GDH, which catalyzes the synthesis and degradation of glutamate using NAD(P)(H), is a widely distributed enzyme among all domains of life. Mammalian GDH is regulated allosterically by multiple metabolites, in which the antenna helix plays a key role to transmit the allosteric signals. In contrast, bacterial GDH was believed not to be regulated allosterically because it lacks the antenna helix. We previously reported that GDH from Thermus thermophilus (TtGDH), which is composed of two heterologous subunits, is activated by leucine. In the present study, we found that AMP activates TtGDH using a catalytically inactive APRTh as the sensory subunit. This suggests that T. thermophilus possesses a complicated regulatory mechanism of GDH to control carbon and nitrogen metabolism.
KEYWORDS: AMP, Thermus thermophilus, adenine phosphoribosyltransferase, allosteric regulation, glutamate dehydrogenase, protein-protein interaction
ABSTRACT
Glutamate dehydrogenase (GDH) from a thermophilic bacterium, Thermus thermophilus, is composed of two heterologous subunits, GdhA and GdhB. In the heterocomplex, GdhB acts as the catalytic subunit, whereas GdhA lacks enzymatic activity and acts as the regulatory subunit for activation by leucine. In the present study, we performed a pulldown assay using recombinant T. thermophilus, producing GdhA fused with a His tag at the N terminus, and found that TTC1249 (APRTh), which is annotated as adenine phosphoribosyltransferase but lacks the enzymatic activity, was copurified with GdhA. When GdhA, GdhB, and APRTh were coproduced in Escherichia coli cells, they were purified as a ternary complex. The ternary complex exhibited GDH activity that was activated by leucine, as observed for the GdhA-GdhB binary complex. Furthermore, AMP activated GDH activity of the ternary complex, whereas such activation was not observed for the GdhA-GdhB binary complex. This suggests that APRTh mediates the allosteric activation of GDH by AMP. The present study demonstrates the presence of complicated regulatory mechanisms of GDH mediated by multiple compounds to control the carbon-nitrogen balance in bacterial cells.
IMPORTANCE GDH, which catalyzes the synthesis and degradation of glutamate using NAD(P)(H), is a widely distributed enzyme among all domains of life. Mammalian GDH is regulated allosterically by multiple metabolites, in which the antenna helix plays a key role to transmit the allosteric signals. In contrast, bacterial GDH was believed not to be regulated allosterically because it lacks the antenna helix. We previously reported that GDH from Thermus thermophilus (TtGDH), which is composed of two heterologous subunits, is activated by leucine. In the present study, we found that AMP activates TtGDH using a catalytically inactive APRTh as the sensory subunit. This suggests that T. thermophilus possesses a complicated regulatory mechanism of GDH to control carbon and nitrogen metabolism.
INTRODUCTION
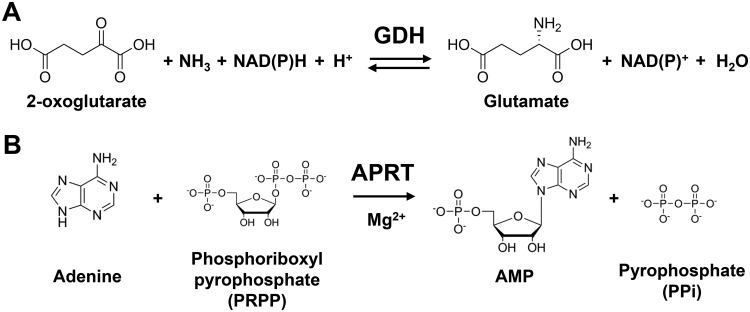
Glutamate dehydrogenase (GDH) catalyzes the reversible conversion between 2-oxoglutarate/ammonium and glutamate using NAD(H) or NADP(H) as a coenzyme (Fig. 1A) (1). Due to its important role in balancing nitrogen consumption in cells, GDH is widely distributed among living organisms. GDH is classified into three types based on the coenzyme specificity: NAD-dependent GDH (EC 1.4.1.2), NAD(P)-dependent GDH (EC 1.4.1.3), and NADP-dependent GDH (EC 1.4.1.4). NAD-dependent GDH is further divided into three types according to the molecular weight of a single subunit: GDH50s, GDH115s, and GDH180s (2). GDH50s are widely distributed among all domains of life and are known to have a homohexameric structure, each with a subunit molecular mass of approximately 50 kDa. In contrast, GDH115s and GDH180s are large GDH composed of the 50-kDa core GDH domain and additional N- and C-terminal domains. GDH115s with a single-subunit molecular mass of approximately 115 kDa are found in several eukaryotic microorganisms, such as Neurospora crassa and Saccharomyces cerevisiae, and have a tetrameric structure (3, 4). GDH180s isolated from several bacteria are the largest type of GDH, with a subunit molecular mass of approximately 180 kDa, and GDH180 from Janthinobacterium lividum has a hexameric structure (2, 5, 6).
FIG 1.
Reactions of GDH and APRT. (A) The GDH reaction. (B) The APRT reaction.
GDH50s are further divided into two subfamilies. One is composed of the 50-kDa core GDH domain, which is widely distributed among all domains of life, and the other is mammalian GDH, possessing an antenna helix (∼50 amino acid residues) inserted in the NAD-binding domain. Mammalian GDH is regulated allosterically in a complicated manner by several metabolites, such as GTP, ADP, leucine, NAD(H), and palmitoyl coenzyme A (palmitoyl-CoA) (7). The antenna helix was also found in GDH from ciliates, and it is controlled allosterically by ADP and palmitoyl-CoA (8). The antenna helix plays roles in intersubunit communication for negative cooperativity and allosteric regulation (9). GTP and NADH inhibit GDH by keeping the active-site cleft in a closed conformation. In contrast, ADP binds to the back side of the NAD-binding domain and activates GDH by keeping the active-site cleft open (10). The allosteric regulation of human GDH is thought to play an important role in insulin and ammonia homeostasis, because the mutation that abrogates GTP inhibition results in dangerously high levels of insulin and ammonium in serum (11, 12). On the other hand, GDH50s from plants, bacteria, or fungi were considered not to be regulated allosterically because they lack the antenna helix (7). However, this is not the case for GDH from T. thermophilus (TtGDH), which is allosterically activated by leucine (13). TtGDH possesses a unique subunit organization. Two GDH homologs, gdhA and gdhB, with 46% amino acid sequence identity, are present in a tandem manner as TTC1212 and TTC1211 on the genome of T. thermophilus. GdhA and GdhB from T. thermophilus form a heterohexamer, in which GdhB acts as the catalytic subunit and GdhA acts as the regulatory subunit to sense leucine. The crystal structure of GdhA/GdhB in a complex with leucine and biochemical analysis revealed that leucine is bound to the pockets at the GdhA-GdhA, GdhA-GdhB, and GdhB-GdhB interfaces. Furthermore, leucine binding increases the turnover of the GDH reaction, possibly through acceleration of the open-close cycle of the active-site cleft between the catalytic domain and NAD-binding domains (14). Of note, comparison of the amino acid sequences and crystal structures between T. thermophilus and mammalian GDH demonstrated that the residues for the recognition of the α-amino and α-carboxyl groups of leucine in TtGDH are conserved in mammalian GDH, whereas they are not conserved in GDH from other organisms except for some bacteria, such as Thermus, Deinococcus, and Thermotoga. We previously performed mutational analysis of human GDH2 (hGDH2) and found that replacement of the residues with alanine abolished the leucine-induced activation, indicating that hGDH2 possesses the same leucine-binding site as TtGDH; however, allosteric regulation by nucleotides was not observed for TtGDH alone.
Here, in order to further investigate the regulatory mechanism of TtGDH, we screened for a partner protein of TtGDH that can alter TtGDH function by binding to TtGDH and found that a homolog of adenine phosphoribosyltransferase (APRTh) formed a stable heterocomplex with TtGDH. TtGDH was also revealed to be activated by AMP in a complex with APRTh where APRTh was necessary for the AMP-mediated allosteric activation of TtGDH. Taken together with the activation by leucine, TtGDH is allosterically regulated in a more complex manner than previously thought.
RESULTS
Screening for a partner protein of TtGDH.
