Abstract
Influenza A virus–specific (IAV-specific) T cell responses are important correlates of protection during primary and subsequent infections. The generation and maintenance of robust IAV-specific T cell responses relies on T cell interactions with dendritic cells (DCs). In this study, we explore the role of the nucleotide-binding domain leucine-rich repeat–containing receptor family member NLRC4 in modulating the DC phenotype during IAV infection. Nlrc4–/– mice had worsened survival and increased viral titers during infection, normal innate immune cell recruitment, and IAV-specific CD8+ T cell responses, but severely blunted IAV-specific CD4+ T cell responses compared with WT mice. The defect in the pulmonary IAV–specific CD4+ T cell response was not a result of defective priming or migration of these cells in Nlrc4–/– mice but was instead due to an increase in FasL+ DCs, resulting in IAV-specific CD4+ T cell death. Together, our data support a role for NLRC4 in regulating the phenotype of lung DCs during a respiratory viral infection and thereby influencing the magnitude of protective T cell responses.
Keywords: Immunology, Virology
Keywords: Adaptive immunity, Dendritic cells, Influenza
Introduction
Influenza A virus (IAV) is a respiratory pathogen responsible for seasonal epidemics that can cause severe disease and death, most commonly in the very young, very old, and immunocompromised individuals in the population (1). Vaccination strategies have traditionally prioritized antibody responses, but there is increasing evidence that IAV-specific T cells confer heterosubtypic protection that correlates with reduced symptom severity during infection (2, 3). During a primary IAV infection, CD8+ T cells are critical for viral clearance, which they accomplish by directly killing infected cells through death receptor interactions and perforin and granzyme (4, 5). IAV-specific CD4+ T cells provide help to both the CD8+ T cell and B cell responses and support the antiviral functions of innate immune cells through production of IFN-γ (6). IAV-specific Th1 cells comprise the majority of the IAV-specific CD4+ T cell response, including the recently described cytotoxic CD4+ T cells that directly kill IAV-infected cells (7). IAV-specific Tregs are also present and help to restrain tissue damage caused by exaggerated nonspecific innate immune responses and targeted cytotoxic CD8+ T cell–mediated killing of infected cells (8–11).
Antigen-presenting cells (APCs) at the site of infection are indispensable for regulating the magnitude and character of T cell responses in diverse inflammatory contexts (12). Depletion of phagocytic cells in the lungs following IAV infection results in premature apoptosis of IAV-specific T cells, due to the loss of critical survival signals normally provided by dendritic cells (DCs) (13, 14). DC interactions with T cells can also result in T cell death, which is exemplified by enhanced killing of IAV-specific CD8+ T cells by plasmacytoid DCs in the lung draining lymph nodes (dLNs) during lethal IAV infection (15–17). Induction of the appropriate phenotype in DCs depends on receipt of proinflammatory signals through cytokine and chemokine receptors and pattern recognition receptors (PRRs).
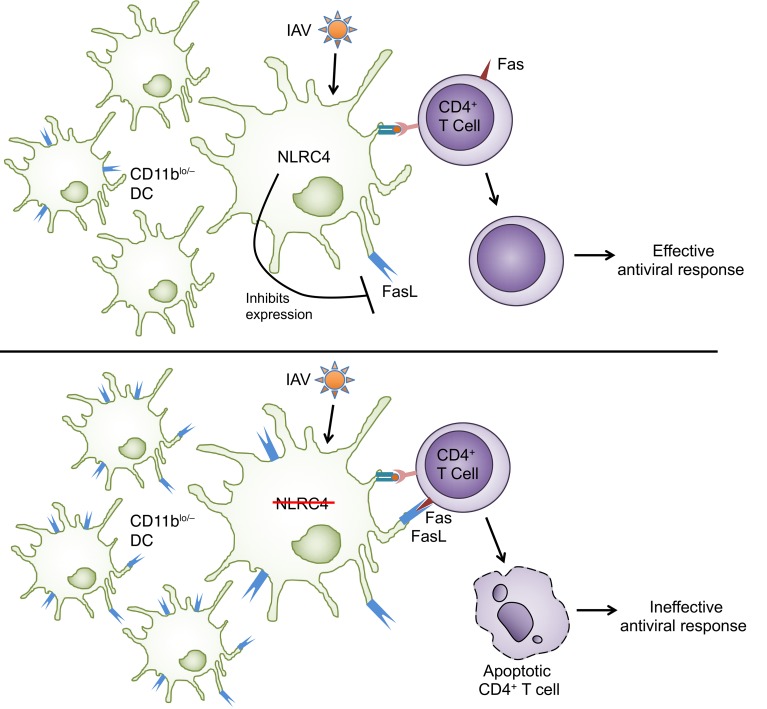
NLRC4 is an intracellular PRR belonging to the nucleotide oligomerization and binding domain and leucine-rich repeat-containing (NLR) family. NLRC4 is best described for its role as part of the multiprotein inflammasome complex that mediates processing and secretion of IL-1β and IL-18 and induces a pyroptotic cell death (18). The NLRC4 inflammasome is activated in response to infection with Gram-negative bacteria, and the resultant IL-18 production in CD8α+ DCs during infection is important for memory CD8+ T cell activation in this context (19). In a syngeneic subcutaneous melanoma model, NLRC4-deficient mice were reported to have defective intratumoral CD4+ and CD8+ T cell responses and increased tumor burdens (20). Considering the importance of T cell responses in viral infection and the evidence supporting a role for NLRC4 in direct or indirect modulation of T cell responses in these different settings, we assessed the role of NLRC4 in the host response to IAV infection. We report that Nlrc4–/– mice had decreased survival following IAV infection with an associated defective IAV-specific CD4+ T cell response. The reduction in the CD4+ T cell response in Nlrc4–/– mice was due to T cell–extrinsic signals that resulted in increased death of IAV-specific CD4+ T cells. We further showed that there was an increase in FasL+ DCs in the lungs of IAV-infected Nlrc4–/– mice and that blocking Fas-FasL interactions in vitro prevented CD4+ T cell killing by NLRC4-deficient DCs. Finally, transfer of NLRC4-deficient DCs into WT mice resulted in both the increased mortality and loss of CD4+ T cells following IAV infection seen in Nlrc4–/– mice. Together, our findings demonstrate a and critical role for NLRC4 in regulating IAV-specific CD4+ T cell responses through FasL expression on DCs.
Results
Nlrc4–/– mice have increased morbidity and mortality during IAV infection.
