Abstract
Schmallenberg virus (SBV), an orthobunyavirus infecting ruminants, emerged in 2011 in Central Europe, spread very rapidly throughout the continent and established an endemic status, thereby representing a constant threat not only to the European livestock population, but also to neighboring countries. Hence, in endemically infected regions, the maintenance and regular verification of diagnostics is needed and in not yet affected regions, suitable diagnostic systems should be established to be prepared for a potential introduction of the disease. In addition, also for the trade of animals into free regions, highly reliable and sensitive diagnostics are of utmost importance. Therefore, a laboratory proficiency trial was initiated to allow for performance evaluations of test systems available for SBV-diagnostics, but also for evaluation of veterinary diagnostic laboratories performing those tests. Ten serum samples (six seropositive, four seronegative) were provided for serological analysis, four of the seropositive samples were provided undiluted, while the remaining samples represented 1/2 and 1/4 dilutions of one of the aforementioned samples in negative serum. Ten further sera (five virus-positive, five negative) were sent to the participants to be analyzed by SBV genome detection methods. A total of 48 diagnostic laboratories from 15 countries of three continents (Europe, Asia, North America) and three kit manufacturers participated in the SBV proficiency test, thereby generating 131 result sets, corresponding to 1310 individual results. The sample panel aimed for serological analysis was tested 72 times; the applied diagnostic methods comprised different commercial ELISAs and standard micro-neutralization tests. The sample set aimed for genome detection was analyzed in 59 approaches by various commercial or in-house (real-time) RT-PCR protocols. Antibody or genome positive samples were correctly identified in every case, independent of the applied diagnostic test system. For seronegative samples, three incorrect, false-positive test results were produced. Virus-negative samples tested false-positive in two cases. Thus, a very high diagnostic accuracy of 99.58% and 99.66% was achieved by the serological and virological methods, respectively. Hence, this ring trial demonstrated that reliable and robust SBV-diagnostics has been established in veterinary diagnostic laboratories in affected and non-affected countries.
Introduction
Schmallenberg virus (SBV), a member of the genus Orthobunyavirus (order Bunyavirales), infects predominantly ruminants, thereby inducing a short-lived viremia of two to six days, in cattle sometimes associated with a mild, transient disease characterized by fever, diarrhea and decreased milk production [1, 2]. However, when pregnant cattle or ewes are infected during a critical period of gestation, SBV may cross the placental barrier and infect the fetus occasionally leading to abortion and/or severe fetal malformation summarized as arthrogryposis-hydranencephaly syndrome [3–5]. Between its mammalian hosts, SBV is transmitted by Culicoides biting midges of various species [6–10].
SBV initially emerged in 2011 in the German/Dutch border region [1]. Thereafter, it spread rapidly throughout the European continent reaching the Scandinavian countries and the British Isles at the North, the Mediterranean region including Spain, Southern France, Italy, Greece and Turkey at the South and Eastern European countries such as Poland or Lithuania and Russia [11–21]. In subsequent years, alternating low-level circulation and re-emergence to a larger extend have been reported from affected regions [22–26]. Therefore, it has to be anticipated that SBV has established an enzootic status in Europe and will re-appear to a larger extent in regular intervals in the future, especially since such patterns of cyclic re-emergence are well-known from closely related viruses [27–30]. Thus, the maintenance and constant verification of reliable diagnostics is needed. Furthermore, diagnostic systems should be established and regularly evaluated in not yet affected countries bordering the endemically infected regions, since a further spread of the disease cannot be excluded, particularly because the insect vectors responsible for virus transmission are wide-spread [31, 32].
The direct detection of SBV is primarily based on RT-PCR systems, either in the form of different commercially available real-time RT-PCR kits or various in-house real-time or conventional RT-PCR protocols (e.g., [1, 33, 34]. In adult animals, the preferred sample material is serum. However, the direct detection of virus or viral genome is time restricted by the short-lived viremia of only a few days. Therefore, the detection of specific antibodies, which are induced between one and three weeks after infection [2, 35–37] and persist for several years [38–40], is more promising for SBV-infection diagnosis. For serological analysis, several commercial or in-house ELISAs, micro-neutralization or indirect immunofluorescence tests are available [36, 41–43].
Here, test systems routinely used for SBV-infection diagnostics have been evaluated in the context of an international interlaboratory proficiency trial. A panel of standardized samples was sent to veterinary diagnostic laboratories across the world with the request to analyze the samples by serological and virological methods routinely applied in the respective institution.
Materials and methods
A total of 20 serum samples were provided, where 10 samples were aimed for the detection of viral genome, and 10 samples for serological analysis. Aliquots of 1ml were prepared in 2-ml injection bottles (Zscheile & Klinger GmbH, Hamburg, Germany) and lyophilized. The bottles were subsequently sealed with rubber plug and flanged caps (both Zscheile & Klinger GmbH) and stored at 4°C until sent to the participating institutions.
The sample panel for serological analysis included two sheep and two cattle sera negative for antibodies against SBV, and six antibody positive sera (1x sheep, 5x cattle), which were collected 3, 4, or 12 weeks after experimental SBV-infection [2, 36, 44]. The sample SBV-S-8 represented a 1/2 dilution of sample SBV-S-10 in SBV antibody negative serum and the sample SBV-S-1 represented a 1/4 dilution of sample SBV-S-10 (Table 1).
Table 1. Status of the samples sent for Schmallenberg virus (SBV) infection diagnosis to the ring trial participants.