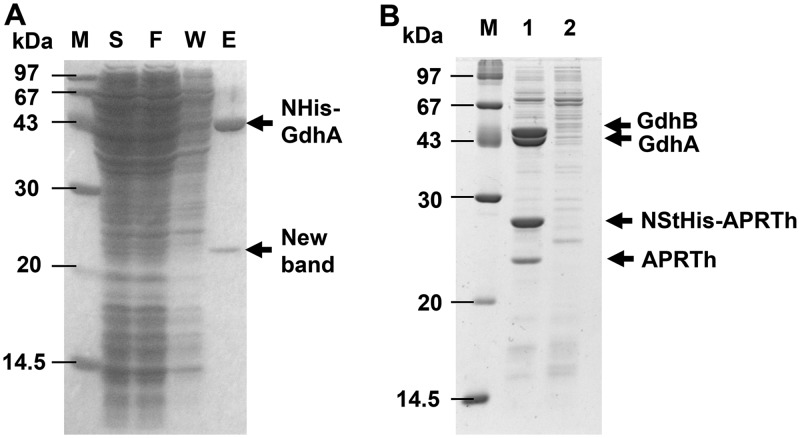
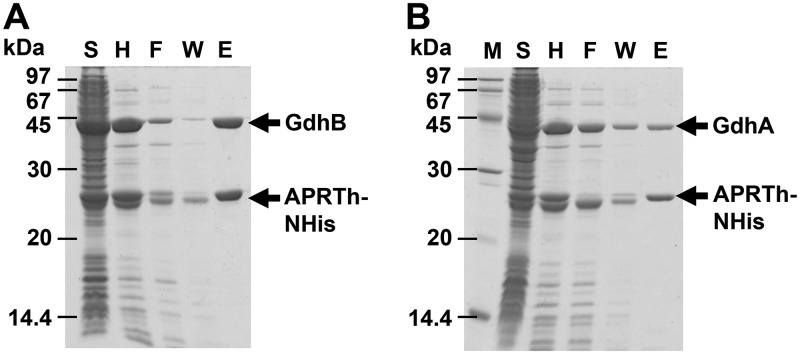
In order to examine partner proteins of TtGDH, we conducted a pulldown assay using the recombinant T. thermophilus, Tt27NHisGdhA, which produces GdhA with a His tag at the N terminus. When cell lysate of the strain was applied to an Ni2+-nitrilotriacetic acid (NTA) column, a unique protein with a molecular weight of 23 kDa on SDS-PAGE was copurified with GdhA (Fig. 2A). As the band of GdhB was not observed on SDS-PAGE, we assumed that GdhA with a His tag possessing a molecular weight of 46,328 Da and GdhB possessing a molecular weight of 46,112 Da overlapped on the SDS-PAGE gel. Using successive matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF-MS) analysis, the copurified protein with a molecular weight of 23 kDa was identified as TTC1249. TTC1249 was annotated as adenine phosphoribosyltransferase (APRT), which is an enzyme that catalyzes the formation of adenine monophosphate (AMP) from adenine and PRPP as the substrates in the purine nucleotide salvage pathway (Fig. 1B). Of note, the TTC1250 gene, which also encodes APRT with 41% amino acid sequence identity to TTC1249, is present upstream of the TTC1249 gene in the genome of T. thermophilus HB27. The tandem coordination of homologous genes is similar to that of the gdhA and gdhB genes.
FIG 2.
Isolation of the partner protein of TtGDH. (A) Pulldown assay for His-tagged GdhA using a Ni2+-NTA column. Lane M, molecular size markers; lane S, supernatant of cell extract of the Tt27NStHisAPRTh strain; lane F, flowthrough fraction from the column; lane W, wash fraction from the column; lane E, elution fraction from the column. (B) Copurification of Strep-tagged ARPTh with TtGDH. The 50-fold concentrated elution fraction from the Strep-Tactin column was applied. Lane 1, Tt27NStHisAPRTh strain; lane 2, wild-type T. thermophilus HB27 strain.
Enzyme assay of TTC1249 and TTC1250.
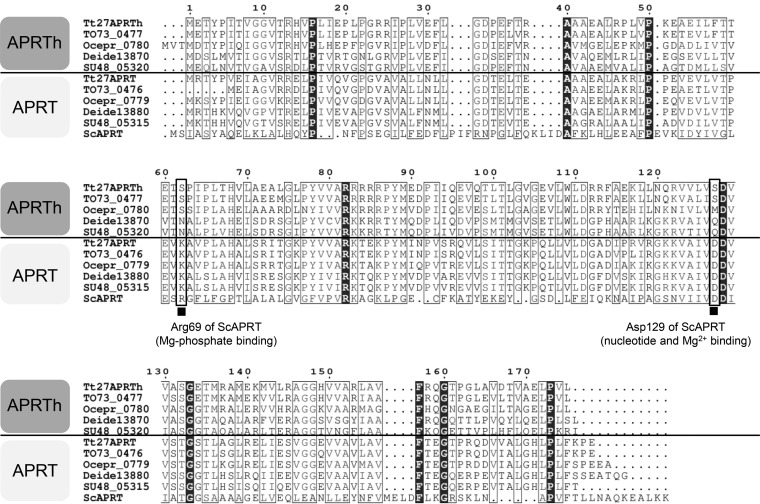
To characterize TTC1249 and TTC1250 as phosphoribosyltransferases (PRT), the recombinant proteins prepared from recombinant Escherichia coli were used for enzyme assay (Table 1). TTC1250 exhibited marked activity when adenine was used as a purine base substrate, whereas TTC1250 exhibited negligible activity toward the other purine bases evaluated. In contrast, TTC1249 exhibited no detectable activity toward adenine, guanine, hypoxanthine, or xanthine. Therefore, here we refer to TTC1250 and TTC1249 as APRT and homolog of APRT (APRTh), respectively. The lack of activity of APRTh may be due to the replacement of the residues involved in binding the monophosphate of nucleotide (Arg69 in Saccharomyces cerevisiae APRT, ScAPRT) and Mg2+ ion (Asp129 in ScAPRT) for catalysis (15) with Ser62 and Ser127 in APRTh, respectively (Fig. 3). In APRT from T. thermophilus, these are conserved as Lys62 and Asp127, respectively.
TABLE 1.
APRT activity of TTC1250 (APRT) and TTC1249 (APRTh)
| Substrate | APRT activity (U/mg) of: |
|
|---|---|---|
| TTC1250 (APRT) | TTC1249 (APRTh) | |
| Adenine | 27 ± 0.17 | <1 |
| Guanine | <1 | <1 |
| Hypoxanthine | <1 | <1 |
| Xanthine | <1 | <1 |
FIG 3.
Amino acid sequence alignment of APRTh and APRT. Tt27APRTh, APRTh from T. thermophilus HB27; TO73_0477, APRTh from T. aquaticus; Ocepr_0780, APRTh from Oceanithermus profundus; Deide13870, APRTh from Deinococcus deserti; SU48_05320, APRTh from Deinococcus puniceus. Tt27APRT, APRT from T. thermophilus HB27; TO73_0476, APRT from T. aquaticus; Ocepr_0779, APRT from Oceanithermus profundus; Deide13880, APRT from Deinococcus deserti; SU48_05315, APRT from Deinococcus puniceus. ScAPRT, APRT from S. cerevisiae. Black boxes at the bottom indicate the residues demonstrated to play important roles in the catalytic reaction of ScAPRT. The corresponding residues in the related proteins are boxed with bold lines.
Complex formation of TtGDH with APRTh.
As we reported previously, GdhA and GdhB form the heterohexamer TtGDH. In order to confirm complex formation between TtGDH and APRTh, a recombinant strain of T. thermophilus producing APRTh with both a Strep-tag and His tag at the N terminus, Tt27NStHisAPRTh, was constructed and the proteins were purified using a Strep-Tactin affinity column. Four unique proteins with molecular weights of 23 kDa, 27 kDa, 44 kDa, and 45 kDa on SDS-PAGE were found (Fig. 2B). Successive MALDI-TOF-MS analysis demonstrated that these proteins were APRTh without a tag (23 kDa protein), NStHis-APRTh (27 kDa protein), GdhA (44 kDa protein), and GdhB (45 kDa protein). In this experiment, APRTh without a tag was copurified unexpectedly. We hypothesized that APRTh without a tag was produced because the original start codon of APRTh remained in the gene encoding APRTh fused with the tags in the recombinant strain. The crystal structure of APRTh from T. thermophilus HB8 has been determined and revealed that the protein, possessing 99% amino acid sequence identity with APRTh from T. thermophilus HB27, forms a dimer (16); therefore, we assumed that a heterocomplex including both APRTh with the tag and APRTh without the tag was purified using the Strep-Tactin affinity column. MALDI-TOF-MS also demonstrated that APRT was not involved in heterocomplex formation.