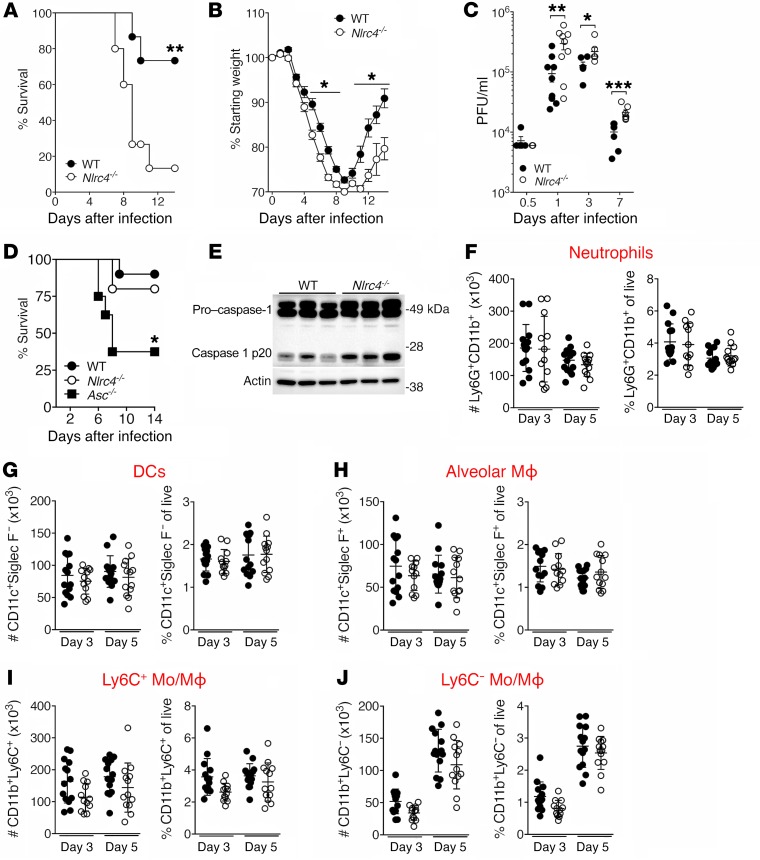
To determine the effect of NLRC4 deficiency on outcome during IAV infection, we compared the morbidity and mortality of WT and Nlrc4–/– mice following infection with IAV. We observed significantly increased morbidity and mortality among the Nlrc4–/– animals compared with WT animals (Figure 1, A and B), accompanied by increased viral titers in the lungs of Nlrc4–/– mice on days 1, 3, and 7 after infection (Figure 1C). The susceptibility of Nlrc4–/– mice to IAV was dose dependent, and infection with a 0.25 median lethal dose (LD50) inoculum of IAV resulted in similar mortality rates between WT and Nlrc4–/– mice (Figure 1D). Consistent with previous studies, Asc–/– mice had increased mortality compared with WT mice (Figure 1D) (21, 22).
Figure 1. Nlrc4–/– mice have reduced survival and viral clearance during IAV infection.
(A–E) Mice were infected with a 0.5 LD50 (A–C and E) or 0.25 LD50 (D) inoculum of IAV. Mortality (A and D) and weight loss (B) were monitored, and pulmonary viral titers (C) were quantified by plaque assay at the indicated time points after infection. (E) Caspase-1 cleavage was assessed in lungs 24 hours after infection with IAV. Each lane represents 1 mouse. (F–J) Innate immune cells in the lungs were quantified at the indicated time points after infection. In addition to the markers shown, dead cells and doublets were excluded, and then cells were gated on CD45.2 expression. Data are from 1 experiment (E, n = 3 per group and D, n = 8–10 per group), or were pooled from 2 (A and B, n = 14 per group, and C, n = 5–9 per group) or 3 (F–J, n = 12–14 per group) separate experiments. *P < 0.05, **P < 0.01, and ***P < 0.001, by Mantel-Cox test (A and D), 1-way ANOVA with Tukey’s post hoc analysis (B), and 2-tailed Student’s t test (C). Mo, monocytes; MΦ, macrophages.
NLRC4 is best known for its role as part of the NLRC4 inflammasome, which is formed upon recognition of bacterial flagellin and components of the type III secretion system by NAIP proteins (23, 24). Activation of the NLRC4 inflammasome results in cleavage of pro–caspase-1 into its active form, which in turn cleaves pro–IL-1β and pro–IL-18 into their mature secreted forms. Formation of the NLRC4 inflammasome within the lungs seemed unlikely in the context of a viral infection, and, indeed, we detected no defect in cleavage of pro–caspase-1 in lung homogenates from Nlrc4–/– mice 24 hours after infection with IAV (Figure 1E). These data suggest an inflammasome-independent role for NLRC4 in the control of IAV infection in vivo.
Nlrc4–/– mice have intact production of inflammatory mediators and innate immune cells in the lungs.
Excessive inflammation is a well-documented cause of pathology during IAV infection (25, 26). Given the increased IAV-induced mortality seen among Nlrc4–/– mice, we compared the production of inflammatory mediators in the lungs of WT and Nlrc4–/– mice following IAV infection and found no significant differences (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI124937DS1). Consistent with the similarity in the levels of innate cell chemoattractants in WT and Nlrc4–/– mice, the number of lung-infiltrating and lung-resident inflammatory cells was similar in WT and Nlrc4–/– mice on days 3 and 5 after infection (Figure 1, F–J).
Defective pulmonary IAV–specific T cell responses in Nlrc4–/– mice.
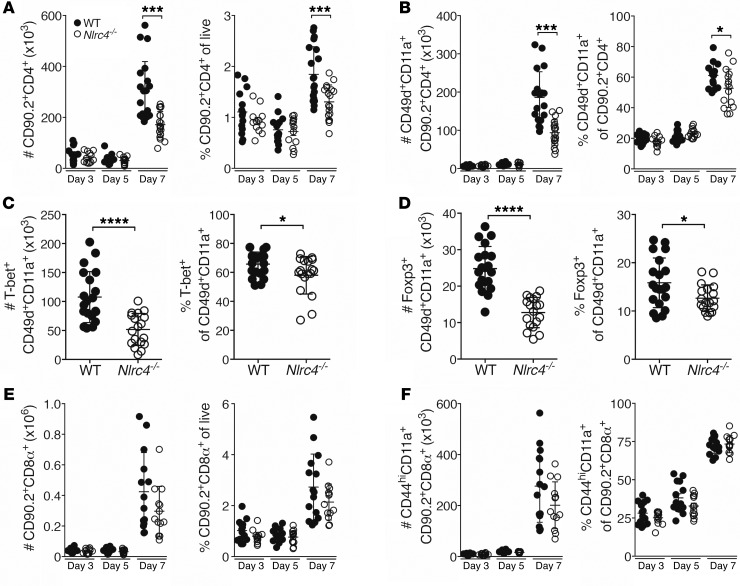
The IAV-specific CD4+ T cell response occurs primarily within the lungs and comprises a wide variety of specificities, with a relatively small proportion of the cells being specific for each antigen (27–29). While useful for the immune response, this breadth of specificities complicates measurement of the total response as the sum of all the individual antigen-specific responses. Hence, we quantified the total IAV-specific CD4+ T cell response using the surrogate markers CD49d and CD11a (30), which are highly expressed on antigen-experienced cells and, in the context of IAV infection, reflect the cells that have been exposed to IAV-specific antigen (31). We detected a significant decrease in the total and IAV-specific CD4+ T cell response in the lungs of Nlrc4–/– mice compared with responses in WT mice on day 7 after infection (Figure 2, A and B). Interestingly, the decrease in IAV-specific CD4+ T cells in Nlrc4–/– mice was not present on day 3 or day 5 after infection in the lungs, dLNs, or spleen (Figure 2, A and B, and Supplemental Figure 2, A–D). Nlrc4+/– and Nlrc4+/+ littermates had similar numbers of pulmonary IAV–specific CD4+ T cells, suggesting that the Nlrc4 gene is haplosufficient (Supplemental Figure 2, E and F).
Figure 2. Decreased IAV-specific CD4+ T cells in the lungs of Nlrc4–/– mice 7 days after infection.
(A–F) Mice were infected with a 0.5 LD50 inoculum of IAV, and lung CD4+ T cell subsets were enumerated by flow cytometry on the indicated days (A, B, E, and F) or on day 7 after infection (C and D). Data are from 3 (C and D, n = 12–16 per group) or 4 (A, B, E, and F, n = 13–20 per group) independent experiments. Error bars represent the SEM. *P < 0.05, ***P < 0.001, and ****P < 0.0001, by 2-tailed Student’s t test.
The IAV-specific CD4+ T cell response is predominantly a Th1-polarized response with a smaller number of IAV-specific Tregs, which are protective during a primary infection (7, 32, 33). Consistent with the significantly decreased number of total IAV-specific CD4+ T cells in Nlrc4–/– mice, we found significant decreases in IAV-specific Th1 (Tbet+) cells and Tregs (Foxp3+) (Figure 2, C and D).
