Abstract
The polarization of macrophages is regulated by transcription factors, such as NF-κB and activator protein 1 (AP-1). In this manuscript, we delineated the role of the transcription factor Fos-related antigen 1 (Fra-1) during macrophage activation and development of arthritis. Network level interaction analysis of microarray data derived from Fra-1– or Fra-2–deficient macrophages revealed a central role of Fra-1, but not of Fra-2, in orchestrating the expression of genes related to wound response, Toll-like receptor activation, and interleukin signaling. ChIP sequencing and standard ChIP analyses of macrophages identified arginase 1 (Arg1) as a target of Fra-1. Luciferase reporter assays revealed that Fra-1 downregulated Arg1 expression by direct binding to the promoter region. Using macrophage-specific Fra-1– or Fra-2–deficient mice, we observed enhanced expression and activity of Arg1 and a reduction of arthritis in the absence of Fra-1, but not of Fra-2. This phenotype was reversed by treatment with the arginase inhibitor Nω-hydroxy-nor-ʟ-arginine, while ʟ-arginine supplementation increased arginase activity and alleviated arthritis, supporting the notion that reduced arthritis in macrophage-specific Fra-1–deficient mice resulted from enhanced Arg1 expression and activity. Moreover, patients with active rheumatoid arthritis (RA) showed increased Fra-1 expression in the peripheral blood and elevated Fra-1 protein in synovial macrophages compared with RA patients in remission. In addition, the Fra-1/ARG1 ratio in synovial macrophages was related to RA disease activity. In conclusion, these data suggest that Fra-1 orchestrates the inflammatory state of macrophages by inhibition of Arg1 expression and thereby impedes the resolution of inflammation.
Keywords: Cell Biology, Inflammation
Keywords: Arthritis, Macrophages
Introduction
Macrophages are remarkably diverse in their plasticity, allowing adaptation to a broad range of environmental stimuli. They participate in various processes essential for homeostatic functions and development, but also in inflammatory and immune responses. To do so, macrophages adapt their responses in a well-orchestrated transcriptional regulatory network, tailoring their response according to the respective stimuli and microenvironmental factors (1). Hence, macrophages generate a large variety of molecules, such as IL-1, IL-6, TNF, and NOS2, that exert proinflammatory effects. Besides their ability to initiate immune responses and eliminate invading pathogens, macrophages also secrete molecules, such as TGF-β or PDGF, that activate fibroblasts and mediate tissue repair and regeneration (2). Thus, macrophages participate in the resolution of inflammation and wound healing, which are indispensable functions for maintaining tissue homeostasis.
During the development of chronic inflammatory disease, such as rheumatoid arthritis (RA), which is characterized by synovial hyperplasia, cartilage degradation, and bone destruction (3), proinflammatory macrophages are deeply involved in the induction phase, whereas antiinflammatory macrophages promote the resolution of inflammation (4, 5). During the initial phase of arthritis, macrophage numbers rise in the synovial membrane, where they exhibit a proinflammatory phenotype characterized by the production of TNF and IL-1, IL-6, and IL-8 (4). In the acute or chronic phases of arthritis, proinflammatory macrophages contribute to the local and systemic inflammation by enhancing monocyte migration to the inflammatory site and by destroying the bone and cartilage through osteoclast activation (4). However, macrophages effectively initiate and promote the resolution of inflammation and thus help to restore tissue homeostasis. Examples of macrophage-derived molecules that are critically involved in tissue repair are TGF-β, VEGF, resistin-like α (Relmα/Retnla/FIZZ1), chitinase-like proteins, and arginase 1 (Arg1), which alter the duration of immune responses, activate fibroblasts, and/or regulate matrix deposition (6).
The cytosolic enzyme Arg1 is a constitutive component of the hepatic urea cycle. Arg1 catalyses the conversion of ʟ-arginine (ʟ-arg) into urea and ʟ-ornithine, which is further converted into polyamines and ʟ-proline. However, Arg1 can also regulate immune responses, mainly by 3 mechanisms. First, polyamines induce cell proliferation and counterregulate proinflammatory cytokine production. For instance, spermine downregulate the monocyte proinflammatory cytokine response of TNF and MIP-1α posttranscriptionally, which prevents local inflammation (7). Aside from that, ʟ-proline produced by the arginase pathway serves as an essential substrate for collagen synthesis. On the one hand, the increased production by Arg1 promotes wound healing and tissue regeneration, but on the other hand, its excessive production can lead to pathological vascular remodelling, fibrosis, and stiffness (8, 9). Second, Arg1-expressing macrophages can also suppress Th2-dependent inflammation by arginine depletion, which impairs T cell activation and proliferation (10–13). Third, Arg1 activity limits the supply of ʟ-arg needed for the formation of cytotoxic levels of NO by iNOS (14–16). Therefore, Arg1 has a regulatory role that seems to be highly dependent on the context of its activation.
The dual role of macrophages, to initiate and resolve immune responses, requires a mechanism for comprehensive reprogramming of macrophage function. This process involves the cooperation of several transcription factors including NF-κB, IFN regulatory factors (IRFs), STATs, and activator protein 1 (AP-1) (17). The AP-1 transcription factor family is composed of homo- and heterodimeric complexes, which consist of JUN and FOS proteins. Several studies have demonstrated the importance of AP-1 signaling during macrophage responses. For example, activation of JUN proteins in macrophages induced the proinflammatory enzyme cyclooxygenase-2, which increased prostaglandin E2 formation and arthritis development (18). On the other hand, FOS proteins, which comprise Fos-related antigen 1 (Fra-1) (Fosl1), Fra-2 (Fosl2), FosB, and c-Fos, interact with JUN, leading to the formation of transactivating or transrepressing complexes (19). c-Fos has been shown to suppress the expression of cytokines (20, 21) and Nos2 as well as the production of NO in macrophages (22). However, c-Jun has also been reported to induce Nos2 expression in hepatocytes (23), indicating that the function of AP-1 members might vary with the cell type and the type and duration of stimulation. Likewise, Fra-1 has been shown to regulate pro- and antiinflammatory cytokine expression, modulating profibrotic responses (24) and promoting LPS-induced injury in mice (25). Notably, the role of Fra-1 in macrophages has mainly been investigated in models of lung inflammation, as it is expressed in alveolar macrophages, where it modulates LPS-stimulated inflammatory cytokine expression, such as IL-10 and IL-1β, during inflammatory lung injury (26, 27). However, how the FRA proteins Fra-1 and Fra-2 influence macrophage functions in other diseases is less well studied. As macrophages are critically involved in many inflammatory and autoimmune diseases, the modulation of their responses might affect not only inflammation, but also tissue and organ homeostasis. Therefore, a comprehensive identification of the role of FRA proteins during macrophage activation could help to delineate new pathways to terminate the acute inflammatory phase and to initiate the resolution phase.
In the present study, we have discovered an important role of Fra-1 for the functional reprogramming of macrophages. Analyses of the K/BxN arthritis mouse model and of tissue sections of patients with active or inactive RA revealed an inverse correlation between Fra-1 and Arg1. Fra-1 directly suppressed Arg1 gene transcription and thereby altered macrophage responses, which impeded the resolution of inflammation.
Results
Fra-1 expression in macrophages is linked to inflammation.
To investigate the role of Fra-1 and Fra-2 in macrophages, Fra-1 or Fra-2 floxed mice were crossed to mice carrying the Cre recombinase controlled by the Mx1 (Fra-1ΔMx) or the Lysozyme2 (Fra-1ΔLysM and Fra-2ΔLysM) promoter, respectively. The regulatory spectrum of Fra-1 and Fra-2 in macrophages was determined through microarray analysis, using Agilent Technologies platforms, performed with thioglycollate-elicited macrophages isolated from Fra-1ΔMx and Fra-2ΔLysM mice and their respective littermate controls. First, the deletion of Fra-2 and Fra-1 in macrophages from each strain was determined by real-time PCR. Both lines showed decreases of gene expression by 85 % when the Fra-deficient cells were compared with their respective controls (Figure 1A and Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI96832DS1). Subsequent microarray analysis and the comparison of each deletion strain to its respective control strain revealed more than 500 genes differentially expressed in Fra-1ΔMx or Fra-2ΔLysM compared with WT macrophages (Figure 1B and data not shown).
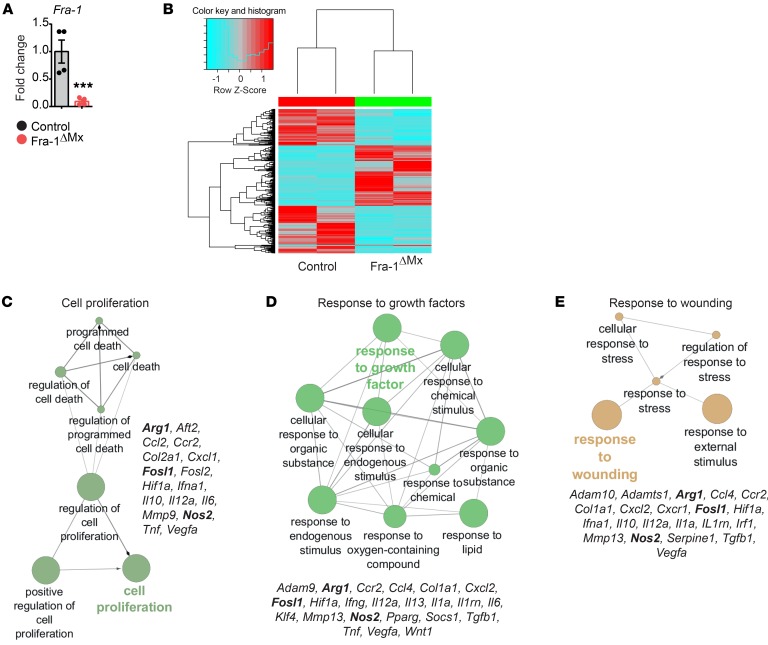
Figure 1. GO enrichment analysis links Fra-1 in macrophages to cell proliferation, response to growth factors, and wounding.
Thioglycollate-elicited macrophages were isolated from Fra-1ΔMx and control mice (n = 2). (A) The deletion efficiency of Fra-1 was quantified by real time PCR (RT-PCR). Data are shown as mean of 2 samples with duplicates and error bars represent SEM. ***P < 0.01, Student’s t test. (B) Heatmap of differentially expressed genes ascertained from microarray analysis. (C–E) GO enrichment analysis of differentially expressed genes found in the microarray analysis (related to Supplemental Figure 2). Depicted are genes associated with the terms in the cluster.
Gene ontology (GO) cluster analyses were performed, defining the molecular pathways associated with the differentially expressed genes. Surprisingly, differentially expressed genes in Fra-2–deficient macrophages were assembled in terms related to developmental functions (Supplemental Figure 1B). This confirms the essential function of Fra-2 during development (28–30). In contrast, GO cluster analysis based on differentially expressed genes in Fra-1–deficient macrophages revealed essential cellular pathways, such as wound response, proliferation, and responses to diverse stimuli (Figure 1, C–E, and Supplemental Figure 2). Interestingly, GO cluster analysis also indicated a dysregulation of Arg1 and Nos2 expression, as both genes were differentially expressed in Fra-1–deficient macrophages (Figure 1, C–E). Considering that the Arg1 and NOS2 pathways can define macrophage responses (16, 31, 32), these data indicate that Fra-1 could shape the pro- and antiinflammatory properties of macrophages.
