Abstract
Nucleic acids from bacteria or viruses induce potent immune responses in infected cells1–4. The detection of pathogen-derived nucleic acids is a central strategy by which the host senses infection and initiates protective immune responses5,6. Cyclic GMP-AMP synthase (cGAS) is a double-stranded DNA sensor7,8. It catalyzes the synthesis of cyclic GMP-AMP (cGAMP)9–12, which stimulates the induction of type I interferons (IFN-Is) through the STING-TBK1-IRF-3 signaling axis13–15. Stimulator of interferon genes (STING) oligomerizes upon cGAMP binding, leading to the recruitment and activation of tank-binding kinase 1 (TBK1)8,16. Interferon regulatory factor 3 (IRF-3) is then recruited to the signaling complex and activated by TBK18,17–20. Phosphorylated IRF-3 translocates to the nucleus and initiates the expression of IFN-Is21. However, the precise mechanisms governing STING activation by cGAMP and subsequent TBK1 activation by STING remained poorly understood. Here we show that a conserved PLPLRT/SD motif within the C-terminal tail of STING mediates the recruitment and activation of TBK1. Crystal structures of TBK1 bound to STING reveal that the PLPLRT/SD motif binds to the dimer interface of TBK1. Cell-based studies confirm that the direct interaction between TBK1 and STING is essential for IFN-β induction upon cGAMP stimulation. Moreover, we show that full-length STING oligomerizes upon cGAMP binding and highlight this as an essential step in the activation of STING-mediated signaling.
Mass spectrometry showed that several residues within STING C-terminal tail (CTT) are phosphorylated by TBK1 (Extended data Fig. 1a, b). To investigate the roles of these residues, we generated several STING mutants and conducted IFN-β luciferase reporter assays. After stimulation with cGAMP, the reporter is activated in cells transfected with wild-type STING (WT), but not in the control cells (Fig.1a). Although the expression of STING alone induces 1–2 fold induction of the reporter, stimulation with cGAMP induces ~10 times higher signals (Fig. 1a, Extended Data Fig. 1c). Consistent with previous studies19, mutation S366A abolishes IFN-β reporter activation and IRF-3 phosphorylation (Fig.1a, b). In contrast, mutations T376A or S379A do not affect TBK1 or IRF-3 activation (Fig. 1b). Strikingly, deletion of the nine C-terminal residues of STING (ΔC9) abolishes the reporter activation and IRF-3 phosphorylation (Fig. 1a, b). Interestingly, deletion of these residues also impairs TBK1 activation (Fig. 1b). Confocal microscopy revealed that truncated STING mutant still translocates to perinuclear punctate structures upon cGAMP stimulation (Fig. 1c, Extended Data Fig. 1d). However, TBK1 does not co-translocate into these puncta with ΔC9 STING (Fig. 1c). Furthermore, immunoprecipitation assays indicate that both WT STING and the S366A mutant bind TBK1; however, deletion of the nine C-terminal residues abolishes TBK1 binding and phosphorylation (Fig. 1d).
Fig. 1. The nine C-terminal residues of STING are critical for STING-mediated signaling and TBK1 binding.
a, IFN-β luciferase reporter assays in HEK293T cells. The cells were transfected with the indicated pcDNA3.1-hSTING plasmid and stimulated with cGAMP. Luciferase signals from stimulated cells are indicated by orange bars and unstimulated controls by blue bars. The data (mean ± S.E.M) are representative of three independent experiments. Each dot represents a technical replicate (n = 3). The p values were calculated by two-tailed Student’s t test: * p < 0.05, ** p < 0.01, *** p < 0.001, NS - not significant.
b, Western blot analyses of cells transfected with STING plasmids containing mutations at the phosphorylation sites and deletion of the nine C-terminal residues.
c, Confocal microscopy images of HEK293T cells transfected with wild type or truncated STING with and without cGAMP stimulation. Localization of STING and TBK1 is shown. Scale bars denote 5 μm.
d, Immunoprecipitation showing interactions between STING mutants and TBK1. HEK293T cells were transfected with mutants of Flag-STING and stimulated with cGAMP. Total and phosphorylated IRF-3 and TBK1 were detected with IRF-3 and TBK1 antibodies, respectively. STING was visualized with FLAG antibody.
e, f, SPR binding studies of phosphorylated human STING C-terminal domain (CTD, residue 155 to 379), ΔC9 (residue 155 to 370), cGAMP-binding domain (residue 155 to 341) and C-terminal tail (residue 342 to 379) with human TBK1 (upper panel) in presence of 1 μM cGAMP. The binding affinity (Kd) was determined by fitting the binding data to a one-site binding model (lower panel).The data of b-f are representative of three independent experiments.
To test if STING binds TBK1 directly, we expressed several biotin-labeled STING truncation mutants (Extended Data Fig. 2a, b) and conducted TBK1 binding studies by SPR (Extended Data Table 1). We observed that unphosphorylated STING binds TBK1 directly with relatively low affinity (Extended Data Fig. 2c, d). The binding affinity does not change significantly in the presence of cGAMP (Extended Data Fig. 2e, f). However, upon phosphorylation, STING binds to TBK1 at almost 20 times higher affinity (Fig. 1e, Extended Data Fig. 2g). The binding affinity does not change significantly in the absence of cGAMP (Extended Data Fig. 2h, i). In addition, phosphorylation of TBK1 does not affect its binding affinity with STING (Extended Data Fig. 2j). To map the TBK1 binding site, we conducted binding studies with several truncated forms of STING (Extended Data Fig. 2a, b). We observed that truncation of the C-terminal 9 or 37 residues of STING abolish TBK1 binding (Fig. 1e, f, Extended Data Fig. 2g, k). In contrast, peptides containing the C-terminal 37 or 14 residues of STING are fully capable of binding TBK1 (Fig. 1f, Extended Data Fig. 2k, l).
To determine the exact STING residues that contribute to TBK1 binding, we generated a number of STING mutants (Fig. 2a, Extended data Fig. 3a) and conducted binding studies (Fig. 2a, Extended Data Fig. 3b to l). These studies showed that mutation L374A disrupts TBK1 binding (Fig. 2a, Extended Data Fig. 3f). However, this mutation does not affect IRF−3 binding (Extended Data Fig. 3m). In addition, mutations of R375A and F378A reduce the binding affinity by 30- and 20-fold, respectively (Fig. 2a, Extended Data Fig. 3g, j). Mutations of L372A and P373A reduce the binding affinity by about 10-fold (Fig. 2a, Extended Data Fig. 3d, e). Mutations P371A, T376A, D377A and S379A only reduce the binding affinity by 3- to 6-fold (Fig. 2a, Extended Data Fig. 3c, h, i, k.). In contrast, mutations K370A and S366A do not affect TBK1 binding (Fig. 2a, Extended Data Fig. 3b, l). Sequence alignment of STING from 60 species (Extended Data Table 2) reveals that the C-terminal residues of STING are highly conserved (Extended Data Fig. 3n). Therefore, these mutagenesis and binding studies identified a highly conserved PLPLRT/SD motif that mediates the recruitment of TBK1 by STING (Extended Data Fig. 3n). Based on these results, we also identified a high affinity phosphomimetic mutant of STING (referred to as EMW, Extended Data Fig. 3o, Table 1).
Fig. 2 A. conserved PLPLRT/SD motif within the C-terminal tail of STING mediates TBK1 recruitment and activation.
a, Binding affinities (Kd) of hSTING mutants to TBK1 determined by SPR.
b, IFN-β luciferase reporter assays using HEK293T cells transfected with STING mutants. The cells were transfected with pcDNA3.1-hSTING (WT or mutants) and stimulated with cGAMP. ΔC9 represents truncation of the nine C-terminal residues. EMW indicates STING with mutations T376E, F378M, and S379W. Luciferase signals from stimulated cells are represented by orange bars and unstimulated controls by blue bars.
c, Western blot analyses of TBK1, STING and IRF-3 phosphorylation in HEK293T cells transfected with WT hSTING or STING mutants and stimulated with cGAMP
d, Immunoprecipitation showing the interactions between STING mutants and TBK1. HEK293T cells were transfected with mutants of Flag-STING and stimulated with cGAMP. STING was detected with an antibody against the FLAG tag.
e, IFN-β luciferase reporter assay of STING containing an insertion of six residues (GSGSGS) between the pLxIS motif and the PLPLRT/SD motif.
f, IFN-β luciferase reporter assays in HEK293T cells expressing hSTING S366A or L374A mutant and co-expressing the S366A and L374A mutants.
g, Western blot showing the phosphorylation of STING, TBK1, and IRF-3 in HEK293T cells expressing hSTING S366A or L374A mutant and co-expressing the S366A and L374A mutants.
