Causal modeling and inference rely on strong assumptions, one of which is conditional exchangeability. Uncontrolled confounding is often seen as if it is the most important threat to conditional exchangeability although collider-stratification bias or selection bias can be just as important.1–4 In this issue of the journal, Flanders and Ye5 (henceforth, F&Y) and Smith and VanderWeele6 (henceforth, S&VW) present their results on new bounds—limits that selection bias would not exceed in any specified context—and accompanying summary measures for the values of the selection bias bounding factors that will be enough to explain away any observed association between the exposure and the outcome on the risk ratio or relative risk scale, with risk difference results given in the appendix of S&VW’s article. These articles on M-bias or selection bias fit into a growing body of work that have renewed researchers’ interests in selection bias including the recent overlapping literature on generalizability and transportability, and bounding factors and related summary measures for bias analysis.1–4,7–13
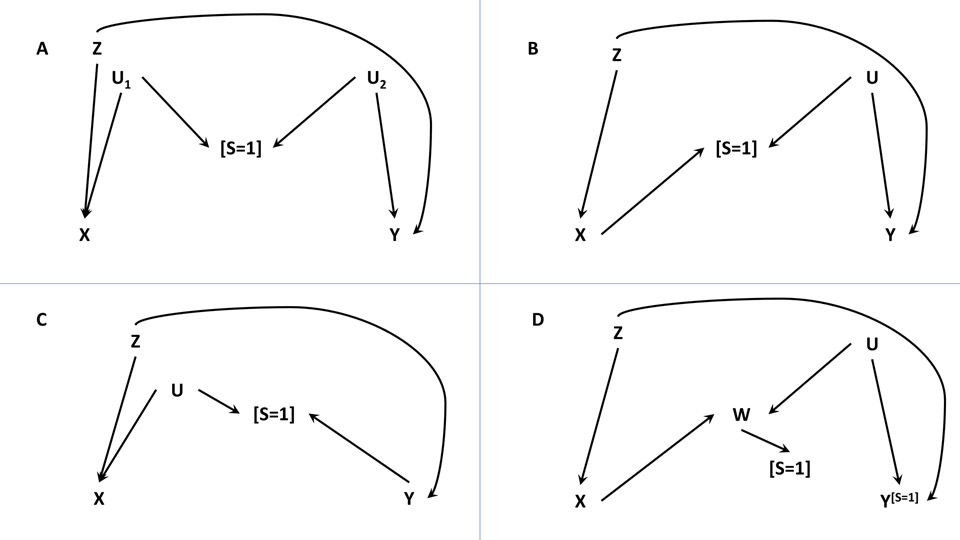
Both F&Y5 and S&VW6 make extensive use of directed acyclic graphs (DAGs) to depict the structure and the mechanism of selection or collider stratification bias. This is not surprising since it would other be difficult to reason about the mechanisms involved in collider-stratification or selection bias without the visual aid provided by DAGs. Common selection bias structures are given in the Figure of this commentary (excluding the special scenario in which the exposure and the outcome jointly lead to selection). Selection bias can be expected when estimating the effect of the exposure (or intervention) X on the outcome Y in the study sample S=1 and when Y is not independent of the selection node or collider S conditional on X and the measured confounder(s) Z (as in DAGs A to C).4,8,10
In their article,5 F&Y derive bounds for M-bias extending previous work.1 They give an expression for the maximum value of M-bias due to conditioning on the collider S in the DAG A (for example) when estimating the effect of the exposure on the outcome among the exposed on the relative risk or risk ratio scale without making a homogeneity assumption.1,5 Their bound is a function of three bias parameters that capture of the strengths of the associations between: (i) the exposure X and the unmeasured cause U1 shared by X and the collider S, (ii) S and the unmeasured causes U1 and U2, (or equivalently, a binary variable N in their Figure 1A) and (iii) U2 (or the authors’ binary N) and the outcome Y. At first sight, the expression for the bound may not look user-friendly to the non-methodologist but can be implemented in a spreadsheet or a general statistical software, for example, by adapting F&Y’s own R codes in their Online Supplement. F&Y also work through several examples showing how to use all or some of the bias parameters in various applications of the bound expressions. As expected, their bound can sometimes be conservative when compared to the actual but unknown bias magnitude. The numeric results in their Table 1 represent increasingly extreme scenarios that demonstrate when their new bound begins to diverge from a previously published bound that assumes homogeneity. However, this divergence occurs at extreme bias parameters that may not be common. Noticeably, F&Y make frequent reference to the mechanism of the bias under consideration using DAGs.