To ascertain whether Fra-1 expression is regulated following macrophage stimulation, mRNA and protein levels of Fra-1 were determined in thioglycollate-elicited macrophages and BM-derived macrophages (BMDMs). Cells were isolated from WT mice and stimulated with IL-4, apoptotic cells (AC), IFN-γ, or LPS. Fra-1 mRNA levels in thioglycollate-elicited macrophages peaked prominently at 1 hour, and protein levels increased 2 hours after AC or LPS challenge in vitro (Figure 2, A and B). Fra-1 expression in BMDMs following LPS stimulation also peaked 1 hour after challenge in a manner similar to that seen in thioglycollate-elicited macrophages (Figure 2C). In cases of AC stimulation, the Fra-1 expression in BMDMs was 4 times increased 30 minutes after stimulation compared with that in unstimulated BMDMs (Figure 2C). However, Fra-1 expression in both types of macrophages remained unaffected by IL-4 or IFN-γ treatment (Figure 2, A and C). These data suggest that Fra-1 might be involved in macrophage immune responses.
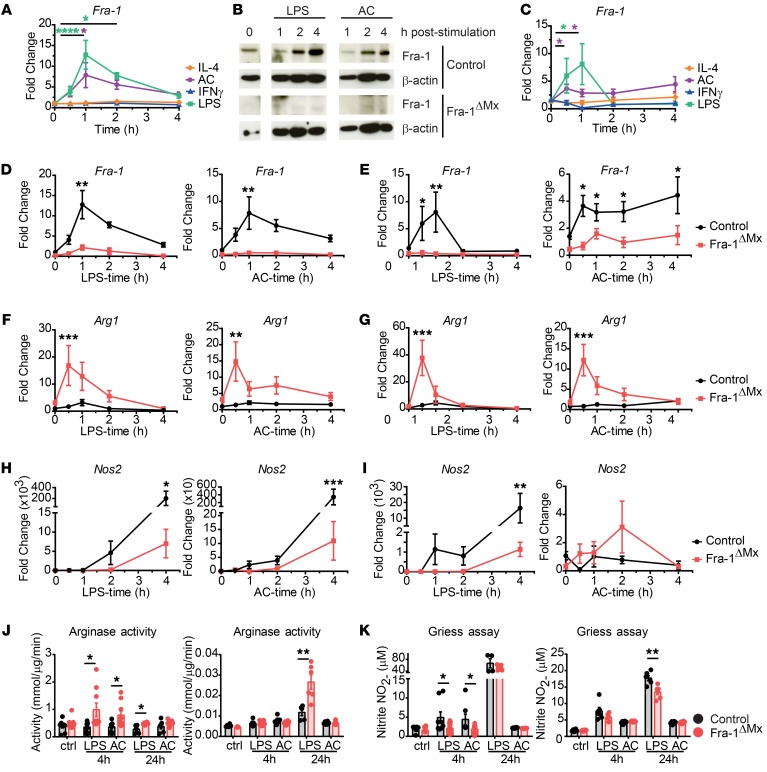
Figure 2. Fra-1 modulates expression and activity of Arg1 and Nos2.
Thioglycollate-elicited macrophages or BMDMs were isolated from Fra-1ΔMx or control littermate mice. 1 × 106 Macrophages were stimulated with 50 ng/ml IFN-γ, 1 μg/ml LPS, 100 ng/ml IL-4, or 5 × 106 ACs for the indicated time points. (A) Fra-1 mRNA levels in WT thioglycollate-elicited macrophages were determined by RT-PCR. (B) Fra-1 protein levels in thioglycollate-elicited macrophages isolated from Fra-1ΔMx or control mice. Shown are representative data from 1 out of 3 experiments. (C) Fra-1 mRNA levels in WT BMDMs after stimulation. (D and E) Fra-1 mRNA levels in WT and mutant (D) thioglycollate-elicited macrophages or (E) BMDMs stimulated with LPS or AC. (F and G) Arg1 mRNA levels in WT and mutant (F) thioglycollate-elicited macrophages or (G) BMDMs stimulated with LPS or AC. (H and I) Nos2 mRNA levels in WT and mutant (H) thioglycollate-elicited macrophages or (I) BMDMs stimulated with LPS or AC. (J) Arginase activity was determined in cell lysates of thioglycollate-elicited macrophages (left) or BMDMs (right). (K) iNOS activity was determined by Griess assay in supernatants of WT and mutant thioglycollate-elicited macrophages (left) or BMDMs (right). Data are shown as mean values of 3 independent experiments, and error bars represent SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, ANOVA.
Fra-1 controls Arg1 and Nos2 expression as well as macrophage activity.
To confirm the deregulation of Arg1 and Nos2 in Fra-1–deficient macrophages (Figure 1, C–E), differentially expressed molecules found in the GO clusters during macrophage responses were profiled. To do so, thioglycollate-elicited macrophages or BMDMs isolated from Fra-1ΔMx mice or littermate controls were stimulated with LPS or AC as stimuli for Fra-1 (Figure 2, A and C). Both types of Fra-1ΔMx macrophages showed a proper deletion of Fra-1 at the protein and mRNA levels in untreated or in AC- and LPS-treated cells when compared with WT cells (Figure 2, B, D, and E). Next, a decreased expression of Il6 in thioglycollate-elicited Fra-1ΔMx macrophages compared with WT macrophages following LPS or AC stimulation was observed, as previously described by Wang et al. (Supplemental Figure 3) (33). No significant difference in Tnf, Il10, Il12b, Il1b, or Retnla levels following AC or LPS stimulation was detected in Fra-1–deficient thioglycollate-elicited macrophages. However, Il1rn expression in these cells was slightly decreased following LPS treatment (Supplemental Figure 3). In control thioglycollate-elicited macrophages and BMDMs, the Arg1 expression following LPS stimulation was 6 times increased 1 hour after stimulation. Interestingly, in thioglycollate-elicited macrophages and BMDMs generated from Fra-1ΔMx mice, the Arg1 mRNA expression was more than 10-fold increased following AC or LPS stimulation compared with WT cells (Figure 2, F and G), while Nos2 expression was reduced following AC and was 40-fold lower following LPS stimulation in Fra-1ΔMx compared with WT macrophages (Figure 2, H and I). Accordingly, an increase of Arg1 enzyme activity was observed in Fra-1–deficient macrophages when compared with control 4 hours after LPS and AC stimulation in thioglycollate-elicited macrophages and 24 hours after LPS stimulation in BMDMs (Figure 2J). A decrease of NO synthesis (measured as nitrite accumulation) was only observed in Fra-1–deficient thioglycollate-elicited macrophages 4 hours after LPS and AC stimulation, but no longer at 24 hours after stimulation, whereas Fra-1–deficient BMDMs still showed decreased NOS2 activity 24 hours after LPS stimulation (Figure 2K). These data suggest that Fra-1 is a key switch in determining the expression of Nos2 and Arg1, shifting macrophages to a proinflammatory activation status.
Since Fra-1 and Fra-2 display similar primary protein structure, we investigated the regulation of Fra-2 expression in thioglycollate-elicited WT macrophages following LPS, AC, IL-4, and IFN-γ stimulation. Similarly to Fra-1 expression, Fra-2 expression was induced following LPS or AC challenge, but in contrast to Fra-1, it was also increased following IL-4 or IFN-γ stimulation (Supplemental Figure 4A). These data suggest that Fra-1 and Fra-2 are differentially regulated depending on the stimulation and the microenvironment of macrophages. Thus, Fra-1 and Fra-2 might possess distinct functions in macrophages. Next, we investigated whether Fra-2 deficiency would affect Arg1 and Nos2 expression in macrophages. Fra-2 deletion in macrophages was confirmed by quantitative PCR analysis in LPS- or AC-stimulated thioglycollate-elicited macrophages isolated from Fra-2ΔLysM compared with WT mice (Supplemental Figure 4B). However, Arg1 and Nos2 mRNA levels, as well as proinflammatory (Il6, Tnf) and antiinflammatory (Il10) cytokines, were similar in Fra-2ΔLysM and WT macrophages following LPS or AC stimulation (Supplemental Figure 4B). These results demonstrate that Fra-2 is unable to regulate these genes under these conditions.
Fra-1 controls Arg1, but not Nos2, expression in macrophages.
The AP-1 transcription factors have been shown to bind nonconsensus tetradecanoyl-phorbol-13-acetate response element (TRE) sequences at promoter levels and regulate the expression of a multitude of genes (34). Therefore, the whole spectrum of Fra-1 target genes in macrophages was determined. To do so, WT thioglycollate-elicited macrophages were left untreated or stimulated with LPS or AC before the cells were subjected to Fra-1 ChIP-Seq analyses. In unstimulated macrophages, more than 3000 genes are predicted to be regulated by Fra-1, according to the peak binding found, and this number doubled after LPS or AC treatment, respectively (Supplemental Table 1). To narrow down each stimulus’s effect, genes that were found under more than one condition were discarded; subsequently, an interaction network for each of the remaining gene lists was constructed (Supplemental Table 1). The genes for which interactors were found were then analyzed using GO enrichment analysis to pinpoint molecular pathways associated with the predicted genes. The GO enrichment confirmed that Fra-1 is a key player in macrophage immune responses (Supplemental Figure 5, A and B). Genes predicted to be regulated by Fra-1 following LPS stimulation were clustered to myeloid differentiation primary response gene 88–dependent (MyD88-dependent) mechanisms, involving Toll-like receptor-3,-4,-5, and -9 pathways as well as IL-6 and IL-7 signaling (Supplemental Figure 5A). Predicted genes regulated by Fra-1 following AC stimulation hinted at new aspects of Fra-1 in myogenesis and apoptosis pathways (Supplemental Figure 5B).
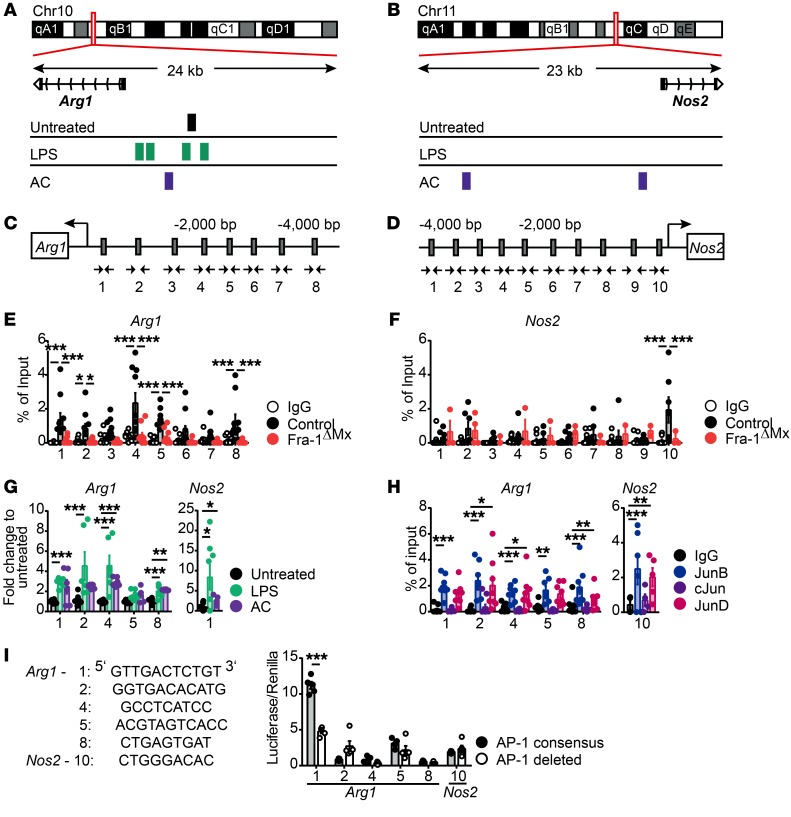
ChIP-Seq analyses suggested that Arg1 and Nos2 might be transcriptionally controlled by Fra-1 in macrophages, according to the binding peaks found in the promoters of these genes (Figure 3, A and B). To pursue the functionality of potential Fra-1 binding on Arg1 and Nos2 promoters, we performed conventional ChIP analyses. Therefore, we first determined the putative AP-1–binding sites up to 4000 bp upstream of the transcription starting site using the online tool TFSEARCH (http://diyhpl.us/~bryan/irc/protocol-online/protocol-cache/TFSEARCH.html). Consistent with the ChIP-Seq data, the promoter regions of Arg1 and Nos2 genes were predicted to harbor numerous TRE consensus sequences for AP-1 proteins (Figure 3, C and D). Fra-1 ChIP analyses in thioglycollate-elicited macrophages from WT or Fra-1ΔMx mice confirmed a Fra-1 binding in the Arg1 promoter region at the 1, 2, 4, 5, and 8 (Figure 3E). However, out of the 10 potential binding sites of the Nos2 promoter, Fra-1 only bound to the consensus sequence located around 500 bp upstream of the Nos2 transcription starting site (Nos2-10) (Figure 3F). Next, we hypothesized that LPS and AC stimulation, which induce Fra-1 expression, might increase its binding. Indeed, ChIP analysis using macrophages stimulated with LPS or AC for 1 hour and precipitating with a Fra-1 antibody showed an increased binding of Fra-1 to the Arg1 promoter region (Figure 3G). Binding of Fra-1 to the Nos2 promoter region was also increased after LPS and AC stimulation compared with unstimulated macrophages, suggesting that Fra-1 binds the Arg1 and Nos2 promoters after macrophage activation and controls its expression (Figure 3G).