The data of b, e, and f (mean ± S.E.M) are representative of three independent experiments. Each dot represents a technical replicate (n = 3). The p values were calculated by two-tailed Student’s t test: * p < 0.05, ** p < 0.01, *** p < 0.001, NS - not significant. All western blot and immunoprecipitation data are representative of three independent experiments.
To determine the functional roles of the nine C-terminal residues of STING, we mutated each of them to alanine and conducted IFN-β reporter assays (Fig. 2b). These assays showed that truncation of the nine C-terminal residues of STING (ΔC9) or mutation L374A disrupts the activation of the luciferase reporter (Fig. 2b). Mutation R375A reduces the reporter activity by about 50% (Fig. 2b). Mutations P371A, T376A, and D377A reduce the reporter signal by about 30% (Fig. 2b). In contrast, mutations K370A, L372A, P373A, F378A and S379A only reduce the signals slightly (Fig. 2b). Interestingly, the high affinity EMW mutant stimulates a similar level of signal as WT STING (Fig. 2b). As a control, mutation S366A disrupts activation of the reporter (Extended Data Fig. 3p). In addition, the reporter assays showed that WT STING induces a faster response to cGAMP compared to the T376A mutant, indicating that phosphorylation of the PLPLRT/SD motif is needed for optimal signaling by STING (Extended Data Fig. 3q).
Next, we tested how these mutations affect STING-mediated signaling in cells. Western blot showed that TBK1, STING, and IRF-3 are phosphorylated upon cGAMP stimulation of cells expressing WT STING (Fig. 2c). Truncation of the nine C-terminal residues or mutation L374A abolished TBK1, STING, and IRF-3 phosphorylation (Fig. 2c). In contrast, mutation S366A only abolished IRF-3 phosphorylation, but did not affect TBK1 activation (Extended Data Fig. 3r). Mutation R375A dramatically reduces the phosphorylation of TBK1, STING, and IRF-3 (Fig. 2c). In contrast, mutations P371A, L372A, D377A, and F378A only reduce TBK1, STING, and IRF-3 phosphorylation slightly (Fig. 2c). Mutations K370A, P373A, T376A, S379A, and the EMW mutation, do not affect the activation of TBK1, IRF-3 or STING (Fig. 2c). Consistent with these results, immunoprecipitation showed that these mutations affect TBK1 binding by STING in cells (Fig. 2d). Moreover, we tested if proximity of the IRF-3 binding pLxIS motif (p, hydrophilic residue, x, any residue, S, phosphorylation site) and the TBK1 binding PLPLRT/SD motif is necessary for STING-mediated signaling. We observed that insertion of six residues (GSGSGS) between these two motifs does not affect IFN-β reporter activation (Fig. 2e). In addition, we tested if the two motifs on separate STING molecules could support signaling. We co-transfected the cells with the S366A and L374A mutants and observed that together, these two mutants support IFN-β reporter activation (Fig. 2f). Western blot showed that TBK1 and IRF-3 are activated in cells co-transfected with the S366A and L374A mutants, but not in cells transfected with either the L374A or the S366A mutant (Fig. 2g).
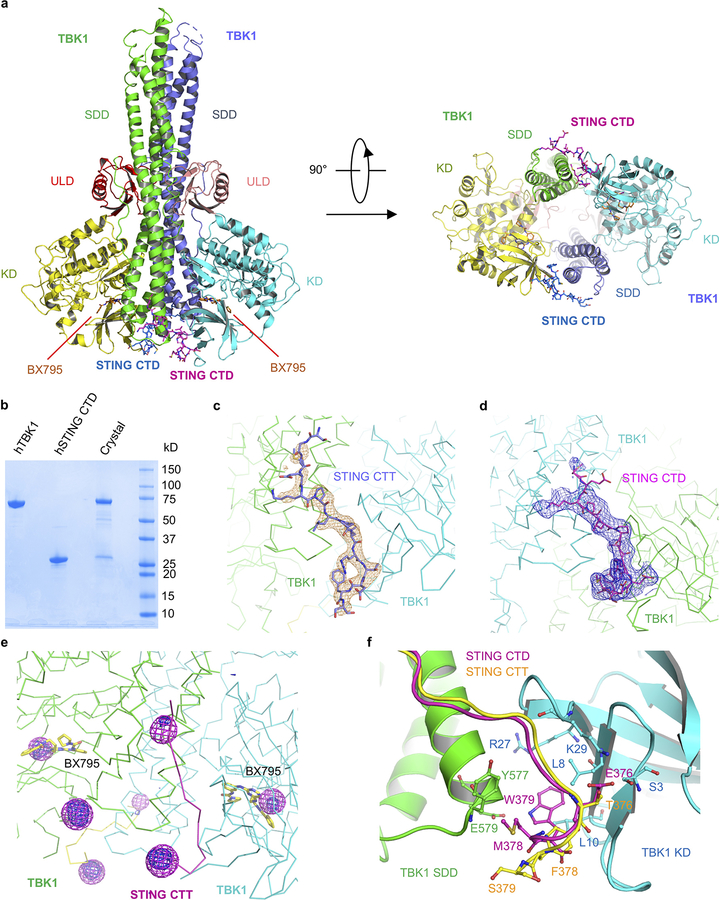
To elucidate the molecular basis of TBK1 recruitment by STING, we determined the crystal structures of TBK1 bound to STING CTT and CTD (Fig. 3, Extended Data Fig. 4 and Table 3). The structure of TBK1 bound to STING CTT showed that a TBK1 dimer binds to two peptides from STING CTT (Fig. 3a). Each STING peptide interacts with two TBK1 molecules simultaneously, forming a 2:2 complex (Fig. 3a). The STING binding sites are located at the interface between the N-lobe of the kinase domain (KD) and the dimerization domain (SDD) of two TBK1 molecules (Fig. 3a, b). STING CTT adopts an extended random coil structure that binds TBK1 mainly through hydrophobic interactions and hydrogen bonds via the PLPLRT/SD motif (Fig. 3c). The mainchain carbonyl group of Lys370, the amide group of Leu372, and the carbonyl group of Pro373, interact with the side chains of Lys584, Gln581 and Tyr577 by hydrogen bonds (Fig. 3c). The side chain of Leu374 reaches into a hydrophobic pocket defined by residues Leu8, Arg27, Lys29, Asn578, Gln581, Ile582, and Phe585, anchoring the PLPLRT/SD motif to its binding groove (Fig. 3c). In addition, the amide group of Leu374 also forms a hydrogen bond with the carbonyl of Lys29. Arg375 forms two hydrogen bonds with the side chain of Asn578 via its backbone amide and carbonyl groups (Fig. 3c). Superposition of the structures of STING CTT bound to mouse TBK1 and STING CTD EMW mutant bound to human TBK1 reveals that the C-terminal residues of the EMW mutant bind to hTBK1 in a similar fashion (Fig. 3d). Glu376, which mimics phosphorylated Thr376, interacts with the side chain of Lys29 through electrostatic interaction and with the side chain of Ser3 through a hydrogen bond (Fig. 3d, Extended Data Fig. 4f), explaining why phosphorylated STING binds TBK1 with higher affinity.
Fig. 3. Crystal structures of STING in complex with TBK1.
a, Ribbon representations of mouse TBK1 in complex with hSTING C-terminal tail (CTT). A side view looking into the active site of TBK1 is shown on the left and a bottom view is shown on the right. The kinase domains (KD) are in yellow and cyan. The ubiquitin-like domains (ULD) are in pink and red. The scaffold and dimerization domains (SDD) are in green and slate. STING CTT are shown by the blue and magenta ball-and-stick models. The TBK1 inhibitor BX795 are shown by the orange stick model.
b, The C-terminal tail of STING binds to the interface between the kinase domain (KD) and the scaffold and dimerization domain (SDD) of a TBK1 dimer. The KD and SDD are shown by the cyan and green surfaces respectively. The C-terminal tail of STING is shown by the magenta ball-and-stick model.
c, Interactions between STING CTT and TBK1. STING CTT is shown by the magenta ball-and-stick model and TBK1 is shown as ribbons. Residues of TBK1 involved in STING binding are shown by the green and cyan ball-and-stick model. The black dashed lines indicate distances less than 3.5 Å.
d, Superposition of the structures of hSTING CTD (yellow ball-and-stick) bound to human TBK1 and hSTING CTT (pink ball-and-stick) bound to mouse TBK1. Mouse TBK1 is shown by the surface representation colored according to surface electrostatic potential with positively charged surface in blue and negatively charged surface in red.