FIGURE 1:
Directed acyclic graphs (DAGs) depicting typical causal scenarios where selection bias can potentially occur (DAGs A-C in particular). X represents the exposure, Y the outcome, Z the measured confounder(s), S the selection (where S = 1 means selected into the study sample), U the unmeasured covariate(s) or confounder(s), and W is a consequence of X that is associated with Y. In all DAGs, X does not cause Y, but they will become non-causally associated if selecting on S = 1. In DAGs A to C, Y is not independent of S conditional on measured variables X and Z; in DAG D, Y is independent of S given X, Z and W. (A) This DAG denotes a selection bias structure, called the M-bias, due to two open backdoors between the selection node S and exposure X and the outcome Y; conditioning on the collider S opens up the path X←U1→[S=1]←U2→Y leading to a type of collider-stratification bias commonly referred to as M-bias. (B) This DAG looks like that in A except X causes S here; conditioning on the collider S opens up the path X→[S=1]←U→Y. (C) Again, this DAG resembles the one in A except Y causes S here; conditioning on the collider S opens up the path X←U→[S=1]←Y leading to selection bias in the effect of X on Y. (D) This DAG is the same as the one in B except W replaces S as the collider between X and Y in the path X→W←U→Y, and W causes S; conditioning on S would appear to induce selection bias between X and Y in the path X→[W]←U→Y because conditioning on the consequence of a collider also leads to collider-stratification bias; nonetheless, the effect of X on Y can still be estimated without selection bias if all variables except Y are fully observed prior to selection and Y is only observed among S=1 (hence, the Y[S=1] notation).
In their article,6 S&VW present several results on how to bound the magnitude of selection bias on the risk ratio and difference scales, using bias parameters that capture the associations between selection S and the unmeasured covariate(s) U and between U and the outcome Y. These parameters reflect the pathways from S to U and from U to Y in common selection bias scenarios such as those depicted in the Figure parts A–C. The authors present different bounds for different target populations, selection mechanisms, and effect measures with some illustrative applications based on examples from the literature. Like F&Y, S&VW make use of DAGs to capture the structure of various selection bias scenarios. Indeed, their bounds are (DAG) structure specific and can be expected to be conservative in that they will provide the maximum bias possible for the assumed bias parameters even when those parameters could be responsible for less bias.6 They also present summary measures (call them ‘selection E-values’ if you will) that quantify how large each of the parameters in the bounding factor will have to be, had they been of equal magnitude, to explain away any empirically observed risk ratio (RRXY+) for the association of the exposure with the outcome. At first look, it appears to me that the summary measure for the general selection bias scenario can be approximated by just the value of observed risk ratio relating X to Y when 1< RRXY+ ≤ 3. Parallel simplifications of the summary measure appear possible for very large RRXY+. The properties and performance of the proposed summary measures will, therefore, benefit from further study.