The IAV-specific CD8+ T cell response is crucial for viral clearance during a primary IAV infection, thus we quantified this response in WT and Nlrc4–/– mice (34). We detected small decreases in the frequency and number of total lung CD8+ T cells that did not rise to the level of statistical significance (Figure 2E), and no significant difference in the frequency or total number of antigen-experienced CD8+ T cells in the lungs was observed (Figure 2F).
Together, these data reveal a defect in the pulmonary IAV–specific CD4+ T cell response in Nlrc4–/– mice. IAV-specific CD4+ T cell responses are important for successful viral clearance and recovery from IAV infection (32, 35), thus these defects probably contribute to the enhanced mortality evident in Nlrc4–/– mice.
DC and T cell accumulation in lung dLNs is intact in Nlrc4–/– mice.
To identify the cause of the blunted IAV-specific CD4+ T cell response, we evaluated key steps in the generation of this response. Initiation of robust IAV-specific T cell responses relies on successful interactions in the secondary lymphoid organs with antigen-bearing APCs, many of which have migrated from the lungs (6). Accelerated respiratory DC migration to the lung dLNs lasts for about 48 hours after IAV infection (36). Twenty-four hours after infection, we noted no significant difference in the proportion of CD11c+ cells originating from the lungs in the lung dLNs of WT or Nlrc4–/– mice (Supplemental Figure 3, A and B), indicating that defective DC migration was not likely to be contributing to the blunted IAV-specific CD4+ T cell response in Nlrc4–/– mice. Further characterization of the DC phenotype in lung dLNs revealed no differences in the abundance or expression of costimulatory molecules on DCs in WT or Nlrc4–/– mice (Supplemental Figure 3, C–H). These findings are consistent with the normal accumulation of total and antigen-experienced CD4+ T cells in the lung dLNs 3 and 5 days after infection (Supplemental Figure 2, C and D). Considered together, these data argue that a defect in activation and expansion of T cells in the dLNs or spleen was not the cause of the blunted CD4+ T cell response in the lungs of Nlrc4–/– mice.
Increased T cell death in the lungs of Nlrc4–/– mice following IAV infection.
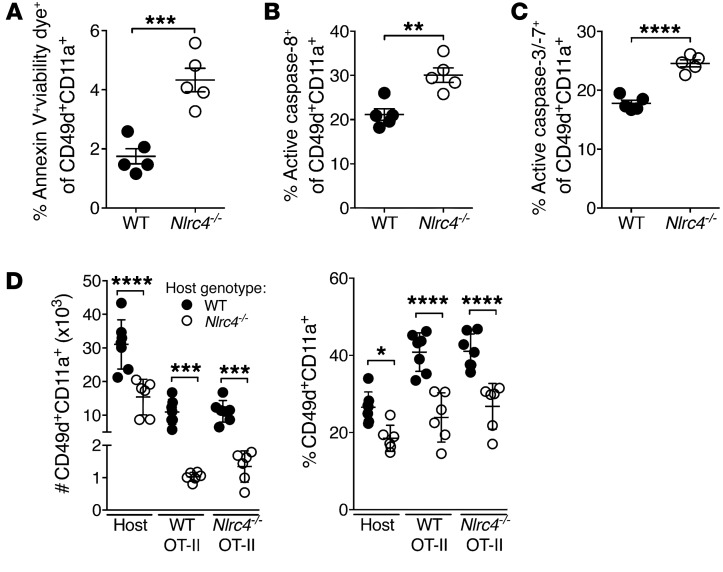
Given that early DC-dependent events in the development of the IAV-specific CD4+ T cell response appeared to proceed normally in Nlrc4–/– mice, we assessed whether the decrease in CD4+ T cells was due to increased death among these cells. Annexin V and viability staining showed increased dead or dying IAV-specific CD4+ T cells in the lungs of Nlrc4–/– mice compared with that seen in WT mice (Figure 3A). Consistently, IAV-specific CD4+ T cells from the lungs of Nlrc4–/– mice had more active caspase-3/-7 and active caspase-8 as measured with a fluorescent inhibitor probe 7 days after infection compared with expression in CD4+ T cells from WT mice (Figure 3, B and C). These data suggest that increased death among IAV-specific CD4+ T cells may be driving their diminished presence in Nlrc4–/– lungs.
Figure 3. Increased death of IAV-specific CD4+ T cells in the lungs of Nlrc4–/– mice 7 days after infection.
(A–C) Mice were infected with a 0.5 LD50 inoculum of IAV, and cells staining positive for the indicated markers and dyes were enumerated in the lungs on day 7 after infection by flow cytometry. (D) CD90.2+ WT and Nlrc4–/– hosts received 1 × 105 naive CD90.1/2+ WT and 1 × 105 CD90.1+ Nlrc4–/– OT-II cells i.v., followed 1 day later by infection with a 0.5 LD50 inoculum of IAV expressing OVA323–339. Seven days after infection, the indicated cells were quantified in the lungs. Data are representative of 2 independent experiments (n = 5–6 per group). Error bars represent the SEM. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001, by 2-tailed Student’s t test.
Increased IAV-specific CD4+ T cell death in Nlrc4–/– mice is T cell extrinsic.
To determine whether the cause of death in Nlrc4–/– T cells was a defect intrinsic or extrinsic to the Nlrc4–/– T cells themselves, we set up a side-by-side comparison of WT and Nlrc4–/– T cells in WT and Nlrc4–/– hosts. We adoptively transferred both WT (CD90.1/2+) and Nlrc4–/– (CD90.1+) OT-II CD4+ T cells, specific for the amino acids 323–339 of chicken OVA (OVA323–339), intravenously into WT and Nlrc4–/– recipients (CD90.2+). One day later, we infected mice with IAV expressing the OVA323–339 epitope and quantified the IAV-specific CD4+ T cell response in the lungs 7 days after infection. We observed significantly more OT-II cells with an antigen-experienced phenotype in the lungs of WT hosts than in Nlrc4–/– hosts, regardless of the OT-II donor genotype (Figure 3D), indicating that the cause of T cell death in the Nlrc4–/– mice was T cell extrinsic.
Increased FasL+ DCs in IAV-infected Nlrc4–/– lungs.
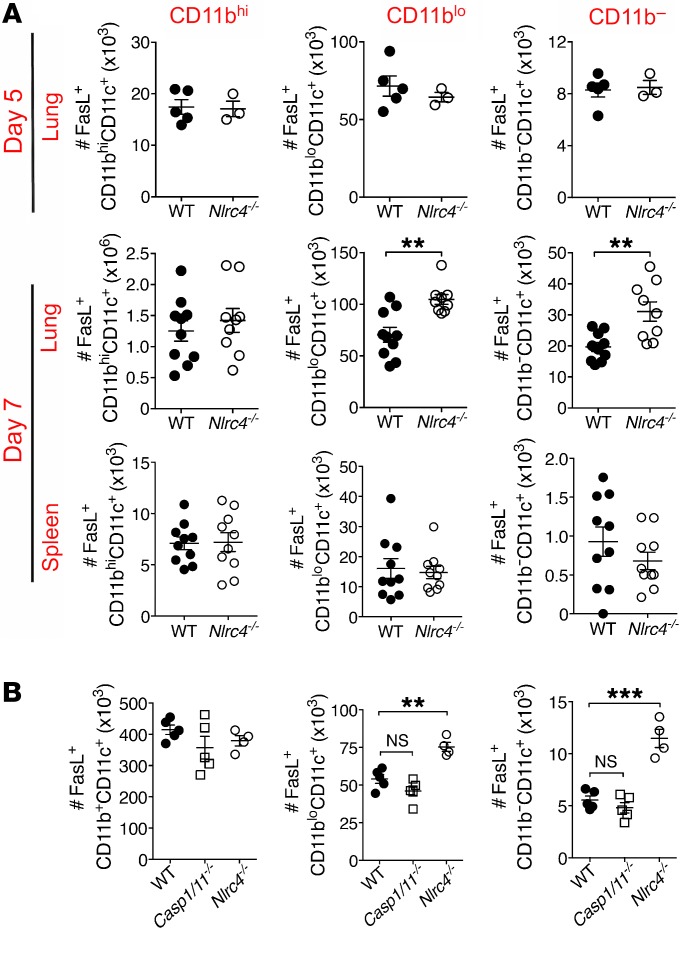
Blunting of T cell responses by FasL+ DCs occurs in diverse inflammatory contexts (15, 16, 37, 38), thus we explored DC expression of FasL as a possible cause of the increased T cell death. We found that there were more FasL+ DCs (Siglec F–CD11c+) in the lungs, but not spleens, of Nlrc4–/– mice 7 days after infection (Figure 4A and Supplemental Figure 4, A and B) specifically within the CD11blo and CD11b– populations of DCs. The increase in FasL+ DCs matched the timing and location of the decrease in IAV-specific CD4+ T cells, as there were no differences in FasL+ DCs on day 5 after infection in the lungs of WT and Nlrc4–/– mice (Figure 4A and Supplemental Figure 4B). The geometric mean fluorescence intensity (GMFI) for FasL was not different between DCs from WT and Nlrc4–/– mice (Supplemental Figure 4C), consistent with more cells expressing FasL rather than a higher expression level per cell.
