls of the SEZ in Notch3-null mice. Notch3 therefore appears to play an important role in the maintenance of qNSCs in an area-specific manner in the adult SEZ and in the production of specific subtypes of neurons.
Materials and Methods
Mice.
Notch3-null mice (Kitamoto et al., 2005) were backcrossed with C57BL/6J mice. Embryonic and postnatal ICR (imprinting control region) mice (Japan SLC) were used for primary neural progenitor (NPC) cultures and in situ hybridization. Adult (2–3 month old) C57BL/6 mice were used for all other experiments. Mice were maintained on a 12 h light/dark cycle with ad libitum access to food and water and cared for according to guidance from the Animal Care and Use Committee of the University of Tokyo.
Expression constructs.
pCAG-IRES-EGFP (pCAGIG) was kindly provided by C.L. Cepko and T. Matsuda. We modified the multicloning site of pCAGIG and made pCAG2-IRES-EGFP (pCAG2IG). 3XFlagNICD1 and 3XFlagNICD3 were purchased from Addgene (plasmids #20183 and #20185, respectively) and inserted into pCAG2IG to generate pCAG2IG-NICD1 and pCAG2IG-NICD3.
Immunofluorescence analysis.
For immunohistofluorescence staining of coronal brain sections, mice were deeply anesthetized and transcardially perfused with ice-cold 4% paraformaldehyde (PFA) in PBS. Brains were postfixed with 4% PFA in PBS at 4°C for 2–3 h. After equilibration with 30% (w/v) sucrose in PBS, the fixed brains were embedded in OCT compound (Tissue TEK) and frozen. Coronal cryosections (12–40 μm thickness) were exposed to TBS containing 0.1% Triton X-100 and 3% BSA (blocking buffer) for 2 h at room temperature (RT) and incubated first overnight at 4°C with primary antibodies in blocking buffer and then 2 h at RT with Alexa Fluor-conjugated secondary antibodies in blocking buffer and mounted in Mowiol (Calbiochem). For staining of lentiviral-infected brains, cryosections were sliced 40 μm thick. TBS containing 0.5% Triton X-100 and 5% BSA was used as blocking buffer. For staining with the antibody to BrdU, CalB, CR, and TH, cryosections were incubated in 0.025N HCl for 30 min at 65°C and rinsed with 0.1 m bolic acid, pH 8.5. We used target-retrieval solution (Dako) for antigen retrieval in staining of iododeoxyuridine (IdU). Antigen retrieval was performed by autoclave treatment of sections for 5–10 min at 105°C. For staining with the antibody to epidermal growth factor receptor (EGFR), we used a tyramide signal amplification kit (Invitrogen) for signal enhancement. For immunocytofluorescence staining, cultured cells were fixed with ice-cold 4% PFA in PBS for 10 min. Cells were exposed to PBS containing 0.1% Triton X-100 for 10 min at RT and then 10 min at RT with PBS containing 3% BSA (blocking buffer). Cells were incubated first overnight at 4°C with primary antibodies in blocking buffer and then 30 min at RT with Alexa Fluor-conjugated secondary antibodies in blocking buffer and mounted in Mowiol (Calbiochem). Antibodies used for immunostaining included mouse monoclonal antibodies to Ascl1 (1:500; BD PharMingen, 556604, RRID:AB_396479), to BrdU (1:500; BD Bioscience, 347580), to CR (1:1000; Millipore, MAB1568, RRID:AB_94259), to GFAP (1:1000, Millipore, MAB360, RRID:AB_2109815), to S100β (1:500; Abcam, ab11179, RRID:AB_297818); and to TH (1:500; Millipore, MAB318, RRID:AB_2313764); rabbit monoclonal antibodies to Ki67 (1:1000; Abcam, ab16667, RRID:AB_302459), to Notch1 (1:200; Cell Signaling Technology, 3608, RRID:AB_2153354), to Sox2 (1:200; Cell Signaling Technology, C70B1, RRID:AB_2194037), and to S100β (1:5; DAKO, IS504); a rabbit polyclonal antibody to CalB (1:500; Millipore Bioscience Research Reagents, AB1778) and to RFP (Medical & Biological Laboratories, PM005, RRID:AB_591279); chicken polyclonal antibody to GFAP (1:500; Abcam, ab4674, RRID:AB_304558) and to GFP (1:1000, Abcam, ab13970, RRID:AB_300798); a rat monoclonal antibody to BrdU (1:200; Abcam, ab6326, RRID:AB_305426); a sheep polyclonal antibody to EGFR (1:5000; Millipore, 06-129); and a goat polyclonal antibody to doublecortin (Dcx, 1:500; Santa Cruz Biotechnology, sc8066, RRID:AB_2088494). Alexa-labeled secondary antibodies and Hoechst 33342 (for nuclear staining) were obtained from Invitrogen. All fluorescence images were obtained with laser confocal microscopes (Leica TCS-SP5 or Olympus FV3000). Images were processed and analyzed using Fiji software (RRID:SCR_002285).
In situ hybridization.
Notch3 cDNA (nucleotides 906–2115, NM_183029.2) used as probe corresponds to nucleotides 4987–6957 (NM_008716.2). For in situ hybridization, brains were fixed with 4% paraformaldehyde in PBS at 4°C for 24 h. After equilibration with 30% (w/v) sucrose in PBS, the fixed brains were embedded in OCT compound (Tissue TEK) and frozen. Coronal cryosections (12 μm thick) were cut on a cryostat, placed on Superfrost microscope slides (Matsunami), and desiccated at RT for 2 h. Then the sections were fixed with cold 4% PFA in PBS for 10 min, washed with PBTw (0.1% Tween 20 in PBS), treated with TEA buffer (100 mm triethanolamine, 0.4% acetic acid) for 15 min, washed with PBTw, prehybridized with hybridization solution (50% formamide, 5 × SSC, 5 × Denhardt's solution, 500 mg ml−1 salmon sperm DNA, 250 mg ml−1 yeast RNA) for 1 h at RT and incubated with hybridization solution containing 5–10 mg ml−1 DIG-labeled riboprobes at 65°C overnight. They were washed with 2 × SSC for 30 min twice at 65°C, 2 × SSC/50% formamide for 30 min twice at 65°C, MABT (1.16% maleic acid, 0.07% NaCl, pH adjusted to 7.5 with Trizma base) for 5 min three times at RT, blocked with blocking solution (10% Tween 20 MABT, 10% donkey serum), and then incubated with alkaline phosphatase-conjugated anti-DIG antibody (1:2000; Roche, blocking solution) at 4°C overnight. The sections were washed with MABT for 5 min twice at RT, with NTMT solution (100 mm NaCl, 10 mm Tris, pH 9.5, 50 mm MgCl2, 1% Tween 20) for 5 min twice at RT, and treated with NBT-BCIP (50 mg ml−1 nitro-blue tetrazolium chloride, 50 mg ml−1 5-bromo-4-chloro-3′-indolylphosphatase p-toluidine salt, NTMT solution) until the color appeared. Then the sections were washed with PBTw for 5 min three times at RT, fixed with cold 4% PFA in PBS, washed with PBS twice, and mounted into Mowiol.
Fluorescence-activated cell sorting.
The SEZs from 2-month-old wild-type C57BL/6J mice were dissected with a scalpel. Brain tissue was then subjected to enzymatic digestion with a papain-based solution (Sumitomo Bakelite) and dissociated single cells were resuspended in PBS. Cells were centrifuged for 10 min at 4°C without brakes in 25% Percoll (Sigma-Aldrich) to remove myelin. After being resuspended in 0.2% BSA PBS, cells were incubated for 10 min with mouse anti-Lex (1:500; Millipore Bioscience Research Reagents, MAB4301), washed by centrifugation, and incubated with Alexa-Fluor-labeled secondary antibody (1:500; Invitrogen, A21042), A555-complexed EGF (1:200; Invitrogen) and APC-conjugated rat anti-mCD24 (1:100; BioLegend, 101814, RRID:AB_439716). All staining and washes were performed on ice in 0.2% BSA PBS. Cells were analyzed or sorted on a AriaIII fluorescence-activated cell sorter (BD Biosciences). Debris and aggregated cells were gated out by forward and side scatter. Gates were set as described previously (Daynac et al., 2013). The data were analyzed using FlowJo software.
Quantitative real-time PCR.
RNA purification from fluorescence-activated cell-sorted populations and cDNA generation were performed as previously described (Sasagawa et al., 2013). The resulting cDNA was subjected to real-time PCR analysis in a Roche LightCycler with a KAPA SYBR Fast qPCR Kit (Kapa Biosystems). The amount of target mRNA was normalized relative to that of β-actin or GAPDH. The sense and antisense primers, respectively, were as follows: β-actin forward, AATAGTCATTCCAAGTATCCATGAAA; β-actin reverse, GCGACCATCCTCCTCTTAG; GAPDH forward, ATGAATACGGCTACAGCAACAGG; GAPDH reverse, CTCTTGCTCAGTGTCCTTGCTG; Notch1 forward, CATCACAGCCACACCTCAGT; Notch1 reverse, CTTTCCTGGGGCAGAATGGT; Notch3 forward, CTCTCAGACTGGTCTG; Notch3 reverse, GGAGGGAGGGAACAGA.
Administration of thymidine analogues.
