Abstract
Mass spectrometry-based peptidomic approaches are powerful techniques to detect and identify the peptide content of biological samples. The present study investigated the limitations of peptidomic approaches using trimethylammonium butyrate isotopic tags to quantify relative peptide levels and Mascot searches to identify peptides. Data were combined from previous studies on human cell lines or mouse tissues. The combined databases contain 2155 unique peptides ranging in mass from 444 to 8765 Da, with the vast majority between 1 and 3 kDa. The amino acid composition of the identified peptides generally reflected the frequency in the Eukaryotic proteome with the exception of Cys, which was not present in any of the identified peptides in the free –SH form but was detected at low frequency as a disulfide with Cys residues, a disulfide with glutathione, or as S-cyanocysteine. To test if the low detection rate of peptides smaller than 500 Da, larger than 3 kDa, or containing Cys was a limitation of the peptidomics procedure, tryptic peptides of known proteins were processed for peptidomics using the same approach used for human cell lines and mouse tissues. The identified tryptic peptides ranged from 516 to 2418 Da, while the theoretical digest ranged from 217 to 7559 Da. Peptides with Cys were rarely detected, and if present the Cys was usually modified S-cyanocysteine. Additionally, peptides with mono- and di-iodo Tyr and His were identified. Taken together, there are limitations of peptidomic techniques and awareness of these limitations is important to properly use and interpret results.
Keywords: Peptidomics, peptides, glutathione, cyanide, cyanocysteine, iodohistidine
Introduction
The development of mass spectrometry-based peptidomic techniques has revolutionized the field of peptides [1–15]. Before development of peptidomics approaches, the detection of peptides was largely performed using radioimmunoassays [16]. While radioimmunoassays are often sensitive, they require antisera to the peptide which take months to generate and characterize, and are limited to known peptides. For the identification of unknown peptides, it was previously necessary to extensively purify each peptide in order for Edman degradation sequencing to be effective. Mass spectrometry-based peptide discovery allowed for identification of peptides in complex mixtures [1–8,11–15]. Quantitative peptidomic approaches facilitated the relative quantification of peptides in mixtures, including both known and previously unknown peptides. Collectively, the various peptidomic approaches led to the discovery of dozens of novel neuropeptides, including some of the most abundant neuropeptides in brain such as peptides named SAAS, PEN, and LEN [17]. Peptidomics also led to the discovery that peptides derived from proteasome-mediated cleavage of intracellular proteins exist within the cell [18–20]. The proteasome cleaves proteins into peptides, and it was previously thought that these peptides are highly unstable due to the activity of intracellular aminopeptidases [21]. This hypothesis was shown to be incorrect based on the discovery that numerous peptides derived from cytosolic, nuclear, and mitochondrial proteins are present in tissues and cell extracts [18–20,22,23]. Many of these peptides require proteasome activity for their production, with levels of peptides affected by treatment of cells with proteasome inhibitors [20,24,25]. These and many other peptidomic studies have led to major advances in our knowledge of peptides in biological samples.
Most of the literature describing peptidomic techniques and their uses has focused on the advantages of these techniques, not on their limitations. In 2000, my laboratory began using peptidomics to detect and identify peptides in mouse extracts that had been purified on affinity columns to isolate neuropeptide processing intermediates [17,26,27]. This affinity procedure was necessary to detect neuropeptides over the background of postmortem protein degradation fragments. In 2003, Svennson and colleagues found that the postmortem changes could be prevented by heating mouse brain with focused microwave irradiation prior to dissection [13]. This allowed peptidomic analysis on mouse brain extracts, without the need for affinity purification. In 2005, my laboratory found that a conventional microwave oven can be used to rapidly heat an undissected mouse head, raising the temperature of the brain to >80°C within seconds of decapitation [28]. Peptidomic analysis showed that the profile of peptides in brains heated with a conventional microwave oven was similar to the peptides detected in brains treated by focused microwave irradiation, and free of the postmortem artifacts that appear 1 minute after death within brains not subjected to heat inactivation either by microwave irradiation or another technique [13,28,29]. Also in 2005, we found that trimethylammonium butyrate (TMAB) isotopic labels were superior to other isotopic tags for the quantification of neuropeptide levels [30]. The TMAB tags were developed by Fred Regnier for proteomic applications, and because they label free amines the TMAB reagents are well suited for peptidomic studies [31]. The TMAB tags can be produced in 5 distinct isotopic forms, allowing for multivariate analysis [32]. In the 10 years since the combination of heat inactivation and TMAB labels were used for quantitative peptidomics, we have performed hundreds of LC/MS runs of samples and MS/MS analyses of peptides [20,22,24,25,28,30,33–48]. In each LC/MS run, up to several hundred peptides were detected and identified. During these analyses some trends were noted such as the relative absence of peptides with free Cys residues, but because Cys is a relatively rare amino acid [49], the sample size of each individual dataset was not sufficient to permit conclusions. For the present study, datasets were combined of all peptidomics analyses performed in my laboratory over the past 10 years using heat inactivation of tissue/cells and TMAB isotopic labels. The combined datasets were then examined for features such as peptide mass, amino acid composition, and the presence of post-translational modifications. This analysis reveals that there are several limitations to quantitative peptidomics techniques, some which are specific to the TMAB-label approach, and others that are likely common to all peptidomic studies.
Methods
Three databases were compiled for the present meta-analysis: mouse, human, and tryptic peptides. These three databases were composed of published studies [20,22,24,25,28,30,33–48] and a few additional studies that have not yet been published; these unpublished studies used identical methods, differing only in the treatment groups (which is not relevant to the present meta-analysis). The mouse database contains all peptidomic results from studies using mouse tissues; mainly brain regions, but also whole brain as well as heart, spleen, and testis. Mouse cell lines from kidney and pancreatic beta cells are also included in this database. The human database contains all peptidomic results with human cell lines: HEK293T, HEK293H, SH-SY5Y, MCF7, and RPMI8226. The third database contains peptides from analysis of proteins digested with trypsin. Five proteins purchased from Sigma were digested with trypsin: bovine serum albumin (P02769); bovine thyroglobulin (P01267); human α-hemoglobin (P69905); human β-hemoglobin (P68871); and bovine α-lactalbumin (P00711).
Prior to extraction of peptides, samples (tissue, cell lines, or tryptic peptides) were heated to 80°C [50–52]. For cell lines, cells were pelleted and resuspended in 80°C water, and then incubated for 20 minutes at 80°C. For analysis of mouse brain, the head of the mouse was placed in a conventional microwave oven for 8 seconds, which raises the temperature to 80°C [28]. After cooling, the brain was removed, dissected into regions (if needed for the experimental protocol) and peptides extracted as described below. Other mouse tissues were rapidly removed from the animals, cut into pieces, and heated in a water bath at 80°C for 20 minutes. In some experiments, cells or tissue were frozen and stored at −70°C prior to peptide extraction. Peptides were extracted by sonication of the tissue in cold water followed by heating to 70°C in a water bath for 20 minutes. Homogenates were cooled in an ice bath and combined with cold 1 M HCl to a final concentration of 10 mM, then centrifuged in a microfuge for 40 minutes. The supernatant was removed and labeled with isotopic labels as described below.
Peptides were labeled with TMAB using the N-hydroxysuccinimide (NHS) ester of the TMAB reagents, prepared as described [32]. In brief, the TMAB-NHS esters are produced from the reaction of methyl iodide with gamma-amino butyric acid in methanol with potassium bicarbonate as a base. Methyl iodide containing 1, 2, or 3 atoms of deuterium were used to create the D3-, D6-, and D9-TMAB forms (three methyl groups react with the amino group). The D12 form was generated from methyl iodide containing 3 deuterium atoms and 13C. After the reaction, hydrochloric acid was added to produce the chloride salt of the quaternary ammonium ion and the product (TMAB) was isolated after precipitation with tetrahydrofuran. TMAB was activated with dicyclohexylcarbodiimide and NHS in acetonitrile, the product precipitated with tetrahydrofuran and purified by recrystallization.
