Abstract
Transfer RNAs (tRNAs) are a major class of noncoding RNA. Stress-induced cleavage of tRNA is highly conserved and results in tRNA fragments. Here, we found that specific tRNA fragments in plasma are associated with epilepsy. Small RNA-Seq of plasma samples collected during video EEG monitoring of patients with focal epilepsy identified significant differences in 3 tRNA fragments (5′GlyGCC, 5′AlaTGC, and 5′GluCTC) compared with samples from healthy controls. The levels of these tRNA fragments were higher in pre-seizure than in post-seizure samples, suggesting that they may serve as biomarkers of seizure risk in patients with epilepsy. In vitro studies confirmed that production and extracellular release of tRNA fragments were lower after epileptiform-like activity in hippocampal neurons. We designed PCR-based assays to quantify tRNA fragments in a cohort of pre- and post-seizure plasma samples from patients with focal epilepsy and from healthy controls. Receiver operating characteristic analysis indicated that tRNA fragments potently distinguished pre- from post-seizure patients. Elevated levels of tRNA fragments were not detected in patients with psychogenic nonepileptic seizures and did not result from medication tapering. This study potentially identifies a new class of epilepsy biomarker and reveals the possible existence of prodromal molecular patterns in blood that could be used to predict seizure risk.
Keywords: Neuroscience
Keywords: Epilepsy, Noncoding RNAs, Seizures
Introduction
Temporal lobe epilepsy (TLE) is the most prevalent form of focal epilepsy in adults, with approximately 0.1% of the world’s population affected. Diagnosis of epilepsy is complex and challenging. Seizures are rarely witnessed by epileptologists, and diagnosis is primarily based on clinical history, supported by EEG and imaging (MRI) data. Accordingly, there is an unmet need for biomarkers of epilepsy. To date, few studies have focused on biomarkers of seizure onset, and seizure prediction studies mainly rely on EEG recordings to provide a patient-specific profile of seizure activity, which has limited predictive power in a subset of patients (1). From a patient’s perspective, the ability to forecast seizures would allow greater control in daily life and improve patient safety.
Circulating blood-based molecules represent an attractive source of epilepsy biomarkers, given the potential for fast analysis of easy-to-collect samples. Serum levels of high-mobility group box 1 (HMGB1) protein have been shown to indicate epileptogenesis in animals and drug-refractory epilepsy in patients (2). Small noncoding RNAs have recently emerged as potential biomarkers, with several studies reporting differences in serum or plasma levels of miRNAs in patients with epilepsy (3). Research into predictive biomarkers for seizure onset or imminence has been limited. Interestingly, a recent study sampling blood from patients with epilepsy in a video EEG monitoring unit identified 4 miRNAs that were significantly elevated immediately after onset of generalized tonic-clonic seizures (GTCSs) (4).
Transfer RNAs (tRNAs) are ubiquitous noncoding RNAs that deliver amino acids to the ribosome during protein synthesis. Cleavage of tRNAs occurs as part of a highly conserved stress response present in single-celled organisms (5). In humans, tRNA fragments are generated by ribonucleases including Dicer and angiogenin (6). Numerous functions have been attributed to tRNA fragments, including inhibition of protein translation (7), initiation of stress granule formation (8), and regulation of gene expression (9, 10). tRNA fragments have also been found circulating in the blood (11), indicating that they are protected from degradation, which makes them ideal candidates for investigation as biomarkers.
Here, we identified plasma tRNA fragments in RNA-Seq data from patients with focal epilepsy (12). We found 3 specific tRNA fragments that were significantly elevated in pre-seizure samples compared with post-seizure samples and those from healthy controls. We performed mechanistic and clinical validation studies to explore the potential of these tRNA fragments as biomarkers of seizure risk. Together, these data suggest that specific tRNA fragments may constitute a novel class of epilepsy biomarker that could support the prediction of seizure risk in patients diagnosed with epilepsy.
