Abstract
Bronchopulmonary dysplasia (BPD) remains a major respiratory illness in extremely premature infants. The biological mechanisms leading to BPD are not fully understood, although an arrest in lung development has been implicated. The current study aimed to investigate the occurrence of autophagy in the developing mouse lung and its regulatory role in airway branching and terminal sacculi formation. We found 2 windows of epithelial autophagy activation in the developing mouse lung, both resulting from AMPK activation. Inhibition of AMPK-mediated autophagy led to reduced lung branching in vitro. Conditional deletion of beclin 1 (Becn1) in mouse lung epithelial cells (Becn1Epi-KO), either at early (E10.5) or late (E16.5) gestation, resulted in lethal respiratory distress at birth or shortly after. E10.5 Becn1Epi-KO lungs displayed reduced airway branching and sacculi formation accompanied by impaired vascularization, excessive epithelial cell death, reduced mesenchymal thinning of the interstitial walls, and delayed epithelial maturation. E16.5 Becn1Epi-KO lungs had reduced terminal air sac formation and vascularization and delayed distal epithelial differentiation, a pathology similar to that seen in infants with BPD. Taken together, our findings demonstrate that intrinsic autophagy is an important regulator of lung development and morphogenesis and may contribute to the BPD phenotype when impaired.
Keywords: Development, Pulmonology
Keywords: Autophagy, Organogenesis, Pulmonary surfactants
Introduction
Bronchopulmonary dysplasia (BPD) continues to be a common and important sequela of extremely preterm infants (1). The exact cause of BPD remains unknown, although arrest of lung development has been implicated (2, 3). Lung development is a well-coordinated process of various cellular events, including proliferation, death, and differentiation (2, 4). It is orchestrated by multiple factors that regulate intracellular signaling networks (2, 5, 6). In mammals, the lung anlage arises by budding from the ventral foregut (7). The lower respiratory tract then develops by progressive branching followed by the division of terminal bronchioles and the formation of terminal saccules (primitive alveoli) and establishment of capillaries until birth (3, 8). Lung development continues after birth through alveolar formation and maturation of the microvasculature (9). The result is an organ with a tremendously large surface area capable of efficient gas exchange across a very thin membrane.
Given the high rates of cell division and differentiation during lung development, energy requirements and metabolic rates tend to be high (10). Thus, the ability of cells to sense and respond to changes in nutrient availability is a fundamental requirement for the survival and maintenance of metabolic homeostasis in the developing lung. Functioning as a major prosurvival mechanism, autophagy is one of the key cellular responses to nutrient deprivation (11). Autophagy can also be rapidly upregulated in response to hypoxia and/or increased cellular energy demands associated with rapid cell proliferation (12, 13). Moreover, autophagy provides energy for the clearance of apoptotic bodies during development (14–16). The requirement for autophagy in maintaining cellular nutrient homeostasis was revealed in Atg5- and Atg7-deficient mice. These autophagy-defective mice died within a 24-hour period after birth as a result of systemic amino acid deficiency and decreased glucose levels (13, 17). Autophagy has been shown to participate in the proper development of many organs (18, 19). In a recent study using Ulk1/2- and Atg5-deficient mouse strains, the lungs of newborn pups were examined and showed reduced airspaces with thickened septae and accumulation of glycogen in the lung (20). As only newborn lungs were analyzed, the occurrence, magnitude, and role of autophagy during lung development are still unknown.
In the present study, we investigated the occurrence of autophagy in the developing mouse lung and examined its regulatory role during airway branching and terminal sacculi formation. Our findings revealed 2 windows of autophagy activation in the airway epithelium, both resulting from AMPK activation. Inhibition of AMPK-mediated autophagy led to impaired lung-branching morphogenesis in vitro. Abrogation of autophagy via deletion of beclin 1 (Becn1) in mouse lung epithelium, either during early (E10.5) or late (E16.5) gestation, resulted in reduced airway branching and impaired sacculi formation as well as delayed distal epithelial differentiation, leading to respiratory distress and death in newborn mice. These results demonstrate that intrinsic autophagy is an important regulator of lung development and morphogenesis.
Results
Autophagy occurrence during embryonic lung development.
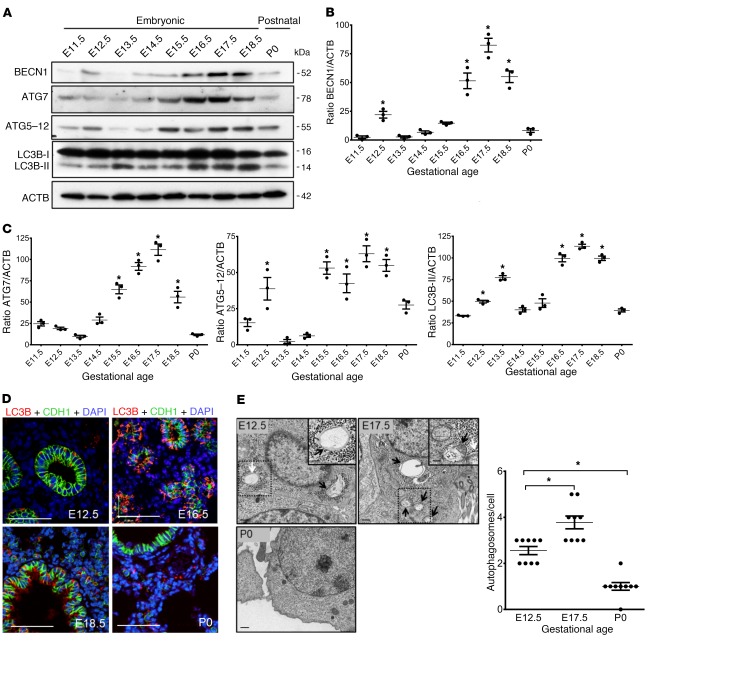
To define the role of autophagy in lung development, we first evaluated the temporal and spatial expression of autophagy-related proteins. Lungs were harvested at various stages of embryonic development (E11.5–E18.5) and at birth (P0) and analyzed by immunoblotting, immunofluorescence (IF), and transmission electron microscopy (TEM). Western blot analysis revealed that several autophagy-related proteins, including BECN1, ATG7, ATG5–12 complex, and LC3B-II, were significantly upregulated at 2 developmental windows of lung development (Figure 1, A–C). Autophagy activation occurred at the early (E12.5) pseudoglandular stage, declined thereafter (E13.5–E15.5), and then peaked again at the canalicular/saccular stages of lung development (E16.5–E18.5), while returning to its basal level at birth.
Figure 1. Autophagy activity during embryonic mouse lung development.
(A) Representative immunoblots of the autophagy proteins ATG7, ATG5–12, BECN1, and LC3B-II (lower band) in mouse lung lysates during lung development. (B and C) Densitometric measurements of ATG7, ATG5–12, BECN1, and LC3B-II proteins at various gestation points relative to E11.5 lung. ACTB was used as a protein loading control. Results are expressed as the mean ± SEM from 3 separate experiments. *P < 0.05 versus E11.5. (D) IF confocal microscopic images showing the autophagosome marker LC3B (red) and the epithelial marker E-cadherin (CDH1, green) during lung development. Nuclei were stained with DAPI (blue). (E) Representative TEM images from embryonic (E12.5 and E17.5) and postnatal (P0) mouse lungs. Arrows indicate autophagosomes present in the epithelial cells of the lungs. Scale bars: 500 nm and 250 nm (insets). Graph shows quantitative analysis of the number of autophagosomes per epithelial cell (≥10 micrographs per gestational age for 2 mice; autophagic vacuoles were counted from 8 to 10 randomly selected fields). Results are expressed as the mean ± SEM. *P < 0.05 versus E12.5. Statistical significance for all data was determined by 1-way ANOVA followed by Tukey’s post hoc test.
To determine the spatial distribution of autophagy activation in the lung, we performed double IF for detection of LC3B and the epithelial cell marker CDH1 (Figure 1D). LC3B predominantly localized to the CDH1+ bronchial and terminal sacculi epithelium of the E12.5–E18.5 lung (Figure 1D). In the newborn lung (P0), we found that LC3B expression in the epithelium was low, whereas positive LC3B signal was detected in the nonepithelial compartment (Figure 1D). TEM, the gold standard for autophagy detection, confirmed the presence of autophagosomes (hallmarks of autophagy) in the epithelium of the E12.5 and E17.5 lungs but were absent in P0 lungs (Figure 1E).
Autophagy inhibition reduces lung branching in vitro.
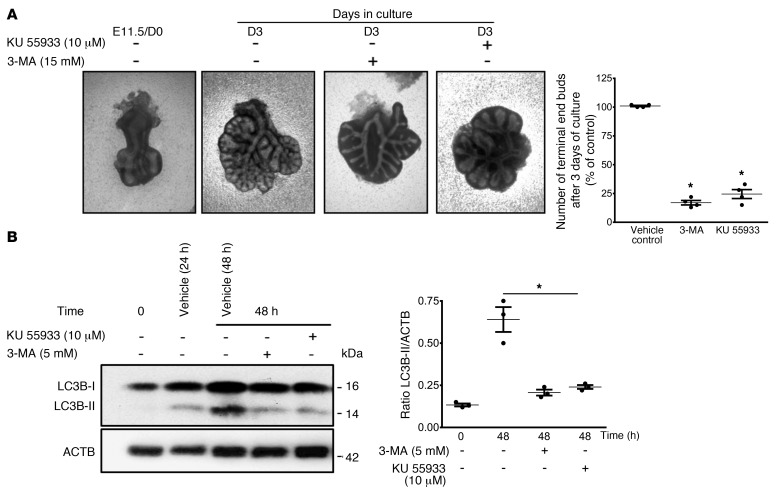
To investigate a potential role for autophagy in lung development, we assessed the effect of autophagy inhibition on lung-branching morphogenesis using ex vivo lung explant cultures. Despite the limitations of long-term culturing, these explant cultures are an attractive tool for studying early lung branching (21). E11.5 lungs were cultured at a liquid-air interface for 72 hours in the presence of PI3K inhibitors (3-methyladenine [3-MA] and KU5593) or vehicle control. Both small-molecule inhibitors block autophagy by inhibiting the initiation of autophagosome formation (22–24). After 72 hours of culture (day 3 [D3]) without inhibitors, lung explants displayed ample branching compared with the starting E11.5 (D0) lung (Figure 2A). Addition of 5 mM 3-MA or 10 μM KU5593 to the culture medium impaired lung branching, as evidenced by the significant reduction in the total number of terminal end buds (Figure 2A). We assessed the efficacy of autophagy inhibition by measuring LC3B-II protein levels, which were indeed significantly reduced after 48 hours of culture with either inhibitor (Figure 2B). Early lung branching was also significantly reduced when the autophagy flux was blocked with 80 nM bafilomycin A1 (Baf A1) (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI127307DS1). Inhibition of autophagolysosome formation by Baf A1 was confirmed by increased LC3B-II levels in lung explants treated with Baf A1 compared with vehicle control lung explants (Supplemental Figure 1, B and C). Consistent with autophagosome-lysosome blockage, we observed an accumulation of enlarged autophagic vacuoles containing undegraded materials in Baf A1–treated versus vehicle control lung explants (Supplemental Figure 1D, white arrow). Together, these results suggest that the autophagy pathway plays an important role in early lung branching.