Overall, F&Y and S&VW should be commended for adding these new tools to our bias analysis toolbox. Whether epidemiologists will rise to the occasion and use them well is another matter. Quantitative bias—and bounding—analyses are infrequently used in the large well-conducted studies for which they might be best suited. Bounding selection bias should be considered in those instances where the adjusting for the selection mechanism using measured covariates is not possible, as in Figure parts A-C. Before resorting to or in addition to bounding, we have several existing options to consider when addressing selection bias. First, bounding may be unnecessary as seen in Figure part D of this commentary and S&VW’s article because appropriate adjustment for W will remove bias due to the selection on S = 1 whenever X, Z, W, and S are fully observed but Y is only observed for S = 1. Since Y is independent of S given X, W, and Z,4,14 there is no selection bias in DAG D. For example, the causal risk ratio relating binary X to Y in DAG D can be obtained using
Where E(Y|X = x, W = w, Z = z, S = 1) can be estimated from the data on S=1 only but P(W = w|X = x)and P(Z = z) can be computed using the full data before selection S=1 occurred. Second, we could use inverse-probability-of-selection-weighting (IPSW)15 even when the selection mechanism involves an unmeasured variable as in DAGs A to C. Unlike in DAG D where we could use the observed data only to estimate and implement IPSW to overcome potential selection bias, we must compute the IPSWExt = 1/[P(S = 1|X = x, Y = y, Z=z)] in DAGs 1A to 1C using externally obtained bias parameters relating S to both X and Y (possibly marginalized over U).10 The numerator can also be replaced with P(S = 1). For example, on the risk ratio scale, IPSWExt could be given by 1/[P(S = 1|X = x, Y = y, Z = z)] = 1/[RS=1|X=0,Y=0·RRxXS|Y=0·RRyYS|X=0·RRRxyXYS] assuming binary variables for illustration, conditioning or stratification on Z = z (for simplicity here), and that RS=1|X=0,Y=0 would represent P(S = 1|X = 0, Y = 0, Z = z), while RRXS|Y=0 and RRYS|X=0 would represent the corresponding risk ratios for the associations of X with S and Y with S, and RRRXYS the product of term between X and Y in the model for S = 1 on the risk ratio scale. Each of the last three bias parameters will be raised to the power X, Y, and XY respectively based on each individual record’s X, Y, and XY values in the sample S = 1. We would then proceed with analyzing the S = 1 population data, weighting each of the individual records weighted by their computed IPSWExt, just as we would in any other selection or censoring weighted analysis.2,7,10 This IPSWExt method is quite general in that it can be applied to different study designs and effect measures.
Third, we could draw the assumed selection bias DAG under no true effect of the exposure on the outcome and use it to simulate the magnitude and direction of the selection bias under specified parameters for relating U, X, Y, and S (given Z) guided by the study under consideration. This bias simulation approach has been described elsewhere for uncontrolled confounding.15 The advantage here is that, unlike in the case of bounding, we actually specify parameters in the direction of the edges or arrows in our DAG, making it easier to reason about them causally and to use the resulting data to compute bounding bias parameters if we want or the selection bias factor for direct use in adjusting for selection bias in empirical data analysis. Finally, we could adapt existing bias formulas16,12 to the selection setting by making the formulas conditional on S = 1 and replacing the backdoor X←U→Y of uncontrolled confounding with X→[S=1]←U→Y. This exploits the known connections between analytical methods for uncontrolled confounding and selection.1,5,11 In all the foregoing approaches, we are guided by the assumed DAG.
Neither of the articles by F&Y5 and S&VW6 aimed to deal with time-varying treatment and confounding, mediation, or multiple-bias settings. It is prudent that further simulation studies be conducted to examine the performance of the proposed bounding factors and summary measures in different selection bias scenarios and for difference effect measures where our heuristics tend to fail. For example, it will be interesting to see how the bounds and summary measures perform when selection bias exists without collider-stratification and when collider-stratification does not result in selection bias in the S = 1 population. More importantly, further work should provide more detailed guidelines for applying, reporting, and interpreting the bounds to avoid their misuse (for example, purportedly as evidence of causality). Guidance on how to think about and obtain the bias parameters needed for these methods is sorely needed. It is also crucial to develop good practices for the use of bounding analysis and quantitative bias analysis more generally.17
In conclusion, quantitative analysis of selection bias using any of the methods described here and by F&Y5 and S&VW6 should be seen as aimed at making causal modeling and inference more credible. Whichever approach is chosen, it is important to think carefully about the assumed selection bias structure and mechanism. As seen in the work of F&Y and S&VW, drawing DAGs augmented with the selection mechanism was helpful, if not indispensable, in visualizing and reasoning about the structure of the bias and the various pathways that should be captured by the bias parameters. Reasoning about selection bias can be daunting and can lead to doubts about the mechanism and the relevant bias parameters. When in doubt, DAG it out.