To confirm that the binding surface of TBK1 identified through the structural analyses is involved in its recruitment by STING, we expressed nine mutants of TBK1 for binding studies (Fig. 4a, Extended Data Fig. 5a to g). These studies showed that mutations L8A, K29A, Y577A, N578A, and I582A abolish the interactions between TBK1 and phosphorylated STING (Fig. 4a, Extended Data Fig. 5a to d, and f). In addition, mutations Q581A and K584A also reduce the binding affinity by almost 20 fold (Fig. 4a, Extended Data Fig. 5e, g). In contrast, mutations P404A and F585A do not affect binding (Fig. 4a, Extended Data Fig. 5c, d). Similar results were obtained using mTBK1 for the binding studies (Fig. 4a). Next, we tested how these mutations of TBK1 affect STING-mediated signaling. We have generated TBK1 knockout (KO) HEK293T cells, which were deficient in STING-mediated signaling, but were responsive after TBK1 transfection (Fig. 4b, Extended Data Fig. 5h to j). Mutations P404A and F585A only have minor effects on the signaling (Fig. 4b). In contrast, mutations R27A, K29A, I582A, and K584A, reduce the signals by about 40% (Fig. 4b). Mutations L8A and Y577A reduce the signals by over 60% (Fig. 4b). Mutations N578A and Q581A reduce the signals to a level similar to that of the kinase inactive S172A mutant (Fig. 4b). Western blot showed that mutations L8A, R27A, Y577A, N578A, Q581A and I582A dramatically reduced TBK1, STING, and IRF-3 phosphorylation (Fig. 4c). Although mutations K29A and K584A also reduced TBK1 and STING phosphorylation, they only have minor effects on IRF-3 phosphorylation (Fig. 4c). In contrast, mutations P404A and F585A only have minor effects on the phosphorylation of TBK1, STING, and IRF-3 (Fig. 4c). Moreover, immunoprecipitation confirmed that these residues are critical in TBK1 recruitment by STING in cells (Extended Data Fig. 5k).
Fig. 4. Mutations at the TBK1/STING interface affect STING binding and signaling.
a, Binding affinities (Kd) of TBK1 mutants to phosphorylated hSTING CTD determined by SPR. n.d., no data.
b, IFN-β luciferase reporter assays using TBK1 knockout HEK293T cells transfected with TBK1 mutants. The cells were transfected with pcDNA3.1-hSTING plasmids and/or pcDNA3.1-hTBK1 plasmids and stimulated with cGAMP. The kinase inactive S172A mutant of TBK1 was used as a negative control. Luciferase signals from stimulated cells are indicated by orange bars and unstimulated controls by blue bars. The data (mean ± S.E.M) are representative of three independent experiments. Each dot represents a technical replicate (n = 3). The p values were calculated by two-tailed Student’s t test: * p < 0.05, ** p < 0.01, *** p < 0.001, NS - not significant.
c, Western blot showing the phosphorylation of TBK1, STING, and IRF-3 in TBK1 knockout cells transfected with TBK1 mutants. The cells were transfected with pcDNA3.1-hSTING plasmid plus pcDNA3.1-hTBK1 plasmids and stimulated with cGAMP. The data are representative of three independent experiments.
Although STING CTT is capable of mediating IRF-3 activation in an in vitro reconstitution system16, expression of STING CTD alone is not sufficient to activate STING-mediated signaling in cells22. To investigate how the transmembrane domain mediates STING activation, we expressed full-length hSTING in Freestyle 293F cells. Intriguingly, in the absence of cGAMP, full-length STING aggregates (Extended Data Fig. 6a, b). However, in the presence of cGAMP, STING forms stable oligomers with an estimated mass of ~400 kD (Extended Data Fig. 6a, b). SDS-PAGE analysis of cross-linked STING oligomers reveals a protein band of ~360 kD (Extended data Fig. 6c, d). In addition, size exclusion chromatography and multiangle light scattering (SEC-MALS) also confirmed the formation of the STING oligomers (Extended Data Fig. 6e). Based on these observations, we estimate that 4 to 5 STING dimers are present in the peak fractions of the STING-cGAMP complex. To provide additional evidence of STING oligomerization upon cGAMP binding, we cross-linked the STING oligomer and imaged the sample by cryo-EM (Extended Data Fig. 6f). Both the raw images and the 2D averages show that the STING-cGAMP complex forms oligomers (Extended Data Fig. 6f, g). We have generated a ~12 Å resolution map of the STING oligomer, which reveals that the STING dimers likely stack against each other side by side to form the oligomers (Extended Data Fig. 6h to j). Based on results from previous mutagenesis studies of mouse STING22, we identified a STING mutant (E68A, E69A, H74A, S75A, R76A) that does not form oligomers in the presence of cGAMP (Extended Data Fig. 6k to m) and is not capable of supporting STING-mediated signaling (Extended Data Fig. 6n, o). Confocal microscopy revealed that this STING mutant does not translocate to the perinuclear punctate structures nor co-localize with TBK1 upon cGAMP stimulation, while other functional mutants behave similarly to WT STING (Extended Data Fig. 6p). Consistent with these findings, a recent study showed that mutations E68A or E69A abolish STING-mediated signaling and STING oligomerization in cells treated with cGAMP23.
Based on the results from this study, we propose a molecular model of STING-mediated signaling (Extended Data Fig. 7). First, cGAMP binding induces the oligomerization of STING. Next, TBK1 is recruited to the STING oligomers via its C-terminal PLPLRT/SD motif and is activated by induced proximity in trans. Activated TBK1 phosphorylates STING at the PLPLRT/SD motif, facilitating further recruitment and activation of TBK1. Phosphorylation of STING at the pLxIS motif allows STING to recruit IRF-3, bringing IRF-3 and TBK1 into proximity and thus facilitates IRF-3 activation. Since simultaneous binding of TBK1 and IRF-3 to a single STING molecule is not required for IRF-3 activation, it is likely that the proximity of IRF-3 and TBK1 induced by adjacent STING molecules in the STING oligomers mediates the activation of IRF-3 by TBK1. This study provides a structural framework for the development of conceptually new approaches to inhibit deleterious STING-mediated immune responses in autoimmune and inflammatory disorders.
METHODS
Mass spectrometry
We expressed a short peptide containing the 22 C-terminal residues human STING as SUMO-fusion. The peptide was purified and phosphorylated with TBK1. The phosphorylated peptide was analyzed using a liquid chromatography electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS) system, consisting of a Dionex Ultimate 3000 LC system (with an Acclaim PepMap 100 C18 column from Thermo) coupled to a Thermo Orbitrap Fusion mass spectrometer. LC system solvents were water with 0.1% FA (mobile phase A) and acetonitrile with 0.1% FA (mobile phase B). The peptides were eluted over a 60-minute gradient from 2% B from 0 to 5 min, 2% to 45% from 5 to 37 min, 45% to 90% from 38 to 46 min, and down to 2% from 46 to 60 min at a flow rate of 0.4 μL/min. The mass spectrometer ion source was set to have a spray voltage of 2.3 kV, ion transfer tube temperature of 275 °C, the scan range was m/z 400–1600 with a resolution of 120,000. MS/MS acquisition was performed with 3 s cycle time. The intensity threshold was set to 5000, Ions with charge states 2+ to 6+ were sequentially fragmented by electron transfer dissociation (ETD) with 100 ms ETD reaction time and 200000 ETD reagent target, followed by high energy collisional dissociation (HCD) with a normalized collision energy (NCE) of 40%. The dynamic exclusion duration was set as 60 s. Raw data were analyzed using the Thermo Proteome Discoverer (v2.1.0.81) software platform. The mass spectrometry data was analyzed using SEQUEST with the following parameters: the protein sequence database contained only the ST358 sequence, dynamic (or variable) modifications included protein N-terminal acetylation, oxidation, serine, threonine and tyrosine phosphorylation. Mass tolerances for precursor ions was set to 10 ppm, and fragment ions set to Δ0.6 Da. Limits for peptide length searched range from 6 to 144, maximum delta Cn is set to 0.05. Maximum number of allowed missed cleavages is 2.
Cell culture
HEK293T cells were obtained from ATCC (CRL-3216), cultured in DMEM (1×) + GlutaMAX medium (Gibco) supplemented with 10% fetal bovine serum (Gibco), penicillin (100 U/ml) and streptomycin (100 μg/ml) at 37 °C in a humidified atmosphere with 5% CO2. TBK1 knockout (KO) HEK293T cells were cultured under the same conditions. Freestyle 293F cells were obtained from Thermo Fisher Scientific (K9000–01) and cultured in Freestyle 293 Expression Medium (Thermo Fisher Scientific) supplemented with penicillin (50 U/ml) and streptomycin (50 μg/ml). The cells were cultured in a 37 °C incubator containing a humidified atmosphere of 8% CO2 on an orbital shaker rotating at 130 rpm.
