Abstract
Recent advances in instrumentation and software for cryoEM have increased the applicability and utility of this method. High levels of automation and faster data acquisition rates require hard decisions to be made regarding data retention. Here we investigate the efficacy of data compression applied to aligned summed movie files. Surprisingly, these images can be compressed using a standard lossy method that reduces file storage by 90–95% and yet can still be processed to provide sub-2Å reconstructed maps. We do not advocate this as an archival method, but it may provide a useful means for retaining images as an historical record, especially at large facilities.
Keywords: cryo electron microscopy (cryoEM), file format, image data, data archiving, structural biology
Introduction
The application of robust automated data acquisition systems and processing pipelines in the field of cryo electron microscopy (cryoEM) has increased throughput and the ability to apply this technique to a greater variety of structural biological applications (Baldwin et al., 2018). Along with advances in detector technology, data generation rates are also growing due to faster frame rates and larger sensor sizes. Meanwhile, the field is still in the process of developing standards as to what forms of the experimental data should be saved and available to the community, adding uncertainty to the scope of data that needs to be archived (Henderson et al., 2012, Patwardhan et al., 2016, Kleywegt et al., 2018).
A data retention and management policy for three dimensional EM has been recognized as an issue that is crucial for the field, scientific computing and future research (Patwardhan et al., 2012). Applying the next generation of computational software to existing data may lead to further gains in resolution and understanding of the underlying biology. In particular, direct detector frame stacks have been reanalyzed using improved motion and dose correction programs (Ripstein and Rubenstein 2016) to improve overall resolution. For example, several researchers have processed publicly available archived datasets to improve 3D reconstructed maps by ~1Å (Tegunov et al., 2018, Heymann et al., 2018).
A concern is that individual researchers may not have the resources to backup and archive all the data generated over the course of an experiment; it is also possible that much of the primary data may not survive the departure of the lead researcher from a lab. Use of publicly available resources for data archiving, such as the Electron Microscopy Public Image Archive (EMPIAR), is far from routine and in any case often only stores a small subset of the data associated with the published map.
At the Simons Electron Microscopy Center (SEMC), a large instrumentation facility based in New York, users are responsible for archiving their own movie frames as long-term storage of these data is beyond our storage capacity. Instead our policy has been to retain the aligned summed images, both as a historical record and against accidental loss of the user’s archival data. However, even this limited data retention policy is no longer sustainable at the current data acquisition rates. We routinely acquire several million high magnification direct detector movies per year and anticipate higher data ingestion rates in the future.
To address this issue, we investigated the effect of using a standard lossy data compression algorithm to reduce the size of the aligned summed images. The compression results in a ~10 fold reduction in storage and, somewhat surprisingly, processing the data after compression resulted in maps at the same resolution and of the same quality as the original images. We absolutely do not advocate using these lossy compression algorithms for archival data but present these results in the spirit of promoting a discussion on data retention policies and potential solutions. This may be of special interest to large facilities in order to extend the time that it is possible to retain images to provide an historical record and cross reference over long periods of time.
Material and Methods
The apoferritin datasets used for this study were downloaded from EMPIAR (EMPIAR-10200 and EMPIAR-10146). The bovine glutamate dehydrogenase (GDH) and Thermoplasma acidophilum 20S (T20S) datasets were collected in house.
Samples:
GDH was purchased from Sigma (G2626) and purified by anion exchange chromatography, followed by size-exclusion chromatography in 150 mM NaOH, 20mM HEPES, pH 7.4. T20S proteasome was a gift from Yifan Cheng, Zanlin Yu, and Kiyoshi Egami. The received stock was separated into small aliquots and stored at −80 °C in 20 mM Tris, pH 8.0, 50 mM NaCl, 0.1 mM EDTA.
Grid Preparation:
3 µl of freshly thawed protein (4 mg/mL for GDH and 0.29 mg/mL for T20S) was applied to plasma-cleaned C-flat 1.2/1.3 400 mesh Cu holey carbon grids (Protochips, Raleigh, North Carolina), blotted for 2.5 s after a 30 s wait time, and then plunge frozen in liquid ethane, cooled by liquid nitrogen, using the Cryoplunge 3 (Gatan) at 75% relative humidity.
Microscopy:
Thermo-Fisher Titan Krios operated at 300 kV and Gatan K2 Summit camera in counting mode were used with a 70 µm C2 aperture, 100 µm objective aperture and calibrated pixel size of 0.66Å.