Preparation of the recombinant TtGDH-APRTh complex using E. coli.
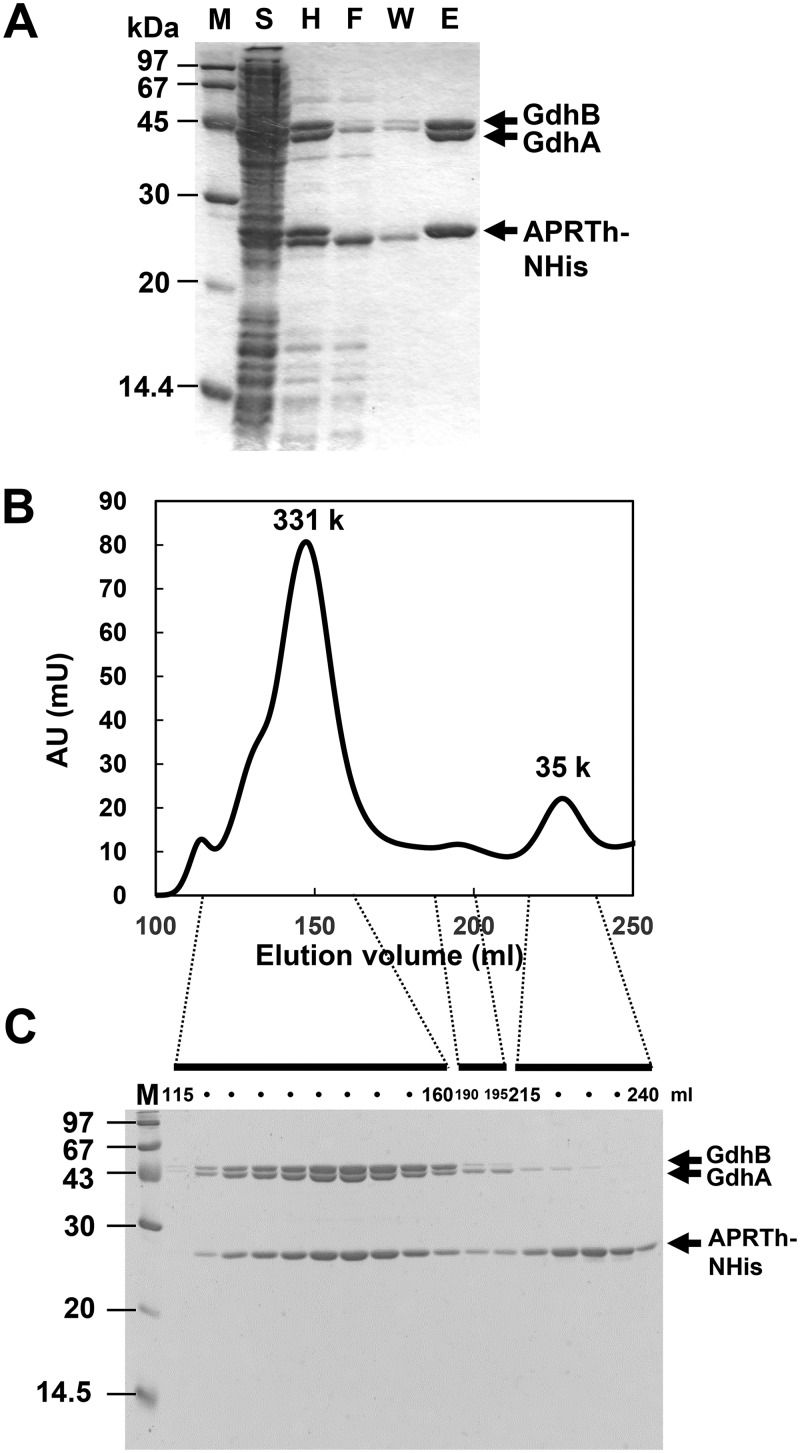
As the yield of the TtGDH·APRTh complex prepared from T. thermophilus was too low for us to perform biochemical characterization, we established E. coli coexpression systems for GdhA, GdhB, and APRTh with a His tag at the N terminus. Ni2+ affinity chromatography successfully confirmed the complex formation of TtGDH with APRTh (Fig. 4A). When the purified fraction was applied to gel filtration chromatography, fractions corresponding to 331 kDa were eluted as the major peak (Fig. 4B). SDS-PAGE of the major fractions demonstrated that the fractions contained GdhA, GdhB, and APRTh (Fig. 4C). In contrast, the molecular weights of TtGDH (GdhA/GdhB) alone and APRTh alone were estimated at 315 kDa and 30 kDa, respectively (see Fig. S3 in the supplemental material). This suggests that the three components formed a stable heterocomplex in the column. In order to further confirm heterocomplex formation, we performed gel filtration analysis and pulldown assay using purified TtGDH without a tag and purified APRTh with a tag (Fig. S4 and S5), which demonstrated the interaction between TtGDH and APRTh in vitro. The band volumes of GdhA-GdhB-APRTh on SDS-PAGE shown in Fig. 4C were quantified to approximate the stoichiometry of complex formation. Taken together with the molecular weight estimated by the gel filtration analysis, we assumed that GdhA-GdhB-APRTh forms a 2:4:2 heterocomplex with a molecular weight of 331 kDa. A minor peak at 35 kDa corresponded to an APRTh dimer that did not form a complex with TtGDH. In order to clarify which subunit of TtGDH interacted with APRTh directly, we next coexpressed gdhA plus aprth or gdhB plus aprth in the same cells and then applied the lysate to Ni2+ chromatography. On SDS-PAGE, GdhB was copurified with APRTh with the His tag with minimal loss into the flowthrough and washing fractions during chromatography (Fig. 5A). In contrast, although a certain amount of GdhA was copurified with APRTh with the His tag, a substantial amount of GdhA was lost into the flowthrough and washing fractions (Fig. 5B), suggesting that the GdhA-APRTh interaction is weaker than that of the GdhB-APRTh interaction. We further conducted gel filtration chromatography of the purified GdhA-APRTh and GdhB-APRTh. As a result, GdhA and APRTh were separately eluted by chromatography as the GdhA dimer (77 kDa) and APRTh dimer (29 kDa) (Fig. S2A and C). In contrast, GdhB and APRTh were mainly eluted forming a complex (Fig. S2B and D). Considering these results together, we consider APRTh to mainly interact with GdhB of the TtGDH heterohexamer.
FIG 4.
Coexpression and purification of TtGDH. (A) Copurification of APRTh with a His tag at the N terminus, GdhA, and GdhB. (B) Gel filtration column chromatography of proteins copurified with His-tagged APRTh. Chromatogram of absorption at 280 nm. AU, arbitrary units. The estimated molecular weights of the peaks are shown. (C) SDS-PAGE of the collected fractions from chromatography. The numbers of corresponding elution volumes are indicated for several fractions.
FIG 5.
Copurification of APRTh with a His tag at the N terminus and Gdh subunits. (A) Copurification of APRTh with a His tag at the N terminus and GdhB. (B) Copurification of APRTh with a His tag at the N terminus and GdhA. Lane M, molecular size markers; lane S, supernatant of cell extract of the strain; lane F, flowthrough fraction from the column; lane W, wash fraction from the column; lane E, elution fraction from the column.
Leucine-induced activation of the TtGDH-APRTh complex.