The results of the pre-testing by real-time RT-PCR [33] or microneutralization test [36] prior to shipment and the time points at which the samples for viral genome detection were taken after experimental infection are given in parenthesis. Cq–quantification cycle value, dpi–days post infection.
| ring trial number | animal species | sample status (neutralizing titer or Cq value) |
|---|---|---|
| SBV-S-1 | cattle | SBV antibody positive, 1/4 dilution of sample SBV-S-10 (1/28) |
| SBV-S-2 | sheep | SBV antibody negative (< 1/5) |
| SBV-S-3 | cattle | SBV antibody positive (1/90) |
| SBV-S-4 | cattle | SBV antibody negative (< 1/5) |
| SBV-S-5 | cattle | SBV antibody positive (1/28) |
| SBV-S-6 | sheep | SBV antibody negative (< 1/5) |
| SBV-S-7 | cattle | SBV antibody negative (< 1/5) |
| SBV-S-8 | cattle | SBV antibody positive, 1/2 dilution of sample SBV-S10 (1/57) |
| SBV-S-9 | sheep | SBV antibody positive (1/22) |
| SBV-S-10 | cattle | SBV antibody positive (1/71) |
| SBV-P-11 | cattle | SBV genome positive (25, 3dpi) |
| SBV-P-12 | cattle | SBV genome positive (26, 3 dpi) |
| SBV-P-13 | sheep | SBV genome negative (no Cq) |
| SBV-P-14 | cattle | SBV genome positive (27, 4dpi) |
| SBV-P-15 | cattle | SBV genome negative, = SBV-S-10 (no Cq) |
| SBV-P-16 | cattle | SBV genome positive (28, 5dpi) |
| SBV-P-17 | sheep | SBV genome negative (no Cq) |
| SBV-P-18 | cattle | SBV genome negative (no Cq) |
| SBV-P-19 | cattle | SBV genome positive (25, 4dpi) |
| SBV-P-20 | cattle | SBV genome negative (no Cq) |
The panel aimed for the detection of viral genome comprised two SBV genome negative sheep sera, three SBV genome negative cattle sera (the antibody positive serum SBV-S-10 and two times fetal calf serum, Biochrom GmbH, Berlin, Germany), and five cattle sera obtained during the viremic phase after experimental infection with SBV [44].
The identifiers and classifications of all samples are given in Table 1.
A total of 48 veterinary diagnostic laboratories from 15 countries (Austria, Belgium, Canada, Denmark, Finland, France, Germany, Hungary, Ireland, Israel, Italy, Netherlands, Poland, Russia, and Switzerland) and three kit manufacturers participated in the SBV proficiency test. Every German state veterinary laboratory had been invited, and every non-German reference center that asked previously whether the authors organize SBV ring trials was allowed to participate. The participants (except the kit manufacturers) perform routine diagnostics for Simbu serogroup viruses in their country and/or pre-export or pre-import investigations.
The participants were asked to analyze the provided samples with the test systems routinely used in their institution. If results were considered “doubtful” by a participant, it was assumed that the necessary clarifications/follow-up analysis would have been initiated in practice and, therefore, that no divergent result was produced.
To assess the discriminative property of the tests, the diagnostic accuracy was calculated by taking the sensitivity and specificity into account. The sensitivity represents the ability to identify a positive sample correctly and is defined as the proportion of true positive results in a set of positive cases (calculated as: true positive results/true positives + false negatives), while the specificity of a test is its ability to determine the negative cases correctly (calculated as: true negative results/true negatives + false positives) [45]. The diagnostic accuracy was finally calculated by using the free statistical calculator MedCalc (MedCalc Software, Ostend, Belgium).
Results
Serology
The sample panel for serological analysis was investigated in 43 laboratories by commercially available antibody ELISA tests, in some cases several test systems were used, whereby 50 result sets were generated (= 500 individual results). The applied test systems included: (I) ID Screen Schmallenberg virus Competition Multi-species, IDvet, Grabels, France (n = 28); (II) ID Screen Schmallenberg virus Indirect Multi-species, IDvet, in either the monophasic (n = 11) or biphasic (n = 5) variant; (III) IDEXX Schmallenberg Ab Test, IDEXX Europe B.V., Hoofddorp, the Netherlands (n = 4); (IV) SVANOVIR SBV-Ab, Boehringer Ingelheim Svanova, Uppsala, Sweden (n = 2). The status of the samples was correctly identified in every case (Table 2).
Table 2. Results of commercially available Schmallenberg virus antibody ELISAs and of the standard microneutralization tests performed by the participating laboratories.