Figure 3. Fra-1 transcriptionally regulates Arg1 at the promoter level.
Thioglycollate-elicited macrophages from Fra-1ΔMx or control littermate mice unstimulated or stimulated with LPS or AC were used for ChIP analysis. Chromatin was precipitated using an anti-mouse Fra-1 antibody or IgG isotype control; the obtained eluate was analyzed by sequencing or real-time PCR. (A and B) Peak of Fra-1 binding on the (A) Arg1 and (B) Nos2 promoters assessed by ChIP-Seq of LPS- or AC-stimulated WT macrophages. (C and D) AP-1 consensus sequences on the (C) Arg1 or the (D) Nos2 promoter were determined by the online tool TF search and are indicated by gray boxes. Additionally, the locations of the primers (arrows) are indicated. (E and F) Fra-1 ChIP analysis by real-time PCR for (E) Arg1 and (F) Nos2 promoters. The eluates arose from Fra-1ΔMx or control macrophages, and the Ct values are normalized to input. (G) Fra-1 ChIP analysis by real-time PCR for Arg1 and Nos2 promoters. The eluates arose from control macrophages stimulated for 1 hour with LPS or AC, and the Ct values normalized to the input were subsequently normalized to Fra-1 binding in unstimulated macrophages. (H) Jun protein ChIP analysis for Arg1 and Nos2 promoters. The eluates arose from control macrophages that were precipitated using anti-mouse JunB, JunD, cJun, or IgG isotype. Ct values were normalized to input. (I) AP-1 consensus sequences (Arg1-1/2/4/5/8 and Nos2-10) were cloned into a luciferase reporter construct and transfected into 293T cells; luciferase activity was determined. Mutated reporter constructs deleted for the respective binding sites were used as negative and renilla as internal control. The luciferase/renilla ratio was normalized to an empty pGL4.23 luciferase construct (n = 3). Data are shown as mean values of 3 independent experiments, and the error bars represent SEM. *P < 0.05; **P < 0.01; ***P < 0.001, ANOVA.
Fra-1 forms transcriptionally active heterodimers with JUN family members; therefore, the binding potential of cJun, JunB, and JunD on the Arg1 and Nos2 promoters was analyzed. To do so, ChIPs for c-Jun, JunB, and JunD proteins were performed using unstimulated thioglycollate-elicited WT macrophages. As shown in Figure 3H, only JunB and JunD bound to Arg1 and Nos2 promoters, whereas cJun showed no binding at the tested TRE elements (Figure 3H).
To determine whether Fra-1 can regulate the transcription of Arg1 and Nos2 expression, the promoter fragments 1, 2, 4, 5, and 8 of the Arg1 and 10 of the Nos2 promoter were cloned into an expression reporter plasmid. The plasmid carried a luciferase gene with a minimal promoter in front of which the respective promoter fragment was inserted. The transfection of 293T cells and subsequent luciferase assay showed that the presence of the Arg1 promoter fragment 1 increased luciferase expression, suggesting Fra-1 activates fragment 1 of the Arg1 promoter, located around 170 bp upstream of the transcription start site of the Arg1 gene (Figure 3I). However, direct regulation of Fra-1 through fragment 10 of the Nos2 promoter was not seen under unstimulated conditions (Figure 3I).
With respect to Fra-1 and Fra-2’s similar protein structures, we aimed to exclude that Fra-2 can replace Fra-1 on the Arg1 promoter in its absence. Therefore, we performed ChIP analyses for Fra-1 and Fra-2 on the AP-1 consensus element found on the Arg1 promoter using thioglycollate-elicited macrophages from control or Fra-1ΔMx mice. The ChIP analyses denied the ability of Fra-2 to bind the Arg1 promoter in control and Fra-1–deleted macrophages when compared with Fra-1 binding (Supplemental Figure 6, A and B).
Together, these data suggest that Fra-1 transcriptionally regulated Arg1 expression in macrophages, likely in cooperation with JunD or JunB, but could not confirm that Fra-1 directly regulates Nos2 expression in macrophages.
Fra-1 in myeloid cells exacerbates arthritis.
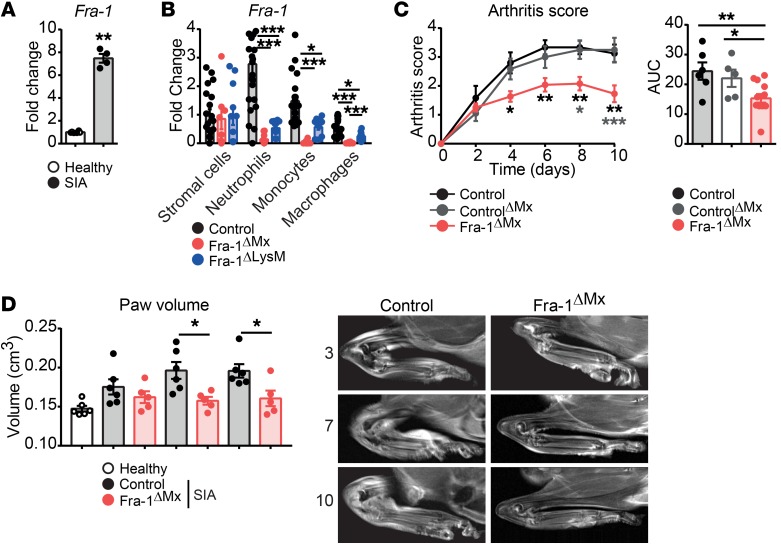
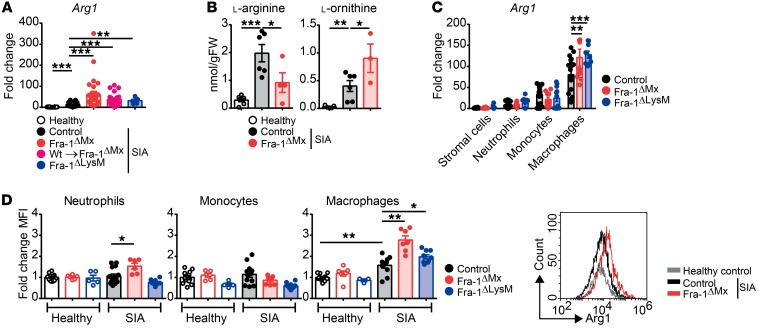
To determine the functional relevance of Fra-1 in macrophages, a KEGG cluster analysis was performed, allowing interpretation of high-level biological functions. This bioinformatic analysis linked macrophage Fra-1 expression to melanoma, prostate cancer, insulin resistance, and rheumatoid arthritis (RA) (Supplemental Figure 7). While the role of Fra-1 has been extensively studied in cancer (35), its function in chronic inflammatory diseases, such as RA, has not yet been determined. To investigate the in vivo relevance of Fra-1 in arthritis, the K/BxN serum-induced arthritis (SIA) model, which is a B cell– and T cell–independent representative model of RA, was used (36). First, SIA in WT mice was induced and Fra-1 mRNA expression in healthy versus arthritic paws 10 days after serum transfer was quantified. Fra-1 mRNA levels were increased in arthritic as compared with normal joints (Figure 4A). Next, the K/BxN model was applied to Fra-1ΔMx and control littermate mice. To quantify the Fra-1 deletion efficiency in different cell types, stromal cells (CD11b–), neutrophils (CD11b+Ly6G+), monocytes (CD11b+Ly6C+), and macrophages (CD11b+F4/80+) were sorted from arthritic paws of control and Fra-1ΔMx mice. First, compared with stromal cells, the expression of Fra-1 in WT neutrophils and monocytes was strongly increased and most abundant in neutrophils (Figure 4B). The expression of Fra-1 was unaltered in Fra-1ΔMx stromal cells compared with control cells, but its expression was absent or very low in Fra-1ΔMx neutrophils, monocytes, and macrophages when compared with control cells in arthritic paws, confirming the proper deletion of Fra-1 in the immune cell populations (Figure 4B).
Figure 4. Ameliorated joint inflammation in Fra-1ΔMx arthritic mice.
K/BxN arthritis was induced in Fra-1 mutant mice and their respective control mice. Healthy or arthritic mice were analyzed 10 days after serum transfer. (A) Fra-1 mRNA levels in paws of healthy and arthritic control mice (n = 4). (B) Fra-1 mRNA levels in stromal cells (CD11b–), neutrophils (CD11b+Ly6G+), monocytes (CD11b+Ly6C+), and macrophages (CD11b+F4/80+) sorted from arthritic Fra-1ΔMx, Fra-1ΔLysM, and control littermate mice. (C) Arthritis scores of control, controlΔMx, and Fra-1ΔMx mice and quantification of AUC. (D) Quantification and representative images of paw volume ascertained from in vivo MRI analysis of healthy, arthritic Fra-1ΔMx and arthritic control mice. Graph points indicate individual mice. Data are shown as mean values, and the error bars represent SEM. *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t test (A and B) or ANOVA (C and D).