Results and Discussion
Blood samples were collected from 16 patients with refractory focal epilepsy (the majority of whom had TLE) upon arrival for video EEG monitoring at the Epilepsy Monitoring Unit (EMU) in the Epilepsy Center Hessen. All patients had drug-refractory focal epilepsy and were on polytherapy. Demographic and medication information is provided in Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI126346DS1 Each patient underwent continuous video EEG monitoring with the 10–20 standard international electrode placement system. Recordings were manually reviewed by a neurologist with special training in epilepsy. A second blood sample was collected 24 hours after a confirmed electroclinical seizure. Plasma samples were obtained at the same site from age- and sex-matched healthy volunteers.
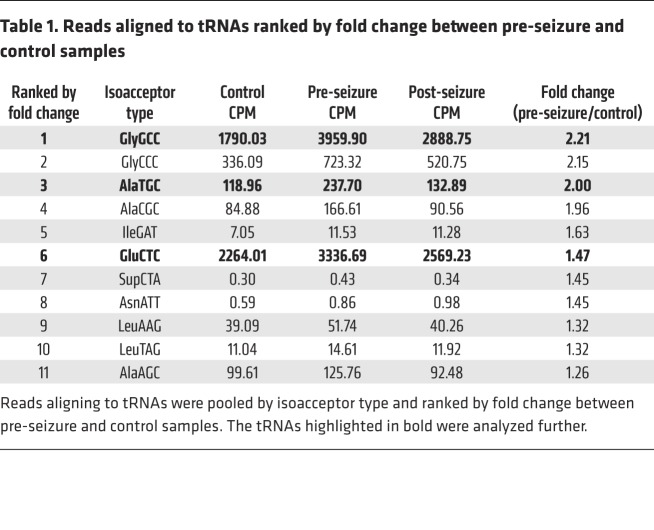
Small RNA-Seq (<50 nt) was performed on pooled samples from 16 controls and pre-seizure or post-seizure samples from 16 patients with focal epilepsy. A custom tRNA library was used to quantify reads aligning to tRNAs, which are summarized in Supplemental Table 2. Reads were pooled for tRNAs of the same isoacceptor type, and tRNAs were ranked by fold change between pre-seizure and control samples (Table 1). Three tRNA fragments were chosen for further investigation on the basis of abundance (counts per million [CPM]) and fold change: glycine GCC, alanine TGC, and glutamic acid CTC (Table 1). Read coverage plots revealed reads aligned to the 5′ end of the tRNA (Figure 1, A, D, and G). The cleavage site was mapped onto the predicted secondary structure of each of the tRNAs (Figure 1, B, E, and H). Predicted secondary structures of the tRNA fragments indicated that they form short hairpin structures (Figure 1, C, F, and I). Of note, tRNA fragment levels were highest in pre-seizure samples and closer to control levels in post-seizure samples (Figure 1, A, D, and G).
Table 1. Reads aligned to tRNAs ranked by fold change between pre-seizure and control samples.
Figure 1. 5′tRNA fragments are differentially expressed in plasma from patients with epilepsy.
Small RNA-Seq of pooled pre- and post-seizure samples from 16 patients with focal epilepsy and samples from 16 healthy controls. Coverage plots show reads aligned to (A) GlyGCC, (D) AlaTCG, and (G) GluCTC tRNAs in CPM (y axis) and tRNA sequences (x axis). (B, E, and H) Cleavage sites are indicated (red triangle) on the mature tRNA structures (downloaded from GtRNAdb 2.0 [ref. 25]). (C, F, and I) Predicted secondary structures of the tRNA fragments.
