Abstract
Background
Giardia lamblia is a very common cause of gastrointestinal symptoms worldwide. There are several methods for the diagnosis of Giardia infection, however none are ideal. We aim to find a new, microRNA-based method that will improve the currently available diagnostic methods for giardiasis.
Methods
Deep-sequence profiling of Giardia small-RNA revealed that miR5 and miR6 are highly expressed in Giardia. These miRNAs were tested by qRT-PCR in duodenal biopsies of patients with giardiasis who were positive by microscopic pathological evaluation. The gastric biopsies of the same patients served as negative control tissues. Additionally, these miRNAs were evaluated in stool samples of patients with proven giardiasis.
Results
All histologically proven duodenal biopsies of patients with Giardia infection were positive for Giardia miR5, with a mean threshold cycle (Ct) of 23.7, as well as for Giardia DNA qPCR (16S-like gene, mean Ct 26.3). Gastric biopsies which were tested as a control all were negative. Stool evaluation of miR6 in patients with giardiasis showed 90% specificity but only 66% sensitivity, and a lower accuracy rate was obtained with miR5.
Conclusion
Giardia miR5 testing in duodenal biopsies may be a new method for the diagnosis of giardiasis. It seems to be more sensitive when compared with testing for Giardia DNA by qPCR in duodenal biopsies. It will be important to investigate the contribution of routine Giardia miRNA testing in duodenal biopsies from patients with persistent abdominal symptoms
Author summary
Giardiasis is a major cause of diarrheal disease throughout the world. It is more common in areas with poor sanitation such as in many low-income countries, but it occurs in high-income countries as well. It is the most commonly identified intestinal parasite in the United States and it is endemic in other industrialized countries. The causative agent is the flagellate protozoan Giardia lamblia, and transmission is mainly by the fecal-oral route. The basic method of diagnosis is stool examination. It is usually found through stool microscopy examination which should be performed on fresh stool and repeated in 3 days. Despite some newer diagnostic methods, Giardia is still difficult to detect, often leading to misdiagnoses.
In this study we show that using Giardia microRNA (miR5) as a marker for Giardia infection in duodenal biopsies may be a new method for diagnosis of giardiasis. It appears to be more sensitive than histological diagnosis and also more sensitive than Giardia DNA testing in duodenal biopsies. Interestingly, in our patients, duodenal biopsies were done for persistent abdominal symptoms and the finding of Giardia in their biopsy was unexpected. Thus, testing duodenal biopsies for Giardia miRNA in patients with persistent abdominal symptoms might contribute to diagnosis and prompt treatment for those with giardiasis.
Introduction
Giardia lamblia (also known as G. intestinalis or G. duodenalis) is the causative agent of giardiasis. Among the intestinal protozoan parasites, it is one of the three most common agents of diarrhea worldwide [1]. Giardia species have two life cycle stages; the flagellated trophozoite that attaches to the intestinal microvilli and an infectious cyst that persists in the environment [2]. Transmission to humans occurs either through direct person-to-person contact in environments with compromised hygiene levels [3] or by the contamination of water or food by the cysts, the resistant form. In industrialized countries, Giardia is the most common parasite identified in stool samples [4, 5]. Symptoms include diarrhea, flatulence, excessive fatigue, nausea, foul smelling stools, abdominal cramps and weight loss. In about 16% of patients the disease may become chronic, with long-term effects such as loose stools, malnutrition, growth delays, cognitive impairment, abdominal pain, malabsorption and malaise [6, 7].
In travelers returning from tropical countries, persistent abdominal symptoms (PAS) including chronic diarrhea, abdominal pain, flatulence and fatigue are common [8]. Unfortunately, in most cases, the etiology of these complaints remains unknown, partially due to the low sensitivity of current tests. In only about a third of the cases pathogens can be identified, the most common being Giardia [9]. Although the majority of returning travelers with chronic complaints remain undiagnosed, many of them nonetheless respond to anti-parasitic treatment [8].
Currently, there are several diagnostic methods for Giardia, but none serves as a real gold- standard. Classically, laboratory diagnosis is performed by microscopic examination of stool samples, ‘‘ova and parasite examination” (O&P) [10]. The recommendation is to collect at least three independent, preferably watery stool specimens to maximize the sensitivity of the detection. Moreover, the stool should be fresh because the trophozoites break down very quickly [10]. Direct fluorescent antibody assay and stool antigen detection using enzyme-linked immunosorbent assay (ELISA) have been accepted as more sensitive tools for diagnosis of giardiasis and have provided a potentially attractive alternative to conventional O&P examinations [11, 12]. However, studies have shown that at least a pair of stool samples is needed for sufficient sensitivity [11]. Furthermore, in recent years, PCR-based nucleic acid detection methods have shown higher sensitivity compared to microscopy and antigen detection tests [13]. Since Giardia parasites reside in the small intestines, searching for it in the duodenum seems reasonable [14]. A prospective study done in Italy has found giardiasis through direct histological examination of duodenal biopsy specimens in 9 of the 137 patients (6.5%) with symptoms consistent with irritable bowel syndrome (IBS) and no alarming signs [15].
miRNAs are small non-coding regulatory RNAs that can direct post-transcriptional repression of protein synthesis from mRNAs containing miRNA binding sites. In animals, miRNAs have diverse biological functions, including regulation of key aspects of development and life cycles [16]. These molecules, are also found in unicellular organisms, including Giardia species [17] (although the existence of canonical miRNA in these primitive eukaryotes is under debate [18], being that their genome lacks orthologs of the miRNA processing genes DROSHA and XPO5 [19]), and as potential markers have the advantage of being in part genus specific [20, 21] and relatively stable. It was shown that miRNAs are resistant to freeze-thaw cycling, RNase A digestion, and treatment with a high pH solution [22]. In addition, miRNAs were shown to be stable in FFPE specimens [23] and in feces [24]. Therefore, we attempted to identify Giardia in positive duodenal biopsies and stool samples of proven giardiasis patients through detecting Giardia miRNAs by quantitative reverse-transcription-PCR.
Materials and methods
Ethics statement
The study was approved by the Sheba Medical Center ethics Committee (Helsinki), protocol number 1335-14-SMC. The FFPE sample were taken from the archives of the Department of Pathology at Sheba Medical Center. The requirement for informed consent was waived due to the archived nature of the study FFPE specimens. The control newborn fecal samples were collected after the mothers were informed and provided consent. Anonymized frozen fecal samples of Giardia-infected patients were provided by the Helsinki Committee-approved repositories of the parasitology laboratory of the Israeli Ministry of Health and the microbiology laboratory at Sheba Medical Center.