Labeling of peptides with TMAB-NHS ester was performed at pH 9.5 in 0.4 M sodium phosphate buffer. The protocol has been described in detail [50–52]. In brief, an excess of TMAB-NHS was dissolved in DMSO and an aliquot added to the peptide at room temperature. After ~10–20 minutes incubation, the pH was adjusted back to 9.5 using dilute NaOH and another aliquot of TMAB-NHS added. This procedure was repeated for a total of 5–7 additions of TMAB-NHS over several hours. Excess TMAB-NHS was quenched by the addition of 2.5 M glycine and incubation at room temperature for 40 minutes. In a typical experiment, 4–5 different samples were labeled with a different isotopic form of the TMAB-NHS reagent. After the reaction was quenched, the samples were combined, treated with hydroxylamine to remove TMAB groups from Tyr residues, filtered on a 10 kDa membrane to remove proteins, and desalted using C18 spin columns, as described [50–52].
LC/MS/MS analysis was performed on a quadrupole time-of-flight (qTOF) mass spectrometer. A variety of qTOF instruments have been used for analysis of the various datasets over the past 10 years: API Q-star Pulsar-i (Applied Biosystems/MDS Sciex); Ultima (Micromass, Manchester, UK); Synapt G1 (Waters Co., EUA); and Synapt G2 (Waters Co., Milford, MA, USA). LC conditions always used 0.1% formic acid in water at a flow rate appropriate for the LC system; peptides were eluted with a gradient of acetonitrile over 20–50 minutes, depending on the experiment. Prior to elution, the sample was desalted online using a C18 trapping column. Data were acquired in data-dependent mode and selected peptides dissociated by collisions with argon. For optimal MS/MS analysis of TMAB-labeled peptides, the collision energy values were higher than those used for non-labeled peptides.
The first phase of the analysis involved searching MS spectra for peak sets that co-eluted and differed in mass by 3 Da or a multiple of 3 Da (after consideration of the charge state); these represent the D0, D3, D6, D9, and/or D12 TMAB-labeled peptides. The m/z values and intensity of the observed ions, the charge state, and the elute times were logged into a spreadsheet. In the second phase of the analysis, peptides were identified by MS/MS analysis, using a combination of Mascot searches and manual validation of the results based on previously defined criteria [50–52]. Mascot searches of the databases always included the modifications of the various isotopic forms of TMAB tags (named GIST in Mascot) on both N-termini and Lys side chains. Mascot searches routinely included commonly found modifications such as methionine oxide and N-terminal acetylation. Additional parameters used include C-terminal amidation and phosphorylation of Ser, Thr, and Tyr. Identification of S-cyanoCys, glutathione-conjugated to Cys, and iodinated His/Tyr were initially made by manual searches of unmatched ions that showed high quality MS/MS spectra. Once these were identified, further Mascot searches include these modifications. Other modifications of Cys were tested for several of the MS/MS data files but did not reveal additional modifications; these included acetyl-Cys, geranyl-geranyl-Cys, farnesyl-Cys, Cys oxidation, Cys methylation, palmitoyl-Cys, palmitoleyl-Cys, phospho-Cys, sulfo-Cys, and Ub-amide on Cys. Mascot searches were performed on either the Swiss Protein or NCBI databases, usually limited by the species of the sample (i.e. human or mouse); the studies on the tryptic proteins involved both human and bovine proteins, so this was analyzed using “mammalian” filter. No enzyme was specified, even for the analysis of tryptic peptides.
In each dataset, peptides that were detected at multiple ionization states are represented by multiple rows, with each row a distinct m/z. Similarly, peptides that incorporate different numbers of isotopic tags also show multiple entries. Analysis of the amino acid composition of each database was performed using DNA Star. For this, the individual amino acid sequences were combined into a single file and the DNA Star Protean program was used to calculate the amino acid composition.
Results
Three master datasets were created by combining datasets from individual studies, each separate study typically consisting of 2–20 LC/MS runs. Most of the individual studies have been published over the past decade [20,22,24,25,28,30,33–48]. Several of the studies have not been published; these unpublished studies were performed using techniques identical to those of the published studies. All of these studies involved TMAB isotopic labels and heat-inactivated samples. In combining the datasets, peptides found in multiple studies were listed multiple times; this was done so that the peptides found most frequently would be highly represented in the database. Each row in the database contains m/z values for each of the TMAB isotopic forms observed in the peak group; for representative data, see Table S1 in [20]. Peptides detected with multiple ionization states are represented by multiple rows, with each row a distinct ionization state reflecting the different m/z. Similarly, peptides that incorporated different numbers of isotopic tags also show multiple entries; in some experiments the peptides were incompletely labeled and lacked the TMAB tag on the N-terminus. In most studies, the number of isotopic tags corresponded to the theoretical number of free amines (peptide N-terminus and unmodified Lys residues).
The combined human cell line database consists of 14,433 total rows of peptides. Of these, 7523 peptides were identified by MS/MS analysis using a combination of Mascot searches and manual validation of the results based on previously defined criteria for excluding false positives [50–52].These 7523 rows of data represent 755 unique peptides, each found on average 10 times (range from 1 to 227 times). The most highly represented peptide in the database is thymosin beta-10 which was typically found in 3–4 different ionization states in each LC/MS run. The combined mouse peptide database consists of 29,050 rows of peptides, of which 17,100 were identified by MS/MS analysis. Only the identified peptides in each database were further considered in the present study. The mouse peptide database was subdivided into peptides derived from secretory pathway proteins (i.e. neuropeptides and related molecules) and those derived from cytosolic, mitochondrial, nuclear, and other non-secreted proteins (termed “intracellular” proteins). There were 7945 rows of peptides in the secretory pathway group (representing 397 unique peptides) and 9155 rows in the intracellular group (1003 unique peptides). The third dataset included in the present study did not reflect the endogenous peptides in a biological sample; instead, this dataset was generated by tryptic digestion of five purified proteins (bovine serum albumin, bovine thyroglobulin, human α-hemoglobin and β-hemoglobin, and bovine α-lactalbumin). This dataset was included in the present analysis so that comparisons between theoretical and observed peptides could be made; this was not possible with peptides isolated from biological samples. The tryptic peptide database consisted of 904 peptides detected by MS analysis, of which 411 peptides were identified by MS/MS analysis. As with the other two databases, tryptic peptides found in multiple LC/MS runs were represented multiple times in the combined dataset.