To label dividing cells, mice were injected once with EdU (Invitrogen; 5 mg kg−1) intraperitoneally 1 h before killing. For identification of label-retaining cells (LRCs), IdU (Sigma-Aldrich) was added to drinking water (1% IdU, 1% sucrose) for 2 weeks. Mice were killed immediately or 3 weeks after stopping IdU administration. For labeling newborn neurons, BrdU (Sigma-Aldrich; 50 mg kg−1) was injected intraperitoneally twice per day for 5 consecutive days and killed 4 weeks after last administration. EdU was detected according to the manufacturer's instructions (Click-iT EdU Imaging Kit, Invitrogen).
Quantification.
For quantitative analysis of immunopositive cells in the SEZ, sections that covered SEZ coronal planes (+0.9 to +0.1 mm, relative to bregma) were examined. Six subregions of the SEZ along the dorsoventral and mediolateral axes were defined as previously described (López-Juárez et al., 2013). We used z stacks of confocal images for cell counting in Figures 1B, 3B,C (five optical planes, 2.03 μm steps, 8.01 μm thickness in total), 5C,E,F (six optical planes, 2.42 μm steps, 12.1 μm thickness in total), and 6D,H. The total number of immunopositive cells per whole OB was determined as previously described (Yamaguchi and Mori, 2005). In brief, coronal sections of the OB from the rostral tip to the caudal end were selected at the rate of one per 10 serial sections and subjected to immunofluorescence analysis. The periglomerular layer (PGL) was cropped by hand and analyzed. The counted numbers were multiplied by 10 to estimate the total number of immunopositive cells per whole OB. To compare PGL size, areas of cropped images were measured. All other quantifications with cryosections were performed with a single z plane. All image processing and analysis were performed using Fiji software.
Figure 1.
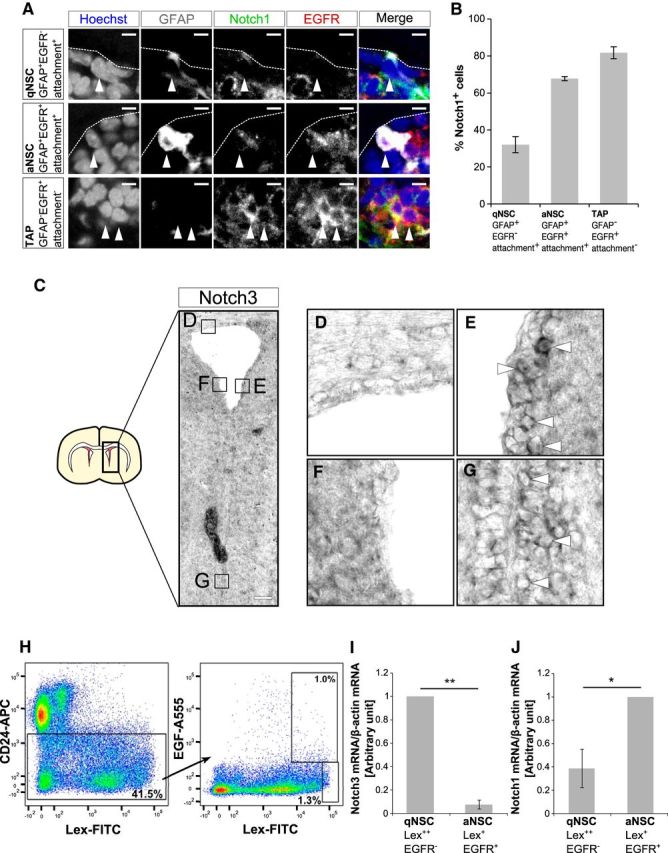
Notch1 and Notch3 expression patterns in the postnatal mouse SEZ. A, Immunostaining of Notch1 (green), GFAP (white), and EGFR (red) in the SEZ of 2-month-old mice. Arrowheads indicate each cell type. Expression levels of Notch1 is higher in TAPs than in qNSCs or aNSCs. B, Quantification of percentage of Notch1-positive cells in indicated cell types. Higher percentages of aNSCs and TAPs express Notch1 than qNSCs. C–G, In situ hybridization for Notch3 at postnatal day 18. Notch3 is expressed in the lateral region (E) and the ventral region (G) but weakly expressed in dorsal region (D) and could not be detected in the medial region (F) of the SEZ. Arrowheads indicate cells with high-level Notch3 expression. H, Representative flow cytometry for sorting SEZ cell types from wild-type adult mice. At first CD24− cells were selected, which were then gated on Lex and EGF-A555. I–J, Quantitative real-time PCR analysis of Notch3 (I) and Notch1 (J) of collected SEZ cells. Scale bars: A, 10 μm; C, 100 μm. Quantitative data are means ± SEM. n = 3 in B, I; n = 4 in J. Nuclei were stained with Hoechst (blue). Broken lines indicate the ventricular surface. Paired t test (I, J); *p < 0.05, **p < 0.01.
Figure 3.
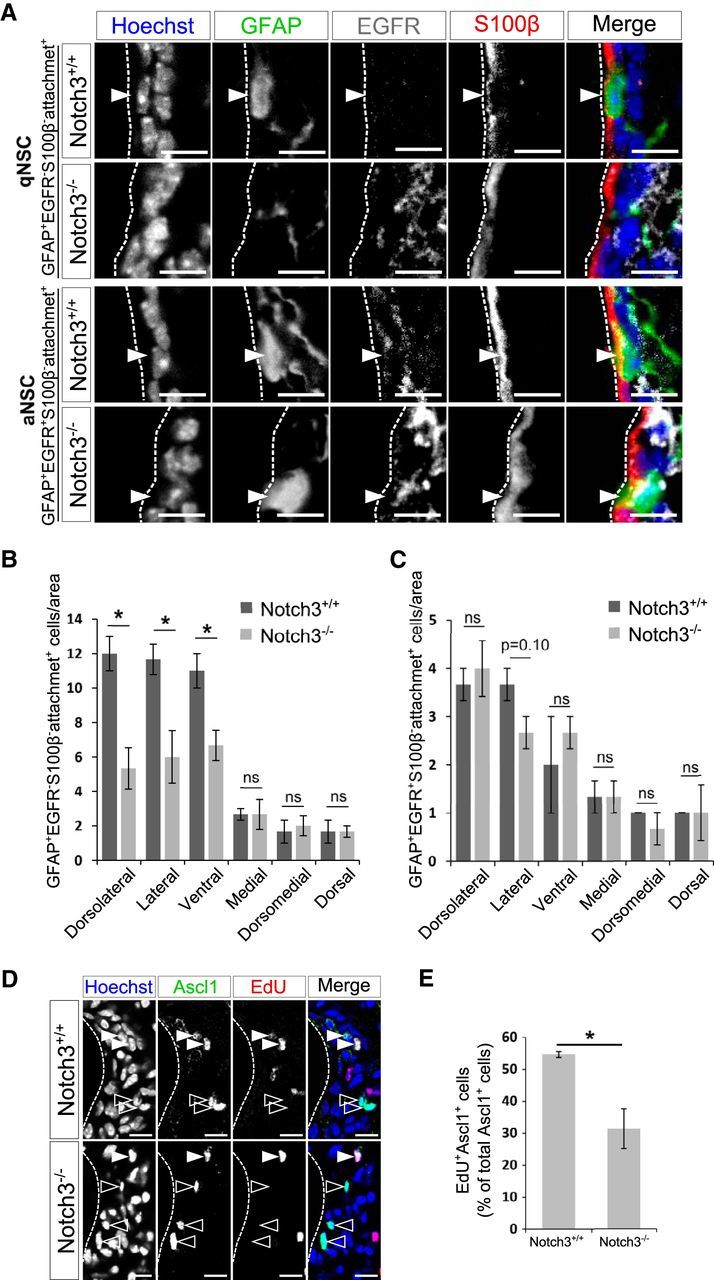
Deletion of Notch3 resulted in domain-specific reduction of qNSCs but not of aNSCs. A, Immunostaining on brain sections of 2-month-old control (Notch3+/+) and Notch3-null (Notch3−/−) mice. GFAP, Green; EGFR, white; S100β, red. qNSCs are defined as GFAP+EGFR−S100β− cells with an apical attachment (attachment+) and aNSCs as GFAP+EGFR+S100β−attachment+ cells. Arrowheads indicate each cell type. B, Quantification of the number of GFAP+EGFR−S100β−attachment+ cells. GFAP+EGFR−S100β−attachment+ cells are decreased in the dorsolateral, lateral, and ventral regions of the SEZ, but not significantly in the medial, dorsomedial, and dorsal regions, which is consistent with other cell types. C, Quantification of the number of GFAP+EGFR+S100β−attachment+ cells. GFAP+EGFR+S100β−attachment+ cells are not significantly reduced in any region of the SEZ. D, Ascl1 (green) and EdU (red). Open and closed arrowheads indicate Ascl1+EdU− cells and Ascl1+EdU+ cells, respectively. E, Percentage of EdU+ cells among Ascl1+ cells. Scale bar, 10 μm. Quantitative data are mean ± SEM. n = 3 and 3. Nuclei were stained with Hoechst (blue). Broken lines indicate the ventricular surface. Two-tailed Student's t test; *p < 0.05, **p < 0.01.
Ara-C treatment.
Ara-C treatment was performed as described previously (Doetsch et al., 1999). In brief, Ara-C (Sigma-Aldrich; 2%) in PBS was infused into the brain for 6 d using a miniature osmotic pump (Alzet, model 1007D). Cannulas (Alzet, Brain Infusion Kit III) were implanted stereotaxically in the left lateral ventricle (anterior, 0 mm; lateral, 1.1 mm; depth, 2 mm relative to bregma and the surface of the brain). Immediately (0 h) or 12 h after pump removal, mice were killed.
