Abstract
Invasive fungal infection is a serious health threat with high morbidity and mortality. Current antifungal drugs only demonstrate partial success in improving prognosis. Furthermore, mechanisms regulating host defense against fungal pathogens remain elusive. Here, we report that the downstream of kinase 3 (Dok3) adaptor negatively regulates antifungal immunity in neutrophils. Our data revealed that Dok3 deficiency increased phagocytosis, proinflammatory cytokine production, and netosis in neutrophils, thereby enhancing mutant mouse survival against systemic infection with a lethal dose of the pathogenic fungus Candida albicans. Biochemically, Dok3 recruited protein phosphatase 1 (PP1) to dephosphorylate Card9, an essential player in innate antifungal defense, to dampen downstream NF-κB and JNK activation and immune responses. Thus, Dok3 suppresses Card9 signaling, and disrupting Dok3-Card9 interaction or inhibiting PP1 activity represents therapeutic opportunities to develop drugs to combat candidaemia.
Keywords: Immunology, Infectious disease
Keywords: Adaptor proteins, Innate immunity
Introduction
Invasive Candida infection is a major public health concern due to its high mortality rates and increased costs of treatment (1). Surprisingly, the incidence of candidiasis rises with the advancement of medicine, mainly because of the emergence of large numbers of invasive procedures and the extensive use of broad-spectrum antibiotics (2). The risk of infection is especially high among the immunocompromised, including patients in intensive care units and people undergoing cancer chemotherapy (3). However, treatment of Candida infection remains challenging. Current antifungal drugs only demonstrate partial success in improving prognosis, and the rapid emergence of drug resistance among Candida species is a growing problem (4). As such, there is a pressing need to develop novel antifungal therapies to improve clinical outcomes. Understanding the mechanistic interaction between host immune cells and fungal pathogens holds the key for uncovering novel immune-based treatments to combat candidiasis (5).
During fungal infection, C-type lectin receptors (CLRs), such as Dectin-1 (6), Dectin-2 (7), and Mincle (8), expressed on innate immune cells play a major role in the recognition of fungal cell wall constituents such as β-glucans, α-mannans, and glycolipids. Upon agonist binding, CLR signaling triggers the phosphorylation of immunoreceptor tyrosine-based activation motif–like (ITAM-like) motifs in the cytoplasmic tail of Dectin-1 or mediates the phosphorylation and recruitment of ITAM-containing adaptor FcRγ to Dectin-2 and Mincle. These events activate tyrosine kinase Syk, which subsequently transduces the signal to phospholipase Cγ2 (PLCγ2) (9, 10), and PKCδ (11), which phosphorylates central adaptor caspase recruitment domain-containing protein 9 (Card9). As a result, Card9 associates with the adaptor B cell lymphoma 10 (Bcl-10) and paracaspase Mucosa-associated lymphoid tissue lymphoma translocation 1 (Malt1) to form a scaffold responsible for the activation of downstream NF-κB and MAPKs (12–15). These signaling pathways turn on a series of effector mechanisms, including the release of proinflammatory cytokines and phagocytosis, ultimately leading to fungi clearance (16, 17).
Although the mechanisms involved in the initiation of fungicidal pathways have been extensively studied, little is known with regard to how CLR signaling is being negatively regulated. Recent studies have identified 2 negative regulators that limit innate antifungal immunity through inhibition of fungal recognition. E3 ubiquitin ligase CBLB targets Dectin-1, -2, and -3 for polyubiquitination and degradation (18–20), while JNK1 suppresses the expression of the CLR CD23 (21). However, it remains to be determined how other downstream components of CLR signaling are being negatively regulated to maintain immune homeostasis during fungal infection. In particular, how Card9 signaling is constrained during antifungal response is poorly understood.
Downstream of kinase 3 (Dok3) is an adaptor molecule preferentially expressed in hematopoietic cells (22). It is known to act as a cell type–specific regulator downstream of various immune receptors, including TLR3, TLR4, and B cell receptor (BCR), during viral infection (23), LPS stimulation (24) as well as plasma cell differentiation (25), respectively. However, the role of Dok3 during fungal infection is completely unknown. Here, we report that Dok3 negatively regulates antifungal immunity in neutrophils by recruiting protein phosphatase 1 (PP1) to suppress Card9 activity. Consequently, loss of Dok3 ameliorates fungal pathology and protects the host from lethal systemic infection with Candida albicans. Hence, this study extends our understanding of the mechanisms regulating host innate antifungal defense and may have translational relevance for the development of antifungal drugs.
Results
Loss of Dok3 ameliorates fungal pathology in vivo.
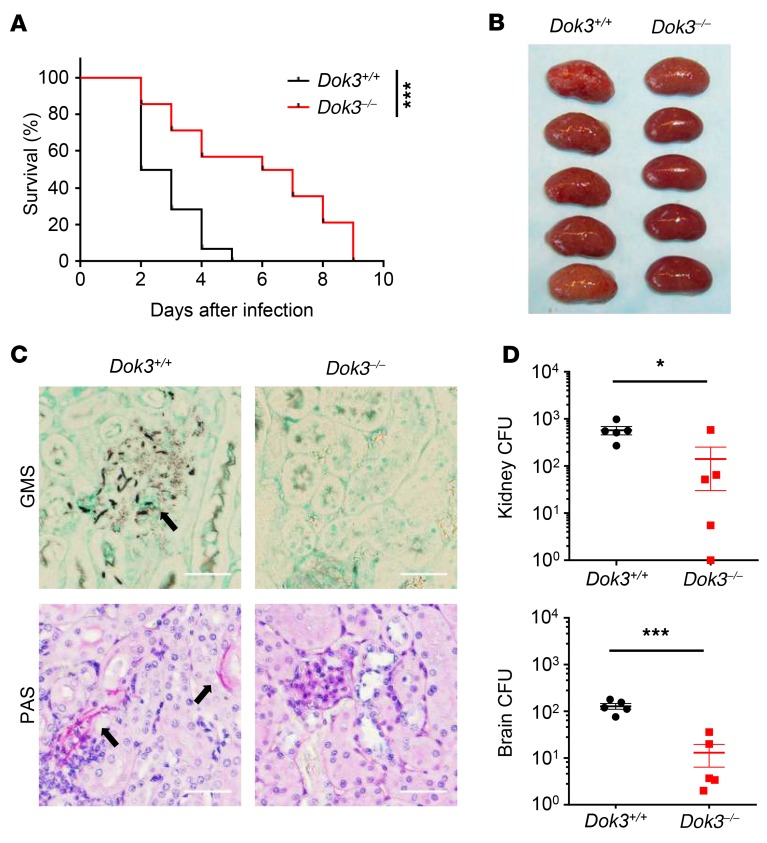
Adaptor proteins such as Card9 and FcRγ serve important roles in CLR signaling by facilitating specific interactions between proteins (12, 26). To test for a potential role of the Dok3 adaptor in antifungal immunity, we challenged WT (Dok3+/+) and Dok3-deficient (Dok3–/–) mice with a lethal dose of C. albicans systemically. Loss of Dok3 in Dok3–/– mice was verified by protein blot analysis (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI126341DS1). Strikingly, Dok3–/– mice demonstrated an improved survival rate as compared with Dok3+/+ controls (Figure 1A). Unlike the kidneys of Dok3+/+ mice, which were enlarged with macroscopically visible fungal colonies, the kidneys of Dok3–/– mice had significantly reduced fungal burden (Figure 1B). Histopathology analysis also revealed reduced numbers of fungal hyphae in the kidneys of Dok3–/– mice (Figure 1C). Quantitatively, fungal titers were approximately 10-fold higher in the kidneys and brains of Dok3+/+ mice as compared with those of Dok3–/– mice (Figure 1D). These findings demonstrated that Dok3 deficiency protects mice from lethal systemic infection with C. albicans.
Figure 1. Dok3 negatively regulates antifungal immunity in vivo.
(A) Dok3+/+ (n = 14) and Dok3–/– (n = 12) mice were infected intravenously with 1 × 106 CFU of C. albicans. Survival was monitored over indicated time periods. Data were pooled from 3 independent experiments. ***P = 0.0007, log-rank test. (B) Kidneys were harvested for macroscopic analysis of fungal burden 2 days after infection (n = 5). (C) Analysis of kidney fungal burden by GMS and PAS staining 1 day after infection with 2.5 × 105 CFU of C. albicans. Arrows indicate fungal hyphae. Scale bars: 50 μm. Six sections per kidney were analyzed (n = 2). (D) Fungal titers of kidneys and brains were determined 2 days after infection. Symbols represent individual mice (n = 5). Data are shown as mean ± SEM. *P = 0.02; ***P = 0.0003, unpaired 2-tailed Student’s t test. One representative experiment out of 3 independent experiments is shown.
Dok3 is dispensable for the development of immune cells.
The innate immune system forms the first and most important line of defense against fungal infection (27). To determine whether there is any developmental defect in innate immune cells that could affect antifungal defense in Dok3–/– mice, we assessed their distribution across various lymphoid organs. However, Dok3 deficiency has no effect on the total cellularity and composition of innate immune cells in the BM, spleen, and lymph nodes. Notably, the percentages and numbers of neutrophils (Ly6G+Ly6C+), monocytes (Ly6G–Ly6C+), macrophages (F4/80+MHC IIhi/lo), and DCs (CD11c+ MHC II+/hi/lo) were similar between Dok3+/+ and Dok3–/– mice. Moreover, peripheral T and B cell populations were also not affected by Dok3 deficiency (Supplemental Figure 1, B–G) (28), suggesting that Dok3 is dispensable for the development of innate and adaptive immune cells.