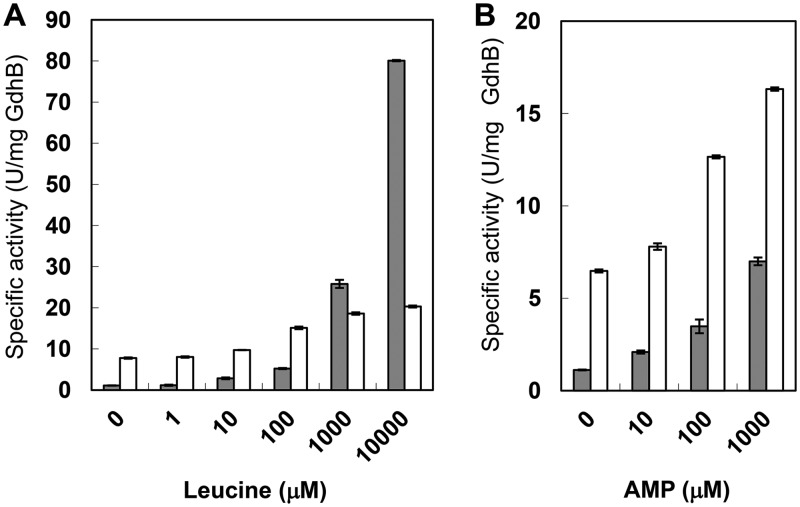
We performed an enzymatic assay for the TtGdh-APRTh complex prepared from E. coli. The TtGDH·APRTh complex exhibited specific activity of 1.1 U/mg (GdhB) and 7.8 U/mg (GdhB) for reductive amination and oxidative deamination, respectively, without leucine (Fig. 6A). The corresponding activity of TtGDH alone was 6.8 U/mg (GdhB) and 21.5 U/mg (GdhB), respectively (13). Thus, the specific activity of TtGDH-APRTh per GdhB subunit was approximately 2.8- to ∼6.2-fold lower than that of TtGDH. We previously reported that TtGDH activity was enhanced by leucine (9.7-fold and 2.6-fold for reductive amination and oxidative deamination, respectively, by 10 mM leucine). Therefore, we examined the effects of leucine on the activity of the TtGDH-APRTh complex. The TtGDH-APRTh complex was also activated by the increased concentration of leucine (Fig. 6A). Activation by 10 mM leucine was reached at 73- and 2.6-fold for reductive amination and oxidative deamination, respectively. These results imply that allosteric regulation by leucine is more effective in the TtGDH-APRTh complex.
FIG 6.
Activation profiles of GDH activity of the TtGDH-APRTh complex by leucine (A) and AMP (B). The concentrations of supplemented effectors are shown at the bottom. The gray and white bars indicate the reductive amination and oxidative deamination reactions, respectively.
Effects of other metabolites on GDH activity.
In order to examine the other roles of APRTh in sensing the allosteric effector, we evaluated the effects of several nucleotide compounds, because mammalian GDH is activated by ADP and inhibited by ATP and GTP. In particular, it was of interest to examine the effects of AMP, because docking modeling of the APRTh ortholog from T. thermophilus HB8 with AMP in a previous report suggested that APRTh can bind AMP at the active-site pocket even though it possesses several amino acid substitutions compared with other types of APRT (16). As expected, reductive amination and oxidative deamination activity was enhanced by AMP (Fig. 6B) by up to 6.3- and 2.5-fold, respectively, by the addition of 1 mM AMP (Table 2). The other candidates also activated the activity to some extent, but they were not as effective as AMP. When a similar assay was performed for TtGDH, only activation by leucine, not AMP, was observed, demonstrating that APRTh mediates the AMP-induced allosteric activation of GDH activity. Of note, the TtGDH-APRTh complex was synergistically activated by the simultaneous addition of AMP and leucine (Table 2), suggesting that they act on GDH independently. To elucidate the mode of activation by AMP, we conducted kinetic analyses in the presence or absence of AMP and/or leucine (Table 3). In reductive amination, the influence of AMP and leucine on the Km for NADH, 2-oxoglutarate (2-OG), and ammonium was not large, although approximately 4.5-, 5.3-, and 10.2-fold increases in Km for 2-OG were observed with AMP, leucine, and the copresence of AMP and leucine, respectively. In contrast, the kcat markedly increased in the presence of AMP (3.5-fold) and leucine (51.6-fold). The cosupplementation of AMP and leucine further increased the kcat by 142-fold. For oxidative deamination, the influence of AMP and leucine on the Km for NAD+ was limited. The kinetics for glutamate when leucine was not added as effector are complicated because there was a break in the Lineweaver-Burk plots for various concentrations of glutamate (Fig. S6). Such complicated kinetic behavior is similar to the negative cooperativity of bovine GDH against NAD(H) (8). In their report, Allen et al. noted a break in the Lineweaver-Burk plot for various concentrations of NAD+, and they estimated the kinetic parameters for the high NAD+ concentration and low NAD+ concentration separately (8). Therefore, we similarly estimated the kinetic parameters for the high glutamate concentration and low glutamate concentration of the oxidative reaction without leucine (Table 3). In contrast, the Km for glutamate was able to be measured with leucine and with AMP plus leucine. As a result, the Km for glutamate with leucine and with leucine plus AMP exhibited a 0.38- and 0.64-fold decrease, respectively, compared with that of low-concentration glutamate without the effector. This clarified that the negative cooperativity for glutamate occurs only in the absence of leucine. In our previous report, we found that glutamate binds to the subunit interface between the two GdhB subunits of the GdhB homohexamer, which corresponds to the leucine-binding site of the GdhA-GdhB heterohexamer (13). Taking this into account, we assumed that glutamate binds the leucine-binding sites of TtGDH in the absence of leucine, which causes the negative cooperativity for glutamate of TtGDH-APRTh, whereas in the presence of leucine, glutamate cannot bind to the effector-binding site occupied by leucine and no longer causes negative cooperativity. When the enzyme reaction with NAD+ as the variable substrate was performed under glutamate-saturating conditions, the kcat was increased in the presence of AMP (3.3-fold) and leucine (1.3-fold) during oxidative deamination. The cosupplementation of AMP and leucine further increased the kcat by 4.3-fold. The degree of increase in the kcat in the presence of AMP and/or leucine correlated with the increase in specific activity. Thus, AMP and leucine may increase the turnover of the TtGDH reaction in both directions but do not greatly affect the Km for the substrates.
TABLE 2.
Effects of metabolic compounds on GDH activity
| Compound | TtGDH APRTha
|
TtGDHb
|
||
|---|---|---|---|---|
| Reductive amination (%) | Oxidative deamination (%) | Reductive amination (%) | Oxidative deamination (%) | |
| None | 100 ± 2 | 100 ± 1 | 100 ± 4 | 100 ± 1 |
| AMP | 626 ± 19 | 252 ± 1 | 98 ± 2 | 104 ± 3 |
| ADP | 152 ± 6 | 120 ± 1 | ND | ND |
| ATP | 124 ± 5 | 111 ± 5 | ND | ND |
| GMP | 133 ± 6 | 101 ± 1 | ND | ND |
| GDP | 113 ± 9 | 103 ± 1 | ND | ND |
| GTP | 108 ± 15 | 105 ± 6 | ND | ND |
| IMP | 116 ± 5 | 100 ± 1 | ND | ND |
| Leu | 2,440 ± 134 | 335 ± 3 | 459 ± 14 | 432 ± 2 |
| Leu/AMP | 5,670 ± 102 | 457 ± 6 | 470 ± 14 | 426 ± 1 |
100% denotes 0.60 U/mg and 3.5 U/mg of TtGDH-APRTh for reductive amination and oxidative deamination, respectively.
100% denotes 3.7 U/mg and 5.4 U/mg of TtGDH for reductive amination and oxidative deamination, respectively. ND, not determined.
TABLE 3.