Lentivirus preparation and stereotaxic injections.
Lentiviral vector (CS-Rfa-CG or CS-RfA-CMV-mRFP1) harboring an shRNA driven by the H1 promoter was transfected with the packaging vectors pCAG-HIV-gp and pCMV-VSV-G-RSV-Rev into 293FT cells using polyethylenimine “MAX” (Polyscience, catalog #24765). For virus mixture preparation, GFP-expressing control virus and RFP-expressing shNotch3 virus were produced separately and mixed just before stereotaxic injections. All plasmids were kindly provided by H. Miyoshi, RIKEN BioResource Center, Tsukuba-shi, Ibaraki, Japan. For stereotaxic injections of lentivirus, mice were deeply anesthetized and injected with 1 μl of viral suspension (anterior, −0.75 mm; lateral, 1.1 mm; depth, 2 mm relative to bregma and the surface of the brain). The shRNA sequences were as follows: shNotch3#1, GTATAGAGACGAGTTCATTTA; shNotch3#2, GCTTGGGAAATCTGTCTTGCA; shControl#1 [shLuc (luciferase) #1]: GTGCGTTGCTAGTATCCGC; shControl#2 (shLuc#2): GATTTCGAGTCGTCTTAATGT.
Primary NPC culture.
Primary NPC culture was prepared from the embryonic ganglionic eminence (GE) at embryonic day 11.5. Dissected GE was subjected to enzymatic digestion with a papain-based solution (Sumitomo Bakelite) and dissociated into single cells. An NPC-enriched cell population (neurospheres) was prepared from dissociated cells after 3 d in vitro as described previously (Hirabayashi et al., 2009). For plasmid transfection, either pCAG2IG, pCAG2IG-NICD1, or pCAG2IG-NICD3 was transfected into dissociated neurospheres with Lipofectamine 2000 (Invitrogen). pCAG was used to keep total DNA concentrations constant between groups. Cells were labeled with 10 μg/ml BrdU for 4 h before fixation. For lentiviral infection, dissociated neurospheres were incubated with viruses for 20 h, followed by incubation without viruses for 2 d.
Experimental design and statistical analysis.
The sample size for each individual experiment is listed in the corresponding figure legend. Quantitative data are presented as means ± SEM. of values. Quantitative real-time PCR data of cells sorted by fluorescence-activated cell-sorting were compared using paired t tests. For quantitative real-time PCR and cell counting of lentivirus-infected cells (see Fig. 6B,D) and BrdU-incorporation assay (see Fig. 6I), data are compared using Dunnett's test. All other data with two groups are compared using unpaired Student's t tests. Two-way ANOVA was used when the data from different SEZ regions met the equal-variance assumption. Dunnett's test, two-way ANOVA, and calculation of effect size (Cohen's d) were performed using R (version 3.3.2, R Project for Statistical Computing, RRID:SCR_001905). p < 0.05 was considered significant.
Figure 6.
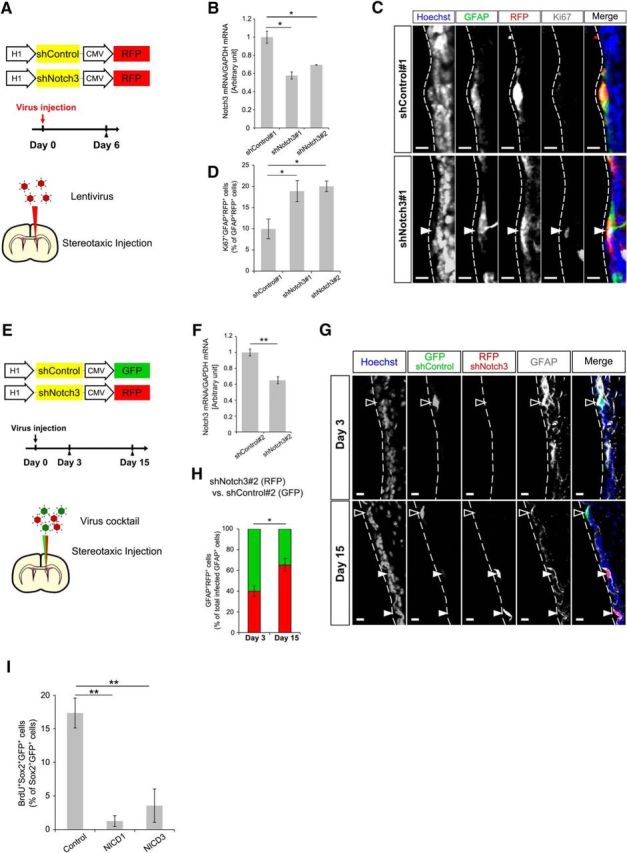
Acute Notch3 knockdown increased proliferation of NSCs. A, Schematic drawing of the lentivirus constructs expressing shRNA and experimental design. Mice were killed 6 d after stereotaxic lentivirus injection into the lateral ventricle. B, Quantitative real-time PCR analysis of Notch3 of the lentivirus-infected cultured NPCs. shRNA against luciferase was used as a negative control. C, Immunostaining of GFAP (green), RFP (red), and Ki67 (white) on the brain sections of the mice 6 d after lentivirus injection. D, Percentage of Ki67+GFAP+RFP+ cells among GFAP+RFP+ cells. E, Schematic drawing of the lentivirus constructs expressing shRNA and experimental design. Lentiviral mixture (shNotch3#2-RFP and shControl#2-GFP lentiviruses) was stereotaxically injected into the lateral ventricle of the 2-month-old mice. Mice were killed 3 or 15 d after lentivirus injection. F, Quantitative real-time PCR analysis of Notch3 of the lentivirus infected cultured NPCs. G, Immunostaining of GFP (green), RFP (red), and GFAP (white) on the brain sections of the mice 3 or 15 d after lentivirus mixture injection. Open and closed arrowheads indicate GFAP+GFP+RFP− cells and GFAP+RFP+ cells, respectively. H, Percentage of GFAP+RFP+ cells among total infected GFAP+ cells 3 and 15 d after lentivirus mixture injection. GFAP+GFP+RFP+ cells were counted as GFAP+RFP+ cells. I, BrdU-incorporation assay of NICD1-overexpressed or NICD3-overexpressed cultured NPCs. Scale bar, 10 μm. Quantitative data are mean ± SEM. n = 3, 3, and 3 in B, D, and I; 3 and 3 in F and H. Nuclei were stained with Hoechst (blue). Dunnett's test in B, D, and I). Two-tailed Student's t test in F and H; *p < 0.05, **p < 0.01.
Results
Notch3 is expressed predominantly in qNSCs in the adult mouse SEZ
A previous report showed that Notch1 is expressed in a subset of glial fibrillary acidic protein (GFAP)+ cells, which include NSCs and parenchymal astrocytes, in addition to TAPs and neuroblasts in the adult mouse SEZ (Basak et al., 2012). Whereas both qNSCs and aNSCs depend upon canonical Notch signaling (Imayoshi et al., 2010), Notch1 deletion results in a selective loss of aNSCs (Basak et al., 2012). This prompted us to investigate in more detail the expression patterns of Notch1 among GFAP+ cells. GFAP+ NSCs can be classified into EGFR− qNSCs and EGFR+ aNSCs (Doetsch et al., 2002; Pastrana et al., 2009). We costained Notch1 with GFAP and EGFR using 8-week-old mouse SEZ. qNSCs were defined as GFAP+EGFR− cells with an apical process attached to the ventricular surface (attachment+) cells and aNSCs were defined as GFAP+EGFR+attachment+ cells (Codega et al., 2014). These GFAP+attachment+ cells also highly expressed Sox2 (Sox22+; data not shown). Notch1 was expressed in fractions of GFAP+EGFR−attachment+ cells (qNSCs), GFAP+EGFR+attachment+ cells (aNSCs), and GFAP−EGFR+attachment− cells (TAPs; Fig. 1A). We found that the percentage of Notch1-expressing cells was greater in aNSCs and TAPs than in qNSCs (Fig. 1A,B): 32 ± 4.4% (mean ± SEM) of GFAP+EGFR−attachment+ cells (qNSCs) were positive for Notch1, whereas 68 ± 1.1% of GFAP+EGFR+attachment+ cells (aNSCs) and 82 ± 3.2% of GFAP−EGFR+attachment− cells (TAPs) were positive for Notch1 (Fig. 1B). The level of Notch1 signal also tended to be greater in GFAP−EGFR+attachment− TAPs than in GFAP+EGFR+attachment+ aNSCs (Fig. 1A). These expression patterns coincide with the notion that aNSCs, but not qNSCs, depend on Notch1.
We then examined the expression patterns of Notch3. Previous reports showed that Notch3 is expressed in radial glial cells in the embryonic mouse forebrain (Dang et al., 2006) and in some GFAP+ cells in the adult mouse SEZ (Basak et al., 2012). By performing an in situ hybridization analysis, we confirmed that Notch3 is expressed in the mouse SEZ at postnatal day 18 (Fig. 1C). Notch3 was expressed in the lateral and ventral walls of the lateral ventricle to a greater extent than in the dorsal or medial walls (Fig. 1C–G). Previous transcriptome analysis has indicated that Notch3 is preferentially expressed in qNSCs compared with aNSCs (Codega et al., 2014). When we isolated qNSCs (Lex2+EGFR−CD24−), aNSCs (Lex+EGFR+CD24−) with cell-surface markers by fluorescence-activated cell-sorting from the SEZ of 8-week-old mice as previously described (Daynac et al., 2013), we found that Notch3 was predominantly expressed in qNSCs compared with aNSCs (Fig. 1H,I). By contrast, Notch1 expression increased more in aNSCs than in qNSCs (Fig. 1J), which is consistent with the immunohistological data (Fig. 1A,B). These results suggested possible roles of Notch3 in qNSCs.