Dok3 negatively regulates fungicidal activity of neutrophils against unopsonized C.
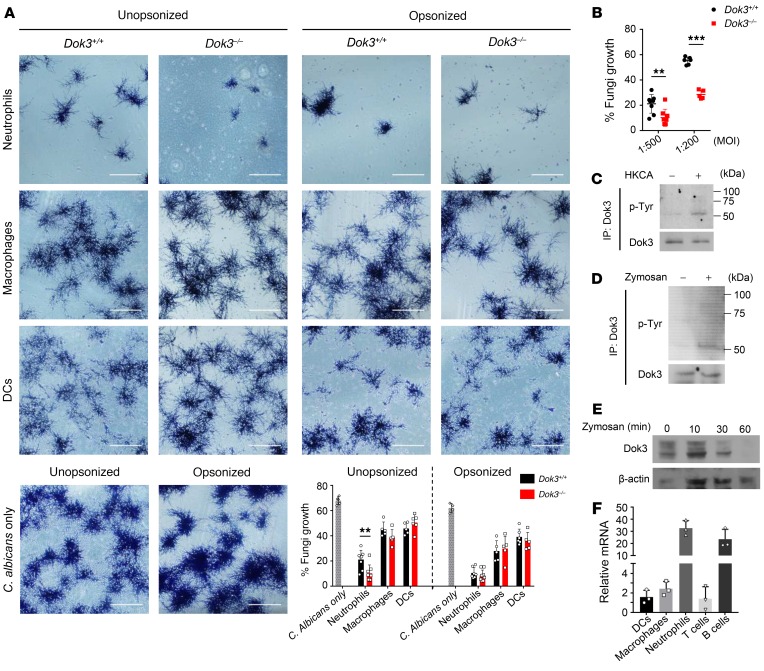
albicans. To determine whether Dok3 is involved in fungi killing in innate cells, we purified bone marrow and splenic neutrophils, macrophages, and DCs from Dok3+/+ and Dok3–/– mice and assessed their ability to restrict germination of C. albicans yeast into clusters of hyphae in vitro after overnight coculture (18, 29). Since adaptive T cell and B cell antibody responses are less relevant during antifungal defense against invasive candidiasis (30), we did not further investigate their fungicidal activity in the absence of Dok3. Strikingly, Dok3–/– neutrophils were highly potent in fungi killing and were able to significantly control the outgrowth of unopsonized C. albicans, whereas no significant differences in fungicidal activity were observed between Dok3+/+ and Dok3–/– macrophages and DCs with unopsonized C. albicans (Figure 2A). On the other hand, although serum opsonization of C. albicans enhances the overall killing effect by innate immune cells, we did not observe any additional advantage in the killing of opsonized C. albicans by Dok3–/– neutrophils (Figure 2A). We further investigated the fungicidal activity of neutrophils by infecting the cells with unopsonized C. albicans at a higher MOI, and similarly, Dok3–/– neutrophils were able to significantly inhibit the germination of unopsonized C. albicans as compared with Dok3+/+ neutrophils (Figure 2B). Hence, these data show that Dok3 deficiency selectively enhances the capacity of neutrophils to prevent outgrowth of unopsonized C. albicans.
Figure 2. Dok3 negatively regulates fungi killing in neutrophils.
(A) Killing capacity of bone marrow neutrophils and splenic macrophages and DCs as assessed by overnight coculture with unopsonized (left) or serum-opsonized (right) C. albicans (MOI 1:500). C. albicans were counterstained with crystal violet. Assays were performed in triplicate; more than 5 fields per group were taken. Scale bars: 200 μm. Data are shown as mean ± SD. **P = 0.008, unpaired 2-tailed Student’s t test. (B) Killing capacity of bone marrow neutrophils against unopsonized C. albicans (MOI 1:500 and 1:200). Assays were performed in triplicates; more than 5 fields per group were taken. Data are shown as mean ± SD. **P = 0.008; ***P = 0.0001, unpaired 2-tailed Student’s t test. (C and D) Purified Dok3+/+ neutrophils were treated with (C) HKCA (MOI 1:1) or (D) zymosan (10 μg/ml) for 10 minutes or 30 minutes, respectively. Cell lysates were IP with a Dok3-specific antibody, and precipitates were probed for p-Tyr and Dok3. Images are representative of 2 independent experiments, respectively. (E) Immunoblot analysis of Dok3 protein levels in purified Dok3+/+ neutrophils stimulated for various times with zymosan (10 μg/ml). β-Actin was used as loading control. Image is representative of 3 independent experiments. (F) Dok3 expression in purified Dok3+/+ splenic DCs, splenic macrophages, bone marrow neutrophils, lymph node T cells, and splenic B cells. Total RNA extracted was used for cDNA synthesis before conducting real-time quantitative reverse-transcription PCR (RT-qPCR). Gene expression was normalized against Gapdh. Data are shown as mean ± SD (n = 3).
Dok3 is tyrosine phosphorylated and degraded upon fungal infection.
To provide direct evidence for the involvement of Dok3 in neutrophil antifungal response, we determined whether the molecule is tyrosine phosphorylated and activated upon neutrophil recognition of fungal components. We stimulated neutrophils with heat-killed C. albicans (HKCA) or zymosan, a Dectin-1 and TLR2 ligand derived from yeast cell wall, and found a robust induction of tyrosine phosphorylation on Dok3 (Figure 2, C and D). In addition, we observed Dok3 to be degraded over time after stimulation of neutrophils with zymosan (Figure 2E), suggesting that Dok3 degradation is necessary to overcome its negative regulatory function during fungal infection. Moreover, in accordance with earlier observations in which Dok3 negatively controls neutrophilic fungal-killing activity, Dok3 mRNA was found to be highly expressed in resting neutrophils in comparison with macrophages, DCs, and T cells (Figure 2F). B cells were also found to express relatively high levels of Dok3 mRNA (Figure 2F), in line with previous studies implicating Dok3 in BCR signaling (25, 28, 31, 32). Collectively, these data point to a role for Dok3 in neutrophil antifungal response.
Dok3 negatively regulates antifungal effector functions in neutrophils.
Recognition of β-glucans and α-mannans on invading fungi via CLRs Dectin-1 and -2, respectively, is crucial for the initiation of antifungal immune responses (15–17). Here, we observed comparable expression of Dectin-1 and -2 on the surfaces of Dok3+/+ and Dok3–/– neutrophils, DCs, and macrophages upon stimulation with various fungal agonists (zymosan, HKCA yeast, and HKCA hyphae) (Supplemental Figure 2, A–C), suggesting that Dok3 does not regulate CLR availability or expression in these innate immune cells.
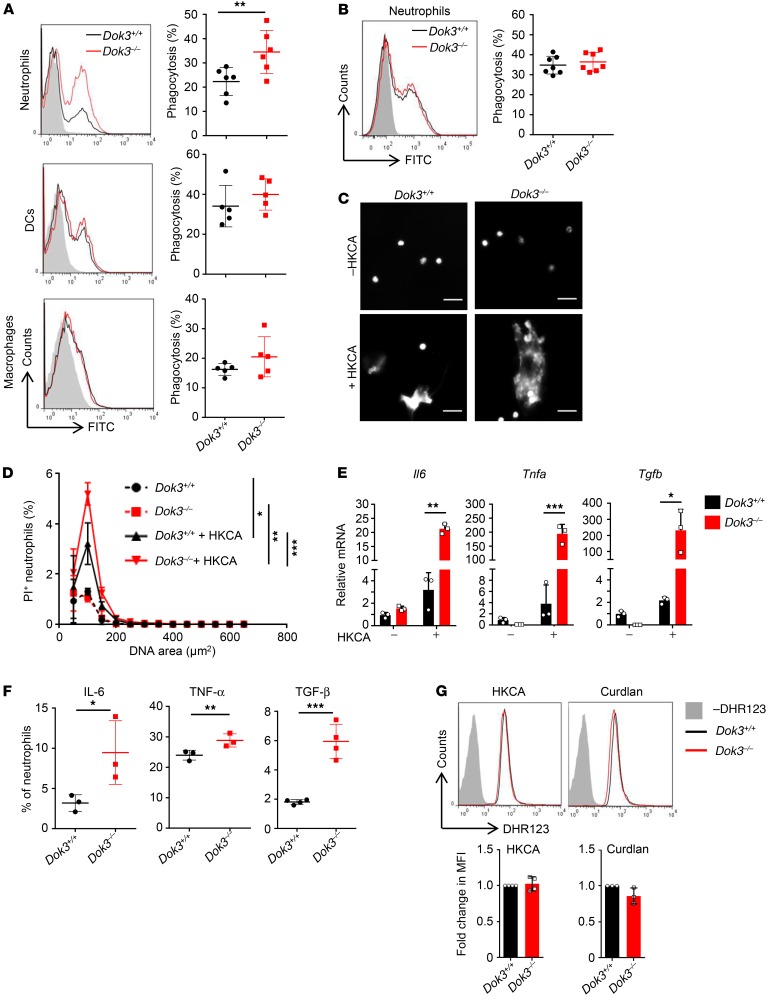
To understand how Dok3 negatively regulates fungicidal activity of neutrophils, we assessed various antifungal and proinflammatory effector functions triggered by CLR signaling. Phagocytosis constitutes the first line of defense against invading fungal pathogens. Indeed, in agreement with their enhanced fungicidal activity against unopsonized C. albicans, we observed higher phagocytic uptake of FITC-labeled unopsonized HKCA yeast by Dok3–/– as compared with Dok3+/+ neutrophils. On the other hand, the extent of phagocytosis was similar between Dok3+/+ and Dok3–/– DCs and macrophages (Figure 3A), consistent with their comparable in vitro fungi-killing capacity against unopsonized C. albicans (Figure 2A). Notably, the uptake of opsonized HKCA was found to be normal in Dok3–/– neutrophils (Figure 3B), which correlates with their intact killing capacity against opsonized C. albicans (Figure 2A).
