Abstract
Purpose
Radiomics is a growing field of image quantification, but it lacks stable and high-quality software systems. We extended the capabilities of the Computational Environment for Radiological Research (CERR) to create a comprehensive, open-source, MATLAB-based software platform with an emphasis on reproducibility, speed, and clinical integration of radiomics research.
Method
The radiomics tools in CERR were designed specifically to quantify medical images in combination with CERR’s core functionalities of radiological data import, transformation, management, image segmentation, and visualization. CERR allows for batch calculation and visualization of radiomics features, and provides a user-friendly data structure for radiomics metadata. All radiomics computations are vectorized for speed. Additionally, a test suite is provided for reconstruction and comparison with radiomics features computed using other software platforms such as the Insight Toolkit (ITK) and PyRadiomics. CERR was evaluated according to the standards defined by the Image Biomarker Standardization Initiative. CERR’s radiomics feature calculation was integrated with the clinically used MIM software using its MATLAB® application programming interface.
Results
CERR provides a comprehensive computational platform for radiomics analysis. Matrix formulations for the compute-intensive Haralick texture resulted in speeds that are superior to the implementation in ITK 4.12. For an image discretized into 32 bins, CERR achieved a speedup of 3.5 times over ITK. The CERR test suite enabled the successful identification of programming errors as well as genuine differences in radiomics definitions and calculations across the software packages tested.
Conclusion
CERR’s radiomics capabilities are comprehensive, open-source, and fast, making it an attractive platform for developing and exploring radiomics signatures across institutions. The ability to both choose from a wide variety of radiomics implementations and to integrate with a clinical workflow makes CERR useful for retrospective as well as prospective research analyses.
Introduction
The concept of “radiomics” in oncology involves identifying quantitative imaging patterns that form the basis of predictive models or diagnostic biomarkers. Radiomics is hypothesized to be related to the underlying tumor biology and response to treatment depending on the timing of image acquisition (1, 2). The number of radiomics studies has greatly increased since the term was introduced by Lambin, et al. (3). Radiomics is by definition quantitative (4), but it is often not reproduced accurately between research groups, even when using the same imaging data (5). This is for various reasons such as different internal parameters used across different software tools, subtle differences in their generation (for example, using physical vs. voxel units), incorrect or insufficient documentation, and/or software defects. Hence, a comprehensive open-source software platform is critical for the development and validation of multi-institutional radiomics-focused research.
Some of the widely used software tools for radiomics include: (i) the Insight ToolKit (ITK; www.itk.org), which is an open-source, BSD copyrighted software developed in C++, with wrappers in commonly used, interpreted, and compiled languages. ITK is a library that is often used in combination with other software tools such as 3D-Slicer (6) for visualization and ITK-SNAP (7) for segmentation. ITK does not provide wrappers for MATLAB® (MathWorks, MA, USA) and includes only a subset of the radiomics features recommended by the Image Biomarker Standardization Initiative (IBSI) (8). (ii) MaZda (9) has been developed in C++, but it is not open-source and is compiled only for Windows operating systems. Similar to ITK, it includes only a subset of the features recommended by IBSI. (iii) PyRadiomics (10) is an open-source, Python-based package to extract radiomics with a plugin for 3D Slicer. It provides a comprehensive set of radiomics in the Python (https://www.python.org) language, but it lacks the calculation of radiomics maps and DICOM-RT input of anatomical structures. Similar to ITK, PyRadiomics is a radiomics library rather than an integrated platform, and it is up to the users to integrate it with their applications that provide bookkeeping to associate radiomics with scans and structures for future use. (iv) The Imaging Biomarker EXplorer (IBEX) (2) is developed in MATLAB and C++, which limits its portability between operating systems and various MATLAB versions. Similar to PyRadiomics, IBEX lacks calculation of radiomics maps and has limited capabilities for data import, export, segmentation, and visualization. Table 1 compares various capabilities of commonly used radiomics software packages.
Table 1. Summary of the main characteristics of available radiomics software.