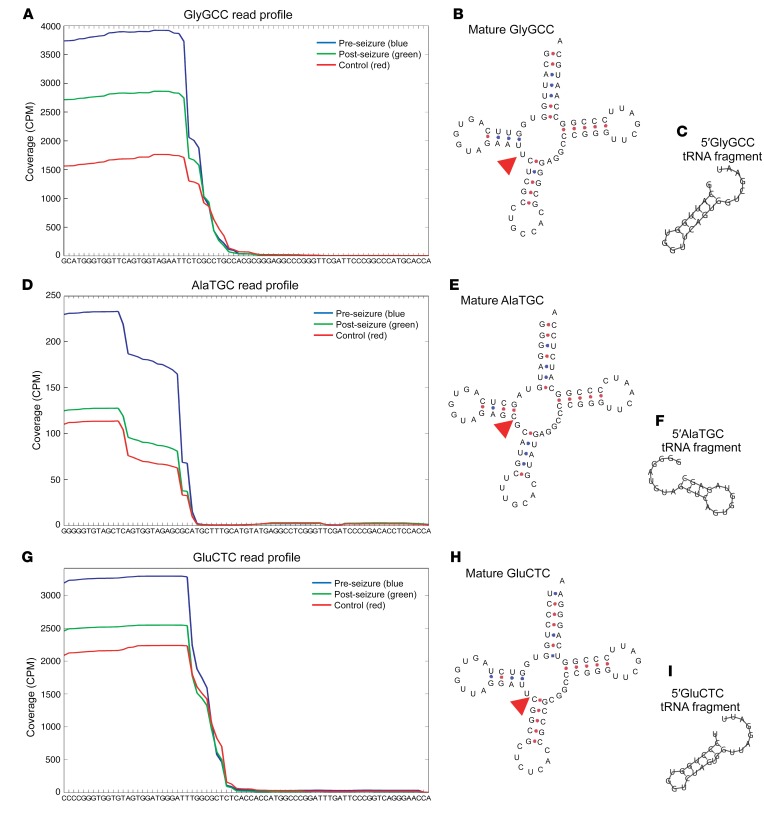
No existing assays were available to quantify tRNA fragments, therefore, we designed custom TaqMan assays. These assays utilize standard quantitative PCR (qPCR) techniques and allow fast, specific quantification of short RNA fragments. As tRNAs are highly conserved, the assays can be used on both mouse and human samples (see Supplemental Figure 1). Next, we aimed to experimentally model the observation that tRNA fragment levels decreased following epileptic seizure. Seizure-like activity was induced in cultured mouse hippocampal neurons by withdrawal of magnesium and assessed by measuring intracellular calcium levels with FLUO4 (Figure 2A) (13). Representative FLUO4 traces in the presence (Mg+: 1 mM MgCl2) and absence (Mg–: 0 mM MgCl2) of magnesium indicated spikes of increased intracellular calcium levels corresponding to increased neuronal activity (Figure 2B). Intracellular 5′GluCTC levels were significantly decreased following epileptiform activity (Figure 2C). As tRNA fragments are secreted from neurons (14), we next quantified tRNA fragments in the extracellular medium and found that the levels of both 5′GlyGCC and 5′GluCTC were significantly reduced following increased neuronal activity (Figure 2D). This confirmed that tRNA fragments are generated in neurons and secreted from neurons and that secretion of tRNA fragments is reduced following neuronal hyperexcitation (mimicking epileptiform activity). tRNA fragment levels were normalized to U6 RNA, which remained unchanged throughout the experiments (Supplemental Figure 2).
Figure 2. tRNA fragments are regulated by epileptiform activity in hippocampal neurons.
(A) Primary mouse hippocampal neurons were incubated with 5 μM FLUO4 for 45 minutes at 37°C and imaged at 5 Hz. Scale bar: 40 μm. (B) Representative FLUO4 traces are shown for neurons in 1 mM Mg (Mg+, upper panel) or 0 mM Mg (Mg–, lower panel). Following 2 hours in Mg+ or Mg– media, the media were collected and total RNA extracted. Both intracellular (C) and extracellular (D) tRNA fragment levels were lower following culture in Mg-free media, in which extracellular 5′GluCTC and 5′GlyGCC levels were significantly lower. *P < 0.05, by Students t test. In C and D, individual data points are plotted, with the mean indicated in red and error bars ± SEM. n = 15–16 replicates from 5 independent preps.
