Abstract
Chronic myeloid leukemia (CML) results from hematopoietic stem cell transformation by the BCR-ABL kinase. Despite the success of BCR-ABL tyrosine kinase inhibitors (TKIs) in treating CML patients, leukemia stem cells (LSCs) resist elimination and persist as a major barrier to cure. Previous studies suggest that overexpression of the sirtuin 1 (SIRT1) deacetylase may contribute to LSC maintenance in CML. Here, by genetically deleting SIRT1 in transgenic CML mice, we definitively demonstrated an important role for SIRT1 in leukemia development. We identified a previously unrecognized role for SIRT1 in mediating increased mitochondrial oxidative phosphorylation in CML LSCs. We showed that mitochondrial alterations were kinase independent and that TKI treatment enhanced inhibition of CML hematopoiesis in SIRT1-deleted mice. We further showed that the SIRT1 substrate PGC-1α contributed to increased oxidative phosphorylation and TKI resistance in CML LSCs. These results reveal an important role for SIRT1 and downstream signaling mechanisms in altered mitochondrial respiration in CML LSCs.
Keywords: Hematology, Oncology
Keywords: Adult stem cells, Leukemias
Introduction
Chronic myeloid leukemia (CML) is a lethal hematological malignancy that results from hematopoietic stem cell (HSC) transformation by the BCR-ABL tyrosine kinase. BCR-ABL expression bestows hematopoietic cells with malignant characteristics, including inappropriate proliferation, reduced apoptosis, and genomic instability. Despite the success of BCR-ABL tyrosine kinase inhibitors (TKIs) in inducing remission and prolonging survival, cures remain elusive because primitive leukemia stem cells (LSCs) are retained, even in patients achieving remission (1, 2). Although a small proportion of CML patients achieving prolonged deep responses can maintain remission after TKI discontinuation, the majority of CML patients require indefinite TKI treatment to prevent disease recurrence, with associated risk of noncompliance, toxicity, and financial burden,(3, 4). Identifying key mechanisms for the persistence of LSCs is critical for the development of effective therapeutic strategies in CML.
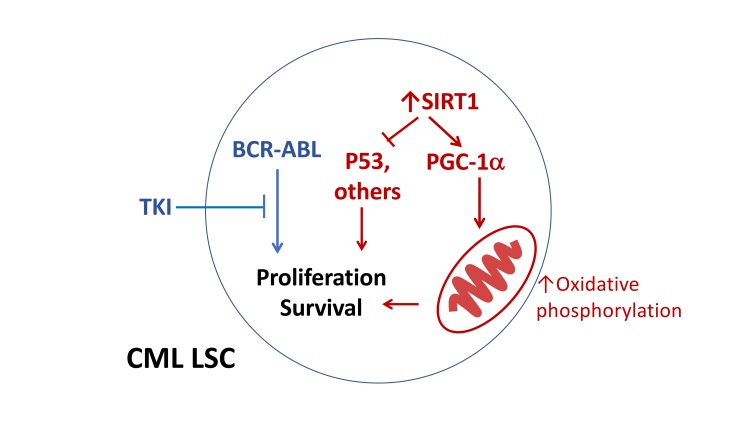
BCR-ABL TKIs effectively inhibit tyrosine kinase activity in CML LSCs, indicating that LSC resistance is related to tyrosine kinase–independent mechanisms (2). We have previously reported that sirtuin 1 (SIRT1), a NAD+ dependent deacetylase, is overexpressed in CML LSCs compared with normal HSCs and that it contributes to CML LSC maintenance and TKI resistance (5, 6). Importantly, SIRT1 also contributes to LSC maintenance and drug resistance in Flt3–ITD+ acute myeloid leukemia (AML), another oncogenic tyrosine kinase–driven malignancy, indicating a broader importance of SIRT1 in diverse leukemias (7, 8). SIRTs regulate many cellular pathways, including energy metabolism and stress response to tumorigenesis and aging. SIRT1 is a nuclear protein expressed in almost all cell types and involved in the deacetylation of histones, transcription factors, and signaling proteins that regulate many metabolic and stress-response pathways (9, 10), including PGC-1α (11), LXR (12), PPARγ (13), FOXO (14), p53 (15), and KU70. Although the role of SIRT1 in longevity and stress responses indicates a role as a tumor suppressor, SIRT1 can also enhance tumorigenesis, depending on cellular context (5–8, 16–19). Indeed, SIRT1 is overexpressed in several cancer types and can deacetylate and repress the activity of tumor suppressor p53 (15).
We have previously shown that SIRT1 inhibition using shRNA or a small molecule SIRT1 inhibitor reduces LSC growth and survival and increases the sensitivity of these cells to TKI (5). Our results indicate that the effects of SIRT1 inhibition were related, at least in part, to increased p53 acetylation and transcriptional activity. However, previous studies evaluating the role of SIRT1 in regulating LSC growth were limited because of possible off-target effects of SIRT1 inhibitors and short duration of exposure as well as lack of an unbiased evaluation of mechanisms underlying SIRT1 effects. Here, we have used a genetic knockout model of SIRT1 to definitively delineate the role of SIRT1 in regulating leukemic and normal stem cell growth and in selectively promoting leukemic hematopoiesis and mediating TKI resistance. These studies reveal a previously unrecognized role for SIRT1 in mediating altered mitochondrial metabolism in CML LSCs in a BCR-ABL kinase–independent manner.
Results
SIRT1 deletion has minimal effects on hematopoiesis in normal mice.
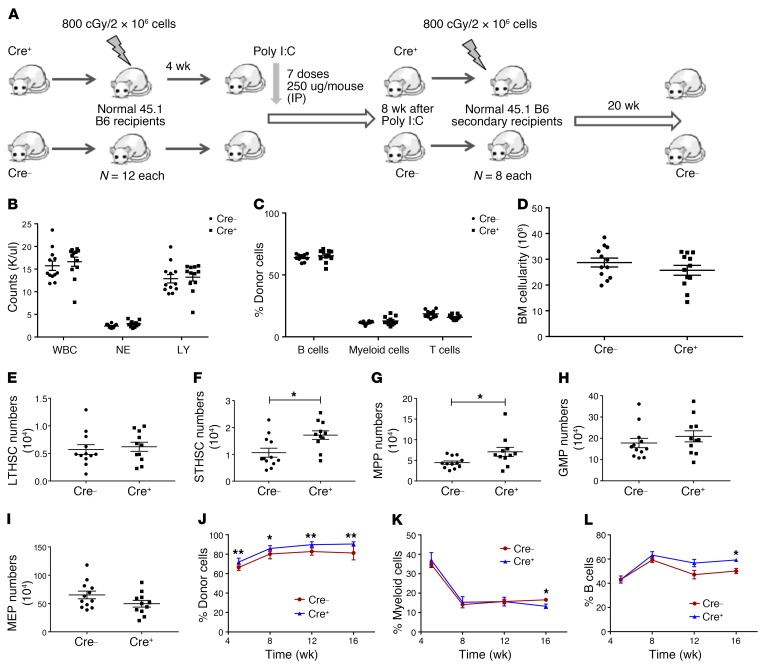
To study the effect of SIRT1 deletion on normal hematopoiesis, we conditionally deleted SIRT1 in murine HSC by crossing SIRT1 exon-4 floxed (SIRT1fl/fl) mice with the Mx1-Cre strain (20). SIRT1 deletion was induced by activating Mx1-Cre by i.p. injections of polyinosine-polycytosine (Poly[I:C]) every other day for 7 doses. SIRT1fl/fl mice without Mx1-Cre were used as controls. BM cells from Mx1-Cre SIRT1fl/fl (Cre+, CD45.2) mice were transplanted into irradiated (800 cGy) CD45.1 congenic recipients to generate a cohort of mice with Mx1-Cre SIRT1fl/fl hematopoietic cells, and BM cells from Cre– mice were transplanted as controls. Mx1-Cre SIRT1fl/fl and control BM recipients received poly(I:C) starting 4 weeks after transplant and were analyzed 8 weeks later (Figure 1A). SIRT1 deletion was confirmed by quantitative reverse-transcriptase PCR (Q-RT-PCR) and genomic D NA PCR (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/JCI127080DS1). SIRT1-deleted mice did not show significant alteration in blood counts (Figure 1B) or in donor-derived mature cell populations compared with controls (Figure 1C). No significant difference in BM cellularity (Figure 1D) or long-term HSCs (LTHSCs) (Lin–Sca1+c-Kit+ [LSK], Flt3–CD150+CD48–) numbers were seen (Figure 1E), but short-term HSCs (STHSCs) (LSK,Flt3–CD150–CD48–) (P = 0.01) (Figure 1F) and multipotent progenitors (MPPs) (LSK, Flt3–CD150–CD48+) (P = 0.03) (Figure 1G) were increased in the BM of SIRT1-deleted mice compared with those in control mice. BM committed progenitor populations, granulocyte-macrophage progenitors (GMPs) (Lin–Sca1–c-Kit+CD34+FcγRII/IIIhi) (Figure 1H), and megakaryocytic-erythrocytic progenitors (MEPs) (Lin–Sca1–c-Kit+CD34–FcγRII/IIIlo) (Figure 1I) remained unaffected upon SIRT1 deletion. Upon secondary transplantation of BM from SIRT1-deleted mice, a modest increase in donor cell engraftment was seen compared with BM from control mice (Figure 1, J–L). Analysis of BM from secondary recipients obtained 20 weeks after transplantation did not show significant change in stem and progenitor populations (Supplemental Figure 1, C–G). Our results are consistent with those of Leko et al., showing that SIRT1 deletion did not affect HSC maintenance and long-term reconstitution in adult mice in the steady state (21), but are in contrast with other studies that show that SIRT1 deletion results in anemia, myeloid expansion, and lymphoid depletion, associated with DNA damage accumulation, gene expression changes associated with aging, and compromised hematopoiesis with increased HSC cycling and exhaustion in response to stress (22–24).
Figure 1. Minimal effects of Mx1-Cre mediated SIRT1 deletion on normal hematopoiesis.