CERR supports all stages of the “radiomics pipeline”.
| Language | IBSI feature defns. | Full OS compatibility | DICOM-RT import | Integrated visualization | Radiomics metadata storage | Built-in segmentation | Radiomics Maps | |
|---|---|---|---|---|---|---|---|---|
| ITK | C++ | ✗ | ✓ | ✓ | ✗ | ✗ | ✗ | ✓ |
| MaZda | C++/Delphi | ✗ | ✗ | ✗ | ✓ | ✗ | ✓ | ✓ |
| PyRadiomics | Python | ✓ | ✓ | ✗ | ✗ | ✗ | ✗ | ✗ |
| IBEX | Matlab/C++ | ✗ | ✗ | ✓ | ✓ | ✓ | ✓ | ✗ |
| CERR | Matlab | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
The Computational Environment for Radiological Research (CERR) (11) was extended to address the shortcomings of the aforementioned software tools. The radiomics functionality in CERR was developed exclusively in the widely used and accessible MATLAB language, but it can also be compiled and distributed without a MATLAB license. The objective was to develop a comprehensive, open-source, MATLAB-based software platform with an emphasis on reproducibility, speed, and clinical integration of radiomics-focused research. The advantage of using CERR for computational radiomics over other software is the availability of a comprehensive and validated pipeline ranging from data import, visualization, segmentation, metadata storage, and feature calculation. Adding computational radiomics to CERR creates a unique research platform capable of combining radiotherapy (RT) treatment planning and outcomes modeling with radiomics. The CERR platform provides a flexible, time-tested data structure to store radiomics metadata and combine it with RT. This further facilitates radiomics-driven longitudinal and multi-modality analysis. CERR is the only open-source platform that provides tests for its radiomics features against other open-source software. It is also the only platform to compute higher-order texture features using vectorized implementations, which results in significant speedups. The computational radiomics codebase is developed purely using MATLAB, making it agnostic to operating system and MATLAB versions.
Description of CERR’s Computational Radiomics
A. Architecture
CERR is a stable and popular platform for developing computational radiomics functionality because it provides extensive visualization, bookkeeping, import, export, image analysis, and transformation functions. CERR has been cited more than 430 times in peer-reviewed literature as of June 2018. Some of the most commonly used CERR plugin modules include positron emission tomography (PET) segmentation (12), the Intensity Modulated Radiotherapy Planning (IMRTP) toolbox (13), and Dose Response Explorer System (DREES) (14). Extending CERR for radiomics analysis provides the ability to combine imaging with CERR’s exhaustive tools for analysis of RT dose and treatment planning data. The critical components of CERR for radiomics include the ability to: (i) import imaging data with standard formats using different modalities, (ii) delineate and import segmentations for radiomics calculation, (iii) define important parameters for radiomics calculation, (iv) visualize and compare the resulting radiomics maps, (v) derive and store radiomics values along with imaging data, and (vi) export the resulting radiomics scalars or maps to any other analysis software (Figure 1). CERR’s data import capabilities (https://github.com/cerr/CERR/wiki/Importing-to-CERR) are vast compared with other radiomics software tools. CERR can import various data formats RTOG, DICOM, MHA, NRRD, NIfTI and XML. It also supports the import of DICOM RTPLAN, RTDOSE, RTSTRUCTS and GSPS in addition to the computed tomography, PET, single photon emission computed tomography, magnetic resonance (dynamic contrast-enhanced and diffusion-weighted imaging), ultrasound, and mammogram modalities. CERR also can import oblique scans along with the segmentations. Data export capabilities in CERR include DICOM export of scan, RTSTRUCT, and RTDOSE. Also, CERR’s contouring tools (https://github.com/cerr/CERR/wiki/Contouring-tools) include pencil, brush, eraser, and active contour-based refinement modes as well as Boolean arithmetic to derive new structures from existing ones. The segment labeler tool (https://github.com/cerr/CERR/wiki/Segment-Labeler) in CERR makes it convenient for users to graphically score different parts of auto-segmentation results, which can then be used for evaluating and improving algorithms. CERR also provides wrappers for Plastimatch (15) for image registration, useful for longitudinal as well as multi-modality analyses.
Figure 1. Flow diagram describing the main components of computational radiomics pipeline in Computational Environment for Radiological Research (CERR).