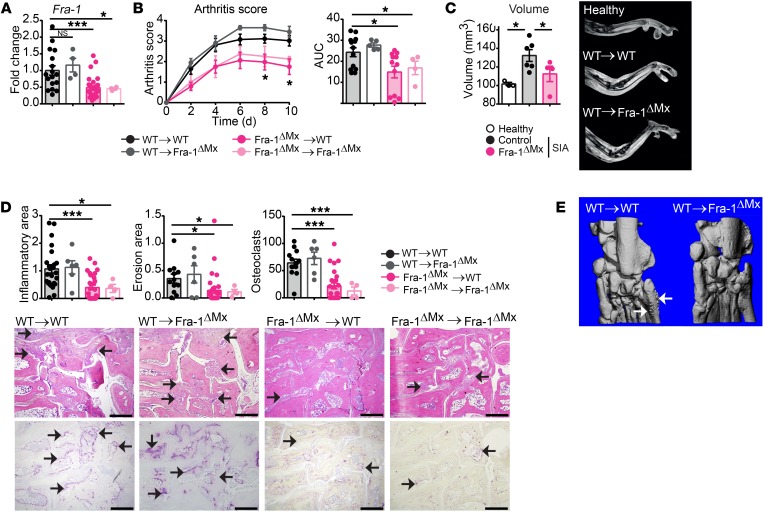
Notably, the severity of arthritis was significantly ameliorated in Fra-1ΔMx compared with both controls, Fra-1WT/fl containing MxCre (controlΔMx), and Fra-1fl/fl (control) mice (Figure 4C). In vivo MRI analysis confirmed that Fra-1–deficient mice had 25 % lower paw volume than WT mice at days 7 and 10 after serum transfer (Figure 4D). Accordingly, flow cytometric analysis revealed reduced neutrophil, monocyte, and macrophage numbers in the inflamed joints of Fra-1ΔMx mice when compared with arthritic WT mice, whereas no difference could be observed in joints of nonarthritic mice (Supplemental Figure 8, A and B). Taking into consideration that the Mx1 promoter can induce a deletion in some nonimmunological cells, such as hepatocytes in the liver (37–40), BM transfer from WT controls or Fra-1ΔMx mice into previously lethally irradiated WT or Fra-1ΔMx recipient mice was performed to restrict the deletion of Fra-1 to the immune cell compartment. Six weeks after BM transfer, arthritis was induced in WT→WT, WT→Fra-1ΔMx, Fra-1ΔMx→WT and Fra-1ΔMx→Fra-1ΔMx mice. A 50% decrease of Fra-1 mRNA level was detected in Fra-1ΔMx→WT and Fra-1ΔMx→Fra-1ΔMx when compared with WT→WT or WT→Fra-1ΔMx mice (Figure 5A). Accordingly, we found ameliorated arthritis in Fra-1ΔMx→WT and Fra-1ΔMx→Fra-1ΔMx mice (Figure 5B), whereas the arthritis in Fra-1ΔMx mice reconstituted with WT BM (WT→Fra-1ΔMx) showed a severity similar to that seen in WT→WT mice (Figure 5B). MRI analysis confirmed the decreased arthritis, showing decreased paw volume in Fra-1ΔMx→WT compared with WT→WT mice (Figure 5C). Additionally, histological analyses showed a reduced inflammatory area, bone erosion, and osteoclast numbers in paws from Fra-1ΔMx→WT and Fra-1ΔMx→Fra-1ΔMx mice (Figure 5D). Moreover, μCT analysis showed less pronounced bone changes in Fra-1ΔMx→WT than in WT→WT mice (Figure 5E). These data suggest that Fra-1 expression in the hematopoietic compartment affects the severity of arthritis and joint destruction.
Figure 5. Fra-1 in myeloid cells exacerbates the development of K/BxN SIA inflammation in joints.
BM from Fra-1ΔMx or control mice was transferred to previously irradiated Fra-1ΔMx or WT mice (WT→WT, WT→Fra-1ΔMx, Fra-1ΔMx→WT, and Fra-1ΔMx→Fra-1ΔMx). Six weeks after BM reconstitution, K/BxN arthritis was induced in recipient mice. (A) Fra-1 expression in arthritic paws. (B) Arthritis score as well as quantification of the AUC. (C) Quantification and representative images of paw volume ascertained by MRI analysis of healthy control paws and arthritic paws from WT→WT and Fra-1ΔMx→WT mice. (D) Quantification of the inflammatory area, erosion area, and number of osteoclast and representative images ascertained from H&E (top) and TRAP (bottom) staining of arthritic paws. The arrows indicate cell infiltrated areas in H&E staining and osteoclasts in TRAP staining, respectively. Scale bars: 500 μm. (E) Representative μCT imaging analysis of arthritic ankles from WT→WT and Fra-1ΔMx→WT. The arrows indicate osteophyte formation. Graph points indicate individual mice. Data are also shown as mean values, and error bars represent SEM. *P < 0.05; ***P < 0.001, Student’s t test (A, C, D) or ANOVA (B).
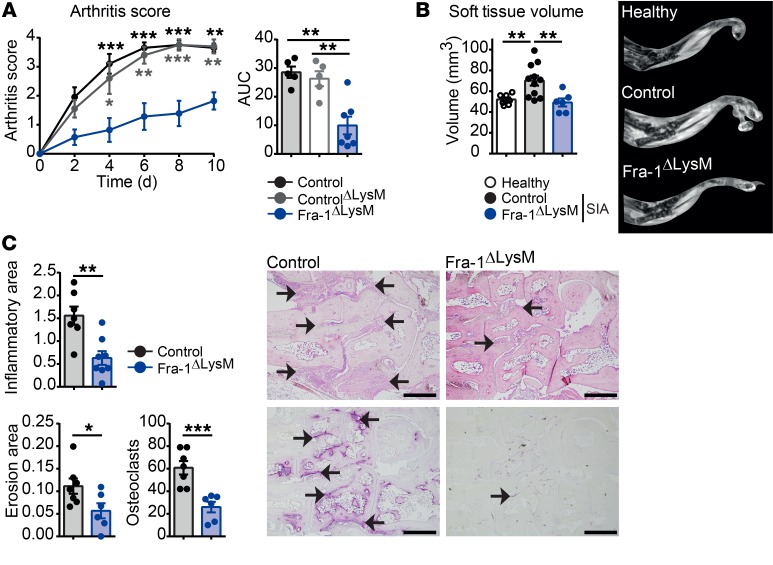
To further restrict Fra-1 deletion, the Fra-1 deletion controlled by the lysozyme promoter (Fra-1ΔLysM) was used. Lysozyme is known to be expressed in neutrophils, monocytes, and macrophages, resulting in a deletion of Fra-1 in these cell types (41). The K/BxN model was applied to Fra-1ΔLysM and control littermate mice. Again, the Fra-1 deletion was controlled in sorted stromal cells, neutrophils, monocytes, and macrophages. Similarly to what occurred in Fra-1ΔMx mice, Fra-1 expression was unchanged in Fra-1ΔLysM stromal cells compared with control cells, but its expression was absent or low in Fra-1ΔLysM neutrophils, monocytes, and macrophages (Figure 4B). Accordingly, the severity of arthritis was ameliorated in Fra-1ΔLysM compared with both controls, Fra-1WT/fl containing LysMCre (controlΔLysM), and Fra-1fl/fl (control) mice (Figure 6A). In addition, MRI analysis confirmed that Fra-1ΔLysM mice had decreased paw volumes compared with WT mice, and histological analysis of inflamed paws showed reduced inflammatory area, bone erosion, and osteoclast numbers in Fra-1ΔLysM compared with control mice (Figure 6, B and C). Despite no differences in myeloid cell numbers in Fra-1 and WT joints in steady state, flow cytometric analysis revealed decreased numbers and percentages of neutrophils, monocytes, and macrophages in the inflamed joints of Fra-1ΔLysM mice compared with arthritic control mice (Supplemental Figure 8, A and B).
Figure 6. Ameliorated joint inflammation in Fra-1ΔLysM arthritic mice.
K/BxN arthritis was induced in Fra-1ΔLysM and control littermate mice. (A) Arthritis score of control, controlΔLysM, and Fra-1ΔLysM mice and quantification of AUC. (B) Quantification of soft tissue volume and representative images ascertained by MRI analysis of healthy control and arthritic control and Fra-1ΔLysM mice. (C) Quantification of the inflammatory area, erosion area, and number of osteoclasts and representative images ascertained from H&E (top) and TRAP (bottom) staining of arthritic paws. The arrows indicate cell infiltrated areas in H&E staining and osteoclasts in TRAP staining, respectively. Scale bars: 500 μm. Graph points indicate individual mice. Data are shown as mean values, and error bars represent SEM. *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t test (B and C) or ANOVA (A).
Next, the role of Fra-2 in the K/BxN arthritis model was investigated. Therefore, K/BxN arthritis was induced in Fra-2ΔLysM mice. Fra-2 mRNA in arthritic Fra-2ΔLysM mice was decreased compared with in control joints (Supplemental Figure 9A). However, deletion of Fra-2 in macrophages, neutrophils, and monocytes did not significantly alter clinical and histological signs of arthritis (Supplemental Figure 9, B and C). This suggests that, in contrast to Fra-1, the expression of Fra-2 in myeloid cells does not exacerbate arthritis.
Increased Fra-1–dependent Arg1 activity ameliorates clinical arthritis.
To investigate whether Arg1 expression is also increased in Fra-1–deficient macrophages in vivo, its expression in normal versus arthritic paws of WT and Fra-1–deficient mutant mice was quantified. Arg1 mRNA levels were upregulated in arthritic compared with healthy paw lysates (Figure 7A). Moreover, Arg1 mRNA levels were more than 30 times increased in all Fra-1–deficient mutant mice (Figure 7A). Consistent with these findings, concentration of ʟ-arg was reduced, while ʟ-ornithine was increased in arthritic paws of Fra-1–deficient mice compared with paws from arthritic WT mice (Figure 7B). These results suggest that Fra-1 represses Arg1 expression in myeloid cells and thereby also prevents resolution of arthritis in vivo. This effect is specific for Fra-1, since Arg1 expression levels were identical in Fra-2ΔLysM compared with WT arthritic paws (Supplemental Figure 9D).
Figure 7. Increased Arg1 in Fra-1–deleted macrophages from arthritic joints.
K/BxN arthritis was induced in Fra-1 mutant mice and their respective control mice. Healthy or arthritic mice were analyzed 10 days after serum transfer. (A) Arg1 mRNA levels were determined in paws of healthy, arthritic control and Fra-1 mutant mice (Fra-1ΔMx, Fra-1ΔMx→WT, and Fra-1ΔLysM). (B) The concentrations of ʟ-arg and ʟ-ornithine in paw lysates of Fra-1ΔMx and control littermate mice were analyzed by HPLC. (C) Arg-1 mRNA levels in stromal cells (CD11b–), neutrophils (CD11b+Ly6G+), monocytes (CD11b+Ly6C+), and macrophages (CD11b+F4/80+) sorted from arthritic Fra-1ΔMx, Fra-1ΔLysM, and control littermate mice. (D) Intracellular Arg1 protein levels observed as MFI in neutrophils, monocytes, and macrophages isolated from healthy control or arthritic joints of WT, Fra-1ΔMx, and Fra-1ΔLysM mice. Shown are ΔMFI as compared with unstained and normalized to healthy controls of each respective cell type. A representative histogram of MFI is shown for macrophages from healthy control mice, arthritic control mice, and arthritic Fra-1ΔMx mice. Graph points indicate individual mice. Data are shown as mean values, and error bars represent SEM. *P < 0.05; **P < 0.01; ***P < 0.001, ANOVA.
To determine the cellular source of Arg1 in the joints, stromal cells (CD11b–), neutrophils (CD11b+Ly6G+), monocytes (CD11b+Ly6C+) and macrophages (CD11b+F4/80+) were sorted from arthritic paws of control, Fra-1ΔMx, and Fra-1ΔLysM mice, and their Arg1 mRNA expression levels were determined. Expression of Arg1 mRNA was around 100-fold higher in macrophages than in stromal cells (Figure 7C). Moreover, Arg1 mRNA expression in macrophages was significantly increased in Fra-1–deleted macrophages compared with control macrophages (Figure 7C). Using flow cytometry, intracellular Arg1 protein levels were compared in neutrophils, monocytes, and macrophages from joints of controls, Fra-1ΔMx mice, and Fra-1ΔLysM mice, either healthy or arthritic, respectively. Increased intracellular Arg1 protein levels were observed in neutrophils and macrophages from Fra-1ΔMx and Fra-1ΔLysM joints compared with arthritic controls, whereas Arg1 levels in monocytes remained unchanged (Figure 7D). The MFI of Arg1 protein in macrophages of arthritic WT paws was 2 times as high as in neutrophils or monocytes (Figure 7D). Notably, Arg1 levels in macrophages of arthritic Fra-1ΔMx mice were 4 times higher than in neutrophils or monocytes of the same mice (Figure 7D). These data indicate that Arg1 in arthritic joints is expressed mainly in macrophages and that its expression is inhibited by Fra-1.