(A) Experimental strategy for studying the role of SIRT1 deletion in normal hematopoiesis. BM cells from Mx1-Cre SIRT1fl/fl mice were transplanted into irradiated (800 cGy) CD45.1 congenic recipients to generate a cohort of mice with Mx1-Cre SIRT1fl/fl hematopoietic cells. BM cells from Cre– SIRT1fl/fl mice were transplanted as controls. Mice were treated with i.p. injections of poly(I:C) starting 4 weeks after transplantation to induce SIRT1 deletion and analyzed 8 weeks later. (B) Peripheral blood WBC, neutrophil (NE), and lymphocyte (LY) counts at 8 weeks after SIRT1 deletion (n = 12 each). (C) Percentages of donor B cells, Gr1+Mac1+ myeloid cells, and T cells assessed by flow cytometry at 8 weeks. (D) BM cellularity at 8 weeks after SIRT1 deletion. (E–I) Effect of SIRT1 deletion on absolute numbers of BM LTHSCs (E), STHSCs (F), MPPs (G), GMPs (H), and MEPs (I) at 8 weeks after SIRT1 deletion. (J–L) Results of transplantation of BM cells into secondary recipients (n = 8 each). Percentages of donor cells (J), myeloid cells (K), and B cells (L) in peripheral blood at 5 through 16 weeks after secondary transplant. Error bars represent mean ± SEM. *P < 0.05; **P < 0.01, t test.
SIRT1 deletion impairs leukemia development in CML mice.
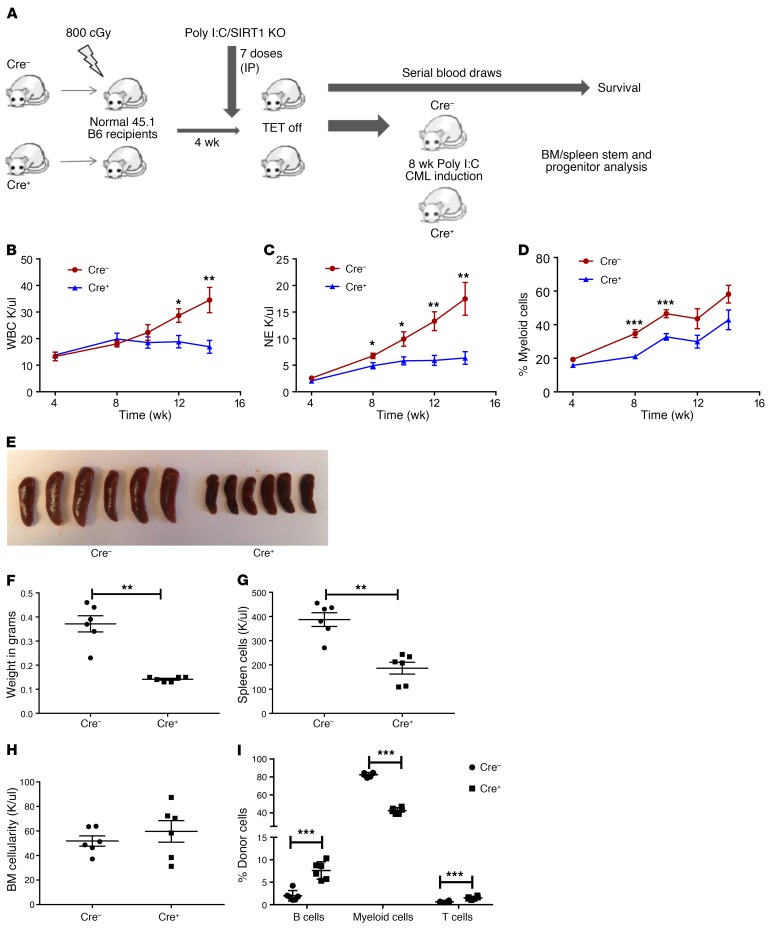
To study the requirement of SIRT1 for CML development, we used a well-characterized and representative SCL-tTA/BCR-ABL transgenic mouse model of chronic-phase CML (25–27). In this model, tetracycline withdrawal leads to BCR-ABL expression in HSCs and development of a CML-like myeloproliferative disorder. SCL-tTA/BCR-ABL mice were crossed with Mx1-Cre SIRT1fl/fl mice to generate SCL-tTA/BCR-ABL Mx1-Cre SIRT1fl/fl mice (BA Mx1-Cre SIRT1fl/fl). BA SIRT1fl/fl mice lacking Mx1-Cre were used as controls. BM cells from BA Mx1-Cre SIRT1fl/fl (Cre+) or control (Cre–) mice were transplanted into irradiated congenic recipients to generate a cohort of mice with a similar time for onset of leukemia (28–30). Cre-mediated deletion of SIRT1 was induced by i.p. poly(I:C) injections, followed by withdrawal of tetracycline to induce BCR-ABL expression (Figure 2A). SIRT1 deletion profoundly inhibited CML development. Control mice developed progressive neutrophilic leukocytosis and increasing morbidity from leukemia after BCR-ABL induction, whereas BA Mx1-Cre SIRT1f/f mice did not develop evidence of morbidity and demonstrated significantly lower WBC (Figure 2B), neutrophil counts (Figure 2C and Supplemental Figure 2A), and Gr1+Mac-1+ myeloid cell frequency at 14 weeks (Figure 2D), with increased lymphocyte frequency (Supplemental Figure 2B).
Figure 2. Mx1-Cre mediated SIRT1 deletion inhibits leukemia development in CML mice.
(A) Experimental strategy for studying the role of SIRT1 deletion on CML hematopoiesis. BM cells from either BA Mx1-Cre SIRT1fl/fl or Cre– controls (both CD45.2) were transplanted into irradiated (800 cGy) CD45.1 congenic recipients (2 × 106 cells/mouse). Cre-mediated deletion of SIRT1 was induced by pIpC injection starting at 4 weeks after transplant, followed by tetracycline withdrawal to induce BCR-ABL expression. Mice were followed for CML and SIRT1 development and analyzed for BM and spleen stem/progenitors (shown in Figure 3). (B–D) Peripheral blood parameters were evaluated over time after SIRT1 deletion. Results for WBCs (B), neutrophils (C), and Gr1+Mac1+ myeloid cells (D) determined by flow cytometry. (E–I) Effect of SIRT1 deletion on spleen and BM parameters at 8 weeks, including spleen size (E), spleen weight (F), spleen cellularity (G), BM cellularity (H), and frequencies of CD45.2 donor cells, B cells, and myeloid cells by flow cytometry in BM (I) (n = 6 each). Error bars represent mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001, t test.
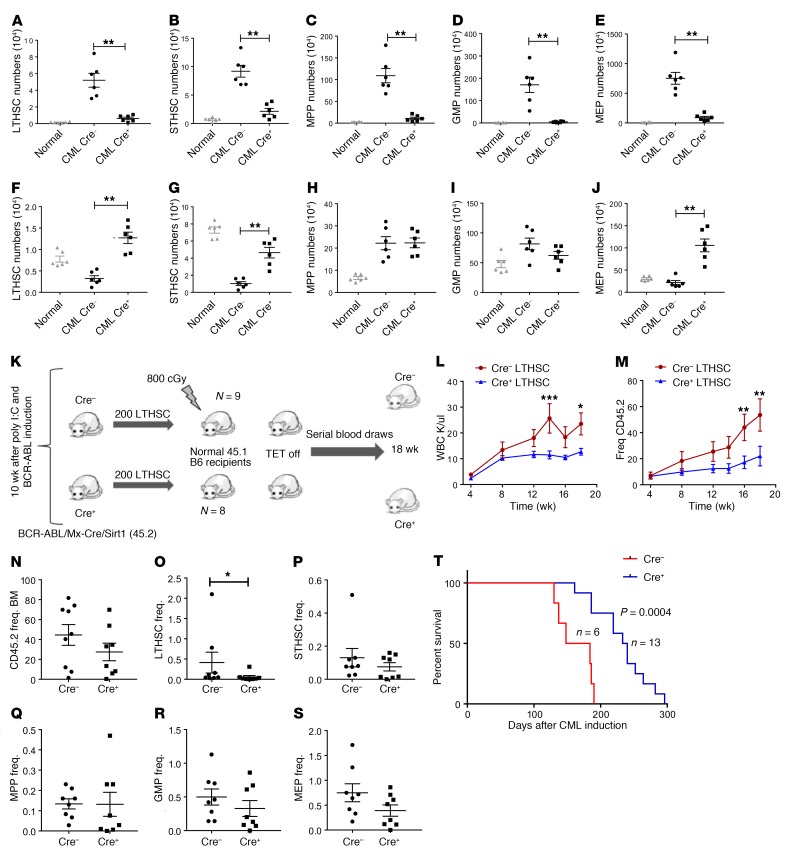
The effect of SIRT1 knockdown on spleen and BM populations was evaluated in BA Mx1-Cre SIRT1fl/fl mice sacrificed at 8 weeks after polyinosinic:polycytidylic acid (pIpC) injection and BCR-ABL induction. SIRT1-deleted mice demonstrated significant reduction in spleen size (Figure 2E), weight (Figure 2F), and cellularity (Figure 2G) compared with controls. There was no significant change in BM cellularity (Figure 2H), but myeloid (Gr1+Mac1+) cells in the BM were significantly reduced compared with those in controls (Figure 2I). As previously shown, CML mice demonstrated reduced LTHSCs, STHSCs, and MEPs as well as increased MPPs and GMPs in the BM and massive expansion in all stem and progenitor populations in the spleen compared with normal mice, consistent with increased egress of stem cells from the BM to the spleen (Figure 3, A–J, and Supplemental Figure 2, C–L) (27). The absolute numbers and frequencies of all splenic primitive stem and progenitor populations were significantly reduced in SIRT1-deleted mice (Figure 3, A–E, and Supplemental Figure 2, C–G), whereas BM LTHSC, STHSC, and MEP populations were significantly increased in SIRT1-deleted mice (Figure 3, F–J, and Supplemental Figure 2, H–L). These observations indicate that SIRT1 deletion reverses several of the hematopoietic alterations observed in the spleen and BM of CML mice.
Figure 3. Mx1-Cre–mediated SIRT1 deletion inhibits CML stem and progenitor cells.
(A–E) Effect of SIRT1 deletion on splenic stem and progenitor subpopulations, including LTHSCs (A), STHSCs (B), MPPs (C), GMPs (D), and MEPs (E), at 8 weeks after SIRT1 deletion (n = 6 each). (F–J) Effect of SIRT1 deletion on BM stem and progenitor subpopulations, including LTHSCs (F), STHSCs (G), MPPs (H), GMPs (I). and MEPs (J). Corresponding stem and progenitor cell populations from normal mice are shown for comparison. (K) Experimental strategy for checking long-term repopulating potential of LTHSCs following SIRT1 deletion. Donor LTHSCs (CD45.2) were selected from SIRT1-deleted and control primary recipient mice by flow cytometry and transplanted to sublethally irradiated (800cGy) secondary recipients (200 cells/mouse), together with 500,000 supporting BM cells (CD45.1) (n = 8–9 each). (L–S) Recipient mice were followed with serial blood counts and subsequently analyzed for BM stem and progenitor cells. Peripheral blood WBC counts and frequency of CD45.2 donor cells (M) in secondary recipients are shown. (N–S) Frequency (Freq.) of donor cells, including total CD45.2 (N), LTHSCs (O), STHSCs (P), MPPs (Q), GMPs (R), and MEPs (S) in secondary recipient BM. (T) Kaplan-Meier analysis of survival of SCL-tTA/BCR-ABL Mx1-Cre SIRT1fl/fl mice compared with Cre controls. Error bars represent mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001, t test, except Kaplan-Meier analysis.