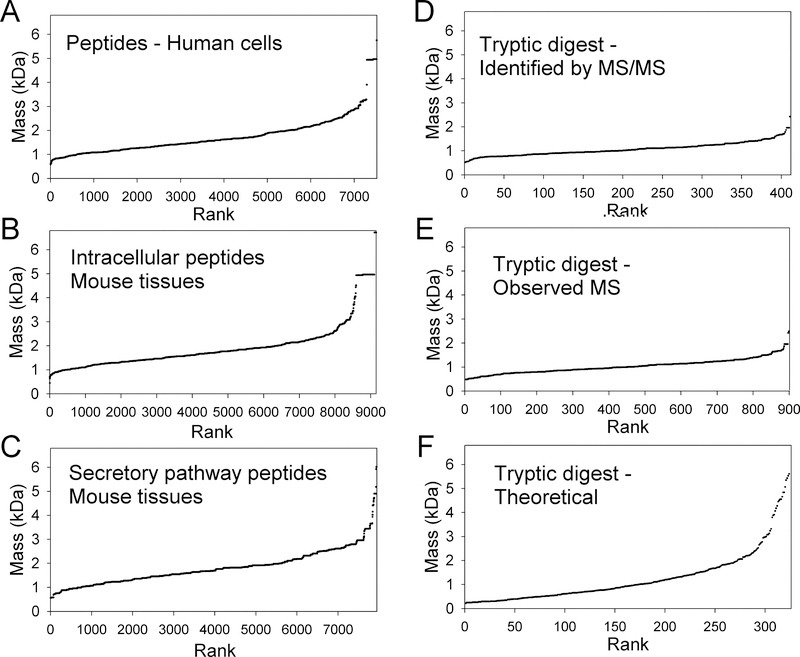
The identified peptides present in the human cell line database range from 578 to 5751 Da, with a median of 1559 Da (Figure 1A). The identified peptides present in the mouse database that were derived from intracellular proteins ranged in size from 444 to 6718 Da with a median of 1693 Da (Figure 1B). Identified mouse peptides derived from secretory pathway proteins ranged from 555 to 8765 Da, with a median of 1694 Da (Figure 1C). The standard procedure used for all of the peptidomic studies employed a 10 kDa microfiltration step to eliminate proteins, and so it was expected that peptides up to, or slightly exceeding 10 kDa would have been detected [50–52]. Although not the focus of the present study, peptides detected by MS but not identified by MS/MS in the mouse tissue and human cell line databases ranged from 224–12,000 Da size, with a median of 1883 Da. Although the mass range of the unidentified peptides is broader than the mass range of the identified peptides, 95% of the unidentified mouse peptides ranged from 625 to 5194 Da and 95% of the unidentified human peptides ranged from 686 to 4430 Da, so the very small and very large peptides represent a minor fraction of the peptides detected by MS analysis. However, it is not clear if the size range of the detected peptides reflects the true size range of the endogenous peptides, or if there is a bias in the detection and identification. To extend this analysis to a well-defined database, peptides derived from tryptic digestion of five proteins were analyzed by the same methods as used for the endogenous peptides. Tryptic peptides whose sequences were identified by MS/MS analysis ranged from 516 to 2418 Da with a median of 1022 Da (Figure 1D). Peptides detected by MS that matched the mass of a predicted tryptic peptide ranged in size from 477 to 2488 Da with a median of 997 Da (Figure 1E). This latter group of peptides did not involve MS/MS analysis and Mascot searches, and in theory should match the theoretical prediction of masses for all tryptic peptides. The theoretical peptide fragments ranged in size from 217 to 7589 Da, with a median of 930 Da (Figure 1F). Because the TMAB isotopic tags add 128–140 Da to the mass of each peptide, even dipeptides of 217 Da would potentially be detectable on qTOF with m/z > 300 if the peptide was labeled with one TMAB tag and present as the 1+ ion. Note that the indicated peptide masses reflect the un-tagged and unprotonated mass, whereas the detected m/z ions reflect the peptide with isotopic tags and often additional protons (the tag contains a quaternary amine and is positively charged without a proton, so protons are only present if the peptide contains an Arg or His residue).
Figure 1.
Size analysis of peptides. A: Peptides identified from MS/MS analysis of human cell lines. B: Peptides identified from MS/MS analysis of mouse tissues and cell lines, and which are derived from proteins expressed in the cytosol, mitochondria, nuclei, and other non-secretory compartments. C: Peptides identified from MS/MS analysis of mouse tissues and cell lines, and which are derived from proteins expressed in the secretory pathway (i.e. neuropeptides, peptide hormones, and related peptides). D: Peptides identified from MS/MS analysis of tryptic digests of five purified human and bovine proteins. E: Peptides observed in MS analysis of tryptic digests of five purified human and bovine proteins; these data include all peptides detected by MS, regardless of whether they were confirmed by MS/MS analysis. F: The predicted peptides resulting from the cleavage of the five human/bovine proteins by trypsin, assuming complete cleavage at every Lys or Arg (except Lys/Arg-Pro). For panels A-E, peptides found multiple times in the various experiments are represented multiple times in the figure, so that the results are weighted for the frequency of the detected peptides. The mass of the identified (A-D), observed (E) or theoretical (F) peptides represents the monoisotopic mass of the peptide in the absence of TMAB tags, not the observed m/z or mass of the TMAB-modified peptide. For all panels, the mass of the peptides were sorted from low to high and plotted by rank order (x-axis).
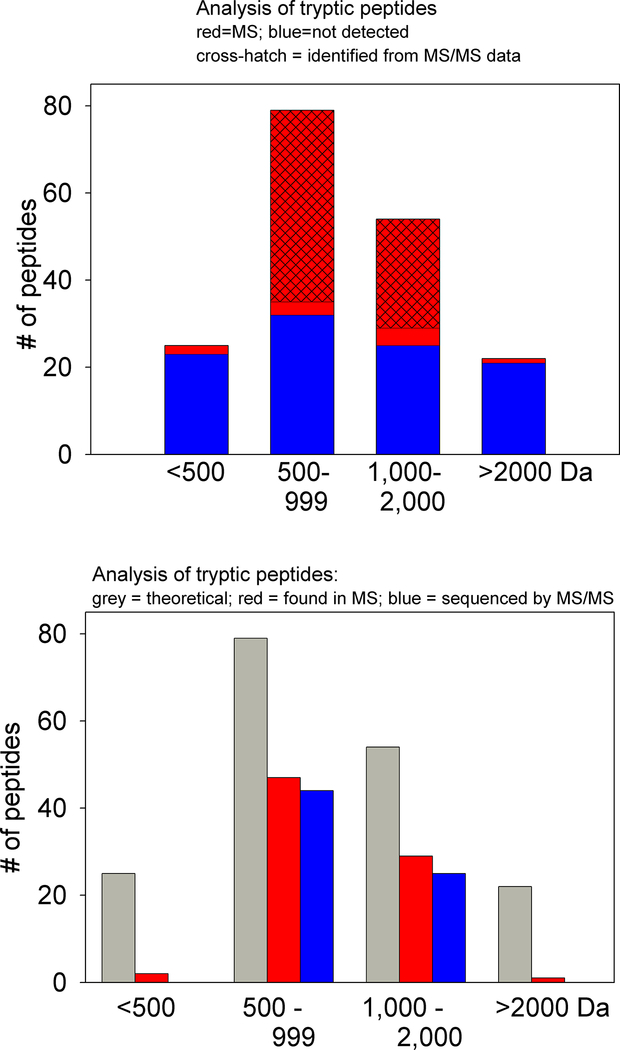
To further explore the difference between theoretical and observed tryptic peptides, the data were further analyzed by comparing the number of theoretical peptides in each size range with the number of peptides detected by MS (Figure 2). For this analysis, only theoretical peptides with mass/charge ratio > 300 were considered, using the expected charge state of 2 for tryptic peptides (3 if His is present) and the additional mass of 128 Da for each D0-TMAB tag incorporated. Also, theoretical peptides with Cys were excluded from the analysis because these peptides may not be easily detected if the Cys is present as a disulfide Cys-Cys with another peptide, or otherwise modified. To eliminate these issues, the theoretical database was filtered to remove all peptides with Cys residues or with an m/z below 300 (for the 2+ ion), leaving 180 peptides that should have been detected. Only 2 of the 25 peptides with mass <500 Da were detected by MS, but neither of these were confirmed by MS/MS sequence either by Mascot or manual sequencing (Figure 2). Similarly, only 1 of the 22 peptides with mass >2000 Da were detected by MS, and this was also not confirmed by MS/MS sequence (Figure 2). In contrast, 47 of the 79 peptides in the 500–999 Da range, and 29 of the 54 peptides in the 1,000–2000 Da range were detected by MS (Figure 2). The vast majority of the peptides detected by MS with mass between 500 and 2000 Da were subsequently confirmed by MS/MS sequencing (Figure 2, cross hatching). Thus, peptides with a mass <500 or >2000 Da are less readily detected by MS than peptides between 500 and 2000 Da using our conditions, which includes labeling with TMAB reagents and analysis on a qTOF. It should be emphasized that in our analysis, all peptides labeled with TMAB tags that are detected in MS spectra are logged into a spreadsheet regardless of whether identified by Mascot from MS/MS data. Thus, the failure to detect most of the theoretical peptides with mass <500 or >2000 Da is unrelated to potential problems with Mascot searches of MS/MS data.
Figure 2.
Comparison of theoretical and observed tryptic peptides. Five proteins were digested with trypsin and the observed peptides compared with the theoretical digest, assuming complete cleavage at Lys or Arg residues except for Lys-Pro and Arg-Pro, which are poorly cleaved by trypsin. The theoretical products containing Cys were excluded from this analysis. Also, any peptide that would have an m/z of <300 was excluded because this was the lower cut-off for the qTOF analysis. The remaining theoretical products were divided into four groups: less than 500 Da, 500–999 Da, 1000–2000 Da, and >2000 Da. These mass ranges represent the uncharged form of the peptide without TMAB tag, not the observed mass of the labeled peptide; all peptides were labeled with at least one tag—those with Lys were labeled with 2 TMAB tags. The peptides detected by mass spectrometry are indicated in red. Cross-hatching indicated the subset of the detected peptides that were confirmed by MS/MS analysis, either by Mascot or by manual sequencing. Blue indicates predicted peptides that were not detected by mass spectrometry.