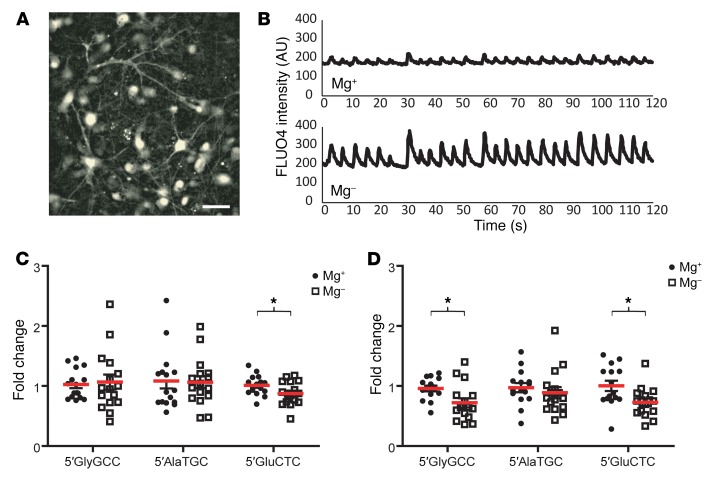
We next used the custom TaqMan assays to validate the RNA-Seq data in the 16 focal epilepsy patients and 16 healthy controls recruited in Marburg, with an additional 16 focal epilepsy patients and 16 healthy controls recruited at the EMU in Beaumont Hospital, Dublin (Supplemental Table 1). We found that the levels of all 3 tRNA fragments were significantly elevated in the pre-seizure samples (Figure 3, A–C). A Wilcoxon signed-rank test indicated that there was a highly significant change in tRNA fragment levels between the pre- and post-seizure samples: 5′GlyGCC, 3.17-fold change (P = 0.0002); 5′AlaTGC, 1.93-fold change (P < 0.0001); and 5′GluCTC, 2.89-fold change (P = 0.0003) (Figure 3, D–F), with the majority of samples showing a decrease in post-seizure tRNA fragment levels. Receiver operating characteristic (ROC) analysis indicated that tRNA fragments could distinguish pre- and post-seizure samples (Figure 3, G–I). 5′GlyGCC had an AUC of 0.816 (P = 0.000027, Figure 3G); 5′AlaTGC had an AUC of 0.916 and was highly significant (P = 1.86 × 10–8, Figure 3H); and 5′GluCTC had an AUC of 0.802 (P = 0.000069, Figure 3I). We applied Youden’s J statistic to determine the optimal cutoff for distinguishing pre- and post-seizure samples, and this indicated that a value of 1.33 was most discriminatory for 5′AlaTGC, with a sensitivity of 96.8% and a specificity of 87.1% (Figure 3H); that a value of 1.36 was optimal for 5′GlyGCC (Figure 3G); and that a 2.13 cutoff for 5′GluCTC performed best (Figure 3I). ROC analyses are summarized in Supplemental Table 3. Analysis of males and females separately revealed that 5′AlaTGC was significantly elevated in pre-seizure samples from both groups (Supplemental Figure 3). Importantly, the levels of 2 other tRNA fragments detected in the plasma RNA-Seq data (5′ValAAC and 5′ProAGG) were validated by custom TaqMan assay and were not elevated in the pre-seizure samples (Supplemental Figure 4). Plasma samples collected at additional time points were available for 24 of the 32 patients (T1: upon arrival at the EMU; T3: 1 hour after seizure). Analysis of extra time points indicated that 5′AlaTGC levels spiked before seizure and fell 1 hour after seizure before rising again by 24 hours after seizure (Supplemental Figure 5). Analysis of the correlation between the time interval (pre-seizure blood collection to seizure onset) and the tRNA fragment level indicated that 5′GluCTC was significantly elevated prior to seizure onset (Spearman’s r = –0.59, P = 0.0035) when the analysis was limited to a time point under 50 hours before onset (Supplemental Figure 6). 5′GlyGCC and 5′AlaTGC showed a weaker correlation that did not reach statistical significance. These analyses indicate that specific tRNA fragments can discriminate between pre- and post-seizure samples and may be of use as epilepsy biomarkers in advance of seizure.