Figure 2. Autophagy inhibition reduces early lung branching in vitro.
(A) Representative micrographs of lung explant tissue cultured in the presence of the autophagy inhibitors 3-MA (5 mM) and KU 55933 (10 mM). E11.5 lung explants (D0) were treated with inhibitors or vehicle control, and terminal buds were counted after 72 hours of culture (D3) for quantitative evaluation of early branching morphogenesis In the graph, the numbers of terminal end buds on D3 are expressed as a percentage of the vehicle control (mean ± SEM, n = 5 separate explant cultures).*P < 0.05 versus vehicle control. (B, left panel) Representative immunoblot for LC3B-II in lysates of lung explants treated with vehicle, 3-MA or KU after 48 hours of culture. (B, right panel) Densitometric analysis of LC3B-II in lysates of lung explants. ACTB was used as a protein loading control. Results are expressed as the mean ± SEM n = 3 separate explant cultures).*P < 0.05 versus 48-hour vehicle control. Statistical significance for all data was determined by 1-way ANOVA followed by Tukey’s post hoc test.
Autophagy activity coincides with AMPK phosphorylation.
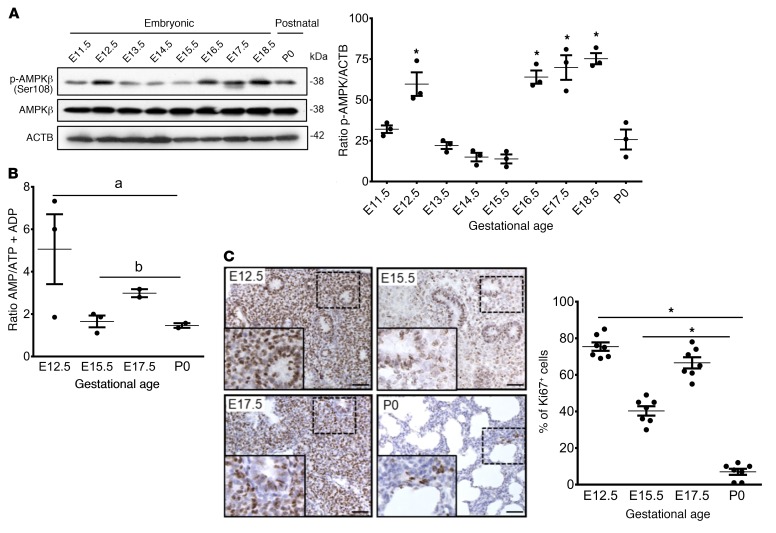
Efficient energy regulation in response to cell growth, differentiation, and survival is a key requirement for cellular homeostasis during development. Failure in the maintenance of ATP demand of a growing tissue will lead to damage of the developing organism. AMPK is one of the major cellular energy sensors found in all eukaryotes (25) that functions as an intracellular low-energy warning system. Recent studies have shown that energy demand triggers autophagy in mammalian cells by activating AMPK (26, 27). To ascertain the underlying mechanisms that may account for autophagy activation during lung development, we examined the expression and phosphorylation of AMPKβ1 (Ser108) at different stages of lung development. Interestingly, we found that AMPK phosphorylation (Figure 3A) occurred during developmental windows similar to those for autophagy activation (Figure 1). Since AMPK is activated by increased AMP and falling ATP levels because of ATP consumption (28), we measured whether AMP, ADP, and ATP levels in mouse fetal lungs were altered during these different stages of lung development. Liquid chromatography–tandem mass spectrometry (LC-MS/MS) revealed significant increases in AMP/ADP plus ATP ratios at gestational time points identical to those for AMPK phosphorylation (Figure 3B). We found that ATP demand was high at these gestational time points because of exuberant cell proliferation in the lung (Figure 3C).
Figure 3. Autophagy is accompanied by AMPK activation during embryonic mouse lung development.
(A) Representative immunoblots of p-AMPKβ (Ser108) and AMPKβ proteins in mouse lung lysates during lung development. Graph shows densitometric analysis of p-AMPKβ (Ser108) during lung development relative to E11.5 lung. ACTB was used as a protein loading control. Results are expressed as the mean ± SEM (n = 3 separate experiments). *P < 0.05 versus E11.5. (B) AMP/ATP plus ADP ratios in embryonic (E12.5, E15.5, and E17.5) and postnatal (P0) lungs. Data show the mean ± SEM (n = 3 separate experiments). aP < 0.05 versus E12.5; bP < 0.05 versus E17.5. (C) Representative IHC images for Ki67 expression in embryonic (E12.5, E15.5, and E17.5) and postnatal (P0) lung tissue. Scale bars: 50 μm. Graph shows quantitative analysis of Ki67+ cells per mm2. Results are expressed as the mean ± SEM (n = 3 separate lungs). *P < 0.05 versus E12.5. Statistical significance for all data was determined by 1-way ANOVA followed by Tukey’s post hoc test.
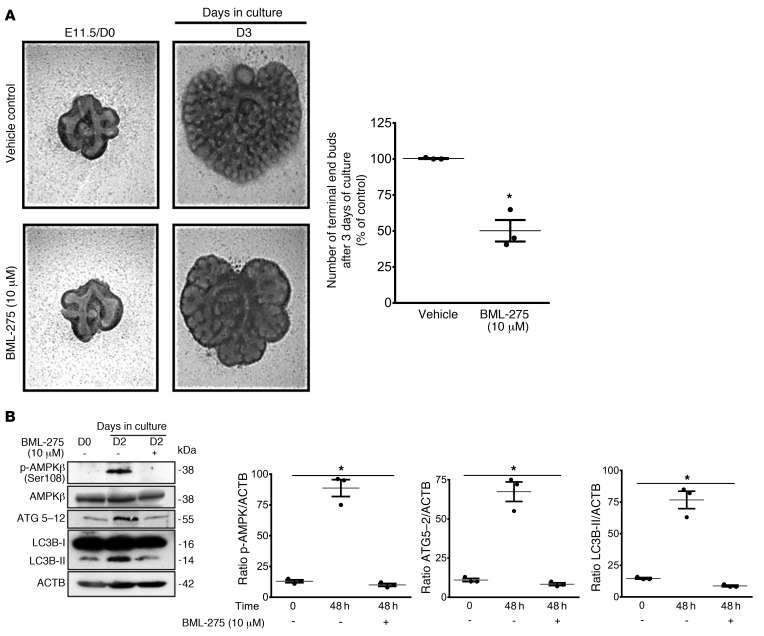
To determine whether the increase in AMPK activity affects lung branching, we blocked AMPK phosphorylation using the specific AMPK inhibitor BML-275 (29). E11.5 lungs cultured for 72 hours in the presence of 10 μM BML-275 exhibited an impaired lung branching (Figure 4A) similar to that seen with autophagy inhibitors (Figure 2). To verify that AMPK activation and downstream autophagy signaling is inhibited by BML-275, we examined the expression of phosphorylated AMPK (p-AMPK), AMPK, and ATG5–12 proteins. Western blot analysis showed that BML-275 effectively blocked AMPK activation in lung explant cultures (Figure 4B). ATG5–12 complex and LC3B-II expression was also inhibited by BML-275 (Figure 4B), observations in line with inhibition of autophagy by the AMPK inhibitor.
Figure 4. Inhibition of AMPK signaling reduces autophagy and early lung branching in vitro.
(A, left panel) Representative micrographs of lung explant tissue cultured with and without the AMPK inhibitor BML-275 (10 mM) for 72 hours (D3). In the graph, the number of terminal end buds on D3 are expressed as a percentage of the vehicle control. Results represent the mean ± SEM from 3 separate experiments. *P < 0.05 versus vehicle control, by Student’s t test). (B) Representative immunoblots of p-AMPKβ (Ser108), AMPKβ, ATG5–12, and LC3B proteins in lysates of lung explants treated with vehicle or BML-275 for 48 hours. Graphs show densitometric analysis of p-AMPKβ (Ser108), ATG5–12, and LC3B-II proteins in lysates of lung explants. ACTB was used as a protein loading control. Results are expressed as the mean ± SEM (n = 3 independent experiments). *P < 0.05 versus 48-hour vehicle control, by 1-way ANOVA followed by Tukey’s post hoc test.
Deletion of epithelial Becn1 at E10.5 reduces airway branching in vivo.
The results shown in Figure 1 revealed the activation of autophagy, including upregulation of BECN1 expression, mainly in the respiratory epithelium of the developing lung. Since constitutive deletion of the Becn1 gene is embryonically lethal, (30), we generated a lung-specific conditional Becn1-KO mouse to study the function of autophagy during lung development. The Becn1 gene was inactivated in the lung epithelium by mating Becn1loxP/loxP mice with an inducible SFTPC-rtTA TetO-Cre–transgenic mouse (31, 32). Cre excision was triggered by doxycycline (Dox) administration via the food and drinking water from E8.5 to E10.5 (Supplemental Figure 2, A and B). We confirmed deletion of Becn1 by PCR genotyping of genomic lung DNA (Supplemental Figure 2C). IF staining for BECN1 on E16.5 lung sections collected from Becn1Epi-KO and control littermate mice confirmed an absence of BECN1 in airway epithelial (CDH1+) cells in Becn1Epi-KO mice (Supplemental Figure 2D). Western blotting of whole-lung tissue lysate from E18.5 Becn1Epi-KO mice showed a significant reduction in protein expression of BECN1, ATG7, and LC3B-II (Supplemental Figure 2, E and F) in comparison with expression levels in control littermate lysate.
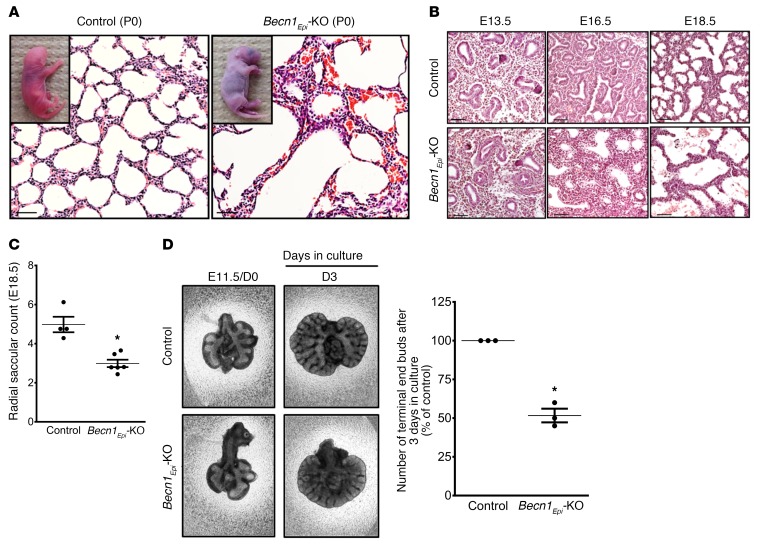
Lung-specific Becn1Epi-KO pups were born alive but died within a couple of hours after birth, whereas littermate control pups survived the perinatal period. The conditional Becn1Epi-KO mice died of respiratory insufficiency, as evidenced by gasping and cyanosis (Figure 5A, inset images). Lungs of WT littermate newborns (P0) had a normal saccular appearance with thin septae (Figure 5A, left panel). By contrast, Becn1Epi-KO newborns had fewer and dilated air sacs with thicker interstitial walls (Figure 5A, right panel). To explore the origin of this morphological defect, we performed macroscopic and histological analyses of the lungs from lung-specific Becn1Epi-KO and littermate control mice at different stages of lung development. Although comparison of lungs from the Becn1Epi-KO and control embryos revealed no structural difference at E13.5 (Figure 5B, left panel), visible defects in branching morphogenesis were evident at E16.5. Transverse or coronal lung sections from Becn1Epi-KO embryos showed fewer but more dilated airway tubules (Figure 5B, middle panel). At E18.5, Becn1Epi-KO lungs had severely dilated distal saccules with thicker septae (Figure 5B, right panel). Morphometric analysis (33) revealed a significant decrease in the number of terminal air sacs in E18.5 Becn1Epi-KO lungs compared with WT littermate lungs (Figure 5C). On the basis of our in vitro data, we postulated that the morphological abnormalities in the conditional Becn1Epi-KO lungs were partially due to diminished airway branching. Thus, we performed explant culturing of whole lungs isolated from E11.5 Becn1Epi-KO and littermate control embryos and, indeed, observed a significant reduction in branching in Becn1Epi-KO lungs after 72 hours of culture compared with littermate controls (Figure 5D).