Giardia parasite strains G. lamblia trophozoite were obtained from the BEI Resources (https://www.beiresources.org/Home.aspx). WB clone 6 (NR-9706, assemblage A [25]), Egypt-4 (NR-9231, assemblage A), Mario (NR-9232, assemblage A), Sug (NR-9233, assemblage not determined) and G2M (NR-9232, assemblage not determined).
Giardia growing conditions
Giardia cells were grown in Keister's Modified TYI-S-33 [26], which contains: 2% casein, 1% yeast extract, 1% glucose, 0.2% NaCl, 0.2% L-Cystein, 0.02% L(+)-ascorbic acid, 0.1% K2HPO4·3H2O, 0.06% KH2PO4, 0.00228% ammonium iron (III) citrate 1% bovine bile solution (all from sigma) and 10% bovine calf serum (Gibco), in double distilled water (DDW). The trophozoites were grown in a polystyrene cell culture tube (Greiner bio-one, Cellstar, Cat. No. 163–160) at 37°C under anaerobic conditions.
RNA extraction from trophozoites and miRNA profiling via small RNA sequencing
Total RNA were extracted from one million trophozoite cells of each Giardia isolate using mirVana miRNA isolation kit (Ambion, AM1561). Barcoded cDNA libraries of small RNA (19–35 nt) were prepared from the total RNA using an in-house method, as previously described [27]. Libraries were deep-sequenced on an Illumina sequencer (HiSeq 2500). Additionally, in order to facilitate miRNA discovery which requires identification of passenger strand reads, that are typically rare compared to the mature strand, we subjected the cDNA libraries to cleavage with a duplex-specific nuclease (DSN, Evrogen, cat. #EA003) [28] and sequenced the cleaved libraries as well. Resulting FASTQ read files were processed as described [29], and demultiplexed data were deposited at NCBI’s gene expression omnibus (GEO record GSE116101).
Bioinformatics analysis
Discovery of miRNA in the demultiplexed libraries was performed using the miRDeep2 algorithm [30]. For this purpose, we combined the intact and DSN-cleaved cDNA library FASTQ files. We allowed reads up to length 29 nt to be included in the analysis, as opposed to the default 25 nt, because a previous investigation of Argonaute-associated small RNAs in Giardia reported a mode length of 26 nt [17], concordant with structural modeling of Giardia Dicer [31]. Previously reported putative Giardia miRNA (S1 Table, adapted from Liao et al [17–19, 32–37]), were specifically sought, by including them as presumed known mature miRNA in the miRDeep2 analysis. For genome mapping, we downloaded the GiardiaDB-37_GintestinalisAssemblageAWB_Genome.fasta file (2018-04-19) from GiardiaDB.
Patient samples
FFPE duodenal biopsy tissue blocks from histopathology proven giardiasis patients were obtained from the Department of Pathology, Sheba Medical Center. In addition, gastric biopsies from the same patients and from unrelated patients were used as negative controls. Fecal samples from microscopy and/or antigen proven giardiasis patients were collected from the Institute of Geographic Medicine and Tropical Diseases, Sheba Medical Center, and from the Parasitology Laboratory of the Israeli Ministry of Health, Jerusalem. Negative control fecal samples were collected from newborns and from infants that had just started eating but are still not attending day-care. Stool samples were stored at -80°C until analysis.
DNA and RNA extraction from FFPE
DNA was extracted from 10–20 slices of FFPE tissue blocks by the QIAamp DNA Mini Kit (Qiagen, 51304) according to the manufacturer’s protocol with minor changes. Proteinase K incubation was performed overnight and then additional proteinase K was added for 1 hour. After adding AL buffer and ethanol, the samples were incubated in -20°C for 1 hour. Subsequent steps were according to protocol. Isolation of RNA from FFPE biopsies was performed using miRNeasy FFPE Kit (Qiagen, 217504) according to the manufacturer’s protocol.
RNA extraction from human stool samples
0.1g of stool was added to 200 μl of 2% polyvinylpolypyrolidone (PVPP, Sigma) suspension and frozen in -20°C overnight. Subsequently, the suspension was heated for 10 min at 100°C [1]. Isolation of miRNA from the suspension was performed using mirVana miRNA isolation kit (Ambion, AM1561) according to the manufacturer’s protocol.
Real time qRT-PCR/qPCR
The ABI Quant Studio6 Flex Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) was used in all qRT-PCR and qPCR experiments.
One step qRT-PCR
Detection of miRNAs, isolated from fecal samples, by Eva Green technology, was performed as previously described [38, 39].
Primers for Giardia miRNA detection in fecal samples
Giardia miR5
RP1-miR5: 5'-GGACGGTAGCAAGCAAAGAGAGAGAAGGCTCGGACAT-3'
RP2-miR5:5'-GGGATTCTGGAAGATGATGATGACGATGCTTCCTTGG-3'
P1: 5'-GGACGGTAGCAAGCAAAGAGAGAG-3'
P2: 5'-GGGATTCTGGAAGATGATGATGAC-3'
Giardia miR6:
RP1-miR6: 5'-GGACGGTAGCAAGCAAAGAGAGAGCAGAATACGACAAA-3'
RP2-miR6: 5'-GGGATTCTGGAAGATGATGATGACGACGCGTGACGAAG'-3'
P1: 5'-GGACGGTAGCAAGCAAAGAGAGAG-3'
P2: 5'-GGGATTCTGGAAGATGATGATGAC-3'
Human RNU6B:
RP1-RNU6B: 5'-GGACGGTAGCAAGCAAAGAGAGAGAAAAATATGGAACGCTTCACGAA-3'
RP2-RNU6B: 5'-GGGATTCTGGAAGATGATGATGACCGCAAGGATGACACGCAAA-3'
P1: 5'-GGACGGTAGCAAGCAAAGAGAGAG-3'
P2: 5'-GGGATTCTGGAAGATGATGATGAC-3'
TaqMan microRNA qRT- PCR
qRT-PCR of miRNAs isolated from FFPE samples was done according to manufacturer’s protocol (Applied Biosystems), using custom primers designed according to the Applied Biosystems miRNA quantification method [40], for either Giardia miR5 (Applied Biosystems Assay-ID 5737335_1) or Giardia miR6 (Applied Biosystems Assay-ID 5710263_1).
TaqMan DNA qPCR
qPCR of DNA isolated from FFPE samples, was done with specific primers to Giardia small subunit ribosomal (16S-like) RNA gene by TaqMan qPCR as described by Verweij et al. [1].