Kinetic parameters of TtGDH-APRTh
| Substrate | Without effector |
AMP |
Leu |
AMP/Leu |
||||
|---|---|---|---|---|---|---|---|---|
| Kmapp (μM) | kcatapp (s−1) | Kmapp (μM) | kcatapp (s−1) | Kmapp (μM) | kcatapp (s−1) | Kmapp (μM) | kcatapp (s−1) | |
| Reductive amination | ||||||||
| NADH | 8.0 ± 2.4 | 0.20 ± 0.017 | 5.8 ± 1.9 | 0.72 ± 0.05 | 3.0 ± 0.7 | 9.1 ± 0.3 | 5.3 ± 1.0 | 29 ± 1.0 |
| 2-OG | 5.1 ± 0.7 | 0.23 ± 0.006 | 22.9 ± 1.1 | 0.66 ± 0.009 | 27 ± 0.1 | 15 ± 1.7 | 52 ± 1.5 | 37 ± 0.4 |
| NH4+ | 29,000 ± 10,000 | 0.31 ± 0.054 | 43,000 ± 11,000 | 1.1 ± 0.15 | 46,000 ± 7,900 | 16 ± 1.6 | 33,000 ± 1,600 | 44 ± 1.0 |
| Oxidative deamination | ||||||||
| NAD+ | 24 ± 0.95 | 2.8 ± 0.05 | 130 ± 22 | 9.1 ± 0.4 | 56 ± 5.2 | 3.7 ± 0.09 | 160 ± 12 | 12 ± 0.20 |
| Glu | 450 ± 26 | 4.5 ± 0.04 | 770 ± 57 | 13 ± 0.24 | ||||
| Glu (high [Glu]) | 52,000 ± 15,000 | 1.5 ± 0.1 | 19,0000 ± 55,000 | 4.4 ± 1.0 | ||||
| Glu (low [Glu]) | 1,200 ± 82 | 0.27 ± 0.01 | 270 ± 50 | 1.2 ± 0.1 | ||||
Growth of the APRTh gene deletion strain.
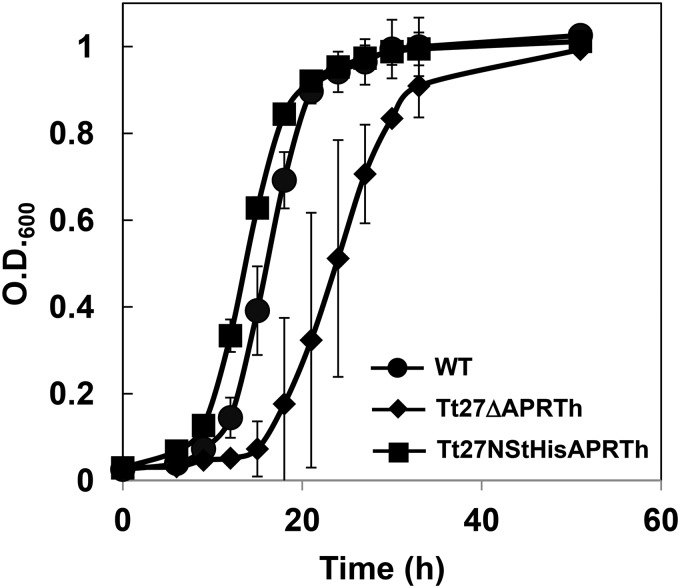
In order to examine the in vivo function of APRTh, we conducted growth analysis of the aprth knockout strain (Tt27ΔAPRTh) and aprth-overexpressing strain (Tt27NStHisAPRTh) of T. thermophilus in minimal medium. The Tt27ΔAPRTh strain exhibited delayed growth and required approximately 36 h to reach the early stationary phase, whereas the wild-type strain reached this phase after 21 h of cultivation (Fig. 7). The Tt27NStHisAPRTh strain exhibited better growth than even the wild-type strain. This suggests that APRTh functions in the cell and supports the optimal growth of T. thermophilus in minimal medium. Although we were unable to clarify the specific function of APRTh with TtGDH, detailed growth analysis with different carbon and nitrogen sources will help to elucidate this point in the future.
FIG 7.
Growth curves of T. thermophilus and the recombinant strains in minimal medium. WT, wild-type strain; Tt27ΔAPRTh, aprth knockout strain; Tt27NStHisAPRTh, aprth overexpression strains. O.D.600, optical density at 600 nm.
DISCUSSION
We previously reported that T. thermophilus GDH was composed of two heterologous subunits, GdhA (regulatory subunit) and GdhB (catalytic subunit). The heterocomplex type GDH is allosterically activated by leucine. In the present study, we found that an APRT homolog, APRTh, without the APRT activity serves as a protein factor that controls the activity of TtGDH, and we further demonstrated that the TtGDH-APRTh complex-type GDH is allosterically activated by AMP in which APRTh acts as the sensor of the signal.
Thus far, allosteric regulation of bacterial GDH is known only for large-type GDH, GDH180s from Streptomyces clavuligerus (2), Pseudomonas aeruginosa (5), Psychrobacter sp. strain TAD1 (17), and Janthinobacterium lividum (6). Large-type GDH is composed of the core 50-kDa GDH domain and additional N- and C-terminal domains. Although two additional domains of the members exhibit amino acid sequence similarity among GDH180s, these domains have no similarity with any other proteins in the database (2). GdhB from P. aeruginosa was found to be subject to allosteric control by arginine and citrate, which function as positive and negative effectors, respectively (5). Recently, GdhZ from Caulobacter crescentus, which is a member of the GDH180s family, was reported to stimulate cell constriction during cytokinesis by directly interfering with Z-ring stability (18). This report also described that the GDH was stimulated by arginine and inhibited by citrate, similar to the GDH from P. aeruginosa. This suggests that the additional domains are involved in the interaction with another protein and allosteric regulation of GDH. In contrast, the allosteric activation of 50-kDa GDH thus far has been observed only for TtGDH, in which leucine is bound to the intersubunit pocket between the GdhA and GdhB subunits to increase the turnover of the catalytic cycle (14). Mammalian GDH, which is also classified into the same 50-kDa GDH family, was reported to share the same mechanism with TtGDH for allosteric regulation by leucine (14).
AMP-mediated activation was also observed for GDH from S. clavurigerus (2). However, the mode of the activation differs between the two types of GDH: AMP is essential for GDH from S. clavurigerus to exhibit activity, whereas T. thermophilus GDH exhibits basal activity even in the absence of AMP. Similar nucleotide-mediated activation is observed for human GDH where reductive amination of 2-OG of housekeeping GDH (GDH1) and tissue-specific GDH (GDH2) was activated by 2.5- and 20-fold, respectively, by 1 mM ADP (19). As the ADP/ATP ratio is high when the intracellular energy level is low, this activation is thought to trigger energy production through the increase in the intracellular level of 2-OG, an intermediate of the tricarboxylic acid (TCA) cycle. We speculate that TtGDH functions via a similar mechanism responsive to the intracellular energy level in T. thermophilus, because AMP is also an indicator of a low energy level in the cell.
In TtGDH, APRTh is essential for allosteric activation by AMP, suggesting that AMP is bound to APRTh and the signal is transmitted to the active site of TtGDH. On the other hand, the crystal structure of bovine GDH complexed with ADP demonstrated that ADP is bound to the pivot helix, which is a rotational axis between the NAD-binding domain and catalytic domain (10). Binding of ADP to the pivot helix was suggested to facilitate the opening of the catalytic cleft between the NAD-binding domain and catalytic domain. As the residues for binding ADP of bovine GDH are not conserved in TtGDH (GdhA and GdhB), the underlying molecular mechanism and evolutionary origin of the activation is different between the two 50-kDa GDH enzymes.
Biological significance of activation of GDH by AMP.
As mentioned above, AMP is a signal of a low energy level in the cell, and GDH activation by AMP may be a response to enhance energy production through replenishment of the intermediates of the TCA cycle. As previously described, AMP is an essential activator of GDH from S. clavuligerus (2); therefore, a similar system to respond to the intracellular energy level may be present in other bacteria. On the other hand, in mammalian cells, AMP activates AMP-activated protein kinase (AMPK), which successively enhances the catabolic pathways to generate ATP (e.g., the uptake and oxidation of glucose and fatty acids and mitochondrial biogenesis) by targeting SREBP1c/ChREBP, TIF-1A, mTORC1, and ACC1 signaling pathways and represses the ATP-consuming anabolic pathways (e.g., the synthesis of lipids, glucose, glycogen, and proteins) by phosphorylating GLUT4/CD36, PGC-1α/SIRT, and ACC2 (20, 21). Although mammalian and T. thermophilus cells differ in quality, GDH activation by AMP may be a common signal transduction pathway to adapt and respond to nutrition or the environment around the cell. Of note, the aprt (TTC1250) gene is present just upstream of the aphth (TTC1249) gene in the genome. We demonstrated that APRT exhibits APRT activity that generates AMP from adenine and PRPP. This suggests that APRT has a functional relationship with APRTh, because APRT itself supplies the effector of the TtGDH-APRTh complex.