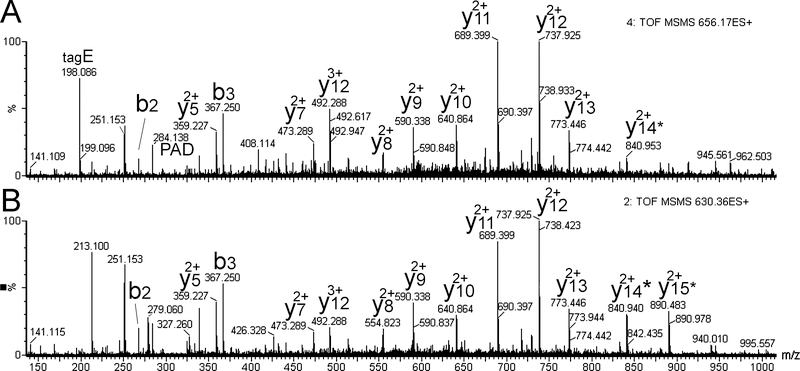
The amino acid composition of the identified peptides in the combined datasets was determined. For this analysis Met and Met-oxide were counted as Met, acetyl groups on the N-terminus of the protein were removed, but all other modifications (phosphorylation, etc) were considered as separate modifications. Except for Cys, which is not detected in the free form within the human cell line peptidome, all of the other amino acids are present at levels ranging from 0.39% (Trp) to 9.2% (Lys) (Table 1). The relative level of each residue other than Cys is generally comparable to the abundance of the amino acid within the Eukaryotic proteome [49]. Cys is present in the Eukaryotic proteome with a frequency around 2% [49], and should have been detected around 2,400 times (out of 120,137 combined amino acids in the human cell line database). Two peptides with modified Cys were detected in multiple experiments, for a total of 24 entries in the human cell line database. Both of these peptides were identical fragments of the protein triosephosphate isomerase, and the difference was the modification. One peptide contained glutathione (GSH) attached to the Cys (mass difference 305.07 Da), the other peptide contained an attachment of 176.03 Da on the Cys which corresponds to the mass of glutathione lacking the Glu residue (Figure 3). Both MS/MS spectra showed a strong 2+ y-series, with y5 to y13 matching the ions predicted by Mascot. The y14 ion includes the conjugated Cys, and this was detected with an additional mass of 31.9 Da, corresponding to the presence of persulfide (Cys-SSH) instead of free Cys-SH. The y15 product ion also had an additional 31.9 Da (Figure 3). These modified y-ions were found for both the glutathione-conjugated peptide and the peptide with an additional mass of 176.03, which strongly suggests that this latter modification is a modified GSH lacking the N-terminal Glu, rather than another modification such as N-glucuronyl, which is also 176.03 Da.
Table 1.
Amino acid composition of identified human and mouse peptides.
| Human cell lines | Mouse intracellular | Mouse secretory pathway | |
|---|---|---|---|
| Amino acid | Number (Frequency) | Number (Frequency) | Number (Frequency) |
| Ala | 10628 (8.9%) | 14366 (9.0%) | 13056 (10.1%) |
| Arg | 4609 (3.8%) | 6561 (4.1%) | 9806 (7.6%) |
| Asn | 3694 (3.1%) | 4393 (2.8%) | 3800 (2.9%) |
| Asp | 7856 (6.5%) | 10166 (6.4%) | 5630 (4.3%) |
| Cys | 0 | 0 | 0 |
| Cys-Cys | 0 | 0 | 50 (<0.1%) |
| Cys-GSH | 13 (<0.1%) | 49 (<0.1%) | 0 |
| Cys-GSH w/o E | 11 (<0.1%) | 0 | 0 |
| CyanoC | 0 | 118 (<0.1%) | 0 |
| Gln | 4831 (4.0%) | 7903 (5.0%) | 5215 (4.0%) |
| Glu | 9187 (7.7%) | 13362 (9.9%) | 12774 (9.8%) |
| Gly | 10646 (8.9%) | 10044 (6.3%) | 8820 (6.8%) |
| His | 1324 (1.1%) | 3262 (2.1%) | 1855 (1.4%) |
| Ile | 6643 (5.5%) | 6824 (4.3%) | 2447 (1.9%) |
| Leu | 10513 (8.8%) | 13363 (8.4%) | 14246 (11.0%) |
| Lys | 11055 (9.2%) | 15351 (9.7%) | 5577 (4.3%) |
| Met | 1984 (1.7%) | 2543 (1.6%) | 2171 (1.7%) |
| Phe | 4430 (3.7%) | 6093 (3.8%) | 4130 (3.2%) |
| Pro | 6842 (5.7%) | 10506 (6.6%) | 12102 (9.3%) |
| Ser | 7821 (6.5%) | 11755 (7.4%) | 11823 (9.1%) |
| Thr | 5758 (4.7%) | 8794 (5.5%) | 4408 (3.4%) |
| Trp | 469 (0.39%) | 819 (0.51%) | 985 (0.76%) |
| Tyr | 1642 (1.4%) | 2650 (1.7%) | 3271 (2.5%) |
| Val | 10181 (8.5%) | 10016 (6.3%) | 7591 (5.9%) |
Figure 3.
MS/MS spectra of Cys-modified peptides identified in extracts of human cell lines. These spectra are from a study of HEK293T cells; similar spectra were obtained from other studies using HEK293T and SH-SY5Y cells. The peptide corresponds to an internal fragment of triosephosphate isomerase 1, sequence AKVPADTEVVCAPPTAYIDFARQK, and with modifications on the Cys residue. A: The 656.17 ion is 5+ and contains 3 D0-TMAB tags (determined from MS spectrum), resulting in a monoisotopic mass of 2894.40 Da for the unprotonated and untagged peptide. The theoretical mass of the peptide without modifications on Cys would be 2589.33 Da, and the mass of the modification is 305.07 Da which matches the mass of glutathione (GluCysGly) attached to the Cys. Because GSH has an amino group, this modified peptide should have 4 TMAB tags, and several times this peptide was observed with 4 tags. However, in this example the peptide lacks a tag on the N-terminal Ala—the b2 and b3 ions match the mass predicted for peptide lacking an N-terminal tag. In addition, a product ion of 198 is observed which matches the predicted mass of Glu (from the GSH moiety) with a tag. Note that the trimethylamine group is lost in MS/MS, and the mass of the tag in MS/MS spectra is 69 Da. The y14 product ion is 31.9 Da heavier than predicted for a peptide containing Cys, which matches the mass of persulfide. B: The 656.17 ion is 5+ and contains 3 D0-TMAB tags (determined from MS spectrum), resulting in a monoisotopic mass of 2765.36 Da; this is 176.03 Da heavier than predicted for the unmodified peptide. Although 176.03 Da corresponds to the mass of the N-glucuronyl group, this modification is not known to target Cys residues. The presence of y14 and y15 product ions that are 31.9 Da heavier than predicted for the peptide with free Cys is consistent with persulfide, strongly suggesting that this peptide is modified with a disulfide bond. One possible candidate is GSH lacking the N-terminal Glu residue; the theoretical mass is 176.03 Da. Asterisks indicate the y14 and y15 ions that contain an additional 31.9 Da.
Cys is present in the human peptidome database well below its typical frequency of 2% in the Eukaryotic proteome [49], even after taking into account the modified Cys residues. The low frequency of Cys in the human peptidome database is not due to the absence of Cys residues within the proteins cleaved to produce the human cell line peptidome. Analysis of the residues immediately upstream or downstream of the observed peptides (i.e. the flanking residues) shows an abundance of Cys residues in these positions. Cys is found in 311 out of 5265 N-terminal flanking-residues (5.9%) and 367 out of 5666 C-terminal flanking residues (6.4%); these are approximately 3-fold higher than the expected frequency of Cys.