Statistics
Statistical significance was evaluated using Student’s t-test or One-way ANOVA. A probability value of p< 0.05 was considered significant. For the comparison of miR5 Ct with DNA Ct, paired student t-test was use. Unpaired t-test and area under the Receiver Operating Characteristic (ROC) curve and the cutoff Ct were computed using GraphPad Prism version 5.00 for Windows, GraphPad Software, San Diego California USA, www.graphpad.com. Briefly, after entering Ct values (infected and healthy control), a series of cutoff values is proposed. The graph plots percentage sensitivity versus percentage of false positive rate (100-specificity) for the different cutoff points. The optimal cutoff is determined as the cutoff with the highest likelihood ratio [defined as %sensitivity / (100-%specificity)]. ROC curve data is presented in S2 and S3 Tables.
Results
Characterization of Giardia miRNAs
Five different Giardia isolates were grown. Total RNA was extracted and small RNA libraries were prepared and deep-sequenced. Results and insights from this small RNA transcriptome analysis are presented in the S1 Appendix. Our deep-sequencing data confirmed the presence of reads arising from two putative miRNAs that have been previously reported as having highest expression levels in Giardia trophozoites, miR5 and miR6 [17], and these were used for subsequent patient-sample analyses. In addition, these two miRNAs are conserved between the three main assemblages infecting humans; A, B and E. miR5 is 26 nt long and 100% identical in all three assemblages. miR6 is 28 nt long and 100% identical in assemblages A and E and 24 out of the 28 are identical in assemblage B (S1 Table).
Detection of Giardia miRNAs in human duodenal biopsies
Eight duodenal biopsies that were determined as Giardia positive by the pathologist were studied. Samples were from patients referred to gastroscopy due to prolonged gastrointestinal system complaints or anemia (Table 1) but had not been diagnosed with giardiasis prior to gastroscopy. RNA and DNA were extracted from biopsy material and analyzed using TaqMan® MicroRNA Assays and TaqMan qPCR DNA Assay, respectively. As can be seen in Table 1, all samples were positive for Giardia miR5 with mean threshold cycle (Ct) value of 23.7. miR6 was mostly undetectable and thus not further examined in duodenal biopsies. We also performed a qPCR test for Giardia DNA using specific primers to Giardia small subunit ribosomal (16S-like) RNA gene by TaqMan qPCR as described by Verweij et al. [1]. Comparison of the Ct levels of Giardia miR5 vs. Giardia DNA on the same samples, using equivalent volumes of the extracted nucleic acid, shows that miR5 yielded lower mean Ct values, 23.7 versus 26.3 (by paired t-test p = 0.004). These findings suggest that the miRNA amplification may be more robust than DNA amplification.
Table 1. miRNA quantification in duodenal and gastric biopsies of positive Giardia patients detected in their duodenum by histological examination.
| Age | Sex | Medical history | Biopsy pathology | Ct miR6 duodenum | Ct miR5 duodenum | Ct DNA | Ct miR5 stomach | |
|---|---|---|---|---|---|---|---|---|
| 1 | 17 | F | Iron deficiency anemia | Positive for Giardia | 33.7 | 22.7 | 26.7 | 39 |
| 2 | 38 | M | Iron deficiency anemia and weight loss in CVID patient | Positive for Giardia | 34.0 | 21.2 | 26.0 | >40 |
| 3 | 46 | M | Iron deficiency and hemorrhoids | Positive for Giardia | 34.0 | 32.9 | 33.8 | - |
| 4 | 56 | M | Iron deficiency in ITP patient | Positive for Giardia | 31.7 | 21.1 | 26.0 | - |
| 5 | 46 | M | Chronic abdominal pain | Positive for Giardia | ND | 25.7 | 27.6 | - |
| 6 | 26 | F | Chronic abdominal pain | Positive for Giardia | ND | 21.5 | 23.3 | - |
| 7 | 8 | F | Chronic abdominal pain | Positive for Giardia | ND | 23.4 | 23.9 | >40 |
| 8 | 5 | M | FTT and iron deficiency anemia | Positive for Giardia | ND | 21.4 | 23.1 | >40 |
| Mean±SD | 33.4±1.1 | 23.7±4 | 26.3±3.4 | |||||
Cutoff for positive Ct for miR5< 33.5. Abbreviations: CVID, common variable immunodeficiency; ITP, immune thrombocytopenic purpura; FTT, failure to thrive.
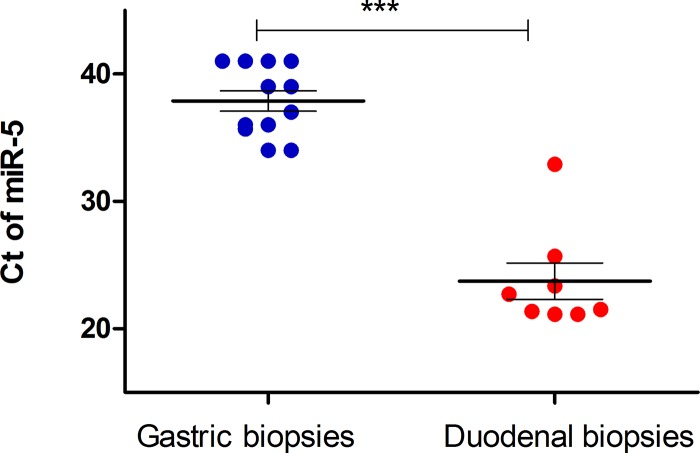
As a negative control, we used gastric biopsies which are not expected to harbor Giardia parasites. Firstly, we used 4 available gastric biopsies from the 8 patients mentioned above with positive duodenal histology. Indeed, no miR5 was detected in gastric biopsies of these patients (Table 1). Six additional gastric biopsies from randomly selected patients who underwent gastroscopy for various reasons showed Ct values above 34. Altogether in all 10 control biopsies miR5 Ct values were above 34 with mean 35.9 (±1.6) compared to 23.7 (±4.0) in the positive cases (p<0.0001) (Fig 1).
Fig 1. Detection of Giardia miR5 in human duodenum biopsies.
RNA was extracted from 8 duodenum biopsies of Giardia positive patients or as control group, from 4 gastric biopsies of Giardia positive patients and from 6 random gastric biopsies. The RNA was subjected to qRT-PCR (see Methods) with specific primers to miR5. Each dot represents patient’s or control individual’s samples. Mean ± SEM are depicted by horizontal lines. ***, P< 0.0001, calculated by unpaired t-test.