Distribution of the GdhA, GdhB, APRTh, and APRT quartet.
In order to examine the distribution of GdhA, GdhB, APRTh, and APRT in nature, we searched the two conserved gene clusters gdhA-gdhB and aprt-aprth using genome databases for several organisms. We found the same gene clusters in several strains belonging to the Thermus-Deinococcus group see (Fig. S7 in the supplemental material). This suggested that a regulatory GDH composed of GdhA, GdhB, APRTh, and maybe APRT as a source of AMP is present in these organisms and allosterically regulated in a manner similar to that in T. thermophilus. This also supports the involvement of APRT in the AMP-mediated regulatory system.
Conclusions.
We demonstrated that an enzymatically inactive paralog of APRT, APRTh, interacts with TtGDH, which is composed of GdhA and GdhB subunits and mediates the allosteric activation of TtGDH by AMP. Taking these results together with our previous reports, we clarified that TtGDH plays an essential role in controlling the carbon-nitrogen balance and intracellular energy level in a complicated allosteric manner, similar to mammalian GDH. Future crystallographic and biochemical analyses of the TtGDH-APRTh complex will lead to the elucidation of the molecular basis and biological importance of this regulation. We expect the results to provide important clues to understand the GDH-mediated metabolic network from different organisms, including mammals.
MATERIALS AND METHODS
Strains, media, and chemicals.
Escherichia coli DH5α was used for DNA manipulation, and E. coli BL21-Codon Plus (DE3)-RIL [F− ompT hsdS(rB− mB−) dcm Tetr gal (DE3) endA hhe (argU ileY leuW Camr)] (Stratagene, La Jolla, CA) was used as the host to express the gdhAB genes. The medium 2× YT (22) was generally used for cultivation of E. coli cells. Antibiotics and isopropyl-β-d-thiogalactopyranoside (IPTG) were added to the medium when required. All chemicals were purchased from Sigma-Aldrich Japan (Tokyo, Japan), Wako Pure Chemical (Osaka, Japan), or Kanto Chemicals (Tokyo, Japan). Enzymes for DNA manipulation were purchased from TaKaRa Bio (Kyoto, Japan) and TOYOBO (Osaka, Japan).
Preparation of the recombinant strains of T. thermophilus.
We constructed several plasmids for transformation of T. thermophilus HB27: pNHis-GdhA for production of GdhA with a His tag at the N terminus, pNStHisAPRTh for production of TTC1249 (APRTh) with a Strep-tag and His tag at the N terminus, and pΔTTC1249 for knockout of the TTC1249 (APRTh) gene. These plasmids were designed as illustrated in Fig. S1 in the supplemental material, and oligonucleotide primers used for their construction are listed in Table S1. pNHis-GdhA was constructed as follows. The first PCR was performed with primers gdhA-up-fw-NotI and gdhA-up-rv-BamHI to amplify the 700-bp upstream region of the gdhA gene. The second PCR was performed with primers hyg10-fw-BamHI and hyg10-rv-PstI to amplify the hyg10 gene encoding thermostable hygromycin B phosphotransferase (23). The third PCR was performed with primers pslpA-fw-PstI and pslpA-rv-ClaI/NdeI to amplify the slpA promoter of T. thermophilus (24). The fourth PCR was performed with primers gdhANHis12-fw-ClaI/NdeI and gdhANHis12-rv-KpnI to amplify a coding region of 700 bp of the gdhA gene. Each of the four amplified fragments was digested with NotI/BamHI, BamHI/PstI, PstI/NdeI, and ClaI/KpnI and cloned separately into pBlueScript II KS(+) for sequence verification. The four fragments with the correct sequences were digested with appropriate enzymes and ligated together with pBlueScript II KS(+) digested with NotI/KpnI to yield the plasmid pNHisGdhA. The resulting plasmid was used for the transformation of T. thermophilus (25) to generate the recombinant strain Tt27NHisGdhA. Colonies that were resistant to 160 μg/ml of hygromycin on TM medium were picked up, and recombination was confirmed by colony PCR using NHisGdhAcheck-fw and NHisGdhAcheck-rv and successive DNA sequencing of the PCR fragment. The other plasmids were constructed in the same manner as pNHisGdhA, using the primers listed in Table S1, and were used for gene integration or knockout of T. thermophilus HB27. The construction of plasmids is illustrated in Fig. S1. T. thermophilus strains for producing APRTh with a Strep-tag and His tag at the N terminus and for the knockout of the APRTh gene were named Tt27NStHisAPRTh and Tt27ΔAPRTh, respectively.
Pulldown assay using His-tagged GdhA as a bait protein.
Tt27NHisGdhA cells were cultured in 800 ml of TM medium supplemented with 40 μg/ml of hygromycin at 70°C for 12 to 14 h. Approximately 10 g of cells was washed and suspended in 16 ml of buffer A (20 mM Tris-HCl, pH 8.0, and 150 mM NaCl). The cells then were disrupted by sonication. The supernatant prepared by centrifugation at 40,000 × g at 4°C for 15 min was loaded onto a column with Ni2+-NTA resin (Novagen) and equilibrated with buffer A supplemented with 20 mM imidazole. After washing with the same buffer, adsorbed proteins were eluted with buffer A supplemented with 500 mM imidazole. The eluates were concentrated to approximately 200 μl with a Vivaspin 20 (molecular weight cutoff [MWCO], 10,000) concentrator (Sartorius Japan, Tokyo, Japan) and subjected to SDS-PAGE. A protein spot that was copurified with GdhA was sliced from the gel, washed twice with distilled water followed by 50% acetonitrile, incubated with 50 mM dithiothreitol in 100 mM NH4HCO3 at 56°C for 30 min, and then incubated with 100 mM iodoacetamide in 100 mM NH4HCO3 at room temperature for 30 min in the dark. The proteins were digested with mass spectrometry-grade trypsin (Promega KK, Tokyo, Japan) at 37°C overnight. The resulting peptides were analyzed by nano-high-pressure liquid chromatography-tandem MS analysis using an Advance nanoLC (Michrom Bioresources, Auburn, CA) system connected to an LTQ linear ion trap mass spectrometer (Thermo Fisher Scientific KK, Tokyo, Japan). Peptides dissolved in solvent A (0.1% formic acid) were separated with a 25-min gradient from 5% to 55% solvent B (acetonitrile containing 0.1% formic acid) at a flow rate of 500 nl/min. Full-scan MS spectra were obtained using the LTQ ion trap mass spectrometer.
Peak lists were searched against the NCBI T. thermophilus HB27 database (2013_07; 5,773 proteins sequences) (26) using the MASCOT server, version 2.4.1 (Matrix Science KK, Tokyo, Japan). The following search parameters in MASCOT were used: peptide mass tolerance, 2 Da; fragment mass tolerance, 1 Da; trypsin cleavage with a maximum of one missed cleavage; fixed modifications, S-carbamidomethylation (Cys); variable modification, oxidation (Met); charge state, +1, +2, or +3; ion score cutoff, 30; and false discovery rate cutoff, <5%. Only peptides with a MASCOT rank of 1 were accepted.
Pulldown assay using Strep-tagged APRTh using T. thermophilus cells.