Figure 5. BECN1 is required for normal lung development and morphogenesis.
(A) Representative light photomicrographs of H&E-stained lung sections from littermate control and Becn1Epi-KO pups after birth. Inserts are representative photographs of littermate control and lung-specific Becn1Epi-KO pups immediately after birth. Scale bars: 70 μm. (B) Representative light photomicrographs of H&E-stained lung sections from littermate control and lung-specific Becn1Epi-KO littermates at different stages of lung development (E13.5, E16.5, and E18.5). Scale bars: 50 μm. (C) Graph shows the radial saccular count for E18.5 lungs from control (WT) mice and Becn1Epi-KO littermates. Data represent the mean ± SEM (n = 5 separate mouse lungs). *P < 0.05 versus control, by Student’s t test. (D) Representative explant cultures of E11.5 control (WT) and Becn1Epi-KO lung tissue that was cultured for 72 hours to evaluate branching morphogenesis. Graph indicates the number of terminal air sacs expressed as the mean ± SEM (n = 5 separate experiments). *P < 0.05 versus control, by Student’s t test.
Deletion of epithelial Becn1 at E10.5 affects proliferation and cell death.
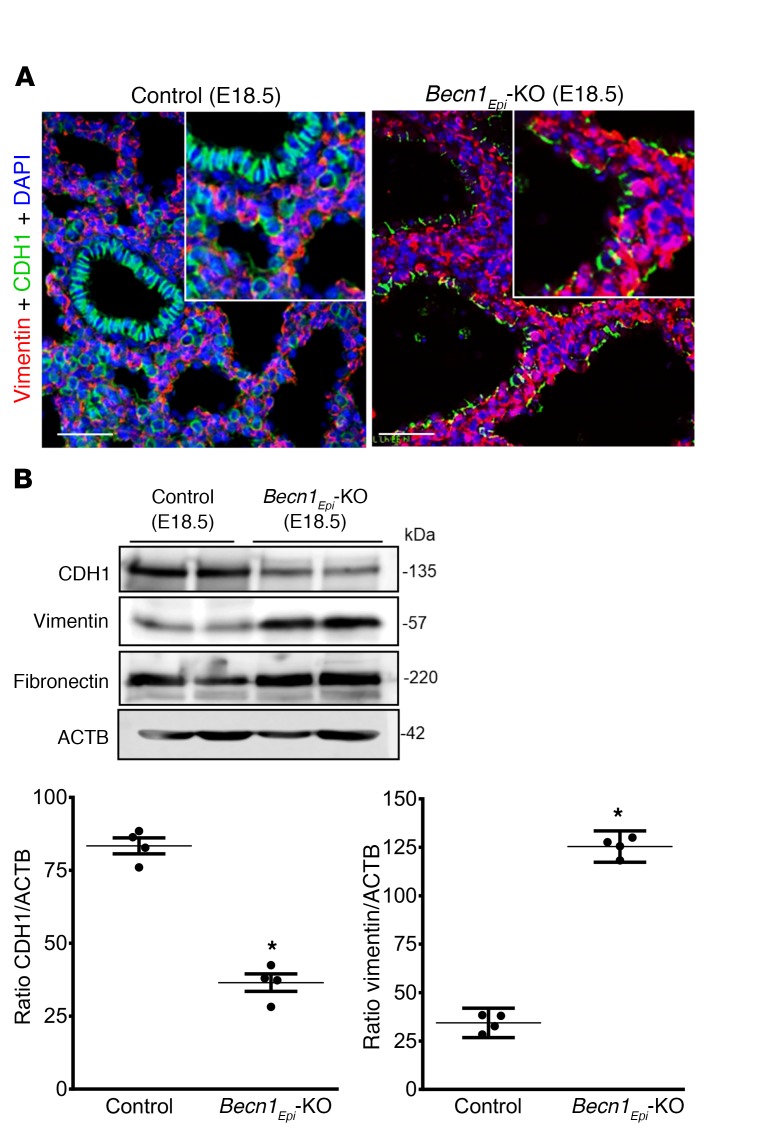
Since E18.5 Becn1Epi-KO lungs had severely dilated terminal sacculi with thicker septae, we first evaluated the mesenchymal and epithelial content using epithelial (CDH1) and mesenchymal (vimentin) markers (Figure 6A). IF revealed markedly more vimentin+ interstitial cells in the thicker septae of the Becn1Epi-KO lungs. In addition, immunoblotting showed reduced epithelial CDH1 levels, whereas expression of mesenchymal markers (vimentin and fibronectin) were increased in Becn1Epi-KO lungs compared with expression in littermate control lungs (Figure 6B), in line with a larger mesenchymal interstitial compartment.
Figure 6. Conditional deletion of epithelial Becn1 affects mesenchymal thinning of the developing lung.
(A) Co-IF staining of CDH1 (green) and vimentin (red) in Becn1Epi-KO and littermate control lungs at E18.5. Nuclei were stained with DAPI (blue). Scale bars: 50 μm; original magnification, ×40 (insets). (B) Representative Western blot for CDH1, vimentin, and fibronectin. Graphs show the densitometric analysis of CDH1 and vimentin expression in whole-lung lysates harvested from Becn1Epi-KO and littermate controls at E18.5. ACTB was used as a protein loading control. Data represent the mean ± SEM (n = 4 separate lungs). *P < 0.05 versus control, by Student’s t test.
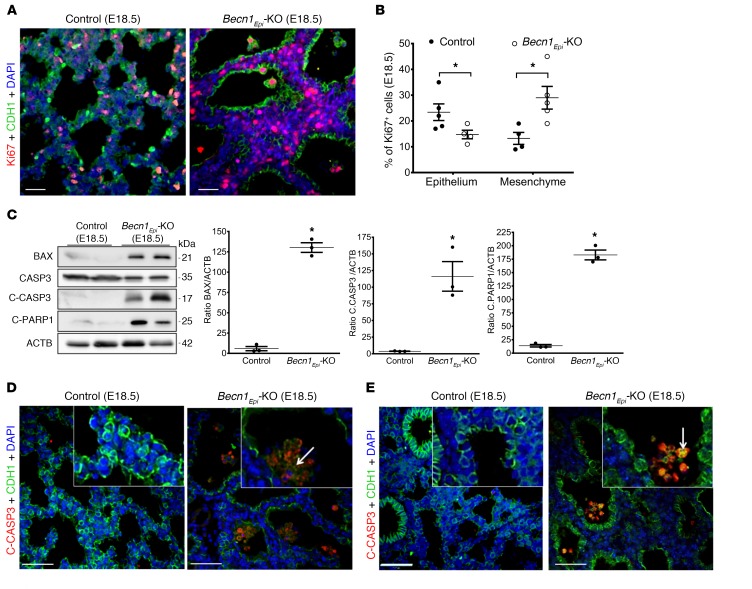
Next, we sought to examine whether the thicker septae and larger airspaces seen in the lungs of conditional Becn1Epi-KO mice were due to changes in cell proliferation or apoptosis. We assessed proliferation and apoptosis in E18.5 Becn1Epi-KO and littermate control lungs by IF using proliferative (Ki67) and apoptotic (cleaved caspase 3 [C-CASP3] and cleaved PARP [C-PARP]) markers. Double-IF staining for Ki67 and CDH1 (Figure 7A) revealed that the number of CDH1+ epithelial cells staining positively for Ki67 was significantly reduced in Becn1Epi-KO lungs compared with that in littermate control lungs (Figure 7B). In contrast, the number of Ki67+ mesenchymal (CDH1–) cells was significantly greater in Becn1Epi-KO lungs (Figure 7B). We then assessed apoptotic cell death in the Becn1Epi-KO lungs. Immunoblotting of E18.5 whole-lung lysates revealed increased BCL-2–associated X (BAX), C-CASP3, and C-PARP protein expression in Becn1Epi-KO lungs compared with expression in littermate control lungs (Figure 7C). Next, we determined which cell type was undergoing apoptosis in the Becn1Epi-KO lungs by double-IF staining for CDH1 and C-CASP3 or C-PARP (Figure 7, D and E, respectively). In contrast to reports showing that most cells undergoing apoptosis during normal lung development are of mesenchymal origin (34–36), we found that apoptotic cells in Becn1Epi-KO lungs were mainly epithelial cells lining the walls of the terminal air sacs or floating in the airspace and dislodged from the walls (Figure 7, D and E).
Figure 7. Conditional deletion of Becn1 perturbs epithelial cell proliferation and apoptosis during lung development.
(A) Co-IF staining of CDH1 (green) and Ki67 (red) in Becn1Epi-KO and littermate control lungs at E18.5. Nuclei were stained with DAPI (blue). Scale bars: 30 μm. (B) Percentage of Ki67+ epithelial and nonepithelial cells in Becn1Epi-KO and littermate control lungs at E18.5. Data represent the mean ± SEM (n = 3 separate lungs). *P < 0.05, by Student’s t test. (C) Representative immunoblots for BAX, CASP3, C-CASP3, and C-PARP in whole-lung lysates harvested from Becn1Epi-KO and littermate control embryos at E18.5. Graphs show densitometric analysis of BAX, C-CASP3, and C-PARP protein expression. ACTB was used as a protein loading control. Data represent the mean ± SEM (n = 4 separate pups). *P < 0.05 versus WT control, by Student’s t test. (D and E) Co-IF staining for E-cadherin (green) with the apoptosis markers C-CASP3 (D, red) and C-PARP (E, red) in Becn1Epi-KO and littermate control lungs at E18.5. Scale bars: 50 μm; original magnification, ×40 (insets).
Deletion of epithelial Becn1 at E10.5 disrupts vascular development.