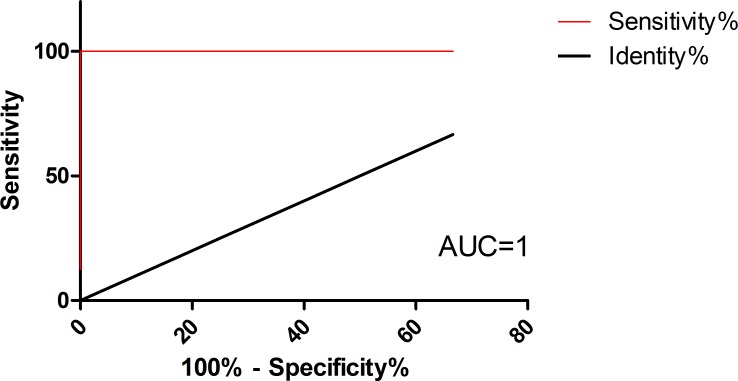
Applying receiver operating characteristic (ROC) curve analysis, we observed highly sensitive and specific results. The calculated area under the curve (AUC) for miR5 was 1, representing a perfect test; [41]. The optimal cutoff is determined as the cutoff with the highest likelihood ratio [likelihood ratio is defined as %sensitivity / (100-%specificity)]. With miR5, Ct <33.5 yielded 100% sensitivity and specificity and therefore likelihood ration of ∞ (95% confidence intervals–sensitivity 63.1% to 100.0%, specificity 59.0% to 100.0%) (Fig 2) and S2 Table.
Fig 2. ROC curve analysis.
Area under ROC curves (AUC) analysis were performed with Ct values of the Giardia miR-5 as predictors for disease category (infected = duodenum positive /control = gastric) as response variable (red lines). P value = 0.0002. ROC curve was fitted using GraphPad Prism version 5.00 for Windows. The diagonal black line reflects the performance of a diagnostic test that is no better than chance level.
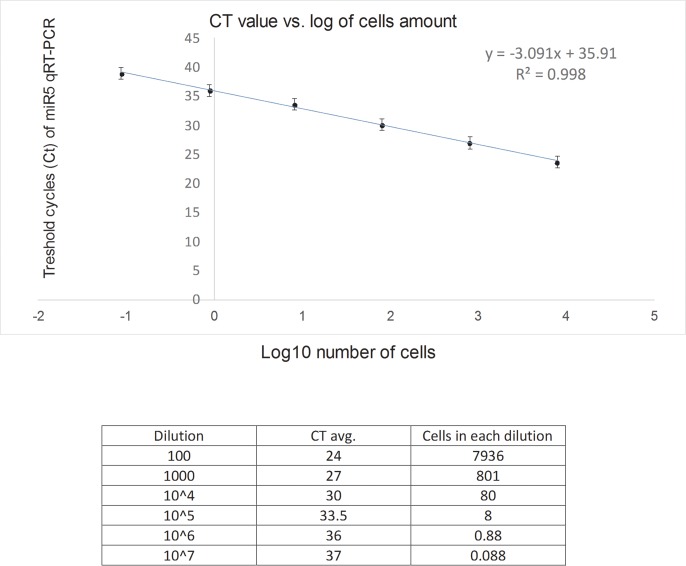
To determine the minimal number of Giardia parasites which can be detected by miR5 qRT-PCR we used an in-vitro model, extracted RNA from parasites, and correlated the number of parasite cells with Ct reading (Fig 3). As can be seen, at 104 dilution we counted ~8 parasites with Ct value of 33.1, slightly below the 33.5 Ct cutoff defined above. Thus, the lower limit of detection of our miR5 assay may be 8 parasite cells.
Fig 3. In-vitro calculation of miR5 qRT-PCR Ct value according to number of parasites.
WB strain Giardia were grown in culture. Number of parasites were counted, and diluted as shown in the table. From each dilution, RNA was extracted and subject to qRT-PCR. The graph represents an average of three repeat, and as the log of parasites concentration versus obtained Ct.
Interestingly, 3 patients with suspected travel-acquired giardiasis without evidence for Giardia by histological examination of duodenum tissue nor by stool microscopy were analyzed for miR5 in the duodenum. In one of these patients the biopsy which was taken before treatment yielded a miR5 Ct value of 31.0, positive for Giardia infection, while the biopsies of the other two patients, which were taken after anti-giardia treatment, yielded borderline Ct levels, 33.8 and 34 (Table 2). All three patients’ symptoms responded to anti Giardia treatment.
Table 2. miR5 detection in duodenal biopsies of patients with travel related persistent abdominal symptoms and no Giardia detection by histological examination.
| Sex | Age | Medical history | Biopsy timing | Improvement after treatment | Biopsy pathology | Ct miR5 duodenum | Ct DNA | |
|---|---|---|---|---|---|---|---|---|
| 1 | M | 34 | Chronic diarrhea | Pre-treatment | Yes | Normal examination | 31.0 | >40 |
| 2 | F | 59 | Chronic flatulence | Post-treatment | Yes | Normal examination | 33.8 | >40 |
| 3 | M | 68 | Chronic diarrhea | Post-treatment | Yes | Normal examination | 34.0 | >40 |
Cutoff Ct for positive miR5: 33.5
Detection of Giardia miRNAs in stool samples
19 stool samples were collected and included in the study. 9 stool samples were taken from patients who suffered from diarrhea with proven giardiasis. In the control group, 10 stool samples were taken from healthy infants; 6 newborns and 4 toddlers, aged 1 to 1.5 years, who have just started eating solid foods and have not yet entered nursery school. Patient samples were tested for the presence of Giardia by three different methods: microscopic examination (O&P), ELISA and DNA PCR. Samples were defined as positive if at least two of the above mentioned diagnostic methods were positive.