IFN-β luciferase reporter assays
The cDNA encoding human STING and human TBK1 were cloned into pcDNA3.1 (−) vector. All mutations were generated by site-directed mutagenesis and confirmed by DNA sequencing. HEK293T cells were seeded in CoStar White 96-well plate (Corning) at 4 × 104 cells per well and cultured at 37 °C with 5% CO2. After 24 hours, the cells were transfected with indicated amounts of pcDNA3.1-hSTING plasmids, IFN-β firefly luciferase reporter plasmid (20 ng per transfection) and phRL-TK–Renilla luciferase plasmid (2 ng per transfection) using the transfection reagent Lipofectamine 2000 (Invitrogen) according to the manufacture’s manual. Empty pcDNA3.1 (−) plasmid was used as transfection control and also added to normalize the amount of DNA in each transfection. The cells were treated with 30 μg/ml cGAMP for 24 hours post transfection. After 16 hours stimulation, the cells were analyzed using the Dual-Glo Luciferase report assay kit (Promega) and detection of the luminescent signals was performed with a BioTek Synergy HTX Multi-Mode microplate reader. IFN-β luciferase reporter assays using the TBK1 KO HEK293T cells were performed in a similar way. The TBK1 KO cells were transfected with indicated amounts of pcDNA3.1-hSTING plasmid and/or pcDNA3.1-hTBK1 plasmid, IFN-β firefly luciferase reporter plasmid (10 ng per transfection) and phRL-TK–Renilla luciferase plasmid (2 ng per transfection). The relative firefly luciferase activity was normalized by the Renilla luciferase activity. The relative IFN-β reporter fold of induction represents the ratio normalized to the negative control plasmid values with the same treatment. Statistical analyses were carried out using Microsoft Excel. All data are presented as mean ± SEM. A two-tailed Student’s t test assuming equal variants was used to compare two groups. The statistical significance between the indicated samples is designated as *P < 0.05, **P < 0.01, ***P < 0.001, or NS (P > 0.05).
Western blot
After transfection with pcDNA3.1-hSTING and/or pcDNA3.1-hTBK1 plasmids and stimulation with cGAMP, wild-type or KO-TBK1 HEK293T cells were washed and suspended in 1×PBS. After centrifugation, the cells were lysed in 200 mM Tris (pH 7.5), 150 mM NaCl, 1 mM EDTA, and 1% Nonidet P-40 supplemented with one complete EDTA-free protease inhibitor mixture tablet (Roche) and one PhosSTOP phosphatase inhibitor mixture tablet (Roche) for each 10 mL of lysis buffer. The proteins were resolved using 10% SDS-PAGE gel and transferred to PVDF membrane for incubating with the primary antibodies. The primary antibodies used in the western blot analysis are as follows, anti-STING (Cell signaling, 13647, 1:1000), anti-STING phosphor-Ser366 (Cell signaling, 85735, 1:1000), anti-TBK1 (Cell signaling, 3013, 1:1000), anti-TBK1 phospho-Ser172 (Cell signaling, 5483, 1:1000), anti–IRF-3 (Cell signaling, 4302, 1:1000), anti-IRF-3 phospho-Ser386 (Abcam, ab76493, 1:2500), anti-Flag M2-Peroxidase (Sigma, A8592, 1:2000) and anti-Actin (Thermo fisher scientific, MA5–11869, 1:4000). After incubation with the primary antibodies overnight at 4°C, the membrane was further incubated with the corresponding HRP-conjugated secondary antibodies at room temperature for 2 hours. Detection of the target proteins was performed with a ChemiDoc Imager (Bio-rad) using the Western Lightening Plus ECL kit (PerkinElmer) according to the manufacturer’s protocol. For immunoprecipitation, the cell lysates were centrifuged at 20,000g for 10 minutes. The supernatant were incubated with anti–FLAG-M2 agarose affinity gel (Sigma, A2220) for 1 hour at 4 °C. The proteins bound to the agarose beads were washed three times with TBS buffer (50 mM Tris, 150 mM NaCl, pH 7.4) and eluted by boiling with 1×SDS-PAGE sample buffer (62.5 mM Tris, pH 6.8, 2% SDS, 10% (v/v) glycerol, and 0.002% bromphenol blue and 10 mM DTT) for 5 minutes. The samples were analyzed using 10% SDS-PAGE gel.
Immunocytochemistry
HEK293T cells grown on poly-L-Lysine coated glass coverslips in 12-well plates for 24 hours and were transfected with equal amount of WT or mutant STING plasmids, respectively. The transfection mixture was combined plasmid DNA with dilution lipofectamine 2000 in Opti-MEM medium (Gibco). After a 24-hour incubation, the medium was replaced with fresh growth medium with or without 30 μg/ml cGAMP. After a 16-hour incubation, cells were washed with PBS, fixed by using 4% paraformaldehyde in PBS for 10 mins at room temperature and permeabilized with PBST (0.5% Triton X-100 in PBS). Cells were washed and blocked with 5% fetal bovine serum in PBST and followed by overnight incubation with primary antibodies, anti-STING (Cell signaling, 13647, 1:200) and anti-TBK1 (GeneTex, GTX12116, 1:200). Cells were then washed three times by PBS and incubated with Alexa 488-conjugated Donkey Anti-Rabbit IgG antibody (Jackson ImmunoResearch, 711-545-152, 1:1000) and Alexa 555-conjugated Donkey Anti-Mouse IgG antibody (Thermo Fisher Scientific, A31570, 1:1000) for 1 hour at room temperature. The coverslips were then washed with PBS, mounted on slides with ProLong Gold antifade reagent with 4′−6-diamidino-2-phenylindole (DAPI) and imaged using a Nikon-Ti Fluorescence microscope.
Protein expression and purification
Constructs of human STING were cloned into a modified pET28(a) vector with an N-terminal Avi-His6-SUMO tag. For protein quantification, the mutation V343W was introduced into human STING C-terminal tail (residue 342 to 379). All the proteins were expressed in BL21 (DE3) in M9 medium. The cDNA encoding human IRF-3 (residue 189–427) was cloned into another modified pET28(a) vector with an N-terminal His6-SUMO tag. IRF-3 was also expressed in BL21 (DE3) in regular LB medium (BD). When OD600 reached 1.0, the cells were induced with 0.4 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and cultured overnight at 16 °C. The proteins were purified using the protocol described previously24. Briefly, the proteins were first purified using a nickel-NTA column. The SUMO tag was then cleaved using SUMO protease and removed using a nickel-NTA column. The proteins were further purified by gel filtration chromatography using Superdex75 or 200 (16/60 GL) columns (GE Healthcare). The Se-methionine substituted human STING was expressed in M9 medium supplemented with selenomethionine (Acros Organics) and purified as described for the wild type protein. Human and mouse TBK1 (residue 1 to 657) were expressed in sf9 insect cells after infection with recombinant baculovirus and purified as described previously25. Biotin labeled-Avi-His6-SUMO proteins and peptides were also expressed in BL21 (DE3) cells in M9 medium, with the exception of the biotin labeled- Avi-His6-SUMO-GFP nanobody, which was expressed in regular LB medium. The cells were co-transformed with the pET28(a) plasmids coding for the target proteins and the pBirAcm plasmid coding for BirA and induced with 0.4 mM IPTG in the presence of 5 μg/mL biotin and cultured at 16°C overnight. The SUMO fusion proteins and peptides were first purified using a nickel-NTA column and further purified on a Superdex75 or 200 column (GE Healthcare).
Full length human STING was cloned into pEGFP-N1 plasmid with a GFP-tag attached to the C-terminus of STING. A thrombin cleavage site was inserted between STING and GFP. FreeStyle 293-F cells were seeded into fresh FreeStyle 293 expression media with a final density of 1.0 × 106 cells/ml and incubated at 37°C, 8% CO2, 130 rpm. After 24 hours, 1 mg pEGFP-N1-STING plasmids, 2 mg of linear PEI25000 (Polysciences, Inc.) were mixed into 100 ml 1×PBS and incubated at room temperature for 20 mins. Then, the mixture was added into 1L FreeStyle 293-F cells. After 2 days, the cells were pelleted at 3000 rpm for 5 mins. The pellets were resuspended in 40 ml pre-chilled lysis buffer (20 mM Tris pH 7.5, 150 mM NaCl, 1% DDM) containing the protease cocktail inhibitors (Roche), sonicated for 3 cycles and shaked gently at 4°C for 2 hours. After centrifugation at 16000 rpm for 30 mins, the supernatant was mixed with 500 μl streptavidin agarose beads (EMD Millipore), which have been coupled with biotin-labeled Avi-SUMO-GFP nanobody. After shaking at 4°C for another 2 hours, the beads were pelleted and washed with pre-chilled washing buffer (20 mM Tris pH 7.5, 150 mM NaCl, 0.1% DDM). Finally, the beads were resuspended in 500 μl washing buffer and SUMO protease was added to cleave the target protein from the beads. The eluted protein was incubated with thrombin at 4°C overnight and further purified using a Superose 6 increase 10/300 GL column (GE Healthcare). All Mutants of STING and TBK1 were generated using the QuikChange sitedirected mutagenesis kit (Agilent) or a PCR-based technique with appropriate primers. The sequences of the plasmids were confirmed by plasmid DNA sequencing.
