Abstract
A deep understanding of the immune landscape in human cancer is essential for guiding the development of immunotherapy to benefit more patients with long-lasting efficacy. Now, two studies from Lavin et al. and Chevrier et al. employ mass cytometry to study immune infiltrates in lung adenocarcinoma and clear cell renal cell carcinoma, respectively.
Immune checkpoint inhibitors such as anti-PD-1 and anti-CTLA4 have revolutionized cancer treatment and reignited the field of tumor immunology. These agents have been shown to invigorate the anti-tumor T cell response, control tumor progression, and in some cases induce complete tumor clearance (Topalian et al., 2015). Despite the success, only a subset of patients responds to immunotherapy, and a better understanding of the tumor microenvironment is essential for identifying biomarkers, designing individualized therapy, and defining new therapeutic targets. Thus, large-scale efforts for in-depth characterization of the immune composition in human tumors, such as the two studies reported in this issue of Cell (Lavin et al., 2017; Chevrier et al., 2017), are of tremendous interest to basic and translational research.
The standard approaches using fluorescent or enzyme-linked antibodies to stain sections or disaggregated cells have identified key cell types associated with immune responses in human cancers (Galon et al., 2013); however, such techniques do not allow simultaneous monitoring of many cell subsets and profiling of complex activation states. In contrast, the Cancer Genome Atlas (TCGA) provides high-dimensional and comprehensive profiles of mutations and gene expression from bulk tumors, revealing valuable insights into the tumor-immune-system relationship (Angelova et al., 2015; Li et al., 2016; Rooney et al., 2015); still, it has been challenging to infer high-resolution cell subsets and functional states from gene expression profiles of bulk tumors.
While flow cytometry typically permits panels of up to 10–15 antibodies, single-cell RNA sequencing (scRNA-seq) and mass cytometry (CyTOF) are evolving technologies for highly multiplexed detection of RNA and protein expression, respectively. CyTOF allows simultaneous detection of > 30 markers with antibodies conjugated to heavy metal ions, while scRNA-seq can detect thousands of transcripts. Combining the two approaches should provide a more comprehensive view of the immune cell types that are present in the tumor microenvironment, as well as their functional features. These data could then be used to associate cell types/states with each other and with variables related to disease or response to immunotherapy (Figure 1).
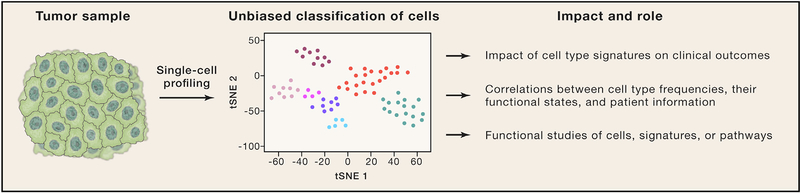
Figure 1. Deconstructing a Human Tumor.
Cells in human tumors are isolated and subjected to single-cell profiling, such as mass cytometry or single-cell RNA sequencing. Cells are then classified into clusters that represent distinct cell types and states, which can be correlated with clinical outcome or other variables. Functional studies are carried out on cells isolated based on markers identified by single-cell profiling.
The work from Lavin et al. (2017) and Chevrier et al. (2017) exploited mass cytometry to map the immune landscape of human cancer. Chevrier et al. (2017) profiled 73 clear cell renal cell carcinoma (cc-RCC) samples and five normal kidneys, whereas Lavin et al. (2017) compared matched primary tumor, non-involved lung, and blood cells isolated from 28 treatment-naive lung adenocarcinoma patients. Both studies characterized the tumors using two panels of > 30 antibodies each: one was tailored to adaptive and the other to innate immune cells. A third panel was used to characterize cytokine expression in a subset of the lung cohort. cc-RCC and lung adenocarcinoma are particularly relevant for immune profiling, as both exhibit substantial cytolytic activity and immunoediting (Rooney et al., 2015) and frequently respond to immunotherapy (Topalian et al., 2015).