Figure 3. tRNA fragments are elevated pre-seizure in plasma from patients with epilepsy.
Blood samples from 32 patients with epilepsy before and after seizure and from 32 healthy controls were analyzed. (A) 5′GlyGCC, (B) 5′AlaTGC, and (C) 5′GluCTC tRNA fragments were significantly elevated in the pre-seizure samples. Kruskal-Wallis test indicated P = 0.0008, P < 0.0001, and P = 0.0001, respectively. *P <0.05 and ***P <0.001. No significant difference was detected between post-seizure samples and those from healthy controls. Data represent the mean ± SEM. Most tRNA fragments decreased significantly after seizure. Wilcoxon signed-rank test was performed for (D) 5′GlyGCC (P = 0.0002), (E) 5′AlaTGC (P < 0.0001), and (F) 5′GluCTC (P = 0.0003). ROC analysis indicated that (G) 5′GlyGCC had an AUC of 0.816 (P = 0.000027), (H) 5′AlaTGC had an AUC of 0.916 (P = 1.86 × 10–8), and (I) 5′GluCTC had an AUC of 0.802 (P = 0.000069).
Having demonstrated that tRNA fragments were elevated in plasma from patients with focal epilepsy in advance of the occurrence of a seizure, we sought to determine whether tRNA fragments were detectable in brain tissues. Five patients from the Dublin cohort had undergone surgical resection in an attempt to alleviate their seizures. Fresh-frozen hippocampal and cortical tissue was cryosectioned, with neighboring sections mounted for histological analysis and collected for RNA analysis. We found that all 3 tRNA fragments were detectable in both hippocampal and cortical tissues (Supplemental Figure 7, A–C) and detected no significant difference in tRNA levels between tissues. Histological analysis indicated that the tissue samples were intact, although components of the hippocampal formation were only visible in 2 samples (Supplemental Figure 7D). Analysis of MRI data revealed that 9 patients with focal epilepsy had no detectable structural lesions (20 patients were positive for lesions, 3 were indeterminate), and, interestingly, tRNA fragment levels were higher in the MRI-negative patients (Supplemental Figure 8), indicating that tRNA fragments are not released as a result of structural damage.
One caveat to interpreting these data is that the patients’ medication was withdrawn upon arrival at the EMU and may therefore have been higher in the pre-seizure than the post-seizure samples; however, we do not believe that medication levels influence tRNA fragment levels for several reasons. First, we noted that 4 patients did not have their medication reduced and went on to experience seizures; however, we found no significant difference in pre-seizure tRNA fragment levels in patients with or without anti-epileptic drug (AED) reduction (Supplemental Figure 9). Second, we analyzed tRNA fragment levels in a cohort of patients for whom AEDs were reduced but who did not experience seizures, and detected no change in tRNA fragment levels (Supplemental Figure 10). Finally, we analyzed plasma tRNA fragment levels in 6 patients who were subsequently diagnosed with psychogenic nonepileptic seizures (PNESs) and found no change in tRNA fragment levels between pre– and post–seizure-like events (Supplemental Figure 11), indicating that elevated tRNA fragments are linked to electroclinical seizures.