To check the effect of SIRT1 deletion on in vivo repopulating ability, LTHSCs purified from BM of mice that received BA-Mx1-Cre SIRT1fl/fl or control cells were transplanted into secondary recipients (Figure 3K). Mice receiving SIRT1-deleted LTHSCs showed significantly reduced WBC counts (Figure 3L) and donor cell engraftment (Figure 3M). BM donor engraftment was not reduced (Figure 3N), but donor LTHSC engraftment in BM was significantly reduced compared with that in controls (Figure 3O), whereas other stem/progenitor populations were not affected (Figure 3, P–S). These results are consistent with the hypothesis that SIRT1 deletion results in reduced CML LSC regenerative potential.
A cohort of primary BA Mx1-Cre SIRT1fl/fl and control mice were followed for survival. SIRT1-deleted mice showed significantly increased survival compared with controls (Figure 3T). Cre+ mice showed significantly reduced percentages and absolute numbers of neutrophils and Gr1+Mac-1+ myeloid cells compared with Cre– controls, but developed neutrophilic leukocytosis with extended follow-up (Supplemental Figure 3, A–D). The percentages of lymphocytes and CD19+B220+ B lymphoid cells relative to myeloid cells was significantly increased in Cre+ mice, but the absolute number of B cells was not increased (Supplemental Figure 3, E–H). Hematopoietic cells in Cre+ mice continued to demonstrate SIRT1 deletion in genomic DNA (Supplemental Figure 4A) and lacked SIRT1 mRNA expression (Supplemental Figure 4B) at the time of development of leukocytosis.
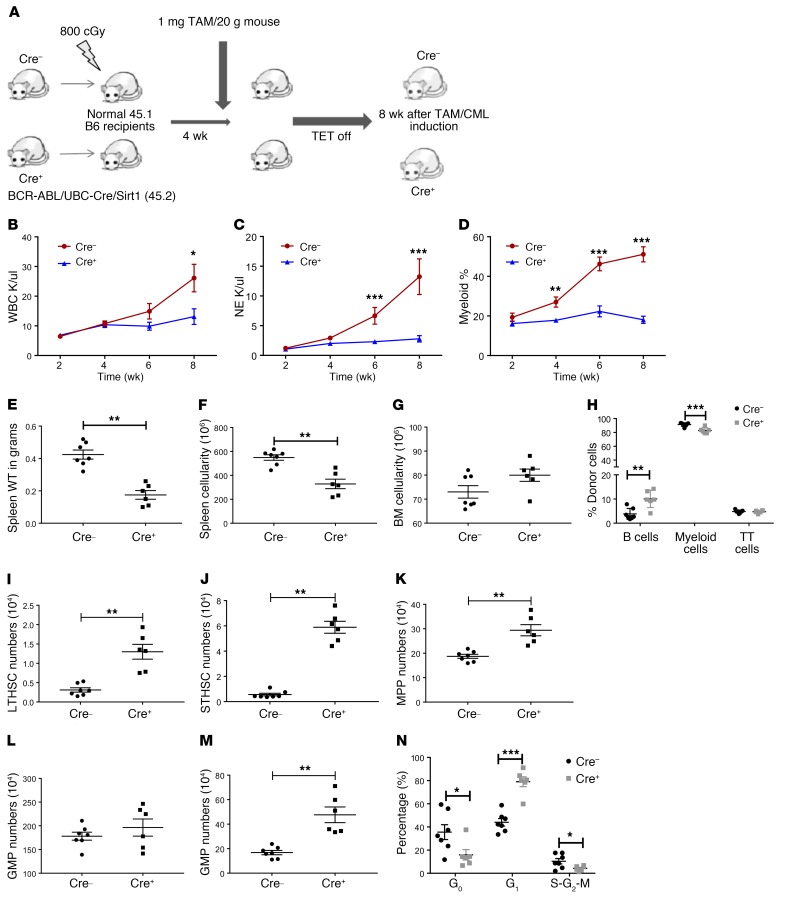
Since poly(I:C) activates Mx1-Cre activity by inducing expression of type 1 interferons, which could also affect HSC cycling and leukemia progression (31), we developed an additional genetic deletion model to study SIRT1’s role in CML by crossing BCR-ABL (BA) mice with SIRT1fl/fl and UBC-Cre-ERT2 strains (tamoxifen inducible). BM cells from BA UBC-Cre-ERT2 SIRT1fl/fl (Cre+) or control (Cre–) mice (both CD45.2) were transplanted into irradiated (800 cGy) CD45.1 congenic recipients (2 × 106 cells/mouse) to generate a cohort of leukemic mice. SIRT1 deletion was induced by i.p. injection of tamoxifen daily for 5 days starting at 4 weeks after transplant, followed by withdrawal of tetracycline to induce BCR-ABL expression (Figure 4A). Effective SIRT1 excision was confirmed by PCR (Supplemental Figure 5A). UBC-Cre-ERT2 SIRT1fl/fl mice demonstrated significant reduction in peripheral blood (PB) WBC counts (Figure 4B), neutrophils (Figure 4C and Supplemental Figure 5B), and Gr1+Mac-1+ myeloid cells (Figure 4D) as well as significantly higher CD19/B200-positive B lymphocytes (Supplemental Figure 5C) compared with controls. Spleen weight (Figure 4E), cellularity (Figure 4F), and splenic stem and progenitor cells (Supplemental Figure 5, D–H) were significantly decreased in SIRT1-deleted mice. BM cellularity was not changed (Figure 4G), and Gr1+Mac-1+ myeloid cells were modestly reduced in the BM of SIRT1-deleted mice (Figure 4H). As in BA Mx1-Cre SIRT1fl/fl mice, SIRT1 deletion from BA UBC-Cre-ERT2 SIRT1f/f mice resulted in increases in BM LTHSCs, STHSCs, and MEP numbers (Figure 4, I–M). Therefore, the effects of SIRT1 deletion were consistent regardless of the model system used and the methods used for Cre induction.
Figure 4. UBC ERT2-Cre–mediated SIRT1 deletion inhibits CML hematopoiesis.
(A) Experimental strategy for studying the effect of SIRT1 deletion on CML development using UBC ERT2-Cre/SIRT1fl/fl/BCR-ABL model: BM cells from either SCL-tTA-BCR-ABL UBC-ERT2-Cre SIRT1fl/fl or Cre– controls (both CD45.2) were transplanted into irradiated (800 cGy) CD45.1 congenic recipients (2 × 106 cells/mouse). Cre-mediated deletion of SIRT1 was induced by tamoxifen (TAM) injection (1 mg/mouse) starting at 4 weeks after transplant, followed by tetracycline withdrawal to induce BCR-ABL expression. Mice were sacrificed and analyzed 8 weeks after CML induction and SIRT1 deletion (n = 10 each). (B–D) Effect of SIRT1 deletion on blood parameters, including WBC (B), neutrophil counts (C), and donor Gr1+Mac1+ myeloid cell frequencies determined by flow cytometry (D) (n = 6–7). Effect of SIRT1 deletion on spleen weight (E), spleen cellularity (F), BM cellularity (G), and frequencies of donor B cells, myeloid cells, and T cells in BM by flow cytometry (H) at 8 weeks. (I–M) Effect of SIRT1 deletion on absolute numbers of BM LTHSCs (I), STHSCs (J), MPPs (K), GMPs (L), and MEPs (M). (N) Cell cycle analysis on LTHSC populations from BM using DAPI and KI67 labeling. Error bars represent mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001, t test.
The cell cycle status of BM LTHSCs was evaluated by Ki-67 and DAPI labeling (BA UBC-Cre SIRT1fl/fl). We observed a significant increase in the proportion of LTHSCs in the G1 phase, but a decrease in LTHSCs in the G0 phase after SIRT1 deletion, indicating that SIRT1 deletion leads to loss of quiescence (Figure 4N). A reduction in cells in the S-G2-M phase was also seen. Loss of quiescence may explain why LTHSC numbers were increased, but their repopulating ability was reduced, following SIRT1 deletion.
TKI treatment leads to additional suppression of CML hematopoiesis in SIRT1-deleted mice.