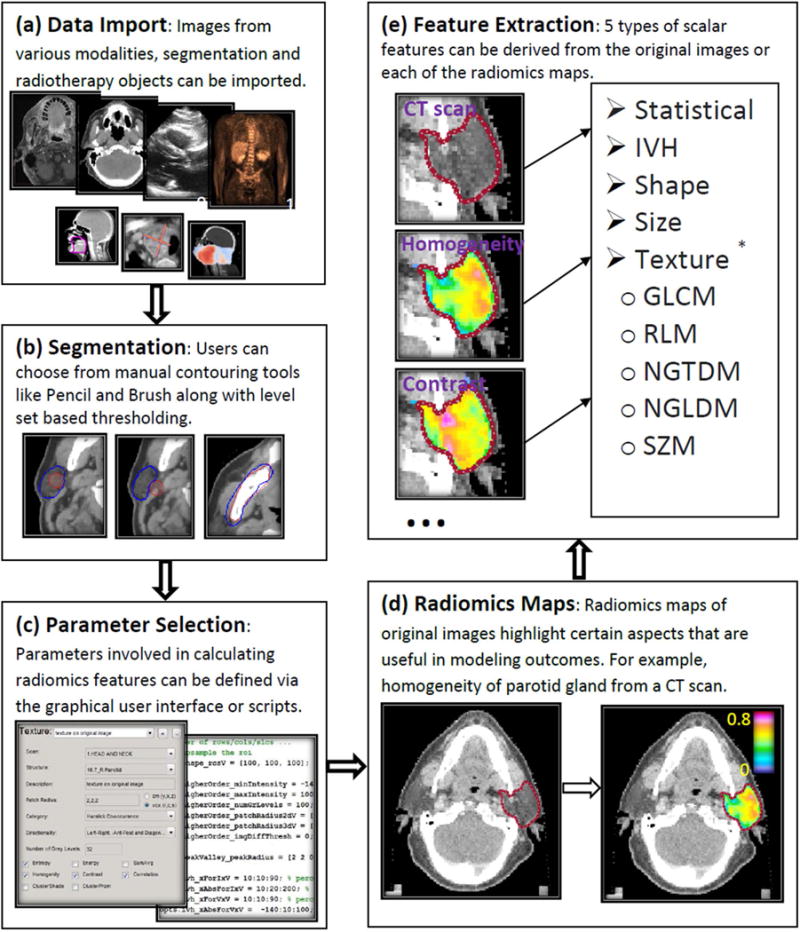
The pipeline consists of (a) data import, (b) segmentation, (c) parameter selection, (d) radiomics map/pre-processing filters, and (e) extraction of scalar radiomics features for further analysis.
* Texture radiomics scalars based on gray level co-occurrence matrix: Gray level co-occurrence (16), Neighborhood gray-tone difference (20), Neighborhood gray-level dependence (21), Run length (22), and Size zone (23) matrices.
Radiomics results are made permanently accessible by extending CERR’s data structure to store radiomics metadata along with the results. This provides a permanent record of calculation parameters, simplifies the bookkeeping for computations across a collection of images, and works seamlessly with longitudinal imaging data. In addition to the native support for CERR’s data structure, the calculation routines were designed to be compatible with MATLAB’s three-dimensional (3D) matrices and logical masks used to define the region of interest (ROI). Hence, CERR’s radiomics can be called from other applications by just passing the matrices for the image and the labels.
B. Radiomics maps and pre-processing filters
Radiomics generated from small neighborhoods around each voxel results in a composite radiomics map, which has the same size as the ROI. These maps, which can be displayed, carry spatial radiomics information and could have implications both in the setting of outcome modeling and image segmentation. The radiomics maps provide another level of image transformations that highlight characteristics of sub-regions within the ROI. CERR allows for the generation of Haralick feature maps (16), Law’s filters maps (17-19), and first order statistics maps in addition to various pre-processing filters (https://github.com/cerr/CERR/wiki/Texture-calculation). Figure 2 illustrates the influence of parameters and methodology in generating radiomics. CERR provides computation of various flavors of the same features for both radiomics maps and scalar radiomics. As described in section D, a unique aspect of CERR is the speedup of radiomics map calculations using a novel matrix formulation. In addition to the computation of Haralick, Law’s and first order statistical radiomics maps, CERR provides various pre-processing filters like Wavelet, Sobel, Gabor, and Laplacian of Gaussian. The parameters for radiomics maps as well as pre-processing filters can be defined in batch mode or through a graphical user interface. The maps can then be visualized side-by-side along with the original image. This is helpful for quality assurance as well as understanding the impact of pre-processing the original image. Figure 3 shows an example of 3D Wavelet pre-processing of CT image. Another pre-processing option is to interpolate the image to a user-defined resolution. This is crucial to standardize heterogeneous datasets where patient scans are acquired at different resolutions. Normalization of image intensities is necessary for images that don’t have standard units. CERR provides tools to compute standard uptake values from FDG PET scans and wrappers for external normalization tools as has been shown in work by Li, et al. (24) for bias field correction in MRI scans. Moreover, CERR’s data structure provides convenient access to images and associated metadata, making it straightforward for users to define custom normalizations or use filters from libraries such as ITK and MATLAB image processing toolbox.
Figure 2. Different approaches to calculating the same feature lead to different radiomics maps.