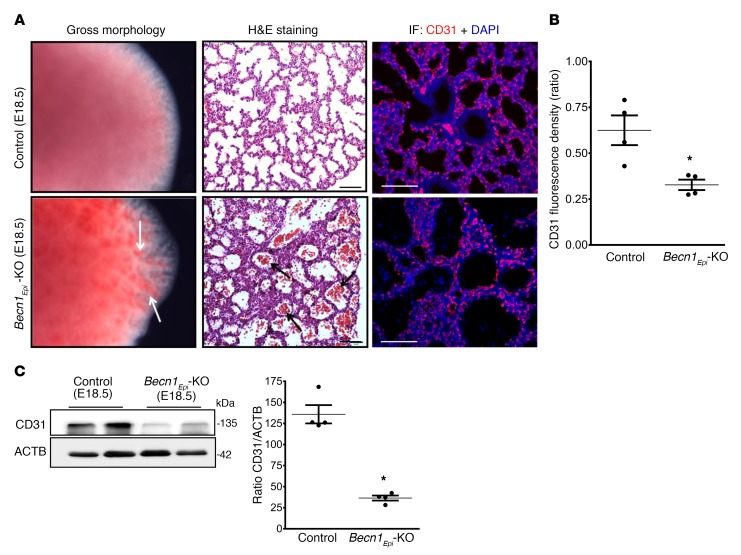
To determine the effect of epithelial Becn1 deletion on vascular development, we examined the vascular morphology of conditional Becn1Epi-KO and littermate control lungs at E18.5. Gross morphology of conditional Becn1Epi-KO lungs showed multiple bleeding spots on the lung periphery, identified as submucosal hemorrhage under a dissection microscope, while littermate control lungs had a normal appearance (Figure 8A, left panel). H&E staining revealed pulmonary hemorrhage with the presence of red blood cells in peripheral air sacs of Becn1Epi-KO lungs (Figure 8A, middle panel). IF staining for the endothelial cell marker CD31 (PECAM) showed that Becn1Epi-KO lungs had fewer CD31+ cells than did littermate control lungs (Figure 8A, right panel and Figure 8B, Supplemental Figure 5). Western blotting corroborated the reduced expression of CD31 in the Becn1Epi-KO lungs compared with the lungs of littermate controls (Figure 8C). Altogether, these results indicate a less developed and leaky pulmonary vasculature in Becn1Epi-KO mice at birth.
Figure 8. Conditional deletion of Becn1 alters proper pulmonary vascular development.
(A) Left panel: Gross morphology of E18.5 lungs from control (WT) and Becn1Epi-KO embryos. White arrows point to hemorrhage regions in Becn1Epi-KO lung. Middle panel: Representative light photomicrographs of H&E-stained lung sections from E18.5 littermate control (WT) and Becn1Epi-KO mice. Note the infiltration of red blood cells in the enlarged air spaces (black arrows) in Becn1Epi-KO lung. Right panel: Confocal IF microscopic images of embryonic lungs (E18.5) stained for the endothelial cell marker CD31 (red). Nuclei were stained with DAPI (blue). Scale bars: 100 μm (middle panel) and 50 μm (right panel). (B) Quantification of the CD31/DAPI fluorescence ratio of E18.5 lungs from control (WT) and Becn1Epi-KO embryos. Data represent the mean ± SEM (n = 3 separate lungs). *P < 0.05 versus WT control, by Student’s t test. (C) Representative immune blot for CD31 on whole E18.5 lung lysates from control (WT) and Becn1Epi-KO embryos. The membrane was re-used in Figure 7C and Figure 8C, which show the same loading control. Graph shows densitometric analysis of CD31 expression. ACTB was used as a protein loading control. Data represent the mean ± SEM (n = 4 separate lungs). *P < 0.05 versus WT control, by Student’s t test.
Deletion of epithelial Becn1 on E10.5 delays distal epithelial differentiation.
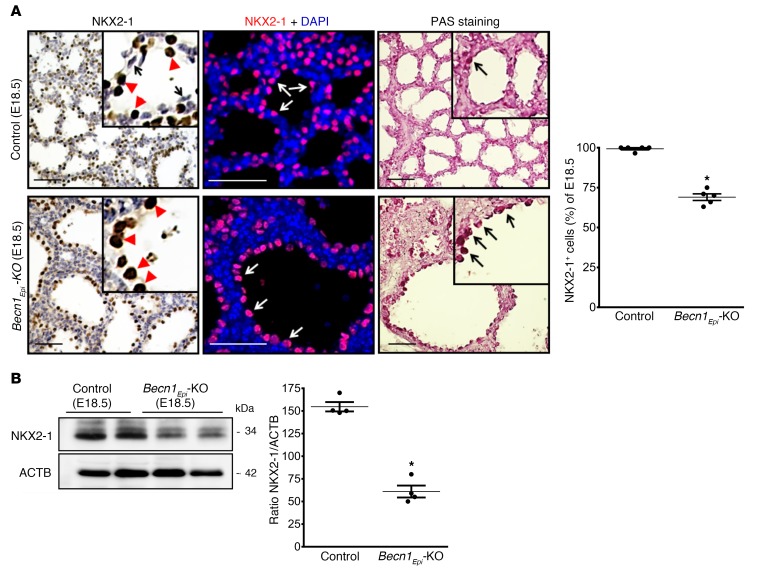
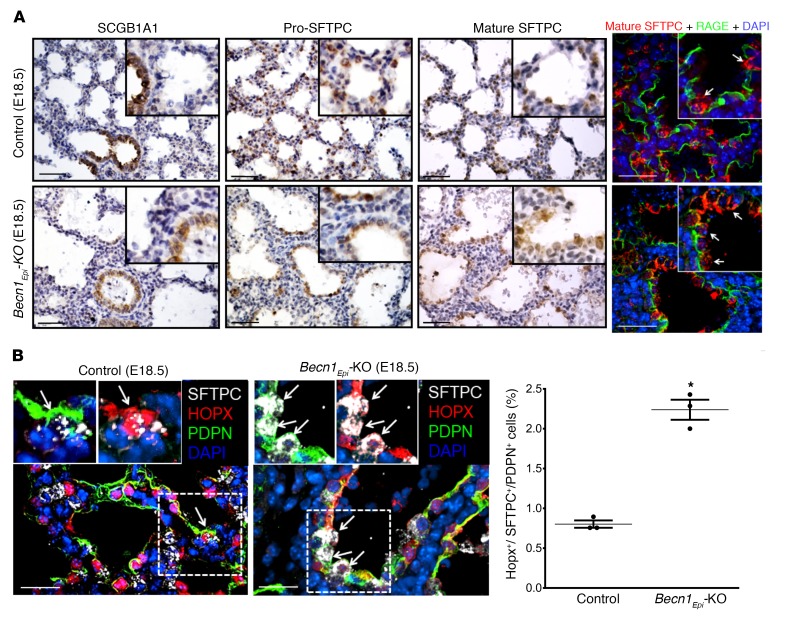
NK2 homeobox 1 (NKX2-1) is a transcription factor expressed in the respiratory epithelium of the lung that is essential for proper lung morphogenesis and formation of the peripheral lung (37–39). In the early lung, NKX2-1 is expressed in a specific subset of cells that includes the progenitor of alveolar type II cells (40). In later gestation and postnatally, NKX2-1 is mainly detected in cuboidal alveolar type II cells and in subsets of nonciliated bronchiolar epithelial cells (40, 41). To determine whether defective branching morphogenesis in Becn1Epi-KO mice alters NKX2-1 expression, we performed IHC and IF on E18.5 lung sections. IHC and IF revealed homogenous nuclear NKX2-1 staining in cuboidal alveolar type II epithelial cells lining the terminal sacculi in littermate control embryos, whereas squamous (flattened) alveolar type I epithelial cells lacked NKX2-1 expression (Figure 9A, left top panels). In Becn1Epi-KO lungs, epithelial cells lining the simple and enlarged sacculi were glycogen filled (defined by periodic acid–Schiff [PAS] staining, Figure 9A, right bottom panels) NKX2-1+ cuboidal cells present in underdeveloped tubules in the thick septae (Figure 9A, left bottom panels). We noted no squamous flattened epithelial cells lining the primitive sacculi of the Becn1Epi-KO lungs. IF staining for NKX2-1 revealed a significant reduction in the number of NKX2-1+ cells in E18.5 Becn1Epi-KO lungs compared with littermate control lungs (Figure 9A, right panel), which was substantiated by immunoblot analysis for NKX2-1 of whole-lung lysates from E18.5 fetuses (Figure 9B). Since NKX2-1 regulates the expression of respiratory epithelium–specific genes, including secretoglobulin 1A member 1 (SCGB1A1), surfactant protein B (SFTPB), and surfactant protein C (SFTPC) (37, 41–45), we next assessed the differentiation of respiratory epithelial cells by performing IHC for club (SCGB1A1) and alveolar type II (pro-SFTPC and mature SFTPC) cell markers on E18.5 lungs from Becn1Epi-KO and littermate control fetuses (Figure 10A). The E18.5 control lungs had a normal appearance, with SCGB1A1+ club cells lining the larger airways. Both cuboidal and squamous epithelial cells lined the terminal sacculi in the E18.5 control lungs, and the cuboidal cells stained positively for pro-SFTPC and mature SFTPC. The presence and distribution of SCGB1A1+ club cells in E18.5 Becn1Epi-KO lungs were similar to that seen in littermate control lungs. However, the larger terminal air sacs of the Becn1Epi-KO lungs contained only cuboidal epithelial cells that were positive for both pro-SFTPC and mature SFTPC (Figure 10A, left bottom panels). Confocal IF of E18.5 lungs for pro-SFTPC, mature SFTPC, and SFTPB corroborated the IHC results (Supplemental Figure 3). IF staining for receptor for advanced glycation end products (RAGE), a marker of alveolar type I cells (46), revealed the presence of squamous flattened alveolar type I cells lining the terminal sacculi in the control lung (Figure 10A, top right panel). Although RAGE was detectable in the primitive air sacs of Becn1Epi-KO lungs, its distribution was confined to the basal side of the SFTPC+ cuboidal cells lining the air sacs (Figure 10A, bottom right panel). These double-positive SFTPC+RAGE+ cells probably represent bipotential alveolar precursor cells (47). To verify this finding, we probed the lungs for HOP homeobox (HOPX), a transcription factor expressed in alveolar precursor cells (47), in combination with the alveolar type I and II cell markers podoplanin (PDPN) and SFTPC, respectively. IF analysis of E18.5 lungs revealed a significant increase in the number of HOPX+SFTPC+PDPN+ cells in the primitive air sacs of Becn1Epi-KO lung (Figure 10B), in line with these cells being bipotential alveolar precursors (47). Thus, distal epithelial differentiation was delayed in Becn1Epi-KO lungs compared with that seen in littermate control lungs.
Figure 9. Conditional deletion of Becn1 reduces NKX2-1 expression.
(A) IHC and confocal IF microscopic images show NKX2-1 and glycogen content (PAS staining) in lung tissue sections from E18.5 Becn1Epi-KO and littermate control (WT) fetuses. In the littermate control, the type II cuboidal alveolar cells stained positive for NKX2-1 (top left inset, red arrowheads), whereas type I squamous alveolar cells lacked NKX2-1 expression (top left inset, black arrows). In the Becn1Epi-KO lung, only NKX2-1+ cuboidal cells were visible (bottom left inset, red arrowheads). In the IF staining (middle panel), NKX2-1+ cells are shown in red, whereas nuclei were stained with DAPI (blue). Scale bars: 50 μm. Graph shows densitometric analysis of NKX2-1 IF results. Data represent the mean ± SEM (n = 4 separate lungs). *P < 0.05 versus control, by Student’s t test. (B) Representative immunoblot for NKX2-1. Graphs shows densitometric analysis of NKX2-1 expression in whole-lung lysate harvested from Becn1Epi-KO and littermate control embryos at E18.5. ACTB was used as a protein loading control. Data represent the mean ± SEM (n = 4 separate lungs). *P < 0.05 versus WT control, by Student’s t test.