Figure 4. Increased FasL+ DCs in the lungs of Nlrc4–/– mice during IAV infection.
(A and B) Mice were infected with a 0.5 LD50 inoculum of IAV, and FasL+ DCs were enumerated by flow cytometry for the indicated organ and time point after infection (A) or in the lung on day 7 after infection (B). Data are from 1 experiment (A, day 5, and B, n = 3–5 per group) or 2 separate experiments (A, day 7, n = 9–10 per group). Error bars show the SEM. **P < 0.01 and ***P < 0.001, by 2-tailed Student’s t test.
Consistent with the normal activation of caspase-1 in lungs from Nlrc4–/– mice following IAV infection, we observed no increase in FasL+CD11blo or FasL+CD11b– DCs in the lungs of Casp1/11–/– mice 7 days after infection (Figure 4B and Supplemental Figure 5A). We also found no difference in the proportion of Fas+ IAV-specific CD4+ T cells in the lungs of WT or Nlrc4–/– mice, in agreement with the data indicating a T cell–extrinsic cause of death (Supplemental Figure 5B).
To determine whether DCs from IAV-infected Nlrc4–/– lungs were killing CD4+ or CD8+ T cells, we modified a previously described DC–T cell coculture assay (15). Seven days after infection, we isolated lung DCs and CD4+ and CD8+ T cells. Lung CD4+ and CD8+ T cell survival was measured following coculture with isolated WT or Nlrc4–/– CD11bhi, CD11blo, or CD11b– lung DCs for 12 hours. We noted similar survival rates of CD4+ T cells following culture alone or following coculture with Nlrc4–/– and WT CD11bhi DCs from the lungs, which had equivalent expression levels of FasL (Figure 5A). However, significantly fewer CD4+ T cells survived culture with CD11blo or CD11b– lung DCs from Nlrc4–/– mice, both of which contain more FasL+ cells than did the corresponding DC populations from WT lungs (Figure 5A). Additionally, we observed no difference in lung CD4+ T cell survival following coculture with splenic DCs isolated from IAV-infected WT and Nlrc4–/– mice, which have similar FasL expression levels (Supplemental Figure 5C). To test whether the increased killing of CD4+ T cells by Nlrc4–/– DCs was dependent on Fas-FasL interactions, we blocked this interaction by adding Fas-Fc to the cocultures. Addition of Fas-Fc to cocultures increased the survival of IAV-specific lung CD4+ T cells cultured with WT and Nlrc4–/– lung DCs to a similar degree (Figure 5A) but had no effect on CD4+ T cells cultured alone (Supplemental Figure 5D), indicating that DC FasL is responsible for killing CD4+ T cells. Consistent with the finding that CD8+ T cell numbers in the lungs of IAV-infected mice were not significantly diminished in the absence of NLRC4 (Figure 2, E and F), we observed similar survival rates of CD8+ T cells following coculture with Nlrc4–/– and WT CD11bhi, CD11blo, or CD11b– lung DCs (Figure 5B).
Figure 5. Increased FasL-mediated killing of CD4+, but not CD8+, T cells by Nlrc4–/– DCs during IAV infection.
Mice were infected with a 0.5 LD50 inoculum of IAV or left uninfected (naive). (A and B) Pulmonary DCs and T cells were purified on day 7 after infection. Pooled WT plus Nlrc4–/– T cells were incubated with the indicated populations of DCs for 12 hours with (+ Fas-Fc) or without (No trx) 2.5 μg/ml Fas-Fc. Live CD4+ and CD8+ T cells (annexin V– viability dye–) were enumerated by flow cytometry. The proportion of live CD4+ and CD8+ T cells cocultured with DCs was normalized to CD4+ and CD8+ T cells cultured alone. (C–G) Five days after infection, WT mice received 5 × 105 WT or Nlrc4–/– BMDCs intranasally, and survival was assessed. (C) Pulmonary CD4+ and CD8+ T cells were quantified on day 7 after infection (C–G). Data are from 1 experiment (B, n = 4 per group) or 2 separate experiments (A, n = 8 per group, and C–G, n = 10 per group). Error bars represent the SEM. *P < 0.05 and **P < 0.01, by 2-tailed Student’s t test (A, D, and E) and Mantel-Cox test (C).
To confirm that the enhanced IAV-induced mortality seen in Nlrc4–/– mice was due to killing of CD4+ T cells by FasL+ DCs, 5 days after infection, WT mice received an intranasal transfer of WT or Nlrc4–/– bone marrow–derived DCs (BMDCs). Like CD11bloCD11c+ and CD11b–CD11c+ lung DCs (Figure 4A), we observed increased FasL expression on BMDCs from Nlrc4–/– mice (Supplemental Figure 6A). Although 40% of the WT mice that received WT BMDCs survived to post-infection day 14, the WT mice that received Nlrc4–/– BMDCs had significantly greater mortality rates (Figure 5C). On day 7 after infection, we enumerated pulmonary CD4+ and CD8+ T cells for a subset of infected WT mice that received WT or Nlrc4–/– BMDCs. We observed a marked loss of total and IAV-specific CD4+ T cells in the WT hosts that received Nlrc4–/– BMDCs in comparison with the WT hosts treated with WT BMDCs (Figure 5, D and E, and Supplemental Figure 6, B and C). In contrast, no difference in IAV-specific CD8+ T cells was observed in mice that received WT or Nlrc4–/– BMDCs (Figure 5, F and G, and Supplemental Figure 6, D and E).
Decreased Akt1 and FoxO3a phosphorylation in NLRC4-deficient BMDCs.
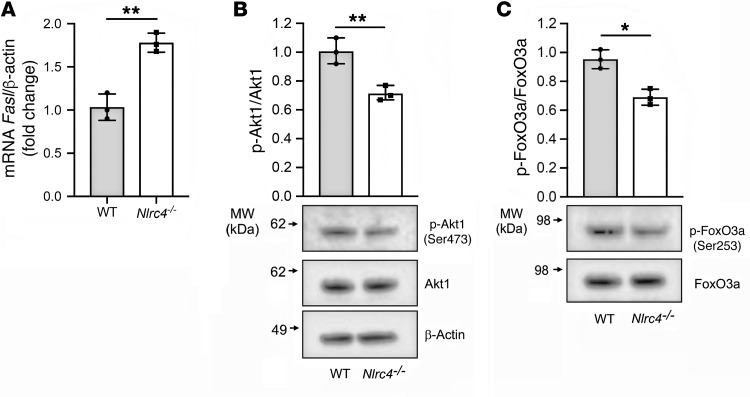
To determine how NLRC4 deficiency regulated FasL expression, we examined mRNA expression of Fasl in WT and Nlrc4–/– BMDCs by quantitative real-time PCR. We found that Nlrc4–/– BMDCs expressed higher levels of Fasl than did WT BMDCs (Figure 6A). To confirm these findings, we assessed FasL on BMDCs from an independently generated Nlrc4–/– mouse line (39). We again observed increased FasL mRNA and protein expression levels in Nlrc4–/– BMDCs compared with levels in WT BMDCs (Supplemental Figure 6, F and G). Consistent with inflammasome-independent regulation of FasL expression, we did not observe any significant difference in FasL expression in Asc–/– or Casp1/11–/– BMDCs compared with WT BMDCs (Supplemental Figure 6, H and I).