Tt27NStHisAPRTh cells were cultured in 800 ml of TM medium supplemented with 50 μg/ml of kanamycin at 70°C for 12 to 14 h. Approximately 10 g of cells was washed and suspended in 16 ml of buffer A. The cells then were disrupted by sonication. The supernatant prepared by centrifugation at 40,000 × g at 4°C for 15 min was loaded onto a column with Strep-Tactin MacroPrep resin (Novagen) that was equilibrated with buffer A. After washing with the same buffer, adsorbed proteins were eluted with buffer A supplemented with 2.5 mM desthiobiotin. The eluates were concentrated to approximately 200 μl using a Vivaspin 20 (MWCO, 10,000) concentrator (Sartorius Japan, Tokyo, Japan) and subjected to SDS-PAGE. In order to identify the protein bands copurified with APRTh, MALDI-MS analysis was performed. Protein bands were excised from the gel and subjected to reduction, alkylation, and hydrolysis with modified trypsin (Promega). Peptides were extracted from gel pieces twice by 5% formic acid and 50% acetonitrile, desalted using ZipTip C18 pipette tips (Millipore), and then eluted by acetonitrile and 0.1% trifluoroacetic acid. Samples for MALDI-TOF analysis were prepared by mixing a small aliquot of the digestion supernatant 1:1 with dihydoxybenzoic acid (10 mg ml−1 in 1:2 solution of acetonitrile–0.1% trifluoroacetic acid). MALDI-TOF MS spectra were acquired and recorded with a mass spectrometer (Bruker). MALDI-TOF MS data were matched against the SWIISSPROT databases using the MS-FIT program (PROTEIN PROSPECTOR package; P. Baker and K. Clauser, http://prospecter.ucsf.edu).
Preparation of recombinant proteins using E. coli cells.
TtGDH was prepared using recombinant E. coli cells by following the procedure of a previous report (13). The recombinant proteins GdhA-GdhB-APRTh (TtGDH complexed with APRTh; TtGDH-APRTh), GdhA-APRTh, GdhB-APRTh, APRT, and APRTh were prepared using E. coli as the host. The construction of plasmids for the production of these proteins is illustrated in Fig. S1, and oligonucleotide primers used for construction are listed in Table S2. The genes were amplified by PCR using T. thermophilus genomic DNA as the template and appropriate oligonucleotide primers. The amplified DNA fragments were digested with appropriate restriction enzymes and ligated into pBlueScript II KS(+) (Novagen), which was prepared by digestion with the same enzymes. In order to use the HindIII site for construction of the coexpression plasmids, the HindIII site in the gdhB gene was mutated by site-directed mutagenesis using the primers listed in Table S2. After DNA sequence verification, these fragments were digested with appropriate restriction enzymes and ligated into the pET-26b(+) vector (Novagen). The resulting plasmids pET-GdhA/GdhB/APRTh, pET-GdhA/APRTh, pET-GdhB/APRTh, pET-APRT, and pET-APRTh were used for the transformation of E. coli BL21-CodonPlus(DE3)-RIL cells.
The TtGDH-APRTh complex was purified as follows. The E. coli transformant harboring pET-GdhA/GdhB/APRTh was inoculated in 2× YT medium supplemented with 50 μg/ml of kanamycin and 30 μg/ml of chloramphenicol and precultured at 37°C for 12 h. An aliquot of 16 ml of the culture broth was transferred into 1.6 liters of the same fresh medium and cultured at 37°C for an additional 3 h. IPTG was supplemented at 0.1 mM, and cultivation was further continued at 25°C for an additional 12 to 14 h. Cells were harvested by centrifugation at 5,000 × g for 10 min and washed with buffer A. The cells were resuspended in the same buffer and lysed by sonication. The supernatant was prepared by centrifugation at 40,000 × g for 15 min and was then heated at 70°C for 30 min. After removal of denatured proteins by centrifugation at 40,000 × g for 15 min, the supernatant was applied to the Ni2+-affinity column preequilibrated with buffer A supplemented with 20 mM imidazole. After washing with the same buffer, the TtGDH-APRTh complex was eluted with buffer A supplemented with 500 mM imidazole. The eluted fraction of GdhA-GdhB-APRTh was subjected to gel filtration chromatography with HiLoad 26/60 Superdex 200 pg (GE HealthCare Japan) equilibrated with buffer A. Purity was evaluated by SDS-PAGE. The proteins were concentrated with a Vivaspin (MWCO, 10,000) concentrator (Sartorius) prior to the enzyme activity assay. The other proteins or complexes, the GdhA-APRTh complex, GdhB-APRTh complex, APRT, and APRTh, were prepared by following the same procedure.
Analysis of subunit assembly of GdhA, GdhB, and APRTh.
In order to analyze interactions among GdhA, GdhB, and APRTh, the purified TtGDH-APRTh complex, GdhA-APRTh complex, and GdhB-APRTh complex were subjected to gel filtration chromatography with HiLoad 26/60 Superdex 200 pg equilibrated with buffer A with the flow rate set at 2.5 ml/min. Protein assembly and protein-protein interactions were analyzed by molecular weight calibration using molecular weight markers, gel filtration calibration kits HMW and LMW (GE HealthCare Japan), and SDS-PAGE of the fractionated eluates.
GDH enzyme assay.
The reaction mixture for oxidative deamination of glutamate contained 100 mM potassium phosphate buffer (pH 7.0), 50 mM KCl, 50 mM glutamate, and 2 mM NAD+. The reaction mixture for reductive amination of 2-oxoglutarate (2-OG) contained 100 mM potassium phosphate buffer (pH 7.0), 50 mM NH4Cl, 50 mM 2-OG, and 150 μM NADH. After the reaction mixture was preheated at 60°C, the reaction was started by adding enzymes, and the reduction of NAD+ to NADH or oxidation of NADH to NAD+ was monitored at 340 nm in a Shimadzu UV2000 spectrophotometer (Kyoto, Japan). One unit of enzyme activity was defined as the amount of enzyme forming 1 μmol of NAD+ or NADH per min. To examine the allosteric regulation of GDH, the GDH activity of TtGDH composed of GdhA and GdhB or the TtGDH-APRTh complex was measured in the reaction mixture containing potential regulatory compounds for GDH, such as 1 mM nucleotides (AMP, ADP, ATP, GMP, GDP, GTP, and IMP) or 1 mM leucine. To examine the sensitivity of the TtGDH-APRTh complex to leucine and AMP, the concentrations of leucine and AMP were varied within the range of 0 to 10,000 μM and 0 to 1,000 μM, respectively.
Steady-state kinetic analyses were performed by measuring the decrease in absorbance at 340 nm in reaction mixtures containing 0.2 to 15 μg/ml of TtGDH-APRTh enzyme in 100 mM sodium phosphate buffer, pH 7.0, at 60°C. For analyses of oxidative deamination, 50 mM KCl was added to the reaction mixture. To determine the Km for NADH, the concentration of NADH was varied in the range of 2 to 100 μM using fixed concentrations of 2-OG (50 mM) and NH4Cl (50 mM). To determine the Km for 2-OG, the concentration of 2-OG was varied in the range of 5 to 200 μM using fixed concentrations of NADH (150 μM) and NH4Cl (50 mM). To determine the Km for ammonia, the concentration of NH4Cl was varied in the range of 2 to 50 mM using fixed concentrations of 2-OG (50 mM) and NADH (150 μM). To determine the Km for NAD+, the concentration of NAD+ was varied in the range of 50 to 2,000 μM using a fixed concentration of Glu (50 mM). To determine the Km for Glu, the concentration of Glu was varied in the range of 1 to 50 mM using a fixed concentration of NAD+ (2 mM). Leu at 1 mM and/or AMP at 1 mM was added to the reaction mixtures when required. Most data were consistent with Michaelis-Menten kinetics; however, we were unable to determine the Km for glutamate without the effector but with AMP because the collected data did not fit Michaelis-Menten or Hill kinetics.
APRT enzyme assay.
APRT catalyzes the condensation of adenine and phosphoribosylpyrophosphate (PRPP) to generate AMP and pyrophosphate. The APRT activity was assayed by measuring released pyrophosphate using a pyrophosphate reagent (Sigma-Aldrich Japan, Tokyo, Japan). After the reaction mixture (792 μl of 50 mM HEPES-NaOH, pH 7.5, containing 1 mM MgSO4, 0.1 mM potential substrates [adenine, guanine, hypoxanthine, or xanthine], 0.1 mM PRPP, and 266 μl of pyrophosphate reagent) was preincubated at 30°C for 5 min, the reaction was started by adding 8 μl of solution containing 1.0 mg/ml of APRT or APRTh. A decrease in the absorbance at 340 nm was monitored with a UV-visible spectrophotometer (UV-2600; Shimadzu, Kyoto, Japan). One unit of the enzyme activity was defined as the amount of enzyme that released 1 μmol of the product per 1 min.