Figure 10. Conditional deletion of Becn1 delays distal epithelial differentiation.
(A) Representative IHC images for Clara cell secretory protein (SCGB1A1), pro-SFTPCC, and mature SFTPC expression in lung tissue sections from E18.5 Becn1Epi-KO and littermate control fetuses. Scale bars: 50 μm; original magnification, ×20 (insets). IF microscopic images show lung tissue sections from E18.5 Becn1Epi-KO and littermate control fetuses stained for mature SFTPC (red) and RAGE (green). The white arrows in the insets point to cuboidal alveolar type II epithelial cells. Scale bars: 25 μm; original magnification, ×40 (insets). (B) Confocal IF microscopic images of E18.5 lung tissue from Becn1Epi-KO mice costained for SFTPC (white), HOPX (red), and PDPN (green). Nuclei were stained with DAPI. Arrows indicate alveolar precursor cells detected by an overlap of all these markers. Scale bars: 25 μm; original magnification, ×40 (insets). Graph indicates the percentage of alveolar precursor cells that stained positive for SFTPC, HOPX, and PDPN in E18.5 lung sections from Becn1Epi-KO and littermate control mice. Data are expressed as the mean ± SEM (n = 3 separate experiments). *P < 0.05 versus WT control, by Student’s t test.
Deletion of epithelial Becn1 at E16.5 of lung development inhibits sacculi formation.
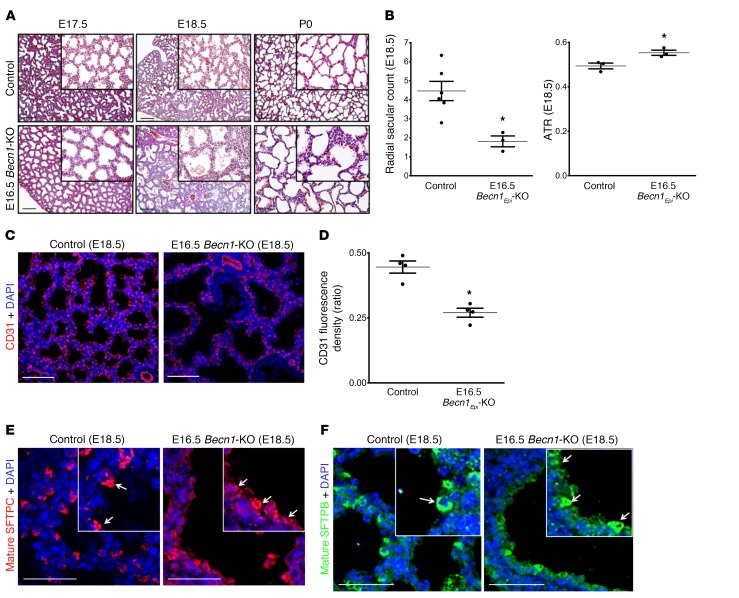
As shown in Figure 1, autophagy activation occurs during 2 distinct periods of lung development. The first activity happens in the early pseudoglandular period, whereas a second activity arises during the canalicular/saccular periods. So far, we have reported findings for mice in which lung epithelial Becn1 was deleted prior to formation of the lungs (E10.5 Becn1Epi-KO mice). This strategy blocks autophagy activation throughout lung development and does not allow for discrimination between the 2 developmental windows of autophagy activity. We therefore took advantage of the Dox-inducible system and deleted the lung epithelial Becn1 gene just prior to the canalicular/saccular periods and investigated the role of autophagy during these periods of lung development. Dox was administered via the food and drinking water from E14.5 to E16.5, and lungs were harvested from fetuses at E17.5, E18.5, and P0 for further analysis (Supplemental Figure 4A). In contrast to the early deletion of lung epithelial Becn1 that resulted in perinatal death within a couple of hours after birth, removal of lung epithelial Becn1 at E16.5 (E16.5 Becn1Epi-KO) did not result in any immediate perinatal lethality. However, all E16.5 Becn1Epi-KO pups died between P2 and P5 as a result of respiratory distress. Histological comparison of lungs from the E16.5 Becn1Epi-KO and littermate control embryos and newborn pups revealed visible defects in terminal air sac formation starting at E17.5 (Figure 11A, left panels). On E18.5, E16.5 Becn1Epi-KO lungs displayed dilated sacculi with slightly thicker septae (Figure 11A, middle panels). P0 lungs of WT littermate newborns had normal saccular appearance, whereas P0 lungs of E16.5 Becn1Epi-KO mice had fewer and dilated air sacs (Figure 11A, right panels). As seen in E10.5 Becn1Epi-KO mice, lung morphometric analysis on E18.5 revealed a significant decrease in the number of terminal air sacs in E16.5 Becn1Epi-KO lungs compared with WT littermate lungs (Figure 11B). Air-to-tissue ratios (ATRs) corroborated that the saccular airspaces were larger in E16.5 Becn1Epi-KO lungs than in WT littermate lungs (Figure 11B). Furthermore, as in E10.5 Becn1Epi-KO mice, IF staining for CD31 showed that E16.5 Becn1Epi-KO lungs at E18.5 had fewer CD31+ cells than did littermate control lungs (Figure 11, C and D). However, in contrast to E10.5 Becn1Epi-KO lungs, we observed no pulmonary hemorrhage. Thus, E16.5 Becn1Epi mice had a less developed, but intact, pulmonary vasculature. Assessment of the differentiation and distribution of respiratory epithelial cells by IHC (Supplemental Figure 4B) and IF (Figure 11, E and F) using SCGB1A1, pro-SFTPC, mature SFTPC, and RAGE antibodies on E18.5 lung tissue from E16.5 Becn1Epi-KO and littermate controls revealed results similar to those observed at E18.5 in E10.5 Becn1Epi-KO lung tissue. Terminal saccules of E16.5 Becn1Epi-KO lungs on E18.5 were lined with pro-SFTPC+, mature SFTPC+, and SFTPB+ epithelial cells that frequently stained positive for RAGE. The presence of bipotential alveolar precursor cells indicates that distal epithelial differentiation was delayed in the E16.5 Becn1Epi-KO lungs.
Figure 11. Pulmonary phenotype following deletion of epithelial Becn1 at canalicular/saccular stages.
(A) Representative H&E-stained images of lungs from littermate control (WT) and E16.5 Becn1Epi-KO mice at E17.5, E18.5, and immediately after birth (P0). Scale bars: 100 μm; original magnification, ×20 (insets). (B) Graph shows the radial saccular count and the ATR for littermate control and E16.5 Becn1Epi-KO mice at E18.5. Data represent the mean ± SEM (n = 5 separate lungs). *P < 0.05 versus WT control, by Student’s t test. (C) Confocal IF microscopic images of embryonic lungs (E18.5) stained for the endothelial cell marker CD31 (red). Nuclei were stained with DAPI (blue). Scale bars: 50 μm. (D) Quantification of the CD31/DAPI fluorescence ratio in E18.5 lung tissue from control (WT) and E16.5 Becn1Epi-KO embryos. Results are shown as the mean ± SEM (n = 3 separate lungs). *P < 0.05 versus WT control, by Student’s t test. (E and F) Representative confocal IF microscopic images of E18.5 lung tissue sections from E16.5 Becn1Epi-KO and littermate control mice stained for mature SFTPC (E, red) and SFTPB (F, green). Nuclei were stained with DAPI. The white arrows in the insets point to cuboidal alveolar type II epithelial cells. Scale bars: 25 μm; original magnification, x40 (insets).
Discussion
Here, we demonstrate that intrinsic autophagy is critical for proper lung development and morphogenesis. Abrogation of intrinsic autophagy in vitro and/or in vivo, either during early or late gestation, disrupts airway branching as well as sacculi formation and delays maturation of the lung. The reduction in branching is accompanied by excessive cell death, diminished and abnormal vascular formation, and delayed differentiation of epithelial cells lining the terminal air sacs into alveolar cells. Together, these developmental impairments contribute to the lethal respiratory distress seen in newborns of Becn1Epi-KO mice.
Despite the importance of autophagy in mammalian development (18, 19), limited information is available regarding its role during lung development. Mice deficient for Ulk1/2 and Atg5 die within 24 hours of birth, and their lungs have reduced airspaces with thickened septae (20), in agreement with our findings. The authors concluded that autophagy is vital for lung maturation, but no mechanisms explaining the reduced airspaces were described. In addition, the gene deletions were not specific to the lung, confounding the interpretation. In the present study, we have extended the findings of these studies by examining the occurrence of autophagy and evaluating its role during murine lung development. Our results revealed that autophagy is present at basal levels throughout the entire period of lung development; however, its activity increases in the early pseudoglandular period, and a second wave of autophagy activity occurs during the canalicular/saccular stages of development. At these developmental stages, fetal lungs undergo active tubular branching and terminal sacculi formation (7, 9, 36). Our data show that autophagy is primarily activated in the epithelium of the developing lungs and that autophagy inhibition markedly reduces airway branching and sacculi formation, resulting in perinatal death. These observations support the idea that intrinsic autophagy may be key to lung development and morphogenesis.
The present study shows that AMPK and autophagy activation occur during similar developmental windows. It is well known that energy homeostasis and maintenance of ATP demand are essential for proper tissue growth and development (48) and are precisely regulated by AMPK (26, 28, 49, 50). Regardless of the stimulus, activated (phosphorylated) AMPK maintains ATP levels by turning on ATP-producing processes and turning off ATP-consuming processes (51). AMPK phosphorylation has been shown to activate autophagy to support cell proliferation and differentiation by maintaining energy homeostasis (26, 49, 50). In line with these findings, we observed activation of AMPK at gestational periods of increased ATP consumption due to exuberant cell growth. Inhibition of AMPK activation prevented autophagy and reduced airway branching, like that seen with autophagy inhibitors, suggesting that AMPK regulates branching morphogenesis via autophagy activation.
BECN1 is a key regulator of autophagosome formation, which acts during the initiation stage of autophagy as part of a complex with ATG14, PIK3C3 (also known as VPS34), and PIK3R4 (also known as p150) (52, 53).
Since BECN1-mediated autophagy activation in the developing lung was dominantly seen in the respiratory epithelium and a ubiquitous deletion of the Becn1 gene is embryonically lethal (30), we conditionally deleted Becn1 in the lung epithelium to establish the function of autophagy during lung development. We found that removal of epithelial Becn1 prior to lung formation severely disrupted airway branching and terminal sacculi formation. The Becn1Epi-KO newborns had fewer and larger distal air sacs with thicker interstitial walls and died at birth of respiratory insufficiency. Perinatal lethality from respiratory insufficiency affecting distal lung development has been reported for several transgenic models (39, 54–59). Mortality in these models was mainly due to defects in surfactant protein metabolism (57–59) or distal epithelial differentiation (54, 56). In the present study, an absence of epithelial Becn1 increased the number of bipotential alveolar progenitor cells lining the primitive air sacs, implying that a delayed maturation of the distal epithelium contributed to the severe respiratory insufficiency.