The sample status is given below the respective sample identifier.
| test system | SBV-S-1 (pos) | SBV-S-2 (neg) | SBV-S-3 (pos) | SBV-S-4 (neg) | SBV-S-5 (pos) | SBV-S-6 (neg) | SBV-S-7 (neg) | SBV-S-8 (pos) | SBV-S-9 (pos) | SBV-S-10 (pos) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ID Screen Schmallenberg virus Competition Multi-species, ID.vet | no. tests | 28 | 28 | 28 | 28 | 28 | 28 | 28 | 28 | 28 | 28 |
| no. positive | 28 | 0 | 28 | 0 | 28 | 0 | 0 | 28 | 28 | 28 | |
| ID Screen Schmallenberg virus Indirect Multi-species (biph.), ID.vet | no. tests | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| no. positive | 5 | 0 | 5 | 0 | 5 | 0 | 0 | 5 | 5 | 5 | |
| ID Screen Schmallenberg virus Indirect Multi-species (monoph.), ID.vet | no. tests | 11 | 11 | 11 | 11 | 11 | 11 | 11 | 11 | 11 | 11 |
| no. positive | 11 | 0 | 11 | 0 | 11 | 0 | 0 | 11 | 11 | 11 | |
| IDEXX Schmallenberg Ab Test, IDEXX | no. tests | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| no. positive | 4 | 0 | 4 | 0 | 4 | 0 | 0 | 4 | 4 | 4 | |
| SVANOVIR SBV-Ab, Svanova | no. tests | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| no. positive | 2 | 0 | 2 | 0 | 2 | 0 | 0 | 2 | 2 | 2 | |
| microneutralization test | no. tests | 22 | 22 | 22 | 22 | 22 | 22 | 22 | 22 | 22 | 22 |
| no. positive | 22 | 2 | 22 | 0 | 22 | 1 | 0 | 22 | 22 | 22 | |
| no. doubtful | 0 | 4 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
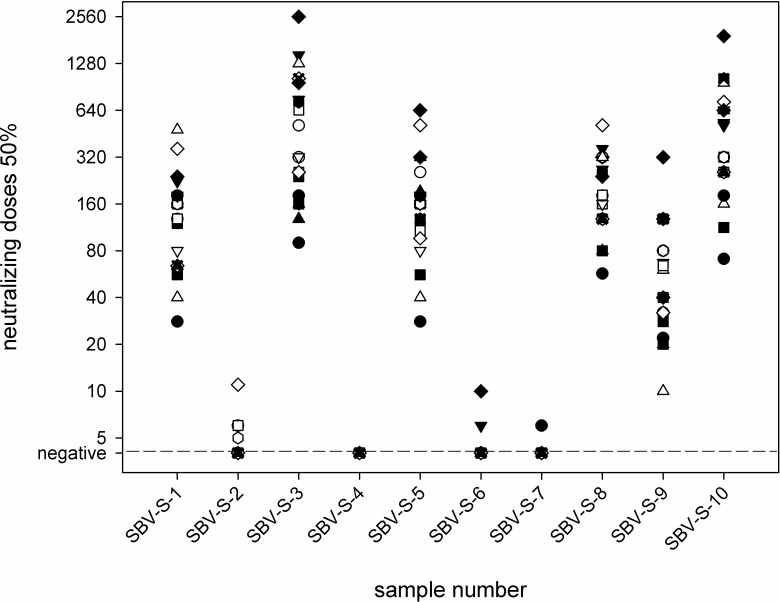
In 22 laboratories, the samples were investigated by standard microneutralization tests in addition or alternatively to the analysis by antibody ELISA. The test was conducted in 17 laboratories according to a previously published protocol [36] using the first SBV-isolate “BH80/11” and a baby hamster kidney (BHK) cell line, and the cytopathogenic effect was assessed after two, three, four or five days. In two further cases, a BHK cell line was used as well, but the specific SBV strain was not given in the results sheet. In the remaining three laboratories, an African green monkey (Vero) cell line was used in combination with a local field strain; the cytopathogenic effect was assessed after four days. Sera containing SBV-specific antibodies (SBV-S-1, SBV-S-3, SBV-S-5, SBV-S-8, SBV-S-9, and SBV-S-10) tested positive as expected in every case, however, the resulting neutralizing titers differed markedly between the laboratories (SBV-S-1: 1/28 to 1/480; SBV-S-3: 1/90 to 1/2560; SBV-S-5: 1/28 to 640; SBV-S-8: 1/57 to 1/513; SBV-S-9: 1/10 to 1/320; SBV-S-10: 1/71 to 1/1920) (Fig 1). Antibody negative sera (SBV-S-2, SBV-S-4, SBV-S-6, and SBV-S-7) were predominantly correctly defined as being negative. The only exceptions were sample SBV-S-2, which scored doubtful in four cases and positive in two laboratories, sample SBV-S-6 that tested once doubtful and once false-positive, and sample SBV-S-7 that tested doubtful in one case (Table 2).
Fig 1. Results of the standard microneutralization tests for the serological panel.
All results of a particular participant are depicted by the identical symbol for each sample.
Considering a total number of 720 individual results produced by serological methods (500 by ELISAs and 220 by neutralization tests) and three false-positive results, a diagnostic accuracy of 99.58% was achieved.