Imaging:
Movies were collected in counting mode using Leginon (Suloway et al., 2005). For the T20S sample, a dose rate of 6 e−/Å2/s was used for an accumulated dose of 62.6 e−/Å2 with a total exposure time of 10 seconds and 0.2 second frame rate. For the GDH sample, a dose rate of 10 e−/Å2/s was used for an accumulated dose of 61.2 e−/Å2 with a total exposure time of 6 seconds and 0.1 second frame rate. Nominal defocus values ranged from 1.0–3.0 µm.
Image Processing:
Movies recorded on the K2 were aligned using MotionCor2 with dose weighting (Zheng et al., 2017) and CTF estimation was performed with CTFFIND4 (Rohou and Grigorieff, 2015) by the Appion (Lander et al., 2009) pre-processing pipeline. For the first 50 images, particles were picked automatically using DoG Picker (Voss et al., 2009), extracted, and subjected to 2D classification in RELION (Zivanov et al., 2018) to create templates for another round of particle picking using FindEM (Roseman, 2004). The picked particles were extracted and subjected to 2D classification and the best classes were selected for ab-initio reconstruction to create an initial model in cryoSPARC (Punjani et al., 2017). This model was used for 3D classification in RELION or heterogeneous refinement in cryoSPARC. For the final reconstruction, particles were selected and subjected to 3D refinement in cryoSPARC, RELION or cisTEM (Grant et al., 2018). The number of micrographs and particles used for each map were as follows: T20S 1173/128,520; GDH 2,491/285,518; apoferritin EMPIAR-10200 1,255/56,990. The T20S and GDH micrographs have been uploaded to EMPIAR and EMDB as EMPIAR-10218 (1,173 micrographs) and EMDB-9233, and EMPIAR-10217 (2,491 micrographs) and EMDB-9203, respectively. Fourier ring correlations utilized the BIOP plugin included with Fiji/ImageJ (Schindelin et al., 2015).
File conversion:
Locally acquired images were recorded as 32-bit MRC files, the EMPIAR datasets were in TIFF format. IMOD (Kremer et al., 1996) was used to convert the images to various JPEG compression formats and EMAN2 (Tang et al., 2007) was used for all other image conversions.
Results and Discussions
Data reduction using lossless formats
The electron microscopy (EM) field uses several file formats during data generation, ranging from vendor specific formats to standard processing formats e.g. MRC and TIFF (Cheng et al., 2015). As an example, the Leginon-Appion workflow allows users the option to save images from electron detectors in the standard 32-bit MRC file format or as TIFF-LZW. Leginon is integrated with a database and web viewer that enables researchers to view these high magnification images through a web browser in context with the parent medium magnification and low magnification images to better track and annotate experiments. Concurrent with data collection on direct detector cameras, motion correction is run on-the-fly and two more images are created in addition to the unaligned sum—an aligned sum and a dose-weighted aligned sum—thereby providing flexibility in the subsequent processing workflow. This leads to significant data expansion because these steps result in an addition of tens of thousands of images generated per microscope per day. We are currently web hosting ~11 million high magnification images consuming ~0.65PB of disk space.
Currently, each new session adds tens of thousands of images (~0.5GB) and several thousand raw direct detector movies (~10TB when stored as uncompressed 16-bit MRCs files). Saving the raw uncompressed movies and gain references for the past 3 years would have required ~12PB disk space, without taking into account needs for backup and disaster recovery. There is every expectation that data ingestion rates will increase well beyond these rates over the next few years (Baldwin et al., 2018).
A common step in the workflow is to compress raw movies after motion correction has been performed. The overall potential compression ratio depends on whether the data has low redundancy to start with. We initially used bzip2, a lossless compression program that uses several layers of compression strategies to reduce redundancy, for storage of our Gatan K2 dark corrected movies because of the reasonably high compression ratio, compatibility of the program across multiple platforms, and the ability to parallelize compression to keep up with data generation. The high redundancy of K2 electron counting images results in compression of ~13–16% of the original file size. For example, an un-gain corrected 50 frame K2 counting movie (3838 × 3710 pixels) in 16-bit MRC is 1,400 MB, but reduces to 221 MB when bzip2 compressed (Table 1).