In the current study, we also observed visible defects in pulmonary vascular development in Becn1Epi-KO mice at birth. It has long been recognized that lung epithelium and vascular morphogenesis are closely linked. Previous studies showed that epithelial-vascular interactions are essential for lung branching, tissue patterning, and primary septa formation during distal lung morphogenesis (60–62). VEGF is produced by the lung epithelium (63) and is known to be important for pulmonary vasculogenesis and morphogenesis (64). Inhibition of VEGF has been shown to impair lung branching (60), and loss of VEGF results in impaired fetal lung maturation (63), like the phenotype seen in our Becn1Epi-KO mice. It is plausible that the higher rate of apoptosis together with the reduced proliferation of the respiratory epithelium in Becn1Epi-KO mice interrupted the desired epithelial-vascular interactions and reduced epithelial VEGF expression, thereby altering pulmonary vascular development.
During lung morphogenesis, the mesenchymal lung compartment “thins” to accommodate the developing airways and air sacs, whereas the epithelium continues to proliferate. Apoptosis plays a role in the removal of excess mesenchymal cells (34, 36). Our finding that apoptotic cells in Becn1Epi-KO lungs were mainly epithelial cells lining the walls of the terminal air sacs contrasts with previous studies reporting that most cells undergoing apoptosis during normal lung development are of mesenchymal origin (34, 36). A possible explanation for this discrepancy is the deletion of Becn1 in the lung epithelium of Becn1Epi-KO mice. BECN1 protein contains a BCL-2–BCL-3 homology (BH2-BH3) domain, which, under normal conditions, inhibits autophagy by interacting with the antiapoptotic protein BCL-2 (30, 52, 65). Disruption of the BECN1–BCL-2 complex has been shown to trigger apoptosis by releasing other proapoptotic BCL-2 family members (65, 66). Here, we found that an absence of BECN1 in the developing lung epithelium of Becn1Epi-KO mice led to an increase in proapoptotic BAX expression as well as enhanced epithelial apoptosis that was so profound that it overwhelmed the low grade of mesenchymal apoptosis. Increased apoptosis has also been reported for other conditional KOs of autophagy (67, 68). Additionally, failure to activate autophagy in response to stimuli is known to lead to apoptotic cell death (69). Thus, the lack of autophagy in the respiratory epithelium of Becn1Epi-KO mice in response to a demand of energy for proliferation may also contribute to apoptosis of the epithelial cells. The lower rate of epithelial proliferation combined with the higher rate of mesenchymal proliferation could explain the reduced lack of mesenchymal thinning normally seen during late fetal lung development and thus resulted in thicker interstitial walls and fewer terminal sacculi in the conditional Becn1Epi-KO lungs at birth.
In the present study, we demonstrated that loss of epithelial Becn1 leads to a reduction in NKX2-1 expression. NKX2-1 plays a key role in regulating the expression of various lung cell–specific genes, including SFTPB, SFTPC, and SCGB1A1 (37, 41–45). Because of this role, it is plausible that a diminished expression of NKX2-1 in Becn1Epi-KO mice led to the lung epithelial phenotype seen in the Becn1Epi-KO mice. To our knowledge, we are the first to show that epithelium-specific deletion of Becn1 in the airways results in decreased NKX2-1 expression. Nuclear translocation of BECN1 indicates a role for BECN1 in regulating DNA damage repair (70) and autophagic growth control (71); however, an interaction between BECN1 and NKX2-1 during lung development remains to be explored. Many of the NKX2-1+ cells lining the primitive and dilated air sacs in E18.5 Becn1Epi-KO mice were bipotential alveolar progenitor cells, based on double-positive SFTPC+RAGE+ and triple-positive HOPX+SFTPC+PNPD+ staining. SCGB1A1 expression in the proximal airways appeared to be unchanged in the Becn1Epi-KO mice, suggesting that specifically distal epithelial differentiation was affected by the absence of epithelial Becn1. Although we did not compare proximal and distal epithelial levels of NKX2-1 expression, we speculate that the delayed distal epithelial differentiation was mainly due to the lower NKX2-1 levels in the cells lining the primitive saccules.
In this study, we also deleted Becn1 in lung epithelial cells during the second developmental window of autophagy activation in late fetal gestation. Most infants who develop BPD are born extremely prematurely (before 28 weeks). At birth, the lungs of these infants reflect the canalicular/saccular stages of lung development, and we were therefore interested in deleting Becn1 during this period. Although the E16.5 Becn1Epi-KO pups survived at birth, they succumbed to respiratory distress between P2 and P5. The morphological phenotype included defects in sacculi formation, vascularization, and epithelial differentiation. The less severe phenotype in E16.5 Becn1Epi-KO lungs compared with that of E10.5 Becn1Epi-KO lungs is likely attributable to the fact that the bronchial tree is completed at E16.5 and that only alveolar sacculi formation and differentiation are affected in the E16.5 Becn1Epi-KO newborns. The lung phenotype of E16.5 Becn1Epi-KO pups, i.e., fewer and dilated immature air sacs, is similar to that of infants with BPD, except that these children receive surfactant, are ventilated, and receive supplemental oxygen (72). It is possible that premature birth affects the intrinsic autophagy occurring at that time, thereby influencing the formation of alveolar sacculi. Ventilation (73) and oxygen (74) have been shown to induce autophagy in newborn rodents but probably at supra-levels that overwhelm the intrinsic autophagy pathway, leading to unwanted cell death.
Collectively, our study demonstrates that the BECN1-mediated autophagy pathway in lung epithelial cells is critical for proper airway branching and terminal sacculi formation. Deficiency of intrinsic autophagy results in structurally abnormal lungs and delayed maturation that lead to respiratory failure.
Methods
Chemicals, reagents, and antibodies.
The following chemicals and reagents were from Gibco (Life Technologies, Thermo Fisher Scientific): PBS (catalog 14190), HBSS (catalog 14175), DMEM/F12 (catalog 11095), penicillin-streptomycin (10,000 U/ml, catalog 15140), TrypLE Express Enzyme Phenol Red (catalog 12605), and FBS (catalog 12483). DAPI was obtained from Invitrogen (Life Technologies, Thermo Fisher Scientific, catalog D1306). Complete protease inhibitor cocktail tablets (catalog 04 693 159001) were obtained from Roche Pharmaceuticals. BSA (catalog A8806), goat serum (catalog G9023), 3-MA (catalog M9281), Baf A1 (catalog B1793), phosphatase inhibitors (catalogs P5726 and P0044), and Schiff’s reagent (catalog S5133) were obtained from Sigma-Aldrich. KU55933 (catalog 3544) and Gö6976 (catalog 2253) were from Tocris Biosciences. Fluorescence mounting medium (catalog S3023) was purchased from Dako. The list of antibodies used in this study is provided in Supplemental Table 1.
Generation of Becn1-KO mice.
Lung epithelium–specific Becn1-KO mice (SFTPC-rtTA TetO-Cre Becn1fl/fl) were generated by crossing Becn1loxP/loxP with SFTPC-rtTA TetO-Cre–transgenic mice. Briefly, cryopreserved Becn1tm1a(KOMP)Wtsi embryos were purchased from the European Mouse Mutant Archive (EMMA) and transferred into pseudopregnant C57BL/6J females at the Toronto Centre for Phenogenomics. Offspring were crossed with FLP deleter mice [B6.129S4–Gt(ROSA)26Sortm1(FLP1)Dym] to generate heterozygous Becn1loxP mice, which were crossed to generate homozygous Becn1loxP/loxP mice. Adult Becn1loxP/loxP mice were healthy and fertile. These animals were then bred with SFTPC-rtTA TetO-Cre–transgenic mice, in which Cre expression is induced in airway epithelial cells by administration of the inducing agent Dox (Supplemental Figure 2B) (31, 32). This enabled us to create triple-transgenic animals in which Becn1 could be deleted in the respiratory epithelial cells upon Dox treatment. Conditional deletion of lung epithelial Becn1 (Becn1Epi-KO) was achieved by administration of Dox via the food (200 mg/kg Dox from BioServ) and water (0.3 mg/ml Dox plus 5% sucrose) to the pregnant mice for 48 hours prior to lung formation (starting at E8.5). In separate experiments, Becn1Epi was conditionally deleted at the canalicular/saccular stage of lung development by administration of Dox in the food and water from E14.5 to E16.5. Lungs were isolated from fetuses at different stages of development for further analysis.
SDS-PAGE and immunoblotting.
Mouse embryonic lung tissues were lysed and homogenized in RIPA lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% [v/v] Triton X-100, 1% [w/v] sodium deoxycholate, and 0.1% [w/v] SDS) supplemented with protease and phosphatase inhibitors and 1 mM PMSF. After centrifugation at 13,000 g for 10 minutes, protein content in the supernatant was determined using a colorimetric assay based on the Bradford method (Bio-Rad Protein Assay Kit, Bio-Rad Laboratories, catalog 5000002). Proteins were then size-fractionated by SDS-PAGE and transferred onto nitrocellulose membranes under reducing conditions. Membranes were blocked with 5% (w/v) nonfat dried milk in 0.01% TBS–Tween-20 for 1 hour, and blots were incubated overnight with the appropriate primary antibodies at 4°C (dilutions are shown in Supplemental Table 1). Bands were visualized using HRP-labeled secondary antibodies and an ECL detection kit from Amersham (GE Healthcare Amersham, catalog 45-002-401) (75). Equal protein loading (50 μg/well unless stated otherwise) was confirmed by immunoblotting for ACTB in the same membrane. Band intensities were quantified using ImageJ (NIH) densitometric software.
TEM analyses.
Mouse embryonic lung tissues were fixed in 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4). Samples were prepared and photographed using a JEOL JSM 6700F transmission electron microscope as previously described (76). A minimum of 10 micrographs from 2 lungs per gestational age were analyzed. Autophagic vacuoles were counted from 8 to 10 randomly selected fields. The number of autophagic vacuoles was counted at a magnification of ×2,500 to ×4,000, and higher-magnification micrographs (×25,000) were taken to confirm the presence of autophagosomes.
Histology, IF, and IHC.
For histology, IHC, and IF confocal microscopy, mouse embryonic lungs were fixed in 4% (v/v) PFA in PBS for 24 hours, dehydrated with ethanol, cleared with xylene, and embedded in paraffin. Sections of 4-μm thickness were prepared from representative paraffin blocks. The sections were then deparaffinized, rehydrated, and used for staining. H&E staining was performed using a standard protocol. The glycogen content of the lung cells was detected by a standardized PAS staining technique using a Schiff’s reagent (Sigma-Aldrich, catalog S5133). Briefly, lung sections were incubated in 1% periodic acid for 5 minutes, rinsed with water, and incubated in Schiff’s reagent for 10 minutes. The slides were then washed with running tap water, counterstained with hematoxylin, rinsed with water, dehydrated, cleared, and permanently mounted. Lung morphometric analysis was performed to assess radial saccular counts as previously described (33). The ATR was determined using 40 × 40 images captured by a Leica DM6000 light microscope equipped with a Q-Imaging Retiga EXi CCD camera (QImaging). The images (n = 3 lungs per group, 25 images each) were processed with MATLAB software (MathWorks). The k-means clustering algorithm was used to separate the air space from the tissue, creating a binarized image. From this binarized image, the ATR was calculated.