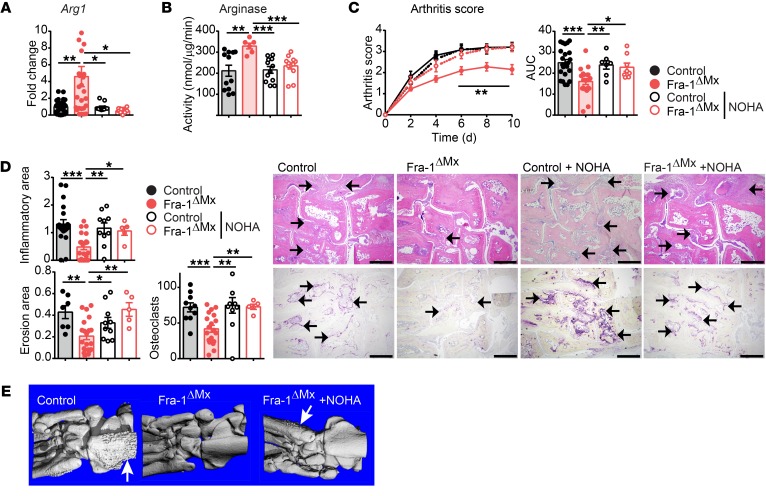
These in vivo and in vitro analyses led us to hypothesize that the increased Arg1 expression in Fra-1–deficient mice potentially accounts for the reduced severity of arthritis. To test this hypothesis, arginase activity was blocked using the inhibitor Nω-hydroxy-nor-l-arg (NOHA) in Fra-1–deficient and WT arthritic mice (42). First, Fra-1 expression was not affected by NOHA treatment (Supplemental Figure 10A). Second, the increased Arg1 mRNA levels and arginase activity in the paws of Fra-1ΔMx arthritic mice returned to basal levels after NOHA treatment (Figure 8, A and B), whereas iNos was not affected, as assessed by the Griess assay (Supplemental Figure 10B). Interestingly, inhibition of arginase in Fra-1–deficient mice was sufficient to aggravate the arthritis, reaching the level observed in WT mice (Figure 8C). This clinical effect of NOHA in Fra-1 mutant mice was paralleled by an increase of inflammation, bone erosion, osteoclast numbers, and osteophyte formation, as determined by μCT and histological analysis of the arthritis paws (Figure 8, D and E). Furthermore, the expression of several cytokines (e.g., Tnf, Il12b, Il1β and Il1rn) that were affected in the arthritic paws of Fra-1–deficient mice was restored after NOHA treatment (Supplemental Figure 10A). Notably, mRNA levels of Reltna, like Arg1, known as a marker for alternatively activated macrophages (43), were also highly increased in Fra-1–deficient arthritic joints, but returned to the low levels seen in WT arthritic mice after NOHA treatment of the Fra-1ΔMx mice (Supplemental Figure 10A). Moreover, arginase activity was blocked in Fra-1ΔLysM mice through NOHA treatment; again, the arthritis score of Fra-1ΔLysM mice treated with NOHA was enhanced to WT levels, as well as the inflammatory area, bone erosion, and the osteoclast numbers (Supplemental Figure 11, A and B). Taken together, these results suggest that Fra-1 expression in myeloid cells inhibits Arg1 expression and thereby promotes arthritis.
Figure 8. Arginase inhibition by NOHA restores arthritis in Fra-1ΔMx mice.
Arthritis was induced by K/BxN serum-transfer in Fra-1ΔMx or control mice. Mice were i.p. injected with NOHA (100 mg/kg body weight, Fra-1ΔMx or control) or PBS (Fra-1ΔMx or control) daily from the day of arthritis induction, and mice were analyzed 10 days after serum transfer. (A) Arg1 mRNA levels were quantified in whole paw. (B) Arg1 enzyme activity in whole-paw lysates was quantified by arginase activity assay. (C) Arthritis scores and AUC. (D) Quantification of the inflammatory area, erosion area, and number of osteoclasts from the histological analysis of H&E (top) and TRAP (bottom) staining and its representative images. The arrows indicate cell infiltrated areas in H&E staining and osteoclasts in TRAP staining, respectively. Scale bars: 500 μm. (E) Representative images of μCT imaging analysis (n = 3). The arrows indicate osteophyte formation. Graph points indicate individual mice. Data are shown as mean values, and error bars represent SEM. *P < 0.05; **P < 0.01; ***P < 0.001, ANOVA.
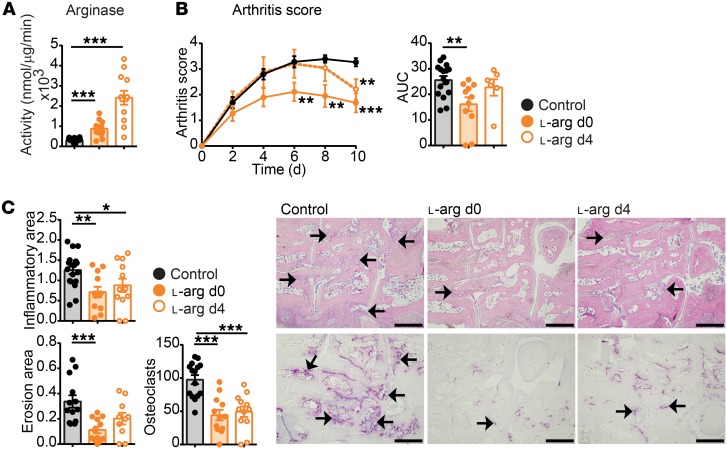
To determine whether the increased arginase activity can be used to inhibit arthritis, WT mice with K/BxN SIA were supplemented with 40 g/l ʟ-arg in drinking water ad libitum, which resulted in an average daily uptake of approximately 100 mg surplus ʟ-arg to increase arginase activity. The mice were either supplemented with ʟ-arg together with the induction of arthritis or in a therapeutic setting at the peak of the inflammation, on day 4 after serum transfer. Indeed, ʟ-arg supplementation led to increased arginase activity in the paws, while iNos activity remained unaffected (Figure 9A and Supplemental Figure 11C). Accordingly, both treatments led to a reduction of arthritis severity and reduced inflammatory areas, bone erosion, and osteoclast numbers (Figure 9, B and C). These data suggest that increased arginase enzyme activity inhibits the severity of arthritis and can be used in a therapeutic setting to alleviate arthritis symptoms and bone erosion.
Figure 9. Therapeutic l-arg supplementation resolves arthritis.
Arthritis was induced by K/BxN serum transfer to WT mice, supplemented with 40 g/l ʟ-arg in the drinking water, either simultaneously with the K/BxN serum transfer (d0) or therapeutically at d4 after serum transfer. Mice were analyzed at day 10 after serum transfer. (A) Arg1 activity in total paw lysates. (B) Arthritis score and its quantification of AUC. (C) Quantification of the inflammatory area, erosion area, and number of osteoclasts from the histological analysis of H&E (top) and TRAP (bottom) staining and its representative images. Scale bars: 500 μm. The arrows indicate cell infiltrated areas in H&E staining and osteoclasts in TRAP staining, respectively. Graph points indicate individual mice. Data are shown as mean values, and error bars represent SEM. *P < 0.05; **P < 0.01; ***P < 0.001, ANOVA.
Human RA is associated with high Fra-1 and low Arg1 expression in synovial macrophages.
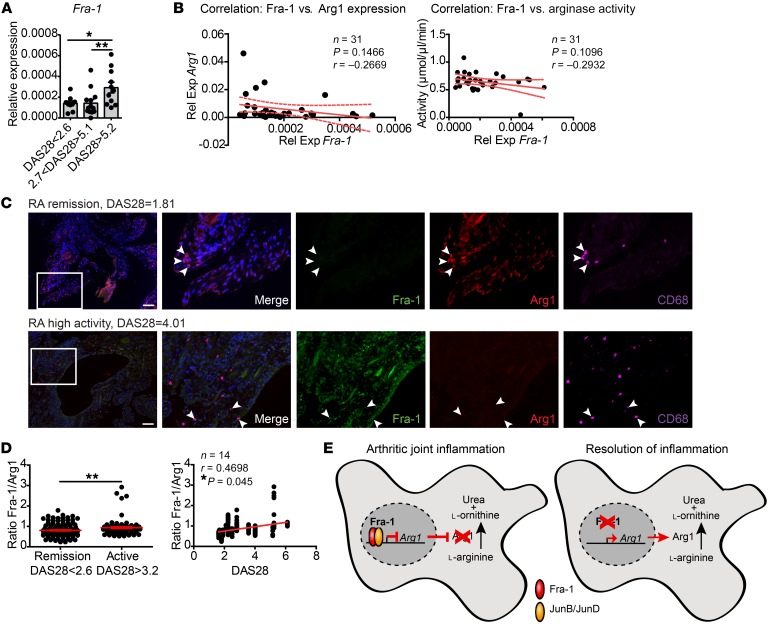
To investigate the relevance of our findings in human RA, we determined Fra-1 expression in the blood and synovial tissue from patients with active RA and RA patients in remission. The expression of Fra-1 in human whole blood from RA patients was increased in active as compared with inactive disease, suggesting that this mechanism might play a role in human RA (Figure 10A). Furthermore, a trend toward an inverse relation between Fra-1 expression in the blood and arginase expression as well as arginase activity was also observed (Figure 10B). Next, the levels of Fra-1 and Arg1 in macrophages locally in the inflamed joints were determined. Therefore, immunofluorescence stainings of Fra-1, Arg1, and CD68 in synovial tissue from patients with active RA and RA in remission were performed and quantified through densitometry. Analyzing the protein levels of Fra-1 and Arg1 in macrophages in the synovial tissue revealed that active RA is characterized by high Fra-1, but low Arg1 expression, while in the synovium of RA patients in remission, Fra-1 levels were low, but Arg1 levels were high in macrophages (Figure 10C). Additionally, the Fra-1/Arg1 protein ratio in the synovial membrane was higher in active than inactive disease and correlated to RA disease activity, as measured by DAS28 score (Figure 10D). In summary, these data led us to assume that Fra-1 effectively inhibits Arg1 expression in arthritis, thereby blocking resolution of inflammation and promoting a more severe clinical disease course (Figure 10E).
Figure 10. Fra-1 and Arg1 expression in the joints of RA patients.
(A) Fra-1 mRNA expression in peripheral blood cells of patients with RA at different levels of inflammatory disease activity measured by DAS28. DAS28 of less than 2.6 indicated low/no disease activity (n = 11), more than 2.7 and less than 5.1 indicated moderate disease activity (n = 11), and more than 5.2 indicated high disease activity (n = 12). (B) Correlation between mRNA expression of Fra-1 and Arg1 (left) or arginase activity (right) in peripheral blood cells or serum, respectively. Rel exp, relative expression. (C) Immunofluorescence image for CD68 (magenta), Fra-1 (green), Arg1 (red), and DAPI (blue) in joint sections from RA patients in remission (DAS28 = 1.81) and moderate/high disease activity (DAS28 = 4.01). Scale bars: 100 μm. Arrowheads point to CD68+ macrophages. White inset boxes are 3 times magnified from the original magnification. (D) CD68+ cells were quantified for their Fra-1/Arg1 ratio by the mean gray value in the synovial tissue of RA patients (n = 14) in remission (DAS28 < 2.6, n = 9) or moderate/high disease activity (DAS28>3.2, n = 5). Each point represents the ratio of the mean gray value (Fra-1/Arg1) per macrophage. (E) Schematic of Fra-1 actions in macrophages: Fra-1 blocks antiinflammatory responses in macrophages by the inhibition of the Arg1 pathways. *P < 0.05; **P < 0.01.
Discussion
The data presented in this study show that the AP-1 transcription factor Fra-1 is a central regulator of macrophage function in arthritis. Fra-1 directly repressed proresolving Arg1 in arthritis and enhanced the clinical course of the disease. We have identified a pathway where Fra-1, but not Fra-2, influences macrophage immune responses and acts as a central transcription factor regulating Arg1 expression. Deletion of Fra-1 mitigated arthritis, suggesting that Fra-1 activation in myeloid cells promotes a proinflammatory activation state. Fra-1 effectively suppressed proresolving Arg1 expression and skewed macrophage function toward a proinflammatory phenotype. Flow cytometric analysis revealed that macrophages are the main source of Fra-1–dependent Arg1 expression in the inflamed joints. The negative correlation between Fra-1 and Arg1 expression represents a molecular mechanism likely conserved in humans, since an inverse ratio of Fra-1 and Arg1 levels was observed in the synovia of the RA patient in clinical remission.