Table 1:
Comparison of image file size in MB for various file formats. In parenthesis is the compression ratio as compared against the original MRC file size.
| File format | K2 EC. 16-bit integer 3838 X 3710 pixels x 50 frames | K2 Summed image 32-bit floating point 3838 X 3710 pixels |
|---|---|---|
| MRC | 1400 | 55 |
| MRC with bz2a | 221 (15.8%) | 42 (76.4%) |
| Tiff | 1400 (100%) | 55 (100%) |
| Tiff ZIP | 282 (20.1%) | 42 (76.4%) |
| Tiff LZW | 286 (20.4%) | 51 (92.7%) |
| PNG (8-bit) | 223 (15.9%) | 12 (21.8%) |
| JPEG000 (most loss) | 8.5 (0.6%) | 0.2 (0.4%) |
| JPEG050 | 188 (13.4%) | 2.7 (4.8%) |
| JPEG075 | 273 (19.5%) | 4.2 (7.6%) |
| JPEG100 (least loss) | 842 (60.1%) | 15 (27.3%) |
The TIFF file format is also commonly used for high quality storage of cryoEM image data. Similar to MRCs, TIFFs tend to be large in size; 16-bit MRC and 16-bit TIFF image are identical in size. For TIFF, disk space may be reduced using native compression. The most common TIFF compressions are the Lempel-Ziv-Welch algorithm, or LZW (Welch, 1984), and ZIP compression. These strategies use codecs, or table-based lookup algorithms, to reduce the size of the original image. Both LZW and ZIP are lossless compression methods and so will not degrade image quality. For non-gain corrected movie stacks, 16-bit compressed TIFFs have no data loss and compress to comparable file sizes as standard bzip2 compression. Using TIFF LZW, the resultant file is 20% of the original file size compared to 16% of the file size using bzip2 (Table 1). Summed images, however, only benefit modestly from these compression algorithms: 93% file size using TIFF LZW and 76% with bzip2.
One advantage for TIFF files over bzip2 compression is that many modern processing packages support TIFF-LZW input files, so that compressed frame stacks may be used directly for refinement (Grant et al., 2018 and Zivanov et al., 2018). While slightly more modest in space savings compared to other lossless compression formats, we have incorporated TIFF-LZW directly into our workflow and this is the format in which the data is handed off to our users for further processing. While data compression substantially reduces the size of the movie files, they nevertheless represent far too large a data storage burden for us to contemplate storing them long term; we estimate that our storage needs would be ~2.5PB if we had retained all K2 movies acquired over the last 3 years in TIF-LZW format. It is thus our current data policy to retain these movie files for one month after data collection, after which they are automatically marked for removal.
While the aligned summed images represent a much smaller data storage burden than the full movies, we nevertheless strongly recommend retention of the raw movie frames, as new algorithms are still being developed that can improve reconstructions based on frame by frame processing and analysis (Ripstein and Rubenstein 2016). For example, several researchers have recently processed publicly available archived datasets to improve 3D reconstructed maps by ~1Å (Tegunov et al., 2018 and Heymann et al., 2018). However, in our experience, the aligned summed images are very valuable as a historical record. In our facility, these images are always viewable using a web-browser interface along with a vast trove of metadata associated with the images (e.g. ice thickness, drift characteristics, resolution limits, number of particles per image etc.) This feedback allows our users to browse and compare previous data collection sessions in order to optimize specimen preparation and data collection parameters. These images also provide a fall back plan should the user fail to archive the raw movie data or lose that data due to some disaster; situations that have occurred several times.
Despite the value of this image record, we have now reached a level of data acquisition where it is not economically feasible to retain all of these aligned summed images indefinitely. Unfortunately, as seen in Table 1, lossless data compression is far less useful for these images and the gains are too small to be useful. For this reason, we have explored the utility of lossy compression for these images.
Data reduction using lossy compression algorithms.
There are a number of additional image file formats often used in photography, but not so far for cryoEM image processing. Two commonly used photography file formats that support up to 24-bit color are PNG (Portable Network Graphics) and JPEG (Joint Photographic Experts Group). PNG is a lossless file format and can utilize LZW compression similar to TIFF images. The disk savings when a 32-bit grayscale TIFF file is converted to an 8-bit grayscale PNG file is comparable with TIFF-LZW with a compression of 16% for K2 counting movies, but there is also a significant 22% compression for the aligned sum image, which corresponds to more than a 4-fold advantage in disk space savings over TIFF-LZW (Table 1).