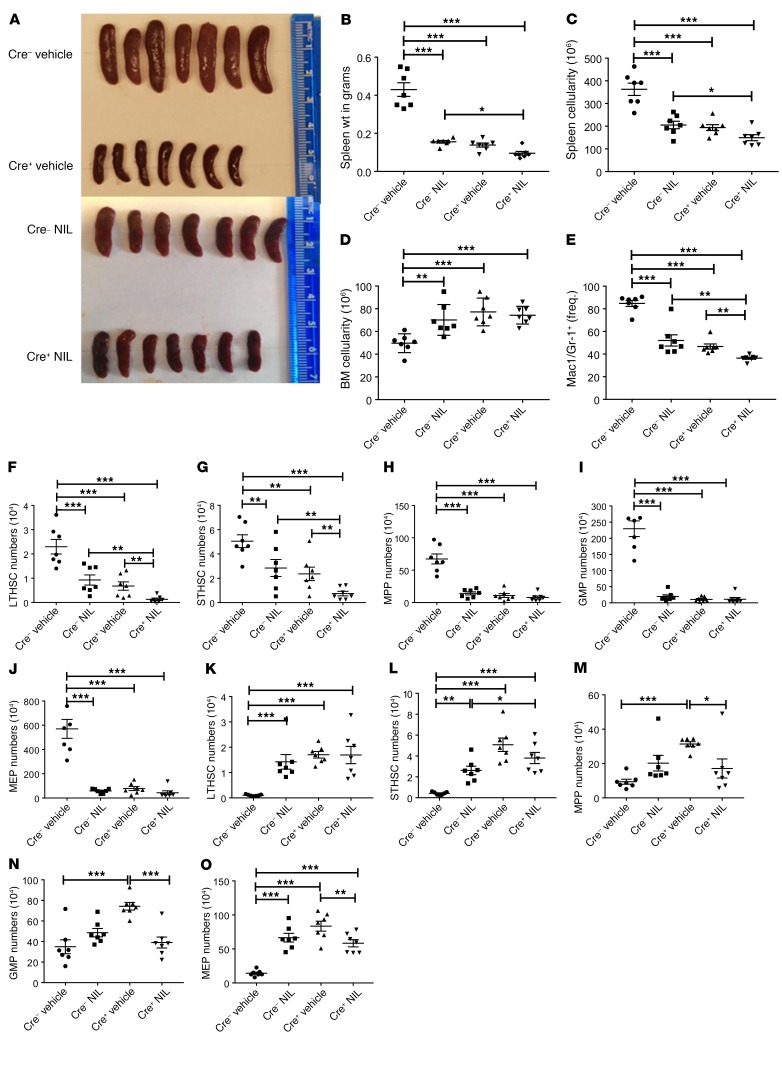
We studied the effect of BCR-ABL TKIs on SIRT1-deleted CML mice. We have previously shown that TKI treatment does not affect SIRT1 expression in CML stem cells (6). SIRT1-deleted (BA Mx1-Cre SIRT1fl/fl) or control CML mice in whom BCR-ABL was induced for 10 weeks were treated with the potent BCR-ABL TKI nilotinib (NIL) (50 mg/kg/d) or vehicle for 4 weeks. Blood counts were monitored during treatment (Supplemental Figure 6, A–C) and mice sacrificed and analyzed after 4 weeks of treatment. NIL treatment resulted in marked reduction in spleen size (Figure 5A), weight (Figure 5B), and cellularity (Figure 5C) in control mice, and further modest reduction in these parameters was seen with NIL treatment of SIRT1-deleted mice. BM cellularity was increased upon NIL treatment in control mice, but not further increased on NIL treatment in SIRT1-deleted mice (Figure 5D). BM myeloid cells were reduced upon NIL treatment in control mice and further reduced in SIRT1-deleted mice (Figure 5E). NIL treatment significantly reduced splenic stem and progenitor cells (Figure 5, F–J), but increased primitive BM stem and progenitor cells (Figure 5, K–O), suggesting sensitivity of splenic stem and progenitor cells, but resistance of BM stem cells to TKI treatment. Treatment of SIRT1-deleted mice with NIL led to further reduction in LTHSCs and STHSCs in the spleen (Figure 5, F–J) and MPPs, MEPs, and GMPs in the BM (Figure 5, K–O) without change in BM LTHSC numbers, indicating that TKI treatment further inhibits primitive CML stem cells in the spleen and committed progenitors in the BM of SIRT1-deleted mice.
Figure 5. TKI treatment leads to enhanced inhibition of CML hematopoiesis in SIRT1-deleted mice.
SIRT1-deleted or control CML mice were treated with NIL or vehicle control for a period of 4 weeks, starting 10 weeks after SIRT1 deletion and CML induction (n = 7 per arm). (A–E) Effects of NIL treatment on spleen size (A), spleen weight (B), spleen cellularity (C), BM cellularity (D), and frequency of Gr1+Mac1+ myeloid cells in BM (E). (F–J) Effect of NIL treatment on absolute numbers of splenic LTHSCs (F), STHSCs (G), MPPs (H), GMPs (I), and MEPs (J). (K–O) Effect of NIL treatment on absolute numbers of BM LTHSCs (K), STHSCs (L), MPPs (M), GMPs (N), and MEPs (O). Error bars represent mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001, ordinary 1-way ANOVA, correcting for multiple comparisons by controlling the FDR using the 2-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli.
SIRT1 deletion inhibits expression of mitochondrial genes in CML stem/progenitor cells.
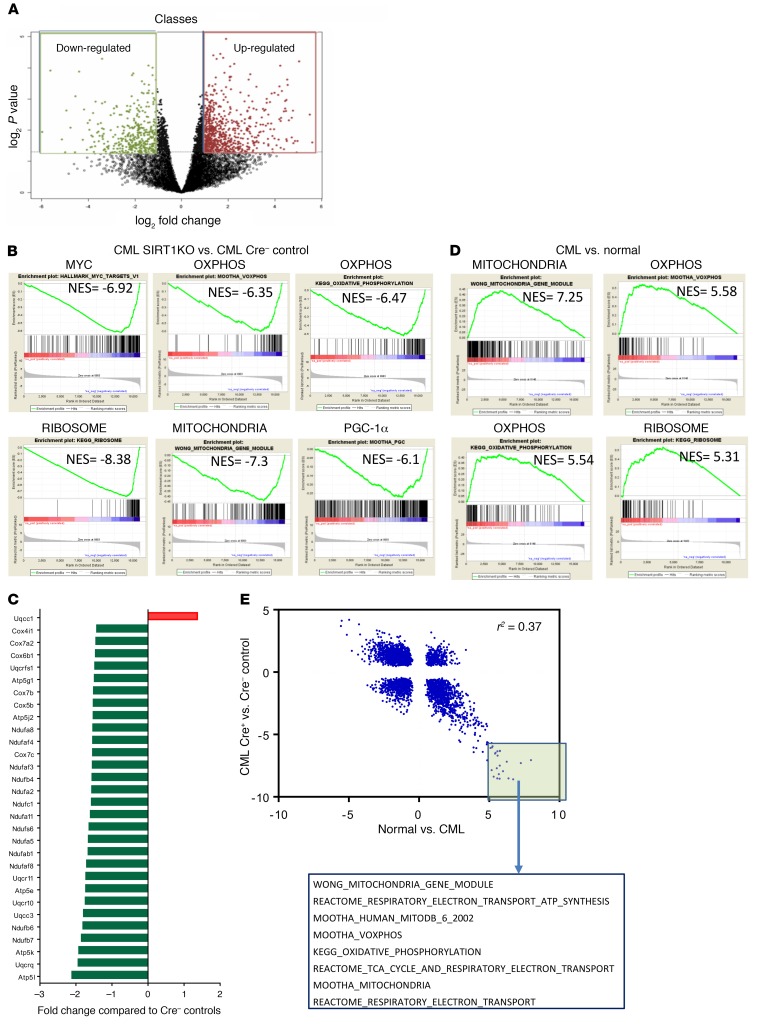
We performed global gene expression analysis to obtain further insights into the mechanisms by which SIRT1 deletion exerts antileukemic effects in BCR-ABL mice. We performed RNA-Seq on BM LSK stem cells from SIRT1-deleted and control CML mice and identified differentially expressed genes (2× fold-change, P < 0.05, n = 1,063) (Figure 6A). Gene Set Enrichment Analysis (GSEA) was performed to identify gene sets enriched in SIRT1-deleted versus control stem cells (32) (http://www.broadinstitute.org/gsea/). Gene sets most positively enriched in SIRT1-deleted stem cells included those related to UV response, HSC signatures, TP53 targets, and WNT and Notch signaling (Supplemental Table 2). The most negatively enriched gene sets in SIRT1-deleted CML stem cells included those related to Myc, mitochondria and oxidative phosphorylation, embryonic stem (ES) cells, ribosomes, proteasomes, and tumor invasiveness (Figure 6B and Supplemental Table 1). Indeed, of the top 25 downregulated gene sets, 10 were related to mitochondria and/or oxidative phosphorylation, and widespread and significant inhibition of expression of mitochondrial electron transport chain–related genes was seen in SIRT1-deleted CML stem cells (Figure 6C). Results from SIRT1-deleted and control CML LSKs were compared with an analysis of LTHSCs from CML and WT mice using RNA-Seq. It was notable that mitochondrial and/or oxidative phosphorylation gene sets were among the top enriched gene sets in CML compared with normal LTHSCs (Figure 6D). Moreover, there was a significant negative correlation (r2 = 0.37, P < 0.0001) between the normalized enrichment scores (NES) of gene sets enriched in CML versus normal stem cells and SIRT1-deleted versus control CML stem cells (Figure 6E), and 61 of the top 100 positively enriched gene sets in CML stem cells were negatively enriched in SIRT1-deleted CML LSCs (Supplemental Table 3). These results suggest that alterations in gene expression programs in CML stem cells are significantly reversed after SIRT1 deletion.
Figure 6. SIRT1 deletion inhibits mitochondrial gene expression in CML and normal hematopoiesis.
(A) Volcano plot showing differentially expressed genes in SIRT1-deleted (SIRT1 KO) compared with control (WT) CML stem cells (LSK) on RNA-Seq analysis. 1,063 Differentially expressed genes (2× fold change and P < 0.05) are highlighted. (B) GSEA analysis of gene expression data showing significant negative enrichment (downregulation) of Myc targets, ribosomal, oxidative phosphorylation, mitochondria, and PGC-1α–related gene sets in SIRT1-KO compared with control CML stem cells. (C) Downregulation of mitochondrial electron transport chain complex genes in SIRT1-deleted compared with control CML stem cells (red signifies upregulation, and green signifies downregulation). (D) GSEA analysis of gene expression data showing significant positive enrichment (upregulation) of mitochondrial, oxidative phosphorylation, and ribosomal gene sets in CML compared with normal stem cells. (E) Pearson’s correlation of NES for the 3567 GSEA signatures derived from analysis of normal versus CML compared with SIRT1-KO versus WT stem cells. The highlighted box indicates gene sets that are most highly enriched in the CML LTHSC versus normal LTHSC analysis and negatively enriched in the SIRT1 WT versus SIRT1-KO LSK analysis, and the mitochondria-related gene sets present among these, are shown.
SIRT1 deletion results in reduced mitochondrial respiration in CML stem/progenitor cells.
