Abstract
Motivation
Genetic analysis of cancer regularly includes two or more samples from the same patient. Somatic copy number alterations leading to allelic imbalance (AI) play a critical role in cancer initiation and progression. Directional analysis and visualization of the alleles in imbalance in multi-sample settings allow for inference of recurrent mutations, providing insights into mutation rates, clonality and the genomic architecture and etiology of cancer.
Results
The REpeat Chromosomal changes Uncovered by Reflection (RECUR) is an R application for the comparative analysis of AI profiles derived from SNP array and next-generation sequencing data. The algorithm accepts genotype calls and ‘B allele’ frequencies (BAFs) from at least two samples derived from the same individual. For a predefined set of genomic regions with AI, RECUR compares BAF values among samples. In the presence of AI, the expected value of a BAF can shift in two possible directions, reflecting an increased or decreased abundance of the maternal haplotype, relative to the paternal. The phenomenon of opposite haplotype shifts, or ‘mirrored subclonal allelic imbalance’, is a form of heterogeneity, and has been linked to clinico-pathological features of cancer. RECUR detects such genomic segments of opposite haplotypes in imbalance and plots BAF values for all samples, using a two-color scheme for intuitive visualization.
Availability and implementation
RECUR is available as an R application. Source code and documentation are available at scheet.org.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Large somatic chromosomal alterations are commonly observed in cancer genomes and appear to have a profound effect on the development and progression of the disease (Ding et al., 2018). Allelic imbalance (AI) resulting from such changes (gain, loss, or copy-neutral loss-of-heterozygosity; cnLOH) is defined as a deviation from the 1:1 ratio of inherited parental haplotypes (Supplementary Fig. S1A). Observed ‘B allele’ frequencies (BAFs) at germline heterozygous loci, from SNP array or next-generation sequencing data, are used to detect AI (Gonzalez et al., 2011; Lamy et al., 2007; Lucas et al., 2016; Olshen et al., 2004, 2011; Staaf et al., 2008; Vattathil et al., 2013). In addition, log R ratio (LRR) or read-depth data, can be analyzed alone or jointly with BAFs to detect gains and losses. When AI is detected in multiple intra-individual samples, spanning similar loci, it is natural to assess whether these signals reflect the same underlying mutation. One way to do this would be to note if the observed AI differs in mutation type (gain, loss), an approach that falls short for events of the same type and subtle AI events that cannot be classified as gains, losses, or cnLOH. In such a case, or even with two mutations of the same type, samples may differ in their maternal/paternal haplotype balance and consequently their alleles will shift in opposite directions (Supplementary Fig. S1B). This phenomenon is indicative of a recurrent or independent mutation (inverse correlation in shifts half the time), or an error in chromosome segregation that generates two ‘mirrored’ clones (would always generate such a pattern), one with a gain and the other with a loss. Information regarding genomic segments with opposite AI haplotypes deviations (‘mirrored AI’), can enrich the analysis of tumor clonal evolution and genomic instability.
Recent studies have used such a directional AI analysis to yield important insights into disease initiation and progression (Jakubek et al., 2016; Jamal-Hanjani et al., 2017; Turajlic et al., 2018). For example, multiple independent chromosome 9 copy number alterations were observed in lung tumor clones and the surrounding normal-appearing tissues, shedding light on particular forms of genomic instability or necessity of mutation in a particular pathway (Jakubek et al., 2016; Jamal-Hanjani et al., 2017). Indeed, studies of intra-tumor heterogeneity of kidney demonstrated selection for 9p loss in clear-cell renal cell carcinoma (Turajlic et al., 2018). Yet, these studies did not provide a formal method for directional AI analysis. While there are methods that test for the presence of specific allele fractions in tumor-normal pairs, these do not explicitly test directionality of AI from multiple samples with overlapping AI segments (Lamy et al., 2007; Olshen et al., 2011).
To address this, we developed REpeat Chromosomal changes Uncovered by Reflection (RECUR) to provide the scientific community with a formal and flexible tool for inference of subclonal or divergent mutations and visualization of directional AI in studies of tumor heterogeneity and other multi-sample studies.
2 Features
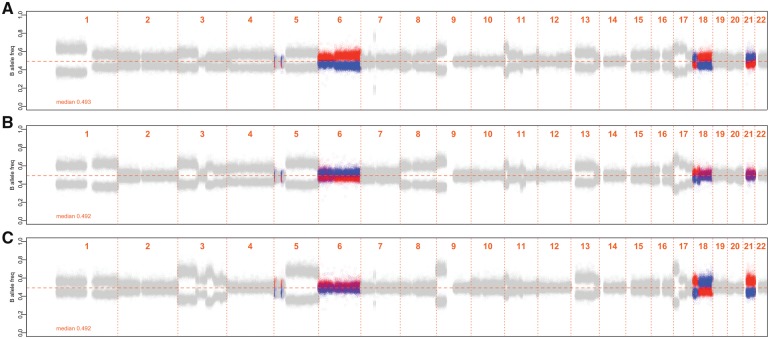
RECUR is an R application that requires genotype calls and BAF data from two or more samples from the same individual. Germline genotypes may be provided directly or by specifying a representative sample (e.g. blood) and must include genomic coordinates. LRR/read-depth data is optional, but can be included in the input files. The application generates two outputs: (i) a list of genomic segments where two or more sample pairs exhibit statistically significant bi-directional AI shifts; and (ii) one graphic display per sample comprised of a BAF plot for bidirectional AI visualization (Fig. 1A). These displays facilitate visual comparison of events across an individual’s samples (Fig. 1).