Phosphorylation of STING
Biotin-labeled Avi-His6-SUMO-STING CTD and CTT peptides were phosphorylated using GST-mTBK1 in a reaction buffer containing 20 mM HEPES (pH 7.5), 10 mM MgCl2, 100 mM NaCl, 5 mM ATP, 0.1 mM Na3VO4, 5 mM NaF, and 5 mM DTT. The target proteins were diluted to 1 mg/ml in the reaction buffer and mixed with GST-mTBK1 in a 10:1 (w/w) ratio. After 24 hours incubation at 27°C, the phosphorylated proteins were purified using a Superdex200 10/300 GL column (GE healthcare) eluted with a buffer containing 20 mM Tris, 150 mM NaCl at pH 7.5.
Surface plasmon resonance (SPR)
The SPR binding studies were performed with a Biacore X100 SPR instrument (GE Healthcare). Biotin CAPture Kit (GE Healthcare) was used for binding analysis in 1× HBS-EP+ buffer (GE Healthcare). Biotin-labeled Avi-His6-SUMO-fusion proteins or peptides were captured on the Sensor Chip CAP using Biotin CAPture reagent. TBK1 and IRF-3 in a series of 2-fold dilutions were flowed through the sensor chip at 30 μl/min. The single-cycle kinetic/affinity protocol was used in all binding studies. After each analysis cycle, the sensor chip was regenerated with a buffer containing 6 M guanidine hydrochloride and 0.25 M NaOH. All measurements were duplicated under the same conditions. The binding affinities (Kd) were determined by fitting the data to a steady-state 1:1 binding model using Biacore X100 Evaluation software version 2.0 (GE Healthcare).
Crystallization, data collection, and structure determination
Purified TBK1 was concentrated to 12 mg/ml and its inhibitor BX795 was added to the samples at a final concentration of 200 μM. Human TBK1 (residue 1–657, S172E, D135N) with human STING CTD (residues 155 to 379, T376E, F378M, S379W) or selenomethionine-labeled human STING CTT (residue 342 to 379, T376E, F378M, S379W) were mixed at 1:1.1 molar ratio. Crystals were obtained by mixing the complexes with equal volume reservoir buffer containing 0.8 M (NH4)2SO4, 0.1 M BIS-TRIS pH 5.5 and 1% PEG3350. Mouse TBK1 (residue 1 to 657, S172A) was mixed human STING CTT (residue 342 to 379, V343W) in a 1:3 molar ratio with a final concentration of TBK1 at ~8 mg/ml. The complex crystals were grown in 18% v/v tacsimate pH 7.0, 0.1 M HEPES pH 7.0, 2% PEG3350. The proteins were crystallized by hanging drop vapor diffusion method at 4 °C. The crystals were flash-frozen in liquid nitrogen in the reservoir solution containing 25% (v/v) glycerol. Diffraction data for the hTBK1/hSTING CTD and mTBK1/hSTING CTT complexes were collected at the Advanced Light Source (ALS) beamline 5.0.2 using a Pilatus3 6M detector. Diffraction data for the hTBK1/hSTING CTT Se-Met derivative complex were collected at ALS beamline 8.2.2 using a Quantum 315R detector. The diffraction data were processed with imosflm26. Both of the structures were determined by molecular replacement using either human (PDB 4IM2) or mouse TBK1 (PDB 4JLC) structures as search models using the Phenix package27. Electron density corresponds to the C-terminal 15 residues of STING are obvious in the difference maps. To facilitate model building, diffraction data at high energy Se wavelength were collected to locate Se atoms in the STING peptide. The heavy atom substructure was determined using SHELXD and the experimental phases calculated with SHELXE28. Three obvious heavy atom peaks were located in the anomalous difference maps calculated with model phases or experimental phases. The STING peptides were modeled with Coot29. The structures were refined using the Phenix package27. All structural figures were generated with PyMOL (https://www.pymol.org).
Generation of TBK1 knockout HEK293T cells
FL-Cas9 lentivirus was produced by transfecting Lenti-X cells (Clontech) with lentiCas9-Blast30, 2nd generation lentiviral packaging plasmids, and PolyJet (SignaGen) according to the manufacturer’s instructions. HEK293T cells were transduced with Cas9 lentivirus with 1:1000 Lipofectamine 2000 (Thermo Fisher Scientific) for two consecutive days and selected with 10 μg/ml blasticidin (Invivogen). FL-Cas9 expression was verified by western blot analysis. To ensure creation of nonsense mutations early in the coding region, TBK1-specific gRNAs were designed against the first four coding exons of TBK1 using the sgRNA Design Tool (Broad Institute): gRNA-TBK1–1: ATCACTTCTTTATTCCTACG; gRNA-TBK1–2: CATAAGCTTCCTTCGTCCAG; gRNA-TBK1–3: GAAGAACCTTCTAATGCCTA; gRNA-TBK1–4: ACAGTTGATCTTTGGAGCAT. GFP-targeting gRNAs were used as negative controls: gRNA-GFP-1: GGGCGAGGAGCTGTTCACCG; gRNA-GFP-2: CAGGGTCAGCTTGCCGTAGG; gRNA-GFP-3: AAGTTCAGCGTGTCCGGCGA. Forward and reverse primers (Integrated DNA Technologies) were cloned into lentiCRISPRv2 as previously described30. gRNA lentivirus was produced as above, and FL-Cas9 HEK293Ts were transduced with gRNA virus and then selected with 0.5 μg/ml puromycin (Invivogen). TBK1 knockout was verified by western blot analysis for endogenous TBK1 (Cell Signaling, 3013). LentiCas9-Blast and lentiCRISPR v2 were gifts from Feng Zhang (Addgene plasmids #52962 and #52961). psPAX2 and pMD2.G (lentiviral packaging plasmids) were gifts from Didier Trono (Addgene plasmids #12260 and #12259).
Electron microscopy
Before cryo-EM imaging, the full-length STING sample in 0.1% DDM was exchanged into Amphipol A8–35. Briefly, STING at ~1 mg/mL was incubated for ~8 hours with 3-fold excess by mass of A8–35. DDM was removed by incubation with Bio-Beads SM-2 (Bio-Rad) overnight at 15 mg Bio-Beads per mL of solution. The sample was filtered to remove Bio-Beads and purified using a Superose 6 increase 10/300 GL column eluted with a buffer containing 20 mM Tris, 150 mM NaCl, and 5 μM cGAMP at pH 7.5. For cryo-EM imaging, 3 μl of STING at a concentration of 0.6 mg/mL stabilized with A8–35 was applied to glow-discharged C-flat holey carbon grids (1.2/1.3, 400 mesh). Girds were blotted for 8s and plunge frozen in liquid ethane using a Vitrobot. Images were recorded by Latitude on a Titan Krios Transmission Electron Microscope operating at 300 kV. A Gatan K3 detector was used in counting mode at a calibrated magnification of ×64,000 (yielding a pixel size of 1.42 Å). The dose rate on the camera was set to be 23.8 electrons per physical pixel per second. Exposure of 8.4 s was dose-fractionated into 84 movie frames, leading to a total accumulated dose of 100 electrons per Å2 on the specimen. Images were recorded with a defocus in the range from 1.0 to 4.0 μm. Movies were collected with 15° and 30° tilt to address the preferred orientation of particle distribution. A total of 3,956 movies were recorded. The cryo-EM images were subjected to MotionCor2 for whole-frame dose-weighted motion correction. Particles picking was performed automatically on the summed images in Gautomatch. A total of 880,703 particles were picked. Per-particle local CTFs were estimated by GCTF. Two rounds of 2D classification were then preformed. After discarding bad class averages, 352,286 particles were re-centered and re-extracted to Relion 3D classification. Initial models for reconstruction were generated from scratch by the EMAN2 e2initialmodel.py program using selected unsupervised 2D averages of good quality based on visual comparison without applying any symmetry. A stack of 47,474 particles was selected to Relion 3D refinement using a 40 Å low-pass filtered reconstruction from the initial model. The final 3D reconstruction of STING oligomer with cGAMP has an overall resolution of 11.6 Å, using gold-standard Fourier shell correlation (FSC) = 0.143 criteria. The cryo-EM maps of STING oligomer was segmented using Segger. We took advantage of the availability of crystal structure of hSTING cytosolic region with cGAMP to guide the decision on the final segmentation choice. After applying the smoothing procedure for four steps to the original EM map, four regions were obtained. STING CTD structures were docked to the segmentation map using Chimera.