Genome detection
To be used for PCR analysis, nucleic acids were extracted from the provided samples either manually by using six different commercial kits or TRIzol, or automated extraction based on a wide range of commercial kits was applied. The extraction kits used by the participants included (sorted alphabetically by manufacturer): (I: manual extraction) NucleoSpin RNA Virus, MACHERY-NAGEL GmbH & Co. KG, Düren, Germany; QIAamp cador Pathogen Mini, Qiagen, Hilden, Germany; QIAamp Viral RNA Mini, Qiagen; RNeasy Mini kit, Qiagen; High Pure Viral RNA Kit, Roche, Basel, Switzerland; Invisorb Spin Virus RNA Mini Kit, Stratec Molecular GmbH, Berlin, Germany; (II: automated extraction) innuPREP Virus DNA/RNA kit, Analytik Jena AG, Jena, Germany; chemagic Viral DNA/RNA Kit PerkinElmer chemagen Technologie GmbH, Baesweiler, Germany; ID Gene Mag Fast Extraction kit, IDvet; ID Gene Mag Universal Extraction kit, IDvet; NucleoMag VET kit, MACHERY-NAGEL GmbH & Co. KG; NucleoSpin RNA Virus, MACHERY-NAGEL GmbH & Co. KG; NucleoSpin Virus kit, MACHERY-NAGEL GmbH &Co. KG; QIAamp cador Pathogen Mini, Qiagen; QIAamp Viral RNA Mini, Qiagen; QIAamp Viral RNA Mini QIAcube kit, Qiagen; MagAttract 96 cador Pathogen kit, Qiagen; MagAttract Virus Mini M48 kit, Qiagen; MagNA Pure 96 DNA and Viral NA Small Volume Kit, Roche; Ribo-Sorb RNA/DNA extraction kit, Sacace Biotechnologies Srl, Caserta, Italy; LSI MagVet Universal Isolation Kit, ThermoFisher Scientific, Waltham, USA; MagMAX CORE Nucleic Acid Purification Kit, ThermoFisher Scientific; MagMAX Pathogen RNA/DNA Kit, ThermoFisher Scientific.
For subsequent PCR analyses, commercially available real-time RT-PCR kits were used in a total of 38 approaches. The applied test systems included: (I) virotype SBV RT-PCR Kit, INDICAL BIOSCIENCE GmbH, Leipzig, Germany (n = 32); (II) virellaSBV real time RT-PCR Kit, gerbion GmbH & Co. KG, Kornwestheim, Germany (n = 3); (III) ADIAVET Schmallenberg virus real time, bioMérieux, Marcy-l’Étoile, Frankreich (n = 1); (IV) ID Gene Schmallenberg Duplex, ID.vet GENETICS, Grabels, France (n = 1); (V) VetMAX Schmallenberg Virus Kit, ThermoFisher Scientific (n = 1). In addition or alternatively to the commercial kits, in-house RT-PCR assays were applied for genome detection in 21 laboratories (16x primers and probe described in [33]; 1x [46]; 3x not further specified real-time RT-PCR protocols, 1x not further specified conventional RT-PCR protocol).
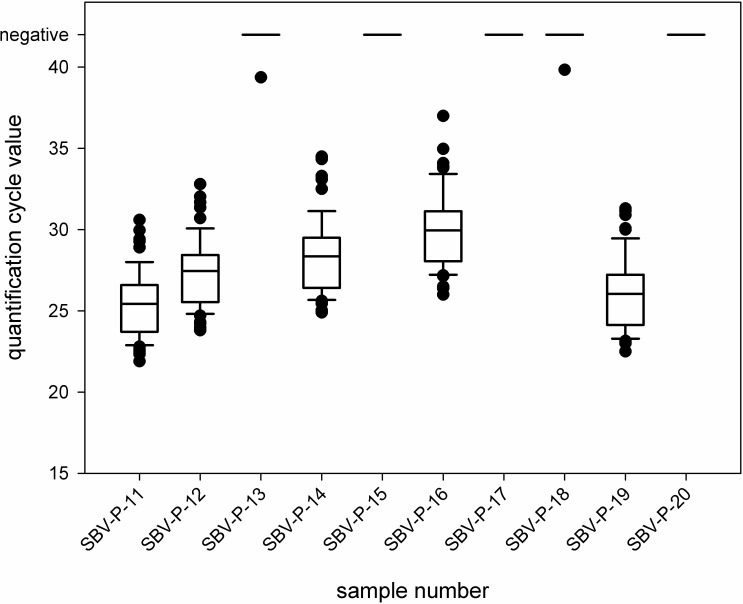
Positive samples (SBV-P-11, SBV-P-12, SBV-P-14, SBV-P-16, and SBV-P-19) were correctly identified in every case independent from the applied test system. The quantification cycle (Cq) values determined for each individual sample are shown in Fig 2 as box plots.
Fig 2. Quantification cycle (Cq) values produced by the ring trial participants using different real-time RT-PCR systems.
Each outlier is depicted by a dot.
The negative samples (SBV-P-13, SBV-P-15, SBV-P-17, SBV-P-18, and SBV-P-20) were tested correctly negative, with the exception of samples SBV-P-13 and SBV-P-18, which tested weak positive in one case each. The Cq values of these incorrect results were 39.4 (sample SBV-P-13) and 39.8 (SBV-P-18), respectively, and therefore very close to the detection limit.
Since the sample panel for genome detection has been tested by 59 extraction/PCR approaches, thereby generating 590 individual results, and only two incorrect results were produced, the sample status was correctly identified in 99.66% of the analyses.
Discussion
Insect-transmitted viruses have been emerging and spreading for centuries, however, their incidence and geographical spread is more rapid and extensive nowadays. Factors that were claimed to be the reasons of this phenomenon range from climate change and the intensive growth of the global transportation systems to an increase in urbanization and conversion of land to agricultural use [47]. An example for such an emerging and intensively circulating virus of veterinary importance represents SBV, which initially emerged in Central Europe and subsequently spread very rapidly throughout the European Union [21]. Thereafter, the virus established an endemic status in Central Europe, thereby presenting a constant threat to the ruminant population, which makes the maintenance of a high quality standard of SBV diagnostics highly important.