Figure 6. NLRC4 regulates FasL expression at the transcriptional level in BMDCs.
(A) Fasl mRNA expression was assessed in WT and Nlrc4–/– BMDCs by quantitative real-time PCR and normalized to β-actin. (B and C) p-Akt1 and p-FoxO3a protein levels in whole-cell lysates of BMDCs from WT and Nlrc4–/– mice were analyzed by immunoblotting. Densitometric analysis was performed with Bio-Rad Image Lab software (version 5.2.1). Total FoxO3a, Akt1, and β-actin were used as loading controls. Data were pooled from 3 independent experiments. *P < 0.05 and **P < 0.01, by 2-tailed Student’s t test.
FoxO3a, a member of the Forkhead family of transcription factors, has been implicated in the regulation of FasL expression. Furthermore, phosphorylation of FoxO3a by the serine/threonine kinase Akt1 prevents FoxO3a-dependent transcription by inhibiting its translocation to the nucleus and preventing it from activating its target genes (40). We observed that BMDCs from Nlrc4–/– mice had diminished Akt1 and FoxO3a phosphorylation compared with WT BMDCs (Figure 6, B and C), suggesting that NLRC4 may regulate FasL expression through Akt1 and FoxO3a phosphorylation.
Discussion
The data presented here show for the first time to our knowledge that Nlrc4–/– mice have a defective immune response during IAV infection. Nlrc4–/– mice show increased morbidity and mortality and impaired viral clearance, but these do not appear to be the result of a defect in cytokine/chemokine production or the innate immune response. Instead, we report a blunted IAV-specific CD4+ T cell response in the lungs of Nlrc4–/– mice. The number of pulmonary IAV-specific CD4+ T cells was dramatically decreased, but CD8+ T cells were largely unaffected. Ultimately, the loss of IAV-specific CD4+ T cells in Nlrc4–/– mice was a result of increased CD4+ T cell death due to Fas-FasL–mediated killing by CD11blo and CD11b– DCs in the lungs.
The increased susceptibility of Nlrc4–/– mice to IAV infection appears to be NLRC4 inflammasome independent, as we observed no changes in the inflammasome-dependent cytokine IL-1β or caspase-1 cleavage in Nlrc4–/– lungs during infection. The NLRP3 and AIM2 inflammasomes are activated during IAV infection (21, 22, 41), thus the caspase-1 activation and IL-1β secretion we observed are expected, given the activity of those inflammasomes. Interestingly, the level of IL-1β was slightly, but not significantly, increased in Nlrc4–/– lungs, as determined by ELISA. This result may indicate increased activation of the NLRP3 or AIM2 inflammasomes, which could be due to the elevated viral titers and therefore more abundant activating signals in Nlrc4–/– mice.
Previous studies focused on the NLRP3 inflammasome showed no effect of NLRC4 deficiency on morbidity or mortality following IAV infection (21, 22). We find these data intriguing in light of the very clear survival defect in Nlrc4–/– mice we report here. The reason for the differences in these studies is probably multifactorial. When we infected mice with a lower inoculum of IAV, we no longer observed an increase in mortality among the Nlrc4–/– mice compared with WT mice, however the Asc–/– mice were more susceptible, even at this lower inoculum, consistent with previously published findings (Figure 1D) (21, 22). Additionally, since those studies were published, we have developed an increased appreciation for the impact that genetic background has on knockout mice. We postulate that differences in the WT substrains used as controls and for backcrossing with Nlrc4–/– mice may further contribute to the discrepancy between the results presented here and previous reports. Of particular note, we and others have recently shown differences between C57BL/6J and C57BL/6N substrains in survival and inflammatory responses to IAV (31, 42). We and others have also reported striking differences in immune responses to additional inflammatory challenges among C57BL/6 substrains (42–44). In the present study, we rigorously tested the specificity of our phenotype to a loss of NLRC4 through the use of littermate controls and by confirmation in an independently generated Nlrc4–/– mouse (Supplemental Figure 2, E and F, and Supplemental Figure 6, F and G).
NLRC4 was recently reported by our laboratory to have an inflammasome-independent role in limiting melanoma progression (20). Although IAV infection and melanoma are considerably different challenges for the host immune system, in both cases, a robust host T cell response is a correlate of protection. During both IAV infection and melanoma challenge, Nlrc4–/– mice exhibit defective T cell responses, and in both models, myeloid cell dysfunction seems to be driving the defect (20). Intriguingly, it was recently shown that FASLG is highly expressed in a large number of human cancers and that engineering tumor-specific T cells to resist Fas-mediated death is a promising strategy for enhancing immunotherapy during cancer (45). It would be of interest to determine whether there is NLRC4-mediated modulation of myeloid cell FasL during melanoma progression and whether the ineffective melanoma-specific T cell responses observed in Nlrc4–/– mice could be rescued by disrupting Fas-FasL interactions or downstream signaling.
An important remaining question to answer is how NLRC4 is acting in these myeloid cells to influence FasL expression. Offering a single explanation that accounts for all of the observations is challenging because of the dynamic nature of immune responses and their regulatory mechanisms, but we speculate that NLRC4 may be involved in differentiation or activation of myeloid cells during an inflammatory insult. That unstimulated Nlrc4–/– BMDCs, but not Casp1/11–/– or Asc–/– BMDCs, have increased FasL mRNA and protein levels is suggestive of an inflammasome-independent function of NLRC4 in either the development and/or differentiation of DCs. Fasl gene expression is controlled by a number of distinct transcription factor interactions at the Fasl promoter (46). FoxO3a can be phosphorylated by Akt1, resulting in its inactivation as a transcription factor. Conversely, dephosphorylation of FoxO3a results in upregulation of FasL and triggers increased apoptosis (40). In the absence of NLRC4, we observed diminished Akt1 and FoxO3a phosphorylation, which may have been driving increased FasL expression. As many signaling pathways involved in metabolism, cytokine sensing, and pathogen-associated molecular pattern (PAMP) and damage-associated molecular pattern (DAMP) sensing converge on Akt (47), it is possible that dysregulated activating signals in the absence of NLRC4 result in increased FasL expression.
In conclusion, the work described here demonstrates a protective role for NLRC4 during IAV infection. Protection is probably NLRC4 inflammasome independent and involved support of the IAV-specific CD4+ T cell response in IAV-infected lungs. In the absence of NLRC4, expression of FasL on CD11blo and CD11b– lung DCs was increased, triggering more CD4+ T cell death in the lungs of IAV-infected animals and the associated increased mortality. These findings are of importance, as they expand our understanding of how the IAV-specific CD4+ T cell response in the lungs is regulated and implicate DC NLRC4 in the regulation of CD4+ T cell responses.
Methods
Mice.