AP-1 transcription factors are environmental biosensors, mediating a broad range of signals, growth factors, cytokines, hormones, and stress responses (44–47). They regulate macrophage responses in conjunction with other key pathways, such as those involving STATs, IRFs, and NF-κB. For instance, LPS-triggered TLR4 activation of macrophages simultaneously activates NF-κB and AP-1 transcription factors (48). In vitro experiments using the monocytic cell line RAW264.7 have shown that activation of AP-1 controls proinflammatory cytokine and chemokine expression (22, 33, 49–52). Here, we found that Fra-1 and Fra-2 expression increased in macrophages following activation of the TLR cascade by LPS. GO analyses clustering the differentially expressed genes of WT and Fra-1–deficient macrophages underscored the important function of Fra-1 in macrophage activation and supported recent findings on Fra-1–mediated control of the cell cycle by regulating P53 expression (35, 53, 54). Furthermore, our Fra-1 ChIP-Seq analysis identified hundreds of genes regulated by Fra-1 that are related to the pattern recognition receptor and interleukin signaling, attributing the downstream role of AP-1 in LPS, TLR, or interleukin signaling (17, 55, 56). Even more importantly, ChIP-Seq and conventional ChIP experiments identified 2 targets of Fra-1 in macrophages following activation: Nos2 and Arg1. Promoter analysis through luciferase assays showed that Fra-1 directly represses Arg1 transcription, but does not directly influence Nos2 expression. Moreover, despite its similar structure, Fra-2 was unable to regulate Arg1 or Nos2 expression in macrophages. ChIP analysis showed no Fra-2 binding on the Arg1 promoter, underlining the functional relevance of Fra-1 in macrophages.
Our previous studies, deleting the AP-1 transcription factor member c-Jun, already revealed dysregulation of Arg1 expression (18). However, c-Jun does not directly control the Arg1 promoter (18). Fra-1 probably acts downstream of c-Jun, as LPS-induced Fra-1 expression is dependent on the collaboration of c-Jun and NF-κB (27). The selective regulation of Arg1 by Fra-1 in macrophages is also highlighted by the fact that other AP-1 members, such as c-Jun or c-Fos, were shown to positively or negatively regulate Arg1 expression in a cell type–dependent manner (22, 23, 57). Arg1 promotes wound healing and tissue remodelling and thereby helps to resolve inflammation and to restore tissue homeostasis, presumably by its generation of ornithine, which is the amino acid precursor for the synthesis of polyamines (via the ornithine decarboxylase pathway) and of ʟ-proline (via the ornithine aminotransferase pathway) (58).
Absence of Fra-1 influenced the inflammatory phase of arthritis, as shown by the reduced disease severity in Fra-1–deficient mice after induction of SIA. Again, the effect of Fra-1 on arthritis was different from that observed with deletion of other AP-1 members. For instance, c-Fos–deficient mice were reported to develop a more severe rather than reduced arthritis (59). Fra-1 deletion in arthritic mice increased Arg1 expression and activity in vivo, ameliorating joint inflammation. Our data showed an increased level of ʟ-ornithine in the paws of Fra-1–deficient arthritic mice, which might entertain an increased synthesis of polyamines. Enhanced production of polyamines is important in initiating resolution mechanisms, such as cell proliferation and growth (60), as well as inhibiting proinflammatory cytokines (7). We suggest that the dominant action of Fra-1 in promoting arthritis is the downregulation of the proresolving action of Arg1. This concept is further supported by the restoration of full-blown arthritis in Fra-1–deficient mice when inhibiting Arg1 by NOHA, as well as by the reduction of arthritis by ʟ-arg supplementation.
In support of our concept, others reported increased arginase activity and protein levels in serum in RA patients compared with healthy controls or patients with systemic lupus erythematosus or osteoarthritis in previous studies (61, 62). Furthermore, evidence suggests that AP-1 transcription factor components are activated in the synovial tissue of RA patients (63). Our data from patients with characterized disease activity score (DAS28) extend the previous findings. Analysis of human RA patients showed an increased expression of Fra-1 in the total blood and the synovium in active, but not inactive, disease. Furthermore, while active RA is characterized by high Fra-1, but low Arg1, expression in synovial macrophages, remission of RA is characterized by the reverse state, high Arg1 but low Fra-1 expression, in the synovium. Our data further extend previous studies and show that Arg1 contributes to entering into the remission phase of RA joint inflammation.
In summary, these data show that Fra-1 orchestrates the polarization of macrophages and thereby modulates the clinical course of arthritis. Fra-1 was shown to directly inhibit Arg1 expression and thus inhibits an essential proresolving mechanism, which leads to excessive inflammation and more severe disease. A selective c-Fos/AP-1 inhibitor has proven in a preclinical model to prevent joint destruction and pannus formation in type II collagen–induced arthritis in rats (64), which also has been investigated in phase II human clinical trials as a therapeutic agent for RA (65). Therefore, targeting Fra-1, but also Arg1, to elevate its enzymatic activities, through ʟ-arg supplementation may provide a therapeutic opportunity to induce resolution of inflammation in arthritis by fostering the transition of macrophages from a proinflammatory to a proresolving state.
Methods
Human samples.
Blood (peripheral blood withdrawal) and synovial samples (ultrasound-guided needle biopsy) were taken from RA patients fulfilling the 2010 American College of Rheumatology/European League Against Rheumatism classification criteria for RA (66). Blood samples for RNA isolation were obtained from RA patients with high (DAS28 > 5.1, n = 12), moderate (DAS28: 3.2–5.1, n = 11), or low/no (DAS28 < 3.2, n = 11) disease activity (67, 68). Blood of RA patients was collected in PAXgene Blood RNA Tubes (QIAGEN), and the RNA was isolated using the PAXgene Blood RNA Kit (QIAGEN) according to the manufacturer’s instructions. Synovial biopsy samples were obtained from patients with active RA (DAS28 > 3.2, n = 5) and patients with RA in remission (DAS28 < 2.6, n = 9).
Animals.
The generation of Fra-1 floxed, Fra-2 floxed, LysMCre, and Mx1Cre mice has been described elsewhere (37, 41, 69, 70). Mice were bred and maintained on a 129/B6 mixed background. The background characterization of these strains through SNPs revealed that their percentage match to the allelic profiles of B6 and 129 was approximately 95 % for B6 and approximately 50% for the 129 strain (Supplemental Table 2). Littermate Fra-1fl/fl and Fra-2fl/fl mice without Cre were used as controls. In some experiments, Fra-1WT/fl and Fra-2WT/fl with Mx-Cre or Lys-MCre, called controlΔMx or controlΔLysM, were used as additional controls. All experiments were performed with 8-week-old male mice. Animals were kept under standardized conditions. A 12-hour light/12-hour dark cycle was maintained, and standard diet and water were provided ad libitum. Detailed information about the time-line of the experiments performed with the Mx-Cre strain is illustrated in Supplemental Figure 12.
KRN TCR-transgenic mouse SIA model.
KRN TCR-transgenic mice were bred with NOD/Lt mice to generate K/BxN mice (71). K/BxN serum was isolated from adult K/BxN arthritic mice for which 150 μl/mouse was used to induce arthritis, as described previously (5). The severity of the arthritis was evaluated using a semiquantitative scoring system, with a score of 0 used for normal mice and a score of 1 given for the swelling of each of the following joints: digits, knuckles, mid-hind paw/mid-forepaw area, and ankle/wrist joint. Each paw was evaluated individually. Thus, the maximum clinical score per leg was 4. The reported clinical score was calculated as the average of all 4 paws (5). Ankle thickness was measured using a digital caliper.
μCT.
All μCT imaging was performed using the cone-beam Desktop Micro Computer Tomograph μCT 40 by SCANCO Medical. The settings were optimized for calcified tissue visualization in murine bones at 55 kVp, with a current of 177 μA, 200 ms integration time for 500 projections per 180°, and an isotropic voxel size of 8.6 μm. The 3D-modeling of the bone was performed with optimized grayscale thresholds of the operating system Open VMS by SCANCO Medical.
MRI.
For determination of soft tissue paw volume ex vivo, excised hind legs were embedded in 4% agarose and placed in a small animal ultra–high-field magnetic resonance scanner (ClinScan 7 Tesla, Bruker). A standard T1-weighted gradient echo sequence was used for segmentation of the soft tissue volume using aycan osirix (aycan Digitalsysteme GmbH and Chimaera GmbH). For in vivo MRI, mice under inhalation anesthesia were imaged using the whole-body mouse coil in an ultra–high-field magnetic resonance scanner (ClinScan 7 Tesla, Bruker). A standard T1-weighted spin echo sequence was used for segmentation of the paw volume. Furthermore, dynamic contrast-enhanced MRI (DCE-MRI) was performed by a 3D flash sequence before, during, and after application of an i.v. contrast agent (0.2 mmol/kg; Gadovist, Bayer).
Generation of macrophages.
Thioglycollate-elicited macrophages were generated by injecting mice i.p. with 2.5 ml of 4 % (w/v) Brewer’s thioglycollate medium (Sigma-Aldrich). Mice were euthanized 72 hours after injection. Peritoneal cavity cells were harvested by lavage, and cells were washed and plated in RPMI-1640 (Gibco, Thermo Fisher Scientific) supplemented with 10 % (v/v) FCS (Gibco, Thermo Fisher Scientific) and 1 % (v/v) penicillin/streptomycin (concentration of the stock solution: 10,000 units/ml penicillin and 10,000 μg/ml streptomycin; Gibco, Thermo Fisher Scientific). Cells were incubated overnight, and adherent cells were stimulated with 50 ng/ml IFN-γ (Peprotech), 1 μg/ml LPS (Sigma-Aldrich), 100 ng/ml IL-4 (Miltenyi Biotec), or ACs (with a 1:5 ratio). BMDMs were generated from BM cells incubated in DMEM (Gibco, Thermo Fisher Scientific) supplemented with 10 % (v/v) FCS (Gibco, Thermo Fisher Scientific), 1 % (v/v) penicillin/streptomycin (concentration of the stock solution: 10,000 Units/ml penicillin and 10,000 μg/ml streptomycin; Gibco, Thermo Fisher Scientific), and 10 % (v/v) L929 conditioned medium for 7 days.
Sequencing.
DNA from the ChIP experiments was sequenced on an Illumina HiSeq 2500 system, using a single end protocol and 100 bp read length. Reads were aligned to the mm10 mouse reference sequence using bwa version 0.7.8-r455 (72). Peak calling was performed with MACS version 1.3.7.1 (73).
Gene expression profiling.
Total RNA was isolated from 1 × 106 thioglycollate-elicited peritoneal macrophages from Fra-1ΔMx or WT littermates, using the RNeasy Mini Kit (QIAGEN). cRNA was synthesized using Cy3 labeling, quantified, and hybridized on 8x60K Arrays (design ID 028005) (Agilent Technologies). Data were extracted by the feature extraction software package (version 11.7.1; Agilent Technologies) using a standard protocol. Text files generated by the feature extraction software were imported into GeneSpring GX v12.5 (Silicon Genetics). Data were log2 transformed, followed by normalization to the 75th percentile, and corrected to the median of all samples. Features passing the quality check (flags detected in at least 1 condition) and showing changes in expression levels equal to or more than 2-fold were selected for further analysis. A volcano plot was applied to identify statistically significant (P < 0.05), more than 2-fold differentially expressed genes between 2 conditions, including the Benjamini-Hochberg multiple test correction. All original microarray data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE128787).
Ontology enrichment analysis.