Supplementary Material
ACKNOWLEDGMENT
This work was supported in part by KAKENHI 24580137 (T.T.) and 15K07382 (T.T.) from the Japan Society for the Promotion of Science.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00710-18.
REFERENCES
- 1.Smith EL, Austin BM, Blumenthal KM, Nyc JF. 1975. Glutamate dehydrogenase, p 293–367. In Boyer PD, Krebs EG (ed), The enzymes. Academic Press, New York, NY. [Google Scholar]
- 2.Miñambres B, Olivera E, Jensen R, Luengo J. 2000. A new class of glutamate dehydrogenases (GDH). Biochemical and genetic characterization of the first member, the AMP-requiring NAD-specific GDH of Streptomyces clavuligerus. J Biol Chem 275:39529–39542. doi: 10.1074/jbc.M005136200. [DOI] [PubMed] [Google Scholar]
- 3.Kinnaird JH, Fincham JR. 1983. The complete nucleotide sequence of the Neurospora crassa am (NADP-specific glutamate dehydrogenase) gene. Gene 26:253–260. doi: 10.1016/0378-1119(83)90195-6. [DOI] [PubMed] [Google Scholar]
- 4.Miller SM, Magasanik B. 1990. Role of NAD-linked glutamate dehydrogenase in nitrogen metabolism in Saccharomyces cerevisiae. J Bacteriol 172:4927–4935. doi: 10.1128/jb.172.9.4927-4935.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu CD, Abdelal AT. 2001. The gdhB gene of Pseudomonas aeruginosa encodes an arginine-inducible NAD(+)-dependent glutamate dehydrogenase which is subject to allosteric regulation. J Bacteriol 183:490–499. doi: 10.1128/JB.183.2.490-499.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawakami R, Sakuraba H, Ohshima T. 2007. Gene cloning and characterization of the very large NAD-dependent L-glutamate dehydrogenase from the psychrophile Janthinobacterium lividum, isolated from cold soil. J Bacteriol 189:5626–5633. doi: 10.1128/JB.00496-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li M, Li C, Allen A, Stanley CA, Smith TJ. 2014. Glutamate dehydrogenase: structure, allosteric regulation, and role in insulin homeostasis. Neurochem Res 39:433–445. doi: 10.1007/s11064-013-1173-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allen A, Kwagh J, Fang J, Stanley CA, Smith TJ. 2004. Evolution of glutamate dehydrogenase regulation of insulin homeostasis is an example of molecular exaptation. Biochemistry 43:14431–14443. doi: 10.1021/bi048817i. [DOI] [PubMed] [Google Scholar]
- 9.Peterson P, Smith T. 1999. The structure of bovine glutamate dehydrogenase provides insights into the mechanism of allostery. Structure 7:769–782. doi: 10.1016/S0969-2126(99)80101-4. [DOI] [PubMed] [Google Scholar]
- 10.Banerjee S, Schmidt T, Fang J, Stanley CA, Smith TJ. 2003. Structural studies on ADP activation of mammalian glutamate dehydrogenase and the evolution of regulation. Biochemistry 42:3446–3456. doi: 10.1021/bi0206917. [DOI] [PubMed] [Google Scholar]
- 11.Li C, Matter A, Kelly A, Petty TJ, Najafi H, MacMullen C, Daikhin Y, Nissim I, Lazarow A, Kwagh J, Collins HW, Hsu BYL, Nissim I, Yudkoff M, Matschinsky FM, Stanley CA. 2006. Effects of a GTP-insensitive mutation of glutamate dehydrogenase on insulin secretion in transgenic mice. J Biol Chem 281:15064–15072. doi: 10.1074/jbc.M600994200. [DOI] [PubMed] [Google Scholar]
- 12.Stanley CA. 2004. Hyperinsulinism/hyperammonemia syndrome: insights into the regulatory role of glutamate dehydrogenase in ammonia metabolism. Mol Genet Metab 81(Suppl 1):S45–S51. doi: 10.1016/j.ymgme.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 13.Tomita T, Miyazaki T, Miyazaki J, Kuzuyama T, Nishiyama M. 2010. Hetero-oligomeric glutamate dehydrogenase from Thermus thermophilus. Microbiology 156:3801–3813. doi: 10.1099/mic.0.042721-0. [DOI] [PubMed] [Google Scholar]
- 14.Tomita T, Kuzuyama T, Nishiyama M. 2011. Structural basis for leucine-induced allosteric activation of glutamate dehydrogenase. J Biol Chem 286:37406–37413. doi: 10.1074/jbc.M111.260265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi W, Tanaka KS, Crother TR, Taylor MW, Almo SC, Schramm VL. 2001. Structural analysis of adenine phosphoribosyltransferase from Saccharomyces cerevisiae. Biochemistry 40:10800–10809. doi: 10.1021/bi010465h. [DOI] [PubMed] [Google Scholar]
- 16.Rehse PH, Tahirov TH. 2005. Crystal structure of a purine/pyrimidine phosphoribosyltransferase-related protein from Thermus thermophilus HB8. Proteins 61:658–665. doi: 10.1002/prot.20624. [DOI] [PubMed] [Google Scholar]
- 17.Camardella L, Di Fraia R, Antignani A, Ciardiello M, di Prisco G, Coleman J, Buchon L, Guespin J, Russell N. 2002. The Antarctic Psychrobacter sp. TAD1 has two cold-active glutamate dehydrogenases with different cofactor specificities. Characterisation of the NAD+-dependent enzyme. Comp Biochem Physiol A Mol Integr Physiol 131:559–567. doi: 10.1016/S1095-6433(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 18.Beaufay F, Coppine J, Mayard A, Laloux G, De Bolle X, Hallez R. 2015. A NAD-dependent glutamate dehydrogenase coordinates metabolism with cell division in Caulobacter crescentus. EMBO J 34:1786–1800. doi: 10.15252/embj.201490730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shashidharan P, Clarke D, Ahmed N, Moschonas N, Plaitakis A. 1997. Nerve tissue-specific human glutamate dehydrogenase that is thermolabile and highly regulated by ADP. J Neurochem 68:1804–1811. [DOI] [PubMed] [Google Scholar]
- 20.Hardie DG. 2011. Sensing of energy and nutrients by AMP-activated protein kinase. Am J Clin Nutr 93:891S–8916. doi: 10.3945/ajcn.110.001925. [DOI] [PubMed] [Google Scholar]
- 21.Ke R, Xu Q, Li C, Luo L, Huang D. 2018. Mechanisms of AMPK in the maintenance of ATP balance during energy metabolism. Cell Biol Int 42:384–392. doi: 10.1002/cbin.10915. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Maniatis T, Fritsch EF. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [Google Scholar]
- 23.Nakamura A, Takakura Y, Kobayashi H, Hoshino T. 2005. In vivo directed evolution for thermostabilization of Escherichia coli hygromycin B phosphotransferase and the use of the gene as a selection marker in the host-vector system of Thermus thermophilus. J Biosci Bioeng 100:158–163. doi: 10.1263/jbb.100.158. [DOI] [PubMed] [Google Scholar]
- 24.Fujita A, Misumi Y, Honda S, Sato T, Koyama Y. 2013. Construction of new cloning vectors that employ the phytoene synthase encoding gene for color screening of cloned DNA inserts in Thermus thermophilus. Gene 527:655–662. doi: 10.1016/j.gene.2013.06.069. [DOI] [PubMed] [Google Scholar]
- 25.Koyama Y, Hoshino T, Tomizuka N, Furukawa K. 1986. Genetic-transformation of the extreme thermophile Thermus thermophilus and of other Thermus spp. J Bacteriol 166:338–340. doi: 10.1128/jb.166.1.338-340.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henne A, Bruggemann H, Raasch C, Wiezer A, Hartsch T, Liesegang H, Johann A, Lienard T, Gohl O, Martinez-Arias R, Jacobi C, Starkuviene V, Schlenczeck S, Dencker S, Huber R, Klenk HP, Kramer W, Merkl R, Gottschalk G, Fritz HJ. 2004. The genome sequence of the extreme thermophile Thermus thermophilus. Nat Biotechnol 22:547–553. doi: 10.1038/nbt956. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.