To investigate the mechanism leading to increased pre-seizure tRNA fragment levels, we assessed whether abnormal interictal neuronal activity may have been occurring, which is not sufficient to trigger an epileptic seizure but may instigate tRNA cleavage and secretion. One study using continuous intracranial EEG monitoring of a small group of patients identified long-term energy bursts up to 7 hours before seizure onset, which increased in frequency in the hours immediately preceding seizure onset (15). To examine this, a neurologist reviewed video EEG recordings made 18 to 24 hours after the patients’ arrival to the EMU, and the patients were then classified into 3 groups on the basis of their video EEG activity: rare, occasional, and frequent. There was no significant difference in pre-seizure tRNA fragment levels between the groups (Supplemental Figure 12). Eight patients developed GTCS; however, no difference in pre-seizure tRNA fragment levels were detected in these patients compared with those who did not develop GTCS (Supplemental Figure 13).
There are currently no reliable biomarkers of seizure onset, and patients with drug-refractory epilepsy consistently report the unpredictable nature of seizures to be a challenging facet of their disease (16). The ability to forecast seizure activity would allow patients to regain control over their condition. Here, we analyzed RNA-Seq data from patients with focal epilepsy and identified 3 tRNA fragments that were elevated in pre-seizure plasma samples. We showed that these fragments are expressed by and secreted from neurons and that tRNA fragment levels are regulated in response to neuronal activity. Finally, we validated the RNA-Seq data results showing that tRNA fragments were elevated in pre-seizure plasma samples from a cohort of patients with focal epilepsy compared with samples from healthy controls. We present a proof-of-concept study indicating that plasma tRNA fragments warrant further investigation as prodromal biomarkers that could be used to predict seizure risk.
The underlying stress that precipitates tRNA cleavage in our study is unknown. Seizures are generally defined as excessive electrical activity in the brain that can lead to involuntary movement, changes in behavior, and loss of consciousness. Many underlying causes, such as infections, tumors, brain trauma, oxygen deprivation, and exposure to drugs or toxins, can lead to seizures. However, for most patients diagnosed with epilepsy, the underlying cause of their seizures is unknown. At the cellular level, numerous changes during epileptogenesis have been identified including neurodegeneration, gliosis, inflammation, neuronal growth, and angiogenesis (reviewed in ref. 17). tRNA cleavage occurs in response to stress in organisms from yeast to humans, indicating that it is a highly conserved process (5, 6). tRNA cleavage can occur at several sites on the tRNA molecule; Dicer cleaves tRNAs between the D-loop and the anticodon loop, whereas angiogenin primarily cleaves tRNAs within the anticodon loop (6). Interestingly, Dicer levels are significantly lower in TLE patients with hippocampal sclerosis (HS) than in those without (18). tRNA cleavage has been identified in response to infection (19) and ischemia (20), indicating that these epileptogenic processes could contribute to the observed tRNA cleavage. A recent study has shown that kainic acid (KA) treatment of synaptosome fractions induced spontaneous release of specific miRNAs, which was inhibited by depolarization, indicating a dynamic regulation of noncoding RNA secretion by neuronal activity (21). An important limitation of the present study is that, despite the detection of tRNA fragments in resected brain tissue and cultured hippocampal neurons, we cannot exclude the possibility that the tRNA fragments we detected in the patients’ plasma may have originated in another tissue, as both acute seizures and chronic epilepsy are system disorders, with physical and biochemical effects on organs and tissues outside the CNS (22).
tRNA fragments share many features with miRNAs that make them both ideal molecules for use as biomarkers. In order to be of use to patients, a biomarker of seizure imminence would require a portable quantification device capable of rapidly assessing tRNA fragment levels in whole blood. Recent advances in this area include development of the TORNADO device (23), which is capable of accurately quantifying miRNA-134 in plasma from patients with focal epilepsy using an electrochemical quantification approach (12). This might complement other new wearable technologies, such as seizure detection watches (24). Our data indicate that tRNA fragments are quantifiable using standard laboratory-based PCR techniques and may provide better specificity than miRNAs as epilepsy biomarkers, which are the current focus of many studies.
Methods
Study approval.
