Abstract
Phosphorylation of dynamin-related protein 1 (Drp1) represents an important regulatory mechanism for mitochondrial fission. Here, we established the role of Drp1 serine 600 (Drp1S600) phosphorylation in mitochondrial fission in vivo and assessed the functional consequences of targeted elimination of the Drp1S600 phosphorylation site in the progression of diabetic nephropathy (DN). We generated a knockin mouse in which S600 was mutated to alanine (Drp1S600A). We found that diabetic Drp1S600A mice exhibited improved biochemical and histological features of DN along with reduced mitochondrial fission and diminished mitochondrial ROS in vivo. Importantly, we observed that the effect of Drp1S600 phosphorylation on mitochondrial fission in the diabetic milieu was stimulus dependent but not cell type dependent. Mechanistically, we show that mitochondrial fission in high-glucose conditions occurs through concomitant binding of phosphorylated Drp1S600 with mitochondrial fission factor (MFF) and actin-related protein 3 (Arp3), ultimately leading to accumulation of F-actin and Drp1 on the mitochondria. Taken together, these findings establish the idea that a single phosphorylation site in Drp1 can regulate mitochondrial fission and progression of DN in vivo and highlight the stimulus-specific consequences of Drp1S600 phosphorylation in mitochondrial dynamics.
Keywords: Metabolism, Nephrology
Keywords: Cytoskeleton, Diabetes, Mitochondria
Introduction
Diabetic kidney disease is a major microvascular complication of both type 1 and type 2 diabetes and the leading cause of end-stage renal disease in the United States (1). Despite significant advances in our understanding of the pathobiology and progression of diabetic nephropathy (DN), current treatment options for DN remain limited, and new and effective therapeutic approaches are urgently needed.
Mitochondrial dysfunction has long been proposed to play a critical role in the pathogenesis of a wide range of microvascular complications of diabetes (2–5). Mitochondria are highly dynamic organelles that undergo remodeling to meet the metabolic demands of the cell. Mitochondrial dynamics encompasses 2 complementary processes: mitochondrial fission and fusion (6–9). Mitochondrial fission is governed primarily by the cytoplasmic dynamin-related protein 1 (Drp1). Drp1 is a conserved dynamin GTPase superfamily protein that is translocated from its cytoplasmic pool to the outer mitochondrial membrane, where Drp1 then assembles into constrictive ring-like multimeric structures, ultimately driving mitochondrial fragmentation through a GTP-dependent mechanism (10–12).
Previously, we have shown that high-glucose (HG) environment leads to an excess of mitochondrial fission in podocytes (13, 14). We found that the enhanced mitochondrial fission in hyperglycemic conditions was due to increased Rho-associated coiled coil–containing protein kinase 1 (ROCK1) activity in podocytes. In support of this observation, we provided evidence indicating that in the diabetic milieu, podocyte-specific ROCK1 activation in mice promotes mitochondrial fission via activation of Drp1 and that Drp1 serves as a direct substrate for ROCK1 in the diabetic milieu (13). Since our published study, others have also established the key contributions of mitochondrial fission in a myriad of kidney diseases (15–20). However, despite recent advances in understanding the role of mitochondrial fission in the kidney, the molecular mechanisms linking Drp1 phosphorylation with progression of DN in vivo remain undefined.
Posttranslational modification of Drp1 represents an important regulatory mechanism for controlling mitochondrial fission. Among a variety of posttranslational modifications, Drp1 phosphorylation at serine 600 (S600, mouse isoform b, NCBI NP_001021118.1; S643, mouse, UniprotKB Q8K1M6) located at the juncture of the variable domain (VD) and N-terminal end of the GTPase effector domain (GED) has been reported to play a critical regulatory function (11, 21, 22). While the importance of Drp1S600 phosphorylation as a key posttranslational modification of Drp1 is well established, the functional impact of this modification on Drp1 activity and mitochondrial network morphology remains controversial. For example, protein kinase A (PKA) phosphorylation of Drp1S600 has been reported to decrease Drp1 GTPase activity in vitro (23, 24), whereas phosphorylation of the same conserved serine residue by Ca2+-calmodulin–dependent protein kinase Iα (CaMKIα) in Drp1 isoform 3 has been reported to cause a significant increase in mitochondrial fission (25). We have also previously reported that ROCK1-mediated Drp1S600 phosphorylation resulted in enhanced mitochondrial fission in podocytes (13). Although several explanations may account for these differences, we speculated that the functional consequences of Drp1 phosphorylation might be highly cell type and/or stimulus dependent.
Given these conflicted roles of Drp1S600 phosphorylation in mitochondrial fission, we sought to investigate its role in mitochondrial fission and progression of DN in vivo. To this end, we generated a knockin mouse model bearing a phosphorylation-deficient serine (S) to alanine (A) mutation at the S600 (S600A) site on a diabetic db/db background. We also developed genetically modified mice in which mitochondrial redox changes within podocytes of diabetic Drp1S600A mice were dynamically monitored in real-time by crossing diabetic Drp1S600A mice with transgenic mice that expressed a redox-sensitive GFP biosensor specifically located in the mitochondrial matrix (mt-roGFP) (26, 27). Our findings now suggest that diabetic mice with the Drp1S600A mutation are protected against progression of DN. Notably, the protective effect of Drp1S600A was associated with decreased mitochondrial fission and reduced mitochondrial ROS (mtROS) in podocytes. Importantly, we found that crosstalk between phosphorylated Drp1S600 (p-Drp1S600) and the actin-binding protein complex Arp2/3 is a required step in mitochondrial Drp1 recruitment and mitochondrial fission under HG conditions. These findings strongly support our hypothesis that Drp1S600 phosphorylation is central in promoting mitochondrial fission and enhanced mtROS in hyperglycemic conditions. Furthermore, our data suggest that Drp1S600 phosphorylation could serve as a new therapeutic target, opening up a new avenue for treatment of patients with DN.
Results
Generation and characterization of Drp1S600A-knockin mice.
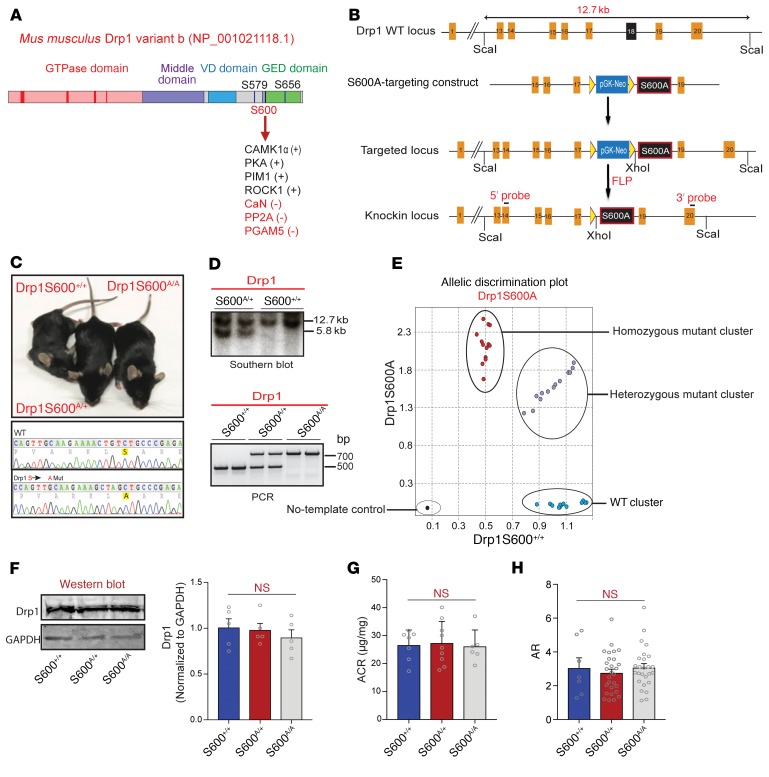
We have previously shown that genetic deletion of Drp1 in diabetic db/db mice, an established model of type 2 diabetes, is associated with improved mitochondrial fitness in podocytes, leading to protection against progression of DN (14). However, the functional consequences of Drp1 phosphorylation for mitochondrial fission in vivo remained unknown. To address this, we generated a Drp1-knockin mutant mouse line on a C57BL/6 background harboring a site-specific serine (S) to alanine (A) mutation at the S600 (S600A) site (Figure 1, A and B). Following electroporation of murine embryonic stem cells and selection and screening of colonies by Southern blot and PCR analysis, we identified several positive clones that we used to generate chimeric Drp1S600A mice. After confirming successful germline transmission of the Drp1S600A mutation in one of the founder lines, we generated heterozygous Drp1S600A/+, homozygous Drp1S600A/A-mutant, and WT Drp1S600+/+ mice (Figure 1, C–E). Heterozygous and homozygous Drp1S600A mutants yielded offspring in a ratio similar to that expected for Mendelian inheritance, suggestive of no significant embryonic lethality. The mutant mice were viable and fertile and showed no gross abnormalities. Analysis of total Drp1 protein levels by Western blotting in podocytes isolated from Drp1S600+/+, Drp1S600A/+, or Drp1S600A/A mice revealed that the Drp1S600A mutation did not lead to changes in Drp1 levels (Figure 1F). Both homozygous and heterozygous Drp1S600A mice showed no significant differences in their mean blood glucose levels, body weight, or the amount of albuminuria (Figure 1G and Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/JCI127277DS1). Notably, we did not observe significant changes in mitochondrial fission in podocytes from mutant mice as assessed by similar aspect ratios (ARs), form factor (FF), length, perimeter, area, and circularity (Figure 1H and Supplemental Figure 1, C–H).
Figure 1. Generation and initial characterization of knockin mice harboring a Drp1S600A mutation.
(A) Diagram of the domain structure of Drp1, illustrating the S600 site at the juncture of the VD and GED. PIM1, serine/threonine protein kinase Pim-1; CaN, calcineurin; PP2A, protein phosphatase 2A; PGAM5, phosphoglycerate mutase family member 5. (B) Structure of the Drp1-targeting locus, the Drp1-targeting construct, and the conditional allele after homologous recombination. The ScaI and XhoI sites used in Southern blot analysis and the location of the 3′ and 5′ probes are indicated. FRT, flippase recognition target (yellow triangles); FLP, flippase. (C) Gross appearance of WT, homozygous, and heterozygous Drp1S600A-knockin mice, and PCR sequence from genomic DNA showing mutation of the allele in the genome. (D) Southern blot analysis of ScaI and XhoI digested genomic DNA from mice of the indicated genotypes, showing the WT (12.7-kb) and mutant (5.8-kb) bands (upper panel). PCR genotyping of Drp1S600A heterozygosity and homozygosity (lower panel). Mutant and WT products are shown. (E) Cartesian allelic discrimination plot shows the relative levels of the Drp1S600A-mutant fluorescence signal for each sample plotted on the y axis and the WT signal on the x axis. Homozygous Drp1S600A (red dots), homozygous WT (blue dots), and heterozygous (violet dots) samples are shown. The no-template control is depicted by the black circle. (F) Western blot of Drp1 protein from mice of the three S600-mutant genotypes and densitometric quantification of Drp1 normalized to GAPDH protein expression. (G) Albumin/creatinine ratio (ACR) analysis of WT, heterozygous, and homozygous Drp1S600A-knockin mice at 20 weeks of age. (H) Mitochondrial AR from podocytes from mice of the three Drp1S600A genotypes as determined from TEM images. P < 0.05, by 1-way ANOVA with Tukey’s multiple comparisons test. Data represent the mean ± standard error of the mean (n = 5–8/group).
Diabetic Drp1S600A mice are protected against progression of DN.