Figure 3. Enhanced antifungal effector functions of Dok3–/– neutrophils.
(A and B) Phagocytosis of FITC-labeled (A) unopsonized or (B) opsonized HKCA by bone marrow neutrophils, splenic DCs, and macrophages (MOI 1:2). Gray histogram represents unstimulated control. Symbols represent individual mice (n = 5–7). Data are shown as mean ± SD. **P = 0.01, unpaired 2-tailed Student’s t test. (C and D) NET release after stimulation of purified Dok3+/+ and Dok3–/– neutrophils with hyphae form HKCA (MOI 1:10). (C) Extracellular DNA stained with DAPI 4 hours following stimulation. Scale bars: 20 μm. (D) Quantification of NET release with PI. Percentage of PI+ neutrophils over total number of neutrophils. *P = 0.0003; **P = 0.0001; ***P = 0.0001, 2-way ANOVA, Sidak’s multiple-comparison post test. Data are pooled from 3 independent experiments (n = 3). (E) mRNA expression levels of indicated cytokines in purified Dok3+/+ and Dok3–/– neutrophils after stimulation with HKCA (MOI 1:2). Data are shown as mean ± SD. (n = 3). *P = 0.03; **P = 0.0002; ***P = 0.0006, unpaired 2-tailed Student’s t test. (F) Flow cytometric analysis of indicated cytokine production by Dok3+/+ and Dok3–/– neutrophils following zymosan stimulation. Representative dot plots are shown in Supplemental Figure 3. Graph indicates percentages of IL-6+, TNF-α+ and TGF-β+ neutrophils. Data are pooled from 3 independent experiments (n = 3–4). Data are shown as mean ± SD. *P = 0.05; **P = 0.009; ***P = 0.0004, unpaired 2-tailed Student’s t test. (G) ROS production by Dok3+/+ and Dok3–/– neutrophils. Neutrophils were labeled with DHR123 before stimulation with HKCA (MOI 1:2) (n = 4) or curdlan (100 μg/ml) (n = 3). Fluorescence was measured by flow cytometry. Histograms were pregated on singlet, live Ly6G+Ly6C+ cells. Gray histograms represent unlabeled control. One representative experiment out of 3 independent experiments is shown. Bar graphs indicate fold change in MFI of DHR123 fluorescence in Dok3–/– neutrophils relative to Dok3+/+ neutrophils. Data are pooled from 3 independent experiments. Data are shown as mean ± SD.
To further address whether Dok3 deficiency contributes to the ability of neutrophils to counter large and invasive fungal hyphae structures that cannot be phagocytosed (33), we examined the release of neutrophil extracellular traps (NETs) following their stimulation with HKCA in preformed hyphae. Interestingly, Dok3–/– neutrophils induced release of more extensive NETs, as measured by the cell-impermeable dyes DAPI and propidium iodide (PI), which stain for extracellular DNA (Figure 3, C and D). Together, these data indicate an important role of Dok3 in suppressing phagocytosis and NETosis in neutrophils, thereby negatively regulating their fungi-killing activity.
Upon fungal infection, CLR signaling triggers the transcription of various genes encoding proinflammatory cytokines (34, 35). We observed increased production of Il6, Tnfa, and Tgfb, cytokines that have been implicated in antifungal defense, by Dok3–/– neutrophils at the mRNA level following treatment with HKCA (Figure 3E). Consistent with the increased transcription, there was increased secretion of these cytokines by Dok3–/– neutrophils upon zymosan stimulation (Figure 3F and Supplemental Figure 3). Hence, Dok3 deficiency enhances the production of proinflammatory cytokines elicited by CLR signaling in neutrophils. We also assessed the contribution of Dok3 to neutrophil respiratory burst, but did not observe any difference in ROS production between Dok3+/+ and Dok3–/– neutrophils upon stimulation with HKCA or curdlan, a β-glucan that is an agonist of Dectin-1 (Figure 3G). Collectively, our data show that loss of Dok3 enhances fungi killing through boosting phagocytosis of unopsonized yeast, netosis, and cytokine production in neutrophils.
Dok3 negatively regulates NF-κB and JNK signaling in neutrophils.
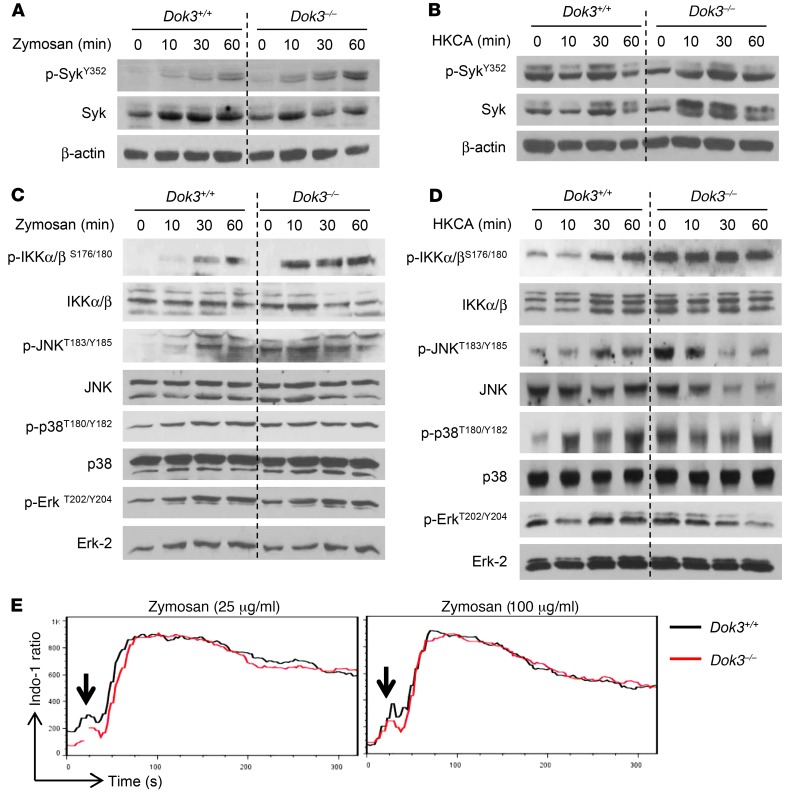
To assess where Dok3 functions in antifungal signaling, we stimulated neutrophils with either zymosan or HKCA, which engages mainly Dectin-1, and analyzed the CLR signaling pathway involving the tyrosine kinase Syk, NF-κB and JNK, p38, and Erk MAP kinases (36). We found no differences in Syk phosphorylation in Dok3+/+ and Dok3–/– neutrophils (Figure 4, A and B, and Supplemental Figure 4A), suggesting that Dok3 is not involved in receptor-proximal signaling. However, Dok3–/– neutrophils exhibited increased levels of phosphorylated IKKα/β (p-IKKα/β) upon stimulation with zymosan or HKCA (Figure 4, C and D, and Supplemental Figure 4B), indicating that Dectin-1–mediated canonical NF-κB activation is enhanced. This is consistent with our earlier findings, in which we observed an upregulation of NF-κB–dependent cytokine transcripts in Dok3–/– neutrophils following treatment with HKCA (Figure 3E). JNK phosphorylation was also observed to be higher in Dok3–/– neutrophils in response to zymosan and HKCA stimulation (Figure 4, C and D, and Supplemental Figure 4C). On the other hand, the phosphorylation levels of Erk and p38 remained similar in Dok3+/+ and Dok3–/– neutrophils (Figure 4, C and D, and Supplemental Figure 4, D and E), suggesting that these signaling pathways are Dok3 independent. We also investigated the calcineurin/NFAT pathway downstream of Syk, but did not observe any difference in calcium flux upon stimulation with both low and high doses of zymosan (Figure 4E). Together, our data show that Dok3 negatively controls NF-κB and JNK signaling downstream of Syk, in response to Dectin-1 engagement.
Figure 4. Dok3 deficiency enhances NF-κB and JNK signaling.