Similar analyses were performed on the mouse intracellular peptidome. All of the identified peptides arising from intracellular proteins collectively constitute 158,928 amino acids. As found with the human cell line data base, there were no peptides containing free Cys in the mouse intracellular peptide database. One peptide, a fragment of tubulin beta 2A found 49 times, contained glutathione-conjugated to a Cys residue on the C-terminus. Another two peptides contained cyanoCys on the N-terminus; these two peptides were found a total of 118 times. Only a few peptides were found with posttranslational modifications on other residues, including one peptide with phosphoThr (found 4 times), one peptide with phosphoSer (found once) and one peptide with N-terminal pyroGlu (found once). Other than the absence of free Cys, the frequency of other amino acids in the mouse intracellular peptide database generally parallels their frequency in the Eukaryotic proteome [49]. Cys is found in 236 out of 5680 N-terminal flanking-residues (4.2%) and 457 out of 6380 C-terminal flanking residues (7.2%); these are higher than the expected frequency of Cys.
No free Cys residues were observed in the mouse secretory pathway peptides. The only Cys-containing peptides detected were oxytocin and vasopressin, both of which contain 2 Cys residues linked by a disulfide bond. No Cys residues were present among the residues flanking the observed peptides, which were primarily (but not exclusively) basic amino acids; the vast majority of secretory pathway peptides are derived from precursors that are cleaved at basic residues by endopeptidases followed by trimming of the basic residues by carboxypeptidases. Of the 7947 peptide entries in the mouse secretory pathway database, 1230 have a posttranslational modification: 690 have a C-terminal amide group, 259 have phosphoSer, 160 have N-terminal pyroGlu, 107 have N-terminal acetyl, 2 have di-acetyl, and 12 have dehydroSer. The overall amino acid composition of the mouse secretory pathway peptides is generally similar to the other databases found in peptidomic analyses (Table 1).
Based on the above analysis, it appears that there is a problem detecting peptides containing Cys. However, it is possible that endogenous peptides containing Cys are present at extremely low abundance in the biological samples used for the peptidomic studies. To resolve these possibilities, the database of TMAB-labeled tryptic peptides was analyzed for amino acid content. Altogether, 127 distinct peptides were identified by MS/MS sequencing, approximately half of which represented predicted tryptic fragments of the five proteins. The remainder represented either missed tryptic cleavage sites (i.e. peptides containing an internal Lys or Arg residue) or cleavage at a non-basic site such as a Cys residue. Twelve peptides contained a cyanoCys residue, 11 of these on the N-terminus (Figure 4). One peptide contained two Cys residues in a disulfide bond; these Cys are known to form a disulfide bond within the protein. Only a single peptide was identified with a free Cys residue. This peptide, a fragment of bovine serum albumin, was also found with a cyanoCys on the N-terminus. Furthermore, the upstream residue was not a basic residue, suggesting it was produced by a non-tryptic mechanism. The five proteins used to generate the tryptic peptide database contain a total of 168 Cys, 100 of which are known to be present in disulfide pairs, leaving 68 free Cys residues in the proteins.
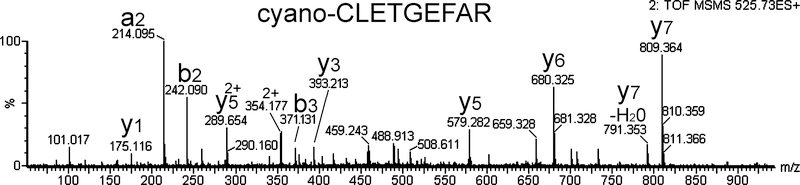
Figure 4.
MS/MS spectrum of a peptide containing S-cyanocysteine from the tryptic database. This peptide is a fragment of thyroglobulin. The peptide was not labeled with TMAB tags, consistent with cyclization of the N-terminal cyano group. The observed ion was 525.73 and was 2+, corresponding to a mass of 1049.46 Da. The theoretical mass of CLETGEFAR is 1024.46, consistent with the addition of cyano on the Cys (25.00 Da). The expected tryptic fragment is DLFIPTCLETGEFAR, indicating cleavage at a non-tryptic site probably due to cyanide-mediated chemical cleavage of the peptide. The source of the cyanide is unknown; none of the reagents are known to contain cyanide.
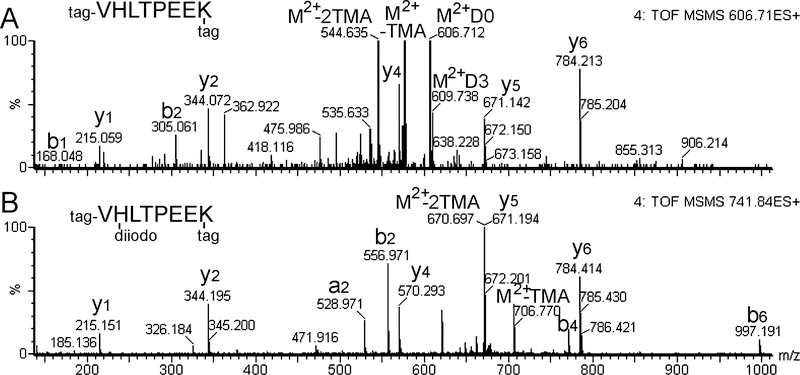
During the analysis of the MS/MS of the tryptic fragments, an effort was made to identify peptides which did not match the mass of expected fragments. Because the input was five purified proteins digested with trypsin, the sequence of each peptide was expected to match one of these proteins (including trypsin) unless the peptide was a contaminant. However, Mascot can only match posttranslational modifications that are defined in the search parameters, and there is a limit to how many modifications can be selected in each Mascot search. In several cases, MS/MS data was sufficient to manually derive a partial sequence, which was then checked against the sequences of the five proteins and trypsin. Several peptides were identified that matched the linear sequence of an expected fragment except that the peptide had an additional mass of 125.91 or 251.81 Da. In all cases, the peptides with the extra mass contained His and/or Tyr. For some peptides the MS/MS data were sufficient to map the extra mass to the His and/or Tyr residue. For example, the beta-hemoglobin-derived fragment VHLTPEEK was found in three forms; the expected form (Figure 5A), the form with one mono-iodoHis (not shown), and the form with di-iodoHis (Figure 5B). Levels of the signals for each of the forms in the MS spectra showed large variation, with the non-iodinated peptide showing strong signals with D0-TMAB, D3-TMAB, and D6-TMAB, and the mono- and di-iodinated peptides showing strong signals with D9-TMAB and D12-TMAB (not shown).
Figure 5.
MS/MS spectra of peptides from the tryptic database with unmodified His and di-iodoHis. A: The peptide VHLTPEEK was identified by Mascot searches of the MS/MS data. The 606.71 ion is 2+ and has 2 D0-TMAB tags (determined from the MS spectrum), and the monoisotopic mass of the untagged peptide is 951.49 Da. This peptide represents the N-terminal fragment of beta-hemoglobin, theoretical mass 951.50 Da. B: The 741.84 ion is 2+ and has 2 D12-TMAB tags (determined from the MS spectrum), and the monoisotopic mass of the untagged peptide is 1203.30. This observed mass is 251.8 Da heavier than the theoretical mass for the VHLTPEEK peptide, which matches the theoretical mass of 125.90 for each iodide. From the MS/MS spectrum, it is clear that this addition occurs on the His residue. Both spectra show 2+ parent ion (M2+) with loss of one and two trimethylamine (TMA) groups.
Discussion
Peptidomic techniques have been used by a large number of laboratories [1–15]. A recent search of PubMed shows over 500 publications on peptidomic(s). While these related techniques are extremely powerful and can detect many of the most abundant peptides in a biological sample, there are several limitations. Some of these limitations are likely to be specific to the technique used; in our case the use of TMAB labels to perform quantification, qTOFs to run the samples, and Mascot to identify the peptide sequence. But some of the limitations noted in the present analysis may represent broader problems common to a variety of peptidomic approaches.