JPEG is a lossy file format that uses discrete cosine transforms (DCT) to express a finite sequence of data points in terms of a sum of cosine functions (Hudson et al., 2017). JPEG format is usually avoided for archiving if images may be used for further processing as the images lose quality when compressed. JPEG has a range of compression ratios ranging from JPEG100 with the least amount of information loss (corresponding to 60% of the original file size for the frame stack and 27% for the aligned sum image) to JPEG000 with the most amount of information loss (corresponding to 0.6% of the original file size for the frame stack and 0.4% for the aligned sum image) (Table 1).
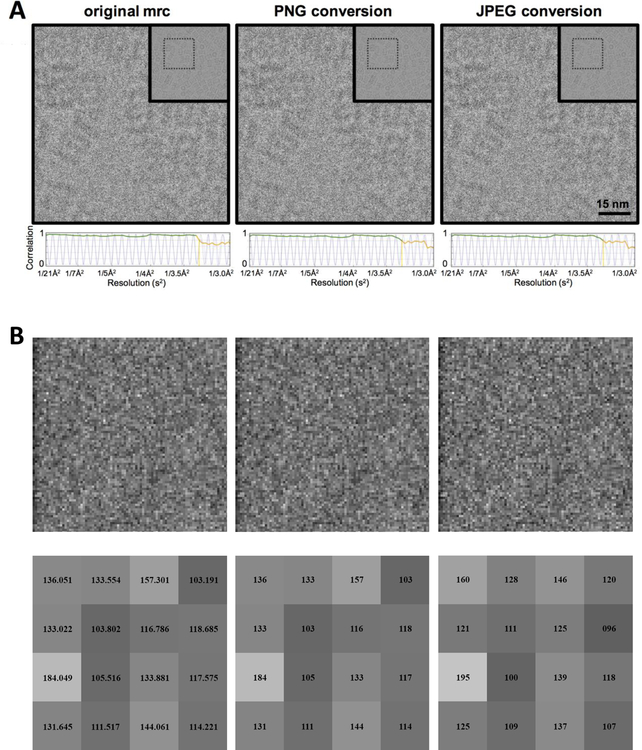
We wished to understand the effect that these compression algorithms have on image evaluation and results obtained from subsequent processing of these compressed images. We chose to explore this using JPEG075 as it is the default compression ratio used in IMOD and compresses images to less than 10% of the original size, thus providing a factor of 10x in disk space savings. In Figure 1, we compare T20S micrographs shown in 32-bit MRC, 8-bit PNG and JPEG075. For feedback on the progress of data collection, manual inspection of the micrograph and evaluation of the CTF estimation are two “best practices” recommended by several centers (Alewijnse et al., 2017). Upon visual inspection, the micrographs in all three formats look qualitatively very similar (Figure 1A) even when examining an enlarged sub-region (Figure 1B, top row). For a more quantitative measure, CTFFind4 was used to estimate the CTF from the three image formats, and all three produced essentially identical values (Figure 1A, bottom row). This observation indicates that using either PNG or JPEG075 images to monitor experiments, would provide sufficient feedback to have confidence in deciding whether to proceed with data collection. Nevertheless, converting a 32-bit image to an 8-bit image certainly results in precision loss. This effect can be seen by examining a sub-region of the image at the individual pixel level; the 32-bit to 8-bit conversion truncates the floating point value to an integer which produces quantization artifacts (Figure 1B, bottom row). Image artifacts are expected for JPEG file formats because the compression method uses a data reduction algorithm that leads to some original image information being lost that cannot be restored. This information loss is distinct from integer truncation and potentially could impact image processing results.
Figure 1. Effect of data reduction treatment on images A.
A. Comparison between original MRC file, PNG lossless compressed image, and JPEG075 lossy compressed image. Inset: The original micrograph indicating region of interest. Below each micrograph is a 1D plot of the CTF estimated by CTFFind4; the green curve tracks the 80% confidence level of the estimation, which is at 3.18Å for each of the images shown.
B. Top: Zoomed in region of the images in 1A. Bottom: 4×4 selected pixel sub region area showing the pixel values before and after conversion.