Extended Data
Extended Data Fig. 1. Potential phosphorylation sites within the STING C-terminal tail.
a, List of phosphorylated STING peptides (residues 358–379, S358W) identified by LC-MS/MS. Phosphorylated residues are underlined and shown in orange.
b, Representative MS/MS spectra of phosphorylated STING peptides (residues 358–379, S358W). b and c fragment ions are shown in red, while y and z fragment ions are shown in blue. Residues phosphorylated are shown in orange. The data are representative of two independent experiments.
c, STING and cGAMP dependent activation of IFN-β luciferase reporter in HEK293T cells. The cells were transfected with indicated amounts of pcDNA3.1-wild-type human STING (STING WT) plasmid and stimulated with cGAMP. Luciferase signals from stimulated cells are indicated by orange bars and unstimulated controls by blue bars. The data (mean ± S.E.M) are representative of three independent experiments. Each dot represents a technical replicate (n = 3). The p values were calculated by two-tailed Student’s t test: * p < 0.05, ** p < 0.01, *** p < 0.001, NS-not significant.
d, Western blot of HEK293T cells transfected with the plasmids of STING WT or deletion of the nine C-terminal residues. The data are representative of three independent experiments.
Extended Data Fig. 2. Surface plasmon resonance (SPR) binding studies of human STING with TBK1.
a, Domain organization of human STING and truncated forms of STING used in this study.
b, SDS-PAGE analyses of human STING (left panel) and TBK1 (right panel) used in the SPR studies.
c to l, SPR binding studies of human STING with TBK1 (upper panels). Experiments without cGAMP in the running buffer are indicated. All others were conducted with 1 μM cGAMP in the running buffer. The binding affinity (Kd) was determined by fitting the binding data to a one-site binding model (lower panels).
The data of b-l are representative of at least two independent experiments.
Extended Data Fig. 3. Binding studies of human STING mutants with TBK1.
a, SDS-PAGE analysis of human STING mutants and human IRF-3 used in the SPR binding studies.
b to l, SPR binding studies of human STING mutants (residue 155 to 379) with human TBK1 (upper panels) in presence of 1 μM cGAMP. The binding affinity (Kd) was determined by fitting the binding data to a one-site binding model (lower panels).
m, SPR binding study of STING L374A mutant with IRF-3.
n, STING C-terminal tail contains a highly conserved PLPLRT/SD motif. The sequence Logo of STING is generated by WebLogo based on the sequence alignment of STING from mammals. The frequency of occurrence of an amino acid is indicated underneath the sequence. The PLPLRT/SD motif is downstream of the pLxIS motif that is involved in IRF-3 binding.
o, SPR binding study of the high affinity phosphomimetic EMW mutant of STING with human TBK1.
p, IFN-β luciferase reporter assays of STING S366A and L374A mutants. For each assay, HEK293T cells were transfected with pcDNA3.1-hSTING variants and stimulated with cGAMP.
q, Time course IFN-β luciferase reporter assays of HEK293T cells transfected with WT and T376A mutant of STING. The cells were stimulated with cGAMP.
r, Western blot showing the phosphorylation of STING, TBK1, and IRF-3 in HEK293T cells transfected with WT STING, the S366A and L374A mutants of STING.
The data of b-m, o, and r are representative of at least two independent experiments. The data of p and q (mean ± S.E.M) are representative of three independent experiments. Each dot represents a technical replicate (n = 3 in p and n = 6 in q). The p values were calculated by two-tailed Student’s t test: * p < 0.05, ** p < 0.01, *** p < 0.001, NS-not significant.
Extended Data Fig. 4. Crystal structures of STING in complex with TBK1.
a, Ribbon representation of the structure of human TBK1 bound to human STING CTD EMW mutant (residue 155 to 379, T376E, F378M, S379W). The kinase domains (KD) are in yellow and cyan, the ubiquitin-like domains (ULD) are in pink and red, the scaffold and dimerization domains (SDD) are in green and slate. The STING CTDs are shown by the blue and magenta ball-and-stick models. The TBK1 inhibitor BX795 is shown by the orange stick models.
b, SDS-PAGE analysis of crystals of human TBK1 in complex with human STING CTD EMW mutant. The data are representative of two independent experiments.
c, Difference map of human STING CTT bound to mouse TBK1 contoured at 2.5 σ. The σA-weighted Fo - Fc map was calculated with STING CTT omitted from the model. STING CTT is shown by the slate ball-and-stick model. TBK1 dimer is shown by the ribbon representation colored in green and cyan.
d, Difference map of human STING CTD bound to human TBK1 contoured at 2.5 σ. The σA-weighted Fo - Fc map was calculated with STING CTD omitted from the model. STING CTD is shown by the purple stick model. The TBK1 dimer is shown by the ribbons colored green and cyan.
e, Anomalous difference maps of Se-Met derivative of human STING CTT EMW mutant bound to human TBK1. The blue map was calculated with model phases (ϕc) and the magenta map was calculated with experimental phases after density modification (ϕdm). The STING peptide is shown by the magenta ribbon and TBK1 shown by the green and cyan ribbons.
f, Superposition of the structures of human STING CTD EMW mutant (magenta) and human STING CTT (yellow) bound to human and mouse TBK1. Mouse TBK1 is shown by the green and cyan cartoon representation. Residues Glu376, Met378, and Trp379 from STING CTD EMW mutant are shown by the magenta ball-and-stick models. Residues Thr376, Phe378, and Ser379 from STING CTT are shown as the yellow ball-and-stick models.
Extended Data Fig. 5. Binding studies of human STING with human TBK1 mutants and characterization of TBK1 knockout HEK293T cells.
a to g, Surface plasmon resonance (SPR) binding studies of phosphorylated human STING C-terminal domain (CTD, residue 155 to 379) with human TBK1 mutants in presence of 1 μM cGAMP. The binding affinity (Kd) was determined by fitting the binding data to a one-site binding model. SDS-PAGE analysis of proteins used in these studies are shown in the inset of panel a.
h, Western blot characterization of TBK1 knockout HEK293T cell lines.
i, IFN-β luciferase reporter assays using TBK1 knockout cells. For each assay, 0.2 ng pcDNA3.1-human STING plasmids or/and 1.0 ng pcDNA3.1-human TBK1 plasmids were transfected into TBK1 knockout cells. Luciferase signals from stimulated cells are indicated by orange bars and unstimulated controls by blue bars. The data (mean ± S.E.M) are representative of three independent experiments. Each dot represents a technical replicate (n = 3). The p values were calculated by two-tailed Student’s t test: * p < 0.05, ** p < 0.01, *** p < 0.001, NS-not significant.
j, Western blot showing the phosphorylation of TBK1, STING, and IRF-3 in TBK1 knockout cells transfected with STING and TBK1 plasmids. TBK1 knockout cells were transfected with 0.2 ng pcDNA3.1-human STING plasmids or/and 1.0 ng pcDNA3.1-human TBK1 plasmids and stimulated with cGAMP.
k, Immunoprecipitation and immunoblot of Flag-STING and TBK1 in TBK1 KO cells. The cells were transfected with Flag-STING and TBK1 mutants and stimulated with cGAMP. Flag-STING and TBK1 in the pull-downs and whole cell lysates (WCL) were analyzed by immunoblotting. STING was visualized with FLAG antibody. TBK1 in the pull-downs were detected with an antibody against TBK1.
The data of a-h are representative of at least two independent experiments. The western blot data in j and k are representative of three independent experiments.