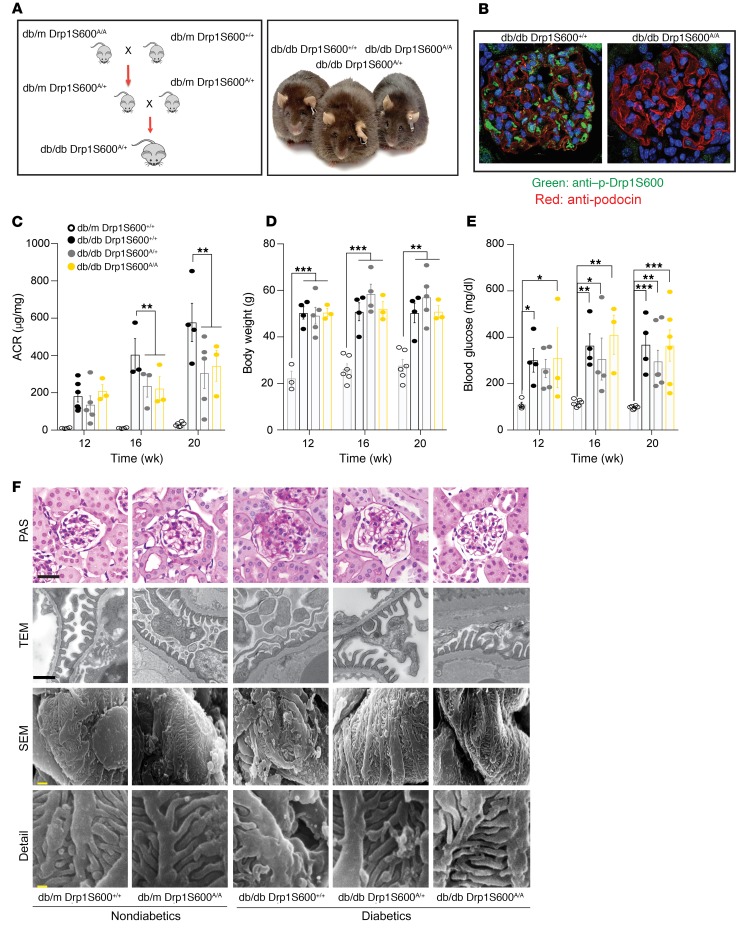
To test the impact of Drp1S600 phosphorylation on the development and progression of DN in vivo, we crossed db/m Drp1S600A/+ mice with db/m Drp1S600A/+ mice to generate diabetic heterozygous Drp1S600A/+ and homozygous Drp1S600A/A mice on a db/db background (refereed to hereafter as db/db Drp1S600A/+ and db/db Drp1S600A/A mice) (Figure 2A). Immunohistochemical staining with an antibody directed against p-Drp1 at the S600 site demonstrated no reactivity in the glomeruli of diabetic Drp1S600A/A mice (Figure 2B and Supplemental Figure 2, A–C). Diabetic Drp1S600-mutant mice were followed over a period of 20 weeks. Although we observed that diabetic Drp1S600A/A-mutant mice did not exhibit any differences in body weight or blood glucose levels when compared with their respective diabetic db/db Drp1S600+/+ controls (hereafter referred to as diabetic control mice), the diabetic Drp1S600A/A mice exhibited significantly reduced albuminuria at 20 weeks of age compared with diabetic controls (345 ± 83 μg/mg vs. 578 ± 103 μg/mg, P < 0.01), despite similar body weights, blood glucose levels, and serum creatinine levels (Figure 2, C–E, and Supplemental Figure 3). Interestingly, heterozygous diabetic Drp1S600A/+ mice exhibited similar reductions in albuminuria compared with diabetic Drp1S600A/A mice, suggesting that the Drp1S600A mutation may exert a dominant effect over WT Drp1 (306 ± 83 μg/mg vs. 345 ± 83 μg/mg, P > 0.05) (Figure 2C). Histological analysis by periodic acid–Schiff (PAS) staining and quantification of glomerular area revealed attenuated mesangial matrix expansion (MME) in the glomeruli of diabetic Drp1S600A/+ and Drp1S600A/A mice compared with diabetic controls (Figure 2F and Supplemental Figure 4A). Similarly, analysis by transmission electron microscopy (TEM) revealed reduced podocyte foot process effacement and improved glomerular basement membrane (GBM) thickening in diabetic, Drp1S600A/+, and Drp1S600A/A mice compared with diabetic controls. Scanning electron microscopy (SEM) revealed preserved podocyte structure in mutant mice (Figure 2F and Supplemental Figure 4B). Analysis of podocyte numbers using the podocyte nuclear marker Wilm’s tumor 1 (WT1) revealed no significant reduction of podocyte numbers in db/db Drp1S600A/+ or db/db Drp1S600A/A mice compared with numbers in diabetic control mice (Supplemental Figure 4C).
Figure 2. Drp1S600A mutation protects against progression of DN.
(A) Breeding scheme to obtain diabetic Drp1S600A-mutant mice and representative images of diabetic mice of each genotype. (B) Immunostaining of glomeruli from diabetic Drp1, WT, and homozygous mutant mice. Glomeruli were stained with an antibody against the p-Drp1 (S637) site (green). Podocytes were identified with an antibody against podocin (red), and nuclei were stained with DAPI (blue). Images were acquired by confocal microscopy (original magnification, ×60). (C) ACR analysis of nondiabetic db/m mice,Drp1 WT mice, and mice of the 3 diabetic genotypes at 12, 16, and 20 weeks demonstrating a significant reduction in albuminuria in 16- and 20-week-old diabetic Drp1S600A heterozygous and homozygous mutant mice compared with diabetic db/db mice. (D) Body weight and (E) blood glucose levels at different time points for the groups described in C. (F) Representative PAS-stained images (scale bar: 50 μm), TEM images (scale bar: 500 nm), SEM images (scale bar: 10 μm), and SEM inset images (scale bar: 1 μm) of kidney glomeruli from mutant allele mice. Images are from a sampling of 3 to 5 animals. *P < 0.05, **P < 0.01, and ***P < 0.001, by 1-way ANOVA followed by Tukey’s multiple comparisons test. Results are presented as the mean ± standard error of the mean (n = 3–8/group).
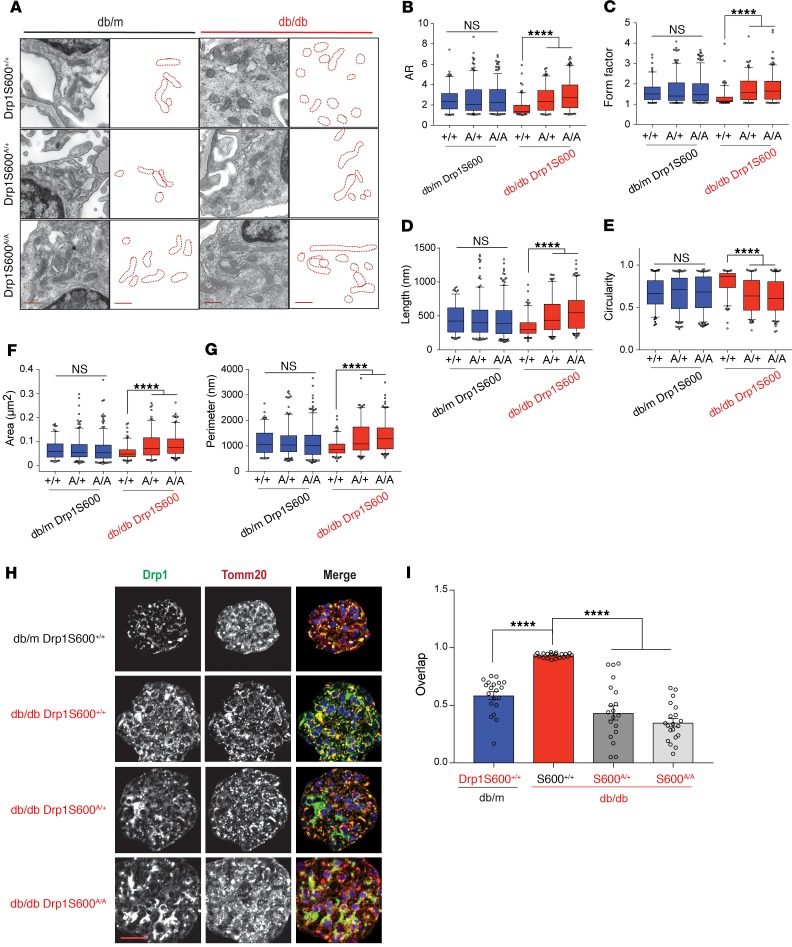
Given the protective effect of Drp1S600A on the progression of DN, we inferred that enhanced mitochondrial fission in the diabetic milieu might have been mitigated in diabetic Drp1S600A mice. Consistent with this expectation, when mitochondria were visualized with TEM, we observed that podocytes from diabetic Drp1S600A/+ and DrpS600A/A mice had more elongated mitochondria with increased AR and FF ratios compared with mitochondria in podocytes obtained from diabetic controls (Figure 3, A–G, and Supplemental Figure 5). Importantly, a careful examination of images from immunostaining experiments showed that mitochondria from glomeruli of Drp1S600A/A and Drp1S600A/+ mice had significantly lower levels of mitochondrial Drp1 recruitment compared with mitochondria from diabetic control mice (Figure 3, H and I). When considered together, these results suggest that Drp1S600 phosphorylation is an important step in the initiation of mitochondrial fission and progression of DN in vivo. However, how increased Drp1S600 phosphorylation promotes mitochondrial fission and the progression of diabetic kidney disease remains largely unknown.
Figure 3. Mitochondrial fission is dependent on Drp1S600 phosphorylation in the diabetic milieu in vivo.
(A) Representative TEM images of mitochondria in podocytes. The 3 possible genotypes at the Drp1S600-knockin alleles — WT, heterozygous, and homozygous — are shown. Scale bars: 500 nm. To the right of each micrograph are tracings of mitochondria from the TEM micrographs. Changes in mitochondrial morphology were quantitated from TEM micrographs as the (B) mitochondria AR, (C) mitochondria form factor, (D) mitochondrial length, (E) mitochondrial circularity, (F) mitochondrial area, and (G) mitochondrial perimeter. (H) Immunofluorescence staining of paraffin-embedded kidney sections. Sections were stained for Drp1 (grayscale or green in merge) and Tomm20 (grayscale, or red in merge), with the merged images shown on the far right. Representative images were cropped to show the glomerular area only. Scale bar: 50 μm. (I) Colocalization of total Drp1 and mitochondria determined from the images represented in H using Pearson’s correlation coefficient. Representative images are from a sampling of 3 to 5 animals. ****P < 0.0001, by 1-way ANOVA followed by Tukey’s multiple comparisons test. Results are presented as the mean ± standard error of the mean (n = 5–8/group).
Assessment of mitochondrial redox status in the kidneys of live Drp1S600-knockin mice.