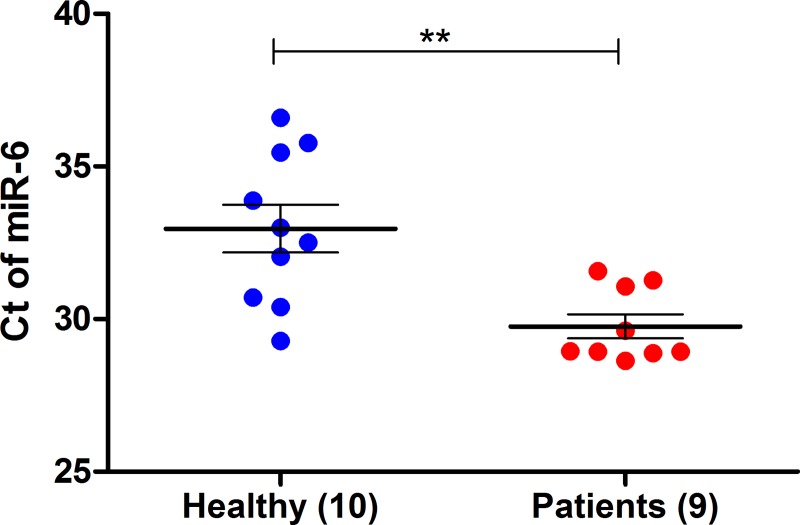
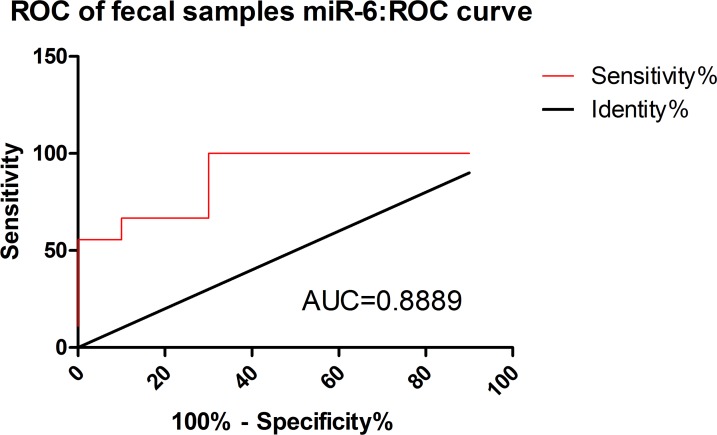
Measurement of miR5 and miR6, in patients’ fecal samples was performed. RNA from stool was extracted and one-step real time RT-PCR analysis was done [39], an assay we have previously used, successfully identifying very small amount of miRNA [38]. As can be seen in Fig 4, a significant difference was found for miR6 between positive patients compared to healthy individuals (p = 0.0025). We applied ROC curve analysis to evaluate the accuracy of our test. As shown in Fig 5, the accuracy of analyzing the presence of Giardia miRNAs extracted from stool samples of infected patients was moderate; with AUC for miR6 is 0.88. The likelihood ratios for miR6, based on, sensitivity of 66.6% and specificity of 90% was Ct of 30.0 (3S Table).
Fig 4. Detection of Giardia miR6 in human feces.
RNA was extracted from the feces and was subjected to one step RT-PCR (see Methods) with specific primers to miR6. Each dot represents patient’s or healthy individual’s samples. Mean ± SEM are denoted by horizontal lines. **, P value, 0.0025, was calculated by unpaired t-test.
Fig 5. ROC curve analysis.
Area under ROC curves (AUC) analysis were performed with Ct values of the Giardia miR6 as predictors for disease category (infected = stool positive /control = stool negative) as response variable (red lines). P value = 0.0043. ROC curve was fitted using GraphPad Prism version 5.00 for Windows. The diagonal black line reflects the performance of a diagnostic test that is no better than chance level.
Unlike miR6, when miR5 was measured, its levels did not differ between the infected and healthy samples (data not shown), implying non-specific (false-positive) amplification.
Discussion
Giardiasis is a significant disease and is underdiagnosed. In this study, our aim was to find a new method that will contribute to the current available diagnostic methods of Giardia infection. In order to do this, we chose to focus on miRNA molecules of Giardia due to their stability, which makes them good candidates to survive complex extraction methods. The fact that miRNA molecules do not exist in the bacteria that constitute most of the gut flora makes them promising in terms of specificity. The use of miRNA for diagnosis of parasitic infections is in its prime [17] and to our knowledge has never been used in the diagnosis of Giardia infection.
We are aware of the discussion of whether Giardia has bona fide miRNA. However, this discussion is less relevant to our work. The precise biochemical definition and function of these small RNAs is of lesser importance with regards to their potential as biomarkers. Nevertheless, since the small-RNA we analyzed were already named as miRNA by others, we maintained this nomenclature (e.g., miR5 and miR6).
We performed deep sequencing analysis of small RNA extracted from 5 different Giardia isolates. Based on their abundance in our data as well as in previous reports by Saraiya et al and Liao et al [17, 18, 36], we decided to focus on two molecules- miR5 and miR6, which by BLAST analysis appear to be non-cognate to the human genome and transcriptome.
Since Giardia resides in the upper gut, we decided to look for the presence of our two chosen molecules in duodenal biopsy specimens of 8 patients who were found to have Giardia parasites on histological examination (Table 1). These 8 patients were referred to gastroscopy due to prolonged gastrointestinal system complaints. Interestingly, none of these patients were suspected of having giardiasis prior to the gastroscopy.
All eight specimens determined as positive by pathology were verified as positive by using a DNA PCR test for Giardia. Applying qRT-PCR for Giardia miR5 was also positive in all samples. Interestingly, miR6 was less efficient, moreover in 4 out of the 8 samples we did not detect it at all (Table 1). This might be because our primers were designed to miR6 of assemblages A and as shown in S1 Table there are 4 nucleotides different between miR6 of assemblage A and assemblage B. Interestingly, although we extracted DNA and RNA from the same amount of FFPE slides, we identified miR5 at lower Ct cycles, that might indicate robustness of miR5 compared to DNA testing. Using gastric biopsies as negative controls, we indeed found that no miR5 was detected even in patients who had Giardia in their duodenal specimen. In addition, all of the control gastric biopsies from non-Giardia patients were negative for miR5.
We had three additional cases with clinically suspected giardiasis but negative Giardia diagnosis by conventional methods and negative duodenal biopsies (Table 2). One of these patients was miR5 positive with Ct of 31 (our calculated cutoff is 33.5). The two other patients were biopsied after empiric treatment and according to the Ct values of 33.8 and 34.0 could be categorized as borderline or negative after treatment, for Giardia miRNA. Moreover, although we do not have any additional supporting evidence that these three patients had giardiasis, all three responded to anti-Giardia treatment. As can be seen in Fig 3A and 3B we generated a calibration-curve of the ratio of Ct obtained in qRT-PCR of Giardia miRNA to the number of parasite cells counted. Based on this curve, Ct of 33.0 indicates 8 parasite cells. Hence, as our calculated cutoff for positive infection is 33.5, it would suggest that qRT-PCR can detect as few as 8 parasites, while based on the mean Ct level (23.7) of histology-positive biopsy there is a need for 100–1000 times more parasites to be detected by histopathology (Fig 3), illustrating the advantage of qRT-PCR miRNA detection in duodenal biopsies. Additionally, these positive Giardia miRNA cases were negative for Giardia DNA which also strengthens the superiority of miRNA on DNA in duodenal biopsies.