For IHC and IF analysis, heat-induced epitope retrieval was performed by placing the slides in a pressure cooker with citrate buffer (0.1 M citric acid, 0.1 M sodium citrate, pH 6.0) in a Coplin Jar for 15 minutes. For IHC, sections were incubated with the assigned primary antibody overnight at 4°C in a humidified chamber, followed by incubation with a biotinylated secondary antibody (1:400, Jackson ImmunoResearch) and visualization with a VECTASTAIN ABC Kit (VECTOR Laboratories, catalog PK-6100). The slides were then counterstained with Mayer’s hematoxylin (Sigma-Aldrich, catalog H9627), dehydrated, and mounted with Permount (Life Technologies, Thermo Fisher Scientific, catalog SP15-500). As a negative control, sections were processed as above, but the addition of a primary antibody was omitted. Digital images were captured using a Leica DM6000 B light microscope equipped with a QImaging Retiga EXi CCD camera (Teledyne QImaging). For IF analysis, 4-μm sections were incubated overnight with primary antibodies at 4°C in a humidified chamber. The next day, the sections were washed 3 times with PBS and then incubated with fluorochrome-conjugated secondary antibodies at room temperature for 1 hour in the dark. After washes with PBS, nuclei were counterstained with DAPI (Invitrogen, Life Technologies, Thermo Fisher Scientific, catalog D1306). The sections were washed twice with PBS and mounted using fluorescence mounting medium (Dako, catalog 3023). Fluorescence images were captured with a Leica CTRMIC 6000 microscope in conjunction with a Hamamatsu C910013 spinning-disc camera (Leica Microsystems) (76). Laser intensity and detector sensitivity settings remained constant for all image acquisitions within a respective experiment. Images were later analyzed using Volocity software (PerkinElmer).
Lung explant cultures.
Timed-pregnant CD1 mice were sacrificed on postcoitum day 11.5 (E11.5), and the embryos were harvested. Lung primordia were isolated from embryos by microdissection under sterile conditions. Isolated lung explants were placed on Nucleopore hydrophobic floating membranes (8-μm pore size, Whatman Nucleopore, Sigma-Aldrich, catalog 110614) and cultured on the surface of DMEM/F12 medium (Gibco, Life Technologies, Thermo Fisher Scientific) supplemented with 200 U/ml penicillin-streptomycin (Gibco, Life Technologies, Thermo Fisher Scientific, catalog 15140) and 0.5% (v/v) FBS (Gibco, Life Technologies, Thermo Fisher Scientific, catalog 12483). The cultures were maintained in an atmosphere of 95% air and 5% CO2 for 3 days. The medium was changed every day. For autophagy inhibition experiments, lung explants were cultured in medium containing either DMSO (vehicle control), 80 nM Baf A1 (Sigma-Aldrich, catalog B1793), or a PI3K inhibitor (5 mM 3-MA; Sigma-Aldrich, catalog M9281) or 10 μM KU55933 (Tocris Biosciences, Bio-Techne Canada, catalog 3544) for 72 hours. At the indicated time points, pictures of the lung explants were taken, and tissues were either processed for immunoblotting or histological analysis.
Mass spectrometric analysis of ATP, ADM, and AMP.
Lung tissue AMP, ADP, and ATP amounts were measured by LC-MS/MS. Briefly, lung tissue samples were harvested and homogenized with 20 nm Tris buffer. Tissue homogenates were then extracted in acetonitrile for several hours at –20°C. The supernatant was then lyophilized, reconstituted in water, and injected for LC-MS/MS analysis (77).
IHC image analysis.
Quantification of apoptosis (C-CASP3+ and C-PARP+ cells) or proliferation (Ki67+ cells) has been described previously (78). In brief, following IHC staining of lung sections obtained from at least 3 different transgenic and littermate control mice, 4–6 areas of lung parenchyma were randomly photographed using a ×20 objective. Images were then analyzed using NIH ImageJ software. Data were then presented as average percentages of positively stained cells in Becn1Epi-KO animals versus littermate controls.
Statistics.
All numerical data are presented as the mean ± SEM of at least 3 separate experiments. Statistical significance was determined by 2-tailed Student’s t test for comparisons of 2 groups or 1-way ANOVA followed by Tukey’s post hoc test for more than 2 groups, using GraphPad Prism, version 6.0 (GraphPad Software). Differences were considered significant at a P value of less than 0.05.
Study approval.
All animal experiments were approved and performed in accordance with the animal care committee guidelines of the Hospital for Sick Children Research Institute (Peter Gilgan Centre for Research and Learning, Toronto, Ontario, Canada).
Author contributions
BY and MP designed the study, wrote the manuscript, and performed experiments. JL and LE performed experiments and analyzed data. IL and CA performed morphometric and electron microscopic analysis.
Supplementary Material
Acknowledgments
The authors would like to thank Angie Griffin (Hospital for Sick Children) for mouse breeding and husbandry; members of the Analytical Facility for Bioactive Molecules at the Hospital for Sick Children for their assistance with LC-MS/MS; and members of the laboratory of Giles Santyr (Hospital for Sick Children) for their technical assistance with lung ATR measurements. This work was supported by grants from the Canadian Institutes of Health Research (FDN-143309, to MP); the Canadian Foundation of Innovation (CFI-12156, to MP); and the Willliam and Meredith Saunderson Family Endowment Fund (to MP). MP holds the Tier 1 Canadian Research Chair in Fetal, Neonatal, and Maternal Health.
Version 1. 06/04/2019
Electronic publication
Version 2. 07/01/2019
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(7):2904–2919.https://doi.org/10.1172/JCI127307.
See the related Commentary at Normal lung development needs self-eating.
Contributor Information
Behzad Yeganeh, Email: behyeganeh@gmail.com.
Joyce Lee, Email: j90lee@gmail.com.
Leonardo Ermini, Email: ermini74@icloud.com.
Cameron Ackerley, Email: cameron.ackerley@sickkids.ca.
Martin Post, Email: martin.post@sickkids.ca.
References
- 1.Edwards MO, Kotecha SJ, Kotecha S. Respiratory distress of the term newborn infant. Paediatr Respir Rev. 2013;14(1):29–36; quiz 36. doi: 10.1016/j.prrv.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Cardoso WV, Lü J. Regulation of early lung morphogenesis: questions, facts and controversies. Development. 2006;133(9):1611–1624. doi: 10.1242/dev.02310. [DOI] [PubMed] [Google Scholar]
- 3.Harding R, Hooper SB. Regulation of lung expansion and lung growth before birth. J Appl Physiol. 1996;81(1):209–224. doi: 10.1152/jappl.1996.81.1.209. [DOI] [PubMed] [Google Scholar]
- 4.Conlon I, Raff M. Size control in animal development. Cell. 1999;96(2):235–244. doi: 10.1016/S0092-8674(00)80563-2. [DOI] [PubMed] [Google Scholar]
- 5.Cardoso WV. Molecular regulation of lung development. Annu Rev Physiol. 2001;63:471–494. doi: 10.1146/annurev.physiol.63.1.471. [DOI] [PubMed] [Google Scholar]
- 6.Maeda Y, Davé V, Whitsett JA. Transcriptional control of lung morphogenesis. Physiol Rev. 2007;87(1):219–244. doi: 10.1152/physrev.00028.2006. [DOI] [PubMed] [Google Scholar]
- 7.Ten Have-Opbroek AA. Lung development in the mouse embryo. Exp Lung Res. 1991;17(2):111–130. doi: 10.3109/01902149109064406. [DOI] [PubMed] [Google Scholar]
- 8.Burri PH, Moschopulos M. Structural analysis of fetal rat lung development. Anat Rec. 1992;234(3):399–418. doi: 10.1002/ar.1092340310. [DOI] [PubMed] [Google Scholar]
- 9. Burri, P. Lung development and pulmonary angiogenesis. In: Gaultier C, Jacques B, Post M eds. Lung Development. New York, New York, USA: Oxford University Press; 1999:122-151. [Google Scholar]
- 10.Davies PS. Energy requirements for growth and development in infancy. Am J Clin Nutr. 1998;68(4):939S–943S. doi: 10.1093/ajcn/68.4.939S. [DOI] [PubMed] [Google Scholar]
- 11.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15(3):1101–1111. doi: 10.1091/mbc.e03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- 13.Kuma A, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432(7020):1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 14.Qu X, et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128(5):931–946. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 15.Mellén MA, de la Rosa EJ, Boya P. The autophagic machinery is necessary for removal of cell corpses from the developing retinal neuroepithelium. Cell Death Differ. 2008;15(8):1279–1290. doi: 10.1038/cdd.2008.40. [DOI] [PubMed] [Google Scholar]
- 16.Aburto MR, Sánchez-Calderón H, Hurlé JM, Varela-Nieto I, Magariños M. Early otic development depends on autophagy for apoptotic cell clearance and neural differentiation. Cell Death Dis. 2012;3:e394. doi: 10.1038/cddis.2012.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Komatsu M, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169(3):425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cecconi F, Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell. 2008;15(3):344–357. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12(9):823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheong H, Wu J, Gonzales LK, Guttentag SH, Thompson CB, Lindsten T. Analysis of a lung defect in autophagy-deficient mouse strains. Autophagy. 2014;10(1):45–56. doi: 10.4161/auto.26505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yeganeh B, Bilodeau C, Post M. Explant culture for studying lung development. Methods Mol Biol. 2018;1752:81–90. doi: 10.1007/978-1-4939-7714-7_8. [DOI] [PubMed] [Google Scholar]
- 22.Farkas T, Daugaard M, Jäättelä M. Identification of small molecule inhibitors of phosphatidylinositol 3-kinase and autophagy. J Biol Chem. 2011;286(45):38904–38912. doi: 10.1074/jbc.M111.269134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci U S A. 1982;79(6):1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu Y, et al. Synthesis and screening of 3-MA derivatives for autophagy inhibitors. Autophagy. 2013;9(4):595–603. doi: 10.4161/auto.23641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hardie DG, Carling D, Carlson M. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu Rev Biochem. 1998;67:821–855. doi: 10.1146/annurev.biochem.67.1.821. [DOI] [PubMed] [Google Scholar]
- 26.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Egan DF, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331(6016):456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kemp BE, et al. AMP-activated protein kinase, super metabolic regulator. Biochem Soc Trans. 2003;31(Pt 1):162–168. doi: 10.1042/bst0310162. [DOI] [PubMed] [Google Scholar]
- 29.Zhou G, et al. Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest. 2001;108(8):1167–1174. doi: 10.1172/JCI13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100(25):15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manwani N, et al. Reduced viability of mice with lung epithelial-specific knockout of glucocorticoid receptor. Am J Respir Cell Mol Biol. 2010;43(5):599–606. doi: 10.1165/rcmb.2009-0263OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perl AK, Tichelaar JW, Whitsett JA. Conditional gene expression in the respiratory epithelium of the mouse. Transgenic Res. 2002;11(1):21–29. doi: 10.1023/A:1013986627504. [DOI] [PubMed] [Google Scholar]