In addition, trade restrictions for live animals and bovine semen have been implemented in several non-affected countries [48], which can be circumvented by appropriate pre-export checks. This further highlights the importance of highly reliable diagnostics that should be established not only in endemically affected countries, but also in areas at risk for disease introduction.
One way of assuring the quality of diagnostics and to independently assess the quality of results produced in diagnostic laboratories represents the participation in interlaboratory proficiency trials. By ring tests, the competency of a laboratory as well as of the applied methods can be demonstrated to accreditation or other regulatory bodies [49]. That is why SBV proficiency trials have been initiated among laboratories located in the initially mostly affected regions shortly after the establishment of first methods for either the serological or virological diagnosis of SBV-infections [50, 51]. However, to ensure a high level of quality, laboratories should participate in ring trials on a regular basis, as has been accomplished in Germany, where an infection with SBV represents a reportable disease (Regulation on Reportable Animal Diseases, http://www.gesetze-im-internet.de/tkrmeldpflv_1983/BJNR010950983.html), and where ring trials are organized regularly [51, 52].
Here, we describe the assessment of the diagnostic capacity of numerous European laboratories and, for the first time, also of laboratories from further continents, i.e. Asia and North America. Serum samples taken during the viremic phase after experimental SBV-infection were provided and viral genome was reliably detected in every case regardless of the applied nucleic acid extraction/RT-PCR assay combination, which is quite considerable when one recalls the very large number of used extraction methods. In terms of specificity, five SBV-negative sera were tested 59 times and false-positive results were only generated in two cases (one incorrect test result each in two different laboratories). Possible explanations for these incorrect test results might be improper PCR conditions or unspecific reactions of the applied PCR reagents [53]. However, different extraction methods (manual vs. automated) and two distinct commercial real-time RT-PCR kits were used and the genome negative samples were tested in both cases in direct proximity to a highly positive sample. Therefore, the most likely explanation is cross-contamination during nucleic acid extraction or PCR preparation, which is a well-known risk in molecular diagnostics, especially when carried out in conjunction with the propagation and application of virus- or plasmid-based positive controls or sequencing [54, 55]. In order to notice such contamination the constant inclusion of proper controls, such as a sufficient number of negative extraction controls and no template controls, is highly recommended [53, 56], thereby preventing incorrect examination reports.
When analyzing the serological sample panel, an excellent sensitivity of 100% was achieved as well. Moreover, despite a previous study reported clear differences in the diagnostic performance of commercial SBV antibody ELISAs [57], this optimal sensitivity of 100% was reached in the present study regardless of whether a commercial kit or a cell-culture based method has been used. However, when comparing the titers measured by the applied microneutralization tests considerable variations are noticeable. This phenomenon has been already described earlier [50, 52], and is most likely caused by the variations in the test protocols of this biological assay, such as different cell lines, virus isolates or viral doses and readout after two, three, four or five days. Nonetheless, the final classifications, i.e. the identification of positive samples, was in full agreement between the participating laboratories. In terms of specificity, three false-positive results were produced leading to the questions as to whether the cut-off should be adjusted or the test protocol slightly modified in the concerned laboratories.
In conclusion, a very high overall diagnostic accuracy of 99.62% was achieved in this SBV ring trial. Hence, the presented interlaboratory proficiency test demonstrated that reliable SBV-infection diagnostics was established and maintained for both, antibody and viral genome detection.
Acknowledgments
We thank Bianka Hillmann and Alrik-Markis Kunisch for excellent technical assistance and especially all the diagnostic laboratories for their efforts in participating in this SBV ring trial.
Data Availability
All relevant data are within the manuscript.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Hoffmann B, Scheuch M, Höper D, Jungblut R, Holsteg M, Schirrmeier H, et al. Novel orthobunyavirus in cattle, Europe, 2011. Emerg Infect Dis. 2012;18(3):469–72. 10.3201/eid1803.111905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wernike K, Hoffmann B, Bréard E, Bøtner A, Ponsart C, Zientara S, et al. Schmallenberg virus experimental infection of sheep. Veterinary microbiology. 2013;166(3–4):461–6. Epub 2013/08/27. 10.1016/j.vetmic.2013.06.030 . [DOI] [PubMed] [Google Scholar]