Sources of financial support:
This work was supported by grant UL1TR001881 from the National Center for Advancing Translational Science, grant R01HD072296 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH, and grant 273823 from the Norwegian Research Council’s BEDREHELSE program. It also benefited from facilities and resources provided by the California Center for Population Research at UCLA (CCPR), which receives core support (R24HD041022) from the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
Footnotes
Conflicts of interest: None declared.
Obtaining data and computing code: Not applicable.
REFERENCES
- 1.Greenland S Quantifying biases in causal models: classical confounding vs collider-stratification bias. Epidemiology. 2003;14:300–306. [PubMed] [Google Scholar]
- 2.Thompson CA, Zhang ZF, Arah OA. Competing risk bias to explain the inverse relationship between smoking and malignant melanoma. Eur J Epidemiol. 2013;28:557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liew Z, Olsen J, Cui X, Ritz B, Arah OA. Bias from conditioning on live birth in pregnancy cohorts: an illustration based on neurodevelopment in children after prenatal exposure to organic pollutants. Int J Epidemiol. 2015;44:345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bareinboim E, Pearl J. Causal inference and the data-fusion problem. Proc Natl Acad Sci. 2016;113:7345–7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flanders WD, Ye D. Limits of the magnitude of M-bias and certain other types of structural bias. Epidemiology. 2019; (in press). [DOI] [PubMed] [Google Scholar]
- 6.Smith LH, VanderWeele TJ. Bounding bias due to selection. Epidemiol. 2019; (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Infante-Rivard C, Cusson A. Reflection on modern methods: selection bias — a review of recent developments. Int J Epidemiol. 2015;44:345–354. [DOI] [PubMed] [Google Scholar]
- 8.Pearl J, Bareinboim E. Note on “Generalizability of Study Results”. Epidemiology. 2019;30:186–188. [DOI] [PubMed] [Google Scholar]
- 9.Bareinboim E, Pearl J. A general algorithm for deciding transportability of experimental results. J Causal Inference. 2013;2:115–129. [Google Scholar]
- 10.Thompson CA, Arah OA. Selection bias modeling using observed data augmented with imputed record-level probabilities. Ann Epidemiol. 2014;24:747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greenland S Bayesian perspectives for epidemiologic research: III. Bias analysis via missing data methods. Int J Epidemiol. 2009;38:1662–1673. [DOI] [PubMed] [Google Scholar]
- 12.Lash TL, Fox MP, Fink AK. Applying Quantitative Bias Analysis to Epidemiologic Data. New York: Springer Science+Business Media, LLC; 2009. [Google Scholar]
- 13.Ding P, VanderWeele TJ. Sensitivity analysis without assumptions. Epidemiology. 2016;27:368–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cinelli C, Pearl J. Re: a practical example demonstrating the utility of single-world intervention graphs. Epidemiology. 2018;29:e50–e51. [DOI] [PubMed] [Google Scholar]
- 15.Arah OA. Bias analysis for uncontrolled confounding in the health sciences. Annu Rev Public Health. 2017;38:23–38. [DOI] [PubMed] [Google Scholar]
- 16.Arah OA, Chiba Y, Greenland S. Bias formulas for external adjustment and sensitivity analysis analysis of unmeasured confounders. Ann Epidemiol. 2008;18:637–646. [DOI] [PubMed] [Google Scholar]
- 17.Lash TL, Fox MP, MacLehose RF, Maldonado G, McCandless LC, Greenland S. Good practices for quantitative bias analysis. Int J Epidemiol. 2014;43:1969–1985. [DOI] [PubMed] [Google Scholar]