In our early studies on mass spectrometry of neuropeptides (prior to our use of TMAB labels), we noted that small and large peptides are difficult to detect and identify by mass spectrometry [27]. A typical neuropeptide precursor is cleaved into a range of products of varying size which should be present in equal levels (unless a site is only partially cleaved). For example the precursor prothyrotropin releasing hormone (proTRH) is cleaved into thyrotropin releasing hormone (TRH), a tripeptide of the sequence pyroGlu-His-Pro-amide (mass 361 Da), and 6 other peptides ranging from 1110 Da to over 5000 Da. The 361 Da peptide was never detected in our peptidomic studies, even though it is present in 5 copies within the precursor and should be 5 times more abundant than any other proTRH-derived peptide. Although the presence of an N-terminal pyroGlu residue would prevent labeling by the TMAB tags, we did not detect this peptide even in studies without isotopic tags. Furthermore, in mice lacking carboxypeptidase E activity there is an accumulation of neuropeptide processing intermediates with C-terminal Lys and Arg, and we should have detected pyroGluHisProGlyLysArg (mass 703 Da) in these animals. We detected all of the predicted Lys/Arg-extended fragments of proTRH ranging from 1482 to 3676 Da in the brains of mice lacking carboxypeptidase E activity, but we did not detect any peptide with a mass of 703 Da [41,45]. We also did not detect either of the two proTRH fragments that are >4 kDa, even after taking into account potential post-translational modifications of these peptides (Cys disulfide bonds, phosphorylation of Ser). This analysis is independent of the identification of the peptide from MS/MS analysis, which can be impacted by the size of the peptide [53]. In our analysis, the detection of peptides depends only on MS data—all observed peptides tagged by the TMAB labels are logged into the database in the first phase of our analyses. In subsequent steps, the peptides are tentatively matched to the size of predicted neuropeptides and most are eventually confirmed by MS/MS analysis. Based on the analysis of the detected proTRH peptides as well as similar analysis of other neuropeptide precursors, there appears to be an optimal size range for detection of peptides by mass spectrometry on qTOF instruments.
The previous studies on neuropeptides detected a relatively small number of peptides, making it difficult to draw firm conclusions regarding the inability to detect particular peptides. The present analysis combined datasets from individual experiments to create very large databases so that trends in the results would be clear. The vast majority of peptides detected in either human cell lines or mouse tissues were between 1 and 3 kDa, consistent with the previously observed analysis of peptides derived from proTRH and other neuropeptide precursors. Another finding was the absence of peptides containing free Cys residues in either the human cell line or mouse tissue databases. Disulfide bonds are rare in cytosolic proteins and it was expected that free Cys would be detected in the human cell line peptides and also in the subset of mouse tissue peptides that are derived from intracellular proteins. Although modified Cys was occasionally detected, peptides with free Cys were generally not found. One of the observed Cys modifications was attachment of glutathione, a known modification that adds 305.07 Da to the mass of the peptide. One peptide modified by attachment of GSH was also found with an attachment of 176.03 Da and the MS/MS spectrum was similar to that of the GSH-modified peptide. The mass difference is 129.04 Da, corresponding to a Glu residue; GSH has a Glu on the N-terminus. It is possible that the 176.03 modification is GSH lacking the Glu, which is a novel post-translational modification. Unimod (www.unimod.org/) lists 176.03 as N-glucuronyl, which can attach to the N-terminus of proteins and also Ser residues, but is not known to modify Cys residues. Based on the MS/MS spectra (Figure 3) that show persulfide in the y14 and y15 product ions, it is likely that the Cys is modified by a disulfide bond and not by glucuronylation. However, further studies are needed to determine whether the 176.03 modification detected in the human cell lines represents de-Glu GSH attached to Cys or another modification involving a disulfide bond.
The trends observed in the analysis of the large combined datasets suggest problems in the detection and identification of small peptides, large peptides, and any peptide containing free Cys. Alternatively, these peptides may not be abundant in the biological samples analyzed by peptidomics. To distinguish these possibilities, a dataset of tryptic peptides was also analyzed in the present study. The tryptic peptides were generated for studies characterizing the specificity of carboxypeptidases; the tryptic peptides were incubated with different carboxypeptidases and then labeled with TMAB isotopic tags and processed for peptidomics using the same techniques used for the human cell line and mouse tissue peptides (a manuscript described the biological results of this study is being prepared). The present analysis of the tryptic peptide dataset showed the same trends that were found for the human and mouse peptide databases. Very few tryptic peptides <500 Da or >2000 Da were detected, and none of the peptides in these ranges were sequenced by MS/MS either manually or by Mascot. In contrast, the majority of the theoretical peptides in the 500–2000 Da range were detected by MS and nearly all of these were subsequently confirmed by MS/MS sequencing. It is not clear if this problem is specific to our approach that uses TMAB reagents and analysis by qTOF instruments. Because the TMAB reagents add 128–140 Da to the mass of the peptide and a quaternary positive charge, the use of this reagent should improve the detection of small peptides.
Analysis of the tryptic peptide database provided solid evidence that free Cys is a rarely detected using our peptidomics approach, and that one problem is the formation of cyanoCys during peptide extraction, labeling, or analysis. Cyanide is known to cleave peptides at Cys residues, forming cyanoCys [54]. Although none of the reagents used in our peptidomic approach are known to contain cyanide, this could be a trace contaminant present in one of the reagents or solvents. An important question is whether formation of cyanoCys is unique to our approach with TMAB labels, or if it is common to other peptidomic approaches. Although cyanoCys has not been reported in peptidomic studies, it is unlikely that anyone previously looked for this modification. Free Cys is rarely detected in other peptidomic studies. A study of human urinary peptides did not detect any peptide containing Cys residues out of 31,296 total amino acids [55]. In a study on mouse brain peptides [56], Cys was detected in only three related peptides derived from actin, representing an abundance of 0.04% which is well below the 2% frequency of Cys in the Eukaryotic proteome. A study of the peptidome of human breast and ovarian cancer found several hundred Cys residues in each sample, but this represented an abundance of 0.66% and 0.28%, respectively [57]. The low frequency of detection of peptides with Cys in these peptidomic studies may be due to post-translational modifications that aren’t routinely included in database searches (i.e. glutathione) or novel ones such as the 176.03 Da modification (Figure 3B). Alternatively, it could reflect a problem like cyanylation, as found for our mouse tissue and tryptic peptide databases. To check this possibility, the MS/MS data from other peptidomics studies need to be searched using cyanoCys as a potential modification.
The finding of mono- and di-iodoHis/Tyr is likely to be a problem only for studies that use TMAB isotopic tags. Iodine is presumably a contaminant of the TMAB reagents, which are synthesized from methyl iodide. In the synthesis, the iodide salt is converted into the chloride salt by the addition of excess hydrochloric acid, but it is likely that traces of iodide remain. The uneven levels of iodinated peptides in the MS spectra indicate that the impurity is variable between batches of TMAB-NHS reagent. For this reason, it is essential that experiments include biological replicates in which the TMAB labels are switched. In spite of this drawback to the TMAB reagents, there are many advantages of this isotopic tag for quantitative peptidomics. The availability of 5 isoforms allows multivariate analysis of multiple groups. The reagents are relatively inexpensive, ranging from $100 to $1000 per gram of reagent. Although they are not commercially available, their synthesis is relatively easy and does not require extensive experience with organic chemistry. The various isotopic forms of TMAB co-elute from LC columns, which is important for accurate quantification of peak intensity. Unlike i-TRAQ reagents that require MS/MS spectra for quantification, the TMAB-labeled peptides are quantified from MS spectra, and many peptides can be quantified in each LC/MS run. Disadvantages of the TMAB reagents include the loss of the TMA moiety in MALDI or if the peptide is exposed to basic conditions. Also, the TMAB-labeled peptides don’t work well with ion trap mass spectrometers, including Orbitraps. Still, despite these limitations and the finding from the present study of iodoTyr/His, the TMAB reagents are effective for quantification of peptides.