- 3.Dominguez M, Gache K, Touratier A, Perrin JB, Fediaevsky A, Collin E, et al. Spread and impact of the Schmallenberg virus epidemic in France in 2012–2013. BMC Vet Res. 2014;10(1):248 10.1186/s12917-014-0248-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lievaart-Peterson K, Luttikholt SJM, Van den Brom R, Vellema P. Schmallenberg virus infection in small ruminants—First review of the situation and prospects in Northern Europe. Small Ruminant Res. 2012;106(2–3):71–6. 10.1016/j.smallrumres.2012.03.006 PubMed PMID: WOS:000307688800001. [DOI] [Google Scholar]
- 5.Wernike K, Elbers A, Beer M. Schmallenberg virus infection. Rev Sci Tech. 2015;34(2):363–73. . [DOI] [PubMed] [Google Scholar]
- 6.De Regge N, Deblauwe I, De Deken R, Vantieghem P, Madder M, Geysen D, et al. Detection of Schmallenberg virus in different Culicoides spp. by real-time RT-PCR. Transboundary and emerging diseases. 2012;59(6):471–5. Epub 2012/10/03. 10.1111/tbed.12000 . [DOI] [PubMed] [Google Scholar]
- 7.Rasmussen LD, Kristensen B, Kirkeby C, Rasmussen TB, Belsham GJ, Bodker R, et al. Culicoids as vectors of Schmallenberg virus. Emerg Infect Dis. 2012;18(7):1204–6. Epub 2012/06/20. 10.3201/eid1807.120385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goffredo M, Monaco F, Capelli G, Quaglia M, Federici V, Catalani M, et al. Schmallenberg virus in Italy: a retrospective survey in Culicoides stored during the bluetongue Italian surveillance program. Prev Vet Med. 2013;111(3–4):230–6. Epub 2013/06/25. 10.1016/j.prevetmed.2013.05.014 . [DOI] [PubMed] [Google Scholar]
- 9.Elbers AR, Meiswinkel R, van Weezep E, Sloet van Oldruitenborgh-Oosterbaan MM, Kooi EA. Schmallenberg virus in Culicoides spp. biting midges, the Netherlands, 2011. Emerg Infect Dis. 2013;19(1):106–9. Epub 2012/12/25. 10.3201/eid1901.121054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larska M, Polak MP, Grochowska M, Lechowski L, Zwiazek JS, Zmudzinski JF. First report of Schmallenberg virus infection in cattle and midges in Poland. Transboundary and emerging diseases. 2013;60(2):97–101. Epub 2013/01/22. 10.1111/tbed.12057 . [DOI] [PubMed] [Google Scholar]
- 11.Larska M, Krzysiak M, Smreczak M, Polak MP, Zmudzinski JF. First detection of Schmallenberg virus in elk (Alces alces) indicating infection of wildlife in Bialowieza National Park in Poland. Vet J. 2013;198(1):279–81. Epub 2013/09/12. 10.1016/j.tvjl.2013.08.013 . [DOI] [PubMed] [Google Scholar]
- 12.Lazutka J, Zvirbliene A, Dalgediene I, Petraityte-Burneikiene R, Spakova A, Sereika V, et al. Generation of recombinant Schmallenberg virus nucleocapsid protein in yeast and development of virus-specific monoclonal antibodies. Journal of immunology research. 2014;2014:160316 10.1155/2014/160316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandez-Aguilar X, Pujols J, Velarde R, Rosell R, Lopez-Olvera JR, Marco I, et al. Schmallenberg virus circulation in high mountain ecosystem, Spain. Emerg Infect Dis. 2014;20(6):1062–4. 10.3201/eid2006.130961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaintoutis SC, Kiossis E, Giadinis ND, Brozos CN, Sailleau C, Viarouge C, et al. Evidence of Schmallenberg virus circulation in ruminants in Greece. Tropical animal health and production. 2014;46(1):251–5. Epub 2013/07/23. 10.1007/s11250-013-0449-5 . [DOI] [PubMed] [Google Scholar]
- 15.Rasmussen LD, Kirkeby C, Bodker R, Kristensen B, Rasmussen TB, Belsham GJ, et al. Rapid spread of Schmallenberg virus-infected biting midges (Culicoides spp.) across Denmark in 2012. Transboundary and emerging diseases. 2014;61(1):12–6. Epub 2013/11/14. 10.1111/tbed.12189 . [DOI] [PubMed] [Google Scholar]
- 16.Wisloff H, Nordvik BS, Sviland S, Tonnessen R. The first documented clinical case of Schmallenberg virus in Norway: fetal malformations in a calf. The Veterinary record. 2014;174(5):120 Epub 2014/01/09. 10.1136/vr.102149 . [DOI] [PubMed] [Google Scholar]
- 17.Mason C, Stevenson H, Carty H, Hosie B, Caldow G, Boyes G. SBV in a dairy herd in Scotland. The Veterinary record. 2013;172(15):403 Epub 2013/04/16. 10.1136/vr.f2256 . [DOI] [PubMed] [Google Scholar]
- 18.Bradshaw B, Mooney J, Ross PJ, Furphy C, O'Donovan J, Sanchez C, et al. Schmallenberg virus cases identified in Ireland. The Veterinary record. 2012;171(21):540–1. Epub 2012/11/28. 10.1136/vr.e7928 . [DOI] [PubMed] [Google Scholar]
- 19.Bouchemla F, Agoltsov VA, Larionov SV, Popova OM, Shvenk EV. Epizootiological study on spatiotemporal clusters of Schmallenberg virus and Lumpy skin diseases: The case of Russia. Veterinary world. 2018;11(9):1229–36. Epub 2018/11/10. 10.14202/vetworld.2018.1229-1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yilmaz H, Hoffmann B, Turan N, Cizmecigil UY, Richt JA, van der Poel WH. Detection and partial sequencing of Schmallenberg virus in cattle and sheep in Turkey. Vector Borne Zoonotic Dis. 2014;14(3):223–5. Epub 2014/03/01. 10.1089/vbz.2013.1451 . [DOI] [PubMed] [Google Scholar]
- 21.EFSA. "Schmallenberg" virus: analysis of the epidemiological data (May 2013). EFSA Supporting Publications 2013 EN-3429 http://wwwefsaeuropaeu/de/supporting/doc/429epdf; accessed 15/07/2013. 2013.