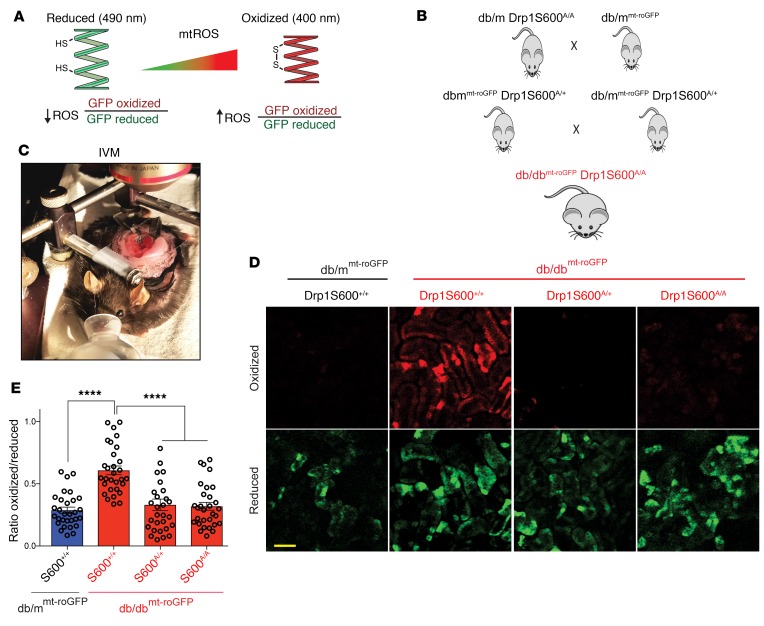
HG-induced mitochondrial fission is associated with increased mtROS (13, 26, 28, 29). So far, our data suggested that preventing Drp1 phosphorylation at the S600 site leads to improvements in several key biochemical and histological features of DN associated with a significant improvement in diabetes-induced mitochondrial fission. However, the role of Drp1S600A in mtROS and potential contribution to the progression of DN in vivo are not known. To determine the impact of Drp1S600A mutation on mtROS, diabetic Drp1S600A/A mice were crossed with mitochondrial matrix–targeted redox-sensitive GFP–transgenic (roGFP-transgenic) mice (26). Mitochondrial roGFP mice express a redox-sensitive mitochondrial matrix–localized GFP that contains a modified fluorophore (27, 30, 31) that, when exposed to changes in the mitochondrial matrix redox pool, undergoes reversible disulfide linkages. The redox changes are detected by probing the biosensor at 2 wavelengths and monitoring emission at a third wavelength in live mice (Figure 4A). We have previously shown that mt-roGFP has the dynamic range to detect changes in the mitochondrial redox state in the kidneys without affecting mitochondrial morphology (26). Thus, to determine the effect of Drp1S600A on mtROS, transgenic mt-roGFP mice were crossed with Drp1S600A-mutant mice to generate diabetic db/dbmt-roGFP Drp1S600A/A mice (Figure 4B). Next, db/dbmt-roGFP Drp1S600A/A mice were subjected to real-time intravital microscopy (IVM) in vivo to monitor the mitochondria-specific redox status of roGFP in the kidneys of live mutant mice as previously described (26) (Figure 4C). Ratiometric analysis of fluorescence emission intensity at oxidized (405 excitation) and reduced (488 excitation) wavelengths confirmed increased levels of mtROS in the kidneys of diabetic db/dbmt-roGFP Drp1S600+/+ compared with nondiabetic controls (Figure 4, D and E). Importantly, we found reduced levels of mtROS in the kidneys of db/dbmt-roGFP DrpS600A/A compared with levels in diabetic db/dbmt-roGFP Drp1S600+/+ kidneys (Figure 4, D and E, and Supplemental Figure 6). Taken together, this powerful new model of live monitoring of mitochondrial redox status in the kidneys of diabetic mice strongly suggests that reduced mtROS are associated with the Drp1S600A mutation and that reduced mtROS might serve as an underlying mechanism by which Drp1S600S mitigates the progression of DN.
Figure 4. Mitochondrial redox–sensitive roGFP indicates that the Drp1S600A attenuates mtROS in the kidneys of live diabetic mice.
(A) Cartoon depicting the principles of ratiometric redox-sensitive mt-roGFP. CMV-driven expression was localized to the mitochondrial matrix using a cytochrome oxidase subunit IV–signaling (COX IV–signaling) sequence. The oxidation state of the engineered thiols determines the fluorescence properties of the sensor, which shifts depending on the oxidized, disulfide-bonded glutathione/reduced glutathione (GSSG/GSH) equilibrium. Cysteine sulfhydryl groups are illustrated, HS, free sulfhydryl; SS, disulfide bonded. (B) Generation of db/dbroGFP Drp1S600A/A mice allows for monitoring of mitochondrial redox status in the kidneys of live Drp1S600-knockin mice. (C) Image of IVM setup. (D) Ratiometric changes in the fluorescence intensity obtained at the oxidized excitation wavelength (red) and the reduced excitation wavelength (green). Scale bar: 100 μm. (E) Ratiometric quantitative analysis from intravital images, in which oxidized fluorescence is placed as the numerator and reduced fluorescence as the denominator. Representative images are from a sampling of 3 animals. ****P < 0.0001, by 1-way ANOVA with Tukey’s multiple comparisons test. Results are presented as the mean ± standard error of the mean (n = 3 mice/group).
Tissue-independent but stimulus-specific effects of Drp1S600 phosphorylation.
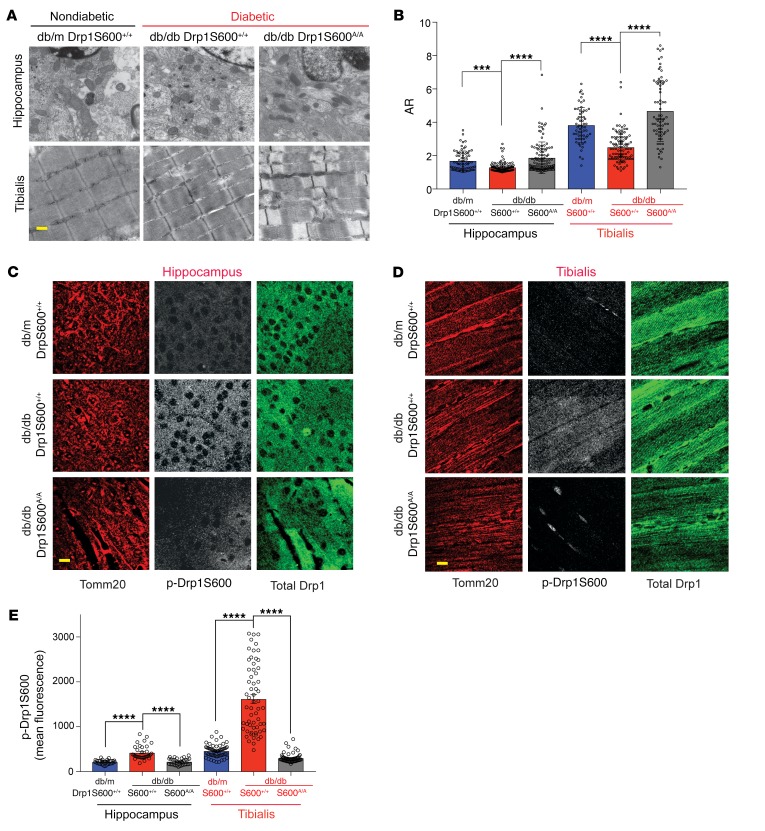
Our previously published work suggested that the consequences of Drp1S600 phosphorylation in mitochondrial fission could be highly cell and/or stimulus dependent (13). This conclusion was based on our initial findings, in which Drp1S600 phosphorylation in podocytes led to enhanced mitochondrial fission under HG conditions, whereas phosphorylation of the same site in other cell lines has been reported to reduce mitochondrial fission (24, 28, 32, 33). Thus, to determine whether the consequences of Drp1S600 phosphorylation in our diabetic model are tissue specific as opposed to stimulus dependent, we evaluated mitochondrial morphology in several tissues, including in hippocampal tissue and tibialis anterior (TA) muscle obtained from our diabetic Drp1S600+/+ and diabetic Drp1S600A/A mice. We found that in contrast to the traditional view regarding the effect of Drp1S600 phosphorylation on mitochondrial fission, especially in neurons (34), mitochondria within hippocampal neurons as well as in TA muscle cells exhibited enhanced mitochondrial fission in diabetic db/db mice compared with nondiabetic db/m mice (Figure 5, A and B). Importantly, we found improved mitochondrial features in hippocampal neurons and skeletal muscle cells in diabetic db/db Drp1S600A/A mice that were consistent with a tissue-independent effect of the Drp1S600A mutation. Histological staining of the tissues revealed increased Drp1S600 phosphorylation, consistent with our previous observations in the kidney (Figure 5, C–E). These findings suggest that the effect of Drp1S600 phosphorylation on mitochondrial fission is stimulus dependent but not tissue specific.
Figure 5. Tissue-independent effect of Drp1S600A on mitochondrial fission in diabetic mice.
Hippocampal neurons and TA muscle cells from mice of different groups were fixed for histology and TEM analysis. (A) Representative TEM images. Scale bar: 500 nm. (B) Quantification of mitochondrial morphology from TEM images. (C) Immunofluorescence staining of paraffin-embedded hippocampal tissue. Left panels show mitochondria stained for Tomm20 (red). Middle panels show gray-scale images of p-Drp1S600 staining. All gray-scale images were captured with identical confocal settings, and images were manipulated simultaneously and equally for each tissue. Right panels show total Drp1 staining (green). Scale bar: 25 μm. (D) Immunofluorescence staining of TA muscle cells as performed in C. Scale bar: 25 μm. (E) Quantification of p-Drp1S600 staining intensity from sections of hippocampus and TA muscle immunostained with anti–p-Drp1S600. Representative images are from a sampling of 3 animals. ***P < 0.001 and ****P < 0.0001, by 1-way ANOVA with Tukey’s multiple comparisons test. Results are presented as the mean ± standard error of the mean (n = 3–5/group).
Interactome analyses identify a role for cytoskeletal remodeling in mitochondrial fission.
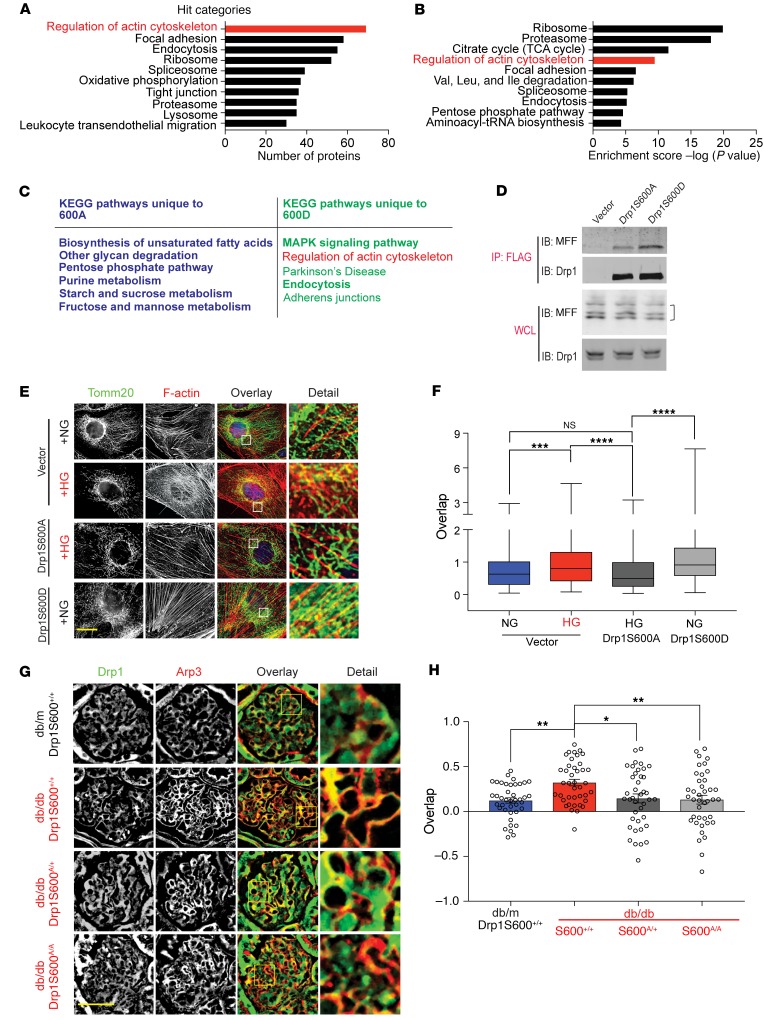
Given our initial results, we explored the underlying molecular mechanisms by which p-Drp1S600 promotes enhanced mitochondrial fission in the diabetic environment. We investigated the possibility that interacting protein partners of p-Drp1S600 in the diabetic milieu might mediate Drp1-induced mitochondrial fission. To characterize highly abundant interactomes, cultured podocytes stably expressing amino terminal FLAG-tagged Drp1S600 (WT) or Drp1S600A were subjected to HG conditions (25 mM, for 48 h), while the phosphomimetic Drp1S600D was maintained in normal glucose (NG) (5 mM) conditions. Anti-FLAG immunoprecipitated fractions of cell lysates were submitted for MS analysis. Comparison of 3 independent experiments led us to identify 1187 proteins that were interacting with both Drp1S600A and S600D mutants. Notably, there were 302 proteins that only interacted with the Drp1S600A mutant and 227 proteins that were exclusively interacting with Drp1S600D. The Gene Ontology (GO) database in conjunction with gene set enrichment analysis (GSEA) revealed an enriched selection of proteins involved in the regulation of the actin cytoskeleton pathway from the Kyoto Encyclopedia of Genes and Genomes (KEGG) (Figure 6, A–C, and Supplemental Figure 7, A–D). Among the proteins enriched using the KEGG database, cofilin 1 and the Arp2/3 complex were highlighted in silico (Supplemental Figure 7, A–D). The STRING protein interaction database also suggested an interaction of Drp1 with Arp3 (Supplemental Figure 7C). Interestingly, a recent body of evidence supports the notion that crosstalk between Drp1 and actin filaments may play an important role in mitochondrial Drp1 recruitment, leading to enhanced mitochondrial fission (35–38). Thus, on the basis of our results and the recent literature, we tested the hypothesis that DrpS600 phosphorylation might lead to enhanced mitochondrial fission in the diabetic milieu through concordance of two p-Drp1S600 interactions, one through facilitating the interaction between Drp1 and mitochondrial fission factor (MFF), a Drp1 receptor, and the other by promoting the interaction between Drp1 and F-actin (39, 40). Since recent studies suggest that MFF is involved in mitochondrial fission and serves as the primary Drp1 receptor (12, 22), we first assessed the functional contribution of the binding of putative Drp1S600 with MFF. We performed IP of cultured podocytes stably expressing FLAG-tagged Drp1S600 mutants and found strong enrichment of recovered MFF with Drp1S600D compared with Drp1S600A (Figure 6D), suggesting that binding with MFF is important for p-Drp1S600 recruitment to mitochondria.