Extended Data Fig. 6. cGAMP binding induces the oligomerization of full-length STING.
a, Gel-filtration chromatography analyses of full length STING in presence and in absence of 1 μM cGAMP using a Superose 6 column.
b, SDS-PAGE analyses of fractions containing full-length STING from gel filtration chromatography.
c, Gel filtration chromatography analyses of full length STING in 0.1% DDM or Amphipol A8–35 using a Superose 6 column in presence of 1 μM cGAMP.
d, SDS-PAGE analysis of cross-linked full length STING.
e, SEC-MALS analysis of full length STING in 0.1% DDM and 1 μM cGAMP.
f, Representative cryo-EM micrograph of full-length STING stabilized with Amphipol A8–35.
g, Representative 2D averages of full-length STING particles in Amphipol A8–35.
h, i, and j, Three views of 12 Å resolution map of STING oligomers. Human STING CTD dimers bound to cGAMP were docked into the map.
k, A list of human STING mutants in the transmembrane domain.
l, Gel-filtration chromatography analyses of wild type and mutants of full length STING in presence of 1 μM cGAMP using a Superose 6 column.
m, SDS-PAGE analyses of fractions of WT STING and STING mutants purified by gel filtration chromatography using a superose 6 column.
n, IFN-β luciferase reporter assays showing that the mutations in N-terminal transmembrane domain affect STING mediated signaling. Indicated amounts of pcDNA3.1-human STING plasmids were transfected into HEK293T cells. Luciferase signals from stimulated cells are indicated by orange bars and unstimulated controls by blue bars. The data (mean ± S.E.M) are representative of three independent experiments. Each dot represents a technical replicate (n = 3). The p values were calculated by two-tailed Student’s t test: * p < 0.05, ** p < 0.01, *** p < 0.001, NS-not significant.
o, Western blot showing that mutations in the transmembrane domain of STING affect the phosphorylation of STING, TBK1, and IRF-3. HEK293T cells were transfected with indicated amounts of pcDNA3.1-human STING plasmids and stimulated with cGAMP.
p, Confocal microscopy images of HEK293T cells transfected with WT STING and STING mutants with or without cGAMP stimulation. Scale bars denote 20 μm.
The data of a-e, l, and m are representative of at least two independent experiments. The data of f, g, o, and p are representative of at least three independent experiments.
Extended Data Fig. 7. Proposed model for the recruitment and activation of TBK1 and IRF-3 through the cGAS-STING pathway.
1) cGAS is activated by dsDNA in the cytosol and catalyzes the synthesis of cGAMP from ATP and GTP. 2) cGAMP binding induces the oligomerization of STING at the ER or Golgi membranes. 3) TBK1 is recruited to the STING oligomers via its C-terminal PLPLRT/SD motif and activated by induced proximity in trans. Phosphorylation of STING by TBK1 increases the binding affinity between TBK1 and STING and facilitates further recruitment and activation of TBK1. 4) Activated TBK1 phosphorylates STING at the pLxIS motif, allowing it to recruit IRF-3 to the signaling complex. 5) The proximity of TBK1 and IRF-3 bound to adjacent STING molecules within the cGAMP-STING oligomer causes the phosphorylation of the pLxIS motif of IRF-3. 6) Phosphorylated IRF-3 dissociates from STING, oligomerizes, translocates to the nucleus, and initiates the transcription of IFN-β gene.
Extended data Table 1.
Binding affinities of human STING mutants with TBK1
| Human STING (hSTING) | Human TBK1 1–657, S172A Kd (μM) |
M0US6 TBK1 1–657, S172A Kd (μM) |
M0US6 TBK1 1–657 Kd (μM) |
|---|---|---|---|
| hSTING 155–379 (no cGAMP) | ~230 | ~260 | |
| hSTING 55–379 | ~290 | ~170 | |
| p-hSTING 155–379 | 11.0 | 9.1 | 13.3 |
| p-hSTING 155.379 (no cGAMP) | 20.0 | 12.5 | |
| p-hSTING 155–341 | no binding | no binding | |
| p-hSTING 155–370 | no binding | no binding | |
| p-hSTING 342–379 | 16.8 | 15.3 | |
| p-hSTING 342–370 | no binding | no binding | |
| p-hSTING 353–379 | 22.6 | 25.1 | |
| p-hSTING 366–379 | 35.6 | 30.8 | |
| hSTING 366–379, F376W | ~190 | ||
| hSTING 155–379, T376E | ~62.0 | ||
| hSTING 356–379, T376E F378W | ~50.0 | ||
| hSTING 366–379, T376E F378W S379E | ~127 | ||
| hSTING 366–379, T376E F378W 3379M | 43.9 | ||
| hSTING 366–379, T376E F378W S379I | ~59.0 | ||
| hSTING 366–379, T376E F378W S379F | 27.1 | ||
| hSTING 356–379, T376E F376M S379W | 2.6 | 2.7 | |
| hSTING 155–379, T376E F378M S379VV | 1.4 | 1.2 |
Extended data Table 2.
Sequences of STING C-terminal region from 60 mammals. The conserved pLxIS motif and PLPLRT/SD motif are highlighted in blue and red.
| Species | NCBI Entry | Sequence |
|---|---|---|
| Acinonyx jubatus | XP_014919539.1 | NLLISGMEQPLPLRTDVF |
| Ailuropoda meianoleuca | XP_002912620.1 | KLLISGLEQPLPLRTDVF |
| Balaenoptera acutorostrata scammoni | XP_007172318.1 | ELLISGMEQPLPLRSDVF |
| Bison bison bison | XP_010848632.1 | ELLISGLEKPLPLRSDVF |
| Bos taurus | NP_001039822.1 | ELLISGLEKPLPLRSDVF |
| Callithrix jacchus | XP_002744307.1 | ELLISGMEKPLPLRSDLF |
| Came/us bactrianus | NP_001306707.1 | ELLISGMEQPLPLRTDVF |
| Canis lupus familiaris | XP_005617314.1 | NLFISGLEQPLPLRTDIF |
| Capra hircus | NP_001306207.1 | ELLISGMEKPLPLRSDVF |
| Cariito syrichta | XP_008046376.1 | ELLISGMEKPLSLHTDFF |
| Castor canadensis | XP_020023516.1 | RLLISDMEQPLPLRTDII |
| Cavia porcellus | XP_003477199.1 | QLLISEMEQPLPLRTDIF |
| Ceratotherium simum simum | XP_014650944.1 | ELLISGTEQPLPLRSDIF |
| Chrysochloris asiatica | XP_006866201.1 | RFLISDIEQPLPLRTDVF |
| Condylura cristata | XP_012583163.1 | QLLISDMDMPLPLRTDLF |
| Dipodomys ordii | XP_012875724.1 | KLLISDMEQPLPLRTDLI |
| Echinops telfairi | XP_004697079.1 | MFLISDMEQPLPLRTDVF |
| Enhydra iutris kenyoni | XP_022349371.1 | KLLISGLEQPLPLRTDLF |
| Equus asinus | XP_014709351.1 | QLLISGMEQPLPLRSDVF |
| Equus caballus | XP_005599422.1 | QLLISGMEQPLPLRSDVF |
| Erinaceus europaeus | XP_007517598.2 | ELLISGMEQPLPLRTDIF |
| Felis catus | XP_023111467.1 | NLLISGMEQPLPLRTDVF |
| Gorilla gorilla | XP_004042660.1 | ELLISGMEKPLPLRTDFS |
| Heterocephalus glaber | XP_021111568.1 | QLLISGMDQPLPLRTDIF |
| Homo sapiens | NP_938023.1 | ELLISGMEKPLPLRTDFS |
| Ictidomys tridecemlineatus | XP_005327332.1 | KLLISDMEQPLPLRTDVF |
| Jaculus jaculus | XP_004652491.1 | KLLISDTDQPLPLRTGFT |
| Leptonychotes weddellii | XP_006730795.1 | KLLISGLEQPLPLRTDVF |
| Lipotes vexillifer | XP_007461503.1 | ELLISGMEQPLPLRSDVF |
| Loxodonta africana | XP_003404845.1 | KLLISGLEQPLPLRSDVF |
| Macaca mulatta | XP_014996496.1 | ELLISGMEKPLPLRTDFS |
| Mandrillus leucophaeus | XP_011852614.1 | ELLISGMEKPLPLRTDFS |
| Microcebus murinus | XP_012604522.1 | ELLISGMEQPLPLRTDIF |
| Monodelphis domestica | XP_016284133.1 | ELLITGMEQPLPLRTDGF |
| Mus musculus | NP_082537.1 | RLLISGMDQPLPLRTDLI |
| Mustela putorius furo | XP_012907883.1 | KLLISGLEQPLPLRTDLF |
| Myotis lucifugus | XP_006086577.1 | QLLISEMDRPLPLRTDIF |
| Neomonachus schauinslandi | XP_021557627.1 | KLLISGLEQPLPLRTDVF |
| Nomascus leucogenys | XP_012360436.1 | ELLISGLEKPLPLRTDFS |
| Odobenus rosmarus divergens | XP_004397863.1 | KLLISGLEQPLPLRTDIS |
| Odocoileus virginianus texanus | XP_020764082.1 | ELLISGMEKPLPLRSDVF |
| Orcinus orca | XP_004280346.1 | ELLISGMEQPLPLRSDIF |
| Orycteropus afer afer | XP_007937166.1 | KFLISGLEQPLPLRTDVF |
| Oryctolagus cuniculus | XP_002710295.1 | QLLISGMEQPLPLRTDVF |
| Otolemur gamettii | XP_012663496.1 | RLLISGMEQPLPLRTDIF |
| Ovis aries | XP_004008906.1 | ELLISGMEKPLPLRSDVF |
| Pan troglodytes | XP_001135484.1 | ELLISGMEKPLPLRTDFS |
| Panthera tigris altaica | XP_007077937.1 | NLLISDMEQPLPLRTDVF |
| Pantholops hodgsonii | XP_005971883.1 | ELLISGMEKPLPLRSDVF |
| Papio anubis | XP_003900232.1 | ELLISGMEKPLPLRTDFS |
| Pongo abelii | XP_002815998.1 | ELLISGMEKPLPLHTDFS |
| Propithecus coquereli | XP_012501803.1 | ELLISGMEQPLPLRTDIF |
| Pteropus vampyrus | XP_011380568.1 | ELLISGMDQPLPLRTDIF |
| Rattus norvegicus | NP_001102592 1 | RLLISGMEQPLPLRTDLI |
| Sarcophilus harrisii | XP_003756672.1 | QLLISGMEQPLSLRTDGF |
| Sus scrofa | NP_001136310.1 | ELLISGMEQPLPLRSDIF |
| Trichechus manatus latirostris | XP_004381119.2 | KLLISGMEQPLPLRTDVF |
| Tursiops truncatus | XP_019780073.1 | ELLISGMEQPLPLRSDIF |
| Ursus maritimus | XP_008689754.1 | KLLISGLEQPLPLRTDVF |
| Vicugna pacos | XP_015094987.1 | ELLISGMEQPLPLRTDVF |
Extended data Table 3.