Domain-specific reduction of qNSCs by Notch3 deletion in the adult SEZ
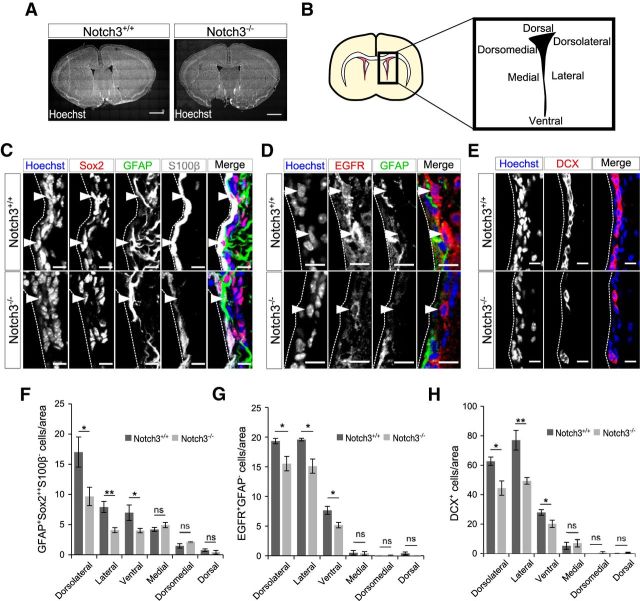
To investigate the role of Notch3 in the adult SEZ, we used Notch3-null mice (Kitamoto et al., 2005). Notch3 is expressed in various cell types, including vascular smooth muscle cells and pericytes, but Notch3-null mice are viable and fertile and develop with no overt abnormalities except for impaired smooth muscle cell differentiation and mild artery abnormality (Domenga et al., 2004; Kitamoto et al., 2005). The gross cytoarchitecture of the brain in these mice appeared normal (Fig. 2A). However, when we examined NSCs as GFAP+Sox22+S100β− cells, the number of NSCs in the SEZ was significantly lower in Notch3−/− mice compared with control mice (Fig. 2B,C,F). Remarkably, the number of these NSCs was greatly reduced by Notch3 deletion in the dorsolateral, lateral, and ventral walls of the SEZ, with this effect being less pronounced in the dorsal and medial walls at least with the sample size used (dorsolateral, p = 0.026, Cohen's d = 2.38; lateral, p = 0.0062, Cohen's d = 3.45; ventral, p = 0.023, Cohen's d = 2.44; medial, p = 0.19, Cohen's d = −1.12; dorsomedial, p = 0.11, Cohen's d = −1.44; dorsal, p = 0.44, Cohen's d = 0.637; two-tailed Student's t tests, n = 3 and 4 animals; Fig. 2F). This tendency was apparently consistent with the expression patterns of Notch3 (Fig. 1). Consistent with this observation, significantly fewer GFAP−EGFR+ TAPs and DCX+ immature neurons, the progeny of NSCs, were in Notch3−/− mice than in control mice (dorsolateral, p = 0.041, Cohen's d = 2.42; lateral, p = 0.021, Cohen's d = 3.01; ventral, p = 0.038, Cohen's d = 2.47; medial, p = 0.87, Cohen's d = 0.140; dorsomedial, p = 0.37, Cohen's d = −0.816; dorsal, p = 0.13, Cohen's d = 1.54; n = 3 and 3 animals; Fig. 2G; dorsolateral, p = 0.017, Cohen's d = 2.29; lateral, p = 0.0077, Cohen's d = 2.77; ventral, p = 0.049, Cohen's d = 1.73; medial, p = 0.62, Cohen's d = −0.359; dorsomedial, p = 0.25, Cohen's d = −0.880; dorsal, p = 0.11, Cohen's d = −1.30; two-tailed Student's t tests, n = 4 and 4 animals; Fig. 2H).
Figure 2.
Deletion of Notch3 resulted in domain-specific reduction of the total number of NSCs and their progenies. A, Hoechst staining of 2-month-old control (Notch3+/+) and Notch3-null (Notch3−/−) mice. The gross cytoarchitecture of the brain in these mice appeared normal. B, Schematic diagram of the SEZ subregions. C–E, Immunostaining of GFAP (green), Sox2 (red), and S100β (white; C); GFAP (green) and EGFR (red; D); and Dcx (red; E) on brain sections of 2-month-old control (Notch3+/+) and Notch3-null (Notch3−/−) mice. Images are from lateral region of the SEZ. Arrowheads indicate GFAP+Sox22+S100β− cells (C) and GFAP−EGFR+ cells (D). F–H, Quantification of GFAP+Sox22+S100β− NSCs (F), EGFR+GFAP− TAPs (G), and Dcx+ immature neurons (H). Scale bars: A, 1 mm; C, 5 μm; D, E, 10 μm. Quantitative data are mean ± SEM. n = 3 and 4 in F; 3 and 3 in G, 4 and 4 in H. Nuclei were stained with Hoechst (blue). Broken lines indicate the ventricular surface. Two-tailed Student's t test; *p < 0.05, **p < 0.01.
We then investigated which populations of GFAP+attachment+ NSCs were reduced in Notch3−/− mice. The number of GFAP+EGFR−attachment+ cells (qNSCs) was lower in Notch3−/− mice compared with control mice in the dorsolateral, lateral, and ventral walls of the SEZ (the interaction of genotypes × domain: F(5,24) = 6.40, p = 0.00065, two-way ANOVA; dorsolateral, p = 0.013, Cohen's d = 3.48; lateral, p = 0.032, Cohen's d = 2.62; ventral, p = 0.031, Cohen's d = 2.65; medial, p = 1, Cohen's d = 0; dorsomedial, p = 0.72, Cohen's d = −0.308; dorsal, p = 1, Cohen's d = 0; two-tailed Student's t tests, n = 3 and 3 animals; Fig. 3A,B). However, the number of GFAP+EGFR+attachment+ cells (aNSCs) was not reduced in Notch3−/− mice (dorsolateral, p = 0.64, Cohen's d = −0.408; lateral, p = 0.10, Cohen's d = 1.73; ventral, p = 0.56, Cohen's d = −0.516; medial, p = 1, Cohen's d = 0; dorsomedial, p = 0.37, Cohen's d = 0.816; dorsal, p = 1, Cohen's d = 0; two-tailed Student's t tests, n = 3 and 3 animals; Fig. 3A,C). Therefore, Notch3 appears to be important for the establishment or maintenance of qNSCs in the dorsolateral, lateral, and ventral parts of the SEZ. Next, we asked why neurogenesis was reduced even though the number of aNSCs did not decrease. One possible explanation is that qNSCs in Notch3−/− mice are apt to undergo abnormal activation but cannot complete the process for producing TAPs. We indeed observed the reduction of TAPs in Notch3−/− mice (Fig. 2G). Moreover, we found that the fraction of dividing cells among Ascl1+ cells (which include aNSCs and TAPs) was smaller in Notch3−/− mice than that in control mice (Fig. 3D,E). This further supports the notion that aberrantly activated NSCs and/or TAPs in Notch3−/− mice have a reduced proliferative capacity to produce TAPs.
Notch3 deletion reduces IdU LRCs in the adult SEZ
To confirm that qNSCs are indeed reduced by Notch3 deletion, we performed thymidine analog labeling of NSCs. Prolonged administration of thymidine analogs, such as IdU, labels NSCs that have passed the S phase. After a long chasing period, NSCs that have remained in a quiescent state are expected to retain IdU labeling and thus can be detected as LRCs (Doetsch et al., 1999). We thus administered IdU through drinking water for 2 weeks. A large proportion of IdU+Sox22+GFAP+ NSCs were distributed to the dorsolateral, lateral, and ventral walls, and fewer of them were found in the dorsal and medial walls of the SEZ. The number of IdU+Sox22+GFAP+ NSCs was lower in Notch3−/− mice compared with control mice in the lateral domain (total of dorsolateral, lateral, and ventral walls) of the SEZ (Fig. 4A–C). This is consistent with the result that the total number of NSCs in these SEZ domains was lower in Notch3−/− mice (Fig. 2F). Three weeks after the end of IdU administration, we detected IdU-retaining Sox22+GFAP+ NSCs in the SEZ and found that Notch3 deletion resulted in a significant decrease in the number of IdU+Sox22+GFAP+ LRCs in the SEZ (Fig. 4D,E). Importantly, although the number of initial IdU+Sox22+GFAP+ NSCs was already lower in Notch3−/− mice than in control mice just after IdU administration (before dilution), the extent of reduction by Notch3 deletion was even greater when IdU+Sox22+GFAP+ NSCs were counted 3 weeks after the end of IdU administration (after dilution; Cohen's d = 6.02 and 2.95, after and before dilution, respectively). This suggests that Notch3 deletion rather reduced qNSCs than aNSCs, which is consistent with data in Figure 3. Since these IdU-retaining cells had undergone ≥1 cell division at the adult stage and became quiescent after that, this result also suggests that Notch3 deletion suppressed the returning step of aNSCs into qNSCs, or enhanced the activation step of qNSCs into aNSCs, or both.
Figure 4.