Figure 6. Actin cytoskeleton interacts with p-Drp1S600 and mitochondria.
Cultured podocytes stably expressing FLAG-tagged versions of Drp1 (WT) and Drp1S600A were treated with HG (25 mM) for 48 hours. FLAG-tagged Drp1S600D was cultured under NG conditions. Cells were cross-linked, immunoprecipitated, and subjected to MS. KEGG Pathway analysis of protein interaction partners was performed. Top interaction pathways for Drp1S600 according to (A) the number of proteins and (B) enrichment score. (C) KEGG pathway analysis of the mutant proteins indicating the top pathways identified for each Drp1 mutant. (D) Podocytes stably expressing FLAG-tagged Drp1S600A or S600D in conjunction with untagged MFF or empty vector were immunoprecipitated with anti-FLAG agarose gel. The top 2 panels show recovery of MFF and Drp1 by immunoblotting (IB) following IP. Immunoblots in the bottom panels are for the WCL starting material. Different isoforms of MFF are indicated by a bracket on the right. (E) Podocytes were immunostained with Tomm20 (green) for mitochondria and rhodamine-phalloidin (red) for actin. From left to right, gray-scale image of mitochondria staining (green in merge), gray-scale images of actin staining (red in merge), and merged image. Scale bar: 25 μm. (F) Quantification of the overlap based on Mander’s coefficient for each condition. (G) Immunofluorescence staining of paraffin-embedded kidney sections. Sections were stained for total Drp1 (grayscale, green in merge) and Arp3 (grayscale, red in merge). Scale bar: 50 μm. (H) Colocalization analysis using Pearson’s correlation analysis of total Drp1 and mitochondria determined from the images represented in F. Representative images are from a sampling of 3 to5 separate cell cultures or animals. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001, by 1-way ANOVA with Tukey’s multiple comparisons test. Results are presented as the mean ± standard error of the mean (n = 5–8/group).
Next, we examined whether key mediators of actin remodeling were also required for Drp1S600-mediated mitochondrial fission. We first assessed whether mitochondrial de novo polymerization of F-actin is required for Drp1S600-mediated mitochondrial fission by incubating cultured podocytes with latrunculin A, an inhibitor of actin polymerization. The addition of latrunculin A mitigated both Drp1S600D as well as HG-mediated mitochondrial fission (Supplemental Figure 8, A and B). We concluded that F-actin polymerization is required for p-Drp1S600–mediated mitochondrial fission. Concurrent analysis of interactions between mitochondria and F-actin in Drp1S600-mutant cells suggested that, although there was no significant colocalization of rhodamine-phalloidin–labeled F-actin and Tomm20-labeled mitochondria in Drp1S600A-transfected podocytes treated with HG, we observed significant colocalization of F-actin and mitochondria in HG-treated cultured podocytes as well as in cultured podocytes transfected with Drp1S600D (Figure 6, E and F). These results suggest that p-Drp1S600 facilitates mitochondrial assembly of F-actin under HG conditions.
We next determined whether cofilin 1 and/or Arp2/3 complex, the main regulatory proteins related to actin remodeling and selected on the basis of our mass spectrometry (MS) experiments, serve as the key elements for Drp1S600-mediated mitochondrial fission. We initially examined this correlation in the glomeruli of diabetic Drp1S600 WT and mutant mice in vivo. We found that, whereas cofilin 1 did not overlap with Drp1 in the glomeruli of diabetic db/db mice, Arp3 showed a significant overlap with Drp1 (Figure 6, G and H, and Supplemental Figure 9) in the kidneys of diabetic mice. Importantly, this colocalization was markedly reduced in the glomeruli of both diabetic Drp1S600A/+ and Drp1S600A/A mice (Figure 6, G and H, Supplemental Figure 5, A and B, and Supplemental Figure 9). These data strongly suggest the possibility that Drp1S600 could play an important role in interactions between the Arp2/3 complex and mitochondria in vivo.
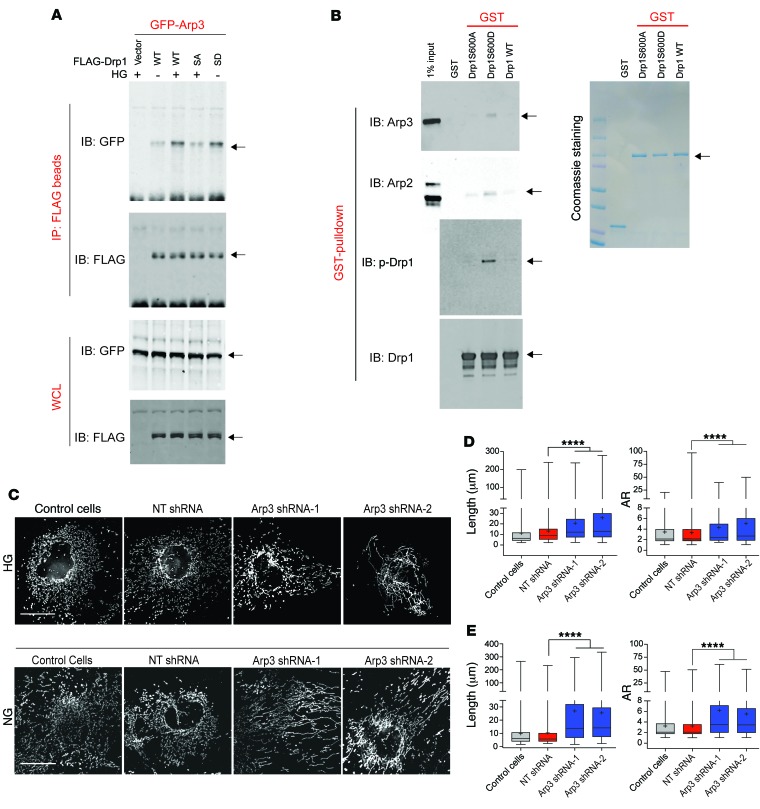
Since we noticed that the relative protein levels of Arp2 and Arp3 were unchanged under HG conditions (Supplemental Figure 10A), we next examined whether the interaction of Drp1 with Arp2/3 was dependent on phosphorylation of Drp1S600. To this end, we transfected GFP-tagged Arp3 into cultured podocytes stably expressing empty vector control, FLAG-tagged WT Drp1S600, or Drp1S600 mutants (S600A or S600D). Western blot detection of the immunoprecipitated FLAG-tagged proteins indicted that the interaction of Drp1 with the GFP-Arp3 complex was markedly increased in cells that were cultured in HG conditions and in Drp1S600D-transfected podocytes (Figure 7A). However, this interaction was markedly reduced in FLAG-Drp1S600A cells under HG conditions, suggesting that, indeed, the binding of Drp1 to the Arp3 complex was enhanced by the phosphorylation of Drp1S600.
Figure 7. Drp1 binds to the Arp2/3 complex in a p-Drp1S600–dependent manner.
(A) Cultured podocytes with empty vector, FLAG-tagged WT Drp1 (WT), FLAG-tagged Drp1S600A (SA), and FLAG-tagged Drp1S600D (SD) were used. Cells were also transiently transfected with GFP-Arp3. Top panels show anti-FLAG IP material and immunoblotting against GFP and FLAG. Bottom panels show the WCLs. (B) Bacterially expressed GST, GST-Drp1S600A, GST-S600D, and GST-S600 WT proteins on GST-sepharose were mixed with purified Arp2/3 complex in the GST-pulldown assay. Coomassie staining of SDS-PAGE gel is shown on the right. Top 2 left blots show recovered materials that were immunoblotted to detect the binding of Arp2 and Arp3 to Drp1. Third blot on the left shows immunoblotting with p-Drp1S600 (p-Drp1), illustrating good mimicry of the phosphorylation epitope by the aspartate mutation. The bottom blot on the left shows immunoblotting for the total level of input Drp1 from the GST-pulldown assay. (C) Top panels show control podocyte cells cultured under HG conditions after being treated with vehicle, nontargeting (NT) shRNA, shRNA-1 against Arp3, or shRNA-2 against Arp3. Cells were fixed and stained for mitochondria with an antibody against Tomm20. Mitochondria are shown in grayscale. Bottom panels show podocytes expressing Drp1S600D cultured under NG conditions after being treated as indicated above and stained for mitochondria as before. Mitochondria are shown in grayscale. Scale bars: 25 μm. (D) Quantification of mitochondrial length and AR for native podocytes for the images shown in C (top). (E) Quantification of mitochondrial length and AR for podocytes stably expressing Drp1S600D for the images shown in C (bottom). Representative images are from a sampling of 3 to 5 separate cell cultures. ****P < 0.0001, by 1-way ANOVA with Tukey’s multiple comparisons test. Results are presented as the mean ± standard error of the mean (n = 5–8/group).
To validate the direct interaction of Drp1 with the Arp2/3 complex, we used glutathione-pulldown assays with purified Arp2/3 complex. Bacterially expressed and purified WT GST-Drp1S600 or GST-Drp1S600 mutants (S600A or S600D) were immobilized on glutathione S-transferase–sepharose (GST-sepharose) beads and incubated with purified Arp2/3 complex in vitro followed by immunoblotting with anti-Arp3 and anti-Arp2 antibodies, respectively. We found enhanced recovery of both Arp2 and Arp3 in GST-Drp1S600D beads but not in GST-Drp1S600A beads (Figure 7B). Together, these data indicate that phosphorylation of Drp1S600 triggers a direct interaction of Drp1 with the Arp2/3 complex.
Finally, to determine whether Arp3 is required for Drp1S600-mediated mitochondrial fission, we designed viral infection vectors containing shRNAs targeted against Arp3 (Supplemental Figure 10, B and C). We found that downregulation of Arp3 by 2 separate shRNAs led to a marked reduction in both HG culture and S600D-mediated mitochondrial fission (Figure 7C). Analysis of the mitochondrial morphology indicated that downregulation of Arp3 restored the tubular morphology of mitochondria, as evidenced by an increase in the AR of mitochondria in podocytes (Figure 7, D and E). Similar experiments were conducted with cofilin 1 (Supplemental Figure 11, A–D), however, downregulation of cofilin 1 did not lead to a significant improvement in mitochondrial fission under these conditions (Supplemental Figure 11D). We believe that these observations suggest that Arp3, but not cofilin 1, is critically involved in Drp1S600-mediated mitochondrial fission under HG conditions.