C. Fourier Ring Correlation of PNG to MRC (solid black line) and JPEG to MRC (dashed grey line) of images shown in 1A.
D. Four Ring Correlations comparing MRC images to various JPEG compression formats.
To further characterize the impact of the lossy compression algorithms on the image quality, we calculated Fourier Ring Correlation (FRC) curves for the images shown in Figure 1A. FRC measures the correlation between the two images at different spatial frequencies (Banterle et al., 2013); the FRC of the MRC image to itself is 1.0 across all spatial frequencies, as expected. Comparing MRC to PNG, which primarily affects precision, results in an FRC envelope that still maintains an FRC value over 0.99 (Figure 1C, solid black line). The MRC to JPEG075 FRC dampens out at high frequencies (Figure 1C, dashed grey line). In Figure 1D, the FRC is shown for a range of JPEG compression formats; as the aggressiveness of the compression is increased (corresponding to an increase in the area over with the DCT is applied), there are increasingly severe effects eventually across the entire frequency range.
CryoEM reconstructions of JPEG images
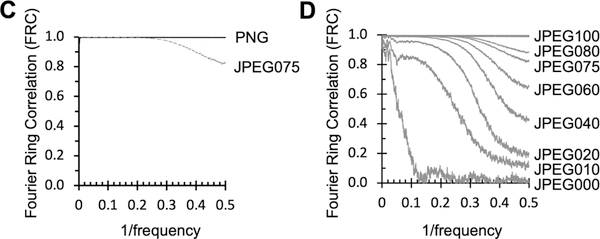
Although there is clearly some loss of data due to JPEG file conversion, it is not obvious what the impact of the data loss is on subsequent processing and analysis. To characterize this impact, we determined 3D reconstructions for test samples of T20S and GDH and a publicly available dataset (EMPIAR-10200) using our standard workflow (Kim et al., 2018).
These results showed that 32-bit MRCs and 32-bit TIFF images may be used interchangeably for data processing as expected. 2D classification followed by 3D refinement in RELION of a selected stack of 32-bit MRC particles provides maps at resolutions of FSC0.143 2.5Å, 2.1Å and 1.8Å for T20S, GDH, and EMPIAR-10200 respectively. When the particles were extracted from JPEG compressed micrographs (generated by IMOD’s default JPEG075) and processed in the same way as the 32-bit MRC particles, we obtained similar reconstructions for the T20S and GDH datasets, with identical FSC0.143 values (Figure 2A) and a somewhat lower resolution for the EMPAIR-10200 maps (2.0Å). As refinement packages do not have support to read JPEG compressed images, images were converted from JPEG back to MRC prior to processing. Comparison of the unsharpened maps of GDH gave a cross correlation of 0.99. Examination of the maps confirms that the overall structures are qualitatively quite similar (Figure 2B).
Figure 2. Comparison between 3D maps generated from MRC and JPEG075 images.
A. Fourier shell correlation of reconstructions between original MRC files (solid line) and lossy compressed JPEG quality 75 files (dashed lines) of T20S (orange, FSC0.143 2.5Å), GDH (green, FSC0.143 2.1Å), and EMPIAR-10200 apoferritin (blue, FSC0.143 1.8Å).
B. 3D reconstruction comparison of selected areas of GDH reconstruction with original MRC (top) and JPEG compressed (bottom) micrographs. Clockwise from the top is a representative α-helix in mesh, and representative parts of the maps: Ile β179, Trp β178 and Phe β399.
C. FRC of JPEG075 to MRC images from T20S (orange), GDH (green) and EMPIAR-10200 apoferritin (blue) datasets.
D. FSC curves for T20S (orange), GDH (green) and EMPIAR-10200 apoferritin (blue) of 3D half maps. Solid lines are from reconstructions of MRC images using Euler angles and CTF information generated from JPEG images. T20S FSC0.143 is 2.5Å, GDH FSC0.143 is 2.1Å and EMPIAR-10200 apoferritin FSC0.143 is 2.0Å. The horizontal dotted lines correspond to the FSC0.143 values for each sample as calculated from 2A.
E. FSC comparing MRC and JPEG compressed unsharpened maps (dashed lines) and sharpened maps (solid lines).
Although these real space comparisons are qualitatively helpful, a more quantitative comparison is provided in Fourier space. The FRC comparison of the analogous MRC and JPEG075 compressed images from each of the datasets in figure 2 show that they follow a similar envelope as noted for the JPEG075 compression in figure 1C; JPEG compression dampens the amplitudes of high resolution frequencies (Figure 2C). The FRC envelope indicates that information is preserved out to ~40% Nyquist, gradually decays ~0.9 at 75% Nyquist and drops to 0.8 at Nyquist frequency.