The generation of Nlrc4–/–, Asc–/–, and Casp1/11–/– mice has been described elsewhere (48–50). Mice were backcrossed with C57BL/6N mice for at least 10 generations and maintained in a specific pathogen–free (SPF) facility. C57BL/6N mice were purchased from Charles River Laboratories and used as WT controls unless otherwise stated; B6.Cg-Tg(TcraTcrb)425Cbn/J (OT-II CD90.2+) mice were purchased from The Jackson Laboratory. Femurs from Nlrc4–/– mice (39) were a gift from Matam Vijay-Kumar (University of Toledo, Toledo, Ohio, USA). Both male and female mice (6–12 weeks of age) were used, however, mice were sex, age, and weight matched for individual experiments.
Virus and in vivo infection.
The mouse-adapted IAV strain A/PR/8/34 (PR/8) was propagated as previously described (16). Recombinant IAV-OT-II was created using standard reverse genetics as previously described (51) and grown in 10-day-old embryonated chicken eggs (Charles River Laboratories). The OT-II epitope (Ova323–339) was inserted into the mRNA nucleotide position 186 encoding the neuraminidase stalk region, which is known to tolerate such insertions (52). Mice were anesthetized with ketamine and xylazine and infected intranasally with 0.5 LD50 virus diluted in 50 μl sterile DMEM. Weight was monitored daily, and the mice were euthanized upon loss of 30% of their starting weight. CFSE labeling of lung cells followed by IAV infection were performed as described previously (36), but CFSE was administered 15 minutes prior to infection.
Lung titers.
To measure virus titers, lungs were homogenized using a Tissue-Tearor (BioSpec), and homogenates were then clarified by centrifugation, snap-frozen in liquid nitrogen, and stored at –80°C. A standard plaque assay on Madin-Darby canine kidney (MDCK) cells was subsequently used to quantify infectious virus (53).
ELISA.
Cytokines and chemokines were quantified in cell culture supernatants and lung homogenate supernatants using DuoSet Mouse ELISA kits from R&D Systems (for CXCL1, CXCL2, CXCL5, CXCL9, CXCL10, IL-1β, CCL2, and CCL5) or ReadySetGo! Mouse ELISA kits from eBioscience (for IL-1α, IL-6, IL-10, and TNF-α) following the manufacturers’ instructions.
Flow cytometry.
Single-cell suspensions were prepared by pressing tissues through wire mesh screens (lungs) or dissociating between frosted ends of glass slides (spleens and LNs). For some experiments, cells were minced and digested in Iscove’s DMEM (Gibco, Thermo Fisher Scientific) containing 1 mg/ml collagenase XI (MilliporeSigma) and 0.02 mg/ml DNase (MilliporeSigma) for 15 minutes at 37°C prior to physical dissociation. Live cells were enumerated by trypan blue exclusion. Cells (1 × 106 cells/well) on a 96-well plate (Corning) were blocked with 2% normal rat serum (The Jackson Laboratory) and anti–mouse CD16 and anti–mouse CD32 (clone 2.4G2, Tonbo Biosciences) in FACS buffer (1× PBS, 2% heat-inactivated FCS [HI-FCS, Atlanta Biologicals]) for 30 minutes at 4°C. Following blocking, cells were stained in FACS buffer with fluorochrome-conjugated antibodies in the dark for 30 minutes at 4°C. Cells were then fixed in FACS lysis buffer (BD) according to the manufacturer’s instructions and resuspended in PBS. For transcription factor staining, cells were fixed, permeabilized, and stained using an eBioscience Transcription Factor Staining Kit according to the manufacturer’s instructions. The following fluorochrome-conjugated antibodies were used: CD4 (clone GK1.5), CD8a (53-6.7), CD11a (M17/4), CD11b (M1/70), CD44 (IM7), CD45.2 (clone 104), CD49d (R1-2), CD80 (16-10A1), CD86 (GL-1), CD90.1 (HIS51), CD90.2 (30-H12), CD95 (15A7), CD178 (MFL3), T-bet (4B10), Foxp3 (FJK16s), Ly6G (1A8), Ly6C (HK1.4 and AL-21), and I-A/I-E (M5/114.15.2) from BioLegend; CD4 (RM4-5) from eBioscience; and Siglec F (E40-2440) and CD11c (HL3) from BD Biosciences. For some experiments, a fixable viability dye (eBioscience, catalog 65-0865) and annexin V (eBioscience, catalog BMS306APC-20) were used according to the manufacturer’s instructions. For detection of active caspases, Vybrant FAM Caspase-3/7 and Caspase-8 Assay kits (Thermo Fisher Scientific, catalogs V35118 and V35119) were used according to the manufacturer’s instructions. Data were acquired on a BD LSR II and analyzed with FlowJo software. Live cells were sorted for in vitro culture experiments on a BD FACSAria II.
DC–T cell coculture.
The DC–T cell coculture assay was adapted from a previously described assay (15). Live CD11bhi/lo/– DCs (CD11c+Siglec F–) and IAV-specific CD4+ T cells (CD90.2+CD4+CD49d+CD11a+) were stained and sorted from lungs or spleens of mice 7 days after infection, as above. DCs (1 × 104) and IAV-specific CD4+ T cells (1 × 104) or CD4+ T cells (2 × 104) alone per well of a 96-well plate were incubated in Iscove’s DMEM (Gibco, Thermo Fisher Scientific) containing 10% HI-FCS (Atlanta Biologicals), 1× β-mercaptoethanol (Gibco, Thermo Fisher Scientific), 1× penicillin-streptomycin (Gibco, Thermo Fisher Scientific), 1× l-glutamine (Gibco, Thermo Fisher Scientific), and 1× sodium pyruvate (Gibco, Thermo Fisher Scientific). For some experiments, 2.5 μg/ml Fas-Fc (R&D Systems) was included. Cells were incubated for 12 hours at 37°C and 5% CO2 and then assessed by flow cytometry, as above.
Isolation of CD4+ T cells.
For adoptive transfer experiments, splenic naive CD4+ T cells were purified by magnetic separation using a negative selection kit (STEMCELL Technologies). Purified cells were suspended in sterile DMEM, and 1 × 105 WT and 1 × 105 Nlrc4–/– cells were injected intravenously 24 hours prior to IAV infection.
Western blotting.
Lysates were prepared in RIPA buffer (Cell Signaling Technology) with 1 mM PMSF according to the manufacturer’s instructions. Lungs were homogenized in RIPA buffer using a Tissue-Tearor (BioSpec Products). A bicinchoninic acid (BCA) assay (Thermo Fisher Scientific) was performed to measure total protein in lung homogenates, and then samples were diluted to the same concentration in RIPA buffer. Lysates were stored at –80°C. Proteins were separated on a NuPAGE gel (Invitrogen, Thermo Fisher Scientific) and transferred onto a PVDF membrane using the XCell II blotting system (Invitrogen, Thermo Fisher Scientific). Membranes were blocked with 5% nonfat milk or BSA and incubated with anti–caspase-1 (p20) antibody (1:1000, AG-20B-0042-C100, Adipogen), β-actin (1:2000, sc-47778, Santa Cruz Biotechnology), phosphorylated Akt1 (p-Akt1) (1:1000, CST 9018, Cell Signaling Technology), Akt1 (1:1000, CST 75692, Cell Signaling Technology), p-FoxO3a (1:1000, CST 9466), or FoxO3a (1:1000, CST 12829, Cell Signaling Technology) overnight at 4°C. Following washing, the membranes were incubated with HRP-tagged anti–mouse IgG (1706516, Bio-Rad) or anti–rabbit IgG (NA934, GE Healthcare) and developed using SuperSignal West Pico or Femto substrate (Thermo Fisher Scientific).
Real-time PCR.