Discussion
The current understanding of the role of Drp1S600 phosphorylation in mitochondrial fission in vivo is limited. Even less is known about the role of Drp1S600 in the progression of DN. The findings in this study provide important insights into the pathobiology of mitochondrial dynamics in the diabetic milieu and the progression of diabetic kidney disease. First, we have defined, for the first time to our knowledge, the in vivo consequences of Drp1S600 phosphorylation for mitochondrial fission and progression of DN. Second, our work highlights a stimulus-dependent effect of Drp1S600 phosphorylation on mitochondrial fission in the diabetic milieu. Third, we show a critical role for mtROS as a key downstream effector of Drp1S600 phosphorylation. Finally, our study reveals that F-actin and cytoskeletal remodeling play a central role in Drp1S600-mediated mitochondrial fission and Drp1 accumulation in the mitochondria.
Posttranslational modifications of Drp1 have emerged as an important regulatory mechanism responsible for Drp1 activation and mitochondrial fission (7, 25, 28, 41, 42). However, despite the central regulatory role of Drp1 phosphorylation in its activation, identification of the specific role of Drp1S600 in Drp1 activation and mitochondrial fission in vivo has been controversial. We and others have previously shown that Drp1S600 phosphorylation by ROCK1 or Ca2+/calmodulin-dependent protein kinase Iα leads to Drp1 activation and its translocation to the mitochondria (13, 14, 25). Paradoxically, other reports have indicated that phosphorylation of Drp1 at the same conserved serine residue, for instance by PKA, leads to reduced Drp1 GTPase activity and decreased recruitment of Drp1 to mitochondria in other cell types (23, 24). Thus, given the central role of Drp1S600 phosphorylation in regulating Drp1 and mitochondrial function, it was essential to address the functional consequences of Drp1S600 phosphorylation for Drp1 activation and mitochondrial fission in vivo. To this end, we used a gene-targeting approach in mice and generated a diabetic knockin mutant Drp1S600A mouse as our experimental model. Although we did not observe significant changes in mitochondrial dynamics in podocytes from nondiabetic Drp1S600A mice, we showed that mitochondrial Drp1 and mitochondrial fission are significantly decreased in the podocytes of diabetic Drp1S600A mice. Consistent with these conclusions, our findings also indicate a link between Drp1S600A and protection from DN progression. Specifically, we report that mice lacking the Drp1S600 phosphorylation site exhibit protection against worsening albuminuria, mesangial matrix expansion, GBM thickness, and podocyte effacement — all key biochemical and histological features of DN.
One unanticipated discovery from this study was that a stimulus-dependent regulatory effect of Drp1S600 phosphorylation on activation and accumulation of Drp1 in the mitochondria ultimately modulates mitochondrial fission. Examination of diabetic hippocampal and TA skeletal muscle cells showed mitochondria that appeared to be protected from the diabetes-induced excessive fragmentation in the Drp1S600A mouse. These findings, however, do not completely exclude the possibility that Drp1S600 phosphorylation might also trigger tissue-dependent effects. It is plausible that specific cellular subtypes with diverse mitochondrial requirements and morphology could exhibit distinct phenotypes in response to Drp1S600 phosphorylation with a variety of stimuli.
Another interesting aspect of Drp1S600 phosphorylation is that Drp1S600A leads to improvement in mtROS. The free radical theory of microvascular complications of diabetes also known as the “unifying hypothesis” proposes that the underlying molecular mechanism for the detrimental effects of diabetes is the increased production of mtROS (43). However, the causes of increased mtROS production in DN are complicated and remain unclear (26, 44, 45). The traditional assumption has been that increased mtROS production in DN is mainly due to an increased flow of glucose as the substrate to oxidative phosphorylation machinery in the mitochondria, but it is still unclear whether and how hyperglycemia is mechanistically linked to mtROS production and progression of DN in vivo. In this work, we uncovered a critical role for Drp1S600 in mediating the hyperglycemia-induced increase in mtROS production. We have previously demonstrated that phosphorylation of Drp1S600 in podocytes increased mtROS and apoptosis in vitro and identified the detrimental effects of mtROS in vivo on the progression of DN (13). In the current study, our results highlight 2 important findings: first, we show that there is enhanced mtROS generation in the diabetic milieu; and second, we provide strong evidence of a direct link between phosphorylation of Drp1 at S600 and mtROS production in vivo.
The underlying molecular mechanisms that regulate Drp1 recruitment to fission sites are not well understood. It is known, however, that downregulation of some of the Drp1 receptors, including MFF, mitigates mitochondrial Drp1 recruitment (46). Several recent studies have also clearly shown that actin polymerization near fission sites stimulates oligomeric maturation of Drp1 on mitochondria (37, 38, 47, 48). Our data suggest that Drp1S600 phosphorylation regulates the interaction between Drp1 and actin on the mitochondrial membrane. We found that podocytes transfected with phosphomimetic Drp1S600D had increased mitochondrial accumulation of F-actin, similar to the effect seen with HG, whereas podocytes transfected with Drp1S600A displayed an opposite effect on F-actin. Furthermore, we provide evidence for a modulatory effect of Drp1S600 on the actin regulatory Arp2/3 complex. Our data indicate that depletion of the Arp2/3 complex blocked HG-induced mitochondrial fission. Furthermore, Drp1S600D-mediated mitochondrial fission was similarly blocked with Arp3 shRNAs, providing evidence for the importance of Drp1-actin interactions leading to mitochondrial fission. Our results are in agreement with the recently proposed model of mitochondrial fission by the Higgs group (49). However, our findings highlight the critical roles of S600 phosphorylation of Drp1 and Arp3 on mitochondrial fission in the diabetic milieu. Since the Arp2/3 complex mediates nucleation and branching of the actin cytoskeleton, one interpretation of our data is that these branch points might serve as key organizing centers for the assembly of mitochondrial dynamic factors involved in diabetes-induced mitochondrial fission in the kidney.
Finally, our results suggest that the effect of p-Drp1S600 on mitochondrial fission is not an all-or-none phenomenon and that a critical amount of p-Drp1S600 attached to mitochondria is required to direct mitochondrial fission. This critical threshold may not be reached in the heterozygous mutant state. We believe our findings have important implications for a wide variety of human pathologies that are associated with mitochondrial dysfunction and pave the way for future studies to identify and target other posttranslational modifications of Drp1, including other phosphorylation sites, to assess for their physiological and mechanistic significance.
Methods
Animal work.
Diabetic db/db, mice and their control littermates db/m were obtained from The Jackson Laboratory (strain: BKS.Cg-Dock7m+/+Leprdb/J) and bred in-house. All animals were maintained on a normal chow diet with free access to water and housed in a room with a 12-hour light/12-hour dark cycle and an ambient temperature of 22°C. Type 1 diabetes was induced in mice as previously described (13)
Drp1S600A-knockin mice.
We designed a Drp1S600A-knockin construct using a replacement targeting strategy that involved 3 cloning steps. First, a 3.5-kb genomic DNA encompassing exons 15–17 and the proximal intronic regions of the Drp1 gene was PCR amplified and subcloned directly into a pLNTK vector. This DNA fragment was sequencing verified and used as the 5′ homologous recombination arm. Second, a neomycin-resistant minigene cassette flanked by flippase recognition target (FRT) sequences was added to the 3′ end of the 5′ arm in the pLNTK backbone. Third, a 5.1-kb genomic fragment encompassing exons 18–19 and the proximal intronic regions was PCR amplified and cloned into a pGEM-T-Easy vector and then sequence verified. A T-to-G transversion in exon 18 (resulting in a corresponding S600A amino acid mutation) was introduced by site-directed mutagenesis (Takara Bio USA). The resulting DNA fragment was used as the 3′ arm and was cloned into the 3′ end of the Neo cassette to complete the knockin construct. The targeting construct was restriction digested, purified, and electroporated into C57BL/6N-Tac ES cells at the BCM Mouse Embryonic Stem Cell Core Facility (https://bcm.corefacilities.org/service_center/show_external/3152/mouse-embryonic-stem-cell-core). Successful gene targeting was determined by Southern blot analysis. Briefly, genomic DNA was digested with the restriction enzymes ScaI and XhoI (New England BioLabs). The WT allele generated a 12.7-kb ScaI DNA fragment, whereas the targeting construct introduced an Xhol I site. The introduced restriction site produced a 5.8-kb fragment after restriction enzyme digestion. A 3′ probe was generated by PCR for Southern blot analysis. Six targeted clones were then further verified by PCR and sequencing for the Drp1S600A mutation. Two of these clones were injected into blastocysts to achieve germline transmission. We generated 16 chimera pups from which targeted founders were established through Southern blot analysis using tail DNA.
Tissue culture.
Conditionally immortalized mouse podocytes were a gift from Jochen Reiser (Rush University, Chicago, Illinois, USA). Briefly, cells were cultured at 33°C in RPMI (Corning, 10-041-CV) containing 10% FBS (FBS Opti-gold, GenDepot, F0900-050) and 20 U/ml mouse recombinant IFN-γ (MilliporeSigma, I4777) to enhance expression of a thermosensitive T antigen. The cells were differentiated at 37oC in DMEM (Corning, 10-014-CV) supplemented with 5% FBS without IFN-γ on 10-cm dishes coated with collagen type I (Gibco, Thermo Fisher Scientific, A10483-01) for 7 to 12 days. For imaging, the differentiated cells were trypsinized, dissociated, and plated onto collagen type I–coated coverslips. Podocytes prepared for experiments involving HG (25 mM) conditions were serum deprived for 24 hours prior to addition of HG. Control cells were cultured with 5 mM NG.
Southern blot analysis and PCR genotyping.
Approximately 10 μg genomic DNA was subjected to restriction enzyme digestion by XhoI and ScaI (New England BioLabs). Digested DNA was separated by gel electrophoresis (0.9% agarose in 1× TAE buffer, 35 V, 8 h). Gels were depurinated, denatured, and neutralized before transfer onto nitrocellulose membranes. Membranes were UV cross-linked and prehybridized in hybridization buffer (PerfectHyb Plus, MilliporeSigma, H7033). PCR was performed to generate a biotinylated 3′ probe. The Southern probe was hybridized and then the membrane washed and subsequently incubated with HRP-labeled streptavidin (Vector Laboratories, SA-5004). Bands were revealed using chemiluminescence detection kits (Thermo Fisher Scientific). Mouse tail DNA was isolated using the REDExtract-N-Amp Tissue PCR Kit (MilliporeSigma Aldrich, XNAT) according to the manufacturer’s instructions. The primers used for PCR genotyping were Drp1 MF and Drp1 MR (CCATCTGCAGGTGGTGGGATTGGAG and GACATCACAAACTCATCCAACA GT). PCR was performed as described by the REDExtract-N-Amp protocol, and the products were resolved on a 2% agarose gel using the TAE buffer system.
Cartesian allelic discrimination plot.
TaqMan genotyping was performed with preoptimized PCR primer pairs (SNP forward/reverse, CCCTCTTTTAGCCAGTTCCAGTTG/CAATAACCTCACAATCTCGCTGTTC) and 2 probes for allelic discrimination (FAM/VIC, AAAACTGGCCGCCCGA/CAAGAAAACTGTCTGCCCGA; Thermo Fisher Scientific, AHZAGZD). Each assay contained a pair of unlabeled primers and 3 TaqMan probes (1 with a FAM dye label and 1 with a VIC dye label). The relative levels of fluorescence from the labeled probes detecting either the mutant or WT sequence for each sample were plotted on the x and y axes, respectively.
Metabolic and physiological parameters.
Mice were individually housed in metabolic cages and fasted overnight but were provided water ad libitum. Blood glucose levels were determined the next morning with a glucose meter (Walgreens True Metrix). Urine was collected and urinary albumin concentration measured with a mouse albumin ELISA kit (Exocell, 1011). Urinary creatinine was measured using the QuantiChrom Creatinine Assay Kit (Bioassay Systems, DICT500).