Fig. 1.
RECUR output for three samples derived from the same patient. One sample is used as the reference at each genomic segment (‘R-HAP’) with red BAF values indicating upwards shifts and blue values indicating downwards BAF shifts in the reference. BAF values for all samples, at the genomic segment are colored using the same directional scheme as the reference. Genomic segments with no detectable mirrored AI segments are gray. Information on the data used to generate this figure is found in the Supplementary Note
2.1 Bidirectional AI shift inference
The user can pre-define genomic segments for bidirectional AI testing, such as detected copy number or AI calls from at least one of the samples; alternatively, the user can provide a list comprised of all chromosome arms. For each segment, the algorithm identifies a ‘reference’ over-represented haplotype (‘R-hap’; Supplementary Fig. S1B). For each heterozygous marker within the event region, the algorithm first determines absolute BAF deviations from the expected value of 0.5, and then calculates the median deviation for the event region. R-hap is then generated from the sample with the largest median BAF deviation. In this approach, we use the BAF values to construct R-hap as the sequence of putatively over-represented alleles (i.e. B for BAF >= 0.5 and A otherwise). Similarly, for the non-reference samples from the same individual, an over-represented haplotype is identified and its alleles are compared to those of R-hap using Pearson’s correlation (e.g. B as 1 and A as 0). By default, events that are significantly anti-correlated (R < 0; P < 1*10−4, which is a parameter) are reported.
2.2 Bidirectional AI shift visualization
One graphic is generated per input sample (Fig. 1). The graphic includes a plot of BAFs ordered by genomic coordinate and an optional LRR/read-depth plot. In segments with statistically significant mirrored AI, markers that are ‘B’ in R-hap are plotted using red points and ‘A’ markers are plotted using blue points (Fig. 1 and Supplementary Fig. S1B). The color scheme for the markers is constant across samples. Therefore, for samples exhibiting mirrored AI, relative to the R-hap sample, blue points will generally be higher than the red points (Fig. 1). The differentiation between red and blue bands depends on the fraction of aberrant cells, sequencing technology, and event type (Supplementary Note).
2.3 Additional features
The application can take output directly from the AI detection algorithms hapLOH and hapLOHseq (Lucas et al., 2016; Vattathil and Scheet, 2013). RECUR can also run with a ‘window’ testing option. In this mode, the algorithm will test windows of N heterozygous markers at each AI segment, where N is user-specified (Supplementary Note). This feature is intended to help detect or refine boundaries of bidirectional AI in genomic regions with complex or multiple/overlapping chromosome aberrations.
3 Concluding remarks
Tumor and non-cancer genomic studies comprised of two or more samples (multiple core-needle biopsies, tumor-metastasis pairs, and single cell methods) have become increasingly common. We developed RECUR as a comprehensive and flexible tool for bidirectional AI profiling in this type of data set. These data are intended to complement genetic heterogeneity studies. The current version of the method is agnostic to event type; however, integration of gain/loss status with directional AI information can help elucidate biological mechanisms driving chromosomal aberrations. It can help identify genomic regions under selective pressure, for example recurrent deletions/gains in the same genomic loci and/or regions with generalized genomic instability. Integration of directional AI, copy number, and somatic mutation data can help build more accurate phylogenetic trees and further illuminate the timing and distribution of somatic chromosomal aberrations to offer insights into disease initiation and progression.
Funding
This work was supported by NIH grants R25CA057730, R01GM081441 and R01HG005855 and by the Cancer Prevention Research Inst. of Texas awards RP150079 and RP160668. Study content is solely the responsibility of the authors and does not necessarily represent the official views of any funding agency.
Conflict of Interest: none declared.
Supplementary Material
References
- Ding L. et al. (2018) Perspective on oncogenic processes at the end of the beginning of cancer genomics. Cell, 173, 305+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez J.R. et al. (2011) A fast and accurate method to detect allelic genomic imbalances underlying mosaic rearrangements using SNP array data. BMC Bioinformatics, 12, 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubek Y. et al. (2016) Genomic landscape established by allelic imbalance in the cancerization field of a normal appearing airway. Cancer Res., 76, 3676–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamal-Hanjani M. et al. (2017) Tracking the evolution of non-small-cell lung cancer. N Engl J Med., 376, 2109–2121. [DOI] [PubMed] [Google Scholar]
- Lamy P. et al. (2007) A hidden Markov model to estimate population mixture and allelic copy-numbers in cancers using affymetrix SNP arrays. BMC Bioinformatics, 8, 434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas F.A.S. et al. (2016) Rapid and powerful detection of subtle allelic imbalance from exome sequencing data with hapLOHseq. Bioinformatics, 32, 3015–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormand J.R., Prince P.J. (1980) A family of embedded Runge–Kutta formulae. J. Comp. Appl. Math., 6, 19–26. [Google Scholar]
- Olshen A.B. et al. (2011) Parent-specific copy number in paired tumor-normal studies using circular binary segmentation. Bioinformatics, 27, 2038–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshen A.B. et al. (2004) Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics, 5, 557–572. [DOI] [PubMed] [Google Scholar]
- Staaf J. et al. (2008) Segmentation-based detection of allelic imbalance and loss-of-heterozygosity in cancer cells using whole genome SNP arrays. Genome Biol., 9, R136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turajlic S. et al. (2018) Tracking cancer evolution reveals constrained routes to metastases: tRACERx renal. Cell, 173, 581+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vattathil S., Scheet P. (2013) Haplotype-based profiling of subtle allelic imbalance with SNP arrays. Genome Res., 23, 152–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.