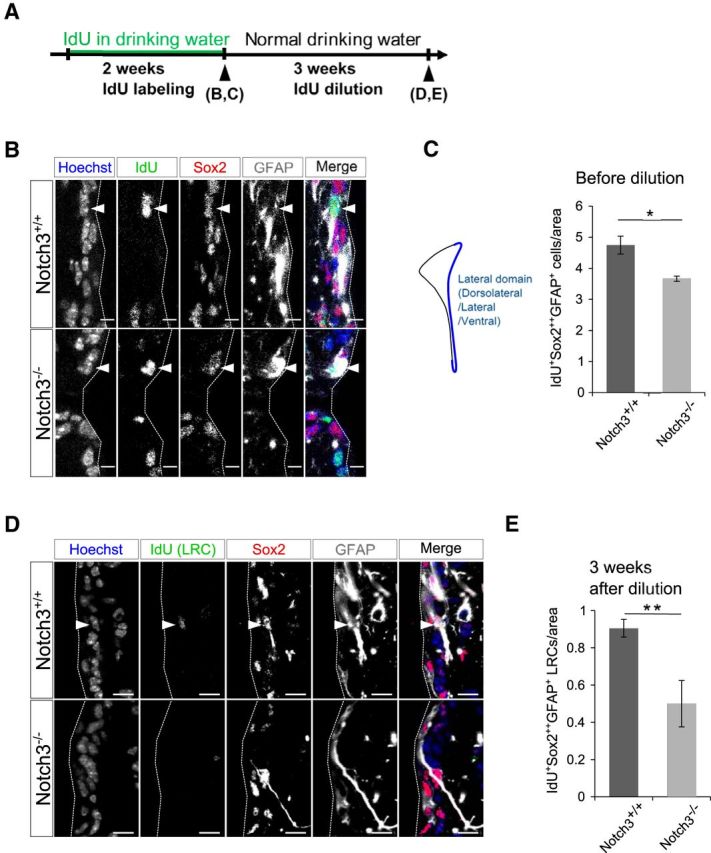
Notch3 deletion reduced LRCs. A, Experimental design. IdU was added to drinking water of 2-month-old control (Notch3+/+) and Notch3-null (Notch3−/−) mice. After 2 weeks, drinking water was changed to normal. Mice were killed just after 2 weeks IdU drinking or 3 weeks after change to normal drinking water. B, Immunostaining on brain sections of adult control (Notch3+/+) and Notch3-null (Notch3−/−) mice after 2 weeks IdU drinking. IdU, Green; Sox2, red; GFAP, white. Arrowheads indicates IdU+Sox22+GFAP+ cells. C, Quantification of IdU+Sox22+GFAP+ cells in dorsolateral, lateral, and ventral SEZ subregions just after 2 weeks of IdU drinking. D, Immunostaining on brain sections of adult control (Notch3+/+) and Notch3-null (Notch3−/−) mice after 3 weeks IdU dilution. IdU, Green; Sox2, red; GFAP, white. Arrowhead indicates IdU+Sox22+GFAP+ LRC. E, Quantification of IdU+Sox22+GFAP+ cells in dorsolateral, lateral, and ventral SEZ subregions after 3 weeks dilution. Scale bars: B, 5 μm; D, 10 μm. Quantitative data are mean ± SEM. n = 3 and 3. Nuclei were stained with Hoechst (blue). Broken lines indicate the ventricular surface. Two-tailed Student's t test; *p < 0.05, **p < 0.01.
Notch3 deletion reduces regenerative qNSCs without affecting their activation rate
To further investigate the effect of Notch3 deletion on qNSCs, we performed Ara-C treatment experiments. While actively dividing cells, such as aNSCs, TAPs, and neuroblasts, are killed by an infusion of the antimitotic drug Ara-C for 6 d, qNSCs survive and repopulate dividing cells after Ara-C treatment (Doetsch et al., 1999). Mice were thus treated with Ara-C for 6 d and then killed immediately after (0 h) or 12 h after Ara-C treatment, with EdU injected 1 h before the animal was killed (Fig. 5A). Almost all rapidly dividing aNSCs and immature precursors were eliminated after Ara-C treatment (0 h). We found that the number of NSCs (GFAP+Sox22+ cells with an apical attachment), which survived Ara-C treatment and therefore were quiescent during the treatment, was significantly reduced in the lateral domain of Notch3−/− mice (Fig. 5B,C). This observation confirmed the notion that Notch3 deletion reduces the number of qNSCs under the homeostatic condition. Twelve hours after Ara-C treatment, newly activated cells were labeled by EdU (Fig. 5D). The number of newly activated NSCs (GFAP+Sox22+ cells with an apical attachment) labeled by EdU was also decreased in Notch3−/− mice, particularly in the lateral domain compared with the dorsal or medial domains in the SEZ (lateral, p = 0.039, Cohen's d = 2.45; medial, p = 0.56, Cohen's d = 0.516; dorsal, p = 0.37, Cohen's d = 0.816; two-tailed Student's t tests, n = 3 and 3 animals; Fig. 5E). Interestingly, Notch3 deletion did not alter the percentage of EdU+ cells among NSCs, suggesting that, while Notch3 deletion decreased the number of qNSCs, it did not affect the activation step of qNSCs after injury (Fig. 5F). Our data thus suggest that Notch3 participates in regulation of the qNSC–aNSC transition under homeostatic but not regenerative conditions, with the reduction in the number of regenerative qNSCs observed in the Notch3 mutant after injury likely being due to a reduction in baseline qNSCs under the homeostatic condition.
Figure 5.
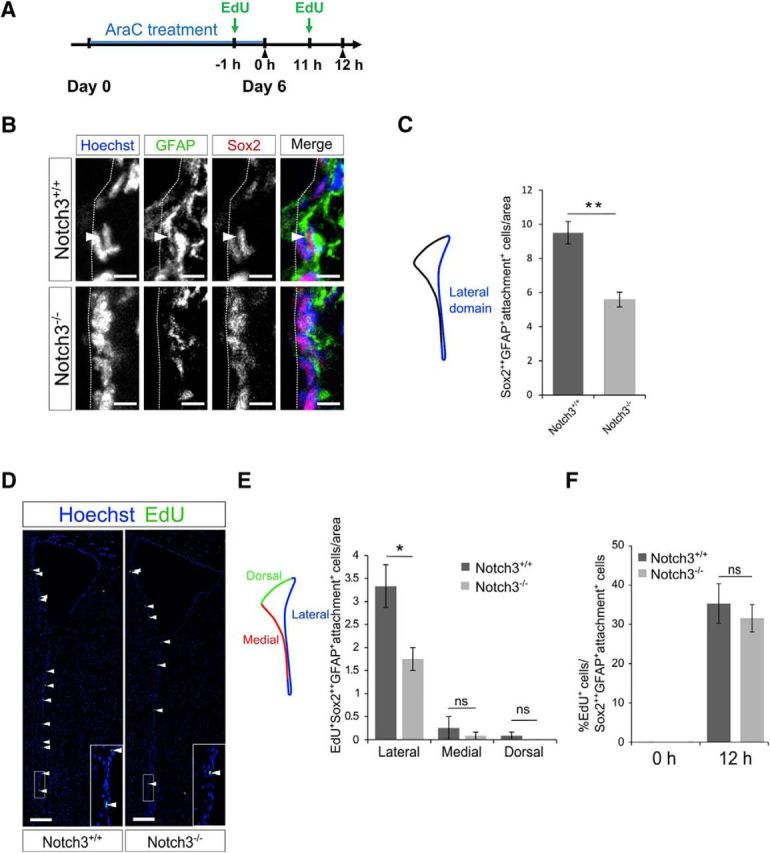
Notch3 deletion decreased qNSCs after Ara-C treatment. A, Experimental design. Ara-C was infused for 6 d into the brains of 2-month-old control (Notch3+/+) and Notch3-null (Notch3−/−) mice. Mice were killed just after (0 h) or 12 h after the pump removal. EdU was injected 1 h before the animals were killed. B, Immunostaining on brain sections of adult control (Notch3+/+) and Notch3-null (Notch3−/−) mice 12 h after the pump removal. GFAP, Green; Sox2, red. Arrowhead indicates Sox22+GFAP+attachment+ cells. C, Quantification of the number of Sox22+GFAP+attachment+ cells in the lateral domain (dorsolateral, lateral, and ventral SEZ subregions). The number of Sox22+GFAP+attachment+ cells was reduced in the Notch3−/− mice 12 h. D, Staining for EdU on the section of the indicated mice brain 12 h after the pump removal. Arrowheads indicate EdU+ cells. Inset is a high-magnification view of the boxed region. E, Quantification of EdU+Sox22+GFAP+attachment+ cells lining indicated domains in the SEZ. F, Percentage of EdU+ cells among Sox22+GFAP+attachment+ cells. Scale bars: B, 5 μm; D, 100 μm. Quantitative data are mean ± SEM. n = 3 and 3. Nuclei were stained with Hoechst (blue). Broken lines indicate the ventricular surface. Two-tailed Student's t test; *p < 0.05, **p < 0.01.