Understanding the limitations of scientific techniques is essential in their effective use. For example, an important technique for the quantification of mRNA levels is that of microarray analysis. On occasion, there was considerable variability between microarray experiments done at different locations, or done at the same location on different days of the year. Careful analysis of the environmental factors found that the dyes used in microarray analysis are differentially sensitive to ozone, with Cy5/Alexa647 affected by 5–10 parts per billion ozone while Cy3/Alexa555 are not affected until ozone levels reach >100 parts per billion [58]. Introduction of air purifiers to reduce ozone levels have improved the consistency of microarray data. Likewise, the reproducibility of peptidomics data will be improved by careful attention to details, such as the purity of the TMAB reagents in order to avoid formation of iodoHis and/or Tyr. The source of the cyanide contamination needs to be determined so the problem with formation of cyanoCys can be eliminated. It is not clear if this problem is unique to our TMAB approach, or common to other approaches. The difficulty in detecting and identifying small and large peptides is a broad problem for peptidomic studies, and is not likely to be limited to studies using TMAB labels. Although part of the problem may be the database searching to identify peptides, there is under-detection of m/z signals for small and large peptides in our studies that is independent of the database searches. Until these problems are resolved, knowledge of the limitations of the peptidomic techniques will help in the interpretation of data. A key point is that the failure to detect a particular peptide in a peptidomic study does not necessary mean that the peptide is absent from the sample, as it may be undetected for a number of different reasons.
Acknowledgments
The study was funded by NIH grant R01-DA004494. Most of the people who performed the work to generate the databases are listed as co-authors of the individual publications describing each database. In addition to the published studies, several recent studies included in the present analysis have not yet been published. Scientists who contributed to these projects include Dr. Sayani Dasgupta, Michael Fishman, Javier Garcia Pardo, Dr. Leandro Castro, Dr. Emer Ferro, and Dr. Alexandre Tashima. Special thanks to Dr. Sayani Dasgupta for helpful comments regarding the preparation of this manuscript.
References
- 1.Schrader M, Schultz-Knappe P, Fricker LD: Historical perspective of peptidomics. EuPA Open Proteomics 3, 171–182 (2014) [Google Scholar]
- 2.Fricker LD: Neuropeptides and other bioactive peptides. Fricker LD, Devi L, (eds) Morgan & Claypool Life Sciences, Charleston, S.C. (2012) [Google Scholar]
- 3.Lone AM, Nolte WM, Tinoco AD, Saghatelian A: Peptidomics of the Prolyl Peptidases. AAPS.J, (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sasaki K, Takahashi N, Satoh M, Yamasaki M, Minamino N: A peptidomics strategy for discovering endogenous bioactive peptides. J Proteome Res 9, 5047–5052 (2010) [DOI] [PubMed] [Google Scholar]
- 5.Boonen K, Landuyt B, Baggerman G, Husson SJ, Huybrechts J, Schoofs L: Peptidomics: the integrated approach of MS, hyphenated techniques and bioinformatics for neuropeptide analysis. J.Sep.Sci 31, 427–445 (2008) [DOI] [PubMed] [Google Scholar]
- 6.Svensson M, Skold K, Nilsson A, Falth M, Nydahl K, Svenningsson P, Andren PE: Neuropeptidomics: MS applied to the discovery of novel peptides from the brain. Anal.Chem 79, 15–21 (2007) [DOI] [PubMed] [Google Scholar]
- 7.Dowell JA, Heyden WV, Li L: Rat Neuropeptidomics by LC-MS/MS and MALDI-FTMS: Enhanced Dissection and Extraction Techniques Coupled with 2D RP-RP HPLC. J.Proteome.Res. 5, 3368–3375 (2006) [DOI] [PubMed] [Google Scholar]
- 8.Fricker LD, Lim J, Pan H, Che FY: Peptidomics: Identification and quantification of endogenous peptides in neuroendocrine tissues. Mass Spectrom.Rev 25, 327–344 (2006) [DOI] [PubMed] [Google Scholar]
- 9.Li L, Sweedler JV: Peptides in the brain: mass spectrometry-based measurement approaches and challenges. Annu Rev Anal Chem (Palo Alto Calif) 1, 451–483 (2008) [DOI] [PubMed] [Google Scholar]
- 10.Hummon AB, Amare A, Sweedler JV: Discovering new invertebrate neuropeptides using mass spectrometry. Mass Spectrom.Rev 25, 77–98 (2006) [DOI] [PubMed] [Google Scholar]
- 11.Baggerman G, Verleyen P, Clynen E, Huybrechts J, De Loof A, Schoofs L: Peptidomics. J.Chromatogr.B Analyt.Technol.Biomed.Life Sci 803, 3–16 (2004) [DOI] [PubMed] [Google Scholar]
- 12.Clynen E, De Loof A, Schoofs L: The use of peptidomics in endocrine research. Gen.Comp Endocrinol 132, 1–9 (2003) [DOI] [PubMed] [Google Scholar]
- 13.Svensson M, Skold K, Svenningsson P, Andren PE: Peptidomics-based discovery of novel neuropeptides. J.Proteome.Res 2, 213–219 (2003) [DOI] [PubMed] [Google Scholar]
- 14.Schrader M, Schulz-Knappe P: Peptidomics technologies for human body fluids. Trends Biotechnol. 19, S55–S60 (2001) [DOI] [PubMed] [Google Scholar]
- 15.Verhaert P, Uttenweiler-Joseph S, de Vries M, Loboda A, Ens W, Standing KG: Matrix-assisted laser desorption/ionization quadrupole time-of-flight mass spectrometry: an elegant tool for peptidomics. Proteomics 1, 118–131 (2001) [DOI] [PubMed] [Google Scholar]
- 16.Chard T: An Introduction to Radioimmunoassay and Related Techniques. Burdon RH, Van Knippenberg PH, (eds) Elsevier, Amsterdam: (1987) [Google Scholar]
- 17.Fricker LD, McKinzie AA, Sun J, Curran E, Qian Y, Yan L, Patterson SD, Courchesne PL, Richards B, Levin N, Mzhavia N, Devi LA, Douglass J: Identification and characterization of proSAAS, a granin-like neuroendocrine peptide precursor that inhibits prohormone processing. J.Neurosci 20, 639–648 (2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferro ES, Rioli V, Castro LM, Fricker LD: Intracellular peptides: From discovery to function. EuPA Open Proteomics 3, 143–151 (2014) [Google Scholar]
- 19.Ferro ES, Hyslop S, Camargo AC: Intracellullar peptides as putative natural regulators of protein interactions. J.Neurochem 91, 769–777 (2004) [DOI] [PubMed] [Google Scholar]
- 20.Fricker LD, Gelman JS, Castro LM, Gozzo FC, Ferro ES: Peptidomic analysis of HEK293T cells: effect of the proteasome inhibitor epoxomicin on intracellular peptides. J Proteome Res 11, 1981–1990 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldberg AL: Protein degradation and protection against misfolded or damaged proteins. Nature 426, 895–899 (2003) [DOI] [PubMed] [Google Scholar]
- 22.Gelman JS, Sironi J, Castro LM, Ferro ES, Fricker LD: Peptidomic analysis of human cell lines. J.Proteome.Res 10, 1583–1592 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fricker LD: Analysis of mouse brain peptides using mass spectrometry-based peptidomics: implications for novel functions ranging from non-classical neuropeptides to microproteins. Mol.Biosyst 6, 1355–1365 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dasgupta S, Castro LM, Dulman R, Yang C, Schmidt M, Ferro ES, Fricker LD: Proteasome inhibitors alter levels of intracellular peptides in HEK293T and SH-SY5Y cells. PloS one 9, e103604 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gelman JS, Sironi J, Berezniuk I, Dasgupta S, Castro LM, Gozzo FC, Ferro ES, Fricker LD: Alterations of the intracellular peptidome in response to the proteasome inhibitor bortezomib. PloS one 8, e53263 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gomes I, Aryal DK, Wardman JH, Gupta A, Gagnidze K, Rodriguiz RM, Kumar S, Wetsel WC, Pintar JE, Fricker LD, Devi LA: GPR171 is a hypothalamic G protein-coupled receptor for BigLEN, a neuropeptide involved in feeding. Proc Natl Acad Sci USA, (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Che FY, Yan L, Li H, Mzhavia N, Devi L, Fricker LD: Identification of peptides from brain and pituitary of Cpe fat /Cpe fat mice Proc.Natl.Acad.Sci.USA 98, 9971–9976 (2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Che FY, Lim J, Biswas R, Pan H, Fricker LD: Quantitative neuropeptidomics of microwave-irradiated mouse brain and pituitary. Mol.Cell.Proteomics 4, 1391–1405 (2005) [DOI] [PubMed] [Google Scholar]