As mentioned, none of the research group patients were suspected to have giardiasis prior to the diagnostic gastroscopy (Table 1). This suggests that there is indeed under-diagnosis of Giardia infections in Israel and probably in the industrialized world in general. In addition, our assumption is that giardiasis is an underdiagnosed illness due to the low sensitivity of current diagnostic tools. This assumption was made from observing patients with chronic gastrointestinal complaints, some being returning travelers from the tropics. These patients had negative stool tests for parasites and were left with a presumable diagnosis of irritable bowel syndrome, however empiric anti-protozoal treatment led to significant improvement in 70% [8].
Early studies showed that a single duodenal biopsy might be insufficient for diagnosis of giardiasis. In one of the studies it was suggested that two samples might be sufficient to diagnose all of the cases probably due to the non-homogenous distribution of the parasites in the duodenum [42].
Comparing duodenal aspirate samples to biopsy samples in giardiasis patients had mixed results in different studies. Some studies have shown superiority of duodenal aspirate samples over duodenal mucosal biopsy [43]. Others showed that duodenal mucosal biopsies were more sensitive compared to duodenal aspirate samples [44].
In both studies the test was histological examination and both suggested that duodenal aspirate samples or duodenal mucosal biopsies were more accurate, compared to stool examination.
Fouad and colleagues subjected stool samples and duodenal aspirates from 120 patients with dyspepsia to PCR analysis for Giardia DNA and searched concurrent duodenal biopsy samples for organisms. Giardia was detected by PCR in duodenal aspirates in 23 cases, but organisms were present in biopsy samples from only 2 of these patients (sensitivity 9%). While a study done in a high prevalence setting showed that as many as 44% (96/220) of patients who underwent gastroduodenoscopy due to dyspeptic symptoms had Giardia on duodenal biopsy [45]. In that study it seemed that duodenal diagnosis is much more sensitive than stool microscopy since in only a minority of them (5/85, 6%) Giardia parasites were found in stool examination.
In the last part of our study we aimed to detect miR5 and miR6 molecules in stool samples of patients with proven giardiasis (who were found to be positive by at least two alternative stool tests). As a control group we chose two populations. The first were newborns, who presumably were not yet to be exposed to the parasite. The second control group was toddlers without siblings, who had not yet started attending nursery school but were no longer breastfed and started eating solid foods. These groups had a relatively small chance of exposure to Giardia. The results of our study showed moderate accuracy with 90% specificity and only 66.7% sensitivity in diagnosing Giardia infection using stool miR6. The results of miR5 quantification in stool were even less accurate.
It seems that identifying Giardia miRNA molecules in duodenal specimens shows more potential than in stool sampling. Extraction of miRNA from paraffin is straightforward and less prone to the background noise that exists in the stool due to heavy bacterial burden. Giardia parasites reside in the duodenum where they reach high cell counts, whereas in the stool their numbers are relatively small and excretion is periodic. The fact that miR5 was identified in duodenal specimens but not stool samples, and vice versa miR6, suggests differences in expression of miRNA molecules between Giardia trophozoites (exist in duodenum) and Giardia cysts (exist in stool). miRNA libraries that we have built were derived from sequencing Giardia cultures in its trophozoite form. That is also the main form of the parasite residing in the duodenum, though in a different environment.
The main limitation of our study is the relatively small sample numbers in both the experimental and the control groups. Another limitation is that we have assessed only two miRNA molecules.
In conclusion, miR5 testing for Giardia infection in duodenal biopsies may be a breakthrough method for diagnosis of giardiasis that has a potential of being more sensitive than current methods. Obviously, we do not suggest duodenal biopsies for each patient suspected of giardiasis. However, we think that qRT-PCR for Giardia miRNAs should be one of the tests in patients undergo endoscopy investigation for undiagnosed persistent abdominal symptoms.
We want to emphasize that fact the duodenal biopsy in our samples were taken from patients that suffer from persistent abdominal symptoms and were not suspected of being infected with Giardia. Only the pathologist, unexpectedly, detect Giardia in there duodenal biopsy. Therefore, we think that qRT-PCR for Giardia miRNAs should be one of the tests such patients undergo. Therefore, it would be important to further investigate the contribution of Giardia miRNA testing in duodenal biopsies and duodenal aspirates from patients with persistent abdominal symptoms.
Supporting information
Included in this appendix are:
(DOCX)
The result_16_06_2018_t_13_52_59.html file is an index, through which pdf plot can be accessed.
(ZIP)
The output shows collapsed read pile-ups for every proposed miRNA precursor and lists reasons for disqualification. The sequences are sorted by abundance and their source library is specified. For example, the sequence “Gi3_8900428_x63185” was found 63185 times in library Gi3
(PDF)
We specifically sought these sequences in the current analysis, by including them as presumed known mature miRNA in the miRDeep2 analysis.
(DOCX)
Calculation of the optimal cutoff as determined as the cutoff with the highest likelihood ratio [defined as %sensitivity / (100-%specificity)].
(DOCX)
Calculation of the optimal cutoff as determined as the cutoff with the highest likelihood ratio [defined as %sensitivity / (100-%specificity)].
(DOCX)
All are lowly expressed, and thus unlikely to exert significant regulation.
(PDF)
The sequences are sorted by abundance and their source library is specified (as in S1 Text).
(PDF)
The sequences are sorted by abundance and their source library is specified (as in S1 Text).
(PDF)
Data Availability
All sequencing data were deposited at NCBI’s gene expression omnibus (GEO record GSE116101).