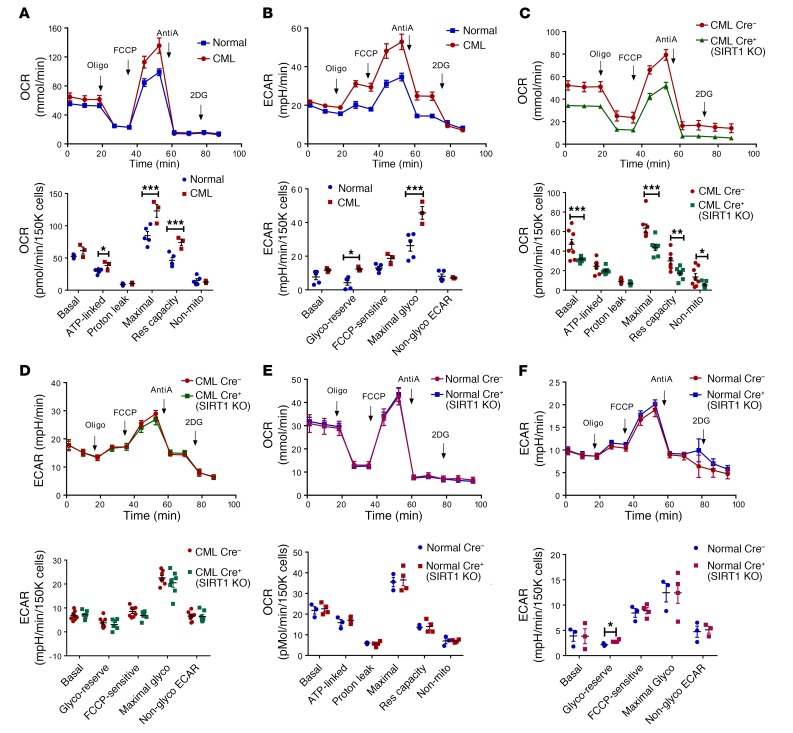
We investigated the effect of SIRT1 deletion on energy metabolism by performing extracellular flux analysis for cellular bioenergetics and glycolysis in c-Kit+ stem/progenitor cells selected from SIRT1-deleted and control CML and normal mice. Comparing CML and normal mice, we found significantly increased ATP-linked respiration, maximal mitochondrial respiration, and mitochondrial reserve capacity (Figure 7A), as well as increased maximal glycolysis and glycolytic reserve (Figure 7B), in c-Kit+ cells from CML mice compared with normal mice, consistent with recent reports that mitochondrial respiration is increased in CML stem cells (33). Next, comparing Cre+ SIRT1-deleted CML mice (BA UBC-Cre-ERT2 SIRT1f/f) and control Cre– CML mice, we observed that SIRT1-deleted CML c-Kit+ cells demonstrated significant reduced basal and maximal mitochondrial respiration and reduced mitochondrial reserve capacity compared with control CML c-Kit+ cells (Figure 7C). In contrast, SIRT1-deleted CML c-Kit+ cells did not demonstrate alteration in extracellular acidification rate (ECAR) parameters, including maximal glycolysis and glycolytic reserve, compared with control CML cells (Figure 7D). These results support an important role for SIRT1 in enhanced oxidative phosphorylation, but not in enhanced glycolysis in CML stem/progenitor cells. Unlike CML cells, c-Kit+ cells from normal SIRT1-deleted (UBC-Cre-ERT2 SIRT1f/f) mice did not demonstrate significant change in mitochondrial respiration (Figure 7E) or ECAR parameters (Figure 7F) compared with c-Kit+ cells from normal Cre– mice. These observations suggest that effects of SIRT1 deletion on mitochondrial respiration seen in CML were not observed in normal stem/progenitor cells in the steady state.
Figure 7. SIRT1 deletion inhibits mitochondrial respiration in CML and normal hematopoiesis.
Analysis of OCR and ECAR was performed using Seahorse XF analyzer to assess mitochondrial respiration and glycolysis. (A) OCR was measured in CML (n = 3) and normal (n = 5) c-Kit+ cells with sequential addition of oligomycin (Oligo, complex V inhibitor), FCCP (a protonophore), and antimycin A (AntiA, complex III inhibitor), to analyze ATP-linked respiration, proton leak respiration, maximal respiratory capacity, mitochondrial reserve capacity, and nonmitochondrial respiration. The upper panel shows the OCR profiles for CML and normal c-Kit+ cells. The lower panel shows mitochondrial bioenergetic parameters calculated from extracellular flux analysis. Mito, mitochondrial, Res, reserve. (B) ECAR was measured in CML (n = 3) and normal (n = 5) c-Kit+ cells with serial addition of oligomycin, FCCP, antimycin A, and 2-DG to measure basal glycolysis, glycolytic reserve, FCCP-sensitive glycolysis, maximal glycolysis, and nonglycolytic ECAR. The upper panel shows the ECAR profiles for CML and normal c-Kit+ cells. The lower panel shows the calculated glycolytic parameters. Glyco, glycolysis. (C and D) OCR and ECAR in CML Cre– (n = 8) and CML Cre+ SIRT1-deleted (SIRT1-KO; n = 7) c-Kit+ cells. (E and F) OCR and ECAR in normal Cre– (n = 8) and normal Cre+ SIRT1-deleted (n = 7) c-Kit+ cells. Error bars represent mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001, 2-way ANOVA correcting for multiple comparisons by controlling the FDR using the 2-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli. 3–6 assay replicate assays were performed for each sample.
TKI treatment does not inhibit mitochondrial respiration in murine and human CML stem/progenitor cells.
To evaluate the effect of TKI on mitochondrial metabolism in LSCs, CML mice were treated with NIL for 2 weeks. c-Kit+ cells obtained from NIL-treated CML mice did not demonstrate significant changes in mitochondrial bioenergetics (Figure 8, A and B) or glycolytic parameters (Supplemental Figure 7A) compared with cells from vehicle-treated mice. These results suggest that increased mitochondrial respiration in murine CML stem/progenitor cells is BCR-ABL kinase independent.
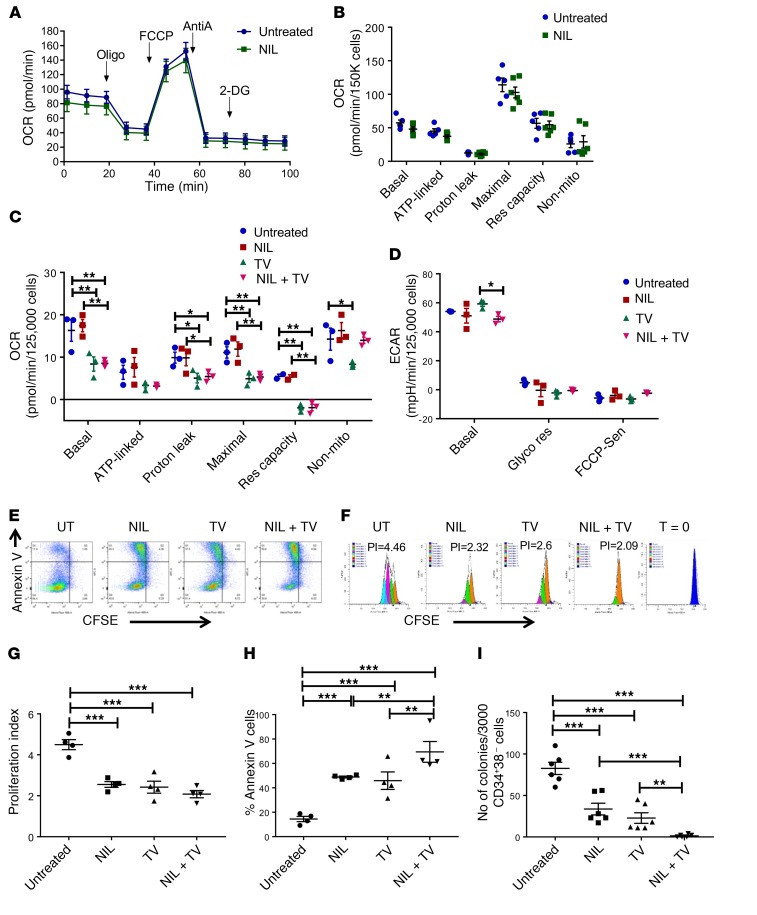
Figure 8. SIRT1 and tyrosine kinase inhibition result in dual inhibition of mitochondrial respiration and glycolysis in human CML stem/progenitor cells.
(A and B) OCR measurements in c-Kit+ cells from CML mice treated with NIL or vehicle (n = 7 each) for 2 weeks. (C and D) OCR (C) and ECAR (D) measurements in CP CML CD34+ cells (n = 4) after exposure to NIL (1 μM), TV39OH (5 μM), combination, or vehicle for 3 hours. (E–G) CML CD34+CD38–CFSEhi cells (n = 4) were exposed to NIL (1 μM) TV39OH (5 μM), combination, or vehicle for 72 hours. (E) Representative flow cytometry plots showing annexin V and CFSE expression. (F) Representative flow cytometry histograms showing CFSE fluorescence in CD34+CD38–CFSEhi cells. (G) Proliferation index measured based on analysis of CFSE fluorescence in CD34+CD38–CFSEhi cells. (H) Apoptosis in CD34+CD38–CFSEhi cells measured by annexin V labeling. (I) CP CML CD34+CD38– cells (n = 3) were exposed to vehicle, NIL (1 μM), TV39OH (5 μM), or combination for 72 hours and plated in methylcellulose progenitor culture. Colonies were enumerated after 14 days. Error bars represent mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001, ordinary 1-way ANOVA (G–I) or 2-way ANOVA (B–D) correcting for multiple comparisons by controlling the FDR using the 2-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli.
GSEA analysis of reported gene expression data for CD34+CD38– stem cells from CML patients with or without treatment with the TKI imatinib showed that TKI-treated CML stem cells did not demonstrate significant enrichment of mitochondrial- or oxidative phosphorylation–related gene sets compared with vehicle-treated controls (not shown) (34). We studied the effects of SIRT1 inhibition and TKI treatment on mitochondrial energetics in human CML CD34+ stem and progenitor cells. Our previous studies used the SIRT1 inhibitor, Tenovin 30 (TV30), which has recently been shown to have additional autophagy inhibitory effects that could affect mitochondrial mass and function independently of SIRT1. Therefore, for the current studies, we used a congener, Tenovin 39OH (TV39OH), which retains SIRT1 inhibitory effects, but does not have autophagy inhibitory properties (35). CML BM CD34+ cells were treated with TV39OH (5 μM), the TKI NIL (1 μM), or a combination. SIRT1 inhibition with TV39OH resulted in significant reduction in oxygen consumption rate (OCR) in CML CD34+ cells compared with vehicle controls (Figure 8C), without significantly affecting ECAR (Figure 8D), consistent with results obtained with SIRT1 deletion in murine CML stem cells. In contrast, NIL treatment did not affect OCR or ECAR in CML CD34+ cells compared with vehicle controls. These results indicate that increased mitochondrial respiration in human CML stem/progenitor cells is also kinase independent.
We evaluated the effect of treatment with NIL, TV39OH, or the combination on CML stem/progenitor cell viability and growth. CML CD34+CD38– stem/progenitor cells and CD34+CD38+ committed progenitors cultured for 72 hours with TV39OH or NIL both showed significant reduction of CML cell proliferation compared with controls, as measured by reduction in CFSE fluorescence (Figure 8, E–G). Treatment with TV39OH or NIL both significantly increased apoptosis of CML CD34+CD38– cells, and the combination resulted in significantly increased apoptosis compared with either agent alone (Figure 8, E and H). Similar results were seen for CML CD34+CD38+ cells (Supplemental Figure 7, B–E). TV39OH and NIL significantly inhibited CML CD34+CD38– cell colony–forming ability, and the combination of NIL and TV39OH led to further reduction in colony-forming capacity (Figure 8I). These results show that combined SIRT1 and BCR-ABL kinase inhibition can enhance suppression of human CML stem/progenitor cells.