Western blot analysis.
Total protein lysates were extracted using RIPA buffer (50 mM Tris-HCl, pH 8.0, with 150 mM NaCl, 1.0% igepal CA-630 [NP-40], 0.5% sodium deoxycholate, and 0.1% SDS). Protein concentration was determined by BCA Protein Assay (Pierce, Thermo Fisher Scientific, 23228). Total protein lysate (20 μg) was diluted in 5× Laemmli buffer, loaded on a 4%–20% gradient SDS-PAGE (Bio-Rad,) and transferred onto PVDF membranes (Roche, 03010-040-001). Membranes were probed with the indicated primary antibody and a fluorescent secondary antibody followed by visualization with a Odyssey Infrared Imaging System (LI-COR Biosciences). The primary antibodies used for Western blotting were: anti-Drp1 (BD, 611113; RRID: AB_398424); anti–FLAG tag (MilliporeSigma, F7425; RRID: AB_439687); anti-MFF1 (Cell Signaling Technology, 86668; RRID: AB_2734126); and anti-GFP (MilliporeSigma, AB3080; RRID: AB_2630379). The secondary antibodies used for Western blotting were: goat anti–mouse Dylight 680 (Thermo Fisher Scientific, 35519; RRID: AB_1965956); goat anti–mouse Dylight 800 (Thermo Fisher Scientific, SA510172; RRID: AB_2556752); goat anti–rabbit Dylight 680 (Thermo Fisher Scientific, 35519; RRID: AB_1965956); and goat anti–rabbit Dylight 800 (Thermo Fisher Scientific, SA535571; RRID: AB_2556775).
Cell and tissue staining.
For cell culture, podocytes were washed with cold PBS, fixed in 4% formaldehyde, and permeabilized with 0.1% Triton X-100 (Acros, 21568-0010). Cells were blocked in 1% BSA (Jackson ImmunoResearch, 001-000-162), 50 mM Tris, pH 7.6, 155 mM sodium chloride (TBS), and 0.1% Triton X-100. The cells were incubated overnight at 4°C with the appropriate primary antibodies in blocking buffer. Coverslips were washed 3 times in TBS and incubated with the appropriate secondary antibodies in blocking buffer for 1 to 2 hours at room temperature (RT). When indicated, the secondary antibody solution had 1:100 rhodamine/phalloidin (Life Technologies, Thermo Fisher Scientific, R415; RRID: AB_2572408) added at the same time to label actin. Coverslips were washed 3 times in TBS and mounted onto slides. Images were obtained using a DeltaVision Elite deconvolution inverted microscope (GE Healthcare, see below). Quantification was performed using ImageJ software (NIH). For tissue sections, formalin-fixed, paraffin-embedded tissue sections were cut at approximately 5-μm thickness and mounted onto slides. The slides were deparaffinized and rehydrated through an ethanol series with a final wash in distilled water. Antigen retrieval was performed by incubating the slides in 20 mM Tris and 1 mM EDTA at 95°C for 1 hour. Sections were then washed twice with 1× TBS, and nonspecific binding was blocked in blocking buffer (see above) for 1 hour at an ambient temperature. After blocking, the sections were incubated overnight at 4°C with 1:100 dilutions of primary antibodies in blocking buffer. The sections were washed 3 times in TBS and incubated with secondary antibodies for 1 to 2 hours at RT in blocking buffer. The sections were washed 3 times with TBS, and nuclei were counterstained with DAPI (Thermo Fisher Scientific, 62248). Finished slides were mounted with ProLong Gold Antifade Mounting Reagent (Molecular Probes, P36934). The antibodies used for immunohistochemistry were: anti-Drp1 (BD, CA, 611113; RRID: AB_398424); anti–p–Drp1 (S637) (Biorbyt, Orb127984; RRID: AB_2734124); anti-Tomm20 (Genetex, GTX32928; RRID: AB_2734125); anti–cofilin 1 (Abcam, ab131519; RRID: AB_11155720); anti-Arp3 (C-term) (ECM Biosciences, AP4581; RRID: AB_2734127); anti-SDHA (Abcam, ab14715; RRID:AB_301433); donkey anti–rabbit Alexa Fluor 488 (Thermo Fisher Scientific, A21206; RRID: AB_2535792); donkey anti–rabbit Alexa Fluor 594 (Thermo Fisher Scientific, A21207; RRID: AB_141637); donkey anti–rabbit Alexa Fluor 647 (Thermo Fisher Scientific, A21244; RRID: AB_141663); donkey anti–mouse Alexa Fluor 488 (Invitrogen, Thermo Fisher Scientific, A21202; RRID: AB_141607); and donkey anti–mouse Alexa Fluor 594 (Thermo Fisher Scientific, A21203; RRID: AB_141633). The sections were examined using a Nikon A1Si inverted confocal microscope. The images were quantified using ImageJ software, version 2.00. Postanalyses of colocalization were performed using ImageJ coloc2.
Mesangial expansion and WT1 staining.
Formalin-fixed, paraffin-embedded tissue sections were rehydrated as described above. The sections were subjected to PAS staining to evaluate the general histologic changes in glomerular and tubular structures. Mesangial fraction/volumes from PAS-stained images were estimated using ImageJ. The glomerular tuft was outlined in the images, simultaneously providing the cross-sectional area of the tuft. ImageJ thresholding was used to highlight the regions of similar intensity of red staining within the region of interest (glomerulus). The fractional area of the glomerulus with positive staining was then calculated. WT1 quantification was achieved using fluorescence immunohistochemistry. Tissue sections were subjected to WT1 staining (Abcam, ab89901, RRID: AB_1965956) and DAPI counterstaining. The acquired images were analyzed using ImageJ software. The images were viewed and the glomerular region of interest outlined to obtain a cross-sectional area. The number of WT1-positive nuclei and the total number of nuclei within the region of interest were determined. The results are presented as either WT1-positive nuclei per mm2 or the percentage of total nuclei observed. Five mice were analyzed per group, and fifty glomeruli were measured per mouse.
Confocal and deconvolution microscopy.
Confocal image collection was performed on a Nikon A1Si inverted confocal microscope equipped with 4 solid-state lasers under acousto-optic tunable filter (AOTF) control. The objective lenses used were either a Nikon CFI60 Plan Fluor ×40 oil (NA 1.3) or a Nikon CFI Apo TIRF ×60 oil. Multicolor channel fluorescence images were captured sequentially using 405-, 488-, 561-, and 640-nm excitation wavelengths and the following emission filters (range limits): 425–475 nm, 500–550 nm, 570–620 nm, and 663–738 nm, respectively. Wide-field and deconvolution images were captured using a DeltaVision Elite deconvolution inverted microscope (GE Healthcare), equipped with a PCO sCMOS camera. The lenses used were a U Plan FLN ×40 (1.3 NA oil) and a Plan Apo N ×60 (1.42 NA oil). Fluorescence images were captured using quadruple polychroic mirrors (DAPI/FITC/TRITC/Cy5 or DAPI/FITC/Alexa 594/Cy5) and the corresponding sets of excitation and emission filter sets mounted in filter wheels. The specifications for each filter set were: DAPI 390/18 nm and 435/48 nm; FITC 475/28 nm and 525/48 nm; TRITC 542/27 nm and 597/45 nm; mCherry 575/25 nm and 625/45 nm; and Cy5 632/22 nm and 679/34 nm (excitation and emission respectively, expressed as peak/bandwidth).
IP for MS.
Cultured podocytes were cross-linked (XL) by adding 37.5% formaldehyde to culture medium with a final concentration of 0.5%, followed by incubation at 37°C for 8 minutes. The cross-linking was quenched by adding glycine to a final concentration of 0.2 M. The cells were collected and washed twice with ice-cold PBS. The cells were lysed in 3 packed cell volumes of NETN buffer (25 mM Tris/pH7.5, 170 mM NaCl, 1 mM EDTA, 0.5% NP-40, 1× phosphatase inhibitor, and 1× protease inhibitor cocktail) by sonication (SONICS Ultrasonic Processors VCX; 30 s on, 1 min rest, 25% amp, 6 cycles). After centrifugation for 20 minutes at 100,000 g, the supernatant was collected as whole-cell lysate (WCL) for IP experiments. For each IP experiment, 12 mg WCL was incubated for 30 minutes at 4°C with 30 μl Protein A–Dynabeads slurry (MilliporeSigma, catalog GE17-0780-01) for preclearance. The resulting supernatant was removed and cleared by ultracentrifugation (44,000 g for 15 min). The supernatants were then incubated with 30 μl M2 antibody–conjugated beads (MilliporeSigma, catalog F1804; RRID: AB_262044) for 1 hour at 4°C. The bead-bound complexes were washed 3 times with NETN buffer (100 mM NaCl, 20 mM Tris, pH 8.0, 0.5 mM EDTA, 0.5% NP-40). The washed beads were boiled with 1× NuPAGE LDS sample buffer with 50 mM 2-mercaptoenthanol and subjected to 4% to 20% Tris-glycine SDS-PAGE (Novex Gel, Invitrogen, Thermo Fisher Scientific). The gels were stained with Coomassie brilliant blue, and visualized proteins bands were excised into 4 gel pieces according to molecular size.
IP validations.
Stable podocytes were generated by subcloning FLAG-tagged Drp1S600A or Drp1S600D into the TRE-responsive vector. The podocytes were then transiently transfected with native MFF1, CFL1-EGFP (gift from James Bamburg, Addgene plasmid no. 50859 [ref. 50]), or Arp3-EGFP (gift from Matthew Welch, Addgene plasmid no. 8462 [ref. 51]) mammalian expression vectors. Drp1 and MFF expression was induced with doxycycline (200 nM) for 72 hours. To study the interaction of Drp1 with MFF1 and CFL1, each group of cells (vector control, S600A, and S600D) were seeded onto 10 collagen I–coated tissue culture dishes (100-mm) at 1 × 105 cells/dish in NC DMEM (5% FBS, 5 mM glucose-DMEM) and differentiated for 7 days at 37°C. Cells were then serum-starved overnight before treatment with HG DMEM (5% FBS, 25 mM glucose) for 72 hours. Cells were washed, scraped into PBS, and cross-linked with 0.5% formaldehyde and PBS for 10 minutes at RT. Glycine was added to 0.2 M to quench the cross-link. Cells were washed 3 times with PBS and lysed by sonication in a 3× packed cell volume of lysis buffer NETN (50 mM Tris, pH 7.3, 170 mM NaCl, 1 mM EDTA, 0.5% NP-40) supplemented with protease inhibitor and phosphatase inhibitor cocktails. Lysates were precleared with Protein A/G PLUS Beads (Santa Cruz Biotechnology) at 4°C for 30 minutes before IP with FLAG-M2 beads (anti-FLAG M2 Affinity Gel, MilliporeSigma, catalog A2220; RRID: AB_10063035) or anti-MFF1 antibody (Cell Signaling Technology, catalog 86668; RRID: AB_2734126) and protein A/G PLUS beads (Santa Cruz Biotechnology). Beads were washed with lysis buffer 5 times and eluted with 0.1 mg/ml 3× FLAG peptide (for FLAG-M2 beads, MilliporeSigma F4799). Samples had cross-links undone by subjection to 65°C for 1 hour in Laemmli buffer (for protein A/G beads). Proteins were subjected to 4% to 20% SDS-PAGE (Bio-Rad) and immunoblotted with antibodies against FLAG (anti–FLAG tag, MilliporeSigma, catalog F7425; RRID: AB_439687); GFP (anti-GFP, MilliporeSigma, catalog AB3080; RRID: AB_2630379); MFF1; and cofilin 1 (anti–cofilin 1, Cytoskeleton, catalog ACFL02-A; RRID: AB_10708808). To study the interaction of Drp1 with Arp3, cells were lysed in M-PER buffer (Thermo Fisher Scientific, catalog 78501), and IP and Western blotting were carried out as above.
GST-pulldown assay.