We also used the final Euler angles and CTF estimations generated from the JPEG compressed images and applied them to the original MRC images for T20s, GDH and EMPIAR-10200. The FSC0.143 of the resultant maps were the same as the JPEG reconstructions to within 0.1Å accuracy (Figure 2D). The main point of divergence in the FSCs between JPEG and MRC reconstructions occurs beyond the FSC0.143 frequency (Figure 2E, dashed and dotted lines). The refinement package used (RELION) masks the volume at FSC0.143 frequency. After that point, the Fourier correlations between the models start to diverge, presumably due to the JPEG compression artifacts, as highlighted in Figure 1B, thereby resulting in a higher correlation due to the noise no longer being random. The FSC curves also show the effect of applying JPEG compression and DCT to the noise. In particular, during sharpening, the maps are masked and filtered at FSC0.143, which boosts the artifacts in the noise beyond the FSC0.143 values (Figure 2E, solid lines).
Considerations when approaching Nyquist
Lossy compression, in addition to resampling, may lead to unrecoverable loss of resolution. In Figure 2A the EMPIAR-10200 dataset is the only example where processing lossy images does not result in the same resolution map as the original images. The final resolutions reached for T20S (2.5Å) and GDH (2.1Å) are respectively about half or two-thirds of Nyquist frequency (pixel size 0.66Å). In contrast, the resolution obtained for EMPIAR-10200 apoferritin is at ~80% of Nyquist (2.0Å) when using JPEG compressed images compared to 90% (1.8Å) of Nyquist when using the MRC images (Figure 2A).
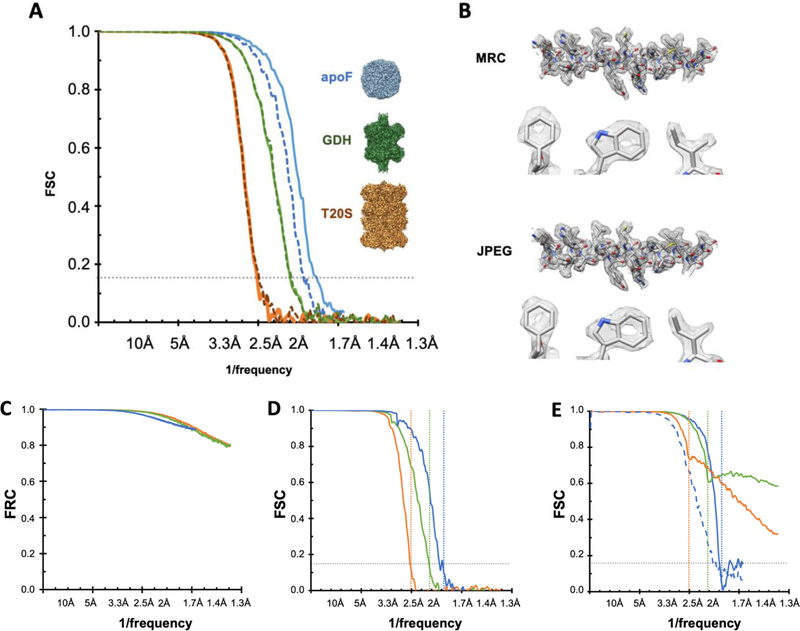
To investigate this further we used another publicly available dataset (EMPIAR 10146 Apoferritin, pixel size 1.5Å) to determine how compression affects FSC0.143 resolution when close to Nyquist frequency. This dataset is used as a test case for the cisTEM program (Grant et al. 2018). Following the cisTEM tutorial, reconstructions at 3.0Å (Nyquist) may be obtained using the raw uncompressed data. If the entire processing workflow (frame alignment, CTF estimation, particle picking, classification and refinement) is completed from PNG-converted movie stacks, the 3D reconstructions are the very similar with little apparent loss of resolution or quality of the resultant maps. Compression using JPEG100 (highest quality, least compression) produced a reconstruction within 0.1Å of the uncompressed data set (FSC0.143 3.1Å). (Figure 3). Increasing the aggressiveness of compression leads to increasing loss of resolution; using JPEG050 there is more than a 10% decrease in resolution (FSC0.143 3.3Å). We also tested these effects on the movie stacks which produced similar results (grey squares in Figure 3)
Figure 3.