The immune compartment in cc-RCC and lung adenocarcinomas was dominated by T cells and monocytes/macrophages (80% of immune cells), with granulocytes, B cells, and natural killer (NK) cells representing the remainder of immune cells. The total number of CD45+ cells per mg of tissue appeared only slightly increased in lung adenocarcinoma versus normal lung. As a fraction of the immune compartment, NK cell numbers were lower in tumor than normal lung, while other immune cell types did not change considerably. Tertiary lymphoid structures (TLS), which have long been observed in cancer and are associated with better prognosis (Palucka and Coussens, 2016), were found in some lung tumor specimens and showed significantly higher fractions of T cells and possibly B cells. Surprisingly, the immune composition was relatively conserved across stages 1, 2, and 3 lung adenocarcinoma, suggesting that immune-targeted therapies may benefit cancer patients starting from the earliest stages. One limitation in interpreting the results of both studies is that non-hematopoietic cells were not assessed, precluding the analysis of the ratio of immune cells to tumor cells (or other cells), a parameter that may impact tumor control.
While at a superficial level, cell populations showed few changes between tumor and normal, both groups leveraged CyTOF multiplex measurements to dig deeper into immune subpopulations. It was here that CyTOF eclipsed what is possible in traditional flow cytometry, especially given the limited cell numbers available from one-of-a-kind human samples. Notably, both reports found changes in T cell subsets, with an increase in Tregs, previously shown to suppress anti-tumor immunity (Palucka and Coussens, 2016), and CD8+PD-1+ T cells. For the lung study, higher percentages of CD8+PD-1+ cells correlated with increased TCR clonality scores in tumors, but not normal lung, suggesting that these CD8+PD-1+ T cells might be tumor specific. CD38 and CD39 were highlighted as potentially targetable activation/exhaustion markers in cc-RCC and lung adenocarcinoma, respectively. CD39 is known to hydrolyze the proinflammatory metabolite ATP and may shift metabolism to shape the immune response (Speiser et al., 2016). The role of CD38 still needs to be determined. Multiplexed staining for surface markers, various cytokines, and the cytolytic effector molecule granzyme B further revealed the functional state of distinct immune cell subsets. CD8+ T cells in lung tumors, for example, expressed less IFNγ and granzyme B compared to normal lung. Additionally, the CD16+ NK cell population was significantly reduced in frequency and IFNγ production in lung adenocarcinomas.
It has been shown that myeloid cells play a critical role in the immune response to cancer (Palucka and Coussens, 2016). Dendritic cells (DCs), particularly the CD141+ DC subset, can promote anti-tumor responses by presenting tumor antigens to CD8+ T cells. The role of monocytes and macrophages is complex, with some subsets suppressing and others promoting anti-tumor immunity, using diverse mechanisms. The limited panel size in traditional flow cytometry has made it difficult to clearly define myeloid cell subsets and assign to them specific functions. Using CyTOF to overcome these limitations, Chevrier et al. (2017) developed a myeloid panel (based on an antibody screen of in-vitro-activated macrophages) and discovered a CD38+ monocyte population with a mature phenotype in cc-RCC tumor specimens. In lung adenocarcinomas, substantial reductions in CD16+ monocytes, CD141+ DCs, basophils, and mast cells were observed, while eosinophil and neutrophil populations remained relatively unchanged compared to normal lung. To some extent, the loss of immune cell types usually patrolling the normal lung parenchyma may simply reflect the loss of healthy tissue; however, the differential loss of some subsets relative to others suggests that tumors might specifically attract some populations, while excluding others, to evade immune-mediated destruction. For example, CD16+ monocytes decreased substantially in comparison to the CD14+ monocyte population. Similarly, the shifts in the DC subsets were intriguing. Whereas the anti-tumor CD141+ DC population decreased, CD1c+ DCs (which are much poorer at cross-presenting tumor antigens to CD8+ T cells) increased in frequency.