A GST-pulldown assay was carried out as previously described (52). Briefly, GST-tagged Drp1 isoforms (WT, S600A, and S600D) or GST vector control (pGEX-2T, GE Healthcare) were transformed into BL21 (DE3) (New England BioLabs) and induced with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) (MilliporeSigma, catalog I6758) at 25°C overnight (18 h). Immobilized GST proteins were purified according to the manufacturer’s protocol. GST proteins (2 μg) were incubated with 5 μg purified Arp2/3 complex (Cytoskeleton Inc., catalog RP01P-A) in 1 ml GST-binding buffer (20 mM Tris-HCl at pH 7.5, 25 mM KCl, 1 mM MgCl2, 50 mM NaCl) at 4°C for 2 hours. Beads were washed 5 times with binding buffer, boiled in SDS sample buffer, and subjected to 4% to 15% PAGE (Bio-Rad, 4561086). Proteins were transferred to a solid support and blotted with Arp3 mAb (1:200, MilliporeSigma; RRID: AB_476749) and Arp2 polyclonal Ab (1:100, ECM Biosciences, RRID: AB_1944438), respectively.
Vectors and recombinant DNA.
Genomic integration of tetracycline-inducible transgenic genes was accomplished using the piggybac transposon system. Cells were transfected with the nonintegratable PiggyBac transposase pCMV-PB to mediate translocation of the transposable vectors into the genomic DNA without extended expression of the transposase. Integration involved 2 vectors in each case. The transposase was transfected along with a transposable Tet-on and transposable transgenic vectors. The transposable vectors had independent selection antibiotics. Vector backbone has been described previously (26). This vector had the TRE-tight promoter. For selection purposes, the vectors contained either puromycin or G418 behind the SV40 promoter. Cells were then selected with G418 and puromycin.
Mitochondrial measurements.
Images were acquired by deconvolution microscopy. ImageJ was used to analyze the resulting images. AR measurements were performed by assigning each mitochondria into an ellipse. The mitochondrial AR was determined as the major and minor axes of the ellipse expressed as a fraction. Length was the major axis. The form factor was defined as (Pm2)/(4pAm), where Pm is the length of the mitochondrial outline, and Am is the area of the mitochondrion. Mitochondrial area was measured as the 2D area (nm2) projected to the focal plane of the image. Circularity was 4π(Am/Pm2). These measurements were made using ImageJ and previously published methods. The morphology of at least 120 mitochondria was determined for each condition.
SEM.
Fixed samples containing 3% glutaraldehyde plus 2% paraformaldehyde in 0.1 M cacodylate buffer, pH 7.3, were washed with 0.1 M cacodylate buffer, pH 7.3, post-fixed with 1% cacodylate-buffered osmium tetroxide, and washed with 0.1 M cacodylate buffer and then in distilled water. Afterwards, the samples were sequentially treated with Millipore-filtered 1% aqueous tannic acid, washed in distilled water, treated with Millipore-filtered 1% aqueous uranyl acetate, and then rinsed thoroughly with distilled water. The samples were dehydrated with a graded series of increasing concentrations of ethanol and then transferred to a graded series of increasing concentrations of hexamethyldisilazane (HMDS) and air-dried overnight. The samples were mounted onto double-stick carbon tabs (Ted Pella) that had been previously mounted onto glass microscope slides. Next, the samples were coated under vacuum using a Balzer MED 010 evaporator (Technotrade International) with platinum alloy for a thickness of 25 nm and then immediately flash carbon-coated under vacuum. The samples were transferred to a desiccator for examination at a later date. The samples were examined and imaged using a JEOL JSM-5910 scanning electron microscope at an accelerating voltage of 5 kV.
TEM.
The samples were fixed with a solution containing 3% glutaraldehyde plus 2% paraformaldehyde in 0.1 M cacodylate buffer, pH 7.3, and then washed in 0.1 M sodium cacodylate buffer and treated with 0.1% Millipore-filtered cacodylate-buffered tannic acid, post-fixed with 1% buffered osmium, and stained en bloc with 1% Millipore-filtered uranyl acetate. The samples were dehydrated in increasing concentrations of ethanol, infiltrated, and embedded in LX-112 medium. The samples were polymerized in a 60°C oven for approximately 3 days. Ultrathin sections were cut in a Leica Ultracut microtome (Leica), stained with uranyl acetate and lead citrate in a Leica EM Stainer, and examined in a JEOL JEM 1010 transmission electron microscope at an accelerating voltage of 80 kV. Digital images were obtained using the AMT Imaging System (Advanced Microscopy Techniques).
MS.
The gel pieces were subject to in-gel digestion using trypsin (GenDepot, T9600). The tryptic peptide was dried under vacuum and then resuspended in 10 μl loading solution (5% methanol containing 0.1% formic acid). One-half of the suspended peptide was injected using the Nano-HPLC 1000 system (Thermo Fisher Scientific) coupled to the Q Exactive Plus mass spectrometer (Thermo Fisher Scientific). The peptides (2 μm) were loaded onto an in-house Reprosil-Pur Basic C18 trap column (Dr. Maisch GmbH), which was 2 cm × 100 μm in size. Then, the trap column was washed with loading solution and switched in-line with an in-house 5-cm × 150-μm column packed with 2 μm Reprosil-Pur Basic C18 beads equilibrated in 0.1% formic acid in water. The peptides were separated with a 75-minute discontinuous gradient of 2% to 24 % acetonitrile and 0.1% formic acid at a flow rate of 800 nl/min. Separated peptides were directly electrosprayed into the mass spectrometer. The instrument was operated in data-dependent mode, with acquisition of fragmentation spectra of the top 35 strongest ions and under the direct control of Xcalibur software (Thermo Fisher Scientific). The parent MS spectrum was acquired in the Orbitrap with the full MS range of 375 to 1300 m/z at a resolution of 140,000. The CID fragmented tandem MS (MS/MS) spectrum was acquired in the ion trap in rapid-scan mode. The MS/MS spectra obtained were searched against the target-decoy Human RefSeq database (released June 2015, containing 73,637 entries; https://www.ncbi.nlm.nih.gov/refseq/) in the Proteome Discoverer 2.1 interface (Thermo Fisher Scientific) with the Mascot algorithm (Mascot 2.4, Matrix Science). Variable modification of oxidation of methionine and protein N-terminal acetylation was allowed. The precursor mass tolerance was confined within 20 ppm, allowing a fragment mass tolerance of 0.02 Dalton and a maximum of 2 missed cleavages. The assigned peptides were filtered with a 1% FDR. The iBAQ algorithm was used to calculate protein abundance to compare relative amounts between different proteins in the sample using an in-house data-processing algorithm (53). Simply, iBAQ was calculated on the basis of normalization of the summed peptide intensity divided by the number of the theoretically observable tryptic peptide of certain proteins.
Latrunculin A and shRNA experiments.
Podocytes were differentiated on collagen-I–coated coverslips. Cells were treated with 50 nM latrunculin A for the duration of the 48-hour HG (25 mM) treatment in 5% FBS and DMEM in normal cells or for 48 hours in 5% FBS and DMEM with NG (5 mM) and 50 nM doxycycline in the case of Drp1S600D-expressing cells. The cells were transduced with lentiviral shRNAs 1 day prior to the start of the glucose treatments. GIPZ mouse CFL1 shRNA (Dharmacon, V3LMM_521443 and V3LMM_449309), GIPZ mouse ACTR3 shRNA (Dharmacon, V3LMM_34878 and V3LMM_444971), and GIPZ nonsilencing control (Dharmacon, RHS4346) were used to prepare lentiviruses at the Baylor College of Medicine Gene Vector Core.
In silico analyses.
Functional protein interaction analysis was performed using STRING (https://string-db.org/). For KEGG pathway analysis, the KEGG Automatic Annotation Server–KEGG (KAAS-KEGG) (http://www.genome.jp/tools /kaas/) was used, and for gene enrichment analysis, GSEA (http://software.broadinstitute.org/gsea/index.jsp) was used.
IVM for live animal imaging.
We monitored the presence of mitochondrial ROS in the kidney in real time in vivo by IVM imaging using diabetic mice that expressed a redox-sensitive GFP biosensor. IVM imaging of the kidney was performed by the Intravital Microscopy Core at the Houston Methodist Research Institute. Kidney imaging was performed by exposing the kidney with a lateral incision. Image acquisition was performed over selected fields of view, with a resolution of 512 × 512 pixels and an optical slice thickness of 7.1 μm. The IVM system uses an upright Nikon A1R laser scanning confocal microscope with a resonance scanner, motorized and heated stage, and Nikon long-working distance 4× and 20× dry plan apochromat objectives. Images were obtained with a 2-channel setup in which fluorescence was collected at 525 nm, exciting sequentially with a 405-nm and 488-nm laser at a rate of 5 fps. Images were analyzed and exported using Nikon Elements. To measure the mitochondrial ROS, the intensity ratio of the areas covered by the biosensor was quantified with Nikon Elements.
Human archival kidney biopsy.
Human kidney biopsy samples were obtained from archival biopsy specimens collected at the Houston Methodist Hospital. Histological sections of formalin-fixed, paraffin-embedded biopsy samples were subjected to immunofluorescence staining with antibodies against synaptopodin (GeneTex, GTX39067, RRID: AB_11161967); p-Drp1 (S637) (Biorbyt, orb127984); Alexa Fluor 647 goat anti-rabbit (Invitrogen, Thermo Fisher Scientific, A21244); and Alexa Fluor 488 donkey anti-mouse (Invitrogen, Thermo Fisher Scientific, A21202).
Statistics.
All group data are expressed as the mean ± standard error of the mean. Comparisons of multiple groups were performed using 1-way ANOVA followed by Tukey’s multiple comparisons test. Comparisons between 2 groups were performed using a 2-tailed Student’s t test, with a P value of less than 0.05 considered statistically significant. Statistical analyses were performed using GraphPad Prism 6.0b (GraphPad Software).
Study approval.
All animal studies were approved and conducted according to the Principles of Laboratory Animal Care (NIH publication no. 85023, revised 1985) and the guidelines of the IACUC at the University of Texas MD Anderson Cancer Center. Animal IVM studies were performed in accordance with the guidelines of the Animal Welfare Act and the Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011) and based on approved protocols by the IACUCs of Baylor College of Medicine and Houston Methodist Research Institute. Human kidney biopsy samples were obtained from archival biopsy specimens collected at the Houston Methodist Hospital under a protocol approved by the hospital’s IRB. Written informed consent was obtained from all patients prior to their inclusion in the study.
Author contributions
DLG and JL performed experiments and analyzed data. NG, BHC, PO, JSL, and PS analyzed data and provided resources and critical review of the manuscript. DLG, NG, BHC, and FRD wrote the manuscript, with critical review from PO and PS. LDT provided human biopsy specimens. FRD conceived and supervised the study and provided funding.
Supplementary Material
Acknowledgments
This work was supported by funding from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), NIH (RO1DK078900 and R01DK091310, to FRD). We thank members of the Electron Microscopy Facility (UTMDACC, Houston, Texas, CCSG grant NIH P30CA016672); the Center for Advanced Imaging (Texas A&M University, Houston, Texas); the Mass Spectrometry Proteomics and Gene Vector Core Facilities at Baylor College of Medicine, Houston, Texas; the National Cancer Institute [NCI], NIH (P30CA125123); and the Houston Methodist Research Institute Intravital Core (HMRIIC). We also thank Enrica De Rosa (HMRIIC) for her assistance with intravital imaging.
Version 1. 05/07/2019
In-Press Preview
Version 2. 06/17/2019
Electronic publication
Version 3. 07/01/2019
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(7):2807–2823.https://doi.org/10.1172/JCI127277.
Contributor Information
Daniel L. Galvan, Email: dlgalvan@mdanderson.org.
Jianyin Long, Email: jlong1@mdanderson.org.
Nathanael Green, Email: nathanag@bcm.edu.
Benny H. Chang, Email: bchang@bcm.edu.
Jamie S. Lin, Email: JLin8@mdanderson.org.
Luan D. Truong, Email: ltruong@houstonmethodist.org.