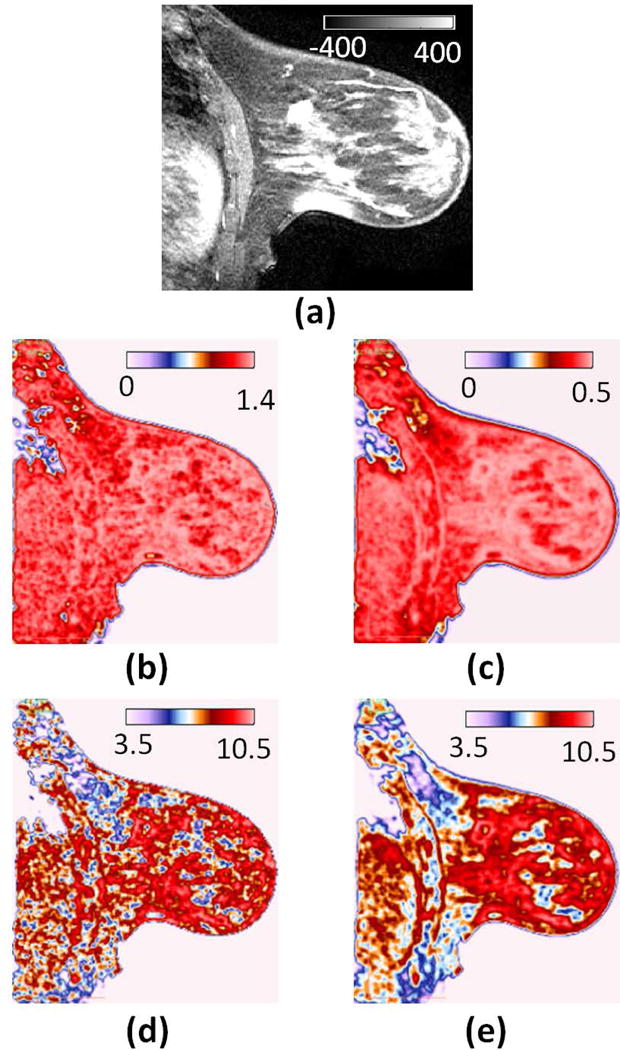
This often ignored aspect is critical when validating radiomics signatures across institutions. (a) T1 post-contrast image from a breast cancer patient. (b) Local gray level co-occurrence matrix (GLCM) Contrast averaged across two-dimensional (2D) directional offsets. (c) Local GLCM Contrast averaged across three-dimensional (3D) directional offsets. (d) Local GLCM Contrast computed by accumulating co-occurrence frequencies from 2D directional offsets into a single co-occurrence matrix. (e) Local GLCM Contrast computed by accumulating co-occurrence frequencies from 3D directional offsets into a single co-occurrence matrix.
Figure 3. Graphical user interface (GUI) to define parameters for pre-processing filters and radiomics maps.
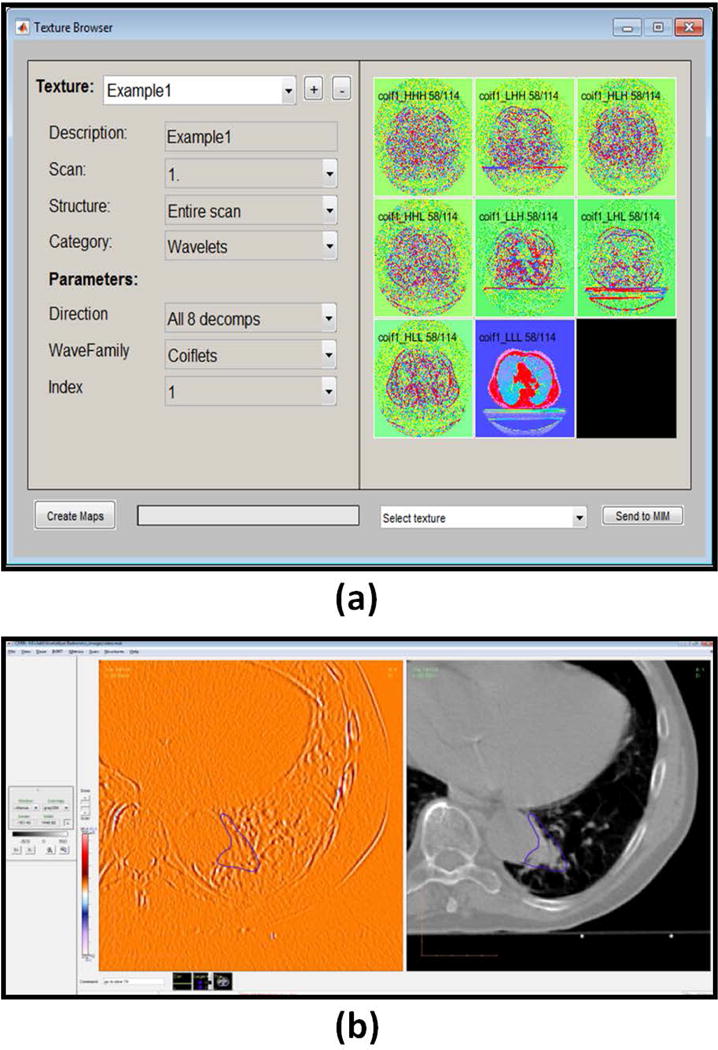
(a) GUI allows user to select a filter and its associated parameters. For example, three-dimensional wavelets filter. (b) The radiomics maps and the pre-processed images can be visualized along with the original image. For example, computed tomography scan and the HLH direction Coiflet1 wavelets (1) pre-processed image.