(A and B) Immunoblot analysis of p-SykY352 and Syk in purified Dok3+/+ and Dok3–/– neutrophils stimulated for various times with (A) zymosan (10 μg/ml) or (B) HKCA (MOI 1:1). β-Actin served as loading control. Images are representative of 3 independent experiments. (C and D) Immunoblot analysis of p-IKKα/βS176/180, IKKα/β, p-JNKT183/Y185, JNK, p-p38T180/Y182, p38, p-ErkT202/Y204, and Erk-2 in purified Dok3+/+ and Dok3–/– neutrophils stimulated with (C) zymosan (10 μg/ml) or (D) HKCA (MOI 1:1) for indicated periods of time. Images are representative of 3 to 4 independent experiments. Quantifications of immunoblots are shown in Supplemental Figure 4. (E) Calcium signaling in Indo-1–loaded Dok3+/+ and Dok3–/– neutrophils after stimulation with either 25 μg/ml or 100 μg/ml of zymosan. Intracellular calcium flux was monitored in real time via flow cytometry. One representative out of 3 independent experiments is shown.
Dok3 suppresses Card9 signaling.
NF-κB and JNK activation have been reported to be regulated by the Syk-dependent Card9/Bcl10/Malt1 (CBM) complex (12, 13, 37). Since both NF-κB and JNK signaling were enhanced in the absence of Dok3, we investigated whether Dok3 functions in the same signaling cascade as the CBM complex. Phosphorylation of adaptor protein Card9 at Thr231 by PKCδ is critical for its activation and signal transduction function (11). To study this, we IP Card9 from neutrophils before immunoblotting it with a phospho-threonine antibody due to the lack of a commercially available phospho-specific antibody against Card9 Thr231. The specificities of various Card9 antibodies used were verified with Card9–/– cells (Supplemental Figure 5). In Dok3+/+ neutrophils, threonine phosphorylation was induced on Card9 upon zymosan stimulation. However, Card9 was observed to be constitutively phosphorylated at higher levels in Dok3–/– neutrophils (Figure 5A). This indicates that Dok3 negatively regulates Card9 activation in neutrophils, thereby suppressing downstream NF-κB and JNK signaling.
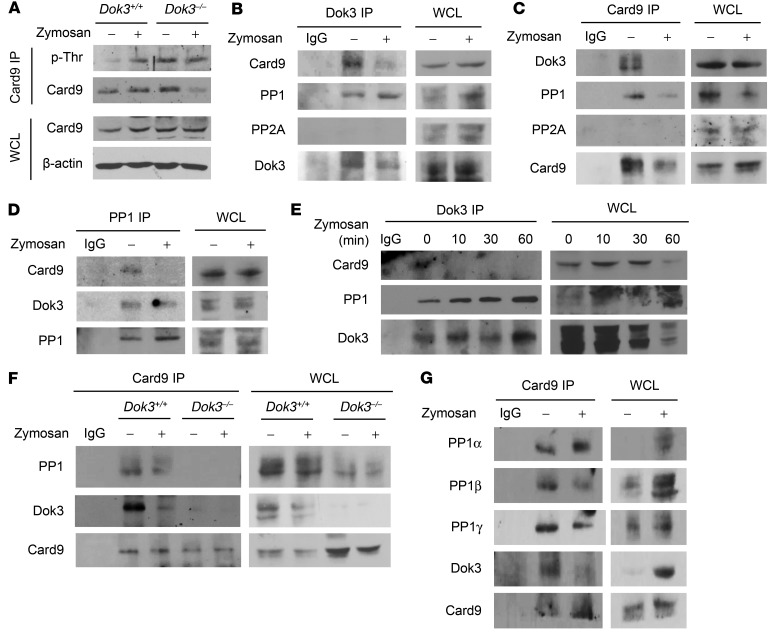
Figure 5. Dok3 mediates Card9 dephosphorylation through PP1 recruitment.
(A) Purified Dok3+/+ and Dok3–/– neutrophils were treated with zymosan (10 μg/ml) for 30 minutes. Whole cell lysates (WCL) were IP with a Card9-specific antibody, and precipitates were probed for p-Thr and Card9. Whole cell lysates were also probed for Card9 and β-actin. (B) Co-IP analysis of cell lysates from purified Dok3+/+ neutrophils treated with zymosan (10 μg/ml) for 30 minutes, and IP with a Dok3-specific antibody or an IgG control. Precipitates and whole cell lysates were probed for Card9, PP1, PP2A, and Dok3. (C) Co-IP analysis of cell lysates from purified Dok3+/+ neutrophils treated for 30 minutes with zymosan (10 μg/ml), and IP with a Card9-specific antibody or an IgG control. Precipitates and whole cell lysates were probed for Dok3, PP1, PP2A, and Card9. (D) Co-IP analysis of cell lysates from purified Dok3+/+ neutrophils treated with zymosan (10 μg/ml) for 30 minutes, and IP with a PP1-specific antibody or an IgG control. Precipitates and whole cell lysates were probed for Card9, Dok3, and PP1. (E) Co-IP analysis of cell lysates from purified Dok3+/+ neutrophils treated with zymosan (10 μg/ml) for indicated periods of time and IP with a Dok3-specific antibody or an IgG control. Precipitates and whole cell lysates were probed for Card9, PP1, and Dok3. (F) Co-IP analysis of cell lysates from purified Dok3+/+ and Dok3–/– neutrophils treated with zymosan (10 μg/ml) for 30 minutes, and IP with a Card9-specific antibody or an IgG control. Precipitates and whole cell lysates were probed for PP1, Dok3, and Card9. (G) Co-IP analysis of cell lysates from purified Dok3+/+ neutrophils treated with zymosan (10 μg/ml) for 30 minutes and IP with a Card9-specific antibody or an IgG control. Precipitates and whole cell lysates were probed for PP1α, PP1β, PP1γ, Dok3, and Card9. All images are representative of 3 independent experiments.
Dok3 recruits PP1 to mediate Card9 dephosphorylation.
To determine whether the hyperphosphorylation of Card9 in Dok3–/– neutrophils is due to increased phosphorylation by PKCδ, we analyzed the phosphorylation status of PKCδ at Tyr311, a residue governing their activation (11). However, we did not observe increased PKCδ phosphorylation in Dok3–/– neutrophils upon zymosan stimulation. On the contrary, lower levels of p-PKCδ were detected (Supplemental Figure 6), possibly as a compensatory mechanism to reduce hyperphosphorylation of Card9 in the absence of Dok3.
To decipher how Dok3 negatively regulates Card9 phosphorylation, we first determined whether Dok3 interacts physically with Card9. Through endogenous co-immunoprecipitation (co-IP) studies with anti-Dok3 antibody, we found Card9 to be associated with Dok3 during steady state in neutrophils, and this binding was diminished upon stimulation with zymosan (Figure 5B). Conversely, anti-Card9 antibody was able to coprecipitate Dok3, and the interaction was abrogated upon neutrophil activation (Figure 5C). Since there was a stimulus-dependent dissociation of Card9 from Dok3 (Figure 5, B and C) and, reciprocally, Card9 was phosphorylated upon zymosan stimulation (Figure 5A), one possibility is that Dok3 recruits a phosphatase to mediate the dephosphorylation of Card9 in neutrophils. PP2A, a serine/threonine phosphatase, has been shown to mediate dephosphorylation of Card11, a homologue of Card9, in T cells (38). However, our endogenous co-IP studies did not reveal any association of PP2A with either Dok3 or Card9 in neutrophils (Figure 5, B and C). Instead, another member of that phosphatase family, PP1, was found to interact with Dok3 and Card9 (Figure 5, B–D). In particular, PP1 complexed with Dok3 constitutively, while the interaction between the Dok3-PP1 complex and Card9 existed predominantly during steady state and was disrupted upon zymosan stimulation (Figure 5, B–E). This inverse correlation between Card9-PP1 interaction and Card9 threonine phosphorylation status suggested that PP1 is responsible for maintaining Card9 in its dephosphorylated state. Crucially, PP1 complexed with Card9 only in Dok3+/+ neutrophils (Figure 5F), indicating that PP1 does not bind directly to Card9, and Dok3 was required to mediate the interaction between Card9 and PP1. In Dok3–/– neutrophils in which PP1 was unable to associate with Card9, markedly enhanced phosphorylation of Card9 threonine residues was detected (Figure 5, A and F). Together, these data indicate that adaptor protein Dok3 recruits PP1 to dephosphorylate Card9 on threonine residues, thereby negatively regulating Card9 activity and, subsequently, downstream NF-κB and JNK signaling.
Mammalian PP1 consists of 3 isoforms, namely PP1α, PP1β, and PP1γ, which are ubiquitously expressed across various cell types (39). To address whether all 3 isoforms are involved in Card9 dephosphorylation, we examined their interaction with Card9. From endogenous co-IP analyses in neutrophils, we observed that anti-Card9 antibody was able to precipitate PP1α, PP1β, and PP1γ together with Dok3 (Figure 5G), suggesting that all 3 isoforms of PP1 are regulators of Card9 dephosphorylation.
Inhibition of PP1 promotes antifungal immunity in neutrophils.