Total RNA was isolated from BMDCs using a RNeasy Mini Kit (QIAGEN), and cDNA was generated with PrimeScript RT Master Mix (Takara Bio) according to the manufacturer’s instructions. Real-time PCR was carried out using the CFX Real-Time PCR System (Bio-Rad) with a SYBR Green molecular probe (Applied Biosystems). mRNA levels were quantitatively analyzed using Bio-Rad CFX Manager 3.1 software. Relative mRNA expression levels were normalized to the housekeeping gene Actb. The primers used were: FasL, forward, 5′-CCTGTGTCACCACTACCACC-3′, reverse, 5′-CCACCGGTAGCCACAGATTT-3′; β-actin, forward, 5′-CGAGGTATCCTGACCCTGAA-3′, reverse, 5′-GGTGTGGTGCCAGATCTTCT-3′.
DC adoptive transfer.
BMDCs were differentiated as previously described (54). BMDCs were analyzed by flow cytometry and/or 5 × 105 BMDCs in 50 μl sterile DMEM were administered intranasally to IAV-infected mice 5 days after infection.
Statistics.
Statistical tests used to determine significance are indicated in the figure legends. Error bars represent the mean ± SEM. Statistical significance was based on the Mantel-Cox test for Figure 1, A and D, and Figure 5C; 1-way ANOVA with Tukey’s post hoc analysis for Figure 1B; and a 2-tailed Student’s t test for all remaining data. P values of 0.05 or less were considered statistically significant. Data were graphed and statistical tests performed using GraphPad Prism (GraphPad Software).
Study approval.
All animal studies were approved by and performed according to the guidelines of the IACUC of the University of Iowa and Cedars-Sinai Medical Center.
Author contributions
EEH, FSS, and SLC conceptualized the project. EEH, KLL, FSS, and SLC developed the methodology. EEH and JD performed the experiments with assistance and/or reagents from ZRZ, AMM, RAL, and KLL. EEH, JD, FSS, and SLC performed formal analysis. EEH, FSS, and SLC wrote the original draft of the manuscript. EEH, JD, ZRZ, AMM, KLL, GAB, PC, FSS, and SLC reviewed and edited the manuscript. FSS and SLC acquired funding. GAB, FSS, and SLC supervised the project.
Supplementary Material
Acknowledgments
We are grateful to members of the Inflammation Program, Eric Elliott, Mary Wilson, Diogo Valadares, and Bruce Hostager (University of Iowa, Iowa City) for valuable discussions and technical assistance. We thank Beng San Yeoh and Matam Vijay-Kumar (University of Toledo) for providing Nlrc4–/– femurs. We thank members of the University of Iowa Flow Cytometry Facility, a Carver College of Medicine/Holden Comprehensive Cancer Center core research facility, for their expertise. NIH grants R01 AI118719 (to FSS), R01 AI104706 (to SLC), and T32 AI007485 (to EEH and AMM) and a grant from the Harry J. Lloyd Charitable Trust (to FSS) supported this work. This material is the result of work supported in part with resources and the use of facilities at the Iowa City Veterans Administration Medical Center. The contents of this report do not represent the views of the Veterans Administration or the United States Government.
Version 1. 04/30/2019
In-Press Preview
Version 2. 06/04/2019
Electronic publication
Version 3. 07/01/2019
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(7):2888–2897.https://doi.org/10.1172/JCI124937.
Contributor Information
Emma E. Hornick, Email: emma-hornick@uiowa.edu.
Jargalsaikhan Dagvadorj, Email: jargalsaikhan.dagvadorj@cshs.org.
Zeb R. Zacharias, Email: zeb-zacharias@uiowa.edu.
Ann M. Miller, Email: ann-janowski@uiowa.edu.
Ryan A. Langlois, Email: langlois@umn.edu.
Peter Chen, Email: peter.chen@cshs.org.
Kevin L. Legge, Email: kevin-legge@uiowa.edu.
Gail A. Bishop, Email: gail-bishop@uiowa.edu.
Fayyaz S. Sutterwala, Email: Fayyaz.Sutterwala@cshs.org.
Suzanne L. Cassel, Email: suzanne.cassel@cshs.org.
References
- 1.Thompson WW, et al. Influenza-associated hospitalizations in the United States. JAMA. 2004;292(11):1333–1340. doi: 10.1001/jama.292.11.1333. [DOI] [PubMed] [Google Scholar]
- 2.Wilkinson TM, et al. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med. 2012;18(2):274–280. doi: 10.1038/nm.2612. [DOI] [PubMed] [Google Scholar]
- 3.Sridhar S, et al. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat Med. 2013;19(10):1305–1312. doi: 10.1038/nm.3350. [DOI] [PubMed] [Google Scholar]
- 4.Topham DJ, Tripp RA, Doherty PC. CD8+ T cells clear influenza virus by perforin or Fas-dependent processes. J Immunol. 1997;159(11):5197–5200. [PubMed] [Google Scholar]
- 5.Brincks EL, Katewa A, Kucaba TA, Griffith TS, Legge KL. CD8 T cells utilize TRAIL to control influenza virus infection. J Immunol. 2008;181(7):4918–4925. doi: 10.4049/jimmunol.181.7.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun J, Braciale TJ. Role of T cell immunity in recovery from influenza virus infection. Curr Opin Virol. 2013;3(4):425–429. doi: 10.1016/j.coviro.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown DM, Lee S, Garcia-Hernandez Mde L, Swain SL. Multifunctional CD4 cells expressing gamma interferon and perforin mediate protection against lethal influenza virus infection. J Virol. 2012;86(12):6792–6803. doi: 10.1128/JVI.07172-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cardani A, Boulton A, Kim TS, Braciale TJ. Alveolar macrophages prevent lethal influenza pneumonia by inhibiting infection of type-1 alveolar epithelial cells. PLoS Pathog. 2017;13(1):e1006140. doi: 10.1371/journal.ppat.1006140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brandes M, Klauschen F, Kuchen S, Germain RN. A systems analysis identifies a feedforward inflammatory circuit leading to lethal influenza infection. Cell. 2013;154(1):197–212. doi: 10.1016/j.cell.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antunes I, Kassiotis G. Suppression of innate immune pathology by regulatory T cells during Influenza A virus infection of immunodeficient mice. J Virol. 2010;84(24):12564–12575. doi: 10.1128/JVI.01559-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mock JR, et al. Foxp3+ regulatory T cells promote lung epithelial proliferation. Mucosal Immunol. 2014;7(6):1440–1451. doi: 10.1038/mi.2014.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hemann EA, Legge KL. Peripheral regulation of T cells by dendritic cells during infection. Immunol Res. 2014;59(1-3):66–72. doi: 10.1007/s12026-014-8530-3. [DOI] [PubMed] [Google Scholar]
- 13.McGill J, Van Rooijen N, Legge KL. IL-15 trans-presentation by pulmonary dendritic cells promotes effector CD8 T cell survival during influenza virus infection. J Exp Med. 2010;207(3):521–534. doi: 10.1084/jem.20091711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGill J, Van Rooijen N, Legge KL. Protective influenza-specific CD8 T cell responses require interactions with dendritic cells in the lungs. J Exp Med. 2008;205(7):1635–1646. doi: 10.1084/jem.20080314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langlois RA, Legge KL. Plasmacytoid dendritic cells enhance mortality during lethal influenza infections by eliminating virus-specific CD8 T cells. J Immunol. 2010;184(8):4440–4446. doi: 10.4049/jimmunol.0902984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Legge KL, Braciale TJ. Lymph node dendritic cells control CD8+ T cell responses through regulated FasL expression. Immunity. 2005;23(6):649–659. doi: 10.1016/j.immuni.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 17.Boonnak K, Vogel L, Feldmann F, Feldmann H, Legge KL, Subbarao K. Lymphopenia associated with highly virulent H5N1 virus infection due to plasmacytoid dendritic cell-mediated apoptosis of T cells. J Immunol. 2014;192(12):5906–5912. doi: 10.4049/jimmunol.1302992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Y, Shao F. The NAIP-NLRC4 inflammasome in innate immune detection of bacterial flagellin and type III secretion apparatus. Immunol Rev. 2015;265(1):85–102. doi: 10.1111/imr.12293. [DOI] [PubMed] [Google Scholar]