C. Scalar radiomics
Scalar radiomics features used to model outcomes can be derived from the original images as well as from the pre-processed images/radiomics maps. CERR provides six classes of scalar radiomics (class definitions according to arXiv:1612.07003 (9): (i) First-order/histogram statistics, (ii) Intensity-volume histogram, (iii) Peak/Valley, (iv) Shape, (v) Size, and (vi) Texture, which refers to the higher-order radiomics where the ROI is reduced to a scalar, as opposed to a voxel-wise, radiomics map. CERR provides the computation of such scalar texture using: (a) Gray level co-occurrence matrix (GLCM) (16), (b) Neighborhood gray tone difference (20), (c) Neighborhood gray level dependence (21), (d) Run length (22), and (e) Size zone (23) matrices. CERR provides the ability to parameterize these radiomics calculations via the graphical interface or batch scripts. CERR also provides the ability to compute gray level co-occurrence and run length features separately for each direction, or by combining frequency contributions from all the directions. Feature calculation can also be parameterized for two-dimensional (2D) or 3D calculation. Such flexibility is useful to accurately reproduce radiomics signatures. The features can be stored within MATLAB’s data structure or output to a CSV file.
D. Speed-up using matrix algebra
Radiomics feature calculation in CERR makes extensive use of matrix algebra, and the code is vectorized for speed. Haralick texture features, commonly used in radiomics, are a prime example of involved computation because they require processing the neighborhood around each voxel in the ROI. We demonstrate the use of matrix algebra and the resulting speedup for Haralick texture calculation. The computation involves counting neighbor pairs with all gray level combinations along a particular direction and within a neighborhood around each voxel. The time complexity of computing such a radiomics map is O(N3) for an image of size N × N × N voxels in the local region. However, the most time-consuming operation occurs at the unit step for each voxel while populating the co-occurrence matrix (25). It involves: (i) determining neighbors in the given direction and offset, (ii) filtering out neighbor-pairs outside the ROI, (iii) determining voxels within the neighborhood around the voxel for the sliding window-based calculation, and (iv) adding entries to the co-occurrence matrix, which has a computational complexity O(NL2), where NL is the number of gray levels. All previously suggested approaches to speed this computation up (25, 26) use parallelization of steps (i)-(iv) across all voxels. Although parallelization reduces the total computational time, it does not address the computational cost per voxel involved in (i)-(iv). Instead, using the proposed matrix approach, repetitive bookkeeping is replaced by fast indexing operations for all the voxels in the concerned ROI (Supplementary material A0; Examples A1 and A2). This eliminates the computational overhead associated with each voxel. For example, computing patch-wise Haralick radiomics features using an image discretized into 32 bins resulted in a speedup of 3.5 times over ITK (Figure 4).
Figure 4. Comparison of runtime between Computational Environment for Radiological Research (CERR) and Insight Toolkit’s Haralick radiomics maps.