Notch3 suppresses the cell division of adult SEZ NSCs
Since Notch3−/− (null) mice were used for the above-mentioned experiments, we wondered whether Notch3 is needed for establishing qNSCs in the embryonic stage or for maintaining qNSCs in the adult stage. To address this, we acutely knocked down Notch3 in NSCs located at the lateral wall of the adult SEZ. Thus, by stereotaxic lentiviral injection to the lateral ventricle of 2-month-old mice, we infected NSCs with a lentivirus expressing an shRNA targeting Notch3 (shNotch3#1 or shNotch3#2) or an shRNA targeting luciferase (shControl#1) as a control (Fig. 6A). We found that, at 6 d after injection of these lentiviruses, the percentage of Ki67+ cells was greater among shNotch3#1-expressing or shNotch3#2-expressing (RFP+) GFAP+ NSCs than shControl#1-expressing (RFP+) GFAP+ NSCs (Fig. 6B–D). This suggests that acute reduction of Notch3 promotes activation of qNSCs in the adult SEZ and, in other words, that Notch3 is required for maintaining the quiescent state of NSCs in the adult stage. Moreover, although Notch3 is expressed also in the vasculature system, this result indicates that Notch3 expressed in nonvasculature SEZ cells (including NSCs) is indeed necessary for suppressing NSC division.
If Notch3 suppresses the division of qNSCs in the adult SEZ, acute knockdown of Notch3 might subsequently increase the number of NSCs. To examine this, we compared the percentage of NSCs expressing a control vector (GFP+) with those expressing a Notch3-knockdown vector (RFP+) 3 and 15 d after infection of a mixture of lentiviral vectors into the lateral ventricle of 2-month-old mice (Fig. 6E). We used two combinations of lentiviral mixtures: a lentivirus expressing shLuc#2 and GFP (shControl#2) and that expressing shNotch3#2 and RFP (shNotch3#2). The percentage of shNotch3-expressing RFP+GFAP+ NSCs among total infected (GFP+ and RFP+) GFAP+ NSCs became greater 15 d after infection compared with that 3 d after infection (Fig. 6F–H). Although we do not fully understand how much Notch3-dependent cell survival and self-renewal contribute to these data, our data suggest that acute reduction of Notch3 in the adult NSCs results in the increase of NSCs, supporting the notion that Notch3 is necessary for the suppression of NSC division in the adult SEZ.
Our results indicate that Notch3 is preferentially expressed in qNSCs and suppresses their division, in contrast to the proposed function of Notch1 in promoting the division of aNSCs (Basak et al., 2012). We then wondered whether Notch1 and Notch3 might have an intrinsic difference in cell-cycle regulation or other factors might cause apparently distinct functions of these Notch receptors. To address this question, we overexpressed the intracellular domain of Notch1 (NICD1) and that of Notch3 (NICD3) together with GFP in NPCs isolated from the embryonic GE. Notch is a transmembrane receptor and, upon activation, the Notch intracellular domain is released from the membrane and translocates into the nucleus, where it associates with RBP-J and induces the transcription of the target genes (Yoon and Gaiano, 2005; Pierfelice et al., 2011). NICD overexpression thus mimics the activation of Notch signaling. We found that overexpression of either NICD1 or NICD3 reduced the percentage of BrdU+ cells among GFP+Sox2+ cells (Fig. 6I). This suggests that a high-level expression of both activated Notch1 and Notch3 is capable of suppressing proliferation of NPCs at least under the condition used. Further investigation is needed to clarify the cause of apparently distinct functions of these Notch receptors.
Neuronal type-specific reduction of neurogenesis by Notch3 deletion
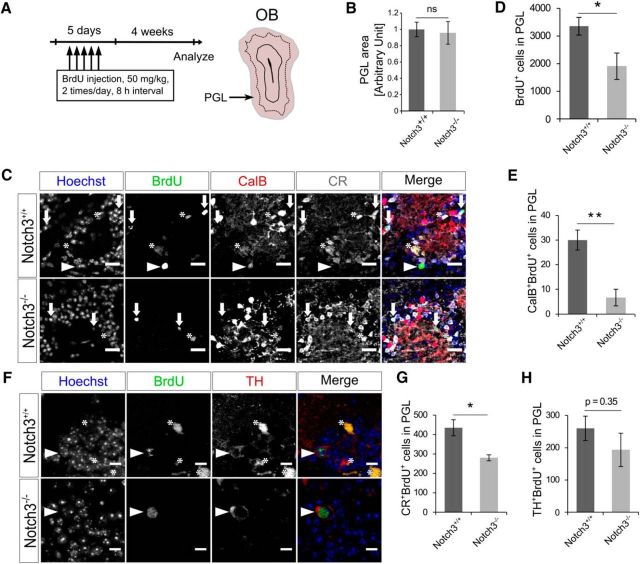
We next investigated how Notch3 deletion affects adult neurogenesis in the OB, which depends on the SEZ NSCs. To label newborn neurons, we injected BrdU twice per day for 5 consecutive days, and analyzed BrdU+ mature neurons in the OB 4 weeks later. Although the size of the PGL was not affected by Notch3 deletion (Fig. 7A,B), the number of BrdU+ PGCs in the OB of Notch3−/− mice was approximately half of that in control mice (Fig. 7C,D). We further examined the numbers of PGC subtypes, which are derived from different regions of the SEZ. We found that the deletion of Notch3 markedly reduced the number of CalB-expressing PGCs, which are preferentially derived from NSCs located in the lateral SEZ (Fig. 7C,E). By contrast, Notch3 deletion only slightly reduced the numbers of CR-expressing PGCs and no statistically significant reduction was observed in the numbers of TH-expressing PGCs (Fig. 7C,F–H), which are preferentially derived from NSCs located in the medial and dorsal SEZ, respectively (Merkle et al., 2007; Young et al., 2007; Fernández et al., 2011). These results indicate that Notch3 plays an important role in promoting adult neurogenesis in a neuronal subtype-specific manner. Although a direct causal link remains to be established, this subtype-specific function of Notch3 may be ascribable to its domain-specific expression patterns in the SEZ.
Figure 7.
Notch3 deletion resulted in neuronal type-specific reduction of newborn neuron in the OB. A, Experimental design. Two-month-old control (Notch3+/+) and Notch3-null (Notch3−/−) mice were injected BrdU twice per day for 5 consecutive days and PGL was analyzed 4 weeks after last injection. B, The size of the PGL of Notch3−/− is not changed from that of Notch3+/+. C, Immunostaining of BrdU (green), CalB (red), and CR (white) on OB sections of Notch3+/+ and Notch3−/− mice. Arrowheads indicate CalB+BrdU+ cells. Arrows indicate CR+BrdU+ cells. Asterisks indicate nonspecific signal of BrdU defined as signal out of nuclei. D, Quantification of BrdU+ cells in the PGL. E, Quantification of CalB+BrdU+ cells in the PGL. F, Immunostaining of BrdU and TH on OB sections. Arrowheads indicate TH+BrdU+ cells. Asterisks indicate nonspecific signals of BrdU defined as signals out of nuclei. G, H, Quantification of CR+BrdU+ cells (G) and TH+BrdU+ cells (H) in the PGL. Scale bars: C, 20 μm; E, 10 μm. Quantitative data are mean ± SEM. n = 4 and 3. Nuclei were stained with Hoechst (blue). Two-tailed Student's t test; *p < 0.05, **p < 0.01.
Discussion
In the adult SEZ, Notch signaling plays a pivotal role in maintaining the undifferentiated state of both qNSCs and aNSCs. However, Notch signaling has additional functions that differ for the two NSC states; it suppresses the cell cycle in qNSCs and promotes the cell cycle in aNSCs. However, it remained unclear how Notch signaling executes these apparently opposite functions in qNSCs and aNSCs. In this study, we demonstrated that Notch3 is preferentially expressed in qNSCs and that Notch3 deletion abrogates their quiescence and thus reduces the number of qNSCs. In contrast, we found that Notch1 is preferentially expressed in aNSCs and TAPs. Given that the loss of Notch1 was reported to reduce the proliferation of NSCs (Basak et al., 2012), the selective expression of different Notch-receptor isoforms in qNSCs and aNSCs appears to explain the differential roles of Notch signaling in these cells, at least in part.
What mechanism explains the differences between Notch1 and Notch3 knock-out phenotypes in NSCs? Although we found that high-level expression of either the activated form of Notch1 or Notch3 can suppress the division of NPCs in vitro, this does not exclude the possibility that Notch1 and Notch3 have distinct intrinsic properties that operate in the proper contexts. Whereas Notch1 and Notch3 activate common downstream effectors, such as RBP-J and Hes/Hey transcription factors, they also show differences in downstream signals. Indeed, the intracellular domains of these Notch isoforms show distinct target sequence preference (Ong et al., 2006). It is also possible that other signaling pathways involved in stem-cell quiescence or activation preferentially cooperate with one of the Notch-receptor isoforms. For example, EGFR and Ascl1 play central roles in promoting proliferation of NSCs in the adult SEZ and have been shown to interact with Notch signaling (Andreu-Agulló et al., 2009; Aguirre et al., 2010; Castro et al., 2011; Kim et al., 2011). Therefore, it would be interesting to investigate whether Notch1 and Notch3 differentially affect EGFR and Ascl1. Similarly, molecules involved in NSC quiescence, such as the cdk inhibitors p57 and p21, Id1, BMPR, PTEN, FoxO3, and NFIX, might also be differentially affected by Notch isoforms in the adult SEZ (Groszer et al., 2006; Renault et al., 2009; Mira et al., 2010; Martynoga et al., 2013; Porlan et al., 2013; Furutachi et al., 2015).
In addition to the qualitative differences between Notch3 and Notch1, differences in their expression levels and dynamics might be important for the distinct outcomes caused by their deletion. Previous reports have shown that, in embryonic NPCs, high Notch signaling suppresses proliferation while low Notch signaling promotes it, that oscillating Hes1 expression maintains proliferation, and that sustained Hes1 expression suppresses proliferation (Guentchev and McKay, 2006; Shimojo et al., 2008; Chapouton et al., 2010; Ninov et al., 2012). Therefore, it would be interesting to investigate the level and dynamics of Notch signaling in qNSCs and aNSCs in the adult SEZ and how different Notch-receptor isoforms contribute to them.