Funding Statement
The research was funded by grant from the The Israel Science Foundation (ISF) I-CORE program number 41/11 to Ben-Dov Z. Iddo. In addition the research was funded by grant from the Sheba Medical Center internal grant, titled: Identifying MicroRNAs in the stool as a diagnostic tool for human Giardiasis to Avni Dror and Schwartz Eli. We declare that the funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Verweij JJ, Blange RA, Templeton K, Schinkel J, Brienen EA, van Rooyen MA, et al. Simultaneous detection of Entamoeba histolytica, Giardia lamblia, and Cryptosporidium parvum in fecal samples by using multiplex real-time PCR. J Clin Microbiol. 2004;42(3):1220–3. Epub 2004/03/09. 10.1128/JCM.42.3.1220-1223.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nosala C, Dawson SC. The Critical Role of the Cytoskeleton in the Pathogenesis of Giardia. Curr Clin Microbiol Rep. 2015;2(4):155–62. Epub 2016/06/28. 10.1007/s40588-015-0026-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colli CM, Bezagio RC, Nishi L, Bignotto TS, Ferreira EC, Falavigna-Guilherme AL, et al. Identical assemblage of Giardia duodenalis in humans, animals and vegetables in an urban area in southern Brazil indicates a relationship among them. PLoS ONE. 2015;10(3):e0118065 Epub 2015/03/12. 10.1371/journal.pone.0118065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silvestri C, Greganti G, Arzeni D, Morciano A, Castelli P, Barchiesi F, et al. Intestinal parasitosis: data analysis 2006–2011 in a teaching hospital of Ancona, Italy. Infez Med. 2013;21(1):34–9. Epub 2013/03/26. . [PubMed] [Google Scholar]
- 5.Ben-Shimol S, Sagi O, Greenberg D. Differences in prevalence of parasites in stool samples between three distinct ethnic pediatric populations in southern Israel, 2007–2011. Parasitol Int. 2014;63(2):456–62. Epub 2013/11/10. 10.1016/j.parint.2013.10.013 . [DOI] [PubMed] [Google Scholar]
- 6.Di Genova BM, Tonelli RR. Infection Strategies of Intestinal Parasite Pathogens and Host Cell Responses. Front Microbiol. 2016;7:256 Epub 2016/03/15. 10.3389/fmicb.2016.00256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanevik K, Hausken T, Morken MH, Strand EA, Morch K, Coll P, et al. Persisting symptoms and duodenal inflammation related to Giardia duodenalis infection. J Infect. 2007;55(6):524–30. Epub 2007/10/30. 10.1016/j.jinf.2007.09.004 . [DOI] [PubMed] [Google Scholar]
- 8.Nissan B, Lachish T, Schwartz E. The effectiveness of empirical anti-parasitic treatment in returning travellers with persistent abdominal symptoms. Journal of travel medicine. 2018;25(1). Epub 2017/12/13. 10.1093/jtm/tax083 . [DOI] [PubMed] [Google Scholar]
- 9.Ross AG, Cripps AW. Enteropathogens and chronic illness in returning travelers. N Engl J Med. 2013;369(8):784 Epub 2013/08/24. 10.1056/NEJMc1308293 . [DOI] [PubMed] [Google Scholar]
- 10.Cartwright CP. Utility of multiple-stool-specimen ova and parasite examinations in a high-prevalence setting. J Clin Microbiol. 1999;37(8):2408–11. Epub 1999/07/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanson KL, Cartwright CP. Use of an enzyme immunoassay does not eliminate the need to analyze multiple stool specimens for sensitive detection of Giardia lamblia. J Clin Microbiol. 2001;39(2):474–7. Epub 2001/02/07. 10.1128/JCM.39.2.474-477.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnston SP, Ballard MM, Beach MJ, Causer L, Wilkins PP. Evaluation of three commercial assays for detection of Giardia and Cryptosporidium organisms in fecal specimens. J Clin Microbiol. 2003;41(2):623–6. Epub 2003/02/08. 10.1128/JCM.41.2.623-626.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zboromyrska Y, Hurtado JC, Salvador P, Alvarez-Martinez MJ, Valls ME, Mas J, et al. Aetiology of traveller's diarrhoea: evaluation of a multiplex PCR tool to detect different enteropathogens. Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2014;20(10):O753–9. Epub 2014/03/14. 10.1111/1469-0691.12621 . [DOI] [PubMed] [Google Scholar]
- 14.Fouad SA, Esmat S, Basyoni MM, Farhan MS, Kobaisi MH. Molecular identification of giardia intestinalis in patients with dyspepsia. Digestion. 2014;90(1):63–71. Epub 2014/09/10. 10.1159/000362644 . [DOI] [PubMed] [Google Scholar]
- 15.Grazioli B, Matera G, Laratta C, Schipani G, Guarnieri G, Spiniello E, et al. Giardia lamblia infection in patients with irritable bowel syndrome and dyspepsia: a prospective study. World J Gastroenterol. 2006;12(12):1941–4. Epub 2006/04/13. 10.3748/wjg.v12.i12.1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bushati N, Cohen SM. microRNA functions. Annual review of cell and developmental biology. 2007;23:175–205. 10.1146/annurev.cellbio.23.090506.123406 . [DOI] [PubMed] [Google Scholar]
- 17.Saraiya AA, Li W, Wu J, Chang CH, Wang CC. The microRNAs in an ancient protist repress the variant-specific surface protein expression by targeting the entire coding sequence. PLoS Pathog. 2014;10(2):e1003791 Epub 2014/03/04. 10.1371/journal.ppat.1003791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao JY, Guo YH, Zheng LL, Li Y, Xu WL, Zhang YC, et al. Both endo-siRNAs and tRNA-derived small RNAs are involved in the differentiation of primitive eukaryote Giardia lamblia. Proc Natl Acad Sci U S A. 2014;111(39):14159–64. 10.1073/pnas.1414394111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li W, Saraiya AA, Wang CC. Gene regulation in Giardia lambia involves a putative microRNA derived from a small nucleolar RNA. PLoS neglected tropical diseases. 2011;5(10):e1338 Epub 2011/10/27. 10.1371/journal.pntd.0001338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36(Database issue):D154–8. 10.1093/nar/gkm952 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li SC, Chan WC, Hu LY, Lai CH, Hsu CN, Lin WC. Identification of homologous microRNAs in 56 animal genomes. Genomics. 2010;96(1):1–9. Epub 2010/03/30. 10.1016/j.ygeno.2010.03.009 . [DOI] [PubMed] [Google Scholar]
- 22.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell research. 2008;18(10):997–1006. Epub 2008/09/04. 10.1038/cr.2008.282 . [DOI] [PubMed] [Google Scholar]