- 33.Emery JL, Mithal A. The number of alveoli in the terminal respiratory unit of man during late intrauterine life and childhood. Arch Dis Child. 1960;35:544–547. doi: 10.1136/adc.35.184.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kresch MJ, Christian C, Wu F, Hussain N. Ontogeny of apoptosis during lung development. Pediatr Res. 1998;43(3):426–431. doi: 10.1203/00006450-199803000-00020. [DOI] [PubMed] [Google Scholar]
- 35.Levesque BM, Vosatka RJ, Nielsen HC. Dihydrotestosterone stimulates branching morphogenesis, cell proliferation, and programmed cell death in mouse embryonic lung explants. Pediatr Res. 2000;47(4 Pt 1):481–491. doi: 10.1203/00006450-200004000-00012. [DOI] [PubMed] [Google Scholar]
- 36.Wongtrakool C, Roman J. Apoptosis of mesenchymal cells during the pseudoglandular stage of lung development affects branching morphogenesis. Exp Lung Res. 2008;34(8):481–499. doi: 10.1080/01902140802271842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan B, Li C, Kimura S, Engelhardt RT, Smith BR, Minoo P. Inhibition of distal lung morphogenesis in Nkx2.1(-/-) embryos. Dev Dyn. 2000;217(2):180–190. doi: 10.1002/(SICI)1097-0177(200002)217:2<180::AID-DVDY5>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 38.Whitsett JA, Glasser SW. Regulation of surfactant protein gene transcription. Biochim Biophys Acta. 1998;1408(2-3):303–311. doi: 10.1016/s0925-4439(98)00076-3. [DOI] [PubMed] [Google Scholar]
- 39.Minoo P, Su G, Drum H, Bringas P, Kimura S. Defects in tracheoesophageal and lung morphogenesis in Nkx2.1(-/-) mouse embryos. Dev Biol. 1999;209(1):60–71. doi: 10.1006/dbio.1999.9234. [DOI] [PubMed] [Google Scholar]
- 40.Minoo P. Transcriptional regulation of lung development: emergence of specificity. Respir Res. 2000;1(2):109–115. doi: 10.1186/rr20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou L, Lim L, Costa RH, Whitsett JA. Thyroid transcription factor-1, hepatocyte nuclear factor-3beta, surfactant protein B, C, and Clara cell secretory protein in developing mouse lung. J Histochem Cytochem. 1996;44(10):1183–1193. doi: 10.1177/44.10.8813084. [DOI] [PubMed] [Google Scholar]
- 42.Kelly SE, Bachurski CJ, Burhans MS, Glasser SW. Transcription of the lung-specific surfactant protein C gene is mediated by thyroid transcription factor 1. J Biol Chem. 1996;271(12):6881–6888. doi: 10.1074/jbc.271.12.6881. [DOI] [PubMed] [Google Scholar]
- 43.Yan C, Sever Z, Whitsett JA. Upstream enhancer activity in the human surfactant protein B gene is mediated by thyroid transcription factor 1. J Biol Chem. 1995;270(42):24852–24857. doi: 10.1074/jbc.270.42.24852. [DOI] [PubMed] [Google Scholar]
- 44.Bohinski RJ, Di Lauro R, Whitsett JA. The lung-specific surfactant protein B gene promoter is a target for thyroid transcription factor 1 and hepatocyte nuclear factor 3, indicating common factors for organ-specific gene expression along the foregut axis. Mol Cell Biol. 1994;14(9):5671–5681. doi: 10.1128/MCB.14.9.5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang L, Whitsett JA, Stripp BR. Regulation of Clara cell secretory protein gene transcription by thyroid transcription factor-1. Biochim Biophys Acta. 1997;1350(3):359–367. doi: 10.1016/s0167-4781(96)00180-7. [DOI] [PubMed] [Google Scholar]
- 46.Demling N, Ehrhardt C, Kasper M, Laue M, Knels L, Rieber EP. Promotion of cell adherence and spreading: a novel function of RAGE, the highly selective differentiation marker of human alveolar epithelial type I cells. Cell Tissue Res. 2006;323(3):475–488. doi: 10.1007/s00441-005-0069-0. [DOI] [PubMed] [Google Scholar]
- 47.Treutlein B, et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509(7500):371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Folmes CD, Terzic A. Metabolic determinants of embryonic development and stem cell fate. Reprod Fertil Dev. 2014;27(1):82–88. doi: 10.1071/RD14383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13(4):251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim KH, Lee MS. Autophagy as a crosstalk mediator of metabolic organs in regulation of energy metabolism. Rev Endocr Metab Disord. 2014;15(1):11–20. doi: 10.1007/s11154-013-9272-6. [DOI] [PubMed] [Google Scholar]
- 51.Towler MC, Hardie DG. AMP-activated protein kinase in metabolic control and insulin signaling. Circ Res. 2007;100(3):328–341. doi: 10.1161/01.RES.0000256090.42690.05. [DOI] [PubMed] [Google Scholar]
- 52.Wirawan E, et al. Beclin1: a role in membrane dynamics and beyond. Autophagy. 2012;8(1):6–17. doi: 10.4161/auto.8.1.16645. [DOI] [PubMed] [Google Scholar]
- 53.Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol. 2009;186(6):773–782. doi: 10.1083/jcb.200907014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dang TP, Eichenberger S, Gonzalez A, Olson S, Carbone DP. Constitutive activation of Notch3 inhibits terminal epithelial differentiation in lungs of transgenic mice. Oncogene. 2003;22(13):1988–1997. doi: 10.1038/sj.onc.1206230. [DOI] [PubMed] [Google Scholar]
- 55.Metzger DE, Stahlman MT, Shannon JM. Misexpression of ELF5 disrupts lung branching and inhibits epithelial differentiation. Dev Biol. 2008;320(1):149–160. doi: 10.1016/j.ydbio.2008.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Brien KB, et al. CARM1 is required for proper control of proliferation and differentiation of pulmonary epithelial cells. Development. 2010;137(13):2147–2156. doi: 10.1242/dev.037150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martis PC, Whitsett JA, Xu Y, Perl AK, Wan H, Ikegami M. C/EBPalpha is required for lung maturation at birth. Development. 2006;133(6):1155–1164. doi: 10.1242/dev.02273. [DOI] [PubMed] [Google Scholar]
- 58.Wan H, et al. Foxa2 regulates alveolarization and goblet cell hyperplasia. Development. 2004;131(4):953–964. doi: 10.1242/dev.00966. [DOI] [PubMed] [Google Scholar]
- 59.Wan H, et al. Kruppel-like factor 5 is required for perinatal lung morphogenesis and function. Development. 2008;135(15):2563–2572. doi: 10.1242/dev.021964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Tuyl M, Liu J, Wang J, Kuliszewski M, Tibboel D, Post M. Role of oxygen and vascular development in epithelial branching morphogenesis of the developing mouse lung. Am J Physiol Lung Cell Mol Physiol. 2005;288(1):L167–L178. doi: 10.1152/ajplung.00185.2004. [DOI] [PubMed] [Google Scholar]
- 61.Healy AM, Morgenthau L, Zhu X, Farber HW, Cardoso WV. VEGF is deposited in the subepithelial matrix at the leading edge of branching airways and stimulates neovascularization in the murine embryonic lung. Dev Dyn. 2000;219(3):341–352. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1061>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 62.Yamamoto H, Yun EJ, Gerber HP, Ferrara N, Whitsett JA, Vu TH. Epithelial-vascular cross talk mediated by VEGF-A and HGF signaling directs primary septae formation during distal lung morphogenesis. Dev Biol. 2007;308(1):44–53. doi: 10.1016/j.ydbio.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 63.Compernolle V, et al. Loss of HIF-2alpha and inhibition of VEGF impair fetal lung maturation, whereas treatment with VEGF prevents fatal respiratory distress in premature mice. Nat Med. 2002;8(7):702–710. doi: 10.1038/nm721. [DOI] [PubMed] [Google Scholar]
- 64.Zeng X, Wert SE, Federici R, Peters KG, Whitsett JA. VEGF enhances pulmonary vasculogenesis and disrupts lung morphogenesis in vivo. Dev Dyn. 1998;211(3):215–227. doi: 10.1002/(SICI)1097-0177(199803)211:3<215::AID-AJA3>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 65.Liang XH, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402(6762):672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 66.Decuypere JP, Parys JB, Bultynck G. Regulation of the autophagic bcl-2/beclin 1 interaction. Cells. 2012;1(3):284–312. doi: 10.3390/cells1030284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jung HS, et al. Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metab. 2008;8(4):318–324. doi: 10.1016/j.cmet.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 68.Hou W, Han J, Lu C, Goldstein LA, Rabinowich H. Autophagic degradation of active caspase-8: a crosstalk mechanism between autophagy and apoptosis. Autophagy. 2010;6(7):891–900. doi: 10.4161/auto.6.7.13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu Y, Levine B. Autosis and autophagic cell death: the dark side of autophagy. Cell Death Differ. 2015;22(3):367–376. doi: 10.1038/cdd.2014.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu F, et al. Nuclear localization of Beclin 1 promotes radiation-induced DNA damage repair independent of autophagy. Sci Rep. 2017;7:45385. doi: 10.1038/srep45385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liang XH, Yu J, Brown K, Levine B. Beclin 1 contains a leucine-rich nuclear export signal that is required for its autophagy and tumor suppressor function. Cancer Res. 2001;61(8):3443–3449. [PubMed] [Google Scholar]
- 72.Abman SH, Bancalari E, Jobe A. The evolution of bronchopulmonary dysplasia after 50 years. Am J Respir Crit Care Med. 2017;195(4):421–424. doi: 10.1164/rccm.201611-2386ED. [DOI] [PubMed] [Google Scholar]
- 73.Yeganeh B, et al. Acid sphingomyelinase inhibition attenuates cell death in mechanically ventilated newborn rat lung. Am J Respir Crit Care Med. 2019;199(6):760–772. doi: 10.1164/rccm.201803-0583OC. [DOI] [PubMed] [Google Scholar]
- 74.Sureshbabu A, et al. Inhibition of regulatory-associated protein of mechanistic target of rapamycin prevents hyperoxia-induced lung injury by enhancing autophagy and reducing apoptosis in neonatal mice. Am J Respir Cell Mol Biol. 2016;55(5):722–735. doi: 10.1165/rcmb.2015-0349OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yeganeh B, Ghavami S, Rahim MN, Klonisch T, Halayko AJ, Coombs KM. Autophagy activation is required for influenza A virus-induced apoptosis and replication. Biochim Biophys Acta Mol Cell Res. 2018;1865(2):364–378. doi: 10.1016/j.bbamcr.2017.10.014. [DOI] [PubMed] [Google Scholar]
- 76.Shojaie S, et al. Acellular lung scaffolds direct differentiation of endoderm to functional airway epithelial cells: requirement of matrix-bound HS proteoglycans. Stem Cell Reports. 2015;4(3):419–430. doi: 10.1016/j.stemcr.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Luo B, Groenke K, Takors R, Wandrey C, Oldiges M. Simultaneous determination of multiple intracellular metabolites in glycolysis, pentose phosphate pathway and tricarboxylic acid cycle by liquid chromatography-mass spectrometry. J Chromatogr A. 2007;1147(2):153–164. doi: 10.1016/j.chroma.2007.02.034. [DOI] [PubMed] [Google Scholar]
- 78.Jensen EC. Quantitative analysis of histological staining and fluorescence using ImageJ. Anat Rec (Hoboken) 2013;296(3):378–381. doi: 10.1002/ar.22641. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.