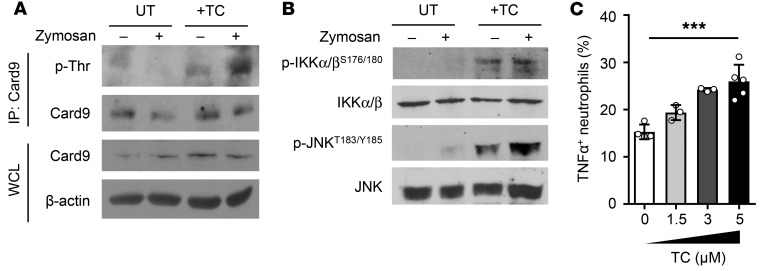
To further validate the role of PP1 as a Card9 threonine phosphatase, we pretreated Dok3+/+ neutrophils with tautomycetin, a specific inhibitor of PP1 (40), prior to stimulation with zymosan. Indeed, inhibition of PP1 activity profoundly enhanced Card9 threonine phosphorylation during steady state as well as upon zymosan stimulation, as compared with untreated controls under similar conditions (Figure 6A). Consequently, NF-κB and JNK signaling, as indicated by the phosphorylation of IKKα/β and JNK, were found to be elevated in tautomycetin-treated, unstimulated, and zymosan-stimulated neutrophils (Figure 6B). These results directly show that PP1 is required for dephosphorylation of threonine residues on Card9, thereby negatively regulating downstream NF-κB and JNK activity.
Figure 6. Inhibition of PP1 enhances Card9 phosphorylation and neutrophil antifungal responses.
Purified Dok3+/+ neutrophils were untreated (UT) or pretreated with tautomycetin (TC) for 2.5 hours. (A and B) Cells were stimulated with zymosan (10 μg/ml) for 30 minutes. (A) Cell lysates were IP with a Card9-specific antibody, and precipitates were probed for p-Thr and Card9. Whole cell lysates were probed for Card9 and β-actin. Images are representative of 3 independent experiments. (B) Immunoblot analysis of cell lysates for p-IKKα/βS176/180, IKKα/β, p-JNKT183/Y185, and JNK. Images are representative of 3 independent experiments. (C) Analysis of TNF-α production by neutrophils pretreated with indicated concentrations of tautomycetin by flow cytometry. Graph indicates percentages of TNF-α+ neutrophils. Data are pooled from 2 independent experiments (n = 3–5). ***P = 0.0002, 1-way ANOVA.
To correlate the biochemical changes associated with PP1 inhibition to the antifungal effects of neutrophils in vitro, we pretreated neutrophils with tautomycetin before infecting the cells with zymosan. Dok3+/+ neutrophils treated with increasing dosages of PP1 inhibitor demonstrated an increasing extent of TNF-α production upon zymosan stimulation (Figure 6C). These molecular and cellular changes induced by PP1 inhibition closely resemble the phenotype of Dok3–/– neutrophils. Together, these data suggest that inhibiting PP1 activity or disrupting Dok3-Card9 interaction represents potential therapeutic strategies for enhancing antifungal immunity.
Loss of Dok3 enhances neutrophil accumulation in Candida-infected brain.
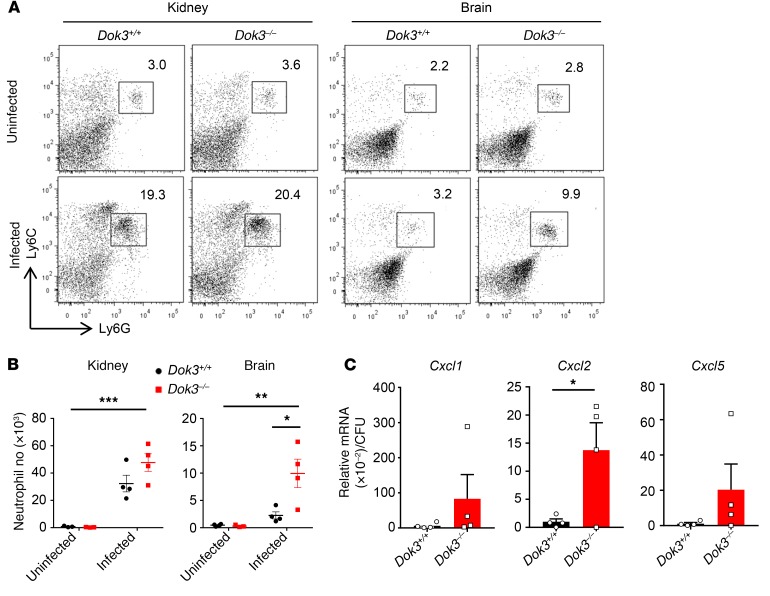
Card9 deficiency has been reported to predispose individuals to C. albicans infection, with preferential targeting of the CNS due to a defect in neutrophil recruitment to the brain, but not the kidneys (41). Since Dok3 negatively regulates Card9 activation, we analyzed the extent of neutrophil infiltration into the kidneys and brains of C. albicans–infected mice to verify the Card9 dependency of the Dok3 pathway in vivo. At 24 hours after infection, there was significant recruitment of neutrophils into the kidneys, with the neutrophil numbers being comparable between Dok3+/+ and Dok3–/– mice (Figure 7, A and B). However, a significant accumulation of neutrophils was observed in the brains of Dok3–/– mice 24 hours after infection, resulting in their lower fungal burden as compared with that of Dok3+/+ mice (Figure 7, A and B). Infiltration of neutrophils into the brain during C. albicans infection depends on various Card9-regulated neutrophil-targeted chemokines, such as Cxcl1, Cxcl2, and Cxcl5, produced by the resident CD45– cells and neutrophils (41). Indeed, Cxcl2 mRNA was found to be significantly upregulated in the brains of Dok3–/– mice, relative to their extent of fungal load (Figure 7C). Thus, Dok3 plays a role in regulating neutrophil infiltration into the brains during candidiasis by limiting Cxcl2 production.
Figure 7. Loss of Dok3 enhances neutrophil accumulation in Candida-infected brain.
(A) Flow cytometric analysis of neutrophils (Ly6G+Ly6C+) in kidneys and brains of Dok3+/+ and Dok3–/– mice 24 hours after infection with C. albicans. Dot plots are pregated on singlet, live CD45+ cells. (B) Total number of neutrophils in kidneys and brains of Dok3+/+ and Dok3–/– mice (n = 3–4). Data are shown as mean ± SEM. Two-way ANOVA demonstrated significant interaction between Dok3 expression and neutrophil numbers in the brain upon C. albicans infection (F[1,11] = 7.2; P = 0.02). *P = 0.02; **P = 0.002; ***P < 0.0001, 2-way ANOVA. (C) mRNA expression of indicated chemokines in homogenized brains expressed relative to the mean brain fungal burden of Dok3+/+ and Dok3–/– mice 24 hours after infection (n = 4). Data are shown as mean ± SEM. *P = 0.04, unpaired 2-tailed Student’s t test.
Discussion
In this study, we identified Dok3 as a critical negative regulator of antifungal immunity in neutrophils. Loss of Dok3 enhances various antifungal effector functions of neutrophils, including phagocytosis, netosis, and production of proinflammatory cytokines, thereby increasing neutrophilic fungicidal activity. Mechanistically, Dok3 mediates the recruitment of PP1 to Card9, thus maintaining Card9 in its dephosphorylated state and subsequently dampening downstream NF-κB and JNK signaling. As such, loss of Dok3 enhances the innate antifungal immune responses and protects mice from morbidity and mortality induced by systemic C. albicans infection.
Dok3 is an adaptor protein known to play important regulatory roles downstream of BCR, including the inhibition of calcium signaling (31) and JNK activation (32) in B cells, as well as promoting the formation of antibody-secreting plasma cells through upregulation of programmed cell death 1 ligands expression (25). In agreement with these reports, we observed Dok3 to be highly expressed in B cells. Since high levels of the Dok3 gene transcript were also found in neutrophils, we hypothesized that Dok3 may be implicated in immune signaling in this cell type. Indeed, our study showed that Dok3 negatively regulates antifungal immunity in neutrophils.
Triggering of CLRs by fungal components elicits various intracellular signaling pathways and activates interactions among adaptors, kinases, phosphatases, and other signaling molecules. Card9 has been reported to play a central role in CLR signaling in innate immune cells (12). Upon fungal sensing, Card9 is activated through phosphorylation by PKCδ in a Syk-dependent manner (11), and this facilitates the assembly of the CBM complex, which subsequently transduces the signal required for NF-κB and JNK activation. These signaling events translate to a series of antifungal killing activities, including the production of proinflammatory cytokines and release of NETs for fungal clearance (15, 16, 36). However, how Card9 signaling is being negatively regulated remains poorly understood. Here, we uncover a signaling mechanism for suppressing Card9 activity in neutrophils. We provide compelling evidence that Dok3 is required to bring PP1 into the vicinity of Card9 to mediate its dephosphorylation, thereby attenuating downstream antifungal immune responses in resting neutrophils. Upon fungal infection, Dok3 will be phosphorylated and subsequently degraded to overcome its negative regulatory effect on Card9, enabling the activation of downstream NF-κB and JNK signaling. These signaling events drive the production of various proinflammatory cytokines and ultimately promote clearance and inhibit growth of fungi in the host. Moreover, ROS production and calcium signaling were largely intact in Dok3-deficient neutrophils, consistent with previous studies that show that these Syk-dependent processes operate in a Card9-independent manner (42, 43) (Figure 8). Intriguingly, while our data demonstrated that Dok3 negatively regulates phagocytosis of unopsonized yeast, phagocytosis has been reported to be a Card9-independent process (41, 42). Given that the differential use of adaptors is a common strategy to orchestrate immune responses, it is likely that, apart from Card9, Dok3 also interacts with other molecules to control phagocytosis during fungal infection. Notably, Rubicon, another known negative regulator of Card9, is able to bind differentially to the NADPH oxidase complex to facilitate phagocytosis during microbial invasion (44, 45). However, it remains to be determined what other proteins are involved in the Dok3-dependent regulation of phagocytosis during C. albicans infection. Nevertheless, in the context of our study, we have demonstrated that Dok3 can negatively regulate innate antifungal immunity in neutrophils largely via a Card9-dependent pathway.