Samples were collected at the Philipps University of Marburg in Marburg, Germany (MAR) and the Department of Neurology of Beaumont Hospital in Dublin, Ireland (DUB). Ethics approval was obtained from the MAR and DUB medical ethics committees (MAR, 17/14; DUB, 13/75), and written informed consent was obtained from all participants according to Declaration of Helsinki principles. The animal work was approved by the Research Ethics Committee of the Royal College of Surgeons in Ireland (REC 1122).
Additional methods are provided in the Supplemental Methods.
Author contributions
MCH, DCH, and JHMP were responsible for the study design. RR, HEN, ND, DOB, SB, and FR performed sample collection and clinical evaluations. MCH, RR, and NM conducted and analyzed experiments. MCH, DCH, and JHMP wrote the manuscript.
Supplementary Material
Acknowledgments
The authors wish to thank the patients who participated in this study, the neuropathologists who assessed surgically resected patient tissue (Jane Cryan, Francesca M. Brett, Alan Beausang, and Michael A. Farrell, all from Beaumont Hospital, Dublin, Ireland), and Heiko Duessman (Department of Physiology and Medical Physics, RCSI) for help with imaging. This publication has emanated from research supported in part by a grant from the Science Foundation Ireland (SFI) under grant number 16/RC/3948 and co-funded under the European Regional Development Fund and by FutureNeuro industry partners. This study was supported by SFI grants SFI/13/IA/1891; the European Union’s ‘Seventh Framework’ Programme (FP7) under grant agreement number 602130 (EpimiRNA); and a fellowship from the Iraqi Ministry of Higher Education and Scientific Research.
Version 1. 04/30/2019
In-Press Preview
Version 2. 06/10/2019
Electronic publication
Version 3. 07/01/2019
Print issue publication
Funding Statement
This publication has emanated from research supported in part by a research grant from Science Foundation Ireland (SFI) under Grant Number 16/RC/3948 and co-funded under the European Regional Development Fund and by FutureNeuro industry partners
This study was also supported by SFI grants SFI/13/IA/1891 and financial support of the European Union’s ‘Seventh Framework’ Programme (FP7) under Grant Agreement No. 602130 (EpimiRNA).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(7):2946–2951.https://doi.org/10.1172/JCI126346.
Contributor Information
Marion C. Hogg, Email: marionhogg@rcsi.ie.
Rana Raoof, Email: ranamraoof@gmail.com.
Hany El Naggar, Email: hanyelnaggar@rcsi.ie.
Naser Monsefi, Email: naser.monsefi@gmail.com.
Norman Delanty, Email: normandelanty@beaumont.ie.
Sebastian Bauer, Email: S.Bauer@med.uni-frankfurt.de.
Felix Rosenow, Email: rosenow@med.uni-frankfurt.de.
David C. Henshall, Email: dhenshall@rcsi.ie.
Jochen H.M. Prehn, Email: prehn@rcsi.ie.