- 23.Kakimoto Y, Tanaka M, Kamiguchi H, Ochiai E, Osawa M. MicroRNA Stability in FFPE Tissue Samples: Dependence on GC Content. PLoS ONE. 2016;11(9):e0163125 10.1371/journal.pone.0163125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Link A, Balaguer F, Shen Y, Nagasaka T, Lozano JJ, Boland CR, et al. Fecal MicroRNAs as novel biomarkers for colon cancer screening. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2010;19(7):1766–74. Epub 2010/06/17. 10.1158/1055-9965.EPI-10-0027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monis PT, Caccio SM, Thompson RC. Variation in Giardia: towards a taxonomic revision of the genus. Trends in parasitology. 2009;25(2):93–100. 10.1016/j.pt.2008.11.006 . [DOI] [PubMed] [Google Scholar]
- 26.Keister DB. Axenic culture of Giardia lamblia in TYI-S-33 medium supplemented with bile. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1983;77(4):487–8. Epub 1983/01/01. 10.1016/0035-9203(83)90120-7 . [DOI] [PubMed] [Google Scholar]
- 27.Shilo V, Mor-Yosef Levi I, Abel R, Mihailovic A, Wasserman G, Naveh-Many T, et al. Let-7 and MicroRNA-148 Regulate Parathyroid Hormone Levels in Secondary Hyperparathyroidism. J Am Soc Nephrol. 2017;28(8):2353–63. 10.1681/ASN.2016050585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qiu X, Zhang H, Yu H, Jiang T, Luo Y. Duplex-specific nuclease-mediated bioanalysis. Trends in biotechnology. 2015;33(3):180–8. 10.1016/j.tibtech.2014.12.008 . [DOI] [PubMed] [Google Scholar]
- 29.Farazi TA, Brown M, Morozov P, Ten Hoeve JJ, Ben-Dov IZ, Hovestadt V, et al. Bioinformatic analysis of barcoded cDNA libraries for small RNA profiling by next-generation sequencing. Methods. 2012;58(2):171–87. 10.1016/j.ymeth.2012.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40(1):37–52. 10.1093/nar/gkr688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, et al. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311(5758):195–8. 10.1126/science.1121638 . [DOI] [PubMed] [Google Scholar]
- 32.Chen XS, Collins LJ, Biggs PJ, Penny D. High throughput genome-wide survey of small RNAs from the parasitic protists Giardia intestinalis and Trichomonas vaginalis. Genome Biol Evol. 2009;1:165–75. 10.1093/gbe/evp017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang PJ, Lin WC, Chen SC, Lin YH, Sun CH, Lyu PC, et al. Identification of putative miRNAs from the deep-branching unicellular flagellates. Genomics. 2012;99(2):101–7. Epub 2011/11/29. 10.1016/j.ygeno.2011.11.002 . [DOI] [PubMed] [Google Scholar]
- 34.Li W, Saraiya AA, Wang CC. The profile of snoRNA-derived microRNAs that regulate expression of variant surface proteins in Giardia lamblia. Cellular microbiology. 2012;14(9):1455–73. Epub 2012/05/10. 10.1111/j.1462-5822.2012.01811.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saraiya AA, Li W, Wang CC. A microRNA derived from an apparent canonical biogenesis pathway regulates variant surface protein gene expression in Giardia lamblia. RNA. 2011;17(12):2152–64. 10.1261/rna.028118.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saraiya AA, Wang CC. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008;4(11):e1000224 Epub 2008/12/02. 10.1371/journal.ppat.1000224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang YQ, Chen DL, Tian HF, Zhang BH, Wen JF. Genome-wide computational identification of microRNAs and their targets in the deep-branching eukaryote Giardia lamblia. Comput Biol Chem. 2009;33(5):391–6. 10.1016/j.compbiolchem.2009.07.013 . [DOI] [PubMed] [Google Scholar]
- 38.Meningher T, Lerman G, Regev-Rudzki N, Gold D, Ben-Dov IZ, Sidi Y, et al. Schistosomal MicroRNAs Isolated From Extracellular Vesicles in Sera of Infected Patients: A New Tool for Diagnosis and Follow-up of Human Schistosomiasis. J Infect Dis. 2017;215(3):378–86. 10.1093/infdis/jiw539 . [DOI] [PubMed] [Google Scholar]
- 39.Yan J, Zhang N, Qi C, Liu X, Shangguan D. One-step real time RT-PCR for detection of microRNAs. Talanta. 2013;110:190–5. Epub 2013/04/27. 10.1016/j.talanta.2013.02.028 . [DOI] [PubMed] [Google Scholar]
- 40.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33(20):e179 Epub 2005/11/30. 10.1093/nar/gni178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hajian-Tilaki K. Receiver Operating Characteristic (ROC) Curve Analysis for Medical Diagnostic Test Evaluation. Caspian journal of internal medicine. 2013;4(2):627–35. [PMC free article] [PubMed] [Google Scholar]
- 42.Oberhuber G, Stolte M. Giardiasis: analysis of histological changes in biopsy specimens of 80 patients. J Clin Pathol. 1990;43(8):641–3. Epub 1990/08/01. 10.1136/jcp.43.8.641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zafar MN, Baqai R, Lodi TZ, Ahmad S, Ahmed W, Qureshi H, et al. Giardia lamblia in patients undergoing upper G.I. endoscopy. J Pak Med Assoc. 1991;41(4):74–5. Epub 1991/04/01. . [PubMed] [Google Scholar]
- 44.Gupta SK, Croffie JM, Pfefferkorn MD, Fitzgerald JF. Diagnostic yield of duodenal aspirate for G. lamblia and comparison to duodenal mucosal biopsies. Dig Dis Sci. 2003;48(3):605–7. Epub 2003/05/22. . [DOI] [PubMed] [Google Scholar]
- 45.Yakoob J, Jafri W, Abid S, Jafri N, Hamid S, Shah HA, et al. Giardiasis in patients with dyspeptic symptoms. World J Gastroenterol. 2005;11(42):6667–70. Epub 2006/01/21. 10.3748/wjg.v11.i42.6667 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Included in this appendix are:
(DOCX)
The result_16_06_2018_t_13_52_59.html file is an index, through which pdf plot can be accessed.
(ZIP)
The output shows collapsed read pile-ups for every proposed miRNA precursor and lists reasons for disqualification. The sequences are sorted by abundance and their source library is specified. For example, the sequence “Gi3_8900428_x63185” was found 63185 times in library Gi3
(PDF)
We specifically sought these sequences in the current analysis, by including them as presumed known mature miRNA in the miRDeep2 analysis.
(DOCX)
Calculation of the optimal cutoff as determined as the cutoff with the highest likelihood ratio [defined as %sensitivity / (100-%specificity)].
(DOCX)
Calculation of the optimal cutoff as determined as the cutoff with the highest likelihood ratio [defined as %sensitivity / (100-%specificity)].
(DOCX)
All are lowly expressed, and thus unlikely to exert significant regulation.
(PDF)
The sequences are sorted by abundance and their source library is specified (as in S1 Text).
(PDF)
The sequences are sorted by abundance and their source library is specified (as in S1 Text).
(PDF)
Data Availability Statement
All sequencing data were deposited at NCBI’s gene expression omnibus (GEO record GSE116101).