Data collection and refinement statistics
| hTBKl/hSTING (155–379, EMW) Complex | mTBK 1/hSTING (WT, 342–379) Complex | hTBKl/Se-Met hSTING (EMW, 342–379) Complex | |
|---|---|---|---|
| Data collection | |||
| Space group | P6522 | P6522 | P6522 |
| Cell dimensions | |||
| a, b, c (Å) | 250.69, 250.69, 239.24 | 249.51,249.51,243.78 | 250.57, 250.57, 236.83 |
| α, β, γ (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 |
| Resolution (Å) | 3.40 (3.49 to 3.40) * | 3.17 (3.24 to 3.17) | 3.40 (3 49 to 3.40) |
| Rmerge | 0.152 (2.0) | 0.184 (2.7) | 0.437 (7.2) |
| I/σI | 7.8 (1.0) | 13.8(1.0) | 8.0 (0.7) |
| Completeness (%) | 99.3 (99.5) | 100.0 (100.0) | 100.0 (100.0) |
| Redundancy | 6.2 (5.9) | 12.3(11.3) | 17.4(17.4) |
| Refinement | |||
| Resolution (Å) | 3.40 | 3.17 | |
| No. reflections | 60195 | 75886 | |
| Rwork/ Rfree | 0.244/0.258 | 0.214/0.230 | |
| No. atoms | |||
| Protein | 10829 | 10538 | |
| Ligand/ion | 68 | 68 | |
| Water | 0 | 0 | |
| B-factors | |||
| Protein | 137,6 | 108.2 | |
| Ligand/ion | 117.8 | 90.6 | |
| Water | |||
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.002 | 0.012 | |
| Bond angles (°) | 0.577 | 1.086 | |
One crystal was used to collect each of the dataset.
Values in parentheses are for the highest-resolution shell.
Supplementary Material
Acknowledgements
The Berkeley Center for Structural Biology is supported in part by the National Institutes of Health, National Institute of General Medical Sciences, and the Howard Hughes Medical Institute. The Advanced Light Source (ALS) is supported by the Director, Office of Science, Office of Basic Energy Sciences, U.S. Department of Energy under Contract No. DE-AC02–05CH11231. We thank Dr. Russell Vance of UC Berkeley and Dr. Craig Kaplan of University of Pittsburgh for valuable discussions. We acknowledge the use of EM facilities at the Biological Science Imaging Resource, which is supported by Florida State University and NIH grants S10 RR025080 and S10 OD018142. We acknowledge TAMU/LBMS for MS/MS analyses. This research was supported in part by the Cancer Prevention and Research Institute of Texas (Grant RP150454 to P.L.), the Welch Foundation (Grant A-1931–20170325 to P.L.) and National Institute of Health (Grant R01 AI145287 to P.L. and R.W.).
Footnotes
Author Information Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing interests. Readers are welcome to comment on the online version of the paper. Correspondence and requests for materials should be addressed to P.L. (pingwei@tamu.edu).
Data Availability Coordinates and structure factors of hTBK1 in complex with hSTING CTD EMW mutant and mTBK1 in complex with hSTING CTT have been deposited in the Protein Data Bank with accession codes 6O8B and 6O8C, respectively.
References
- 1.Chen Q, Sun L & Chen ZJ Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol 17, 1142–1149, doi: 10.1038/ni.3558 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Roers A, Hiller B & Hornung V Recognition of Endogenous Nucleic Acids by the Innate Immune System. Immunity 44, 739–754, doi: 10.1016/j.immuni.2016.04.002 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Barber GN Innate immune DNA sensing pathways: STING, AIMII and the regulation of interferon production and inflammatory responses. Curr Opin Immunol 23, 10–20, doi: 10.1016/j.coi.2010.12.015 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kato H, Takahasi K & Fujita T RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol Rev 243, 91–98, doi: 10.1111/j.1600-065X.2011.01052.x (2011). [DOI] [PubMed] [Google Scholar]
- 5.Paludan SR & Bowie AG Immune sensing of DNA. Immunity 38, 870–880, doi: 10.1016/j.immuni.2013.05.004 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burdette DL & Vance RE STING and the innate immune response to nucleic acids in the cytosol. Nat Immunol 14, 19–26, doi: 10.1038/ni.2491 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Sun L, Wu J, Du F, Chen X & Chen ZJ Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791, doi: 10.1126/science.1232458 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu J & Chen ZJ Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol 32, 461–488, doi: 10.1146/annurev-immunol-032713-120156 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Zhang X et al. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell 51, 226–235, doi: 10.1016/j.molcel.2013.05.022 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ablasser A et al. cGAS produces a 2’−5’-linked cyclic dinucleotide second messenger that activates STING. Nature 498, 380–384, doi: 10.1038/nature12306 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diner EJ et al. The Innate Immune DNA Sensor cGAS Produces a Noncanonical Cyclic Dinucleotide that Activates Human STING. Cell Rep 3, 1355–1361, doi: 10.1016/j.celrep.2013.05.009 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao P et al. Cyclic [G(2’,5’)pA(3’,5’)p] Is the Metazoan Second Messenger Produced by DNA-Activated Cyclic GMP-AMP Synthase. Cell 153, 1094–1107, doi: 10.1016/j.cell.2013.04.046 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu J et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830, doi: 10.1126/science.1229963 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishikawa H & Barber GN STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678, doi: 10.1038/nature07317 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishikawa H, Ma Z & Barber GN STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461, 788–792, doi: 10.1038/nature08476 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka Y & Chen ZJ STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal 5, ra20, doi: 10.1126/scisignal.2002521 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma S et al. Triggering the interferon antiviral response through an IKK-related pathway. Science 300, 1148–1151, doi: 10.1126/science.1081315 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Fitzgerald KA et al. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol 4, 491–496, doi: 10.1038/ni921 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Liu S et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347, aaa2630, doi: 10.1126/science.aaa2630 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Zhao B et al. Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins. Proc Natl Acad Sci U S A 113, E3403–3412, doi: 10.1073/pnas.1603269113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin R, Heylbroeck C, Pitha PM & Hiscott J Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol 18, 2986–2996 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burdette DL et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518, doi: 10.1038/nature10429 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shang G, Zhang C, Chen ZJ, Bai XC & Zhang X Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP. Nature 567, 389–393, doi: 10.1038/s41586-019-0998-5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shu C, Yi G, Watts T, Kao CC & Li P Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat Struct Mol Biol 19, 722–724, doi: 10.1038/nsmb.2331 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shu C et al. Structural insights into the functions of TBK1 in innate antimicrobial immunity. Structure 21, 1137–1148, doi: 10.1016/j.str.2013.04.025 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Powell HR, Battye TGG, Kontogiannis L, Johnson O & Leslie AGW Integrating macromolecular X-ray diffraction data with the graphical user interface iMosflm. Nat Protoc 12, 1310–1325, doi: 10.1038/nprot.2017.037 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221, doi: 10.1107/S0907444909052925 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheldrick GM Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr D Biol Crystallogr 66, 479–485, doi: 10.1107/S0907444909038360 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emsley P & Cowtan K Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132, doi: 10.1107/S0907444904019158 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Shalem O et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87, doi: 10.1126/science.1247005 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.