- 19.Kupz A, et al. NLRC4 inflammasomes in dendritic cells regulate noncognate effector function by memory CD8+ T cells. Nat Immunol. 2012;13(2):162–169. doi: 10.1038/ni.2195. [DOI] [PubMed] [Google Scholar]
- 20.Janowski AM, et al. NLRC4 suppresses melanoma tumor progression independently of inflammasome activation. J Clin Invest. 2016;126(10):3917–3928. doi: 10.1172/JCI86953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allen IC, et al. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity. 2009;30(4):556–565. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas PG, et al. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity. 2009;30(4):566–575. doi: 10.1016/j.immuni.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tenthorey JL, et al. The structural basis of flagellin detection by NAIP5: A strategy to limit pathogen immune evasion. Science. 2017;358(6365):888–893. doi: 10.1126/science.aao1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poyet JL, Srinivasula SM, Tnani M, Razmara M, Fernandes-Alnemri T, Alnemri ES. Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J Biol Chem. 2001;276(30):28309–28313. doi: 10.1074/jbc.C100250200. [DOI] [PubMed] [Google Scholar]
- 25.Tumpey TM, et al. Pathogenicity of influenza viruses with genes from the 1918 pandemic virus: functional roles of alveolar macrophages and neutrophils in limiting virus replication and mortality in mice. J Virol. 2005;79(23):14933–14944. doi: 10.1128/JVI.79.23.14933-14944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramos I, Fernandez-Sesma A. Modulating the innate immune response to influenza A virus: potential therapeutic use of anti-inflammatory drugs. Front Immunol. 2015;6:361. doi: 10.3389/fimmu.2015.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richards KA, Chaves FA, Krafcik FR, Topham DJ, Lazarski CA, Sant AJ. Direct ex vivo analyses of HLA-DR1 transgenic mice reveal an exceptionally broad pattern of immunodominance in the primary HLA-DR1-restricted CD4 T-cell response to influenza virus hemagglutinin. J Virol. 2007;81(14):7608–7619. doi: 10.1128/JVI.02834-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sant AJ, et al. Immunodominance in CD4 T-cell responses: implications for immune responses to influenza virus and for vaccine design. Expert Rev Vaccines. 2007;6(3):357–368. doi: 10.1586/14760584.6.3.357. [DOI] [PubMed] [Google Scholar]
- 29.Crowe SR, et al. Uneven distribution of MHC class II epitopes within the influenza virus. Vaccine. 2006;24(4):457–467. doi: 10.1016/j.vaccine.2005.07.096. [DOI] [PubMed] [Google Scholar]
- 30.McDermott DS, Varga SM. Quantifying antigen-specific CD4 T cells during a viral infection: CD4 T cell responses are larger than we think. J Immunol. 2011;187(11):5568–5576. doi: 10.4049/jimmunol.1102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hornick EE, et al. Nlrp12 mediates adverse neutrophil recruitment during influenza virus infection. J Immunol. 2018;200(3):1188–1197. doi: 10.4049/jimmunol.1700999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown DM, Román E, Swain SL. CD4 T cell responses to influenza infection. Semin Immunol. 2004;16(3):171–177. doi: 10.1016/j.smim.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 33.León B, Bradley JE, Lund FE, Randall TD, Ballesteros-Tato A. FoxP3+ regulatory T cells promote influenza-specific Tfh responses by controlling IL-2 availability. Nat Commun. 2014;5:3495. doi: 10.1038/ncomms4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tripp RA, Sarawar SR, Doherty PC. Characteristics of the influenza virus-specific CD8+ T cell response in mice homozygous for disruption of the H-2lAb gene. J Immunol. 1995;155(6):2955–2959. [PubMed] [Google Scholar]
- 35.Sant AJ, Richards KA, Nayak J. Distinct and complementary roles of CD4 T cells in protective immunity to influenza virus. Curr Opin Immunol. 2018;53:13–21. doi: 10.1016/j.coi.2018.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Legge KL, Braciale TJ. Accelerated migration of respiratory dendritic cells to the regional lymph nodes is limited to the early phase of pulmonary infection. Immunity. 2003;18(2):265–277. doi: 10.1016/S1074-7613(03)00023-2. [DOI] [PubMed] [Google Scholar]
- 37.Latchman Y, et al. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat Immunol. 2001;2(3):261–268. doi: 10.1038/85330. [DOI] [PubMed] [Google Scholar]
- 38.Süss G, Shortman K. A subclass of dendritic cells kills CD4 T cells via Fas/Fas-ligand-induced apoptosis. J Exp Med. 1996;183(4):1789–1796. doi: 10.1084/jem.183.4.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mariathasan S, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430(6996):213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- 40.Brunet A, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96(6):857–868. doi: 10.1016/S0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 41.Zhang H, et al. AIM2 inflammasome is critical for influenza-induced lung injury and mortality. J Immunol. 2017;198(11):4383–4393. doi: 10.4049/jimmunol.1600714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eisfeld AJ, Gasper DJ, Suresh M, Kawaoka Y. C57BL/6J and C57BL/6NJ Mice are differentially susceptible to inflammation-associated disease caused by influenza A virus. Front Microbiol. 2018;9:3307. doi: 10.3389/fmicb.2018.03307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vanden Berghe T, et al. Passenger mutations confound interpretation of all genetically modified congenic mice. Immunity. 2015;43(1):200–209. doi: 10.1016/j.immuni.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ulland TK, et al. Nlrp12 mutation causes C57BL/6J strain-specific defect in neutrophil recruitment. Nat Commun. 2016;7:13180. doi: 10.1038/ncomms13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamamoto TN, et al. T cells genetically engineered to overcome death signaling enhance adoptive cancer immunotherapy. J Clin Invest. 2019;129(4):1551–1565. doi: 10.1172/JCI121491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kavurma MM, Khachigian LM. Signaling and transcriptional control of Fas ligand gene expression. Cell Death Differ. 2003;10(1):36–44. doi: 10.1038/sj.cdd.4401179. [DOI] [PubMed] [Google Scholar]
- 47.Manning BD, Toker A. AKT/PKB signaling: navigating the network. Cell. 2017;169(3):381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lara-Tejero M, et al. Role of the caspase-1 inflammasome in Salmonella typhimurium pathogenesis. J Exp Med. 2006;203(6):1407–1412. doi: 10.1084/jem.20060206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sutterwala FS, et al. Critical role for NALP3/CIAS1/Cryopyrin in innate and adaptive immunity through its regulation of caspase-1. Immunity. 2006;24(3):317–327. doi: 10.1016/j.immuni.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 50.Kuida K, et al. Altered cytokine export and apoptosis in mice deficient in interleukin-1 beta converting enzyme. Science. 1995;267(5206):2000–2003. doi: 10.1126/science.7535475. [DOI] [PubMed] [Google Scholar]
- 51.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci U S A. 2000;97(11):6108–6113. doi: 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heaton NS, Sachs D, Chen CJ, Hai R, Palese P. Genome-wide mutagenesis of influenza virus reveals unique plasticity of the hemagglutinin and NS1 proteins. Proc Natl Acad Sci U S A. 2013;110(50):20248–20253. doi: 10.1073/pnas.1320524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Szretter KJ, Balish AL, Katz JM. Influenza: propagation, quantification, and storage. Curr Protoc Microbiol. 2006;Chapter 15:Unit 15G.1. doi: 10.1002/0471729256.mc15g01s3. [DOI] [PubMed] [Google Scholar]
- 54.Helft J, et al. GM-CSF Mouse bone marrow cultures comprise a heterogeneous population of CD11c(+)MHCII(+) macrophages and dendritic cells. Immunity. 2015;42(6):1197–1211. doi: 10.1016/j.immuni.2015.05.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.