Both groups mined their rich datasets for interesting connections between cell types. In lung adenocarcinomas, the percentage of CD16+ NK cells correlated with the percentage of CD16+ monocytes, consistent with the idea that CD16+ monocytes might recruit NK cells to the lung, as previously shown in a mouse model of lung cancer (Palucka and Coussens, 2016). The newly identified CD38+ macrophage population in cc-RCC correlated with the CD8+PD-1+ T cell population, a potentially important finding that warrants further investigation. Such associations are unlikely to be discovered using traditional approaches and illustrate the power of CyTOF.
Many challenges remain for mass cytometry, especially the selection of optimal marker panels that capture the relevant biology (even with panels of more than 30 markers), as well as the functional characterization of the identified cell clusters. scRNA-seq is poised to resolve some of these issues, enabling relatively unbiased classification of cell types based on the transcriptional programs of thousands of cells. The cell-type-specific markers derived from scRNAseq could be used to develop antibody panels for monitoring and isolating specific cell populations and carrying out functional studies.
The technological developments leveraged by these two studies offer a relatively unbiased analysis of immunity in cancer specimens, which should lead to mechanistic hypotheses and potentially new therapeutic targets. To realize the potential of these technologies, several aspects of experimental design must be considered. First, the patient cohort has to be carefully selected, with medical record review, to answer well-posed questions. Second, decisions must be made regarding tissue sampling: Is the tissue obtained from the primary tumor or a metastasis? What part of the tumor is sampled? Is it important to collect adjacent non-tumor tissue and/or matched blood? Are pre- and post-treatment biopsies available? Third, interdisciplinary teams comprised of pathologists, oncologists, surgeons, clinical research coordinators, technologists, computational biologists, and immunologists are necessary to ensure the quality of both the scientific study and patient care.
The integration of layers of information at the single-cell level—including genetic, epigenetic, proteomics, and, importantly, the spatial context of the cell types and states in the tissue—represents the next generation of multi-dimensional analyses that will more deeply define the tumor and its microenvironment. Such complex datasets also call for creation of web-based, interactive data-sharing platforms such as Morpheus and FireCloud. Our toolbox to study human tumor immunology continues to grow at a fast pace and promises to define the contribution of specific cell types and pathways in natural and therapy-induced tumor immunity.
ACKNOWLEDGMENTS
J.H.C. is supported by NIH/NCI training grant T32CA207021. K.P. and N.H. are supported by NIH/NCI 1R01CA208756.
REFERENCES
- Angelova M, Charoentong P, Hackl H, Fischer ML, Snajder R, Krogsdam AM, Waldner MJ, Bindea G, Mlecnik B, Galon J, and Trajanoski Z (2015). Genome Biol. 16, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevrier S, Levine JH, Zanotelli V, Silina K, Schulz D, Bacac M, Ries C, Ailles L, Jewett MAS, Moch H, et al. (2017). Cell 169, this issue, 736–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galon J, Angell HK, Bedognetti D, and Marin-cola FM (2013). Immunity 39, 11–26. [DOI] [PubMed] [Google Scholar]
- Lavin Y, Kobayashi S, Leader A, Amir ED, Elefant N, Bigenwald C, Remark R, Sweeney R, Becker CD, Levine JH, et al. (2017). Cell 169, this issue, 750–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, Jiang P, Shen H, Aster JC, Rodig S, et al. (2016). Genome Biol. 17, 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palucka AK, and Coussens LM (2016). Cell 164, 1233–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney MS, Shukla SA, Wu CJ, Getz G, and Hacohen N (2015). Cell 160, 48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speiser DE, Ho PC, and Verdeil G (2016). Nat. Rev. Immunol 16, 599–611. [DOI] [PubMed] [Google Scholar]
- Topalian SL, Drake CG, and Pardoll DM (2015). Cancer Cell 27, 450–461. [DOI] [PMC free article] [PubMed] [Google Scholar]