- 22.Wernike K, Hoffmann B, Conraths FJ, Beer M. Schmallenberg Virus Recurrence, Germany, 2014. Emerg Infect Dis. 2015;21(7):1202–4. 10.3201/eid2107.150180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delooz L, Saegerman C, Quinet C, Petitjean T, De Regge N, Cay B. Resurgence of Schmallenberg Virus in Belgium after 3 Years of Epidemiological Silence. Transboundary and emerging diseases. 2017;64(5):1641–2. 10.1111/tbed.12552 . [DOI] [PubMed] [Google Scholar]
- 24.Wernike K, Beer M. Schmallenberg virus: a novel virus of veterinary importance. Adv Virus Res. 2017;99:39–60. 10.1016/bs.aivir.2017.07.001 . [DOI] [PubMed] [Google Scholar]
- 25.Sohier C, Deblauwe I, Van Loo T, Hanon JB, Cay AB, De Regge N. Evidence of extensive renewed Schmallenberg virus circulation in Belgium during summer of 2016—increase in arthrogryposis-hydranencephaly cases expected. Transboundary and emerging diseases. 2017;64(4):1015–9. 10.1111/tbed.12655 . [DOI] [PubMed] [Google Scholar]
- 26.Gache K, Zientara S, Collin E, Authie E, Dion F, Garin E, et al. Spatial and temporal patterns of Schmallenberg virus in France in 2016. The Veterinary record. 2018;182(20):575 Epub 2018/02/16. 10.1136/vr.104769 . [DOI] [PubMed] [Google Scholar]
- 27.Kato T, Yanase T, Suzuki M, Katagiri Y, Ikemiyagi K, Takayoshi K, et al. Monitoring for bovine arboviruses in the most southwestern islands in Japan between 1994 and 2014. BMC Vet Res. 2016;12(1):125 10.1186/s12917-016-0747-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayama Y, Yanase T, Suzuki M, Unten K, Tomochi H, Kakehi M, et al. Meteorological factors affecting seroconversion of Akabane disease in sentinel calves in the subtropical Okinawa Islands of Japan. Tropical animal health and production. 2018;50(1):209–15. 10.1007/s11250-017-1404-7 . [DOI] [PubMed] [Google Scholar]
- 29.Kato T, Shirafuji H, Tanaka S, Sato M, Yamakawa M, Tsuda T, et al. Bovine Arboviruses in Culicoides Biting Midges and Sentinel Cattle in Southern Japan from 2003 to 2013. Transboundary and emerging diseases. 2016;63(6):e160–e72. 10.1111/tbed.12324 . [DOI] [PubMed] [Google Scholar]
- 30.Geoghegan JL, Walker PJ, Duchemin JB, Jeanne I, Holmes EC. Seasonal drivers of the epidemiology of arthropod-borne viruses in Australia. PLoS Negl Trop Dis. 2014;8(11):e3325 10.1371/journal.pntd.0003325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carpenter S, Groschup MH, Garros C, Felippe-Bauer ML, Purse BV. Culicoides biting midges, arboviruses and public health in Europe. Antiviral Res. 2013;100(1):102–13. Epub 2013/08/13. 10.1016/j.antiviral.2013.07.020 . [DOI] [PubMed] [Google Scholar]
- 32.Elbers AR, Koenraadt CJ, Meiswinkel R. Mosquitoes and Culicoides biting midges: vector range and the influence of climate change. Rev Sci Tech. 2015;34(1):123–37. . [DOI] [PubMed] [Google Scholar]
- 33.Bilk S, Schulze C, Fischer M, Beer M, Hlinak A, Hoffmann B. Organ distribution of Schmallenberg virus RNA in malformed newborns. Veterinary microbiology. 2012;159(1–2):236–8. Epub 2012/04/21. 10.1016/j.vetmic.2012.03.035 . [DOI] [PubMed] [Google Scholar]
- 34.Fischer M, Schirrmeier H, Wernike K, Wegelt A, Beer M, Hoffmann B. Development of a pan-Simbu real-time reverse transcriptase PCR for the detection of Simbu serogroup viruses and comparison with SBV diagnostic PCR systems. Virology journal. 2013;10(1):327 Epub 2013/11/06. 10.1186/1743-422X-10-327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wernike K, Silaghi C, Nieder M, Pfeffer M, Beer M. Dynamics of Schmallenberg virus infection within a cattle herd in Germany, 2011. Epidemiol Infect. 2014;142(7):1501–4. Epub 2013/10/17. 10.1017/S0950268813002525 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wernike K, Eschbaumer M, Schirrmeier H, Blohm U, Breithaupt A, Hoffmann B, et al. Oral exposure, reinfection and cellular immunity to Schmallenberg virus in cattle. Veterinary microbiology. 2013;165(1–2):155–9. Epub 2013/03/05. 10.1016/j.vetmic.2013.01.040 . [DOI] [PubMed] [Google Scholar]
- 37.Laloy E, Riou M, Barc C, Belbis G, Bréard E, Breton S, et al. Schmallenberg virus: experimental infection in goats and bucks. BMC Vet Res. 2015;11(1):221 10.1186/s12917-015-0516-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elbers AR, Stockhofe-Zurwieden N, van der Poel WH. Schmallenberg virus antibody persistence in adult cattle after natural infection and decay of maternal antibodies in calves. BMC Vet Res. 2014;10(1):103 10.1186/1746-6148-10-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wernike K, Holsteg M, Szillat KP, Beer M. Development of within-herd immunity and long-term persistence of antibodies against Schmallenberg virus in naturally infected cattle. BMC Vet Res. 2018;14(1):368 10.1186/s12917-018-1702-y . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Claine F, Coupeau D, Wiggers L, Muylkens B, Kirschvink N. Modelling the evolution of Schmallenberg virus seroprevalence in a sheep flock after natural infection. Prev Vet Med. 2018;154:132–8. Epub 2018/04/25. 10.1016/j.prevetmed.2018.03.024 . [DOI] [PubMed] [Google Scholar]