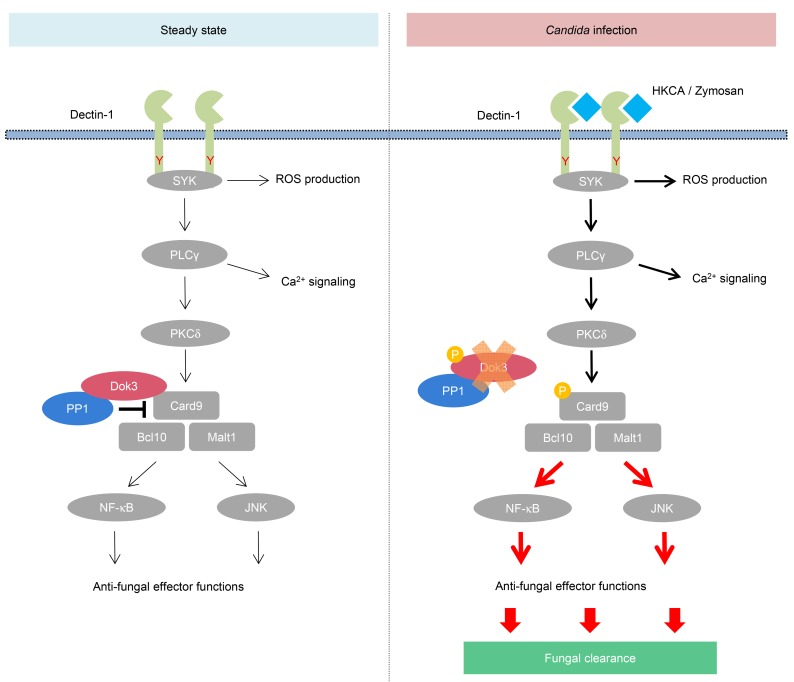
Figure 8. Proposed model of Dok3 regulation of Card9 during antifungal immune signaling.
In resting neutrophils, Dok3 recruits PP1 to maintain Card9 in its dephosphorylated and inactive state. Upon Candida infection, Dok3 is phosphorylated, the Dok3-PP1 complex dissociates from Card9, and Dok3 is also subsequently degraded. This enables Card9 to be phosphorylated by PKCδ, thereby activating downstream NF-κB and JNK signaling, leading to fungal clearance.
Several genetic studies in humans have demonstrated the pivotal role of Card9 in antifungal host defense, in which loss-of-function mutations in Card9 render individuals susceptible to Candida infection, especially affecting the CNS (41, 46–49). This mirrors our findings in Dok3–/– mice, where the release of Card9 from its negative regulation heightens immune responses and promotes the specific infiltration of neutrophils into infected brain, thus conferring protection against systemic candidiasis. Although negative regulation of Card9 by Dok3 and PP1 serves to prevent hyperinflammation and tissue damage, it could, at the same time, dampen essential host immune responses required for fungal killing. Hence, targeting negative regulators of Card9 could be a potential strategy to boost innate immunity for antifungal therapy. In our study, we have demonstrated the ability of the PP1 inhibitor tautomycetin to enhance Card9 activity and antifungal immune responses in neutrophils. However, tautomycetin is reported to be highly toxic (50), possibly because PP1 is ubiquitously expressed across many cell types and is involved in the regulation of diverse cellular processes ranging from muscle contraction to cell-cycle progression (51–53). In order for PP1 to exhibit substrate specificity and selectivity, it needs to work in combination with other protein regulators (54). In this study, we showed that Dok3 acts as a regulator of PP1 specificity by directing it to Card9 during steady state in neutrophils. Since Dok3 expression is limited to mainly immune cells, it may be more feasible to target Dok3 for antifungal therapy. Even though inhibition of immune negative regulator Dok3 could trigger excessive inflammation, which may have detrimental consequences, through limiting the dosage and period of inhibition, we can achieve optimal host immune responses required for antifungal defense while minimizing undesirable side effects. Moreover, we do not observe any signs of autoimmunity in Dok3–/– mice, further supporting the utility of Dok3 inhibitors to fight fungal infections.
In this study, we did not further evaluate whether PP1–/– mice harbor the same antifungal phenotype as Dok3–/– mice, as our data suggest that Dok3 and Card9 associate with all 3 isoforms of PP1 and it will be time-consuming to generate mice lacking one or more of these PP1 isoforms to determine which of them is directly involved in antifungal response. Moreover, reports have shown that PP1β–/– mice are embryonic lethal, while PP1γ–/– male mice are sterile due to a defect in spermatogenesis (55). Hence, given the promiscuous nature of protein phosphatases, it will be difficult to study the effects of PP1 deficiency on antifungal defense in vivo.
In conclusion, our study uncovers Dok3 and PP1 as critical negative regulators of Card9 signaling during antifungal immunity in neutrophils and raises the potential utility of Dok3 inhibitors as therapies for fungal infection. Such an immune-based approach could act as an adjunctive therapy to current antifungal drugs for further improvement of outcomes in Candida-infected patients.
Methods
Mice.
C57BL/6 and Card9–/– mice were purchased from The Jackson Laboratory. Dok3–/– mice were generated as described previously (28). Male and female mice were used at 8 to 10 weeks of age unless otherwise stated. All mice were maintained under specific pathogen–free conditions at A*STAR Biological Resource Centre (BRC).
Systemic C.
albicans infection. Mice were infected intravenously with 2.5 × 105 or 1 × 106 CFU of C. albicans (SC5314) and monitored daily for survival. For flow cytometry analysis and histology, kidneys and brains were harvested 1 day after infection. To determine fungal titers, kidneys and brains were harvested and homogenized 2 days after infection.
Histology.
Kidneys were harvested from C. albicans–infected mice and fixed in 10% neutral buffered formalin for 24 hours before embedding in paraffin wax. Sections were stained with periodic acid–Schiff (PAS) and Grocott’s methenamine silver (GMS).
Isolation of neutrophils, macrophages, and DCs.
Neutrophils were isolated from tibias and femurs of mice using anti–Ly-6G magnetic beads (Miltenyi Biotec). DCs and macrophages were isolated from spleens of mice with CD11c and F4/80 magnetic beads, respectively (Miltenyi Biotec). Purity of isolated cells was confirmed by flow cytometry.
Flow cytometry.
Cell suspensions were surface labeled with fluorochrome-conjugated antibodies for 10 minutes at 4°C in staining buffer (PBS containing 1% BSA). For intracellular labeling, cells were fixed and permeabilized using the Cytofix/Cytoperm kit (BD) according to manufacturer’s protocol before staining for 1 hour at room temperature. DAPI was used to exclude dead cells. Data were acquired using LSRII (BD Biosciences) and analyzed using FlowJo software (Tree Star). The following antibodies were used for flow cytometry analysis: anti-CD11c PerCP (clone N418; BioLegend), anti–Dectin-1 PE (clone 218820; R&D Systems), anti–Dectin-2 APC (catalog FAB1525A; R&D Systems), anti-F4/80 PE (clone BM8; eBioscience), anti-Ly6C FITC (clone HK1.4; BioLegend), anti-Ly6G PE (clone 1A8; BD), anti–I-A/I-E FITC (clone M5/114.152; BioLegend), anti–TNF-α APC and PE (clone MP6-XT22; BioLegend), and anti–TGF-β1 APC (clone TW7-16B4; BioLegend).
Neutrophil stimulation.
Purified neutrophils were rested at 37°C for 2 hours before stimulation with HKCA in cell or hyphae forms, zymosan (Invivogen), or curdlan (Invivogen). For PP1 inhibition experiments, cells were pretreated with indicated concentrations of tautomycetin (Tocris) for 2.5 hours before stimulation. For intracellular cytokine staining, cells were stimulated with zymosan for 3 hours in the presence of GolgiPlug (BD).
In vitro killing assay.
Purified immune cells were plated in triplicate (1 × 105 cells/well) in 96-well plates and incubated overnight at 37°C with unopsonized or serum-opsonized C. albicans (MOI 1:200 or 1:500) before fixing with 2% paraformaldehyde. C. albicans were stained with crystal violet (MilliporeSigma). Percentages of fungal growth were quantified using ImageJ software (NIH).
Phagocytosis assay.
HKCAs were labeled with FITC (Sigma-Aldrich) in 100 mM HEPES buffer (pH 7.5) for 10 minutes. For opsonization, HKCAs were resuspended in 20% mouse serum for 15 minutes at 37°C. Subsequently, purified immune cells were cocultured with FITC-labeled unopsonized or serum-opsonized HKCAs (MOI 1:2) at 37°C for 45 minutes. Adherent fungal cells were quenched with trypan blue, and the extent of phagocytosis measured by flow cytometry.
Netosis assay.
Purified neutrophils were plated (5 × 104 cells/well) on a 24-well plate before stimulation with HKCA hyphae (MOI 1:10). Four hours later, PI or DAPI was added and cells were analyzed for NET release by microscopy. NET release was scored using ImageJ, as previously described (33).
ROS production.