References
- 1.Kuhlmann L, Lehnertz K, Richardson MP, Schelter B, Zaveri HP. Seizure prediction - ready for a new era. Nat Rev Neurol. 2018;14(10):618–630. doi: 10.1038/s41582-018-0055-2. [DOI] [PubMed] [Google Scholar]
- 2.Walker LE, et al. Molecular isoforms of high-mobility group box 1 are mechanistic biomarkers for epilepsy. J Clin Invest. 2017;127(6):2118–2132. doi: 10.1172/JCI92001. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Enright N, Simonato M, Henshall DC. Discovery and validation of blood microRNAs as molecular biomarkers of epilepsy: Ways to close current knowledge gaps. Epilepsia Open. 2018;3(4):427–436. doi: 10.1002/epi4.12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun J, et al. Identification of serum miRNAs differentially expressed in human epilepsy at seizure onset and post-seizure. Mol Med Rep. 2016;14(6):5318–5324. doi: 10.3892/mmr.2016.5906. [DOI] [PubMed] [Google Scholar]
- 5.Lee SR, Collins K. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J Biol Chem. 2005;280(52):42744–42749. doi: 10.1074/jbc.M510356200. [DOI] [PubMed] [Google Scholar]
- 6.Li S, Xu Z, Sheng J. tRNA-derived small RNA: A novel regulatory small non-coding RNA. Genes (Basel) 2018;9(5):E246. doi: 10.3390/genes9050246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43(4):613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emara MM, et al. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem. 2010;285(14):10959–10968. doi: 10.1074/jbc.M109.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Q, et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351(6271):397–400. doi: 10.1126/science.aad7977. [DOI] [PubMed] [Google Scholar]
- 10.Sharma U, et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016;351(6271):391–396. doi: 10.1126/science.aad6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dhahbi JM, et al. 5′ tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. BMC Genomics. 2013;14:298. doi: 10.1186/1471-2164-14-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raoof R, et al. Dual-center, dual-platform microRNA profiling identifies potential plasma biomarkers of adult temporal lobe epilepsy. EBioMedicine. 2018;38:127–141. doi: 10.1016/j.ebiom.2018.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weisová P, et al. Latrepirdine is a potent activator of AMP-activated protein kinase and reduces neuronal excitability. Transl Psychiatry. 2013;3:e317. doi: 10.1038/tp.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, Wu C, Aramayo R, Sachs MS, Harlow ML. Synaptic vesicles contain small ribonucleic acids (sRNAs) including transfer RNA fragments (trfRNA) and microRNAs (miRNA) Sci Rep. 2015;5:14918. doi: 10.1038/srep14918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Litt B, et al. Epileptic seizures may begin hours in advance of clinical onset: a report of five patients. Neuron. 2001;30(1):51–64. doi: 10.1016/S0896-6273(01)00262-8. [DOI] [PubMed] [Google Scholar]
- 16.Dalic L, Cook MJ. Managing drug-resistant epilepsy: challenges and solutions. Neuropsychiatr Dis Treat. 2016;12:2605–2616. doi: 10.2147/NDT.S84852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pitkänen A, Lukasiuk K. Mechanisms of epileptogenesis and potential treatment targets. Lancet Neurol. 2011;10(2):173–186. doi: 10.1016/S1474-4422(10)70310-0. [DOI] [PubMed] [Google Scholar]
- 18.McKiernan RC, et al. Reduced mature microRNA levels in association with dicer loss in human temporal lobe epilepsy with hippocampal sclerosis. PLoS ONE. 2012;7(5):e35921. doi: 10.1371/journal.pone.0035921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Q, Lee I, Ren J, Ajay SS, Lee YS, Bao X. Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol Ther. 2013;21(2):368–379. doi: 10.1038/mt.2012.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Q, et al. tRNA-derived small non-coding RNAs in response to ischemia inhibit angiogenesis. Sci Rep. 2016;6:20850. doi: 10.1038/srep20850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu J, Chen Q, Zen K, Zhang C, Zhang Q. Synaptosomes secrete and uptake functionally active microRNAs via exocytosis and endocytosis pathways. J Neurochem. 2013;124(1):15–25. doi: 10.1111/jnc.12057. [DOI] [PubMed] [Google Scholar]
- 22.Devinsky O. Effects of seizures on autonomic and cardiovascular function. Epilepsy Curr. 2004;4(2):43–46. doi: 10.1111/j.1535-7597.2004.42001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McArdle H, et al. “TORNADO” - Theranostic One-Step RNA Detector; microfluidic disc for the direct detection of microRNA-134 in plasma and cerebrospinal fluid. Sci Rep. 2017;7(1):1750. doi: 10.1038/s41598-017-01947-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bruno E, et al. Wearable technology in epilepsy: The views of patients, caregivers, and healthcare professionals. Epilepsy Behav. 2018;85:141–149. doi: 10.1016/j.yebeh.2018.05.044. [DOI] [PubMed] [Google Scholar]
- 25.Chan PP, Lowe TM. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2016;44(D1):D184–D189. doi: 10.1093/nar/gkv1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.