For the biological interpretation of selected differentially expressed genes, the Cytoscape (74) plug-in ClueGO (75) was used. It visualizes the nonredundant biological terms for large clusters of genes in a functionally grouped network. Related terms that share a high percentage of associated genes can be fused to reduce redundancy; those terms are connected and have the same color code. The ClueGO network was created with κ statistics and reflects the relationships between the terms based on the similarity of their associated genes. A 2-tailed hypergeometric test for enrichment/depletion was used with a subsequent Bonferroni’s step-down or Benjamini-Hochberg correction for multiple testing of P values. The size of the nodes in the network represents the significance level of that term. The largest node in the cluster is the most significant term and is regarded as the cluster representative. For the current analysis, the P value threshold was set to 0.05, and the minimum number of genes associated with a term was set to 3. The ClueGO plugin was used to do both GO enrichment analysis and pathway enrichment analysis using KEGG (76) or Reactome (77) databases.
Amino acid determination.
Supernatants from paws were extracted as described previously (78). Briefly, paw samples were flash frozen in liquid nitrogen and the fresh weight (FW) was determined. Paws were homogenized in ethanol using a Precellys Ceramic Kit on a Precellys 24 tissue grinder (Bertin Instruments) under the conditions 2 × 30 seconds at 6,500 rpm; subsequently, samples were sonicated on ice (settings: cycle 5, power 50%, 40 seconds) and centrifuged at 23,000 g for 10 minutes. For their determination, amino acids in supernatants were derivatized using the fluoropohore 6-aminoquinolyl-N-hydroxysuccimidyl carbamate (AccQ Taq) and separated at a flow rate of 1 ml/min at 37°C on a Thermo (Dionex) Ultimate 3000 HPLC system essentially as described previously (79), using the eluents A (140 mm sodium acetate, pH 5.8; 7 mm triethanolamine), B (acetonitrile), and C (water) and fluorescence detection (excitation at 300 nm and detection at 400 nm) as previously described (79).
For additional information, see Supplemental Methods.
Statistics.
All experiments were repeated at least 3 times. Statistical analysis was performed using GraphPad PRISM software (version 5.03). One-tailed Student’s t test was used for comparison of 2 groups. For multiple comparisons, 2-way ANOVA with a Bonferroni’s corrected after test was used. P < 0.05 was considered significant. Data are shown as means, and the error bars represent SEM.
Study approval.
All analyses of human material were performed in accordance with the institutional guidelines and with the approval of the ethics committee of the Universitätsklinikum Erlangen. All experiments with animals were authorized by the ethics committee of the Government of Lower Franconia. Animals were kept following the guidelines of the German Animal Welfare Act.
Author contributions
NH and AB designed the study and wrote the manuscript. JJ and TB performed imaging analyses. SU and AE performed sequencing analysis. ME and JV contributed network reconstruction. NH and ME carried out enrichment analyses. NH performed in vitro and in vitro experiments. SC and XC cloned the luciferase plasmids and performed luciferase assays. DE provided technical assistance on cloning and edited the manuscript. NH and AS analyzed human samples. JR, AR, XC, and JDC collected human samples. US and CB provided technical assistance and expertise on the NOS2 and Arg1 pathway and edited the manuscript. GS and AB supervised the study and edited the manuscript.
Supplementary Material
Acknowledgments
We thank Wolfgang Baum, Christine Zech, Dana Weidner, Barbara Happich, and Hedwig Symowski for great technical assistance. We thank the staff of the Core Unit Cell Sorting and Immunomonitoring facility, especially Uwe Appelt for his technical support in flow cytometry and for cell sorting. We also thank the staff of the Division of Biochemistry, FAU, especially Stephen Reid and Sophia Sonnewald, for microarray analysis and Jörg Hofmann for amino acid determination. This study was supported by the Deutsche Forschungsgemeinschaft (grant CRC1181 project A01 to AB and GS; grant CRC1181 project C04 to CB and US; BO3811/1-6, BO3811/1-5, BO3811/1-7 to AB; Emmy Noether grant BO3811/1-1 to AB), the Interdisciplinary Center for Clinical Research (IZKF) of the Universitätsklinikum Erlangen (grant D23 to AB and DE; grant A63 to CB), and the German Federal Ministry of Education and Research (BMBF) (e:Bio MelEVIR [031L0073A] to JV).
Version 1. 04/16/2019
In-Press Preview
Version 2. 05/28/2019
Electronic publication
Version 3. 07/01/2019
Print issue publication
Funding Statement
grant CRC1181 project A01 to A.B. and G.S., grant CRC1181 project C04 to C.B. and U.S., BO3811/1-6 to A.B., Emmy Noether grant BO3811/1-1 to A.B.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(7):2669–2684.https://doi.org/10.1172/JCI96832.
Contributor Information
Nicole Hannemann, Email: Nicole.hannemann@uk-erlangen.de.
Shan Cao, Email: Shan.Cao@uk-erlangen.de.
Daniel Eriksson, Email: Daniel.Eriksson@uk-erlangen.de.
Anne Schnelzer, Email: anne_schnelzer@hotmail.de.
Jutta Jordan, Email: jutta-janke@web.de.
Martin Eberhardt, Email: martin.eberhardt@uk-erlangen.de.
Ulrike Schleicher, Email: ulrike.schleicher@uk-erlangen.de.
Jürgen Rech, Email: juergen.rech@uk-erlangen.de.
Andreas Ramming, Email: andreas.ramming@uk-erlangen.de.
Steffen Uebe, Email: steffen.uebe@uk-erlangen.de.
Arif Ekici, Email: Arif.Ekici@uk-erlangen.de.
Juan D. Cañete, Email: JCANETE@clinic.ub.es.
Xiaoxiang Chen, Email: xiaoxiang0721@gmail.com.
Tobias Bäuerle, Email: tobias.baeuerle@uk-erlangen.de.
Julio Vera, Email: Julio.Vera-Gonzalez@uk-erlangen.de.
Christian Bogdan, Email: christian.bogdan@uk-erlangen.de.
Georg Schett, Email: georg.schett@uk-erlangen.de.
Aline Bozec, Email: aline.bozec@uk-erlangen.de.
References
- 1.Xue J, et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 2014;40(2):274–288. doi: 10.1016/j.immuni.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diegelmann RF, Evans MC. Wound healing: an overview of acute, fibrotic and delayed healing. Front Biosci. 2004;9:283–289. doi: 10.2741/1184. [DOI] [PubMed] [Google Scholar]
- 3.Picerno V, Ferro F, Adinolfi A, Valentini E, Tani C, Alunno A. One year in review: the pathogenesis of rheumatoid arthritis. Clin Exp Rheumatol. 2015;33(4):551–558. [PubMed] [Google Scholar]
- 4.Kinne RW, Bräuer R, Stuhlmüller B, Palombo-Kinne E, Burmester GR. Macrophages in rheumatoid arthritis. Arthritis Res. 2000;2(3):189–202. doi: 10.1186/ar86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Z, et al. Th2 and eosinophil responses suppress inflammatory arthritis. Nat Commun. 2016;7:11596. doi: 10.1038/ncomms11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wynn TA, Barron L. Macrophages: master regulators of inflammation and fibrosis. Semin Liver Dis. 2010;30(3):245–257. doi: 10.1055/s-0030-1255354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang M, et al. Spermine inhibits proinflammatory cytokine synthesis in human mononuclear cells: a counterregulatory mechanism that restrains the immune response. J Exp Med. 1997;185(10):1759–1768. doi: 10.1084/jem.185.10.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell L, Saville CR, Murray PJ, Cruickshank SM, Hardman MJ. Local arginase 1 activity is required for cutaneous wound healing. J Invest Dermatol. 2013;133(10):2461–2470. doi: 10.1038/jid.2013.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caldwell RB, Toque HA, Narayanan SP, Caldwell RW. Arginase: an old enzyme with new tricks. Trends Pharmacol Sci. 2015;36(6):395–405. doi: 10.1016/j.tips.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pesce JT, et al. Arginase-1-expressing macrophages suppress Th2 cytokine-driven inflammation and fibrosis. PLoS Pathog. 2009;5(4):e1000371. doi: 10.1371/journal.ppat.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munder M, Choi BS, Rogers M, Kropf P. L-arginine deprivation impairs Leishmania major-specific T-cell responses. Eur J Immunol. 2009;39(8):2161–2172. doi: 10.1002/eji.200839041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monin L, et al. Helminth-induced arginase-1 exacerbates lung inflammation and disease severity in tuberculosis. J Clin Invest. 2015;125(12):4699–4713. doi: 10.1172/JCI77378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mondanelli G, Ugel S, Grohmann U, Bronte V. The immune regulation in cancer by the amino acid metabolizing enzymes ARG and IDO. Curr Opin Pharmacol. 2017;35:30–39. doi: 10.1016/j.coph.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 14.Rutschman R, Lang R, Hesse M, Ihle JN, Wynn TA, Murray PJ. Cutting edge: Stat6-dependent substrate depletion regulates nitric oxide production. J Immunol. 2001;166(4):2173–2177. doi: 10.4049/jimmunol.166.4.2173. [DOI] [PubMed] [Google Scholar]
- 15.Kropf P, et al. Arginase and polyamine synthesis are key factors in the regulation of experimental leishmaniasis in vivo. FASEB J. 2005;19(8):1000–1002. doi: 10.1096/fj.04-3416fje. [DOI] [PubMed] [Google Scholar]
- 16.Schleicher U, et al. TNF-mediated restriction of arginase 1 expression in myeloid cells triggers type 2 NO synthase activity at the site of infection. Cell Rep. 2016;15(5):1062–1075. doi: 10.1016/j.celrep.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tugal D, Liao X, Jain MK. Transcriptional control of macrophage polarization. Arterioscler Thromb Vasc Biol. 2013;33(6):1135–1144. doi: 10.1161/ATVBAHA.113.301453. [DOI] [PubMed] [Google Scholar]
- 18.Hannemann N, et al. The AP-1 Transcription Factor c-Jun Promotes Arthritis by Regulating Cyclooxygenase-2 and Arginase-1 Expression in Macrophages. J Immunol. 2017;198(9):3605–3614. doi: 10.4049/jimmunol.1601330. [DOI] [PubMed] [Google Scholar]
- 19.Zenz R, et al. Activator protein 1 (Fos/Jun) functions in inflammatory bone and skin disease. Arthritis Res Ther. 2008;10(1):201. doi: 10.1186/ar2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ray N, et al. c-Fos suppresses systemic inflammatory response to endotoxin. Int Immunol. 2006;18(5):671–677. doi: 10.1093/intimm/dxl004. [DOI] [PubMed] [Google Scholar]
- 21.Koga K, et al. Cyclic adenosine monophosphate suppresses the transcription of proinflammatory cytokines via the phosphorylated c-Fos protein. Immunity. 2009;30(3):372–383. doi: 10.1016/j.immuni.2008.12.021. [DOI] [PubMed] [Google Scholar]
- 22.Okada S, Obata S, Hatano M, Tokuhisa T. Dominant-negative effect of the c-fos family gene products on inducible NO synthase expression in macrophages. Int Immunol. 2003;15(11):1275–1282. doi: 10.1093/intimm/dxg126. [DOI] [PubMed] [Google Scholar]