Purified neutrophils were labeled with Dihydrorhodamine 123 (Thermo Fisher Scientific) for 5 minutes at 37°C before stimulation with HKCAs (MOI 1:2) or curdlan (100 μg/ml) for 1 hour at 37°C. The amount of ROS produced by neutrophils, as determined by the fluorescence signal, was measured by flow cytometry.
Quantitative PCR.
Purified neutrophils were lysed with TRIzol (Gibco, Thermo Fisher Scientific), and RNA was purified using phenol/chloroform extraction. Complementary DNA was reversed transcribed using RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). The following primers were used: Il6 (forward): ACGGCCTTCCCTACTTCACA; Il6 (reverse): CATTTCCACGATTTCCCAGA; Tnfa (forward): GCCTCTTCTCATTCCTGCTTG; Tnfa (reverse): CTGATGAGAGGGAGGCCATT; Tgfb (forward): CACCGGAGAGCCCTGGATA; Tgfb (reverse): TGTACAGCTGCCGCACACA; Cxcl1 (forward): GGCGCCTATCGCCAATG; Cxcl1 (reverse): CTGGATGTTCTTGAGGTGAATCC; Cxcl2 (forward): AAGTTTGCCTTGACCCTGAA; Cxcl2 (reverse): AGGCACATCAGGTACGATCC; Cxcl5 (forward): GCATTTCTGTTGCTGTTCACGCTG; and Cxcl5 (reverse): CCTCCTTCTGGTTTTTCAGTTTAGC.
IP and Western blotting.
For endogenous co-IP, purified neutrophils were stimulated with HKCAs (MOI 1:1) or zymosan (10 μg/ml) for indicated time periods before lysis with 1% NP40 buffer containing protease and phosphatase inhibitors (Roche). Cell lysates were IP overnight and pulled down using Protein A/G Plus Agarose beads (Santa Cruz Biotechnology Inc.). Precipitates were analyzed by Western blotting according to standard protocol using the indicated antibodies: anti-Card9 (catalog 12283, Cell Signaling Technology; clone A-8, Santa Cruz Biotechnology Inc.), anti-Dok3 (clone E-17, M-214, H-5; Santa Cruz Biotechnology Inc.), anti–p-Erk1/2T202/Y204 (catalog 4370; Cell Signaling Technology), anti-Erk2 (clone C-14; Santa Cruz Biotechnology Inc.), anti–p-IKKα/βS176/180 (catalog 2697; Cell Signaling Technology), anti-IKKα/β (clone H-470; Santa Cruz Biotechnology Inc.), anti–p-JNKT183/Y185 (catalog 9255; Cell Signaling Technology), anti-JNK (catalog 9252; Cell Signaling Technology), anti–p-p38T180/Y182 (clone D-8; Santa Cruz Biotechnology Inc.), anti-p38 (clone N-20; Santa Cruz Biotechnology Inc.), anti–p-SykY352 (catalog 2717; Cell Signaling Technology), anti-Syk (catalog 80460; Cell Signaling Technology), anti–p-Thr (catalog 9381; Cell Signaling Technology), anti–p-Tyr (clone 4G10; Millipore), anti–β-actin (clone ACTBD11B7; Santa Cruz Biotechnology Inc.), anti-PP1 (clone E-9; Santa Cruz Biotechnology Inc.), anti-PP2AC (catalog 2259; Cell Signaling Technology), anti-PP1α (clone G-4; Santa Cruz Biotechnology Inc.), anti-PP1β (clone A-6; Santa Cruz Biotechnology Inc.), anti-PP1γ (clone E-4; Santa Cruz Biotechnology Inc.), anti–p-PKCδY311 (clone A-8;. Santa Cruz Biotechnology Inc.), and anti-PKCδ (clone G-9; Santa Cruz Biotechnology Inc.).
Statistics.
Figures and statistical analyses were generated using GraphPad Prism software. Mice were allocated to experimental groups based on their genotypes and were randomized within their sex- and age-matched groups. Data were tested for normality using the Kolmogorov-Smirnov test. No mouse was excluded from the analyses. For survival analyses, log-rank test was performed. For netosis assays and analyses of neutrophil infiltration into brain and kidneys upon C. albicans infection, 2-way ANOVA was performed. For tautomycetin treatment, 1-way ANOVA was performed. For other analyses, unpaired 2-tailed Student’s t test was performed. A P value of less than 0.05 was considered significant.
Study approval.
All mouse protocols were conducted in accordance with guidelines from and approved by the A*STAR BRC Institutional Animal Care and Use Committee.
Author contributions
JTL designed and conducted most of the experiments, interpreted data, and wrote the manuscript. SX and JXH performed in vivo infection experiments. SSYK helped with co-IP experiments. YW provided Candida isolates. KPL conceived the research and helped in data interpretation and manuscript preparation.
Supplementary Material
Acknowledgments
This work is supported by the Singapore Ministry of Health’s National Medical Research Council under its Open Fund–Young Individual Research Grant (NMRC/OFYIRG/083/2018 to JTL) and an A*STAR core grant (to Bioprocessing Technology Institute).
Version 1. 06/10/2019
Electronic publication
Version 2. 07/01/2019
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(7):2717–2729.https://doi.org/10.1172/JCI126341.
Contributor Information
Jia Tong Loh, Email: loh_jia_tong@bti.a-star.edu.sg.
Shengli Xu, Email: xu_shengli@bti.a-star.edu.sg.
Jian Xin Huo, Email: huo_jianxin@bti.a-star.edu.sg.
Susana Soo-Yeon Kim, Email: susana_kim@bti.a-star.edu.sg.
Yue Wang, Email: mcbwangy@imcb.a-star.edu.sg.
Kong-Peng Lam, Email: lam_kong_peng@bti.a-star.edu.sg.
References
- 1.Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. Hidden killers: human fungal infections. Sci Transl Med. 2012;4(165):165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 2.Edwards JE. Invasive candida infections--evolution of a fungal pathogen. N Engl J Med. 1991;324(15):1060–1062. doi: 10.1056/NEJM199104113241511. [DOI] [PubMed] [Google Scholar]
- 3.Ravikumar S, Win MS, Chai LY. Optimizing outcomes in immunocompromised hosts: understanding the role of immunotherapy in invasive fungal diseases. Front Microbiol. 2015;6:1322. doi: 10.3389/fmicb.2015.01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cowen LE, Anderson JB, Kohn LM. Evolution of drug resistance in Candida albicans. Annu Rev Microbiol. 2002;56:139–165. doi: 10.1146/annurev.micro.56.012302.160907. [DOI] [PubMed] [Google Scholar]
- 5.Underhill DM, Pearlman E. Immune interactions with pathogenic and commensal fungi: a two-way street. Immunity. 2015;43(5):845–858. doi: 10.1016/j.immuni.2015.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor PR, et al. Dectin-1 is required for beta-glucan recognition and control of fungal infection. Nat Immunol. 2007;8(1):31–38. doi: 10.1038/ni1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saijo S, et al. Dectin-2 recognition of alpha-mannans and induction of Th17 cell differentiation is essential for host defense against Candida albicans. Immunity. 2010;32(5):681–691. doi: 10.1016/j.immuni.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Wells CA, et al. The macrophage-inducible C-type lectin, mincle, is an essential component of the innate immune response to Candida albicans. J Immunol. 2008;180(11):7404–7413. doi: 10.4049/jimmunol.180.11.7404. [DOI] [PubMed] [Google Scholar]
- 9.Xu S, Huo J, Lee KG, Kurosaki T, Lam KP. Phospholipase Cgamma2 is critical for Dectin-1-mediated Ca2+ flux and cytokine production in dendritic cells. J Biol Chem. 2009;284(11):7038–7046. doi: 10.1074/jbc.M806650200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorjestani S, Yu M, Tang B, Zhang D, Wang D, Lin X. Phospholipase Cγ2 (PLCγ2) is key component in Dectin-2 signaling pathway, mediating anti-fungal innate immune responses. J Biol Chem. 2011;286(51):43651–43659. doi: 10.1074/jbc.M111.307389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strasser D, et al. Syk kinase-coupled C-type lectin receptors engage protein kinase C-σ to elicit Card9 adaptor-mediated innate immunity. Immunity. 2012;36(1):32–42. doi: 10.1016/j.immuni.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gross O, et al. Card9 controls a non-TLR signalling pathway for innate anti-fungal immunity. Nature. 2006;442(7103):651–656. doi: 10.1038/nature04926. [DOI] [PubMed] [Google Scholar]
- 13.Roth S, et al. Vav proteins are key regulators of Card9 signaling for innate antifungal immunity. Cell Rep. 2016;17(10):2572–2583. doi: 10.1016/j.celrep.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hara H, et al. The adaptor protein CARD9 is essential for the activation of myeloid cells through ITAM-associated and Toll-like receptors. Nat Immunol. 2007;8(6):619–629. doi: 10.1038/ni1466. [DOI] [PubMed] [Google Scholar]
- 15.Romani L. Immunity to fungal infections. Nat Rev Immunol. 2011;11(4):275–288. doi: 10.1038/nri2939. [DOI] [PubMed] [Google Scholar]
- 16.Netea MG, Brown GD, Kullberg BJ, Gow NA. An integrated model of the recognition of Candida albicans by the innate immune system. Nat Rev Microbiol. 2008;6(1):67–78. doi: 10.1038/nrmicro1815. [DOI] [PubMed] [Google Scholar]