PGC-1α regulates mitochondrial metabolism and contributes to TKI resistance in CML stem/progenitor cells.
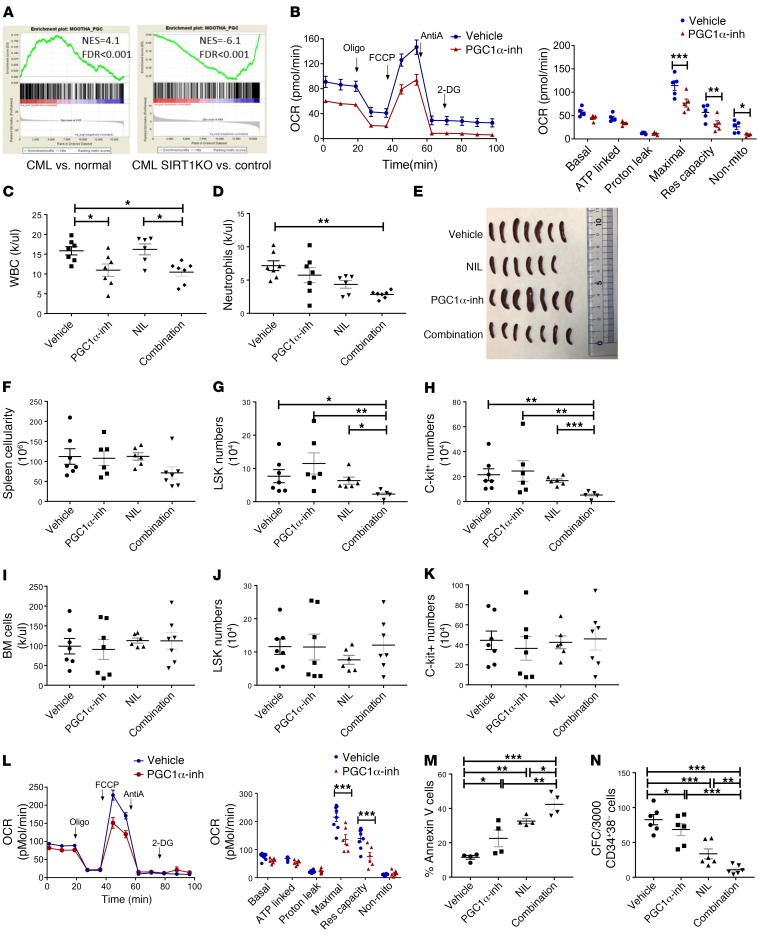
SIRT1 can deacetylate and activate the transcriptional coactivator PGC-1α to enhance mitochondrial DNA replication and gene expression and promote mitochondrial activity in response to energy needs (36). A PGC-1α–related gene set was enriched in stem/progenitor cells from CML compared with normal mice and downregulated in stem/progenitor cells from SIRT1-deleted compared with control CML mice (Figure 9A). Extracellular flux analysis of c-Kit+ cells isolated from CML mice following treatment with the PGC-1α inhibitor SR-18292 (45 mg/kg i.p.) for 14 days showed reduced OCR (Figure 9, B and C) without change in ECAR (Supplemental Figure 8, A and B). Although the PGC-1α inhibitor did not significantly affect CML hematopoiesis by itself, combined treatment with the PGC-1α inhibitor and NIL (50 mg/kg oral gavage) significantly reduced peripheral blood counts, donor engraftment, LSK and c-Kit+ cells in spleen, and donor LTHSCs in BM compared with NIL alone (Figure 9, C–K, and Supplemental Figure 8, B–M). We also evaluated the effect of in vitro treatment of CML CD34+ cells with the PGC-1α inhibitor SR-18292 (10–40 μM). We confirmed that this range of inhibitor concentrations was effective in increasing PGC-1α acetylation in K562 cells (not shown). Extracellular flux analysis demonstrated that treatment with the PGC-1α inhibitor reduced OCR in CML CD34+ cells without an effect on ECAR (Figure 9L and Supplemental Figure 8N). The PGC-1α inhibitor only modestly enhanced apoptosis and reduced colony-forming cell (CFC) potential of CML CD34+CD38– cells by itself, but in combination with NIL, significantly increased apoptosis and reduced CFC numbers compared with NIL alone (Figure 9, M and N). These results support an important role for PGC-1α in enhanced mitochondrial respiration in CML stem/progenitor cells. They also indicate that PGC-1α inhibition does not directly suppress CML hematopoiesis, but inhibits leukemic hematopoiesis in combination with TKI treatment.
Figure 9. PGC-1α regulates mitochondrial metabolism and contributes to TKI resistance in CML stem/progenitor cells.
(A) GSEA analysis of gene expression data shows enrichment of the PGC-1α–related gene set in LTHSCs obtained from CML mice compared with normal mice and downregulation in LSK cells from SIRT1-deleted compared with control Cre– CML mice. (B) OCR measurements in c-Kit+ cells from CML mice treated with PGC-1α inhibitor (PGC-1α inh) (SR18292) or vehicle (n = 7 each) for 2 weeks. (C–E) CML mice were treated with vehicle, NIL, PGC-1α inhibitor, or combination (n = 7–8) for 2 weeks. The effect of treatment on WBC (C), neutrophil counts (D), spleen size (E), spleen cellularity (F), splenic LSK cells (G) splenic c-Kit+ cells (H), BM cellularity (I), BM LSK cells (J), and BM c-Kit+ cells (K) are shown. (L) Measurement of OCR in CML CD34+ cells (n = 3) treated with PGC-1α inhibitor or vehicle overnight. (M) Apoptosis of CML CD34+CD38– cells (n = 3) treated with vehicle, PGC-1α inhibitor, NIL, or combination for 72 hours, measured by annexin V labeling. (N) Colony formation from CML CD34+CD38– cells (n = 3) exposed to vehicle, PGC-1α inhibitor, NIL, or combination for 48 hours and plated in methylcellulose progenitor culture. Error bars represent mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001, ordinary 1-way ANOVA (M and N), Kruskal-Wallis 1-way ANOVA (C–K), or 2-way ANOVA (B and L), correcting for multiple comparisons by controlling the FDR using the 2-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli.
Discussion
In this study, we demonstrate that the SIRT1 deacetylase plays an important role in maintaining the regenerative potential of CML LSCs and promoting leukemia development in CML. Our studies provide a conceptual advance and biological insights regarding the activity of SIRT1 and its role in CML LSCs. It has been reported that CML LSCs rely on upregulated mitochondrial oxidative metabolism for survival (33). However, the underlying mechanisms are not known. Our studies reveal an important role for SIRT1 and the SIRT1 substrate PGC-1α in mediating enhanced mitochondrial oxidative phosphorylation in LSCs. Increased mitochondrial metabolism in LSCs is BCR-ABL kinase independent, and TKI treatment of SIRT1-deleted mice results in further suppression of leukemic hematopoiesis. Identification and targeting of SIRT1-regulated mechanisms responsible for mitochondrial alterations may facilitate selective targeting of mitochondrial abnormalities in CML LSCs.
SIRT1 knockout impaired development of CML in transgenic BCR-ABL mice, as mice showed substantial delays in development of leukocytosis, neutrophilia, splenomegaly, morbidity, and death. SIRT1 deletion also reversed redistribution of CML stem cells from the BM to the spleen, reduced CML LSC quiescence, resulting in fewer quiescent G0 phase cells and increased G1 phase cells, and reduced regenerative capacity of LSCs after transplantation. These results definitively demonstrate an important role for SIRT1 in myeloproliferation and extramedullary hematopoiesis that is characteristic of CML. However, the lack of elimination of LSCs in the BM may contribute to delayed development of leukemia in SIRT1-deleted mice.
Quiescent HSCs reside in hypoxic niches in the BM cavity and rely on glycolysis, rather than oxidative phosphorylation, to support ATP production (37, 38). Self-renewing HSCs limit mitochondrial respiration to remain in a quiescent state, but switch to oxidative phosphorylation on activation and differentiation (39). In contrast to normal HSCs and bulk tumor cells, cancer stem cells show distinct metabolic phenotypes and, depending on cancer type, are variably dependent on oxidative phosphorylation or on glycolysis for growth and maintenance (40). CML and AML LSCs are reported to upregulate oxidative phosphorylation and be selectively targeted by inhibitors of mitochondrial translation, such as tigecycline (33, 41, 42). However, previous studies have not addressed the mechanisms of altered mitochondrial metabolism in leukemia versus normal stem cells.
Our results suggest that increased mitochondrial respiration in CML LSCs is tyrosine kinase independent and SIRT1 dependent. SIRT1 is a NAD+-dependent enzyme that is a cellular metabolic sensor with multiple regulatory effects on cellular metabolism (43). Extracellular flux analysis revealed that increased mitochondrial respiration in CML LSCs is markedly reduced upon SIRT1 deletion. In contrast, SIRT1 deletion did not affect mitochondrial respiration in normal HSCs. These results support an important role for SIRT1 overexpression in mediating enhanced oxidative phosphorylation in LSCs compared with HSCs. SIRT1 can deacetylate and activate the transcriptional coactivator PGC-1α to enhance mitochondrial DNA replication and gene expression and promote mitochondrial biogenesis and activity (44, 45). SIRT1 can also lead to PGC-1α activation by modulating LKB1, leading to increased AMPK activation and PGC-1α phosphorylation (36, 46). PGC-1α gene sets were upregulated in CML stem/progenitor cells and markedly downregulated upon SIRT1 deletion. PGC-1α inhibition substantially reduced mitochondrial oxygen consumption, supporting an important role in regulation of mitochondrial metabolism in CML stem/progenitor cells. Interestingly, PGC-1α inhibition did not directly inhibit CML hematopoiesis, suggesting that additional SIRT1-related mechanisms besides regulation of mitochondrial respiration are also involved in promoting CML stem/progenitor cell growth. SIRT1 overexpression is known to lead to p53 deacetylation and inhibition of p53 activity in CP CML LSCs, and effects of SIRT1 inhibition on leukemic hematopoiesis are mediated at least in part to increased p53 acetylation and transcriptional activity (5, 7, 47). Indeed, p53-related gene signatures were substantially enriched in SIRT1-deleted CML stem cells. In the future, it will be of interest to explore interactions between SIRT1 regulation of metabolism and its regulation of p53 in the effects of SIRT1 inhibition on LSCs.