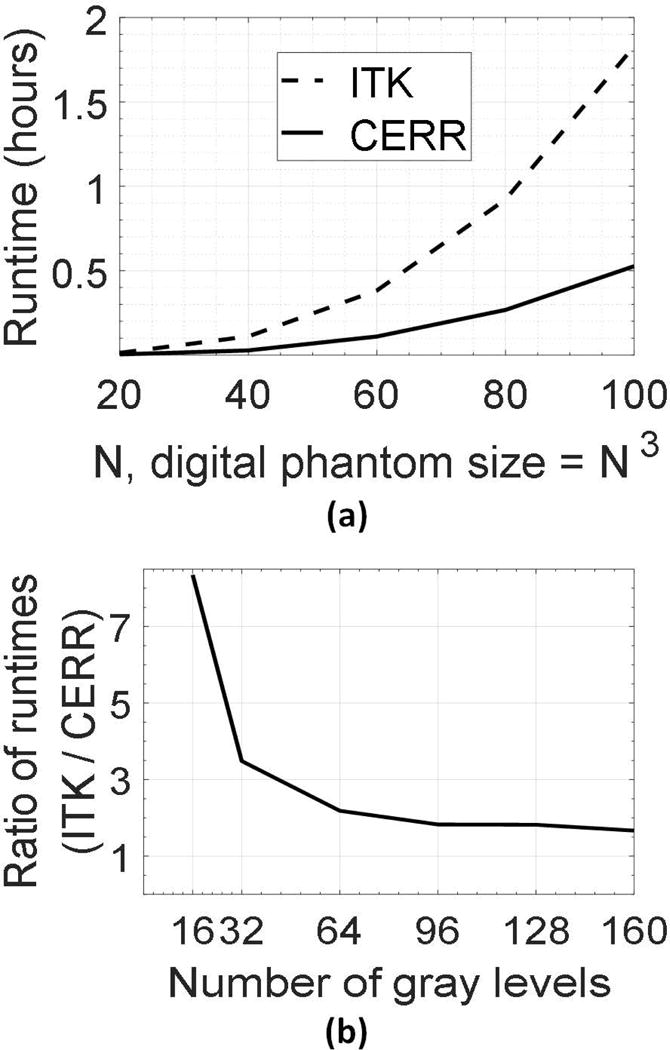
(a) Runtime as a function of digital phantom size when the image is discretized into 32 gray levels. CERR is about 3.5 times faster compared to Insight Toolkit (ITK). (b) The ratio between runtimes of ITK and CERR as a function of the number of gray levels. As the number of gray levels increases, CERR loses some of its speed advantages. This is because the time required to accumulate the co-occurrence frequencies ( ) dominates the gains from indexing and bookkeeping in the matrix-based approach.
E. Testing and reproducibility between software implementations
Differences in radiomics between software systems arise from incorrect/inconsistent definitions or programming errors. Professionally engineered software like ITK provides good coverage with their unit tests. However, such testing may not uncover subtle differences in radiomics definitions. Hence, developing tests that compare different software systems is the only way to address the problem of reproducibility in radiomics. CERR’s radiomics was tested by matching results from the digital phantom provided by IBSI. Additionally, CERR provides tests for its radiomics features to ensure reproducibility with other software systems. This “test suite” compares CERR generated radiomics with those computed from ITK and PyRadiomics. The tests between CERR and ITK have involved GLCM (scalar and patch-wise) and run length matrix (RLM) features whereas the tests between CERR and PyRadiomics have involved the first order, shape and higher order (texture) features. In addition to testing feature calculation, tests have also been developed to evaluate pre-processing filters. The eight Wavelet decompositions used in (1) have been tested between CERR and PyRadiomics. In all, the tests covered 1024 radiomics features that were computed from original and pre-processed images and 9 patch-wise Haralick radiomics maps. Although all the tests between CERR and PyRadiomics have indicated agreement, the following tests between CERR and ITK have failed. The next sections provide details of subtle discrepancies with ITK that have been uncovered by this inter-software testing:
Correlation and Haralick correlation (15) from ITK
The ITK documentation as well as the code use the formula for “Correlation” feature as , where is the weighted pixel-mean, is the weighted pixel variance and g represents the co-occurrence matrix. The correct formula has as the standard deviation ( ) instead of the variance ( ), as has been coded and documented in ITK.
Investigation of “Haralick correlation” calculation from ITK has revealed that the levels run from 0 to the maximum gray level minus 1. This is different from the definition in Haralick’s original paper where the levels ran from 1 to the maximum gray level (16).
Run length matrix (RLM) (21) from ITK
The RLM computed in ITK is designed to be a square matrix, and the maximum number of run length bins can be at most the number of gray levels into which the image has been discretized. This leads to a loss in resolution in cases with relatively less gray levels. Moreover, ITK computes run lengths in physical units, which are accumulated into the specified number of bins whereas most other radiomics software compute the run lengths in units of voxel lengths, as defined and suggested by Galloway (21). Within CERR, the computation of the RLM can be performed using either physical or voxel units.