In addition to the differential expression of Notch3 depending on cell type and proliferative state, we observed regional differences in Notch3 expression. Notch3 is preferentially expressed in NSCs located in the lateral and ventral walls and is required for their maintenance. As NSCs are more abundant in the lateral and ventral walls than in the dorsal and medial walls of the adult SEZ (Fiorelli et al., 2015; Fig. 2), it is possible that the higher expression of Notch3 in the lateral and ventral walls may in part explain the increased abundance of NSCs between the SEZ domains.
NSCs reside in all domains—the dorsal, medial, and lateral walls—of the SEZ, but the subtypes of neurons they produce depend on their location. Our results indicate that Notch3 deletion dramatically reduces CalB+ PGCs but only mildly affects CR+ PGCs and has no statistically significant effect on TH+ PGCs (Fig. 7F–H). This is consistent with previous studies that show that CalB+ PGCs are preferentially produced in the lateral SEZ, TH+ PGCs in the dorsal SEZ, and CR+ PGCs in the medial SEZ (Merkle et al., 2007; Young et al., 2007; Fernández et al., 2011). Therefore, our results suggest that different Notch isoforms might be used to specify different neuronal subtypes that constitute the OB. Moreover, SEZ-derived neurons have also been reported to migrate to other brain regions, such as the striatum and the neocortex, under normal and regenerative conditions (Dayer et al., 2005; Ernst et al., 2014). It is possible that different Notch isoforms also differentially regulate the production of other neuron subtypes of other brain areas in addition to OBs.
Even in the lateral and ventral wall of the SEZ, where we observed the largest effects, the reduction of qNSCs by Notch3 deletion was only partial. Given that the deletion of RBP-J, a common effector of Notch signaling, caused a greater reduction of qNSCs in the same regions, it is possible that the various Notch isoforms have some redundancy in maintaining qNSCs.
We observed that Notch3 deletion caused reduction of the number of qNSCs and overall neurogenesis while the number of aNSCs was not significantly reduced (Figs. 3A–C, 7). One possible reason for this is that qNSCs in Notch3 knock-out mice are prone to undergo abnormal activation, but fewer TAPs are produced. We indeed observed the reduction of the number of TAPs in Notch3 knock-out mice (Fig. 2G). Moreover, we found that the fraction of dividing cells among Ascl1+ cells (which include aNSCs and TAPs) was smaller in Notch3 knock-out mice than that in control mice (Fig. 3D,E). These observations suggest that aberrantly activated NSCs and/or TAPs in Notch3 knock-out mice have a reduced proliferative capacity to produce TAPs. Therefore, reduction of neurogenesis may be caused in part by decreased proliferation of aNSCs and/or TAPs.
Our results shown here are consistent with those of a previous study (Alunni et al., 2013), which used zebrafish as a model system and elegantly demonstrated that Notch3 suppresses the cell cycle of adult NSCs and that Notch1 promotes their proliferation upon activation. Therefore, the regulatory mechanisms of different NSC pools by distinct Notch receptors appear to be evolutionally conserved.
In conclusion, we have provided evidence suggesting that Notch3 is preferentially expressed in qNSCs and plays a critical role in maintaining the appropriate number of qNSCs in the adult SEZ and in regulating genesis of a specific subset of neurons in the OB. Further characterization of the regulation of expression and activation of Notch-receptor isoforms may provide insights into fundamental aspects of adult NSCs, such as the balance between the maintenance and activation of NSCs that allows neurogenesis to continue for a long period into adulthood, and the domain-specific regulation of NSCs within the SEZ.
Footnotes
This work was supported by the Japan Society for the Promotion of Science (Grant #JP25-10011 and Grants-In-Aid for Scientific Research #JP15H05773 and #JP16H06481); the Japan Agency for Medical Research and Development (Advanced Research and Development Program for Medical Innovation); World Premier International Research Center Initiative (WPI), MEXT, Japan. We thank M. Okajima for assistance, and T. Koumura and members of the Gotoh laboratory for helpful discussions. We thank K. Inamori and I. Nikaido for technical advice, A. Miyajima and members of the Miyajima laboratory for their kind support.
The authors declare no competing financial interests.
References
- Aguirre A, Rubio ME, Gallo V (2010) Notch and EGFR pathway interaction regulates neural stem cell number and self-renewal. Nature 467:323–327. 10.1038/nature09347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alunni A, Krecsmarik M, Bosco A, Galant S, Pan L, Moens CB, Bally-Cuif L (2013) Notch3 signaling gates cell cycle entry and limits neural stem cell amplification in the adult pallium. Development 140:3335–3347. 10.1242/dev.095018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreu-Agulló C, Morante-Redolat JM, Delgado AC, Fariñas I (2009) Vascular niche factor PEDF modulates Notch-dependent stemness in the adult subependymal zone. Nat Neurosci 12:1514–1523. 10.1038/nn.2437 [DOI] [PubMed] [Google Scholar]
- Androutsellis-Theotokis A, Leker RR, Soldner F, Hoeppner DJ, Ravin R, Poser SW, Rueger MA, Bae SK, Kittappa R, McKay RD (2006) Notch signalling regulates stem cell numbers in vitro and in vivo. Nature 442:823–826. 10.1038/nature04940 [DOI] [PubMed] [Google Scholar]
- Basak O, Giachino C, Fiorini E, Macdonald HR, Taylor V (2012) Neurogenic subventricular zone stem/progenitor cells are Notch1-dependent in their active but not quiescent state. J Neurosci 32:5654–5666. 10.1523/JNEUROSCI.0455-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro DS, Martynoga B, Parras C, Ramesh V, Pacary E, Johnston C, Drechsel D, Lebel-Potter M, Garcia LG, Hunt C, Dolle D, Bithell A, Ettwiller L, Buckley N, Guillemot F (2011) A novel function of the proneural factor Ascl1 in progenitor proliferation identified by genome-wide characterization of its targets. Genes Dev 25:930–945. 10.1101/gad.627811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaker Z, Codega P, Doetsch F (2016) A mosaic world: puzzles revealed by adult neural stem cell heterogeneity. Wiley Interdiscip Rev Dev Biol 5:640–658. 10.1002/wdev.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapouton P, Skupien P, Hesl B, Coolen M, Moore JC, Madelaine R, Kremmer E, Faus-Kessler T, Blader P, Lawson ND, Bally-Cuif L (2010) Notch activity levels control the balance between quiescence and recruitment of adult neural stem cells. J Neurosci 30:7961–7974. 10.1523/JNEUROSCI.6170-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codega P, Silva-Vargas V, Paul A, Maldonado-Soto AR, Deleo AM, Pastrana E, Doetsch F (2014) Prospective identification and purification of quiescent adult neural stem cells from their in vivo niche. Neuron 82:545–559. 10.1016/j.neuron.2014.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, Yoon K, Wang M, Gaiano N (2006) Notch3 signaling promotes radial glial/progenitor character in the mammalian telencephalon. Dev Neurosci 28:58–69. 10.1159/000090753 [DOI] [PubMed] [Google Scholar]
- Dayer AG, Cleaver KM, Abouantoun T, Cameron HA (2005) New GABAergic interneurons in the adult neocortex and striatum are generated from different precursors. J Cell Biol 168:415–427. 10.1083/jcb.200407053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daynac M, Chicheportiche A, Pineda JR, Gauthier LR, Boussin FD, Mouthon MA (2013) Quiescent neural stem cells exit dormancy upon alteration of GABAAR signaling following radiation damage. Stem Cell Res 11:516–528. 10.1016/j.scr.2013.02.008 [DOI] [PubMed] [Google Scholar]
- Doetsch F, Caillé I, Lim DA, García-Verdugo JM, Alvarez-Buylla A (1999) Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell 97:703–716. 10.1016/S0092-8674(00)80783-7 [DOI] [PubMed] [Google Scholar]
- Doetsch F, Petreanu L, Caille I, Garcia-Verdugo JM, Alvarez-Buylla A (2002) EGF converts transit-amplifying neurogenic precursors in the adult brain into multipotent stem cells. Neuron 36:1021–1034. 10.1016/S0896-6273(02)01133-9 [DOI] [PubMed] [Google Scholar]
- Domenga V, Fardoux P, Lacombe P, Monet M, Maciazek J, Krebs LT, Klonjkowski B, Berrou E, Mericskay M, Li Z, Tournier-Lasserve E, Gridley T, Joutel A (2004) Notch3 is required for arterial identity and maturation of vascular smooth muscle cells. Genes Dev 18:2730–2735. 10.1101/gad.308904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehret F, Vogler S, Pojar S, Elliott DA, Bradke F, Steiner B, Kempermann G (2015) Mouse model of CADASIL reveals novel insights into Notch3 function in adult hippocampal neurogenesis. Neurobiol Dis 75:131–141. 10.1016/j.nbd.2014.12.018 [DOI] [PubMed] [Google Scholar]
- Ernst A, Alkass K, Bernard S, Salehpour M, Perl S, Tisdale J, Possnert G, Druid H, Frisén J (2014) Neurogenesis in the striatum of the adult human brain. Cell 156:1072–1083. 10.1016/j.cell.2014.01.044 [DOI] [PubMed] [Google Scholar]