Gene expression and extracellular flux analyses indicate that TKI treatment does not significantly alter mitochondrial oxidative phosphorylation in human or murine CML LSCs, suggesting that altered oxidative phosphorylation in CML LSCs may be TKI independent. This is consistent with previous observations that TKI treatment does not inhibit SIRT1 activity in CML LSCs (5). TKI treatment increased suppression of leukemic hematopoiesis in SIRT1-deleted mice compared with control CML mice. Similarly, treatment of human CML stem cells with a SIRT1 inhibitor in combination with TKI resulted in substantially greater apoptosis and proliferation inhibition than either agent alone. Interestingly, PGC-1α inhibition resulted in marked suppression of leukemic hematopoiesis in combination with TKI, compared with TKI alone, suggesting that PGC-1α, although not essential for leukemia development, may contribute to maintenance of CML stem/progenitor cells following TKI treatment. However, our studies suggest that SIRT1 and PGC-1α inhibition, although further suppressing residual CML stem/progenitor cells, may not result in their elimination.
Development of effective approaches for targeting CML LSCs that persist despite TKI treatment without toxicity to normal HSCs has been challenging. In previous studies, SIRT1 inhibition by shRNA or pharmacological means did not affect normal human stem or progenitor cell survival (5). In our studies, SIRT1 loss from normal HSCs did not inhibit steady-state normal hematopoiesis. However, we cannot rule out a requirement for SIRT1 in HSC maintenance under chronic stress conditions and with aging, since the effects of SIRT1 deletion on normal hematopoiesis appear to differ depending on the specific models and context.
The current studies focus on CML, which is an outstanding disease model that has contributed greatly to advances in leukemia pathogenesis and treatment. However, their impact extends to other hematological malignancies, including AML, myelodysplastic syndromes, and myeloproliferative neoplasms. The proposed research reveals knowledge and concepts regarding the role of SIRT1 in metabolic regulation of HSC and LSC maintenance, growth, and resistance and raises the possibility of developing improved strategies to target kinase-independent metabolic alterations.
Methods
Animal studies.
SIRT1 exon-4 floxed (SIRT1fl/fl) mice (20) were a gift from Frederick Alt (Harvard Medical School) and were crossed with Mx1-Cre mice (Jackson Laboratories) to generate Mx1-Cre SIRT1fl/fl) mice. Mx1-Cre SIRT1fl/fl) mice were crossed with SCL-tTa/BCR-ABL mice (BA mice) to generate SCL-tTa/BCR-ABL SIRT1fl/fl mice (BA Mx1-Cre SIRT1fl/fl). The mice were maintained on tetracycline till CML induction. Cre activity was induced by i.p. injections of poly(I:C) (12.5 mg/kg) every other day for 7 doses. SIRT1fl/fl mice were also crossed with C57BL/6 UBC-Cre-ERT2 mice (Jackson Laboratories) and SCL-tTa/BCR-ABL mice to generate SCL-tTa/BCR-ABL UBC-Cre-ERT2 SIRT1fl/fl mice (BA UBC-Cre-ERT2 SIRT1fl/fl). Cre activity was induced by i.p. injections of tamoxifen (50 mg/kg) daily for a total of 5 doses. SIRT1 exon4 deletion was confirmed using genomic DNA PCR. Genotypes and age at the time of experiment for these mice are as shown in Table 1.
Table 1. Mouse strains used for experiments with genotypes.
Human samples.
BM samples were obtained from untreated CP CML patients seen at UAB. CD34+ cell isolation was performed using magnetic beads (STEMCELL Technologies). CD34+CD38– and CD34+CD38+ cells were obtained by flow cytometry sorting.
BM transplantation.
BM cells from Cre+ or Cre– BA Mx1-Cre SIRT1fl/fl or BA UBC-Cre-ERT2 SIRT1fl/fl mice were injected into the tail vein (2 × 106 cells/mouse) of sublethally irradiated (400 cGy × 2, 4 hours apart) CD45.1 congenic recipients (C57BL6-Ly5.1/Cr; Charles River laboratories). The mice were maintained on a tetracycline diet for 4 weeks until the induction of SIRT1 deletion by either poly(I:C) or tamoxifen, followed by induction of BCR-ABL transgene by withdrawing the tetracycline diet. Donor LTHSCs from mice engrafted with BM cells from BA Mx1-Cre SIRT1fl/fl and Cre– controls were selected by flow cytometry sorting (BD FACSAria III cell sorter) and transplanted (200 cells/mouse) into secondary congenic CD45.1 recipients together with 500,000 CD45.1 BM cells.
Flow cytometry.
BM, PB, or spleen cells were stained with anti-CD45.1 and CD45.2 antibodies to distinguish donor from host-derived cells and with lineage-specific antibodies Mac1, Gr1, CD19, B220, and CD3 to identify myeloid, B, and T lineages. LTHSCs (LSK, Flt3–CD150+CD48–), STHSCs (LSK,Flt3–CD150–CD48–), MPPs (LSK, Flt3–CD150–CD48+), GMPs (Lin–Sca1–c-Kit+CD34+FcγRII/IIIhi), and MEPs (Lin–Sca1–c-Kit+CD34–FcγRII/IIIlo) were also analyzed. All analyses were performed on BD LSR Fortessa flow cytometer (BD). Details of antibodies used are as shown in Table 2.
Table 2. Antibodies used for flow cytometry analysis.
Cell cycle analysis.
For cell cycle analysis, BM cells were stained for stem and progenitor markers, followed by fixing using BD Cytofix/Cytoperm. Fixed cells were then stained with KI67 and DAPI and analyzed using flow cytometry.
TKI treatment.
Whole BM cells from both BA Mx1-Cre SIRT1fl/fl and controls were transplanted into sublethally irradiated CD45.1 recipients. Ten weeks after CML induction and SIRT1 deletion, mice were randomized and treated with vehicle or NIL (50 mg/kg once daily) by oral gavage for a period of 4 weeks. Human CML CD34+ cells were labeled with CFSE (Molecular Probes), CD34–PE–Cy7 and CD38– APC, and CD34+CD38– and CD34+CD38+ cells with uniform CFSE labeling were selected by flow cytometry (BD FACS Aria III). Cells were cultured with low concentrations of growth factors at 37°C in normoxic conditions. Effects of NIL, the SIRT1 inhibitor TV39OH, or a combination on cell proliferation was evaluated by analyzing CFSE by flow cytometry after 72 hours. Apoptosis was assessed by labeling cells with annexin V (BD Biosciences — Pharmingen) followed by flow cytometry analysis. CFC assays were performed by plating cells in methylcellulose progenitor culture (STEMCELL Technologies) after 72 hours of drug treatment. Colony-forming unit granulocyte, macrophage, and megakaryocyte and colony-forming unit granulocyte and macrophage were counted after 2 weeks.
PGC-1α inhibitor treatment.
For in vivo treatment, whole BM cells from both BA Mx1-Cre SIRT1fl/fl and controls were transplanted into sublethally irradiated CD45.1 recipients. Ten weeks after CML induction, mice were randomized and treated with the PGC-1α inhibitor SR18292 (45 mg/kg once daily) by i.p. injection, NIL (50 mg/kg once daily), or a combination for 2 weeks. Human CML CD34+ cells were treated in vitro with SR18292 (10–40 μM), NIL, or a combination overnight to measure OCR/ECAR and for 3 days to measure apoptosis and colony formation.
RNA-Seq analysis.
LSK cells were sorted from BM of BA UBC-Cre-ERT2 SIRT1fl/fl and controls and RNA extracted using RNeasy Plus Micro Kit (QIAGEN), with 4 biological replicates from each group. Sequencing libraries were prepared with the SMARTer Ultra Low Input RNA Kit for Sequencing (v4, TaKaRa Clontech, catalog 634891) and the Nextera XT DNA Library Preparation Kit (96 samples, Illumina, catalog FC-131-1096). Sequencing was performed using the HiSeq 2500 platform with the HiSeq SBS Kit V4 (Illumina). STAR (version 2.5.3a) was used to align raw RNA-Seq FASTQ reads to the mouse reference genome (Gencode Release M11) (48), and number of reads mapping to each gene was enumerated using HTSeq-count (49). Normalization and differential expression was calculated using DESeq2 (50). Pathway analysis was performed using GSEA. The RNA sequencing data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE129158).
Extracellular flux analysis.
OCR was measured using the Seahorse XFe96 analyzer (Agilent). c-Kit–enriched cells were suspended in XF Assay Medium supplemented with 1 mM pyruvate, 5.5 mM d-glucose, and 4 mM l-glutamine (pH 7.4). 150,000 cells were seeded per well of a Seahorse XF96 cell culture plate precoated with Cell-Tak (BD Biosciences) as described earlier (51). Cells were allowed to adhere for 30 minutes in a CO2- free incubator at 37°C, after which XF Assay Medium was added. The plate was equilibrated for 10 minutes in the CO2-free incubator before being transferred to the Seahorse XFe96 analyzer. Measurement of OCR and ECAR was done at baseline and following sequential injections of (i) oligomycin (1 μM), an ATP synthase inhibitor, (ii) carbonyl cyanide-4-(trifluoromethoxy) phenyl hydrazone (FCCP) (1.75 μM for mice c-Kit cells, and 1.5 μM for human CD34+ cells), a mitochondrial uncoupler, and (iii) antimycin A (10 μM), a complex III inhibitor, respectively. Briefly, oligomycin inhibits mitochondrial ATP synthase, and the resulting drop in OCR and rise in ECAR are attributed to ATP-linked OCR and the compensation of glycolysis for the loss of mitochondrial ATP production. FCCP uncouples the mitochondrial proton gradient and oxygen consumption from ATP synthase and drives maximal OCR. Antimycin A inhibits complex III of the electron transport chain and suppresses all mitochondrial oxygen consumption, and the residual OCR is considered nonmitochondrial. For ECAR measurements, 2-deoxy-glucose (2DG) (100 mM), a competitive inhibitor of hexokinase, was added at the end, so that basal glycolysis, maximum glycolysis, and nonglycolysis ECAR were calculated.
Statistics.
Results are displayed as mean ± SEM. Significance values were calculated using Prism version 5.0 software (GraphPad Prism) using unpaired, nonparametric t tests (Mann-Whitney U test) or 1- or 2-way ANOVA as appropriate. Survival was analyzed using Kaplan-Meier analysis. A P value of less than 0.05 was considered significant.
Study approval.
All human subjects signed an informed consent form. Sample acquisition was approved by the Institutional Review Board at UAB in accordance with assurances filed with the Department of Health and Human Services and met all requirements of the Declaration of Helsinki. Mouse care and experimental procedures were performed in accordance with federal guidelines and protocols approved by the IACUC at the UAB.