F. Integration with clinical software
Software tools, such as MIM (MIMvista, MIM software Inc., Cleveland, OH; https://www.mimsoftware.com/), Eclipse (Varian Medical Systems, Palo Alto, CA; https://www.varian.com/), and RayStation (RaySearch Laboratories, Stockholm, Sweden; https://www.raysearchlabs.com/), provide application programming interfaces (APIs) for data access. Such APIs provide integration of site- and organ-specific radiomics, and thus allow for the use of radiomics for clinical investigations. Radiomics extension was developed using MIM’s MATLAB API in which users can pass images and ROIs from MIM to CERR and export the derived radiomics map back to MIM. Figure 5 demonstrates CERR radiomics Extension’s workflow to generate and display the radiomics maps within MIM. The CERR radiomics Extension provides options for setting parameters for generating radiomics maps. Compiling CERR code is independent of the operating system because it is purely MATLAB-based. CERR can therefore be easily integrated with clinical software that does not provide MATLAB APIs. It is emphasized, of course, that CERR is not Food and Drug Administration- approved software, and can only be used to derive research data with appropriate safeguards.
Figure 5. Computational Environment for Radiological Research (CERR) radiomics extension as integrated into the Food and Drug Administration-approved MIM software.
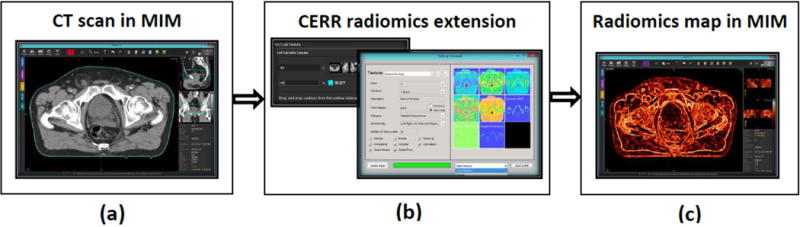
(a) computed tomography scan from a prostate cancer patient in MIM software. (b) The Extension presents users with options to select parameters for radiomics calculation and displays thumbnails for radiomics maps. (c) The resulting radiomics map (correlation from Haralick gray level co-occurrence) for the selected scan and the structure is displayed in MIM.
Discussion
The capabilities of CERR covered in this work include key aspects of accurate radiomics representation and associated research: data import, transformation, segmentation, visualization, radiomics calculation, and bookkeeping in a user-friendly MATLAB environment (Figure 1). CERR is distributed on gitHub (http://www.github.com/cerr/CERR), which provides an extremely stable platform for CERR releases and information related to various modules. Each software change is tested for integrity using the Jenkins framework (http://jenkins.io). Extensive documentation is provided via gitHub Wiki (https://github.com/cerr/CERR/wiki/Radiomics). CERR’s user group (https://groups.google.com/forum/#!forum/cerr-forum) has 536 members as of June 2018.
CERR offers the ability to choose from a wide range of radiomics implementations and parameters, and thus also makes this platform useful to validate radiomics-based models across institutions as exemplified for Haralick contrast in Figure 2. It addresses the lack of reproducibility in generated radiomics, which is critical for deriving radiomics-based models. CERR provides a wide range of radiomics features, and an extensible data structure to add new ones. The role of CERR as a radiomics platform includes sharing and reproducing radiomics results across institutions as well as across software tools, e.g., for external validation of generated radiomics models.
CERR provides a computational speedup of Haralick radiomics calculation over other commonly used implementations such as the C++-based ITK version 4.12. This is crucial for clinical implementation of developed radiomics. The matrix formulation for speeding up Haralick texture calculations can be easily translated into other programming languages, and on-going work focuses on such implementations both for Julia (http://julialang.org) and Python (https://www.python.org).
A further step towards clinical implementation of radiomics is the integration of CERR with the Food and Drug Administration-approved MIM software. The MIM Extension for CERR’s computational radiomics (Figure 5) is distributed along with the source code, which makes it possible for MIM users to readily use it as a template for research purposes.
Supplementary Material
Acknowledgments
This research was partially funded by NIH grant 1R01CA198121 and NIH/NCI Cancer Center Support grant P30 CA008748.
Footnotes
Disclosure of Conflicts of Interest
The authors have no relevant conflicts of interest to disclose.