- Fernández ME, Croce S, Boutin C, Cremer H, Raineteau O (2011) Targeted electroporation of defined lateral ventricular walls: a novel and rapid method to study fate specification during postnatal forebrain neurogenesis. Neural Dev 6:13. 10.1186/1749-8104-6-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorelli R, Azim K, Fischer B, Raineteau O (2015) Adding a spatial dimension to postnatal ventricular-subventricular zone neurogenesis. Development 142:2109–2120. 10.1242/dev.119966 [DOI] [PubMed] [Google Scholar]
- Furutachi S, Miya H, Watanabe T, Kawai H, Yamasaki N, Harada Y, Imayoshi I, Nelson M, Nakayama KI, Hirabayashi Y, Gotoh Y (2015) Slowly dividing neural progenitors are an embryonic origin of adult neural stem cells. Nat Neurosci 18:657–665. 10.1038/nn.3989 [DOI] [PubMed] [Google Scholar]
- Groszer M, Erickson R, Scripture-Adams DD, Dougherty JD, Le Belle J, Zack JA, Geschwind DH, Liu X, Kornblum HI, Wu H (2006) PTEN negatively regulates neural stem cell self-renewal by modulating G0–G1 cell cycle entry. Proc Natl Acad Sci U S A 103:111–116. 10.1073/pnas.0509939103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guentchev M, McKay RD (2006) Notch controls proliferation and differentiation of stem cells in a dose-dependent manner. Eur J Neurosci 23:2289–2296. 10.1111/j.1460-9568.2006.04766.x [DOI] [PubMed] [Google Scholar]
- Hirabayashi Y, Suzki N, Tsuboi M, Endo TA, Toyoda T, Shinga J, Koseki H, Vidal M, Gotoh Y (2009) Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron 63:600–613. 10.1016/j.neuron.2009.08.021 [DOI] [PubMed] [Google Scholar]
- Ihrie RA, Shah JK, Harwell CC, Levine JH, Guinto CD, Lezameta M, Kriegstein AR, Alvarez-Buylla A (2011) Persistent sonic hedgehog signaling in adult brain determines neural stem cell positional identity. Neuron 71:250–262. 10.1016/j.neuron.2011.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imayoshi I, Sakamoto M, Yamaguchi M, Mori K, Kageyama R (2010) Essential roles of Notch signaling in maintenance of neural stem cells in developing and adult brains. J Neurosci 30:3489–3498. 10.1523/JNEUROSCI.4987-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi D, Furutachi S, Kawai H, Hozumi K, Gotoh Y (2013) Dll1 maintains quiescence of adult neural stem cells and segregates asymmetrically during mitosis. Nat Commun 4:1880. 10.1038/ncomms2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EJ, Ables JL, Dickel LK, Eisch AJ, Johnson JE (2011) Ascl1 (Mash1) defines cells with long-term neurogenic potential in subgranular and subventricular zones in adult mouse brain. PLoS One 6:e18472. 10.1371/journal.pone.0018472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamoto T, Hanaoka K (2010) Notch3 null mutation in mice causes muscle hyperplasia by repetitive muscle regeneration. Stem Cells 28:2205–2216. 10.1002/stem.547 [DOI] [PubMed] [Google Scholar]
- Kitamoto T, Takahashi K, Takimoto H, Tomizuka K, Hayasaka M, Tabira T, Hanaoka K (2005) Functional redundancy of the Notch gene family during mouse embryogenesis: analysis of Notch gene expression in Notch3-deficient mice. Biochem Biophys Res Commun 331:1154–1162. 10.1016/j.bbrc.2005.03.241 [DOI] [PubMed] [Google Scholar]
- Kriegstein A, Alvarez-Buylla A (2009) The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci 32:149–184. 10.1146/annurev.neuro.051508.135600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafkas D, Rodilla V, Huyghe M, Mourao L, Kiaris H, Fre S (2013) Notch3 marks clonogenic mammary luminal progenitor cells in vivo. J Cell Biol 203:47–56. 10.1083/jcb.201307046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Juárez A, Howard J, Ullom K, Howard L, Grande A, Pardo A, Waclaw R, Sun YY, Yang D, Kuan CY, Campbell K, Nakafuku M (2013) Gsx2 controls region-specific activation of neural stem cells and injury-induced neurogenesis in the adult subventricular zone. Genes Dev 27:1272–1287. 10.1101/gad.217539.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martynoga B, Mateo JL, Zhou B, Andersen J, Achimastou A, Urbán N, van den Berg D, Georgopoulou D, Hadjur S, Wittbrodt J, Ettwiller L, Piper M, Gronostajski RM, Guillemot F (2013) Epigenomic enhancer annotation reveals a key role for NFIX in neural stem cell quiescence. Genes Dev 27:1769–1786. 10.1101/gad.216804.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle FT, Mirzadeh Z, Alvarez-Buylla A (2007) Mosaic organization of neural stem cells in the adult brain. Science 317:381–384. 10.1126/science.1144914 [DOI] [PubMed] [Google Scholar]
- Mira H, Andreu Z, Suh H, Lie DC, Jessberger S, Consiglio A, San Emeterio J, Hortigüela R, Marqués-Torrejón MA, Nakashima K, Colak D, Götz M, Fariñas I, Gage FH (2010) Signaling through BMPR-IA regulates quiescence and long-term activity of neural stem cells in the adult hippocampus. Cell Stem Cell 7:78–89. 10.1016/j.stem.2010.04.016 [DOI] [PubMed] [Google Scholar]
- Nam HS, Benezra R (2009) High levels of Id1 expression define B1 type adult neural stem cells. Cell Stem Cell 5:515–526. 10.1016/j.stem.2009.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninov N, Borius M, Stainier DY (2012) Different levels of Notch signaling regulate quiescence, renewal and differentiation in pancreatic endocrine progenitors. Development 139:1557–1567. 10.1242/dev.076000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyfeler Y, Kirch RD, Mantei N, Leone DP, Radtke F, Suter U, Taylor V (2005) Jagged1 signals in the postnatal subventricular zone are required for neural stem cell self-renewal. EMBO J 24:3504–3515. 10.1038/sj.emboj.7600816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong CT, Cheng HT, Chang LW, Ohtsuka T, Kageyama R, Stormo GD, Kopan R (2006) Target selectivity of vertebrate notch proteins: collaboration between discrete domains and CSL-binding site architecture determines activation probability. J Biol Chem 281:5106–5119. 10.1074/jbc.M506108200 [DOI] [PubMed] [Google Scholar]
- Pastrana E, Cheng LC, Doetsch F (2009) Simultaneous prospective purification of adult subventricular zone neural stem cells and their progeny. Proc Natl Acad Sci U S A 106:6387–6392. 10.1073/pnas.0810407106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul A, Chaker Z, Doetsch F (2017) Hypothalamic regulation of regionally distinct adult neural stem cells and neurogenesis. Science 356:1383–1386. 10.1126/science.aal3839 [DOI] [PubMed] [Google Scholar]
- Pierfelice T, Alberi L, Gaiano N (2011) Notch in the vertebrate nervous system: an old dog with new tricks. Neuron 69:840–855. 10.1016/j.neuron.2011.02.031 [DOI] [PubMed] [Google Scholar]
- Porlan E, Morante-Redolat JM, Marqués-Torrejón MÁ, Andreu-Agulló C, Carneiro C, Gómez-Ibarlucea E, Soto A, Vidal A, Ferrón SR, Fariñas I (2013) Transcriptional repression of Bmp2 by p21(Waf1/Cip1) links quiescence to neural stem cell maintenance. Nat Neurosci 16:1567–1575. 10.1038/nn.3545 [DOI] [PubMed] [Google Scholar]
- Renault VM, Rafalski VA, Morgan AA, Salih DA, Brett JO, Webb AE, Villeda SA, Thekkat PU, Guillerey C, Denko NC, Palmer TD, Butte AJ, Brunet A (2009) FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell 5:527–539. 10.1016/j.stem.2009.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto M, Imayoshi I, Ohtsuka T, Yamaguchi M, Mori K, Kageyama R (2011) Continuous neurogenesis in the adult forebrain is required for innate olfactory responses. Proc Natl Acad Sci U S A 108:8479–8484. 10.1073/pnas.1018782108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto M, Ieki N, Miyoshi G, Mochimaru D, Miyachi H, Imura T, Yamaguchi M, Fishell G, Mori K, Kageyama R, Imayoshi I (2014) Continuous postnatal neurogenesis contributes to formation of the olfactory bulb neural circuits and flexible olfactory associative learning. J Neurosci 34:5788–5799. 10.1523/JNEUROSCI.0674-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasagawa Y, Nikaido I, Hayashi T, Danno H, Uno KD, Imai T, Ueda HR (2013) Quartz-Seq: a highly reproducible and sensitive single-cell RNA-Seq reveals non-genetic gene expression heterogeneity. Genome Biol 14:R31. 10.1186/gb-2013-14-4-r31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojo H, Ohtsuka T, Kageyama R (2008) Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron 58:52–64. 10.1016/j.neuron.2008.02.014 [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Mori K (2005) Critical period for sensory experience-dependent survival of newly generated granule cells in the adult mouse olfactory bulb. Proc Natl Acad Sci U S A 102:9697–9702. 10.1073/pnas.0406082102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon K, Gaiano N (2005) Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat Neurosci 8:709–715. 10.1038/nn1475 [DOI] [PubMed] [Google Scholar]
- Young KM, Fogarty M, Kessaris N, Richardson WD (2007) Subventricular zone stem cells are heterogeneous with respect to their embryonic origins and neurogenic fates in the adult olfactory bulb. J Neurosci 27:8286–8296. 10.1523/JNEUROSCI.0476-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]