- 17.Hardison SE, Brown GD. C-type lectin receptors orchestrate antifungal immunity. Nat Immunol. 2012;13(9):817–822. doi: 10.1038/ni.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wirnsberger G, et al. Inhibition of CBLB protects from lethal Candida albicans sepsis. Nat Med. 2016;22(8):915–923. doi: 10.1038/nm.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao Y, et al. Targeting CBLB as a potential therapeutic approach for disseminated candidiasis. Nat Med. 2016;22(8):906–914. doi: 10.1038/nm.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu LL, et al. E3 ubiquitin ligase Cbl-b negatively regulates C-type lectin receptor-mediated antifungal innate immunity. J Exp Med. 2016;213(8):1555–1570. doi: 10.1084/jem.20151932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao X, et al. JNK1 negatively controls antifungal innate immunity by suppressing CD23 expression. Nat Med. 2017;23(3):337–346. doi: 10.1038/nm.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemay S, Davidson D, Latour S, Veillette A. Dok-3, a novel adapter molecule involved in the negative regulation of immunoreceptor signaling. Mol Cell Biol. 2000;20(8):2743–2754. doi: 10.1128/MCB.20.8.2743-2754.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim SS, et al. DOK3 is required for IFN-β production by enabling TRAF3/TBK1 complex formation and IRF3 activation. J Immunol. 2014;193(2):840–848. doi: 10.4049/jimmunol.1301601. [DOI] [PubMed] [Google Scholar]
- 24.Peng Q, O’Loughlin JL, Humphrey MB. DOK3 negatively regulates LPS responses and endotoxin tolerance. PLoS ONE. 2012;7(6):e39967. doi: 10.1371/journal.pone.0039967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ou X, Xu S, Li YF, Lam KP. Adaptor protein DOK3 promotes plasma cell differentiation by regulating the expression of programmed cell death 1 ligands. Proc Natl Acad Sci USA. 2014;111(31):11431–11436. doi: 10.1073/pnas.1400539111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sancho D, Reis e Sousa C. Signaling by myeloid C-type lectin receptors in immunity and homeostasis. Annu Rev Immunol. 2012;30:491–529. doi: 10.1146/annurev-immunol-031210-101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drummond RA, Gaffen SL, Hise AG, Brown GD. Innate defense against fungal pathogens. Cold Spring Harb Perspect Med. 2014;5(6):a019620. doi: 10.1101/cshperspect.a019620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng CH, Xu S, Lam KP. Dok-3 plays a nonredundant role in negative regulation of B-cell activation. Blood. 2007;110(1):259–266. doi: 10.1182/blood-2006-10-055194. [DOI] [PubMed] [Google Scholar]
- 29.Wirnsberger G, et al. Jagunal homolog 1 is a critical regulator of neutrophil function in fungal host defense. Nat Genet. 2014;46(9):1028–1033. doi: 10.1038/ng.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cassone A. Development of vaccines for Candida albicans: fighting a skilled transformer. Nat Rev Microbiol. 2013;11(12):884–891. doi: 10.1038/nrmicro3156. [DOI] [PubMed] [Google Scholar]
- 31.Stork B, et al. Subcellular localization of Grb2 by the adaptor protein Dok-3 restricts the intensity of Ca2+ signaling in B cells. EMBO J. 2007;26(4):1140–1149. doi: 10.1038/sj.emboj.7601557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robson JD, Davidson D, Veillette A. Inhibition of the Jun N-terminal protein kinase pathway by SHIP-1, a lipid phosphatase that interacts with the adaptor molecule Dok-3. Mol Cell Biol. 2004;24(6):2332–2343. doi: 10.1128/MCB.24.6.2332-2343.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Branzk N, et al. Neutrophils sense microbe size and selectively release neutrophil extracellular traps in response to large pathogens. Nat Immunol. 2014;15(11):1017–1025. doi: 10.1038/ni.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stockinger B, Veldhoen M. Differentiation and function of Th17 T cells. Curr Opin Immunol. 2007;19(3):281–286. doi: 10.1016/j.coi.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 35.van Enckevort FH, et al. Increased susceptibility to systemic candidiasis in interleukin-6 deficient mice. Med Mycol. 1999;37(6):419–426. doi: 10.1046/j.1365-280X.1999.00247.x. [DOI] [PubMed] [Google Scholar]
- 36.Netea MG, Joosten LA, van der Meer JW, Kullberg BJ, van de Veerdonk FL. Immune defence against Candida fungal infections. Nat Rev Immunol. 2015;15(10):630–642. doi: 10.1038/nri3897. [DOI] [PubMed] [Google Scholar]
- 37.Hsu YM, et al. The adaptor protein CARD9 is required for innate immune responses to intracellular pathogens. Nat Immunol. 2007;8(2):198–205. doi: 10.1038/ni1426. [DOI] [PubMed] [Google Scholar]
- 38.Eitelhuber AC, et al. Dephosphorylation of Carma1 by PP2A negatively regulates T-cell activation. EMBO J. 2011;30(3):594–605. doi: 10.1038/emboj.2010.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cohen PT. Protein phosphatase 1--targeted in many directions. J Cell Sci. 2002;115(Pt 2):241–256. doi: 10.1242/jcs.115.2.241. [DOI] [PubMed] [Google Scholar]
- 40.Mitsuhashi S, Matsuura N, Ubukata M, Oikawa H, Shima H, Kikuchi K. Tautomycetin is a novel and specific inhibitor of serine/threonine protein phosphatase type 1, PP1. Biochem Biophys Res Commun. 2001;287(2):328–331. doi: 10.1006/bbrc.2001.5596. [DOI] [PubMed] [Google Scholar]
- 41.Drummond RA, et al. CARD9-dependent neutrophil recruitment protects against fungal invasion of the central nervous system. PLoS Pathog. 2015;11(12):e1005293. doi: 10.1371/journal.ppat.1005293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drewniak A, et al. Invasive fungal infection and impaired neutrophil killing in human CARD9 deficiency. Blood. 2013;121(13):2385–2392. doi: 10.1182/blood-2012-08-450551. [DOI] [PubMed] [Google Scholar]
- 43.Németh T, Futosi K, Sitaru C, Ruland J, Mócsai A. Neutrophil-specific deletion of the CARD9 gene expression regulator suppresses autoantibody-induced inflammation in vivo. Nat Commun. 2016;7:11004. doi: 10.1038/ncomms11004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang CS, et al. Autophagy protein Rubicon mediates phagocytic NADPH oxidase activation in response to microbial infection or TLR stimulation. Cell Host Microbe. 2012;11(3):264–276. doi: 10.1016/j.chom.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang CS, et al. The autophagy regulator Rubicon is a feedback inhibitor of CARD9-mediated host innate immunity. Cell Host Microbe. 2012;11(3):277–289. doi: 10.1016/j.chom.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glocker EO, et al. A homozygous CARD9 mutation in a family with susceptibility to fungal infections. N Engl J Med. 2009;361(18):1727–1735. doi: 10.1056/NEJMoa0810719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grumach AS, et al. A homozygous CARD9 mutation in a Brazilian patient with deep dermatophytosis. J Clin Immunol. 2015;35(5):486–490. doi: 10.1007/s10875-015-0170-4. [DOI] [PubMed] [Google Scholar]
- 48.Alves de Medeiros AK, et al. Chronic and invasive fungal infections in a family with CARD9 deficiency. J Clin Immunol. 2016;36(3):204–209. doi: 10.1007/s10875-016-0255-8. [DOI] [PubMed] [Google Scholar]
- 49.Pérez de Diego R, et al. Genetic errors of the human caspase recruitment domain-B-cell lymphoma 10-mucosa-associated lymphoid tissue lymphoma-translocation gene 1 (CBM) complex: Molecular, immunologic, and clinical heterogeneity. J Allergy Clin Immunol. 2015;136(5):1139–1149. doi: 10.1016/j.jaci.2015.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen X, Zhu X, Ding Y, Shen Y. Antifungal activity of tautomycin and related compounds against Sclerotinia sclerotiorum. J Antibiot. 2011;64(8):563–569. doi: 10.1038/ja.2011.55. [DOI] [PubMed] [Google Scholar]
- 51.Winkler C, et al. The selective inhibition of protein phosphatase-1 results in mitotic catastrophe and impaired tumor growth. J Cell Sci. 2015;128(24):4526–4537. doi: 10.1242/jcs.175588. [DOI] [PubMed] [Google Scholar]
- 52.Toole BJ, Cohen PT. The skeletal muscle-specific glycogen-targeted protein phosphatase 1 plays a major role in the regulation of glycogen metabolism by adrenaline in vivo. Cell Signal. 2007;19(5):1044–1055. doi: 10.1016/j.cellsig.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 53.Peti W, Nairn AC, Page R. Structural basis for protein phosphatase 1 regulation and specificity. FEBS J. 2013;280(2):596–611. doi: 10.1111/j.1742-4658.2012.08509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Virshup DM, Shenolikar S. From promiscuity to precision: protein phosphatases get a makeover. Mol Cell. 2009;33(5):537–545. doi: 10.1016/j.molcel.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 55.Li J, Sinha N, Vijayaraghavan S, Chen Y. Expression of protein phosphatase PP1 isoforms in developing mouse embryos. Biology of Reproduction. 2011;85(Suppl_1):255. doi: 10.1093/biolreprod/85.s1.255. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.