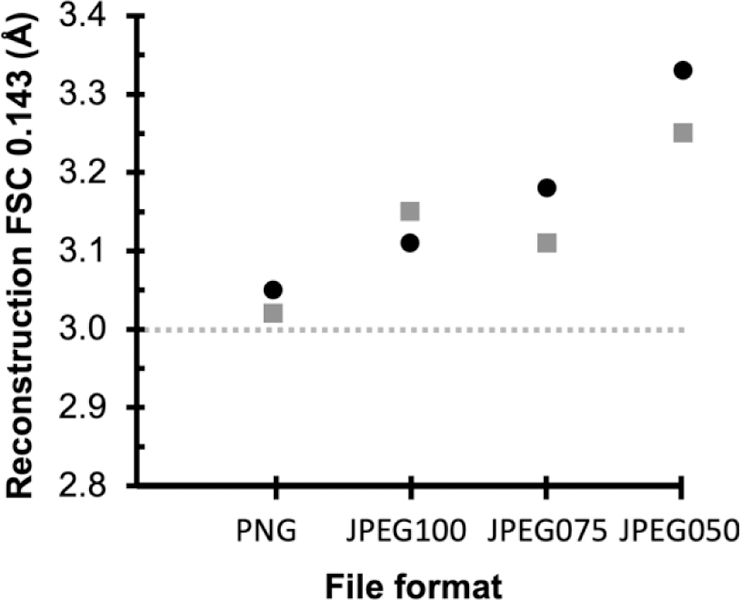
FSC0.143 estimates of resolution from reconstructions of Apoferritin EMPIAR-10146 using various file formats. Black circles are aligned summed images, grey squares are movie stacks. The dotted horizontal line indicates the FSC0.143 of 3.0Å from the original MRC files; this represents Nyquist as the pixel size is 1.5Å.
Conclusions
Data compression is an essential element of data storage for cryoEM given the increasingly larger volume of data being produced. Best practices dictate archiving the original raw movies files so that these can be reprocessed as improved algorithms are developed. Storage of these movie files is beyond the capacity of many EM centers and is often the responsibility of the users. At our center we have opted to store the aligned, summed movie files, to provide a historical record and cross reference for users who return to the center over the course of many months or even years. Nevertheless, even this level of file preservation is rapidly becoming too costly. As a result, we explored the use of various compression algorithms to reduce the file storage burden. A commonly used compression method, JPEG075, reduces file sizes to less than 10% of the original, but still provides an excellent visual record of the image and can be used to accurately estimate the CTF. More surprisingly, these JPEG compressed images can be used to compute a 3D map that is essentially the same as the one computed form the original MRC files, except in the case where the resolution of the reconstruction is very close to Nyquist.
Acknowledgements
The work presented here was performed at the Simons Electron Microscopy Center and National Resource for Automated Molecular Microscopy located at the New York Structural Biology Center, supported by grants from the Simons Foundation (SF349247), NYSTAR, and the NIH National Institute of General Medical Sciences (GM103310) with additional support from Agouron Institute (F00316) and NIH (OD019994).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alewijnse B, Ashton AW, Chambers MG, Chen S, Cheng A, Ebrahim M, Eng ET, Hagen WJH, Koster AJ, López CS, Lukoyanova N, Ortega J, Renault L, Reyntjens S, Rice WJ, Scapin G, Schrijver R, Siebert A, Stagg SM, Grum-Tokars V, Wright ER, Wu S, Yu Z, Zhou ZH, Carragher B, Potter CS. (2017) Best practices for managing large CryoEM facilities. J Struct Biol September;199(3):225–236. 10.1016/j.jsb.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin PR, Tan YZ, Eng ET, Rice WJ, Noble AJ, Negro CJ, Cianfrocco MA, Potter CS, Carragher B. (2018) Big data in cryoEM: automated collection, processing and accessibility of EM data. Curr Opin Microbiol June;43:1–8. 10.1016/j.mib.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banterle N, Bui KH, Lemke EA, & Beck M. (2013). Fourier ring correlation as a resolution criterion for super-resolution microscopy. Journal of Structural Biology, 183(3), 363–367. 10.1016/j.jsb.2013.05.004. [DOI] [PubMed] [Google Scholar]
- Cheng A, Henderson R, Mastronarde D, Ludtke SJ, Schoenmakers RH, Short J, Marabini R, Dallakyan S, Agard D, Winn M. MRC2014: Extensions to the MRC format header for electron cryo-microscopy and tomography. J Struct Biol (2015) 192(2):146–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitry T, Patrick C (2018) Real-time cryo-EM data pre-processing with Warp bioRxiv 338558; 10.1101/338558 [DOI]