Paul Overbeek, Email: overbeek@bcm.edu.
Farhad R. Danesh, Email: danesh@bcm.edu.
References
- 1. United States Renal Data System. 2018 USRDS annual data report: Epidemiology of kidney disease in the United States. Bethesda, Maryland, USA: NIH, National Institute of Diabetes and Digestive and Kidney Diseases; 2018. [Google Scholar]
- 2.Badal SS, Danesh FR. New insights into molecular mechanisms of diabetic kidney disease. Am J Kidney Dis. 2014;63(2 Suppl 2):S63–S83. doi: 10.1053/j.ajkd.2013.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhargava P, Schnellmann RG. Mitochondrial energetics in the kidney. Nat Rev Nephrol. 2017;13(10):629–646. doi: 10.1038/nrneph.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galvan DL, Green NH, Danesh FR. The hallmarks of mitochondrial dysfunction in chronic kidney disease. Kidney Int. 2017;92(5):1051–1057. doi: 10.1016/j.kint.2017.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forbes JM, Thorburn DR. Mitochondrial dysfunction in diabetic kidney disease. Nat Rev Nephrol. 2018;14(5):291–312. doi: 10.1038/nrneph.2018.9. [DOI] [PubMed] [Google Scholar]
- 6.Eisner V, Picard M, Hajnóczky G. Mitochondrial dynamics in adaptive and maladaptive cellular stress responses. Nat Cell Biol. 2018;20(7):755–765. doi: 10.1038/s41556-018-0133-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmitt K, et al. Circadian control of DRP1 activity regulates mitochondrial dynamics and bioenergetics. Cell Metab. 2018;27(3):657–666.e5. doi: 10.1016/j.cmet.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 8.Ramírez S, et al. Mitochondrial dynamics mediated by mitofusin 1 is required for POMC neuron glucose-sensing and insulin release control. Cell Metab. 2017;25(6):1390–1399.e6. doi: 10.1016/j.cmet.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 9.Giacomello M, Scorrano L. The INs and OUTs of mitofusins. J Cell Biol. 2018;217(2):439–440. doi: 10.1083/jcb.201801042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kraus F, Ryan MT. The constriction and scission machineries involved in mitochondrial fission. J Cell Sci. 2017;130(18):2953–2960. doi: 10.1242/jcs.199562. [DOI] [PubMed] [Google Scholar]
- 11.Kalia R, et al. Structural basis of mitochondrial receptor binding and constriction by DRP1. Nature. 2018;558(7710):401–405. doi: 10.1038/s41586-018-0211-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen H, Chan DC. Mitochondrial dynamics in regulating the unique phenotypes of cancer and stemcells. Cell Metab. 2017;26(1):39–48. doi: 10.1016/j.cmet.2017.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W, et al. Mitochondrial fission triggered by hyperglycemia is mediated by ROCK1 activation in podocytes and endothelial cells. Cell Metab. 2012;15(2):186–200. doi: 10.1016/j.cmet.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ayanga BA, et al. Dynamin-related protein 1 deficiency improves mitochondrial fitness and protects against progression of diabetic nephropathy. J Am Soc Nephrol. 2016;27(9):2733–2747. doi: 10.1681/ASN.2015101096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng J, et al. Hyperglycemia, p53, and mitochondrial pathway of apoptosis are involved in the susceptibility of diabetic models to ischemic acute kidney injury. Kidney Int. 2015;87(1):137–150. doi: 10.1038/ki.2014.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao X, Hu Y, Quirós PM, Wei Q, López-Otín C, Dong Z. OMA1 mediates OPA1 proteolysis and mitochondrial fragmentation in experimental models of ischemic kidney injury. Am J Physiol Renal Physiol. 2014;306(11):F1318–F1326. doi: 10.1152/ajprenal.00036.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brooks C, Cho SG, Wang CY, Yang T, Dong Z. Fragmented mitochondria are sensitized to Bax insertion and activation during apoptosis. Am J Physiol, Cell Physiol. 2011;300(3):C447–C455. doi: 10.1152/ajpcell.00402.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He L, et al. AKI on CKD: heightened injury, suppressed repair, and the underlying mechanisms. Kidney Int. 2017;92(5):1071–1083. doi: 10.1016/j.kint.2017.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eirin A, Lerman A, Lerman LO. The emerging role of mitochondrial targeting in kidney disease. Handb Exp Pharmacol. 2017;240:229–250. doi: 10.1007/164_2016_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sumida M, et al. Regulation of mitochondrial dynamics by Dynamin-related protein-1 in acute cardiorenal syndrome. J Am Soc Nephrol. 2015;26(10):2378–2387. doi: 10.1681/ASN.2014080750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Francy CA, Alvarez FJ, Zhou L, Ramachandran R, Mears JA. The mechanoenzymatic core of dynamin-related protein 1 comprises the minimal machinery required for membrane constriction. J Biol Chem. 2015;290(18):11692–11703. doi: 10.1074/jbc.M114.610881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clinton RW, Francy CA, Ramachandran R, Qi X, Mears JA. Dynamin-related protein 1 oligomerization in solution impairs functional interactions with membrane-anchored mitochondrial fission factor. J Biol Chem. 2016;291(1):478–492. doi: 10.1074/jbc.M115.680025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang CR, Blackstone C. Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J Biol Chem. 2007;282(30):21583–21587. doi: 10.1074/jbc.C700083200. [DOI] [PubMed] [Google Scholar]
- 24.Cribbs JT, Strack S. Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep. 2007;8(10):939–944. doi: 10.1038/sj.embor.7401062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han XJ, et al. CaM kinase I alpha-induced phosphorylation of Drp1 regulates mitochondrial morphology. J Cell Biol. 2008;182(3):573–585. doi: 10.1083/jcb.200802164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galvan DL, et al. Real-time in vivo mitochondrial redox assessment confirms enhanced mitochondrial reactive oxygen species in diabetic nephropathy. Kidney Int. 2017;92(5):1282–1287. doi: 10.1016/j.kint.2017.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guzman JN, et al. Corrigendum: Oxidant stress evoked by pacemaking in dopaminergic neurons is attenuated by DJ-1. Nature. 2015;521(7552):380. doi: 10.1038/nature14487. [DOI] [PubMed] [Google Scholar]
- 28.Willems PH, Rossignol R, Dieteren CE, Murphy MP, Koopman WJ. Redox homeostasis and mitochondrial dynamics. Cell Metab. 2015;22(2):207–218. doi: 10.1016/j.cmet.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 29.Yu T, Robotham JL, Yoon Y. Increased production of reactive oxygen species in hyperglycemic conditions requires dynamic change of mitochondrial morphology. Proc Natl Acad Sci U S A. 2006;103(8):2653–2658. doi: 10.1073/pnas.0511154103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Breckwoldt MO, et al. Multiparametric optical analysis of mitochondrial redox signals during neuronal physiology and pathology in vivo. Nat Med. 2014;20(5):555–560. doi: 10.1038/nm.3520. [DOI] [PubMed] [Google Scholar]
- 31.Dooley CT, Dore TM, Hanson GT, Jackson WC, Remington SJ, Tsien RY. Imaging dynamic redox changes in mammalian cells with green fluorescent protein indicators. J Biol Chem. 2004;279(21):22284–22293. doi: 10.1074/jbc.M312847200. [DOI] [PubMed] [Google Scholar]
- 32.Cereghetti GM, et al. Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc Natl Acad Sci U S A. 2008;105(41):15803–15808. doi: 10.1073/pnas.0808249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flippo KH, et al. AKAP1 Protects from cerebral ischemic stroke by inhibiting Drp1-dependent mitochondrial fission. J Neurosci. 2018;38(38):8233–8242. doi: 10.1523/JNEUROSCI.0649-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dickey AS, Strack S. PKA/AKAP1 and PP2A/Bβ2 regulate neuronal morphogenesis via Drp1 phosphorylation and mitochondrial bioenergetics. J Neurosci. 2011;31(44):15716–15726. doi: 10.1523/JNEUROSCI.3159-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calì T, Szabadkai G. Organelles: the emerging signalling chart of mitochondrial dynamics. Curr Biol. 2018;28(2):R73–R75. doi: 10.1016/j.cub.2017.11.040. [DOI] [PubMed] [Google Scholar]
- 36.Hatch AL, Ji WK, Merrill RA, Strack S, Higgs HN. Actin filaments as dynamic reservoirs for Drp1 recruitment. Mol Biol Cell. 2016;27(20):3109–3121. doi: 10.1091/mbc.e16-03-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li S, et al. Transient assembly of F-actin on the outer mitochondrial membrane contributes to mitochondrial fission. J Cell Biol. 2015;208(1):109–123. doi: 10.1083/jcb.201404050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ji WK, Hatch AL, Merrill RA, Strack S, Higgs HN. Actin filaments target the oligomeric maturation of the dynamin GTPase Drp1 to mitochondrial fission sites. Elife. 2015;4:e11553. doi: 10.7554/eLife.11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science. 2011;334(6054):358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friedman JR, Nunnari J. Mitochondrial form and function. Nature. 2014;505(7483):335–343. doi: 10.1038/nature12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strack S, Wilson TJ, Cribbs JT. Cyclin-dependent kinases regulate splice-specific targeting of dynamin-related protein 1 to microtubules. J Cell Biol. 2013;201(7):1037–1051. doi: 10.1083/jcb.201210045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson TJ, Slupe AM, Strack S. Cell signaling and mitochondrial dynamics: Implications for neuronal function and neurodegenerative disease. Neurobiol Dis. 2013;51:13–26. doi: 10.1016/j.nbd.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brownlee M. Biochemistry and molecular cell biology of diabetic complications. Nature. 2001;414(6865):813–820. doi: 10.1038/414813a. [DOI] [PubMed] [Google Scholar]
- 44.Coughlan MT, Sharma K. Challenging the dogma of mitochondrial reactive oxygen species overproduction in diabetic kidney disease. Kidney Int. 2016;90(2):272–279. doi: 10.1016/j.kint.2016.02.043. [DOI] [PubMed] [Google Scholar]
- 45.Flemming NB, Gallo LA, Forbes JM. Mitochondrial Dysfunction and Signaling in Diabetic Kidney Disease: Oxidative Stress and Beyond. Semin Nephrol. 2018;38(2):101–110. doi: 10.1016/j.semnephrol.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 46.Losón OC, Song Z, Chen H, Chan DC. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol Biol Cell. 2013;24(5):659–667. doi: 10.1091/mbc.e12-10-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DuBoff B, Götz J, Feany MB. Tau promotes neurodegeneration via DRP1 mislocalization in vivo. Neuron. 2012;75(4):618–632. doi: 10.1016/j.neuron.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manor U, et al. A mitochondria-anchored isoform of the actin-nucleating spire protein regulates mitochondrial division. Elife. 2015;4:e08828. doi: 10.7554/eLife.08828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hatch AL, Gurel PS, Higgs HN. Novel roles for actin in mitochondrial fission. J Cell Sci. 2014;127(Pt 21):4549–4560. doi: 10.1242/jcs.153791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garvalov BK, et al. Cdc42 regulates cofilin during the establishment of neuronal polarity. J Neurosci. 2007;27(48):13117–13129. doi: 10.1523/JNEUROSCI.3322-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Welch MD, DePace AH, Verma S, Iwamatsu A, Mitchison TJ. The human Arp2/3 complex is composed of evolutionarily conserved subunits and is localized to cellular regions of dynamic actin filament assembly. J Cell Biol. 1997;138(2):375–384. doi: 10.1083/jcb.138.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Serrels B, et al. Focal adhesion kinase controls actin assembly via a FERM-mediated interaction with the Arp2/3 complex. Nat Cell Biol. 2007;9(9):1046–1056. doi: 10.1038/ncb1626. [DOI] [PubMed] [Google Scholar]
- 53.Jung SY, et al. An anatomically resolved mouse brain proteome reveals parkinson disease-relevant pathways. Mol Cell Proteomics. 2017;16(4):581–593. doi: 10.1074/mcp.M116.061440. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.