- 23.Hasselblatt P, Rath M, Komnenovic V, Zatloukal K, Wagner EF. Hepatocyte survival in acute hepatitis is due to c-Jun/AP-1-dependent expression of inducible nitric oxide synthase. Proc Natl Acad Sci USA. 2007;104(43):17105–17110. doi: 10.1073/pnas.0706272104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajasekaran S, Reddy NM, Zhang W, Reddy SP. Expression profiling of genes regulated by Fra-1/AP-1 transcription factor during bleomycin-induced pulmonary fibrosis. BMC Genomics. 2013;14:381. doi: 10.1186/1471-2164-14-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaz M, Reddy NM, Rajasekaran S, Reddy SP. Genetic disruption of Fra-1 decreases susceptibility to endotoxin-induced acute lung injury and mortality in mice. Am J Respir Cell Mol Biol. 2012;46(1):55–62. doi: 10.1165/rcmb.2011-0169OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajasekaran S, et al. Visualization of Fra-1/AP-1 activation during LPS-induced inflammatory lung injury using fluorescence optical imaging. Am J Physiol Lung Cell Mol Physiol. 2015;309(4):L414–L424. doi: 10.1152/ajplung.00315.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mishra RK, Potteti HR, Tamatam CR, Elangovan I, Reddy SP. c-Jun is required for nuclear factor-κB-dependent, LPS-stimulated Fos-related antigen-1 transcription in alveolar macrophages. Am J Respir Cell Mol Biol. 2016;55(5):667–674. doi: 10.1165/rcmb.2016-0028OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ubieta K, et al. Fra-2 regulates B cell development by enhancing IRF4 and Foxo1 transcription. J Exp Med. 2017;214(7):2059–2071. doi: 10.1084/jem.20160514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luther J, et al. Fra-2/AP-1 controls adipocyte differentiation and survival by regulating PPARγ and hypoxia. Cell Death Differ. 2014;21(4):655–664. doi: 10.1038/cdd.2013.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karreth F, Hoebertz A, Scheuch H, Eferl R, Wagner EF. The AP1 transcription factor Fra2 is required for efficient cartilage development. Development. 2004;131(22):5717–5725. doi: 10.1242/dev.01414. [DOI] [PubMed] [Google Scholar]
- 31.Bogdan C. Nitric oxide synthase in innate and adaptive immunity: an update. Trends Immunol. 2015;36(3):161–178. doi: 10.1016/j.it.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 32.Murray PJ, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41(1):14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Q, Ni H, Lan L, Wei X, Xiang R, Wang Y. Fra-1 protooncogene regulates IL-6 expression in macrophages and promotes the generation of M2d macrophages. Cell Res. 2010;20(6):701–712. doi: 10.1038/cr.2010.52. [DOI] [PubMed] [Google Scholar]
- 34.Chinenov Y, Kerppola TK. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene. 2001;20(19):2438–2452. doi: 10.1038/sj.onc.1204385. [DOI] [PubMed] [Google Scholar]
- 35.Zhong G, Chen X, Fang X, Wang D, Xie M, Chen Q. Fra-1 is upregulated in lung cancer tissues and inhibits the apoptosis of lung cancer cells by the P53 signaling pathway. Oncol Rep. 2016;35(1):447–453. doi: 10.3892/or.2015.4395. [DOI] [PubMed] [Google Scholar]
- 36.Korganow AS, et al. From systemic T cell self-reactivity to organ-specific autoimmune disease via immunoglobulins. Immunity. 1999;10(4):451–461. doi: 10.1016/S1074-7613(00)80045-X. [DOI] [PubMed] [Google Scholar]
- 37.Kühn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269(5229):1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 38.Radtke F, et al. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10(5):547–558. doi: 10.1016/S1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 39.Lam KP, Rajewsky K. Rapid elimination of mature autoreactive B cells demonstrated by Cre-induced change in B cell antigen receptor specificity in vivo. Proc Natl Acad Sci U S A. 1998;95(22):13171–13175. doi: 10.1073/pnas.95.22.13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wells CM, Walmsley M, Ooi S, Tybulewicz V, Ridley AJ. Rac1-deficient macrophages exhibit defects in cell spreading and membrane ruffling but not migration. J Cell Sci. 2004;117(Pt 7):1259–1268. doi: 10.1242/jcs.00997. [DOI] [PubMed] [Google Scholar]
- 41.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Förster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8(4):265–277. doi: 10.1023/A:1008942828960. [DOI] [PubMed] [Google Scholar]
- 42.Rodriguez PC, et al. Arginase I production in the tumor microenvironment by mature myeloid cells inhibits T-cell receptor expression and antigen-specific T-cell responses. Cancer Res. 2004;64(16):5839–5849. doi: 10.1158/0008-5472.CAN-04-0465. [DOI] [PubMed] [Google Scholar]
- 43.Raes G, Noël W, Beschin A, Brys L, de Baetselier P, Hassanzadeh GH. FIZZ1 and Ym as tools to discriminate between differentially activated macrophages. Dev Immunol. 2002;9(3):151–159. doi: 10.1080/1044667031000137629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hill CS, Treisman R. Differential activation of c-fos promoter elements by serum, lysophosphatidic acid, G proteins and polypeptide growth factors. EMBO J. 1995;14(20):5037–5047. doi: 10.1002/j.1460-2075.1995.tb00186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410(6824):37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 46.Hill CS, Wynne J, Treisman R. Serum-regulated transcription by serum response factor (SRF): a novel role for the DNA binding domain. EMBO J. 1994;13(22):5421–5432. doi: 10.1002/j.1460-2075.1994.tb06877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gruda MC, Kovary K, Metz R, Bravo R. Regulation of Fra-1 and Fra-2 phosphorylation differs during the cell cycle of fibroblasts and phosphorylation in vitro by MAP kinase affects DNA binding activity. Oncogene. 1994;9(9):2537–2547. [PubMed] [Google Scholar]
- 48.Guha M, Mackman N. LPS induction of gene expression in human monocytes. Cell Signal. 2001;13(2):85–94. doi: 10.1016/S0898-6568(00)00149-2. [DOI] [PubMed] [Google Scholar]
- 49.Swantek JL, Cobb MH, Geppert TD. Jun N-terminal kinase/stress-activated protein kinase (JNK/SAPK) is required for lipopolysaccharide stimulation of tumor necrosis factor alpha (TNF-alpha) translation: glucocorticoids inhibit TNF-alpha translation by blocking JNK/SAPK. Mol Cell Biol. 1997;17(11):6274–6282. doi: 10.1128/MCB.17.11.6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim DS, Han JH, Kwon HJ. NF-kappaB and c-Jun-dependent regulation of macrophage inflammatory protein-2 gene expression in response to lipopolysaccharide in RAW 264.7 cells. Mol Immunol. 2003;40(9):633–643. doi: 10.1016/j.molimm.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 51.Matsumoto M, Einhaus D, Gold ES, Aderem A. Simvastatin augments lipopolysaccharide-induced proinflammatory responses in macrophages by differential regulation of the c-Fos and c-Jun transcription factors. J Immunol. 2004;172(12):7377–7384. doi: 10.4049/jimmunol.172.12.7377. [DOI] [PubMed] [Google Scholar]
- 52.Chen J, et al. IL-17A induces pro-inflammatory cytokines production in macrophages via MAPKinases, NF-κB and AP-1. Cell Physiol Biochem. 2013;32(5):1265–1274. doi: 10.1159/000354525. [DOI] [PubMed] [Google Scholar]
- 53.Xiao S, et al. Fra-1 is downregulated in cervical cancer tissues and promotes cervical cancer cell apoptosis by p53 signaling pathway in vitro. Int J Oncol. 2015;46(4):1677–1684. doi: 10.3892/ijo.2015.2873. [DOI] [PubMed] [Google Scholar]
- 54.Zhu W, et al. FOS-like antigen 1 is highly expressed in human psoriasis tissues and promotes the growth of HaCaT cells in vitro. Mol Med Rep. 2014;10(5):2489–2494. doi: 10.3892/mmr.2014.2509. [DOI] [PubMed] [Google Scholar]
- 55.Takada Y, et al. Interstitial lung disease induced by gefitinib and toll-like receptor ligands is mediated by Fra-1. Oncogene. 2011;30(36):3821–3832. doi: 10.1038/onc.2011.101. [DOI] [PubMed] [Google Scholar]
- 56.Hagemann T, Biswas SK, Lawrence T, Sica A, Lewis CE. Regulation of macrophage function in tumors: the multifaceted role of NF-kappaB. Blood. 2009;113(14):3139–3146. doi: 10.1182/blood-2008-12-172825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharda DR, et al. Regulation of macrophage arginase expression and tumor growth by the Ron receptor tyrosine kinase. J Immunol. 2011;187(5):2181–2192. doi: 10.4049/jimmunol.1003460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu G, Morris SM. Arginine metabolism: nitric oxide and beyond. Biochem J. 1998;336(Pt 1):1–17. doi: 10.1042/bj3360001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Redlich K, et al. Osteoclasts are essential for TNF-alpha-mediated joint destruction. J Clin Invest. 2002;110(10):1419–1427. doi: 10.1172/JCI15582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pegg AE. Mammalian polyamine metabolism and function. IUBMB Life. 2009;61(9):880–894. doi: 10.1002/iub.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang LW, Chang KL, Chen CJ, Liu HW. Arginase levels are increased in patients with rheumatoid arthritis. Kaohsiung J Med Sci. 2001;17(7):358–363. [PubMed] [Google Scholar]
- 62.Jiao Z, Hua S, Wang W, Wang H, Gao J, Wang X. Increased circulating myeloid-derived suppressor cells correlated negatively with Th17 cells in patients with rheumatoid arthritis. Scand J Rheumatol. 2013;42(2):85–90. doi: 10.3109/03009742.2012.716450. [DOI] [PubMed] [Google Scholar]
- 63.Asahara H, et al. Direct evidence of high DNA binding activity of transcription factor AP-1 in rheumatoid arthritis synovium. Arthritis Rheum. 1997;40(5):912–918. doi: 10.1002/art.1780400520. [DOI] [PubMed] [Google Scholar]
- 64.Aikawa Y, et al. Treatment of arthritis with a selective inhibitor of c-Fos/activator protein-1. Nature Biotechnology. 2008;26(7):817–823. doi: 10.1038/nbt1412. [DOI] [PubMed] [Google Scholar]
- 65.Ye N, Ding Y, Wild C, Shen Q, Zhou J. Small molecule inhibitors targeting activator protein 1 (AP-1) J Med Chem. 2014;57(16):6930–6948. doi: 10.1021/jm5004733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aletaha D, et al. 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum. 2010;62(9):2569–2581. doi: 10.1002/art.27584. [DOI] [PubMed] [Google Scholar]
- 67.Aletaha D, et al. 2010 rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Ann Rheum Dis. 2010;69(9):1580–1588. doi: 10.1136/ard.2010.138461. [DOI] [PubMed] [Google Scholar]
- 68.Prevoo ML, van ‘t Hof MA, Kuper HH, van Leeuwen MA, van de Putte LB, van Riel PL. Modified disease activity scores that include twenty-eight-joint counts. Development and validation in a prospective longitudinal study of patients with rheumatoid arthritis. Arthritis Rheum. 1995;38(1):44–48. doi: 10.1002/art.1780380107. [DOI] [PubMed] [Google Scholar]
- 69.Eferl R, et al. The Fos-related antigen Fra-1 is an activator of bone matrix formation. EMBO J. 2004;23(14):2789–2799. doi: 10.1038/sj.emboj.7600282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eferl R, Zenz R, Theussl HC, Wagner EF. Simultaneous generation of fra-2 conditional and fra-2 knock-out mice. Genesis. 2007;45(7):447–451. doi: 10.1002/dvg.20311. [DOI] [PubMed] [Google Scholar]
- 71.Monach PA, Mathis D, Benoist C. The K/BxN arthritis model. Curr Protoc Immunol. 2008;Chapter 15:Unit 15.22. doi: 10.1002/0471142735.im1522s81. [DOI] [PubMed] [Google Scholar]
- 72.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bindea G, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Croft D, et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42(Database issue):D472–D477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Heussner K, et al. Species differences of 11beta-hydroxysteroid dehydrogenase type 2 function in human and rat term placenta determined via LC-MS/MS. Placenta. 2016;37:79–84. doi: 10.1016/j.placenta.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 79.van Wandelen C, Cohen SA. Using quaternary high-performance liquid chromatography eluent systems for separating 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate-derivatized amino acid mixtures. J Chromatogr A. 1997;763(1):11–22. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.