Author contributions
RB, AA, and SQ conceived the project. AA, BKC, AP, JH, MS, RW, SQ, VMDU and RB developed the methods. AA, SQ, BKC, HL, JH, PA, MS, VMDU, and RB performed experiments. AA, JH, BKC, and RB analyzed the data. RB and AA wrote the original draft. RB, AA, SQ, AP, RW, BKC, and VMDU reviewed and edited the manuscript. RB acquired funding. RB and VMDU supervised the project.
Supplementary Material
Acknowledgments
This work was supported by NIH grant R01 CA95684 (to RB). We acknowledge the assistance provided by David Crossman in the Genomics Core, Vidya Sagar Hanumanthu in the Flow Cytometry Core, and the staff of the UAB Animal Resources Program. We thank Fredrick Alt, Harvard Medical School, for providing the SIRT1f/f mice. We are grateful to Maya Robinson and Amanda Mullens for procurement and processing of human samples and Mason Harris for assistance with animal studies.
Version 1. 06/10/2019
Electronic publication
Version 2. 07/01/2019
Print issue publication
Footnotes
AA’s present address is: Department of Anatomy and Cell Biology, University of Florida College of Medicine, Gainesville, Florida, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(7):2685–2701.https://doi.org/10.1172/JCI127080.
Contributor Information
Ajay Abraham, Email: aabraham@uab.edu.
Shaowei Qiu, Email: sqiu@uabmc.edu.
Balu K. Chacko, Email: bchacko@uab.edu.
Hui Li, Email: huili@uabmc.edu.
Andrew Paterson, Email: andrewpaterson@uabmc.edu.
Jianbo He, Email: jianbo@uab.edu.
Puneet Agarwal, Email: pagarwal@uab.edu.
Mansi Shah, Email: mmshah@uab.edu.
Ravi Bhatia, Email: rbhatia@uabmc.edu.
References
- 1.Quintás-Cardama A, Kantarjian H, Cortes J. Imatinib and beyond--exploring the full potential of targeted therapy for CML. Nat Rev Clin Oncol. 2009;6(9):535–543. doi: 10.1038/nrclinonc.2009.112. [DOI] [PubMed] [Google Scholar]
- 2.Chu S, et al. Persistence of leukemia stem cells in chronic myelogenous leukemia patients in prolonged remission with imatinib treatment. Blood. 2011;118(20):5565–5572. doi: 10.1182/blood-2010-12-327437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang X, Cortes J, Kantarjian H. Estimations of the increasing prevalence and plateau prevalence of chronic myeloid leukemia in the era of tyrosine kinase inhibitor therapy. Cancer. 2012;118(12):3123–3127. doi: 10.1002/cncr.26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahon FX, et al. Discontinuation of imatinib in patients with chronic myeloid leukaemia who have maintained complete molecular remission for at least 2 years: the prospective, multicentre Stop Imatinib (STIM) trial. Lancet Oncol. 2010;11(11):1029–1035. doi: 10.1016/S1470-2045(10)70233-3. [DOI] [PubMed] [Google Scholar]
- 5.Li L, et al. Activation of p53 by SIRT1 inhibition enhances elimination of CML leukemia stem cells in combination with imatinib. Cancer Cell. 2012;21(2):266–281. doi: 10.1016/j.ccr.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan H, et al. Activation of stress response gene SIRT1 by BCR-ABL promotes leukemogenesis. Blood. 2012;119(8):1904–1914. doi: 10.1182/blood-2011-06-361691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li L, et al. SIRT1 activation by a c-MYC oncogenic network promotes the maintenance and drug resistance of human Flt3-ITD acute myeloid leukemia stem cells. Cell Stem Cell. 2014;15(4):431–446. doi: 10.1016/j.stem.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sasca D, et al. SIRT1 prevents genotoxic stress-induced p53 activation in acute myeloid leukemia. Blood. 2014;124(1):121–133. doi: 10.1182/blood-2013-11-538819. [DOI] [PubMed] [Google Scholar]
- 9.Chalkiadaki A, Guarente L. The multifaceted functions of sirtuins in cancer. Nat Rev Cancer. 2015;15(10):608–624. doi: 10.1038/nrc3985. [DOI] [PubMed] [Google Scholar]
- 10.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434(7029):113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 12.Li X, Zhang S, Blander G, Tse JG, Krieger M, Guarente L. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol Cell. 2007;28(1):91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 13.Picard F, et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429(6993):771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hariharan N, Maejima Y, Nakae J, Paik J, Depinho RA, Sadoshima J. Deacetylation of FoxO by Sirt1 plays an essential role in mediating starvation-induced autophagy in cardiac myocytes. Circ Res. 2010;107(12):1470–1482. doi: 10.1161/CIRCRESAHA.110.227371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaziri H, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107(2):149–159. doi: 10.1016/S0092-8674(01)00527-X. [DOI] [PubMed] [Google Scholar]
- 16.Jang KY, et al. SIRT1 expression is associated with poor prognosis of diffuse large B-cell lymphoma. Am J Surg Pathol. 2008;32(10):1523–1531. doi: 10.1097/PAS.0b013e31816b6478. [DOI] [PubMed] [Google Scholar]
- 17.Chen X, et al. High levels of SIRT1 expression enhance tumorigenesis and associate with a poor prognosis of colorectal carcinoma patients. Sci Rep. 2014;4:7481. doi: 10.1038/srep07481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jang KY, et al. SIRT1 and c-Myc promote liver tumor cell survival and predict poor survival of human hepatocellular carcinomas. PLoS ONE. 2012;7(9):e45119. doi: 10.1371/journal.pone.0045119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang JC, et al. Histone deacetylase in chronic lymphocytic leukemia. Oncology. 2011;81(5-6):325–329. doi: 10.1159/000334577. [DOI] [PubMed] [Google Scholar]
- 20.Cheng HL, et al. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci U S A. 2003;100(19):10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leko V, et al. SIRT1 is dispensable for function of hematopoietic stem cells in adult mice. Blood. 2012;119(8):1856–1860. doi: 10.1182/blood-2011-09-377077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ou X, et al. SIRT1 deficiency compromises mouse embryonic stem cell hematopoietic differentiation, and embryonic and adult hematopoiesis in the mouse. Blood. 2011;117(2):440–450. doi: 10.1182/blood-2010-03-273011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rimmelé P, et al. Aging-like phenotype and defective lineage specification in SIRT1-deleted hematopoietic stem and progenitor cells. Stem Cell Reports. 2014;3(1):44–59. doi: 10.1016/j.stemcr.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh SK, Williams CA, Klarmann K, Burkett SS, Keller JR, Oberdoerffer P. Sirt1 ablation promotes stress-induced loss of epigenetic and genomic hematopoietic stem and progenitor cell maintenance. J Exp Med. 2013;210(5):987–1001. doi: 10.1084/jem.20121608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koschmieder S, et al. Inducible chronic phase of myeloid leukemia with expansion of hematopoietic stem cells in a transgenic model of BCR-ABL leukemogenesis. Blood. 2005;105(1):324–334. doi: 10.1182/blood-2003-12-4369. [DOI] [PubMed] [Google Scholar]
- 26.Schemionek M, et al. BCR-ABL enhances differentiation of long-term repopulating hematopoietic stem cells. Blood. 2010;115(16):3185–3195. doi: 10.1182/blood-2009-04-215376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang B, et al. Altered microenvironmental regulation of leukemic and normal stem cells in chronic myelogenous leukemia. Cancer Cell. 2012;21(4):577–592. doi: 10.1016/j.ccr.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamilton A, et al. Chronic myeloid leukemia stem cells are not dependent on Bcr-Abl kinase activity for their survival. Blood. 2012;119(6):1501–1510. doi: 10.1182/blood-2010-12-326843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang B, et al. Effective targeting of quiescent chronic myelogenous leukemia stem cells by histone deacetylase inhibitors in combination with imatinib mesylate. Cancer Cell. 2010;17(5):427–442. doi: 10.1016/j.ccr.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang B, et al. Inhibition of interleukin-1 signaling enhances elimination of tyrosine kinase inhibitor-treated CML stem cells. Blood. 2016;128(23):2671–2682. doi: 10.1182/blood-2015-11-679928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Essers MA, et al. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 2009;458(7240):904–908. doi: 10.1038/nature07815. [DOI] [PubMed] [Google Scholar]
- 32.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuntz EM, et al. Targeting mitochondrial oxidative phosphorylation eradicates therapy-resistant chronic myeloid leukemia stem cells. Nat Med. 2017;23(10):1234–1240. doi: 10.1038/nm.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang B, et al. Microenvironmental protection of CML stem and progenitor cells from tyrosine kinase inhibitors through N-cadherin and Wnt-β-catenin signaling. Blood. 2013;121(10):1824–1838. doi: 10.1182/blood-2012-02-412890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ladds MJGW, et al. Autophagic flux blockage by accumulation of weakly basic tenovins leads to elimination of B-Raf mutant tumour cells that survive vemurafenib. PLoS ONE. 2018;13(4):e0195956. doi: 10.1371/journal.pone.0195956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cantó C, Auwerx J. PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr Opin Lipidol. 2009;20(2):98–105. doi: 10.1097/MOL.0b013e328328d0a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simsek T, et al. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell. 2010;7(3):380–390. doi: 10.1016/j.stem.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suda T, Takubo K, Semenza GL. Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell. 2011;9(4):298–310. doi: 10.1016/j.stem.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 39.Ito K, Suda T. Metabolic requirements for the maintenance of self-renewing stem cells. Nat Rev Mol Cell Biol. 2014;15(4):243–256. doi: 10.1038/nrm3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farge T, et al. Chemotherapy-resistant human acute myeloid leukemia cells are not enriched for leukemic stem cells but require oxidative metabolism. Cancer Discov. 2017;7(7):716–735. doi: 10.1158/2159-8290.CD-16-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lagadinou ED, et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell. 2013;12(3):329–341. doi: 10.1016/j.stem.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boily G, et al. SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS ONE. 2008;3(3):e1759. doi: 10.1371/journal.pone.0001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lagouge M, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127(6):1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 45.LeBleu VS, et al. PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol. 2014;16(10):992–1003. doi: 10.1038/ncb3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Price NL, et al. SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metab. 2012;15(5):675–690. doi: 10.1016/j.cmet.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prokocimer M, Rotter V. Structure and function of p53 in normal cells and their aberrations in cancer cells: projection on the hematologic cell lineages. Blood. 1994;84(8):2391–2411. [PubMed] [Google Scholar]
- 48.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chacko BK, et al. Methods for defining distinct bioenergetic profiles in platelets, lymphocytes, monocytes, and neutrophils, and the oxidative burst from human blood. Lab Invest. 2013;93(6):690–700. doi: 10.1038/labinvest.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.