References
- 1.Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang L, Fried DV, Fave XJ, Hunter LA, Yang J, Court LE. IBEX: an open infrastructure software platform to facilitate collaborative work in radiomics. Med Phys. 2015;42(3):1341–1353. doi: 10.1118/1.4908210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: Extracting more information from medical images using advanced feature analysis. European Journal of Cancer. 2012;48(4):441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology. 2016;278(2):563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Limkin EJ, Sun R, Dercle L, et al. Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Annals of Oncology. 2017;28(6):1191–1206. doi: 10.1093/annonc/mdx034. [DOI] [PubMed] [Google Scholar]
- 6.Fedorov A, Beichel R, Kalpathy-Cramer J, et al. Magnetic Resonance Imaging: 3D Slicer as an image computing platform for the Quantitative Imaging Network. 2012;30(9):1323–1341. doi: 10.1016/j.mri.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yushkevich PA, Piven J, Hazlett HC, et al. User-guided 3D active contour segmentation of anatomical structures significantly improved efficiency and reliability. Neuroimage. 2006;31(3):1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 8.Zwanenburg A, Leger S, Vallières M, Löck S. Image biomarker standardisation initiative - feature definitions. CoRR. 2016 abs/161207003. [Google Scholar]
- 9.Szczypiński PM, Strzelecki M, Materka A, Klepaczko A. MaZda–a software package for image texture analysis. Comput Methods Programs Biomed. 2009;94(1):66–76. doi: 10.1016/j.cmpb.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 10.Griethuysen J, Fedorov A, Parmar C, et al. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Research. 2017;77(21):e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deasy JO, Blanco AI, Clark VH. CERR: a computational environment for radiotherapy research. Med Phys. 2003;30(5):979–985. doi: 10.1118/1.1568978. [DOI] [PubMed] [Google Scholar]
- 12.Berthon B, Spezi E, Galavis P, et al. Toward a standard for the evaluation of PET-Auto-Segmentation methods following the recommendations of AAPM task group No. 211: Requirements and implementation. Med Phys. 2017;44(8):4098–4111. doi: 10.1002/mp.12312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deasy J, Lee EK, Bortfeld T, et al. A collaboratory for radiation therapy treatment planning optimization research. Annals of Operations Research. 2006;148(1):55–63. [Google Scholar]
- 14.El Naqa I, Suneja G, Lindsay PE, et al. Dose response explorer: an integrated open-source tool for exploring and modelling radiotherapy dose-volume outcome relationships. Phys Med Biol. 2006;51(22):5719–5735. doi: 10.1088/0031-9155/51/22/001. [DOI] [PubMed] [Google Scholar]
- 15.Shackleford JA, Kandasamy N, Sharp GC. On developing B-spline registration algorithms for multi-core processors. Physics in Medicine & Biology. 2010;55(21):6329–6351. doi: 10.1088/0031-9155/55/21/001. [DOI] [PubMed] [Google Scholar]
- 16.Haralick RM, Shanmugam K. Textural features for image classification. IEEE Transactions on systems, man, and cybernetics. 1973;3(6):610–621. [Google Scholar]
- 17.Laws K. Rapid Texture Identification. SPIE. 1980;0238:376–380. [Google Scholar]
- 18.Suzuki MT, Yaginuma Y, Yamada T, Shimizu Y. A shape feature extraction method based on 3D convolution masks. IEEE International Symposium on Multimedia. 2006:837–844. [Google Scholar]
- 19.Suzuki MT, Yaginuma Y, Kodama H. A 2D Texture Image Retrieval Technique based on Texture Energy Filters. IMAGAPP. 2009:145–151. [Google Scholar]
- 20.Amadasun M, King R. Textural features corresponding to textural properties. IEEE Transactions on Systems, Man, and Cybernetics. 1989;19(5):1264–1274. [Google Scholar]
- 21.Sun C, Wee WG. Neighboring gray level dependence matrix for texture classification. Computer Vision, Graphics, and Image Processing. 1983;23(3):341–352. [Google Scholar]
- 22.Galloway MM. Texture analysis using gray level run lengths. Computer Graphics and Image Processing. 1975;4(2):172–179. [Google Scholar]
- 23.Thibault GFB, Navarro C, Pereira S, et al. Shape and texture indexes application to cell nuclei classification. International Journal of Pattern Recognition and Artificial Intelligence. 2013;27(1):1357002. [Google Scholar]
- 24.Li C, Gore JC, Davatzikos C. Multiplicative intrinsic component optimization (MICO) for MRI bias field estimation and tissue segmentation. Magnetic resonance imaging. 2014;32(7):913–923. doi: 10.1016/j.mri.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lakovidis DK, Maroulis DE, Bariamis DG. FPGA architecture for fast parallel computation of co-occurrence matrices. Microprocessors and Microsystems. 2007;31(2):160–165. [Google Scholar]
- 26.Asadollah S, Pham TA, Bertels K. Parallel implementation of Gray Level Co-occurrence Matrices and Haralick texture features on cell architecture. The Journal of Supercomputing. 2012;59(3):1455–1477. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