- 29.Skold K, Svensson M, Norrman M, Sjogren B, Svenningsson P, Andren PE: The significance of biochemical and molecular sample integrity in brain proteomics and peptidomics: stathmin 2–20 and peptides as sample quality indicators. Proteomics. 7, 4445–4456 (2007) [DOI] [PubMed] [Google Scholar]
- 30.Che FY, Fricker LD: Quantitative peptidomics of mouse pituitary: Comparison of different stable isotopic tags. J.Mass Spectrom 40 238–249 (2005) [DOI] [PubMed] [Google Scholar]
- 31.Zhang R, Sioma CS, Thompson RA, Xiong L, Regnier FE: Controlling deuterium isotope effects in comparative proteomics. Anal.Chem 74, 3662–3669 (2002) [DOI] [PubMed] [Google Scholar]
- 32.Morano C, Zhang X, Fricker LD: Multiple isotopic labels for quantitative mass spectrometry. Anal.Chem 80, 9298–9309 (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berezniuk I, Sironi JJ, Wardman J, Pasek RC, Berbari NF, Yoder BK, Fricker LD: Quantitative peptidomics of Purkinje cell degeneration mice. PloS one 8, e60981 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gelman JS, Dasgupta S, Berezniuk I, Fricker LD: Analysis of peptides secreted from cultured mouse brain tissue. Biochim Biophys Acta 1834, 2408–2417 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berezniuk I, Sironi J, Callaway MB, Castro LM, Hirata IY, Ferro ES, Fricker LD: CCP1/Nna1 functions in protein turnover in mouse brain: Implications for cell death in Purkinje cell degeneration mice. FASEB J. 24, 1813–1823 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lyons PJ, Fricker LD: Substrate specificity of human carboxypeptidase A6. J.Biol.Chem 285:, 38234–38242 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanco S, Zhang X, Morano C, Aviles FX, Lorenzo J, Fricker LD: Human carboxypeptidase A4: Characterization of the substrate specificity and implications for a role in extracellular peptide processing. J.Biol.Chem 285, 18385–18396 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wardman JH, Zhang X, Gagnon S, Castro LM, Zhu X, Steiner DF, Day R, Fricker LD: Analysis of peptides in prohormone convertase 1/3 null mouse brain using quantitative peptidomics. J.Neurochem 114, 215–225 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X, Pan H, Peng B, Steiner DF, Pintar JE, Fricker LD: Neuropeptidomic analysis establishes a major role for prohormone convertase-2 in neuropeptide biosynthesis. J.Neurochem 112, 1168–1179 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berti DA, Morano C, Russo LC, Castro LM, Cunha FM, Zhang X, Sironi J, Klitzke CF, Ferro ES, Fricker LD: Analysis of intracellular substrates and products of thimet oligopeptidase (EC 3.4.24.15) in human embryonic kidney 293 cells. J.Biol.Chem 284, 14105–14116 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X, Che FY, Berezniuk I, Sonmez K, Toll L, Fricker LD: Peptidomics of Cpe(fat/fat) mouse brain regions: implications for neuropeptide processing. J.Neurochem 107, 1596–1613 (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Che FY, Zhang X, Berezniuk I, Callaway M, Lim J, Fricker LD: Optimization of neuropeptide extraction from the mouse hypothalamus. J.Proteome.Res 6, 4667–4676 (2007) [DOI] [PubMed] [Google Scholar]
- 43.Che FY, Vathy I, Fricker LD: Quantitative peptidomics in mice: Effect of cocaine treatment. J.Mol.Neurosci 28, 265–275 (2006) [DOI] [PubMed] [Google Scholar]
- 44.Decaillot FM, Che FY, Fricker LD, Devi LA: Peptidomics of Cpefat/fat mouse hypothalamus and striatum: Effect of chronic morphine administration. J.Mol.Neurosci 28 277–284 (2006) [DOI] [PubMed] [Google Scholar]
- 45.Lim J, Berezniuk I, Che FY, Parikh R, Biswas R, Pan H, Fricker LD: Altered neuropeptide processing in prefrontal cortex of Cpe<fat/fat> mice: Implications for neuropeptide discovery. J.Neurochem 96, 1169–1181 (2006) [DOI] [PubMed] [Google Scholar]
- 46.Pan H, Che FY, Peng B, Steiner DF, Pintar JE, Fricker LD: The role of prohormone convertase-2 in hypothalamic neuropeptide processing: a quantitative neuropeptidomic study. J.Neurochem 98, 1763–1777 (2006) [DOI] [PubMed] [Google Scholar]
- 47.Che FY, Biswas R, Fricker LD: Relative quantitation of peptides in wild type and Cpe<fat/fat> mouse pituitary using stable isotopic tags and mass spectrometry. J.Mass Spectrom 40 227–237 (2005) [DOI] [PubMed] [Google Scholar]
- 48.Pan H, Nanno D, Che FY, Zhu X, Salton SR, Steiner DF, Fricker LD, Devi LA: Neuropeptide processing profile in mice lacking prohormone convertase-1. Biochemistry 44, 4939–4948 (2005) [DOI] [PubMed] [Google Scholar]
- 49.Pe’er I, Felder CE, Man O, Silman I, Sussman JL, Beckmann JS: Proteomic signatures: amino acid and oligopeptide compositions differentiate among phyla. Proteins 54, 20–40 (2004) [DOI] [PubMed] [Google Scholar]
- 50.Gelman JS, Wardman JH, Bhat VB, Gozzo FC, Fricker LD: Quantitative peptidomics to measure neuropeptide levels in animal models relevant to psychiatric disorders. Methods Mol.Biol 829, 487–503 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wardman J, Fricker LD: Quantitative peptidomics of mice lacking Peptide-processing enzymes. Methods Mol.Biol 768, 307–323 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lyons PJ, Fricker LD: Peptidomic approaches to study proteolytic activity. Curr Protoc Protein Sci Chapter 18, Unit18 13 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Akhtar MN, Southey BR, Andren PE, Sweedler JV, Rodriguez-Zas SL: Evaluation of database search programs for accurate detection of neuropeptides in tandem mass spectrometry experiments. J Proteome Res 11, 6044–6055 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Catsimpoolas N, Wood JL: Specific cleavage of cystine peptides by cyanide. J Biol Chem 241, 1790–1796 (1966) [PubMed] [Google Scholar]
- 55.Frantzi M, Zoidakis J, Papadopoulos T, Zurbig P, Katafigiotis I, Stravodimos K, Lazaris A, Giannopoulou I, Ploumidis A, Mischak H, Mullen W, Vlahou A: IMAC fractionation in combination with LC-MS reveals H2B and NIF-1 peptides as potential bladder cancer biomarkers. J Proteome Res 12, 3969–3979 (2013) [DOI] [PubMed] [Google Scholar]
- 56.Van Dijck A, Hayakawa E, Landuyt B, Baggerman G, Van Dam D, Luyten W, Schoofs L, De Deyn PP: Comparison of extraction methods for peptidomics analysis of mouse brain tissue. J Neurosci Meth 197, 231–237 (2011) [DOI] [PubMed] [Google Scholar]
- 57.Xu Z, Wu C, Xie F, Slysz GW, Tolic N, Monroe ME, Petyuk VA, Payne SH, Fujimoto GM, Moore RJ, Fillmore TL, Schepmoes AA, Levine DA, Townsend RR, Davies SR, Li S, Ellis M, Boja E, Rivers R, Rodriguez H, Rodland KD, Liu T, Smith RD: Comprehensive Quantitative Analysis of Ovarian and Breast Cancer Tumor Peptidomes. J Proteome Res, (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fare TL, Coffey EM, Dai H, He YD, Kessler DA, Kilian KA, Koch JE, LeProust E, Marton MJ, Meyer MR, Stoughton RB, Tokiwa GY, Wang Y: Effects of atmospheric ozone on microarray data quality. Anal Chem 75, 4672–4675 (2003) [DOI] [PubMed] [Google Scholar]