- 41.Bréard E, Lara E, Comtet L, Viarouge C, Doceul V, Desprat A, et al. Validation of a commercially available indirect ELISA using a nucleocapside recombinant protein for detection of Schmallenberg virus antibodies. PLoS One. 2013;8(1):e53446 Epub 2013/01/22. 10.1371/journal.pone.0053446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Humphries D, Burr P. Schmallenberg virus milk antibody ELISA. The Veterinary record. 2012;171(20):511–2. Epub 2012/11/20. 10.1136/vr.e7739 . [DOI] [PubMed] [Google Scholar]
- 43.Loeffen W, Quak S, de Boer-Luijtze E, Hulst M, van der Poel W, Bouwstra R, et al. Development of a virus neutralisation test to detect antibodies against Schmallenberg virus and serological results in suspect and infected herds. Acta Vet Scand. 2012;54(1):44 Epub 2012/08/09. 10.1186/1751-0147-54-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wernike K, Mundt A, Link EK, Aebischer A, Schlotthauer F, Sutter G, et al. N-terminal domain of Schmallenberg virus envelope protein Gc delivered by recombinant equine herpesvirus type 1 and modified vaccinia virus Ankara: Immunogenicity and protective efficacy in cattle. Vaccine. 2018;36(34):5116–23. Epub 2018/07/28. 10.1016/j.vaccine.2018.07.047 . [DOI] [PubMed] [Google Scholar]
- 45.Šimundić AM. Measures of Diagnostic Accuracy: Basic Definitions. EJIFCC. 2009;19(4):203–11. [PMC free article] [PubMed] [Google Scholar]
- 46.Golender N, Bumbarov VY, Erster O, Beer M, Khinich Y, Wernike K. Development and validation of a universal S-segment-based real-time RT-PCR assay for the detection of Simbu serogroup viruses. J Virol Methods. 2018;261:80–5. Epub 2018/08/12. 10.1016/j.jviromet.2018.08.008 . [DOI] [PubMed] [Google Scholar]
- 47.Gould E, Pettersson J, Higgs S, Charrel R, de Lamballerie X. Emerging arboviruses: Why today? One Health. 2017;4:1–13. 10.1016/j.onehlt.2017.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.EFSA. Schmallenberg virus: State of Art. EFSA Journal. 2014;12(5):54 10.2903/j.efsa.2014.3681 [DOI] [Google Scholar]
- 49.FAO. Things to know about the ring test. Food and Agriculture Organization of the United Nations Available online: http://wwwfaoorg/ag/againfo/home/documents/2015_Announcement_Ring_testpdf. 2015.
- 50.van der Poel WH, Cay B, Zientara S, Steinbach F, Valarcher JF, Bøtner A, et al. Limited interlaboratory comparison of Schmallenberg virus antibody detection in serum samples. The Veterinary record. 2014;174(15):380 Epub 2014/03/05. 10.1136/vr.102180 . [DOI] [PubMed] [Google Scholar]
- 51.Schulz C, van der Poel WH, Ponsart C, Cay AB, Steinbach F, Zientara S, et al. European interlaboratory comparison of Schmallenberg virus (SBV) real-time RT-PCR detection in experimental and field samples: The method of extraction is critical for SBV RNA detection in semen. J Vet Diagn Invest. 2015;27(4):422–30. 10.1177/1040638715593798 . [DOI] [PubMed] [Google Scholar]
- 52.Wernike K, Beer M, Hoffmann B. Schmallenberg Virus Infection Diagnosis: Results of a German Proficiency Trial. Transboundary and emerging diseases. 2017;64(5):1405–10. 10.1111/tbed.12517 . [DOI] [PubMed] [Google Scholar]
- 53.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–22. Epub 2009/02/28. 10.1373/clinchem.2008.112797 . [DOI] [PubMed] [Google Scholar]
- 54.Hoffmann B, Beer M, Reid SM, Mertens P, Oura CA, van Rijn PA, et al. A review of RT-PCR technologies used in veterinary virology and disease control: sensitive and specific diagnosis of five livestock diseases notifiable to the World Organisation for Animal Health. Veterinary microbiology. 2009;139(1–2):1–23. Epub 2009/06/06. 10.1016/j.vetmic.2009.04.034 . [DOI] [PubMed] [Google Scholar]
- 55.Fischer M, Renevey N, Thür B, Hoffmann D, Beer M, Hoffmann B. Efficacy Assessment of Nucleic Acid Decontamination Reagents Used in Molecular Diagnostic Laboratories. PLoS One. 2016;11(7):e0159274 10.1371/journal.pone.0159274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bustin SA. Why the need for qPCR publication guidelines?—The case for MIQE. Methods. 2010;50(4):217–26. Epub 2009/12/23. 10.1016/j.ymeth.2009.12.006 . [DOI] [PubMed] [Google Scholar]
- 57.Pejaković S, Wiggers L, Coupeau D, Kirschvink N, Mason J, Muylkens B. Test selection for antibody detection according to the seroprevalence level of Schmallenberg virus in sheep. PLoS One. 2018;13(4):e0196532 Epub 2018/04/28. 10.1371/journal.pone.0196532 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the manuscript.