- Grant T, Rohou A, Grigorieff N. cisTEM, user-friendly software for single-particle image processing. Elife 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R, Sali A, Baker ML, Carragher B, Devkota B, Downing KH, Egelman EH, Feng Z, Frank J, Grigorieff N, Jiang W, Ludtke SJ, Medalia O, Penczek PA, Rosenthal PB, Rossmann MG, Schmid MF, Schröder GF, Steven AC, Stokes DL, Westbrook JD, Wriggers W, Yang H, Young J, Berman HM, Chiu W, Kleywegt GJ, Lawson CL. Outcome of the first electron microscopy validation task force meeting. Structure (2012) 20(2):205–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann JB (2018) Map Challenge assessment: Fair comparison of single particle cryoEM reconstructions 10.1016/j.jsb.2018.07.012 [DOI] [PubMed]
- Hudson G, Léger A, Niss B, Sebestyén I, (2017) JPEG at 25: Still Going Strong, IEEE MultiMedia, Volume: 24, Issue: 2, Apr-Jun 10.1109/MMUL.2017.38 [DOI] [Google Scholar]
- Kim LY, Rice WJ, Eng ET, Kopylov M, Cheng A, Raczkowski AM, Jordan KD, Bobe D, Potter CS, Carragher B. (2018) Benchmarking cryo-EM Single Particle Analysis Workflow. Front Mol Biosci June 4;5:50 doi: 10.3389/fmolb.2018.00050 . eCollection 2018. doi: 10.3389/fmolb.2018.0005010.3389/fmolb.2018.00050. eCollection 2018. doi: 10.3389/fmolb.2018.00050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleywegt GJ, Velankar S, Patwardhan A. Structural biology data archiving - where we are and what lies ahead. FEBS Lett (2018) 592(12):2153–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN and McIntosh JR (1996) Computer visualization of three-dimensional image data using IMOD J. Struct. Biol 116:71–76. [DOI] [PubMed] [Google Scholar]
- Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW. (2009). Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol 166: 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwardhan A, Carazo JM, Carragher B, Henderson R, Heymann JB, Hill E, Jensen GJ, Lagerstedt I, Lawson CL, Ludtke SJ, Mastronarde D, Moore WJ, Roseman A, Rosenthal P, Sorzano CO, Sanz-García E, Scheres SH, Subramaniam S, Westbrook J, Winn M, Swedlow JR, Kleywegt GJ. (2012) Data management challenges in three-dimensional EM. Nat Struct Mol Biol 19(12):1203–7. 10.1038/nsmb.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwardhan A, Lawson CL. Databases and Archiving for CryoEM. Methods Enzymol (2016) 579:393–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. (2017) cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. Mar;14(3):290–296. 10.1038/nmeth.4169. Epub 2017 Feb 6. [DOI] [PubMed] [Google Scholar]
- Ripstein ZA, Rubinstein JL (2016) Processing of cryo-EM movie data Methods Enzymol, 579, pp. 103–124 [DOI] [PubMed] [Google Scholar]
- Rohou A, Grigorieff N. (2015) CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol 192, 216–221. 10.1016/j.jsb.2015.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman AM. (2004) FindEM--a fast, efficient program for automatic selection of particles from electron micrographs. J Struct Biol 145, 91–99. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Rueden CT, Hiner MC. (2015), “The ImageJ ecosystem: An open platform for biomedical image analysis”, Molecular Reproduction and Development, [DOI] [PMC free article] [PubMed]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. (2005) Automated molecular microscopy: the new Leginon system. J Struct Biol 151, 41–60. 10.1016/j.jsb.2005.03.010 [DOI] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. (2007) EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157, 38–46. [DOI] [PubMed] [Google Scholar]
- Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. (2009) DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J Struct Biol 166, 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch T. (1984) A Technique for High-Performance Data Compression. Computer 17(6), 8–19 10.1109/MC.1984.1659158 [DOI] [Google Scholar]
- Zheng SQ, Palovcak E, Armache J-P, Verba KA, Cheng Y, Agard DA. (2017) MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, Scheres SH. (2018) New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]